Background: The interferon-stimulated gene (ISG) IFITM3 restricts endosomal entry of enveloped viruses.
Results: IFITM3 also restricts entry of reovirus, a nonenveloped virus.
Conclusion: IFITM3 alters endosomal function, either by delaying acidification or modulating proteolytic activity.
Significance: IFITM3 may restrict other clinically relevant nonenveloped viruses that require endosomes for entry.
Keywords: Endosomes, Innate Immunity, Interferon, Reovirus, Virus Entry, Interferon-stimulated Gene
Abstract
Reoviruses are double-stranded RNA viruses that infect the mammalian respiratory and gastrointestinal tract. Reovirus infection elicits production of type I interferons (IFNs), which trigger antiviral pathways through the induction of interferon-stimulated genes (ISGs). Although hundreds of ISGs have been identified, the functions of many of these genes are unknown. The interferon-inducible transmembrane (IFITM) proteins are one class of ISGs that restrict the cell entry of some enveloped viruses, including influenza A virus. One family member, IFITM3, localizes to late endosomes, where reoviruses undergo proteolytic disassembly; therefore, we sought to determine whether IFITM3 also restricts reovirus entry. IFITM3-expressing cell lines were less susceptible to infection by reovirus, as they exhibited significantly lower percentages of infected cells in comparison to control cells. Reovirus replication was also significantly reduced in IFITM3-expressing cells. Additionally, cells expressing an shRNA targeting IFITM3 exhibited a smaller decrease in infection after IFN treatment than the control cells, indicating that endogenous IFITM3 restricts reovirus infection. However, IFITM3 did not restrict entry of reovirus infectious subvirion particles (ISVPs), which do not require endosomal proteolysis, indicating that restriction occurs in the endocytic pathway. Proteolysis of outer capsid protein μ1 was delayed in IFITM3-expressing cells in comparison to control cells, suggesting that IFITM3 modulates the function of late endosomal compartments either by reducing the activity of endosomal proteases or delaying the proteolytic processing of virions. These data provide the first evidence that IFITM3 restricts infection by a nonenveloped virus and suggest that IFITM3 targets an increasing number of viruses through a shared requirement for endosomes during cell entry.
Introduction
The innate immune system must recognize signatures of pathogen infection (such as the presence of double-stranded RNA (dsRNA) for RNA viruses) and then initiate mechanisms that hinder the ability of the pathogen to replicate (1). Innate detection mechanisms must, practically, be nonspecific. However, after initial detection, the restriction mechanisms, exemplified by the type I interferon (IFN) system, need not be general. Initiating a wide variety of anti-pathogen responses may give an organism the greatest chance to contain an infection whether or not each individual mechanism is effective against the specific pathogen. Type I IFNs (IFN-α/β) are major antiviral cytokines and can be induced by several different pattern recognition receptors, including Toll-like receptors and cytoplasmic RIG-I-like helicases (2). After induction, type I IFNs signal through the type I IFN receptor (IFNAR) to induce a number of ISGs.2 Hundreds of ISGs have been identified, of which only a handful have a characterized function (3). Some induce broad antiviral responses, such as inhibition of cellular protein synthesis or degradation of RNA (4, 5). However, other ISGs, such as TRIM79α, target specific viruses (6). Understanding how individual ISGs restrict virus infection is of broad interest for designing antiviral therapeutics that can be targeted to specific or more general classes of pathogens.
One family of ISGs that is known to restrict specific classes of virus is the interferon-inducible transmembrane proteins (IFITM), which are conserved across numerous vertebrate species. In humans, the IFITM1, -2, -3, and -5 genes are located on chromosome 11 and have previously been shown to function in cell signaling, adhesion, and bone mineralization (7–10). These proteins contain two intramembrane domains linked by a highly conserved intracellular loop, features shared among the more than 200 CD225 protein family members. Family members IFITM1, -2, and -3 restrict the cell entry of influenza A, flaviviruses, dengue virus, West Nile virus, severe acute respiratory syndrome coronavirus, and filoviruses (11–13). These viruses are enveloped and enter cells via membrane fusion in endosomal compartments; indeed, the IFITM proteins restrict viruses independent of receptor usage but are dependent upon processes that occur in late endosomes (12, 14). IFITM3 is the most potent IFITM family member in restricting influenza A replication in cell culture (11), and recently it has been shown to reduce the pathogenesis of influenza in mice (15, 16) and in infected humans (16). Our recent data suggest a model where IFITM3 prevents influenza A virus fusion, thereby trapping virions within the endocytic pathway ultimately leading to their destruction in lysosomes and autolysosomes (14). Whether IFITM3 restricts non-enveloped viruses that also require endosomal access for cell entry is not known.
Mammalian orthoreoviruses (reoviruses) are non-enveloped viruses containing a segmented genome composed of dsRNA (17). Reoviruses utilize a multistep entry process, first binding to cell surface carbohydrates before engagement with the proteinaceous receptor junction adhesion molecule-A (JAM-A) (18). Subsequent interactions with β1-integrins target virions to late endosomal compartments (19), where the actions of acid-dependent cathepsin proteases are required for uncoating (20). These proteases cleave outer capsid protein σ3, revealing the membrane penetration protein μ1 (21–25). Cleavages of μ1 into the particle-associated fragments δ and φ and the dissociated fragment μ1N allow for conformational changes and subsequent membrane association of the viral capsid that mediates penetration of the viral core into the cytoplasm (23, 26). Although reovirus particles traverse a number of endocytic compartments, correct targeting to late endosomes containing Rab7 and Rab9 is crucial for establishing a productive infection (27). Because IFITM3 also localizes to late endosomes, we sought to test whether IFITM3 restricts reovirus infection.
In this study we found that IFITM3 expression significantly restricts infection and replication by reovirus. IFITM3-mediated restriction occurs at the level of endosomal penetration, as neither receptor binding nor RNA synthesis was affected. Targeting of reovirus particles to Rab7-containing late endosomes occurred with similar kinetics in the presence or absence of IFITM3. However, the kinetics of acidification and subsequent proteolysis of the reovirus capsid were delayed in IFITM3-expressing cells. Thus, IFITM3 likely alters the dynamics of endosomal uncoating, either leading to inefficient membrane penetration or lysosomal degradation of viral particles. This represents the first evidence that IFITM3 can restrict the infection of non-enveloped viruses that utilize endosome-dependent cell entry mechanisms and provides further mechanistic evidence for how IFITM3 restricts an increasing number of viruses by targeting a shared requirement for endosomes for cell entry.
EXPERIMENTAL PROCEDURES
Cells, Viruses, and Reagents
HeLa, U2OS, and A549 vector control and IFITM3- and shIFITM3-expressing cells were characterized previously (11, 14) and were maintained in Dulbecco's modified Eagle's medium supplemented to contain 10% fetal bovine serum, 2 mm l-glutamine, 100 units/ml penicillin,100 μg/ml streptomycin, and 250 ng/ml amphotericin B (Sigma). L929 cells were maintained in Joklik's minimum essential medium supplemented to contain 5% fetal bovine serum, 2 mm l-glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin, and 25 ng/ml amphotericin B.
Reovirus strains type 1 Lang (T1L) and type 3 Dearing (T3D) are laboratory stock. Reovirus strain rsT3D-σ1T249I was recovered by plasmid rescue (28). rsT3D-σ1T249I is isogenic to T3D with the exception of a single amino acid substitution in the σ1 protein rendering it insensitive to protease cleavage and, therefore, capable of generating infectious subvirion particles (ISVPs) with infectivity equivalent to that of T3D virions (29).
Purified reovirus virions were generated using second or third passage L-cell lysates of twice-plaque-purified reovirus as described (30). Viral particles were Freon-extracted from infected cell lysates, layered onto 1.2–1.4-g/cm3 CsCl gradients, and centrifuged at 62,000 × g for 18 h. Bands corresponding to virions (1.36 g/cm3) were collected and dialyzed in virion storage buffer (150 mm NaCl, 15 mm MgCl2, 10 mm Tris-HCl, pH 7.4). Concentrations of reovirus virions in purified preparations were determined from an equivalence of 1 absorbance unit at 260 nm equals 2.1 × 1012 virions (31). Viral titer was determined by a plaque assay using murine L929 cells, and all indications of m.o.i. are based on L929 cell titer (32). ISVPs were generated as described (33) and confirmed by SDS-PAGE and Coomassie Brilliant Blue staining. Guinea pig anti-reovirus σNS and rabbit anti-T3D antisera were generously provided by Dr. Terence S. Dermody (Vanderbilt University Medical Center). Plasmids encoding Rab7-EGFP and Rab-interacting lysosomal protein (RILP)-EGFP (27) and guinea pig anti-reovirus σNS antiserum were obtained from Dr. Terence S. Dermody (Vanderbilt University Medical Center).
Succinimidyl Ester Labeling
Reovirus virions were labeled with succinimidyl ester Alexa Fluor 546 (A546) or pHrodo S.E. (pHrodo; Invitrogen) as described previously (27, 34). Succinimidyl esters preferentially label reovirus proteins λ2, μ1, σ2, and σ3 (34). Reovirus particles (3 × 1012) were diluted into fresh 0.05 m sodium bicarbonate, pH 8.5, and incubated with 10 μm succinimidyl ester A546 or pHrodo at room temperature for 90 min in the dark. Virus particles were dialyzed against phosphate-buffered saline (PBS) at 4 °C overnight and stored at 4 °C.
Fluorescent Focus Assay
Cells (4 × 104) were grown in 24-well tissue culture plates and adsorbed with reovirus strains at various m.o.i. for 1 h at 4 °C. After adsorption, 0.5 ml of fresh medium was added, and the cells were incubated at 37 °C for 18 h. Cells were fixed with 2% paraformaldehyde for 30 min, washed twice with PBS, and permeabilized and blocked in PBS + 2% bovine serum albumin + 0.1% Triton-X100 (PBS-T) for at least 1 h at 4 °C. Cells were then incubated with guinea pig anti-reovirus σNS antiserum (1:1000) in PBS-T for at least 1 h at 4 °C, washed 3× with PBS-T, and incubated with an anti-guinea pig Alexa 568-conjugated antibody (1:2000) in PBS-T for at least 1 h at 4 °C. Cells were washed 3× with PBS and visualized using fluorescence microscopy. Reovirus antigen-positive cells were quantified by counting fluorescent cells in at least two random fields of view in triplicate wells at a magnification of 113×. Total cell number was quantified using background fluorescence.
Assessment of Virus Replication by Plaque Assay
Cells (4 × 104) grown in 24-well tissue culture plates were adsorbed with reovirus strain T3D at an m.o.i. of 1 or 100 pfu/cell for 1 h at 4 °C. After adsorption, cells were washed 2× with PBS and incubated in 0.5 ml of fresh medium at 37 °C. After different time intervals, cells were freeze-thawed twice, and viral titer was determined by plaque assay. The viral yields were calculated by dividing viral titers at the indicated times by the viral titer at 0 h.
Analysis of Interferon Sensitivity
Cells (4 × 104) grown in 24-well tissue culture plates were either mock-treated or treated with IFN-α (100 IU/ml) for 6 h before inoculation with either T1L or T3D at an m.o.i. of 10 pfu/cell for 1 h at 4 °C. After inoculation, 0.5 ml of fresh medium was added, and the cells were incubated at 37 °C for 18 h before performing a fluorescent focus assay. Alternatively, viral titers at various times were determined by plaque assay.
Endosomal Acidification Inhibition
Cells (4 × 104) grown in 24-well tissue culture plates were adsorbed with either rsT3D-σ1T249I virions or ISVPs at the indicated m.o.i. for 45 min at 4 °C. After adsorption, cells were either mock-treated or treated with 10 mm ammonium chloride and incubated at 37 °C for 18 h and analyzed by fluorescent focus assay.
Kinetic Ammonium Chloride Protection Assay
Cells (4 × 104) grown in 24-well tissue culture plates were inoculated with rsT3D at an m.o.i. of 25 pfu/cell for 1 h at 4 °C. The inoculum was removed, and the cells were washed 2× with PBS and incubated in fresh prewarmed media at 37 °C. At various times after adsorption, ammonium chloride was added to the medium for a final concentration of 25 mm. Cells were then incubated for 18 h post-adsorption, fixed with 2% paraformaldehyde, and stained for a fluorescent focus assay.
Analysis of Viral Disassembly
Cells (5 × 105) grown in 60-mm dishes were inoculated with T3D at an m.o.i. of 100 pfu/cell for 1 h at 4 °C. The inoculum was removed, and the cells were washed 2× with PBS and incubated in fresh prewarmed media at 37 °C. At various times after adsorption, cells were scraped into 1 ml of ice-cold PBS and pelleted at 3000 × g for 5 min at 4 °C. The supernatant was aspirated, and the pellet was resuspended in 100 μl of ice-cold modified radioimmunoprecipitation assay buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA, 1% sodium deoxycholate, 1% IGEPAL CA-630, 1 mm PMSF) supplemented with protease inhibitor mixture (Roche Applied Science). The lysate was clarified by centrifugation at 13,000 × g for 10 min at 4 °C, and the supernatant was removed to a fresh tube and frozen at −20 °C. Extracts (10 μg of total protein) were resolved by electrophoresis in 4–12% Bis-Tris gels and transferred to PVDF membranes. The membranes were blocked overnight at room temperature in PBS + 1% Tween 20 (PBS-T) containing 5% milk and incubated with a rabbit anti-T3D antiserum (1:500) in PBS-T plus milk at room temperature for 3 h. The membranes were washed 3× for 10 min with PBS-T and incubated with an alkaline phosphatase-conjugated goat anti-rabbit antibody (Bio-Rad) diluted 1:2000 for 3 h. After 3 washes with PBS-T, the membranes were incubated for 5 min with chemiluminescent alkaline phosphate substrate (Bio-Rad) and visualized using a ChemiDock XRS+ molecular imager (Bio-Rad). Band densities were analyzed using Image J software.
Quantitative Reverse Transcriptase Polymerase Chain Reaction (qPCR)
Cells (5 × 105) grown in 60-mm dishes were adsorbed with T3D in PBS at an m.o.i. of 100 pfu/cell at 4 °C for 1 h. Cells were incubated in medium at 37 °C for various intervals, removed from plates with a scraper, washed once with PBS, and centrifuged at 500 × g for 5 min. The supernatant was removed, and the cell pellet was frozen at −20 °C. RNA was extracted by using an RNeasy Plus RNA extraction minikit (Qiagen) according to the manufacturer's instructions. RNA was converted to cDNA by using an Omniscript RT cDNA synthesis kit (Qiagen) with an oligo(dT) primer according to the manufacturer's instructions. qPCR was performed using the Express SYBR Green ER system (Invitrogen). Primers specific for human GAPDH (forward primer 5′-GATCATCAGCAATGCCTCCT-3′ and reverse primer 5′-TGTGGTCATGAGTCCTTCCA-3′) or human IFITM3 (forward primer 5′-ATGTCGTCTGGTCCCTGTTC-3′ and reverse primer 5′-GTCATGAGGATGCCCAGAAT-3′) were used at a final concentration of 0.2 μm. Quantification and melt curve analyses were performed according to the manufacturer's protocol. For each sample, the CT (threshold cycle) for the RNA of interest was normalized to that for GAPDH. -Fold induction was calculated by comparing normalized CT values (ΔΔCT) of duplicate cDNA synthesis reactions to those of samples taken at the time of infection (T = 0) for two independent experiments.
Confocal Microscopy of Reovirus Internalization
Cells were plated on glass coverslips (#1.5; Thermo Scientific) in 24-well plates at 37 °C overnight. Cells were transfected with plasmids using Lipofectamine 2000 (Life Sciences) according to the manufacturer's instructions. After incubation at 37 °C for 18 h, the cells were chilled at 4 °C for 1 h. Cells were then adsorbed with 10,000 Alexa546-labeled reovirus particles/cell at 4 °C for 1 h. The inoculum was removed, and the cells were washed 3 times with PBS and either fixed with 2% paraformaldehyde or supplemented with complete medium and incubated at 37 °C for various intervals. Cells were washed once with PBS and fixed for 20 min with 2% paraformaldehyde, quenched with 0.1 m glycine, and washed 3 times with PBS. Coverslips were removed from wells and placed on slides using ProLong Gold mounting medium (Life Sciences). Images were captured using a Zeiss LSM710 laser-scanning confocal microscope using a 63× Plan-Apochromat objective lens. Images were thresholded for pixel intensity, and the pinhole size used was identical for all fluorophores. All images represent single sections and were adjusted for brightness and contrast to the same extent. Colocalization analysis was performed using the colocalization function of Zeiss LSM image software (Zeiss) taking into consideration endosomal vesicle size to try to isolate individual endosomes. Virions within the boundary of single cells were quantified.
Assessment of pHrodo Fluorescence Intensity by Flow Cytometry
The kinetics of reovirus access to acidic compartments was performed as described previously (27). HeLa cells were chilled at 4 °C for 1 h and adsorbed with pHrodo-labeled reovirus (1 × 104 particles/cell) at 4 °C for 1 h. The inoculum was removed, and the cells were washed 3 times with cold PBS to remove unbound virus and supplemented with warm Opti-MEM-I (Invitrogen) for various intervals. The cells were washed once with PBS, detached with Cellstripper (Corning CellGro), washed once with PBS, and analyzed with a BD Accuri C6 flow cytometer. Cell staining was quantified using BD Accuri C6 software.
RESULTS
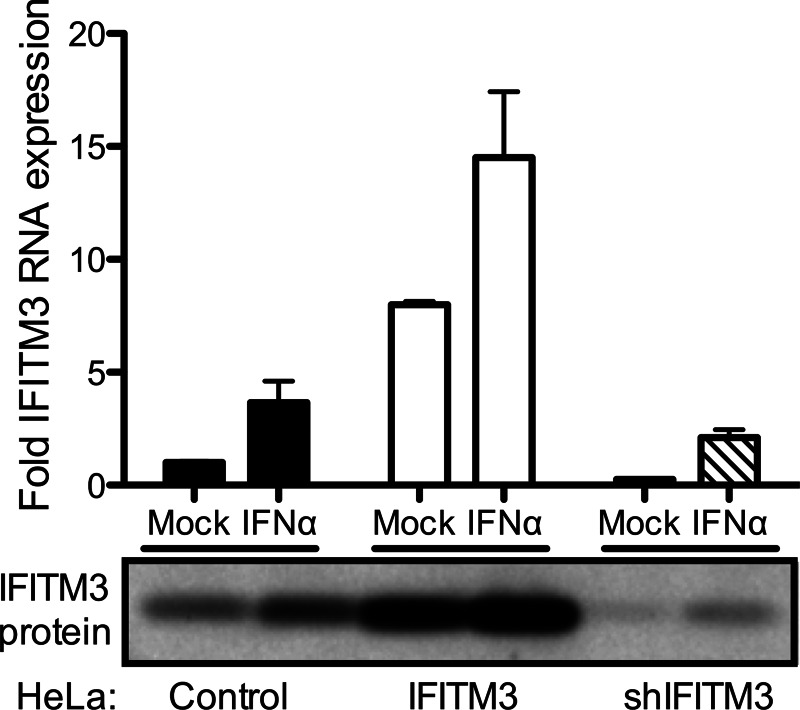
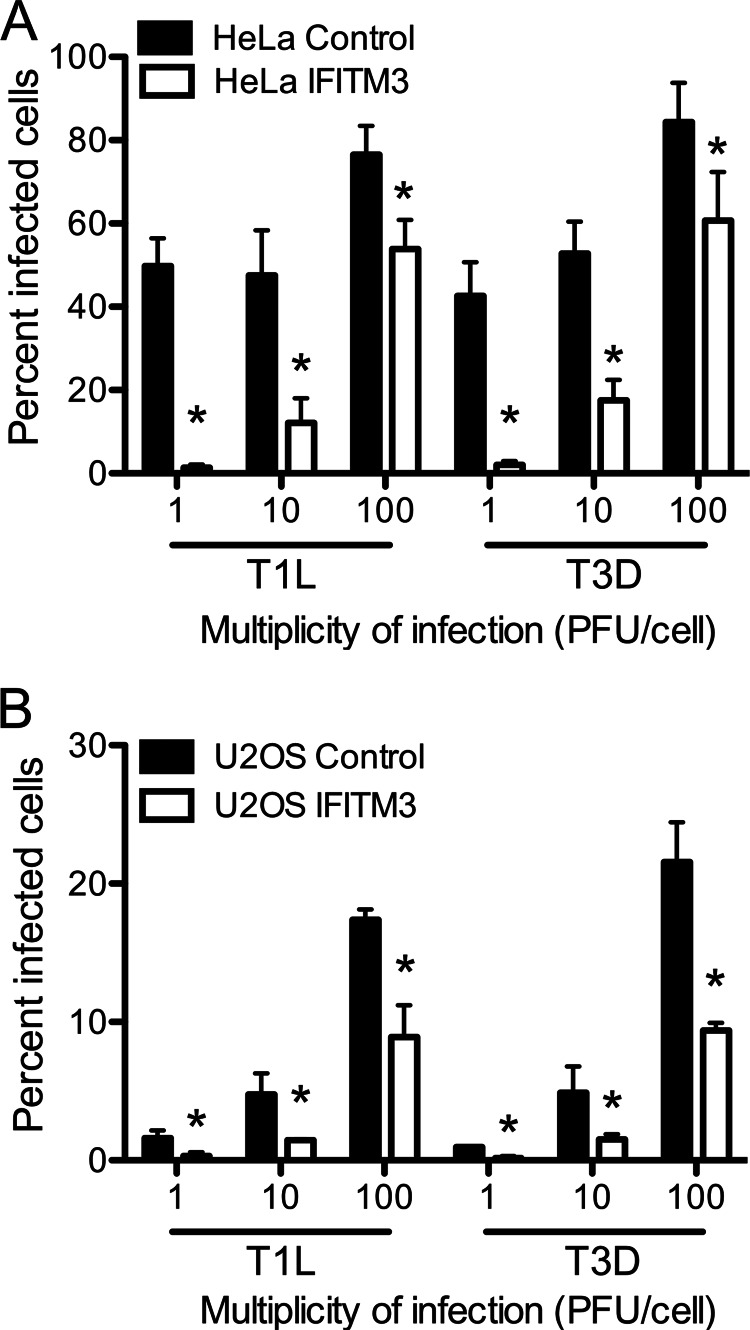
IFITM3 restricts entry of enveloped viruses that require acid pH or endosomal components for membrane fusion (12, 14). As the mechanism for this restriction is not defined, we tested whether IFITM3 also restricts entry of a non-enveloped virus with similar membrane penetration requirements, mammalian orthoreovirus. We utilized previously developed HeLa and U2OS cell lines that were engineered to stably express IFITM3 (11). Empty vector-transduced cell lines served as controls. Expression levels of IFITM3 in mock- and IFN-α-treated cells were determined by qPCR and by immunoblotting (Fig. 1 and data not shown). As previously observed, control HeLa cells express basal levels of IFITM3 in the absence of IFN treatment (11, 16). IFITM3-expressing cells produced ∼4-fold more IFITM3 than mock-treated cells and ∼2-fold more IFITM3 than IFN-α-treated cells. IFITM3-expressing and control cell lines were adsorbed with reovirus strains T1L and T3D at various m.o.i. (as determined using L929 cells), and the percentage of infected cells was determined at 18 h post-infection by immunostaining using an anti-reovirus σNS antiserum. Reovirus infection was significantly reduced in cells expressing IFITM3 (Fig. 2). The reduction in infection was greater at lower m.o.i., suggesting that the restriction mediated by IFITM3 could be saturated at high m.o.i. Similar results were obtained using an A549 cell line engineered to express IFITM3 (data not shown).
FIGURE 1.
IFITM3 expression in HeLa cell lines. Vector control, IFITM-3-expressing, and shIFITM3-expressing HeLa cell lines were mock-treated or treated with 100 IU/ml IFN-α for 6 h. Cultures were scraped into PBS and pelleted. RNA was extracted from cultures, converted into cDNA, and used for qPCR analysis using primers specific for IFITM3 (top panel). The level of IFITM3 RNA was normalized to that of GAPDH in each culture. Alternatively, cultures were lysed with radioimmunoprecipitation buffer and subjected to SDS-PAGE electrophoresis followed by immunoblotting using antisera against IFITM3 (bottom panel).
FIGURE 2.
IFITM3 restricts reovirus infection. HeLa (A and B) or U2OS (C) vector control- or IFITM3-expressing cells were infected with reovirus strains T1L and T3D at the indicated m.o.i. (pfu/cell). The percentage of infected cells was determined by fluorescent focus assay 18 h post-infection. The results are presented as the means of triplicate samples. Error bars indicate S.D. *, p < 0.05 by Student's t test in comparison with vector control cells.
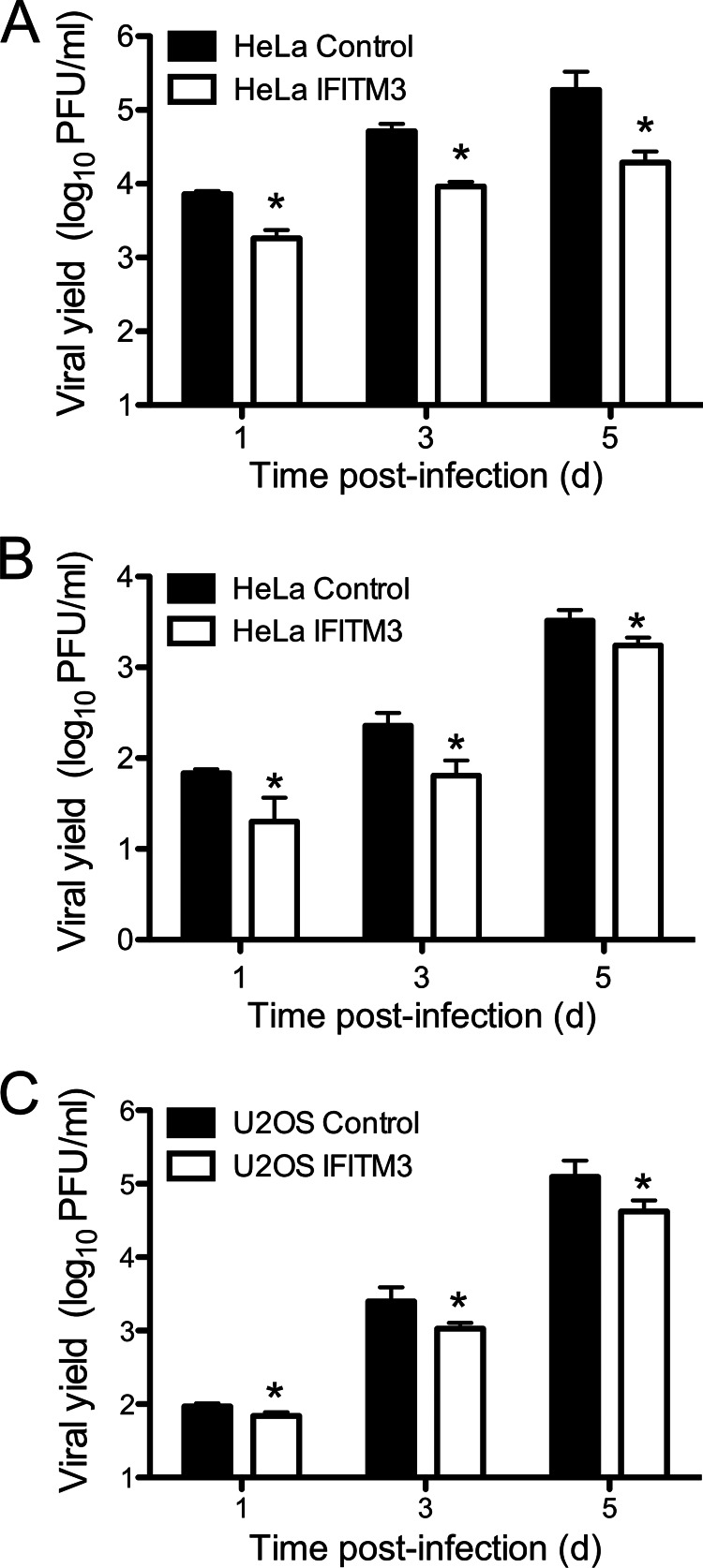
We next tested whether the decrease in the percentage of infected cells mediated by IFITM3 would result in decreased viral replication. IFITM3-expressing and control cell lines were infected with T3D at an m.o.i. of 1 pfu/cell, which allows for the initially infected cells to produce type I IFNs and prime uninfected cells for increased IFITM3 expression. Viral yield was then determined at various times post-infection. IFITM3 expression in HeLa and U2OS cell lines resulted in a 5-to-10-fold decrease (p < 0.05) in viral yield in comparison to vector control cells at all times examined (Fig. 3, A and C). Decreased yields were also observed in IFITM3-expressing HeLa cells infected at an m.o.i. of 100 pfu/cell, which would not allow for IFN-mediated priming, indicating that basal levels of IFITM3 can still restrict infection at high infectious doses of reovirus (Fig. 3B). These data suggest that IFITM3 overexpression restricts, but does completely inhibit, reovirus infection.
FIGURE 3.
IFITM3 restricts reovirus replication. HeLa (A and B) or U2OS (C) vector control- or IFITM3-expressing cells were infected with reovirus strain T3D at an m.o.i. of 1 pfu/cell (A and C) or 100 pfu/cell (B). Viral titers at the indicated times were determined by plaque assay. The results are presented as the mean viral yields, calculated by dividing titer at the indicated time by titer at 0 h for triplicate samples. Error bars indicate S.D. *, p < 0.05 by Student's t test in comparison with vector control cells.
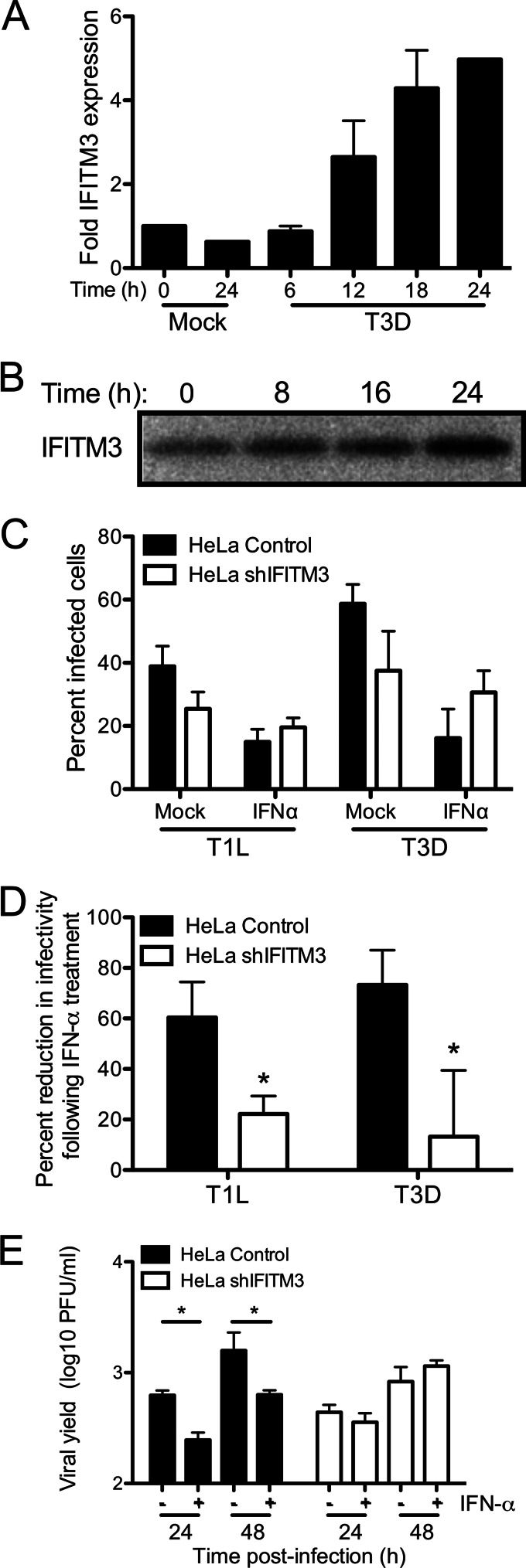
To determine whether IFITM3 is induced by reovirus infection, we first measured IFITM3 RNA levels in HeLa cells after infection. Cells were mock-infected or infected with T3D at an m.o.i. of 100 pfu/cell, and RNA was extracted from cells at various times post-infection. Levels of IFITM3 mRNA were quantified by using qPCR. We observed a continuous, time-dependent increase in IFITM3 expression after reovirus infection, with a maximum of a ∼5-fold increase over base-line expression after 24 h (Fig. 4A). Increased IFITM3 expression was first observed at 12 h post-infection, a result that is consistent with its known dependence on IFN for expression. Similar results were observed at the level of protein expression, as determined by immunoblot (Fig. 4B). We then utilized HeLa cells constitutively expressing a short hairpin RNA targeting IFITM3 (shIFITM3) to determine if IFITM3 expression could restrict reovirus infection in IFN-treated cells. HeLa cells expressing shIFITM3 exhibited reduced IFITM3 expression by ∼4–5-fold at both the RNA and protein level (Fig. 1). Control and shIFITM3-expressing HeLa cells were treated with 100 IU of IFN-α for 6 h before infection with T1L or T3D. This level of interferon is similar to that produced by reovirus infection of cultured fibroblasts (35, 36). The percentage of infected cells was determined by immunostaining at 18 h post-infection. Cells expressing shIFITM3 were significantly less sensitive to the antiviral effects of IFN in comparison to control cells (Fig. 3, C and D), as IFN treatment reduced the percent of infected cells by 60–80% in control cells and by 10–20% in shIFITM3-expressing cells (p < 0.05). In productively infected cells, however, no differences were observed in the number or size of σNS-containing inclusions (data not shown). No significant difference was observed in the percentage of infected control- and shIFITM3-expressing cells in the absence of IFN (p = 0.12). We also examined reovirus replication in mock- and IFN-α-treated control cells and shIFITM3-expressing cells. IFN-α treatment significantly decreased reovirus replication in control cells (Fig. 4E, p < 0.05) but had no effect on replication in shIFITM3-expressing cells, suggesting that IFITM3 expression can restrict reovirus replication in cells responding to type I IFN.
FIGURE 4.
Knockdown of IFITM3 expression decreases reovirus sensitivity to type I interferon. A, HeLa cells were mock-infected or infected with T3D at an m.o.i. of 100 pfu/cell. RNA was isolated from cells at the indicated times post-infection, and IFITM3 expression was quantified by qPCR relative to GAPDH expression. Results represent the means of duplicate cDNA syntheses for two independent experiments. Error bars indicate S.D. B, HeLa cells were infected with T3D at an m.o.i. of 100 pfu/cell. IFITM3 protein levels in cell lysates at the indicated times post-infection were determined by immunoblot using an antibody specific for IFITM3. C, HeLa vector control- or shIFITM3-expressing cells were either mock-treated or treated with IFN-α (100 IU/ml) for 6 h. Cells were then infected with TIL or T3D at an m.o.i. of 10 pfu/cell. The percentage of infected cells was determined by fluorescent focus assay 18 h post-infection. The results are presented as the means of triplicate samples. Error bars indicate S.D. D, values from C were normalized to show a decrease in reovirus infectivity after treatment with IFN-α in HeLa shIFITM3 and vector control cells. E, HeLa vector control- or shIFITM3-expressing cells were either mock-treated or treated with IFN-α (100 IU/ml) for 6 h. Cells were then infected with T3D at an m.o.i. of 1 pfu/cell. Viral titers at the indicated times were determined by plaque assay. The results are presented as the mean viral yields, calculated by dividing titer at the indicated time by titer at 0 h for triplicate samples. Error bars indicate S.D. *, p < 0.05 by Student's t test in comparison with mock treated cells.
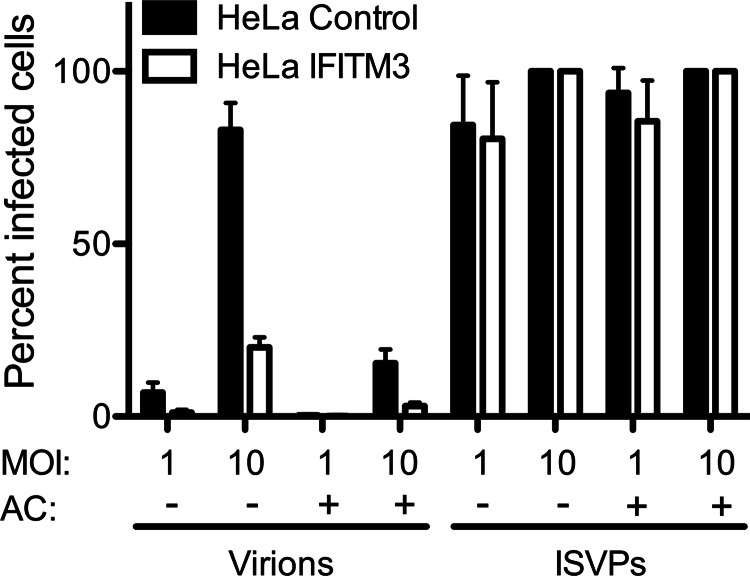
We next sought to determine at which stage of the reovirus replication cycle IFITM3 mediates its effects. Because IFITM3 localizes to endosomal compartments, we hypothesized that it would act at the virion disassembly stage in late endosomes and not at the level of receptor engagement and target cell binding. To test this, we utilized ISVPs, which bypass the requirement for endosomal proteases for cell entry (21). Therefore, ISVPs, unlike virions, are not sensitive to the weak base ammonium chloride, which prevents endosomal acidification. Control- or IFITM3-expressing cells were adsorbed with rsT3D-σ1T249I virions or ISVPs and then incubated in the presence or absence of ammonium chloride for 18 h, at which time the percentage of infected cells was determined by immunostaining. Ammonium chloride treatment reduced the infectivity of virions by ∼80%, with IFITM3-expressing cells exhibiting lower levels of infection than control cells (Fig. 5). In contrast, ammonium chloride had no effect on the entry of ISVPs, and no differences were observed in the percentage of infected cells in the presence or absence of IFITM3. Similar results were observed in control- and IFITM3-expressing U2OS cell lines (data not shown). Additionally, no differences were observed in the initial binding of virions to control- versus IFITM3-expressing cells as determined by flow cytometry using fluorescently tagged virions (not shown). Together, these data indicate that IFITM3 acts at a stage subsequent to initial receptor binding on target cells, likely during endosomal trafficking.
FIGURE 5.
IFITM3 does not restrict entry of reovirus ISVPs. HeLa vector control- or IFITM3-expressing cells were infected with either rsT3D-σ1T249I virions or T3D-σ1T249I ISVPs at the indicated m.o.i. After a 45-min adsorption, cells were either mock-treated or treated with 10 mm ammonium chloride (AC) and incubated at 37 °C for 18 h. The percentage of infected cells was determined by fluorescent focus assay. The results are presented as the means of triplicate samples. Error bars indicate S.D.
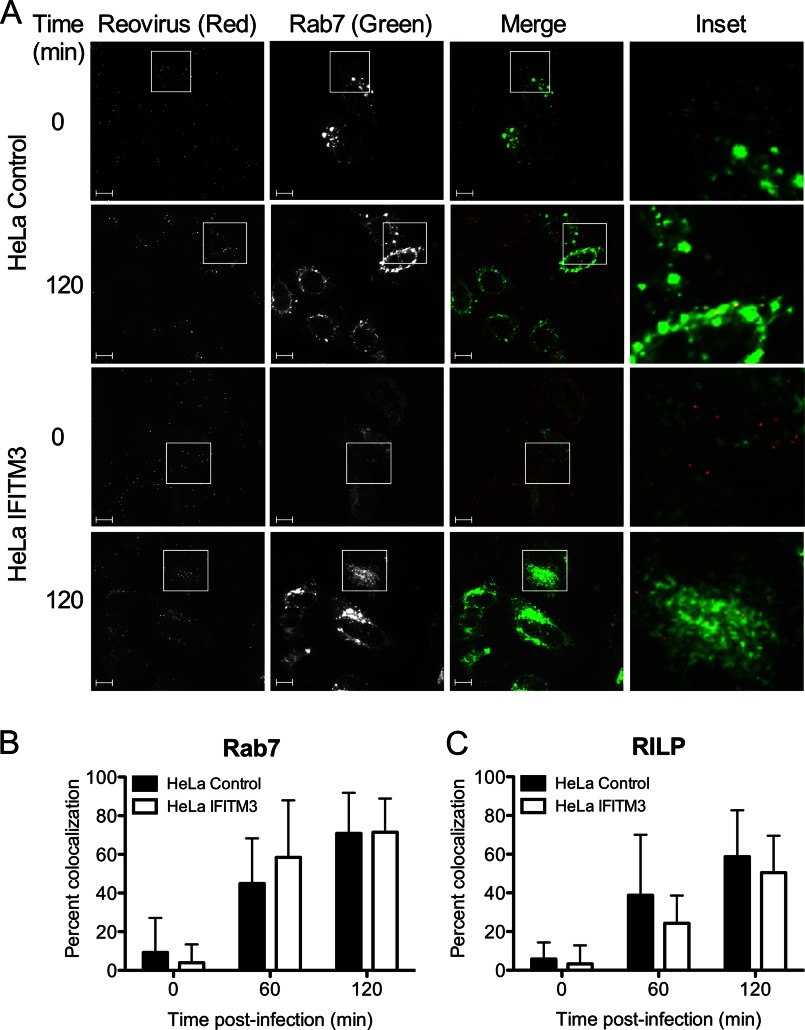
IFITM3 may function in endosomes to restrict reovirus entry via several possible mechanisms. One possibility may be that IFITM3 directs internalized virions to cellular compartments that are non-productive for the uncoating process, as incorrectly targeted virions are incapable of mediating infection (37). Productive reovirus entry occurs in late endosomes that colocalize with specific Rab GTPases such as Rab7 and RILP (27). We examined whether IFITM3 altered reovirus targeting to late endosomes by utilizing GFP-tagged endosomal markers and fluorescently tagged virions. Control- or IFITM3-expressing HeLa cells were transfected with Rab7-EGFP or RILP-EGFP, which localize to late endosomes (38, 39). At 18 h post-transfection, cells were chilled at 4 °C for 1 h, adsorbed with A546-labeled reovirus particles at 4 °C for 1 h, and incubated at 37 °C for 0, 60, or 120 min. At these times, cells were fixed with paraformaldehyde and visualized by confocal microscopy (Fig. 6A). Quantification of colocalization between reovirus particles and either Rab7-EGFP or RILP-EGFP showed no significant differences between control- and IFITM3-expressing cells (Fig. 6, B and C). These data suggest that IFITM3 does not alter virion targeting to late endosomal compartments.
FIGURE 6.
IFITM3 does not alter reovirus trafficking to late endosomes. A, HeLa vector control or IFITM3-expressing cells were transfected with EGFP-Rab7 (green) 18 h before infection. Cells were chilled at 4 °C for 1 h and then adsorbed with 10,000 particles/cell of reovirus-A546 (red) at 4 °C for 1 h. The inoculum was removed, and cells were washed to remove unbound virus and then either fixed with 2% paraformaldehyde or supplemented with complete medium and incubated at 37 °C for the times shown before fixing. Cells were imaged by confocal microscopy. Insets depict enlarged areas of boxed regions. Scale bars, 10 μm. B and C, shown is quantification of colocalization of reovirus-A546 particles with EGFP-tagged versions of Rab7 (B) and RILP (C) after adsorption of reovirus for the times shown. Data are presented as the percent of virus particles exhibiting spectral overlap with EGFP expression (n = 12–18 cells per time point, average of 322 particles per time point). Error bars indicate S.D.
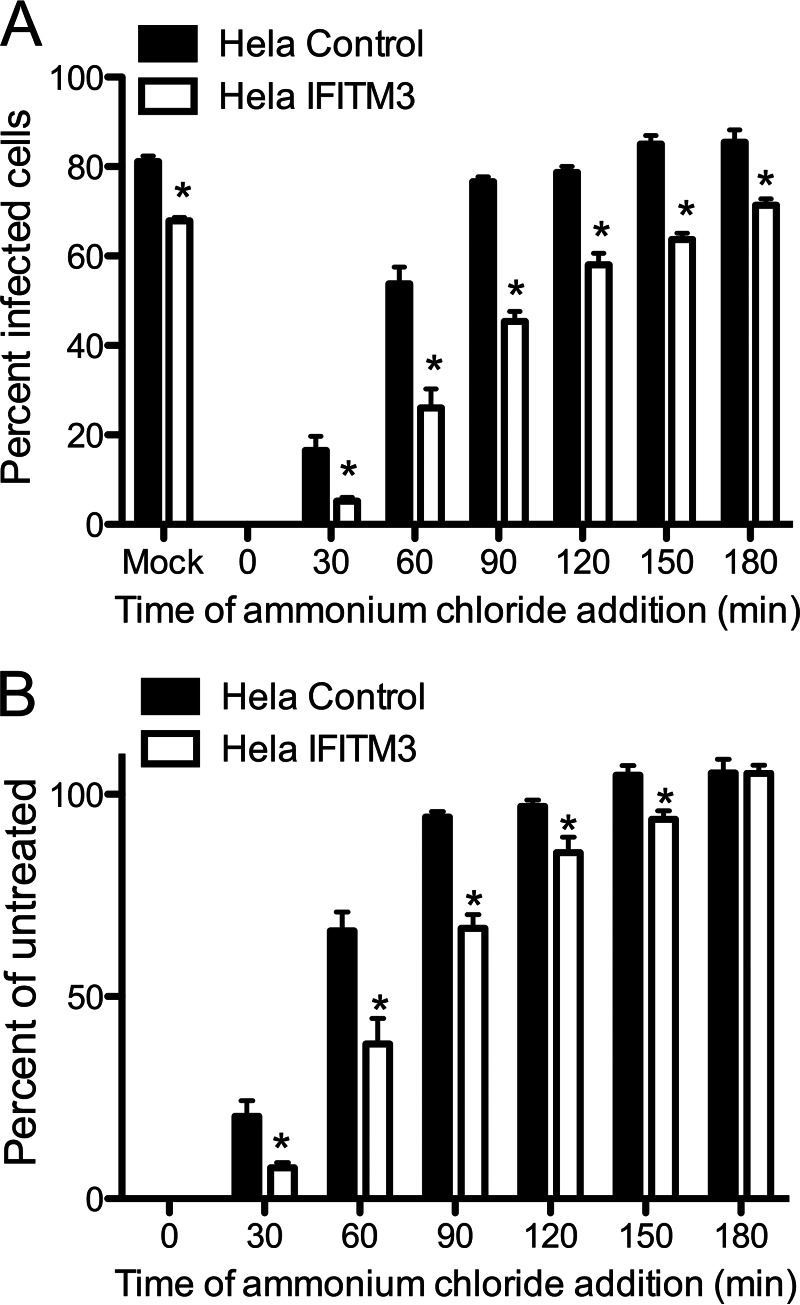
Because endosomal targeting is not affected, we hypothesized that IFITM3 alters the efficiency and/or timing of virion disassembly. To better gauge the efficiency of entry events, we took several approaches. First, we determined whether IFITM3 alters the kinetics with which reovirus particles escape host cell endosomes using an ammonium chloride bypass assay. This assay is predicated upon the observation that reovirus disassembly is prevented by inhibiting the pH decrease required for efficient cathepsin cleavage, whereas intact particles are still resident in endosomes (21, 40). We adsorbed monolayers of control- and IFITM3-expressing HeLa cells with rsT3D at 4 °C for 1 h, washed the cells with PBS, and added prewarmed DMEM. Cells were incubated at 37 °C and ammonium chloride was added at various intervals to prevent endosome acidification. Cells were incubated overnight and scored for the percentage of infected cells by indirect immunofluorescence (Fig. 7). rsT3D escaped ammonium chloride blockade around 30 min earlier (p < 0.05) in control cells than in IFITM3-expressing cells, which indicates that IFITM3 delays the kinetics of endosomal escape.
FIGURE 7.
IFITM3 expression delays the kinetics of reovirus escape from ammonium chloride treatment. A, HeLa vector control- or IFITM3-expressing cells were adsorbed with rsT3D at an m.o.i. of 25 pfu/cell for 1 h at 4 °C. Cells were washed 3 times with PBS and incubated with prewarmed media at 37 °C. At the times shown after adsorption, ammonium chloride was added to a final concentration of 25 mm. The percentage of infected cells was determined by fluorescent focus assay 18 h post-infection. The results are presented as the percentage of cells infected for triplicate samples. B, the data shown in panel A were normalized to the percentage of infected cells in untreated wells. Error bars indicate S.D. *, p < 0.05 by Student's t test versus vector control cells.
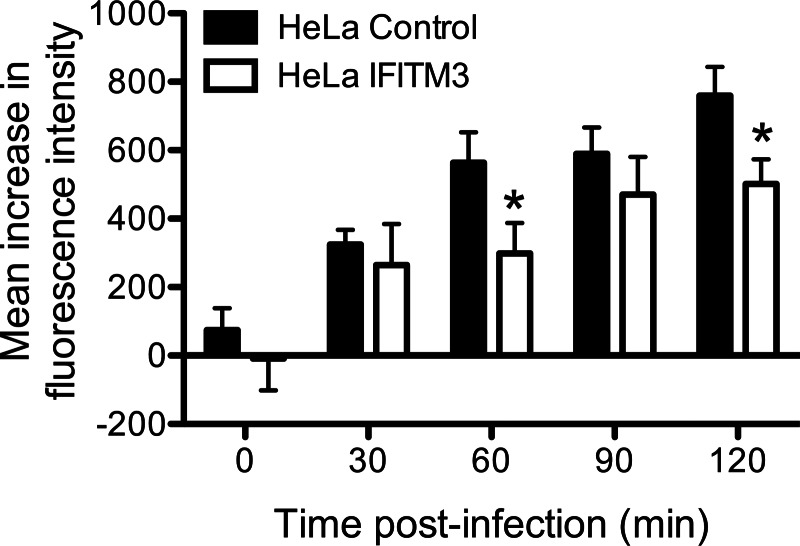
Next, we determined the kinetics of reovirus particles entering into acidic endosomal compartments. Virus particles were labeled with pHrodo, a fluorophore that fluoresces at a pH less than 5. Control- or IFITM3-expressing HeLa cells were chilled for 1 h at 4 °C, adsorbed with pHrodo-labeled particles for 1 h at 4 °C, incubated in prewarmed media at 37 °C for various times, and then assayed for fluorescence intensity by flow cytometry. Fluorescence intensity increased in both control- and IFITM3-expressing cells over time; however, the kinetics of increase in fluorescence intensity was slower in the presence of IFITM3 (p < 0.05; Fig. 8). Because colocalization between virus particles and late endosomes occurred with similar kinetics in control- and IFITM3-expressing cells, these data suggest that IFITM3 may function to alter the rate of acidification of late endosomal compartments.
FIGURE 8.
IFITM3 delays the kinetics of reovirus particle entry into acidified endosomal compartments. HeLa vector control- or IFITM3-expressing cells were chilled at 4 °C for 1 h and adsorbed with 10,000 particles/cell of reovirus-pHrodo at 4 °C for 1 h. The inoculum was removed, the unbound virus was washed away, and the cells were supplemented with prewarmed complete medium and incubated at 37 °C for the times shown. The cells were analyzed by flow cytometry. The data are shown as the mean fluorescence intensity from triplicate samples. Error bars indicate S.D. *, p < 0.05 by Student's t test in comparison with vector control cells.
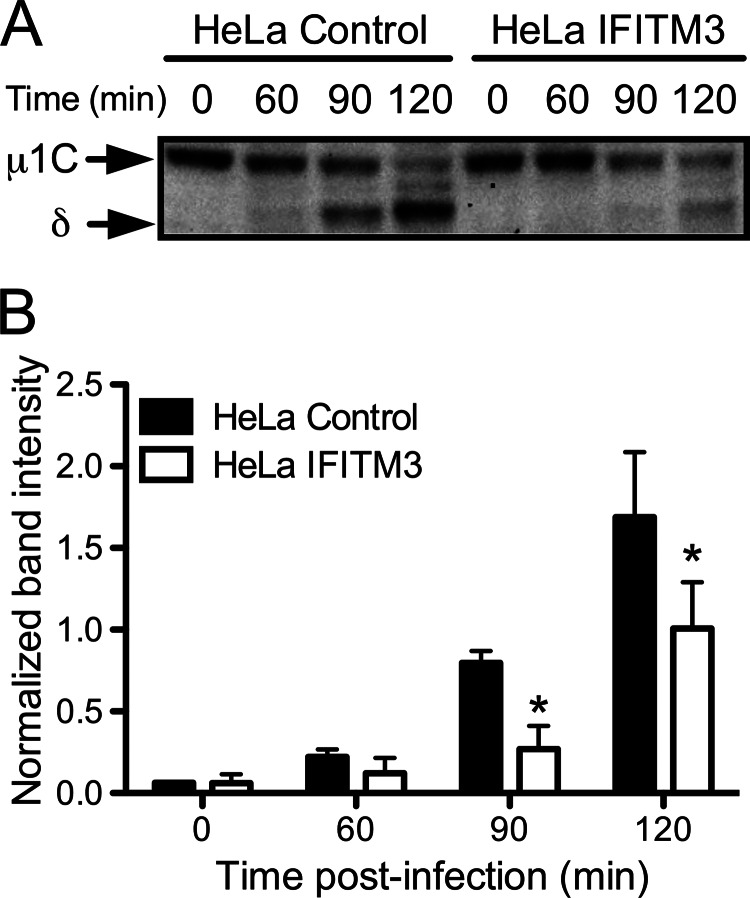
Finally, we examined the kinetics of outer capsid proteolysis in control- and IFITM3-expressing HeLa cells. Cells were adsorbed with virions for 1 h at 4 °C and then incubated at 37 °C for 0, 60, 90, or 120 min. At these time points, cells were lysed in radioimmunoprecipitation buffer, and the lysates were analyzed by SDS-PAGE electrophoresis followed by immunoblotting using a T3D-specific antiserum. Cells expressing IFITM3 exhibited delayed cleavage of μ1 into μ1C and δ in comparison to control cells (Fig. 9), with initial cleavage products detected in control cells by 60 min post-infection and in IFITM3-expressing cells by 90 min post-infection. Together with data regarding endosome acidification and the kinetics of ammonium chloride bypass, these data indicate that IFITM3 restricts reovirus entry by altering the efficiency of endosomal acidification and subsequent protease activity, which serves to alter the kinetics of outer capsid proteolysis and membrane penetration.
FIGURE 9.
IFITM3 expression delays reovirus outer capsid cleavage. A, HeLa vector control- or IFITM3-expressing cells were adsorbed with T3D at an m.o.i. of 100 pfu/cell for 1 h at 4 °C. Cells were washed 2 times with PBS and incubated with prewarmed media at 37 °C. At the times shown after adsorption, cells were scraped into PBS, pelleted at 3000 × g, and resuspended in radioimmunoprecipitation buffer. Lysates were clarified by centrifugation at 13,000 × g, resolved in 4–12% polyacrylamide gels, and transferred to PVDF membranes. The membranes were probed with a rabbit polyclonal anti-T3D antiserum and an anti-rabbit AP-conjugated secondary antibody and visualized using chemiluminescence. B, bands corresponding to μ1C and δ from three individual experiments were quantified using ImageJ software and normalized using nonspecific background bands. *, p < 0.05 by Student's t test in comparison with vector control cells.
DISCUSSION
The type I IFN system works to restrict viral infection through many mechanisms. Requirements shared by many viruses in their replication cycle provide advantageous targets for cellular antiviral mechanisms, as a single protein or family of proteins may prove efficacious against a large number of potential pathogens. Cell entry represents a very attractive target, as all viruses require some mechanism to penetrate into a host cell either at the cell surface, via endosomes, or via other cell fusion or cell transport events. However, because viruses use a wide variety of different receptors, targeting receptor binding or adhesion may not be effective against a broad range of pathogens. In contrast, many viruses, both enveloped and non-enveloped, require the use of endosomal compartments either for proteolytic cleavage or pH-dependent conformational changes necessary for membrane fusion or penetration (41, 42). Hence, evolving the capacity to prevent viruses from penetrating into the cytoplasm from endosomes represents a key antiviral response. The IFITM proteins, and in particular IFITM3, fulfill this function by restricting endosomal viral entry mechanisms.
IFITM3 is a 133-amino acid membrane-associated protein, and its expression is increased 4–40-fold by type I IFN (Refs. 11, 43, and 44; Fig. 1). The topology of IFITM3 in membranes is not fully understood. It was originally characterized as having two transmembrane domains, with the N and C termini facing the lumen of the ER and endosomes (10, 11, 46). However, recent evidence suggests that it may be an intramembrane protein, with the protein termini oriented toward the cytoplasm (47). IFITM3 is S-palmitoylated (47, 48), and palmitoylation enhances the antiviral activity of the protein. Recent mechanistic data indicate that IFITM3 may localize to endosomes containing the vacuolar ATPase proton pump ATP6v0b, leading to alterations in the endosomal compartments including increased association with clathrin (49).
The precise mechanism of action for IFITM3 is not fully understood. Several possible mechanisms have been proposed previously. IFN treatment or IFITM3 expression expands late endosomal and autolysosomal compartments (14, 49), indicative of changes in the activity of these structures. These changes could alter the rate of acidification of endosomes before delivery of the components to lysosomes, influence cathepsin activity (either due to changes in the pH or by directly affecting catalytic activity), or modify endosomal membrane characteristics, such as elasticity or curvature, to prevent membrane fusion. The latter hypothesis was strengthened by recent evidence that IFITM3 alters membrane fluidity to prevent hemifusion mediated by viral fusion proteins (50).
Our data indicate that IFITM3 acts in late endosomes to restrict reovirus cell entry. Although differences in m.o.i. and detection mechanisms make direct comparisons difficult, the level of restriction mediated by IFITM3 against reovirus is similar to that observed for enveloped viruses, including influenza (11), HIV-1 (51), and West Nile Virus (13). Reovirus particles colocalize with late endosomal markers with the same kinetics in the presence or absence of IFITM3, indicating that IFITM3 does not alter entry into endosomal compartments. Rather, particles access an acidic environment later in IFITM3-expressing cells, suggesting that IFITM3 alters the kinetics of endosomal acidification. This is consistent with the observation that ISVPs are not sensitive to IFITM3; ISVPs require endosomes for entry but do not require endosomal acidification (21, 52). Thus, uncoating is delayed, and virions may be delivered to lysosomes or other degradative compartments before they can complete membrane penetration. Accordingly, a previous study indicated that particles that do not efficiently penetrate endosomes localize to lysosomes (23). We cannot exclude the possibility that IFITM3 also alters the properties of endosomal membranes to prevent membrane penetration. Although reovirus does not possess a fusion protein like enveloped viruses, it does form pores in membranes to facilitate entry, and alterations to membrane curvature, elasticity, or bending modulus may alter the dynamics of pore formation and prevent entry.
We observed that IFITM3 expression accounts for between 60 and 80% of the decrease in the percentage of reovirus-infected cells mediated by IFN-α treatment. The fluorescent focus assay used for this experiment primarily assesses how many cells become productively infected, and not other aspects of the viral replication cycle, such as levels of RNA or protein synthesis. Therefore, these data suggest that other ISGs, including possibly other members of the IFITM protein family, may restrict reovirus entry. Other ISGs, including MxA (53) and PKR (protein kinase R) (54, 55) can restrict reovirus infection but primarily act on post-entry events such as protein synthesis. The relative efficacy of each individual ISG in restricting reovirus replication remains to be determined.
Inhibiting expression of IFITM3 after IFN treatment had a relatively greater effect in allowing T3D infection in comparison to T1L. Therefore, IFITM3 could at least partially account for the known differences in IFN sensitivity between T3D and T1L (56, 57). T3D ISVPs are less stable than T1L ISVPs (58) and, therefore, may be more sensitive to changes in the endosomal compartment during uncoating. This phenotype is linked to the M2 gene, particularly a Ala/Val change at position 305 in the T3D μ1 protein (59, 60). This residue is located in the δ region of μ1, which forms the central region of the protein and undergoes conformational changes during uncoating (23). Whether alterations in this region of the protein also affect sensitivity to IFITM3 is unknown.
Perhaps the most intriguing implication of our investigation is that restriction by IFITM3 is not limited to enveloped viruses that penetrate via endosomes but may include nonenveloped viruses with similar requirements for entry as well. Many nonenveloped viruses require access to endosomal compartments for membrane penetration, including parvoviruses (61, 62), adenoviruses (63, 64), and some rotaviruses (65). However, the precise endosomal dynamics required for these viruses to penetrate membranes differ. Rhesus rotavirus only requires early endosomes for cell entry (66), whereas adenovirus and canine parvovirus require acid-dependent activities in late endosomes (45, 62, 67, 68). Thus, determining the full range of viruses that are sensitive to IFITM3 expression may provide further insight into its precise mechanism and location of action and provide useful information in developing broad-spectrum antiviral therapies.
The IFITM protein family, and IFITM3 in particular, represent an important interferon-inducible antiviral response. Recent reports demonstrating that IFITM3 limits the pathogenesis of influenza infection in mice (15, 16) and in humans (16) highlight the relevance of this pathway for innate immunity. Our data suggest that IFITM3 may play a significant role in modulating the pathogenesis of nonenveloped viruses as well, which could inform novel therapies for an expanded range of clinically relevant pathogens.
Acknowledgments
We thank members of our laboratories for many helpful discussions and Walter Atwood, Karl Boehme, Jenna Daly, Pranav Danthi, Terence Dermody, Alexis Griffiths, Jonathan Knowlton, Bernardo Mainou, John Parker, Andrea Pruijssers, Lauren Richardson, and Yevangelina Rybak for technical assistance, reagents, and advice. The confocal microscope was purchased with support from National Science Foundation Grant DBI-0923310.
This work was supported, in whole or in part, by National Institutes of Health Public Health Service Awards R15 AI094440 (to G. H. H.) and R01 AI091786 (to A. L. B.). This work was also supported by the Charles H. Hood Foundation, the Bill and Melinda Gates Foundation, the Phillip T. and Susan M. Ragon Foundation, and the University of Massachusetts Medical School Center for AIDS Research. Additional support was provided by the Colgate University Core Incentive Program and Student Diversity Research Initiative.
- ISG
- interferon-stimulated gene
- IFITM
- interferon-inducible transmembrane protein
- shIFITM3
- short hairpin RNA targeting IFITM3
- T1L
- reovirus type 1 Lang
- T3D
- reovirus type 3 Dearing
- rsT3D
- T3D reovirus strain
- ISVP
- infectious subvirion particle
- EGFP
- enhanced GFP
- RILP
- Rab-interacting lysosomal protein
- m.o.i.
- multiplicity(s) of infection
- pfu
- plaque forming units
- qPCR
- quantitative reverse transcriptase PCR
- A546
- Alexa Fluor 546
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1. Mogensen T. H. (2009) Pathogen recognition and inflammatory signaling in innate immune defenses. Clin. Micro. Rev. 22, 240–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jensen S., Thomsen A. R. (2012) Sensing of RNA viruses. A review of innate immune receptors involved in recognizing RNA virus invasion. J. Virol. 86, 2900–2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Borden E. C., Williams B. R. (2011) Interferon-stimulated genes and their protein products. what and how? J. Interferon Cytokine Res. 31, 1–4 [DOI] [PubMed] [Google Scholar]
- 4. Farrell P. J., Balkow K., Hunt T., Jackson R. J., Trachsel H. (1977) Phosphorylation of initiation factor eIF-2 and the control of reticulocyte protein synthesis. Cell 11, 187–200 [DOI] [PubMed] [Google Scholar]
- 5. Ratner L., Wiegand R. C., Farrell P. J., Sen G. C., Cabrer B., Lengyel P. (1978) Interferon, double-stranded RNA, and RNA degradation. Fractionation of the endonuclease INT system into two macromolecular components. Role of a small molecule in nuclease activation. Biochem. Biophys. Res. Commun. 81, 947–954 [DOI] [PubMed] [Google Scholar]
- 6. Taylor R. T., Lubick K. J., Robertson S. J., Broughton J. P., Bloom M. E., Bresnahan W. A., Best S. M. (2011) TRIM79α, an interferon-stimulated gene product, restricts tick-borne encephalitis virus replication by degrading the viral RNA polymerase. Cell Host Microbe 10, 185–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Evans S. S., Collea R. P., Leasure J. A., Lee D. B. (1993) IFN-α induces homotypic adhesion and Leu-13 expression in human B lymphoid cells. J. Immunol. 150, 736–747 [PubMed] [Google Scholar]
- 8. Imai T., Yoshie O. (1993) C33 antigen and M38 antigen recognized by monoclonal antibodies inhibitory to syncytium formation by human T cell leukemia virus type 1 are both members of the transmembrane 4 superfamily and associate with each other and with CD4 or CD8 in T cells. J. Immunol. 151, 6470–6481 [PubMed] [Google Scholar]
- 9. Lange U. C., Saitou M., Western P. S., Barton S. C., Surani M. A. (2003) The fragilis interferon-inducible gene family of transmembrane proteins is associated with germ cell specification in mice. BMC Dev. Biol. 3, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Smith R. A., Young J., Weis J. J., Weis J. H. (2006) Expression of the mouse fragilis gene products in immune cells and association with receptor signaling complexes. Genes Immun. 7, 113–121 [DOI] [PubMed] [Google Scholar]
- 11. Brass A. L., Huang I.-C., Benita Y., John S. P., Krishnan M. N., Feeley E. M., Ryan B. J., Weyer J. L., van der Weyden L., Fikrig E., Adams D. J., Xavier R. J., Farzan M., Elledge S. J. (2009) The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139, 1243–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huang I.-C., Bailey C. C., Weyer J. L., Radoshitzky S. R., Becker M. M., Chiang J. J., Brass A. L., Ahmed A. A, Chi X., Dong L., Longobardi L. E., Boltz D., Kuhn J. H., Elledge S. J., Bavari S., Denison M. R., Choe H., Farzan M. (2011) Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Pathog. 7, e1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jiang D., Weidner J. M., Qing M., Pan X.-B., Guo H., Xu C., Zhang X., Birk A., Chang J., Shi P.-Y., Block T. M., Guo J.-T. (2010) Identification of five interferon-induced cellular proteins that inhibit west nile virus and dengue virus infections. J. Virol. 84, 8332–8341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Feeley E. M., Sims J. S., John S. P., Chin C. R., Pertel T., Chen L.-M., Gaiha G. D., Ryan B. J., Donis R. O., Elledge S. J., Brass A. L. (2011) IFITM3 inhibits influenza A virus infection by preventing cytosolic entry. PLoS Pathog. 7, e1002337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bailey C. C., Huang I.-C., Kam C., Farzan M. (2012) Ifitm3 limits the severity of acute influenza in mice. PLoS Pathogens 8, e1002909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Everitt A. R., Clare S., Pertel T., John S. P., Wash R. S., Smith S. E., Chin C. R., Feeley E. M., Sims J. S., Adams D. J., Wise H. M., Kane L., Goulding D., Digard P., Anttila V., Baillie J. K., Walsh T. S., Hume D. A., Palotie A., Xue Y., Colonna V., Tyler-Smith C., Dunning J., Gordon S. B., GenISIS Investigators, MOSAIC Investigators, Smyth R. L., Openshaw P. J., Dougan G., Brass A. L., Kellam P. (2012) IFITM3 restricts the morbidity and mortality associated with influenza. Nature 484, 519–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schiff L. A., Nibert M. L., Tyler K. L. in Field's Virology (Knipe D. M., Howley P. M., eds.) 5th Ed., pp. 1853–1915, Lippincott Williams and Wilkins ET, Philadelphia [Google Scholar]
- 18. Barton E. S., Forrest J. C., Connolly J. L., Chappell J. D., Liu Y., Schnell F. J., Nusrat A., Parkos C. A., Dermody T. S. (2001) Junction adhesion molecule is a receptor for reovirus. Cell 104, 441–451 [DOI] [PubMed] [Google Scholar]
- 19. Maginnis M. S., Forrest J. C., Kopecky-Bromberg S. A., Dickeson S. K., Santoro S. A., Zutter M. M., Nemerow G. R., Bergelson J. M., Dermody T. S. (2006) β1 integrin mediates internalization of mammalian reovirus. J. Virol. 80, 2760–2770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ebert D. H., Deussing J., Peters C., Dermody T. S. (2002) Cathepsin L and cathepsin B mediate reovirus disassembly in murine fibroblast cells. J. Biol. Chem. 277, 24609–24617 [DOI] [PubMed] [Google Scholar]
- 21. Sturzenbecker L. J., Nibert M., Furlong D., Fields B. N. (1987) Intracellular digestion of reovirus particles requires a low pH and is an essential step in the viral infectious cycle. J. Virol. 61, 2351–2361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chandran K., Farsetta D. L., Nibert M. L. (2002) Strategy for nonenveloped virus entry. A hydrophobic conformer of the reovirus membrane penetration protein m1 mediates membrane disruption. J. Virol. 76, 9920–9933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chandran K., Parker J. S., Ehrlich M., Kirchhausen T., Nibert M. L. (2003) The delta region of outer-capsid protein m1 undergoes conformational change and release from reovirus particles during cell entry. J. Virol. 77, 13361–13375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nibert M. L., Odegard A. L., Agosto M. A., Chandran K., Schiff L. A. (2005) Putative autocleavage of reovirus m1 protein in concert with outer-capsid disassembly and activation for membrane permeabilization. J. Mol. Biol. 345, 461–474 [DOI] [PubMed] [Google Scholar]
- 25. Odegard A. L., Chandran K., Zhang X., Parker J. S., Baker T. S., Nibert M. L. (2004) Putative autocleavage of outer capsid protein m1, allowing release of myristoylated peptide μ1N during particle uncoating, is critical for cell entry by reovirus. J. Virol. 78, 8732–8745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nibert M. L., Fields B. N. (1992) A carboxyl-terminal fragment of protein m1/m1C is present in infectious subvirion particles of mammalian reoviruses and is proposed to have a role in penetration. J. Virol. 66, 6408–6418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mainou B. A., Dermody T. S. (2012) Transport to late endosomes is required for efficient reovirus infection. J. Virol. 86, 8346–8358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kobayashi T., Ooms L. S., Ikizler M., Chappell J. D., Dermody T. S. (2010) An improved reverse genetics system for mammalian orthoreoviruses. Virology 398, 194–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kobayashi T., Antar A. A., Boehme K. W., Danthi P., Eby E. A., Guglielmi K. M., Holm G. H., Johnson E. M., Maginnis M. S., Naik S., Skelton W. B., Wetzel J. D., Wilson G. J., Chappell J. D., Dermody T. S. (2007) A plasmid-based reverse genetics system for animal double-stranded RNA viruses. Cell Host Microbe 1, 147–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Furlong D. B., Nibert M. L., Fields B. N. (1988) Sigma 1 protein of mammalian reoviruses extends from the surfaces of viral particles. J. Virol. 62, 246–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Smith R. E., Zweerink H. J., Joklik W. K. (1969) Polypeptide components of virions, top component and cores of reovirus type 3. Virology 39, 791–810 [DOI] [PubMed] [Google Scholar]
- 32. Virgin H. W., 4th, Bassel-Duby R., Fields B. N., Tyler K. L. (1988) Antibody protects against lethal infection with the neurally spreading reovirus type 3 (Dearing). J. Virol. 62, 4594–4604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Connolly J. L., Dermody T. S. (2002) Virion disassembly is required for apoptosis induced by reovirus. J. Virol. 76, 1632–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fecek R. J., Busch R., Lin H., Pal K., Cunningham C. A., Cuff C. F. (2006) Production of Alexa Fluor 488-labeled reovirus and characterization of target cell binding, competence, and immunogenicity of labeled virions. J. Immunol. Methods 314, 30–37 [DOI] [PubMed] [Google Scholar]
- 35. Gauntt C. J. (1973) Induction of interferon in L cells by reoviruses. Infect. Immun. 7, 711–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Holm G. H., Zurney J., Tumilasci V., Leveille S., Danthi P., Hiscott J., Sherry B., Dermody T. S. (2007) Retinoic acid-inducible gene-I and interferon-β promoter stimulator-1 augment proapoptotic responses after mammalian reovirus infection via interferon regulatory factor-3. J. Biol. Chem. 282, 21953–21961 [DOI] [PubMed] [Google Scholar]
- 37. Maginnis M. S., Mainou B. A., Derdowski A., Johnson E. M., Zent R., Dermody T. S. (2008) NPXY motifs in the β1 integrin cytoplasmic tail are required for functional reovirus entry. J. Virol. 82, 3181–3191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jordens I., Fernandez-Borja M., Marsman M., Dusseljee S., Janssen L., Calafat J., Janssen H., Wubbolts R., Neefjes J. (2001) The Rab7 effector protein RILP controls lysosomal transport by inducing the recruitment of dynein-dynactin motors. Curr. Biol. 11, 1680–1685 [DOI] [PubMed] [Google Scholar]
- 39. Barbero P., Bittova L., Pfeffer S. R. (2002) Visualization of Rab9-mediated vesicle transport from endosomes to the trans-Golgi in living cells. J. Cell Biol. 156, 511–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dermody T. S., Nibert M. L., Wetzel J. D., Tong X., Fields B. N. (1993) Cells and viruses with mutations affecting viral entry are selected during persistent infections of L cells with mammalian reoviruses. J. Virol. 67, 2055–2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tsai B. (2007) Penetration of nonenveloped viruses into the cytoplasm. Annu. Rev. Cell Dev. Biol. 23, 23–43 [DOI] [PubMed] [Google Scholar]
- 42. Harrison S. C. (2008) Viral membrane fusion. Nat. Struct. Mol. Biol. 15, 690–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Friedman R. L., Manly S. P., McMahon M., Kerr I. M., Stark G. R. (1984) Transcriptional and posttranscriptional regulation of interferon-induced gene expression in human cells. Cell 38, 745–755 [DOI] [PubMed] [Google Scholar]
- 44. Lewin A. R., Reid L. E., McMahon M., Stark G. R., Kerr I. M. (1991) Molecular analysis of a human interferon-inducible gene family. Eur. J. Biochem. 199, 417–423 [DOI] [PubMed] [Google Scholar]
- 45. Seth P. (1994) Mechanism of adenovirus-mediated endosome lysis. role of the intact adenovirus capsid structure. Biochem. Biophys. Res. Commun. 205, 1318–1324 [DOI] [PubMed] [Google Scholar]
- 46. Weidner J. M., Jiang D., Pan X.-B., Chang J., Block T. M., Guo J.-T. (2010) Interferon-induced cell membrane proteins, IFITM3 and tetherin, inhibit vesicular stomatitis virus infection via distinct mechanisms. J. Virol. 84, 12646–12657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yount J. S., Karssemeijer R. A., Hang H. C. (2012) S-Palmitoylation and ubiquitination differentially regulate interferon-induced transmembrane protein 3 (IFITM3)-mediated resistance to influenza virus. J. Biol. Chem. 287, 19631–19641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yount J. S., Moltedo B., Yang Y.-Y., Charron G., Moran T. M., López C. B., Hang H. C. (2010) Palmitoylome profiling reveals S-palmitoylation-dependent antiviral activity of IFITM3. Nat. Chem. Biol. 6, 610–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wee Y. S., Roundy K. M., Weis J. J., Weis J. H. (2012) Interferon-inducible transmembrane proteins of the innate immune response act as membrane organizers by influencing clathrin and v-ATPase localization and function. Innate immun. 18, 834–845 [DOI] [PubMed] [Google Scholar]
- 50. Li K., Markosyan R. M., Zheng Y.-M., Golfetto O., Bungart B., Li M., Ding S., He Y., Liang C., Lee J. C., Gratton E., Cohen F. S., Liu S.-L. (2013) IFITM proteins restrict viral membrane hemifusion. PLoS Pathog. 9, e1003124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lu J., Pan Q., Rong L., He W., Liu S.-L., Liang C. (2011) The IFITM proteins inhibit HIV-1 infection. J. Virol. 85, 2126–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Schulz W. L., Haj A. K., Schiff L. A. (2012) Reovirus uses multiple endocytic pathways for cell entry. J. Virol. 86, 12665–12675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mundt E. (2007) Human MxA protein confers resistance to double-stranded RNA viruses of two virus families. J. Gen. Virol. 88, 1319–1323 [DOI] [PubMed] [Google Scholar]
- 54. Stewart M. J., Blum M. A., Sherry B. (2003) PKR's protective role in viral myocarditis. Virology 314, 92–100 [DOI] [PubMed] [Google Scholar]
- 55. Nilsen T. W., Maroney P. A., Baglioni C. (1982) Inhibition of protein synthesis in reovirus-infected HeLa cells with elevated levels of interferon-induced protein kinase activity. J. Biol. Chem. 257, 14593–14596 [PubMed] [Google Scholar]
- 56. Jacobs B. L., Ferguson R. E. (1991) The Lang strain of reovirus serotype 1 and the Dearing strain of reovirus serotype 3 differ in their sensitivities to b interferon. J. Virol. 65, 5102–5104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sherry B., Torres J., Blum M. A. (1998) Reovirus induction of and sensitivity to β interferon in cardiac myocyte cultures correlate with induction of myocarditis and are determined by viral core proteins. J. Virol. 72, 1314–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Middleton J. K., Agosto M. A., Severson T. F., Yin J., Nibert M. L. (2007) Thermostabilizing mutations in reovirus outer-capsid protein m1 selected by heat inactivation of infectious subvirion particles. Virology 361, 412–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sarkar P., Danthi P. (2010) Determinants of strain-specific differences in efficiency of reovirus entry. J. Virol. 84, 12723–12732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Madren J. A., Sarkar P., Danthi P. (2012) Cell entry-associated conformational changes in reovirus particles are controlled by host protease activity. J. Virol. 86, 3466–3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Parker J. S., Murphy W. J., Wang D., O'Brien S. J., Parrish C. R. (2001) Canine and feline parvoviruses can use human or feline transferrin receptors to bind, enter, and infect cells. J. Virol. 75, 3896–3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Parker J. S., Parrish C. R. (2000) Cellular uptake and infection by canine parvovirus involves rapid dynamin-regulated clathrin-mediated endocytosis, followed by slower intracellular trafficking. J. Virol. 74, 1919–1930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Chardonnet Y., Dales S. (1970) Early events in the interaction of adenoviruses with HeLa cells. II. Comparative observations on the penetration of types 1, 5, 7, and 12. Virology 40, 478–485 [DOI] [PubMed] [Google Scholar]
- 64. FitzGerald D. J., Trowbridge I. S., Pastan I., Willingham M. C. (1983) Enhancement of toxicity of antitransferrin receptor antibody-Pseudomonas exotoxin conjugates by adenovirus. Proc. Natl. Acad. Sci. U.S.A. 80, 4134–4138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wolf M., Vo P. T., Greenberg H. B. (2011) Rhesus rotavirus entry into a polarized epithelium is endocytosis-dependent and involves sequential VP4 conformational changes. J. Virol. 85, 2492–2503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wolf M., Deal E. M., Greenberg H. B. (2012) Rhesus rotavirus trafficking during entry into MA104 cells is restricted to the early endosome compartment. J. Virol. 86, 4009–4013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Blumenthal R., Seth P., Willingham M. C., Pastan I. (1986) pH-dependent lysis of liposomes by adenovirus. Biochemistry 25, 2231–2237 [DOI] [PubMed] [Google Scholar]
- 68. Greber U. F., Willetts M., Webster P., Helenius A. (1993) Stepwise dismantling of adenovirus 2 during entry into cells. Cell 75, 477–486 [DOI] [PubMed] [Google Scholar]