Background: Regulation of Kir2.1 by WT caveolin 3 and LQT9-causing mutants of caveolin3 is unknown.
Results: WT caveolin 3 and mutants of caveolin 3 associate with Kir2.1. Caveolin 3 mutants reduce Kir2.1 current density. Surface expression of Kir2.1 is decreased by caveolin 3 mutations.
Conclusion: Caveolin 3 mutations affect Kir2.1 current density by decreasing cell surface expression of Kir2.1.
Significance: Kir2.1 loss of function may contribute to the mechanism of arrhythmia generation in caveolin 3-mediated LQT9.
Keywords: Cardiomyopathy; Caveolin; Electrophysiology; Ion Channels; Potassium Channels; Long QT Syndrome (LQTS); Arrhythmias, Basic Studies
Abstract
Mutations in CAV3 cause LQT syndrome 9 (LQT9). A previously reported LQT9 patient had prominent U waves on ECG, a feature that has been correlated with Kir2.1 loss of function. Our objective was to determine whether caveolin 3 (Cav3) associates with Kir2.1 and whether LQT9-associated CAV3 mutations affect the biophysical properties of Kir2.1. Kir2.1 current (IK1) density was measured using the whole-cell voltage clamp technique. WT-Cav3 did not affect IK1. However, F97C-Cav3 and T78M-Cav3 decreased IK1 density significantly by ∼60%, and P104L-Cav3 decreased IK1 density significantly by ∼30% at −60 mV. Immunostained rat heart cryosections and HEK293 cells cotransfected with Kir2.1 and WT-Cav3 both demonstrated colocalization of Kir2.1 and WT-Cav3 by confocal imaging. Cav3 coimmunoprecipitated with Kir2.1 in human ventricular myocytes and in heterologous expression systems. Additionally, FRET efficiency was highly specific, with a molecular distance of 5.6 ± 0.4 nm, indicating close protein location. Colocalization experiments found that Cav3 and Kir2.1 accumulated in the Golgi compartment. On-cell Western blot analysis showed decreased Kir2.1 cell surface expression by 60% when expressed with F97C-Cav3 and by 20% when expressed with P104L-Cav3 compared with WT-Cav3. This is the first report of an association between Cav3 and Kir2.1. The Cav3 mutations F97C-Cav3, P104L-Cav3, and T78M-Cav3 decreased IK1 density significantly. This effect was related to a reduced cell surface expression of Kir2.1. Kir2.1 loss of function is additive to the increase described previously in late INa, prolonging repolarization and leading to arrhythmia generation in Cav3-mediated LQT9.
Introduction
Long QT Syndrome (LQTS)3 is characterized by a prolongation of the QT interval on the electrocardiogram in individuals with structurally normal hearts, predisposing them to polymorphic ventricular tachycardia, syncope, and sudden death. Ion channel dysfunction is thought to underlie the pathogenesis of LQTS, either directly from abnormal ion channel function or indirectly through abnormal accessory protein function, and is classified as LQT1–13 according to genetically identified abnormalities (1).
The caveolin family of proteins assists in formation of specialized lipid domains or rafts called caveolae. Three different caveolin genes have been identified (CAV1, CAV2, and CAV3). CAV3 encodes caveolin 3 (Cav3), which is exclusively expressed in heart and skeletal muscle (2). Functionally, Cav3 is both an anchoring protein for molecules within caveolae and a regulatory element for protein signaling activity (3). Mutations in CAV3 have been clinically linked to autosomal dominant skeletal muscle diseases and cardiac disease (4). More recently, our group and colleagues showed that CAV3 mutations are associated with the LQT syndrome phenotype (5). From 905 unrelated LQTS subjects, six non-synonymous nucleotide changes in highly conserved residues, resulting in the discovery of four heterozygous missense mutations (T78M, A85T, F97C, and S141R). These CAV3 mutants, caused a pathologic increase in late sodium current (late INa), which is known to prolong the action potential and cause LQT syndrome (5). On the basis of those results, the syndrome is now referred to as LQT9. However, because Cav3 is known to associate with other cardiac ion channels, we hypothesized that CAV3 mutations are likely to affect other ion currents in addition to late INa.
In the heart, IK1 is thought to be predominantly carried by channels composed of Kir2.1, encoded by KCNJ2 (6, 7). IK1 maintains the resting membrane potential close to the potassium equilibrium potential and contributes to final phase 3 repolarization (8). Mutations in KCNJ2 resulting in a loss of Kir2.1 function are implicated in Andersen-Tawil syndrome (9), also referred to as LQT7. By ECG, loss of Kir2.1 function can cause a prolonged QTu with a dominant U wave with only mild QTc prolongation (9, 10). Kir2.1 has been shown to associate with a variety of scaffolding proteins and signaling molecules, such as SAP97, syntrophin, β 2-adrenergic receptor, and G proteins (11, 12). Nevertheless, the macromolecular complex associated with Kir2.1 in the heart is not known, and, to date, no β subunits have been identified (7). We hypothesized that Cav3 and Kir2.1 may be part of a macromolecular signaling complex. Our interest was heightened by the observation of a prominent U wave in the ECG of a LQT9 patient (T78M-Cav3) (5), suggestive of possible IK1 involvement in this syndrome. In this manuscript, we have explored the association, functional consequences, and mechanisms of LQT9 CAV3 mutations and WT-CAV3 with the inward rectifier potassium channel (Kir2.1).
EXPERIMENTAL PROCEDURES
Ethical Approval
All experiments were performed in accordance with the Institutional Animal Care and Use Committee at the University of Wisconsin, Madison and conform to National Institutes of Health guidelines. Experiments using human tissue were approved by the institutional review board, University of Wisconsin, Madison (protocol number 2011-0194).
Cav3 and KCNJ2 Construction and Mutagenesis
The WT human CAV3 456-bp coding sequence was cloned from human cardiac cDNA as reported (5). The human KCNJ2 clone was created previously and reported in this laboratory (13). Site-directed mutagenesis was performed with the QuikChange site-directed mutagenesis kit (Stratagene) using the vector containing the WT-Cav3 inserts as a template. The primers used for mutagenesis are available on request. PCR and bacterial transformation were performed according to the instructions of the manufacturer.
Cell Culture and Transfections
HEK293 or Cos1 cells were cultured in DMEM (Invitrogen) with 10% FBS. For electrophysiological experiments, HEK293 cells were transiently transfected using FuGENE 6 (Promega) according to the specifications of the manufacturer. For each set of transfections, an equal amount of WT and/or mutant Cav3 (Pires-EGFP, Clontech, Palo Alto, CA) along with hKir2.1 (Clontech) were used. For on-cell Western blot analysis, immunoprecipitation, and cell surface localization of Kir2.1, cells were cotransfected with HA-Kir2.1 (a gift from Dr. A. George) and WT-Cav3 or F97C-Cav3 or T78M-Cav3 or P104L-Cav3. Kir2.1-NT-GFP was constructed as described previously by Eckhardt et al. (13). The Cav3-mCherry construct was a gift from Dr. T. Kamp.
Immunostaining
Immunostaining of tissue and single cells was performed as described previously (28). The rabbit anti-Kir2.1 antibody was generated by using a peptide sequence (388–401, DDSENGVPESTSTD) from the C terminus of hKir2.1. The antibodies were raised in rabbits and purified as described previously (15). Results from in-house anti-Kir2.1 antibody were confirmed by using commercially available Santa Cruz rabbit anti-Kir2.1 antibody (data not shown), which was shown previously to detect Kir2.1 in rat tissue (28). Other antibodies included mouse anti-Cav3 antibody (BD Biosciences), rabbit anti-HA (Applied Biological Materials, Inc.) and mouse anti-HA (Covance). For cell surface expression measurement, cells were fixed with 4% paraformaldehyde in PBS but not permeabilized. To detect extra versus intracellular Kir2.1, fixed cells were treated with mouse anti-HA antibody, washed with PBS, and then permeabilized and incubated with rabbit anti-HA antibody. Slides were washed with PBS with 0.1% Tween 20 (PBS-T) and then incubated in goat secondary antibodies conjugated to Alexa Fluor 488 and/or Alexa Fluor 568 and/or Alexa Fluor 660 (Invitrogen). DAPI staining was used to detect the nucleus. Samples were then washed in PBS-T and mounted using a ProLong Gold anti-fade mounting kit (Invitrogen). Fluorescence was analyzed, and images were acquired on a Nikon A1 or Leica SP5 confocal microscope. ImageJ software was used to investigate colocalization. Analysis by measuring Pearson's correlation coefficient was performed. Tissue from at least three different rat hearts was sectioned and analyzed to confirm results.
Coimmunoprecipitation
Human tissue samples or transfected Cos1 cells were homogenized in radioimmune precipitation assay buffer containing 25 mm Tris-HCl (pH 7.4), 150 mm NaCl, 60 mm n-octyl-D-glucoside, 1% Triton X-100, 2 mm PMSF, and a protease inhibitor tablet/50 ml. The homogenate was centrifuged to remove insoluble debris. 400 μg of supernatant was used for each reaction and was precleared with protein A/G-Sepharose beads (Pierce). The precleared supernatant was incubated in primary antibody overnight at 4 °C, and protein-antibody complexes were recovered using recombinant protein A/G-Sepharose (Pierce). As a negative control, isotype control IgG was used. Cell lysates were separated on precast 10–20% SDS-PAGE gels and immunoblotted as described previously (28).
FRET
The acceptor photobleaching approach was performed as described previously (16). Briefly, images were acquired using the Leica confocal scanning system (Exton, PA) TCS SP5 outfitted with a Leica inverted microscope, an argon laser, and ×60 1.4 numerical aperture plan apochromatic oil immersion objective. EGFP and mCherry fluorescence were excited with 488- and 561-nm laser light, respectively. A dichroic (DD488/561) mirror in the excitation path was used for dual excitation. The emissions for EGFP and mCherry were collected using 492- to 505-nm and 570- to 696-nm band pass windows, respectively. Live samples were imaged within 48 h after transfection. The regions of interest in cells expressing both fluorophores were photobleached using 561-nm laser light at 100% intensity. EGFP and mCherry images were taken both before and after acceptor photobleaching. FRET efficiency (E) was calculated using Leica software: E = Dpost-Dpre/Dpost, where Dpre and Dpost are EGFP emissions before and after regional photobleaching. To calculate the distance between molecules (r) we used the following formula: Fret efficiency = (ro)6/((ro)6+(r)6) where ro = 5.24.
Electrophysiology
Membrane current recordings were carried out 24 h after transfection using the ruptured patch whole cell technique (17) at room temperature and recorded with an Axiopatch-200B amplifier. Borcillica glass pipettes were pulled to resistances of 2–4 MΩ. Cells were identified by GFP fluorescence under fluorescent microscopy (Nikon). The bath solution contained 140 mm NaCl, 5.4 mm KCl, 1.8 mm CaCl2, 0.5 mm MgCl2, 5 mm HEPES, 0.33 mm NaH2PO4, 5.5 mm D-glucose, and NaOH (pH 7.4). The pipette solution contained 30 mm KCl, 85 mm K aspartate, 5 mm MgCl2, 10 mm KH2PO4, 2 mm K2EGTA, 2 mm K2ATP, 5 mm HEPES, and KOH (pH 7.2). Control experiments in the WT were done contemporaneously with the mutations. From a holding potential of −60 mV, voltages were applied from −120 to 30 mV in 10-mV increments for 200 ms. Data were filtered at 10 kHz and digitized using a Digidata 1200 (Axon Instruments). Analysis of data was done using pClamp10 (Axon Instruments) and Origin 7 software (OriginLab Corp.).
On-cell-Western Blot Analysis
Monolayers of HEK293 cells transiently cotransfected with HA-Kir2.1 and WT-Cav3 or F97C-Cav3 were fixed with 4% paraformaldehyde 22–24 h after plating. Cells were washed with 1× PBS and then blocked with 5% normal goat serum made in 1× PBS for 1 h and then incubated with mouse anti-HA antibody for 90 min. Cells were incubated in IRDye 680 goat anti-mouse secondary antibody for 1 h. Cells marked as control were only incubated with secondary antibodies and no primary antibody. Cells were imaged according to the specification of the manufacturer on the Li-Cor Odyssey infrared imager (Li-Cor Biosciences, Lincoln, NE). Integrated intensities of the 700-nm infrared signal for each well were calculated using the Li-Cor odyssey infrared imaging system software. Background intensity was subtracted from the experiment, and results are reported as normalized data (18).
Statistical Analysis
Data are expressed as mean ± S.E. unless specified otherwise. Data were analyzed using either an unpaired Student's t test or analysis of variance using Microsoft Excel. Values of p ≤ 0.05 were considered significant.
RESULTS
Cav3 Mutations Decreased Kir2.1 Current Density Significantly
We studied the effect on IK1 of three different mutations in the membrane domain of Cav3: LQT9-causing mutations F97C-Cav3 and T78M-Cav3, and P104L-Cav3 causing limb-girdle muscular dystrophy. No endogenous caveolins have been found in HEK293 or COS1 cells. Thus, they were chosen for our modeling system. Representative IK1 traces are illustrated in Fig. 1, left panel, Kir2.1 alone (top panel), with WT-Cav3 (center panel), or with F97C-Cav3 (bottom panel). Summary data of the Kir2.1 current-voltage relationships (IV) are shown in the right panel. Kir2.1 channels expressed by itself (■), coexpressed with WT-Cav3 (●), F97C-Cav3 (♦), T78N-Cav3 (▾) and P104L-Cav3 (▴). IV relationships from all three Cav3 mutations compared with Kir2.1 alone or with WT-Cav3 showed a significant decrease in IK1 density. The average peak inward current at −120 mV was −97.44 ± 20.99 pA/pF (n = 9) for F97C-Cav3, −98.46 ± 8.04 pA/pF (n = 6) for T78M-Cav3, and −157.25 ± 14.28 pA/pF (n = 11) for P104L-Cav3, whereas compared with WT-Cav3 and Kir2.1 alone, the peak inward current was −235.68 ± 22.97 pA/pF (n = 12) and −237.32 ± 19.51 pA/pF (n = 9), respectively. The average peak outward current at −60 mV for F97C-Cav3 was −0.42 ± 1.30 pA/pF (n = 9), whereas T78M-Cav3 was 0.31 ± 1.80 pA/pF (n = 6) and P104L-Cav3 was 1.77 ± 0.81 pA/pF (n = 11) as compared with WT-Cav3, which was 9.12 ± 2.23 pA/pF (n = 12), and Kir2.1 alone, which was 7.37 ± 2.30 pA/pF (n = 9). We have shown a significant, ∼60% decrease in Kir2.1 current density with F97C-Cav3 (-120 mV, p < 0.001 and −60 mV, p < 0.01) and T78M-Cav3 (-120 mV, p < 0.001 and − 60 mV, p = 0.05) when compared with Kir2.1 expressed by itself and Kir2.1 coexpressed with WT-Cav3. P104L-Cav3 caused a significant, 30% (-120 mV, p < 0.01 and −60 mV, p = 0.02) decrease in IK1 density compared with Kir2.1 expressed by itself and Kir2.1 coexpressed with WT-Cav3.
FIGURE 1.
Cav3 mutants decrease Kir2. 1 current density significantly compared with Kir2.1 cotransfected with vector (■) only or WT-Cav3 (●). Representative Kir2.1 current density traces are illustrated in the left panel in the absence (top) or presence of WT-Cav3 (center), or F97C-Cav3 (bottom). Scale bar = 50 ms and 50 pA/pF. The dotted line indicates 0 pA. Summary data of the current-voltage relationships of Kir2.1 channels expressed by itself (■, n = 9) and coexpressed with WT-Cav3 (●, n = 12), F97C-Cav3 (♦, n = 9), T78N-Cav3 (▾, n = 6), and P104L-Cav3 (▴, n = 11) are shown in the right panel. The current-voltage relationship from all three mutations of Cav3, but not WT-Cav3, showed a significant decrease in Kir2.1 current density. *, p ≤ 0.05). The inset highlights the current at depolarized potentials.
Kir2.1 and Cav3 Coimmunolocalize in Ventricular Cells and Coimmunoprecipitate in Human Ventricular Myocytes and Heterologous Cells
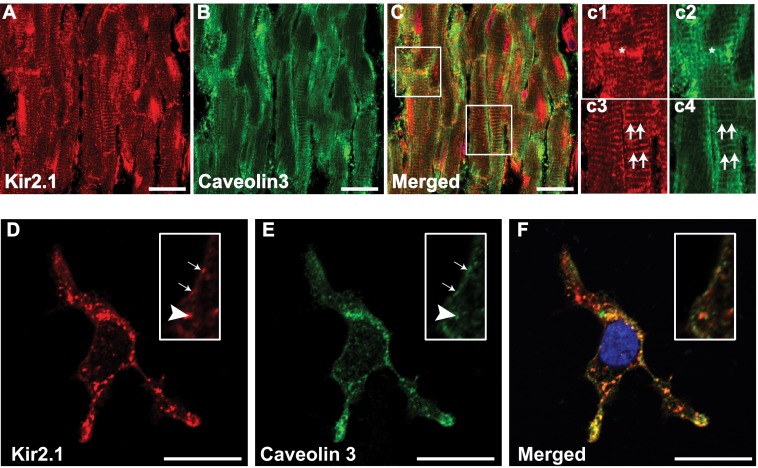
Immunostaining of rat cardiac ventricular cryosections (Fig. 2, A--C) and HEK cells (D–F) was performed. Kir2.1 (Fig. 2, A and D, red) and Cav3 (B and E, green) coimmunolocalize as demonstrated in the merged images (C and F). Fig. 2C, c1–c4, and the insets in D--F are higher magnifications of the areas identified as boxes in C, D, E, and F, respectively. Colocalizations at cellular locations are indicated at intercalated discs by asterisks and at T-tubules by double arrows in Fig. 2C, c1–c4. Two different Kir2.1 and Cav3 antibodies confirmed these results (data not shown). Colocalization was analyzed by Pearson's correlation coefficient (significance of interaction is at ≥ 0.5). For rat cardiac tissue, the coefficient was 0.76 ± 0.03 (three hearts, n = 10), and for HEK293 cells it was 0.87 ± 0.03 (n = 7).
FIGURE 2.
Coimmunolocalization of Kir2. 1 and Caveolin3. Shown is coimmunolocalization of Kir2.1 (A, c1, c3, and D, red) and Cav3 (B, c2, c4, and E, green) in rat cardiac ventricular tissue (A–C) and heterologous cells (D–F). c1–c4 and the insets in D–F are higher magnifications of the areas identified with boxes. Double arrows (c3 and c4) denote T-tubular staining, asterisks (c1 and c2) denote an intercalated disk, single arrows denote membrane localization, and arrowheads represent a punctuate pattern of staining. Nuclei (C and F) are stained blue with DAPI. Scale bar = 20 μm.
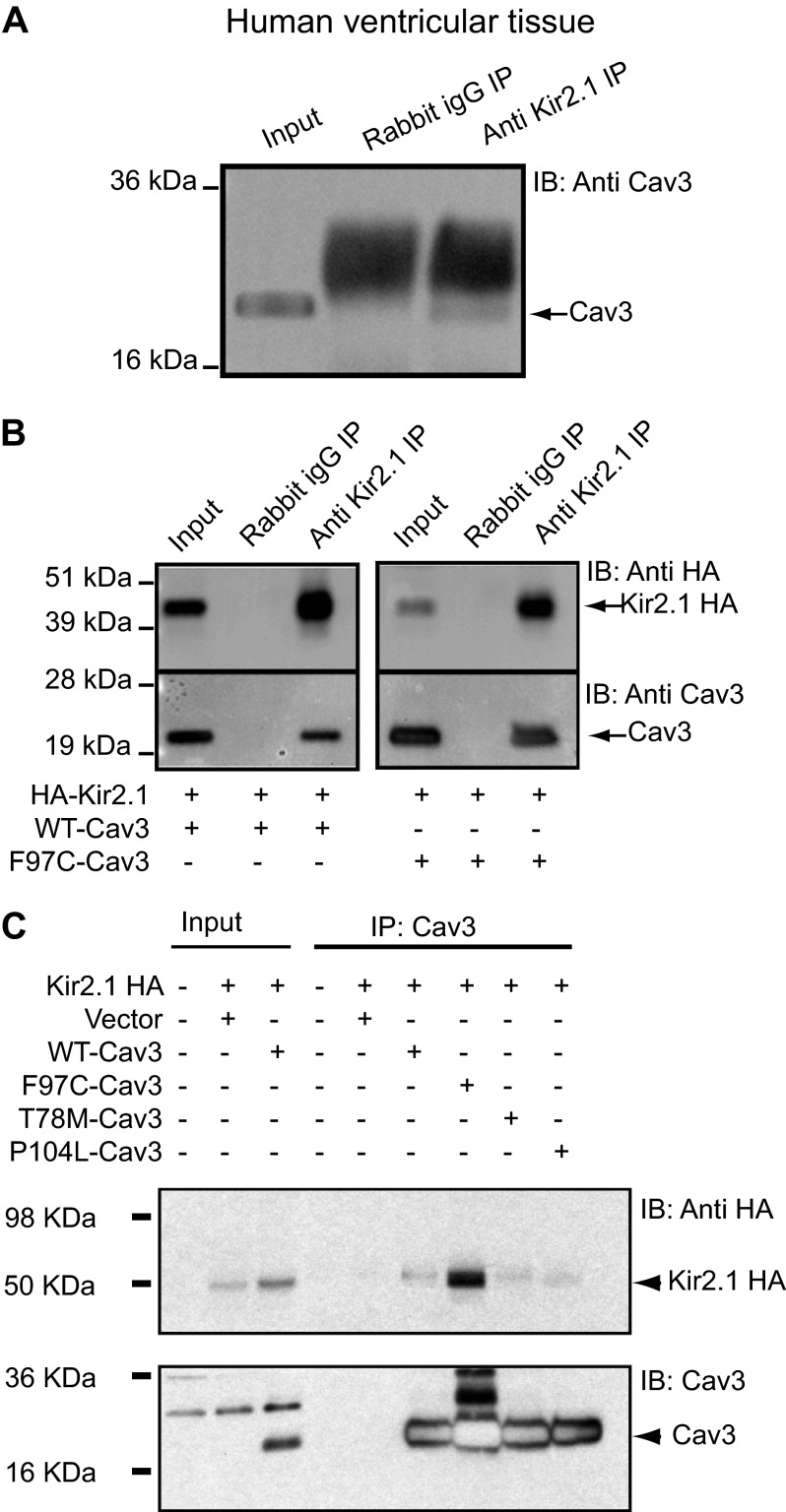
The immunoblot analysis of a coimmunoprecipitation experiment shows that, in human ventricular lysate, Kir2.1 and Cav3 associate, directly or indirectly (Fig. 3A). Anti-Kir2.1 antibody was used to immunoprecipitate Cav3. Rabbit IgG was used as a negative control. COS1 cells, like HEK293 cells, do not express any endogenous caveolins. Thus, we performed the next experiments in COS1 cells to determine whether proteins specific to HEK293 cells (used for the electrophysiology experiments) mediated the association between Cav3 and Kir2.1 or not. Coimmunoprecipitation and Western blot analysis of COS-1 cells coexpressing extracellular hemagglutinin-Kir2.1 (HA-Kir2.1) and WT-Cav3 or F97C-Cav3 are shown in Fig. 3B. Cell lysates were incubated with anti-Kir2.1 antibody or rabbit IgG and analyzed by Western blot analysis with anti-HA antibody (upper panel) or anti-Cav3 antibody (lower panel), Fig. 3B). Rabbit IgG was used as a negative control. Kir2.1 and WT-Cav3 and F97C-Cav3 associate as indicated by arrows. We also investigated coimmunoprecipitation of T78M-Cav3, P104L-Cav3, and F97C-Cav3 with Kir2.1 in HEK293 cells. Fig. 3C demonstrates that all three tested mutants of Cav3 coimmunoprecipitate with Kir2.1. Cav3 antibody failed to pull down Kir2.1 when the vector alone was used as a negative control. The reverse experiment was also performed by immunoprecipitating HA-Kir2.1 with HA antibody and blotting for Cav3 (data not shown). We had similar results with all of the Cav3 mutations, so that all of the mutations in Cav3 associate with Kir2.1. The darker appearance of the Cav3 band for the F97C is related to an increased plasmid protein expression (a variable in heterologous expression) rather than a true differential biophysical relationship. We verified that this was because of variable plasmid expression by performing repeat coimmunoprecipitation and Western blot experiments using tubulin as an internal control of protein expression (data not shown).
FIGURE 3.
Kir2.1 coimmunoprecipitates with WT-Cav3 and Cav3 mutants. Kir2.1 coimmunoprecipitates with WT-Cav3 in human ventricular tissue lysate (A) and in Cos1 cells transiently cotransfected with HA-Kir2.1 and WT-Cav3 (B, left panel) or F97C-Cav3 (B, right panel). IB, immunoblot. Anti-Kir2.1 antibody was used to immunoprecipitate (IP) WT-Cav3 in A and B, left panel, and Cav3F97C (right panel). Rabbit IgG was used as a negative control and failed to immunoprecipitate Cav3. Tissue and cell lysates were used as positive controls. C, WT-Cav3, F97C-Cav3, T78M-Cav3, and P104L-Cav3 were immunoprecipitated with Cav3 antibody (reciprocal immunoprecipitation) and blotted for Kir2.1. Lysate from untransfected cells or cells transfected with Kir2.1 and vector were used as negative controls.
Cav3 Association with Kir2.1 Revealed by FRET
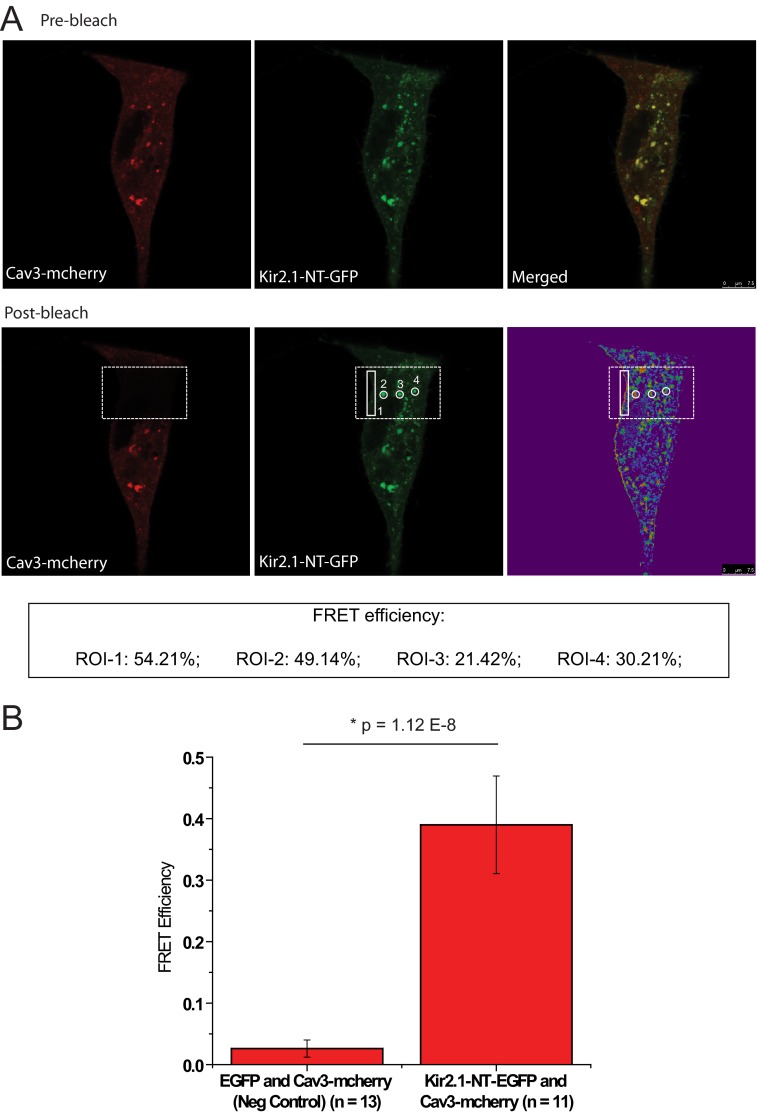
Given that channels localize in caveolar domains, we designed the FRET experiments to have Cav3 as the acceptor and Kir2.1 as the donor. Fluorophores were placed strategically at the N-terminal of both Kir2.1 and Cav3, with Kir2.1 fused to EGFP (donor) and Cav3 fused to mCherry (acceptor). HEK293 cells were transfected with these two constructs, and only cells expressing both the fluorophores were included. Fig. 4A illustrates a representative FRET experiment. Live, prebleached cells expressing both fluorophores were scanned sequentially (Fig. 4A, top panel). Following acceptor bleaching in the area identified by the box, the same cell was scanned again sequentially, and FRET efficiency was calculated as described under “Experimental Procedures” in identified regions of interest. Regions of interest were selected at the edge of the cell and also at punctuate patterns on the cell, as identified previously (Fig. 2, D–F). FRET efficiency is reported in the inset in Fig. 4A. The bar graph in Fig. 4B exhibits summary data from multiple control experiments, where EGFP transfected along with Cav3-mCherry demonstrated no significant FRET efficiency, whereas Cav3-mCherry and Kir2.1-EGFP demonstrated significant FRET efficiency. From the average FRET efficiency, we calculated the distance between the fluorophores and, thus, a measure of distance between the proteins of interest. Using the formula depicted under “Experimental Procedures,” with FRET efficiency as 0.40 and ro as 5.24, the distance between proteins (r) is 5.6 ± 0.4 nm. These results confirm our previous observation of association between Cav3 and Kir2.1 by immunostaining and coimmunoprecipitation (Figs. 2 and 3).
FIGURE 4.
Association between Kir2. 1 and Cav3 identified by FRET. A, representative FRET experiment where HEK293 cells were dual-transfected with Kir2.1-EGFP and Cav3-mCherry and scanned before (top panel, prebleach) and after (bottom panel, post-bleach) photo bleaching of the acceptor. The region identified by the large rectangular box represents the area of photobleach. FRET efficiency (E) was calculated in the regions of interest (ROI) identified in the images. The inset reveals the FRET efficiency of the regions of interest. Scale bar = 7.5 μm. B, the bar graph describes the summary data from multiple FRET experiments, showing that the average FRET efficiency is ∼40% as compared with negative control, which showed negligible or no FRET efficiency. The equation described under “Experimental Procedures” was used to calculate the distance between the molecules, which is 5.6 ± 0.4 nm.
Cav3 Mutants Are Retained in the Golgi Compartment
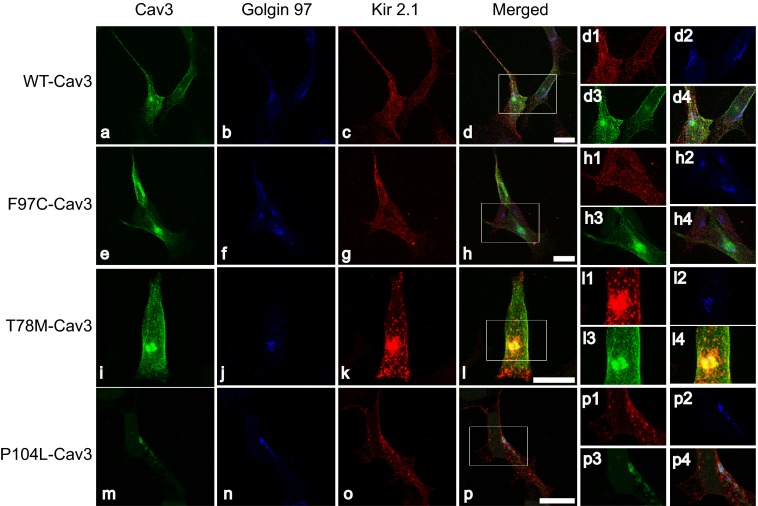
We have now shown that Cav3 and Kir2 associate (directly or indirectly) and that Cav3 mutations have functional consequences on Kir2.1 current density. In this section, we investigated the intracellular localization of Cav3 and Kir2.1 in a heterologous expression system. Initial experiments suggested that LQT9 Cav3 mutations caused Kir2.1 accumulation in intracellular compartments (data not shown). P104L-Cav3 has been shown previously to accumulate in the Golgi (19). Hence we investigated if the other two Cav3 mutations, F97C-Cav3 and T78M-Cav3, also accumulate in the Golgi, along with Kir2.1. Fig. 5, c, g, k, and o, illustrates immunostained HEK293 cells cotransfected with Kir2.1 and WT-Cav3 (a) or F97C-Cav3 (e) or T78M-Cav3 (i) or P104L-Cav3 (m). WT-Cav3 did not accumulate in the Golgi as identified by Golgin97 antibody (Fig. 5, b, f, j, and n), in contrast to the tested Cav3 mutants along with Kir2.1. The data from Fig. 5 suggest that Cav3 mutants affect cell surface expression of Kir2.1 channels. Interestingly, we saw some P104L-Cav3 at the membrane evident in Fig. 5m.
FIGURE 5.
Cav3 mutants accumulate in the Golgi compartment of HEK293 cells in the presence of Kir2. 1. Shown is the accumulation of Cav3 mutants and Kir2.1 in the Golgi compartment of HEK293 cells. Representative photomicrographs of HEK293 cells cotransfected with WT-Cav3 (a), F97C-Cav3 (e), T78M-Cav3 (i), P104L-Cav3 (m), and HA-Kir2.1 (c, g, k, and o), which were triple-stained to detect Kir2.1 (c, g, k, and o, red), Cav3 (a, e, i, and m, green) and Golgin97 (b, f, j, and n, blue). Merged images in H, L, and P suggest that there is accumulation of Cav3 mutants and Kir2.1 in the Golgi as compared with WT-Cav3 and Kir2.1 (d), which do not seem to accumulate in the Golgi. Scale bars = 20 μm. The insets show a magnification of the region highlighted by the white box in the adjacent merged panels.
Cav3 Mutants Affect Kir2.1 Surface Membrane Expression
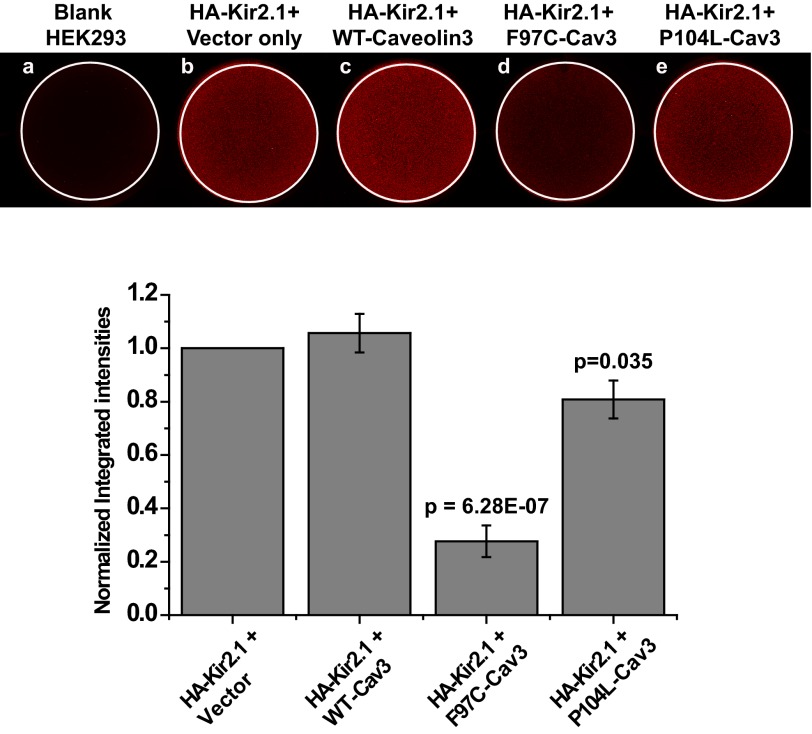
The data in Fig. 5 indicate that the mechanism of the effect of Cav3 mutants on Kir2.1 current density is related to a decrease in the surface expression. We evaluated this observation by analyzing the cell surface expression of Kir2.1 by on-cell Western blotting technique using transiently transfected HEK293 cells with HA-tagged Kir2.1 (HA-Kir2.1) and WT-Cav3/F97C-Cav3. Fig. 6A is a representative on-cell Western experiment illustrating the intensity levels of HA-Kir2.1 surface expression in cells cotransfected with either HA-Kir2.1 and vector (top panel, center), HA-Kir2.1 and WT-Cav3 (top panel, right), HA-Kir2.1 and F97C-Cav3 (bottom panel, left), or HA-Kir2.1 and P104L-Cav3 (bottom panel, right). Blank HEK cells (top panel, left) were used as negative control. F97C-Cav3 decreased the intensity level of Kir2.1 significantly by ∼60% as compared with positive control experiments (p ≪ 0.001, n = 3; Fig. 6B). P104L-Cav3 had a ∼20% decrease as compared with control experiment (p = 0.035, n = 3).
FIGURE 6.
Cav3 mutants affect Kir2. 1 current density by significantly reducing surface expression of Kir2.1 channels. A, representative on-cell Western blot analysis illustrating intensity levels of extracellular HA-tagged Kir2.1 surface expression in wells cotransfected with HA-Kir2.1 and vector only (b), WT-Cav3 (c), F97C-Cav3 (d), and P104L-Cav3 (e). a shows untreated cells stained with both primary and secondary antibody. B, the graph displays quantitative results of three independent experiments.
DISCUSSION
In this study, we present the first report of a Kir2.1 and Cav3 association and the pathological consequences of this interaction. As demonstrated by multiple complimentary techniques, our data showed a physical correlation between Kir2.1 and Cav3 in human ventricular myocytes, rat ventricular myocytes, and by heterologous expression by using coimmunoprecipitation, immunostaining, and FRET. We investigated the pathophysiological effect of this association by studying CAV3 mutations that cause LQT9 (T78M and F97C) and, for comparison, a CAV3 mutation, P104L, associated with limb girdle muscular dystrophy. These mutations induced decreased Kir2.1 current density, compared with Kir2.1, when expressed alone or when coexpressed with WT-Cav3. Taken together, our data support a relevant association and pathophysiologic interaction between Kir2.1 and Cav3. Given the importance of Kir2.1 in normal cardiac physiology, loss of Kir2.1 function in the presence of LQT9 CAV3 mutations is additive to the sodium current in the pathological effects, resulting in LQTS.
Association of Cav3 and Kir2.1
We hypothesized that Cav3 may be a channel-interacting protein of Kir2.1 because of a prominent U wave noted on the ECG of a LQT9 patient with a T78M CAV3 mutation. Prominent U waves are a feature described in patients with LQT7/Anderson's syndrome because of KCNJ2 mutations causing loss of function in Kir2.1 channels (10). Given that LQT9 patients had tested negative for KCNJ2 or other ion channel mutations, we speculated that mutations in Cav3 could also affect IK1. The first step to answer the above question was to investigate whether these two proteins colocalize and/or associate with each other. In Fig. 2, we demonstrate that Cav3 and Kir2.1 colocalize in native tissue and in a heterologous expression system as shown by immunostaining. WT-Cav3 as well as Cav3 mutations coimmunoprecipitate with Kir2.1 in human tissue lysate and by heterologous expression (Fig. 3). We complemented this data by performing FRET on the fluorophore-tagged constructs of Cav3 and Kir2.1. Our FRET results illustrated that Kir2.1 and Cav3 are in close proximity, indicating an association between proteins. These results demonstrate that the effect of Cav3 mutants on Kir2.1 is by association and not because of a lack or loss of association.
Functional Effect of CAV3 Mutations and Kir2.1
To test the hypothesis that the Cav3 and Kir2.1 physical association has a functional consequence, we investigated the coexpression of Kir2.1 with WT-Cav3 and LQT9 Cav3 mutations. As shown in Fig. 1, our results demonstrate that although Kir2.1 does not depend on the presence of WT-Cav3, Cav3 mutations decreased Kir2.1 current density significantly as compared with WT-Cav3. This finding broadens the scope of arrhythmia generation related to loss of IK1, to include not only Kir2.1 protein abnormalities but also interactions with channel-interacting and regulatory proteins such as Cav3. The complement of components in the Kir2.1 macromolecular complex is not fully known but is important, given that interactions with accessory intracellular proteins influence trafficking and function (20). Kir2.1 interaction with SAP97 may be important for targeting newly synthesized Kir2 proteins to specific microdomains, such as caveolar lipid rafts (12). The relationship between Cav3 and Kir2.1 is a crucial step in understanding the components of the Kir2.1 macromolecular complex, regulation, and modification.
Tetramers of Kir2.1 make up the predominant component of IK1 (6). However, Kir2.2 and Kir2.3 also contribute to IK1 and can create homotetramers or heterotramers with another Kir2.X (21). It is possible that Cav3 associates with Kir2.2 or Kir2.3. This association, as well as the functional effects, are part of our ongoing study.
Mechanism of the Effect of LQT9 Cav3 Mutations on Kir2.1
In our study, Cav3 mutants and Kir2.1 had a different cellular localization pattern compared with Kir2.1 with WT-Cav3. Cav3 mutants and Kir2.1 accumulate in the Golgi compartment, as identified by Golgin-97. This was not surprising for P104L-Cav3 because it has been published previously that P104L-Cav3 is trafficking-defective and accumulates in the Golgi compartment of cells (19). We chose to include this particular mutation in our study to serve as a positive control with respect to trafficking and cell surface expression. However, the LQT9 CAV3 mutations are thought to express and exhibit normal membrane trafficking (22). It was surprising that F97C-Cav3 accumulated intracellularly in the Golgi, suggesting that the major effect of Cav3 on Kir2.1 is due to its surface membrane expression. We investigated this further utilizing on-cell Western blotting (Fig. 6). F97C-Cav3 showed an ∼60% decrease in cell surface expression, and P104L-Cav3 decreased cell surface expression of Kir2.1 by ∼30%. This percentage change in surface expression is parallel to the decrease in current density (electrophysiology data, Fig. 1).
Cav3 has been shown to associate with nNos, Nedd4-2, and PIP2 (23, 24). Thus, an alternative mechanism by which Cav3 could affect Kir2.1 includes channel nitrosylation, rapid degradation induced by Nedd4-2, or by changing the gating properties by interaction with PIP2. Our results indicate that the effect of Cav3 mutations on current density parallels the effect on channel trafficking. This indicates that the important physiologic mechanism occurs before the channel reaches the membrane, and it is thus unlikely that any of the aforementioned membrane-related mechanisms play a role in modulating the function of Kir2.1. Our group has shown that the sodium channel is nitrosylated in the presence of Cav3 mutations, which increases the late sodium current (5, 21). For Kir2.1, channel nitrosylation occurs at Cys-76 and results in increased Kir2.1 current (25). Thus, if the same mechanism as the sodium channel were applied to Kir2.1, the opposite of our observed effect and of the presumed arrhythmia mechanism for loss of Kir2.1 function would occur.
Kir2.1 Cellular Localization
Our results in Figs. 1, 2, and 5 suggest that there are at least two populations of Kir2 channels in our model cell system: Kir2.1 by itself and Kir2.1 that associates with WT/mutant-Cav3. In addition, on the basis of our results, there are at least two populations of Cav3: Cav3 by itself and Cav3 associated with Kir2.1. This explains why WT-Cav3 does not affect IK1 density. Cav3 is not required for all proper membrane trafficking, but mutations in Cav3 can disrupt normal membrane trafficking, thus causing a decrease in IK1 density. Further, given that F97C-Cav3 and T78M-Cav3 are not trafficking-defective alone, it is safe to speculate that they accumulate in the Golgi only in the presence of Kir2.1. However, P104L-Cav3, which is known to be a trafficking-defective mutant, does not affect IK1 density as drastically as the other mutants. Our data suggest that P104L-Cav3 in association with Kir2.1 is somewhat rescued to the surface membrane (Fig. 5). This is consistent with the clinical presentation of P104L-Cav3 patients who do not suffer from any cardiac ailments and only from limb girdle muscular dystrophy (26). The possibility that P104L could be partially rescued by Kir2.1 requires additional experiments, which are outside the scope of this study.
On the basis of these results and reports from our laboratory and others, we propose that Kir2.1 is part of a “caveolar microdomain” that includes Nav1.5 and three scaffolding proteins (Cav3, SAP97, and syntrophin). This is derived from this work and recent work by Abriel and colleagues (27), who demonstrated that NaV1.5 associates with syntrophin and SAP97, Makielski and colleagues (5, 23), who established that NaV1.5 coimmunoprecipitates with Cav3 and syntrophin, Vandenberg and colleagues, who showed that Kir2.1 coimmunoprecipitates with syntrophin (11), and Anumonwo and colleagues (28), who assert that Kir2.1 associates with SAP97. This is also in agreement with a recent study by Jalife and colleagues (29), who propose reciprocal modulation of Kir2.1 and NaV1.5 and suggest that these ion channels are in the same macromolecular complex. Although these protein interactions have been shown separately, our findings corroborate these studies, and we demonstrate here pathophysiologic consequences of the association of Cav3 with Kir2.1 and NaV1.5.
Clinical Implications
LQTS causes syncope and sudden cardiac death because of a prolongation of the cardiac action potential and development of Early After Depolarizations (EADs), which can subsequently lead to malignant ventricular arrhythmias such as Torsades de Pointes. IK1 outward current contributes to the final phase 3 repolarization (8), and loss of function has been associated with LQT7 (10). In this report, we have shown that loss of Kir2.1 function, instead, relates to an association with a dysfunctional channel-interacting protein, Cav3. We propose that, in addition to an increase in late-INa, the loss of Kir2.1 function may also be responsible for the pathophysiology of LQT9. More broadly, both Kir2.1 and Cav3 have been shown to be down-regulated in heart failure (30–32). Thus, the clinical implications of the association of these two important cardiac proteins may extend beyond more rare inherited arrhythmic conditions.
Summary
Here we have demonstrated for the first time that Cav3 associates with Kir2.1. This association, in addition to sodium channels, is part of the pathologic mechanism of LQT9. Cav3 and Kir2.1 association is an entirely new protein-protein interaction and has important implications for the pathogenesis of LQTS and other disease states.
Limitations
The overexpression of the proteins of interest a heterologous expression system is useful for studying electrophysiologic properties but may not recapitulate a physiological environment and is an inherent limitation. Overexpression of CAV3 mutations in cultured ventricular myocytes was intentionally not utilized because of the problematic Kir2.1 run-down in cultured myocytes. We anticipate that this issue may be ameliorated with the use of induced pluripotent-derived cardiomyocytes with and without Cav3 mutations to investigate our hypotheses. The use of induced pluripotent-derived cardiomyocytes for this study is a current and ongoing project in our laboratory.
Acknowledgments
We thank Dr. Ravi Balijepalli for valuable discussion and caveolin 3 expertise. We also thank Dr. A. George for providing the HA-Kir2.1 construct, Dr. Takushi Kohmoto for supplying the human heart tissue, and Sara Abozeid and Karen Albright for technical assistance.
This work was supported by the University of Wisconsin Cellular and Molecular Arrhythmia Research Program, by American Heart Association National Scientist Development Grant 11SDG5000003 (to L. E.), and by the University of Wisconsin School of Medicine and Public Health.
- LQTS
- long QT syndrome
- EGFP
- enhanced GFP
- pA
- picoamperes
- pF
- picofarads.
REFERENCES
- 1. Schwartz P. J., Crotti L., Insolia R. (2012) Long-QT syndrome. From genetics to management. Circ. Arrhythm. Electrophysiol. 5, 868–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Song K. S., Scherer P. E., Tang Z., Okamoto T., Li S., Chafel M., Chu C., Kohtz D. S., Lisanti M. P. (1996) Expression of caveolin-3 in skeletal, cardiac, and smooth muscle cells. Caveolin-3 is a component of the sarcolemma and co-fractionates with dystrophin and dystrophin-associated glycoproteins. J. Biol. Chem. 271, 15160–15165 [DOI] [PubMed] [Google Scholar]
- 3. Cohen A. W., Hnasko R., Schubert W., Lisanti M. P. (2004) Role of caveolae and caveolins in health and disease. Physiol. Rev. 84, 1341–1379 [DOI] [PubMed] [Google Scholar]
- 4. Balijepalli R. C., Kamp T. J. (2008) Caveolae, ion channels and cardiac arrhythmias. Prog. Biophys. Mol. Biol. 98, 149–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vatta M., Ackerman M. J., Ye B., Makielski J. C., Ughanze E. E., Taylor E. W., Tester D. J., Balijepalli R. C., Foell J. D., Li Z., Kamp T. J., Towbin J. A. (2006) Mutant caveolin-3 induces persistent late sodium current and is associated with long-QT syndrome. Circulation 114, 2104–2112 [DOI] [PubMed] [Google Scholar]
- 6. Leonoudakis D., Conti L. R., Anderson S., Radeke C. M., McGuire L. M., Adams M. E., Froehner S. C., Yates J. R., 3rd, Vandenberg C. A. (2004) Protein trafficking and anchoring complexes revealed by proteomic analysis of inward rectifier potassium channel (Kir2.x)-associated proteins. J. Biol. Chem. 279, 22331–22346 [DOI] [PubMed] [Google Scholar]
- 7. Anumonwo J. M., Lopatin A. N. (2010) Cardiac strong inward rectifier potassium channels. J. Mol. Cell Cardiol. 48, 45–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Miake J., Marbán E., Nuss H. B. (2003) Functional role of inward rectifier current in heart probed by Kir2.1 overexpression and dominant-negative suppression. J. Clin. Invest. 111, 1529–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Plaster N. M., Tawil R., Tristani-Firouzi M., Canún S., Bendahhou S., Tsunoda A., Donaldson M. R., Iannaccone S. T., Brunt E., Barohn R., Clark J., Deymeer F., George A. L., Jr., Fish F. A., Hahn A., Nitu A., Ozdemir C., Serdaroglu P., Subramony S. H., Wolfe G., Fu Y. H., Ptácek L. J. (2001) Mutations in Kir2.1 cause the developmental and episodic electrical phenotypes of Andersen's syndrome. Cell 105, 511–519 [DOI] [PubMed] [Google Scholar]
- 10. Tristani-Firouzi M., Jensen J. L., Donaldson M. R., Sansone V., Meola G., Hahn A., Bendahhou S., Kwiecinski H., Fidzianska A., Plaster N., Fu Y. H., Ptacek L. J., Tawil R. (2002) Functional and clinical characterization of KCNJ2 mutations associated with LQT7 (Andersen syndrome). J. Clin. Invest. 110, 381–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Leonoudakis D., Conti L. R., Radeke C. M., McGuire L. M., Vandenberg C. A. (2004) A multiprotein trafficking complex composed of SAP97, CASK, Veli, and Mint1 is associated with inward rectifier Kir2 potassium channels. J. Biol. Chem. 279, 19051–19063 [DOI] [PubMed] [Google Scholar]
- 12. Leonoudakis D., Mailliard W., Wingerd K., Clegg D., Vandenberg C. (2001) Inward rectifier potassium channel Kir2.2 is associated with synapse-associated protein SAP97. J. Cell Sci. 114, 987–998 [DOI] [PubMed] [Google Scholar]
- 13. Eckhardt L. L., Farley A. L., Rodriguez E., Ruwaldt K., Hammill D., Tester D. J., Ackerman M. J., Makielski J. C. (2007) KCNJ2 mutations in arrhythmia patients referred for LQT testing. A mutation T305A with novel effect on rectification properties. Heart Rhythm 4, 323–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deleted in proof
- 15. Pu J. L., Ye B., Kroboth S. L., McNally E. M., Makielski J. C., Shi N. Q. (2008) Cardiac sulfonylurea receptor short form-based channels confer a glibenclamide-insensitive KATP activity. J. Mol. Cell Cardiol. 44, 188–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sheridan J. T., Worthington E. N., Yu K., Gabriel S. E., Hartzell H. C., Tarran R. (2011) Characterization of the oligomeric structure of the Ca(2+)-activated Cl-channel Ano1/TMEM16A. J. Biol. Chem. 286, 1381–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hamill O. P., Marty A., Neher E., Sakmann B., Sigworth F. J. (1981) Improved patch-clamp techniques for high-resolution current recording from cells and cell-free membrane patches. Pflugers Arch. 391, 85–100 [DOI] [PubMed] [Google Scholar]
- 18. Delisle B. P., Anderson C. L., Balijepalli R. C., Anson B. D., Kamp T. J., January C. T. (2003) Thapsigargin selectively rescues the trafficking defective LQT2 channels G601S and F805C. J. Biol. Chem. 278, 35749–35754 [DOI] [PubMed] [Google Scholar]
- 19. Galbiati F., Volonte D., Minetti C., Chu J. B., Lisanti M. P. (1999) Phenotypic behavior of caveolin-3 mutations that cause autosomal dominant limb girdle muscular dystrophy (LGMD-1C). Retention of LGMD-1C caveolin-3 mutants within the Golgi complex. J. Biol. Chem. 274, 25632–25641 [DOI] [PubMed] [Google Scholar]
- 20. Romanenko V. G., Rothblat G. H., Levitan I. (2002) Modulation of endothelial inward-rectifier K+ current by optical isomers of cholesterol. Biophys. J. 83, 3211–3222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang Z., Yue L., White M., Pelletier G., Nattel S. (1998) Differential distribution of inward rectifier potassium channel transcripts in human atrium versus ventricle. Circulation 98, 2422–2428 [DOI] [PubMed] [Google Scholar]
- 22. Cheng J., PhD, Valdivia C. M., Vaiyanathan R. P., Balejepalli R. P., Ackerman M., Makielski J. M. (2013) J. Mol. Cell. Cardiol., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ueda K., Valdivia C., Medeiros-Domingo A., Tester D. J., Vatta M., Farrugia G., Ackerman M. J., Makielski J. C. (2008) Syntrophin mutation associated with long QT syndrome through activation of the nNOS-SCN5A macromolecular complex. Proc. Natl. Acad. Sci. U.S.A. 105, 9355–9360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cui S., Ho W. K., Kim S. T., Cho H. (2010) Agonist-induced localization of Gq-coupled receptors and G protein-gated inwardly rectifying K+ (GIRK) channels to caveolae determines receptor specificity of phosphatidylinositol 4,5-bisphosphate signaling. J. Biol. Chem. 285, 41732–41739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gómez R., Caballero R., Barana A., Amorós I., Calvo E., López J. A., Klein H., Vaquero M., Osuna L., Atienza F., Almendral J., Pinto A., Tamargo J., Delpón E. (2009) Nitric oxide increases cardiac IK1 by nitrosylation of cysteine 76 of Kir2.1 channels. Circ. Res. 105, 383–392 [DOI] [PubMed] [Google Scholar]
- 26. Couchoux H., Allard B., Legrand C., Jacquemond V., Berthier C. (2007) Loss of caveolin-3 induced by the dystrophy-associated P104L mutation impairs l-type calcium channel function in mouse skeletal muscle cells. J. Physiol. 580, 745–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Petitprez S., Zmoos A. F., Ogrodnik J., Balse E., Raad N., El-Haou S., Albesa M., Bittihn P., Luther S., Lehnart S. E., Hatem S. N., Coulombe A., Abriel H. (2011) SAP97 and dystrophin macromolecular complexes determine two pools of cardiac sodium channels Nav1.5 in cardiomyocytes. Circ. Res. 108, 294–304 [DOI] [PubMed] [Google Scholar]
- 28. Vaidyanathan R., Taffet S. M., Vikstrom K. L., Anumonwo J. M. (2010) Regulation of cardiac inward rectifier potassium current (I(K1)) by synapse-associated protein-97. J. Biol. Chem. 285, 28000–28009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Milstein M. L., Musa H., Balbuena D. P., Anumonwo J. M., Auerbach D. S., Furspan P. B., Hou L., Hu B., Schumacher S. M., Vaidyanathan R., Martens J. R., Jalife J. (2012) Dynamic reciprocity of sodium and potassium channel expression in a macromolecular complex controls cardiac excitability and arrhythmia. Proc. Natl. Acad. Sci. U.S.A. 109, E2134–2143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tomaselli G. F., Zipes D. P. (2004) What causes sudden death in heart failure? Circ. Res. 95, 754–763 [DOI] [PubMed] [Google Scholar]
- 31. Beuckelmann D. J., Näbauer M., Erdmann E. (1993) Alterations of K+ currents in isolated human ventricular myocytes from patients with terminal heart failure. Circ. Res. 73, 379–385 [DOI] [PubMed] [Google Scholar]
- 32. Hare J. M., Lofthouse R. A., Juang G. J., Colman L., Ricker K. M., Kim B., Senzaki H., Cao S., Tunin R. S., Kass D. A. (2000) Contribution of caveolin protein abundance to augmented nitric oxide signaling in conscious dogs with pacing-induced heart failure. Circ. Res. 86, 1085–1092 [DOI] [PubMed] [Google Scholar]