Background: Saccharomyces cerevisiae and Candida albicans use different transcription factors to regulate ribosomal protein transcription.
Results: The Ifh1-Fhl1 heteromer interacts with Tbf1 through the Fhl1 protein and Rap1 through Ifh1.
Conclusion: Despite the rewiring of the DNA-binding module, the protein-protein interactions reconfigure to maintain the response to external signals.
Significance: Studying the rewiring of transcription regulation will help understand the laws underlying evolutionary variation.
Keywords: Candida albicans, Protein Evolution, Protein-Protein Interactions, Transcription Regulation, Yeast Transcription, Rewiring
Abstract
The genes encoding the ribosomal proteins of fungi form a regulon whose expression is enhanced under good growth conditions and down-regulated under starvation conditions. The fungal pathogen Candida albicans contains an evolutionarily ancient control circuit for this regulon where a heteromer made up of the transcription regulators Ifh1 (interacts with Forkhead 1) and Fhl1 (Forkhead-like 1) is targeted to the ribosomal protein genes by the DNA binding factor Tbf1. In the more recently evolved circuit in the model yeast Saccharomyces cerevisiae (Sc), the generalist repressor-activator protein Rap1 now directs the Ifh1-Fhl1 module to the ribosomal protein genes. Even though overall sequence similarity is low for the respective Fhl1 and Ifh1 subunits, in both species, the Ifh1 protein links to the Forkhead-associated domain of Fhl1 through its FHB domain. Intriguingly, correlated with the transition to the Rap1-regulated circuit, the Sc-Ifh1 contains a Rap1 binding domain that is not present in the C. albicans protein. Because no extensive common sequences are found in Tbf1 and Rap1, it appears that these targeting proteins must connect to the Ifh1-Fhl1 module in distinct ways. Two-hybrid and co-immunoprecipitation analysis has been used to show that in C. albicans Tbf1 is linked to the heterodimer through direct association with Fhl1. By contrast, in S. cerevisiae, the linkage of the heteromer to Rap1 occurs through Ifh1. Thus, in the ascomycetes, the Ifh1-Fhl1 heterodimer has reconfigured its protein associations to remain connected to the ribosomal protein regulon despite rewiring of the targeting transcription factor from Tbf1 to Rap1.
Introduction
Proteins represent a key component of the biomass of cells, and so an actively growing cell invests a great deal of effort in making ribosomes, the machinery necessary to produce these proteins. For a Saccharomyces cerevisiae cell growing in log phase, this process involves coordination of the three RNA polymerases in making proper amounts of the four ribosomal RNAs and the transcripts for around 80 ribosomal proteins. Up to 50% of RNA polymerase II transcription initiations are for ribosomal proteins, and 80% of all nucleic acid in the cell comprises rRNA (1, 2), so this represents a major cellular endeavor. Overall, the formation of ribosomes involves two main elements: the transcription of the ribosomal proteins and the transcription and processing of the ribosomal DNA. The regulatory pathways controlling the expression of these components are conserved across a wide variety of fungi and plants (3, 4).
Recent work has shown that the transcription of ribosomal proteins in Candida albicans and in the majority of Hemiascomycota is controlled by a complex of proteins, including the specialist activators Ifh12 and Fhl1 and the DNA-binding protein Tbf1 (3, 5). Intriguingly, in S. cerevisiae and its close relatives, this critical circuit has been rewired to replace the DNA-binding protein Tbf1 with the general repressor-activator protein Rap1 (3, 5, 6). This rewiring has required the complete restructuring of the promoters of the ribosomal protein genes to replace the palindromic Tbf1 binding site with the Rap1 binding site. However, other elements of the complex involved in the control of RP transcription, in particular Ifh1 and Fhl1, appear to function in the pathways of both the Rap1- and Tbf1-specified regulons (5–9). Because the transcription of RP-encoding genes in a growing cell must remain responsive to an array of nutritional and external environment signals for the cells to modulate the production of ribosomes (10–12), it appears essential that any rewiring event not disconnect the gene expression control from the upstream pathways communicating the environmental signals. Thus, switching from using Tbf1 to regulate RP gene expression to using Rap1 presents the cell with the engineering challenge not just of reconfiguring the promoters of a large regulon but also of maintaining the connectivity to the environmental sensing circuits while the switch is made.
Overall, evolutionary changes in complexes controlling basic transcriptional networks can involve changes in cis-regulatory DNA sequences and trans-acting protein factors either together or separately (13). Examples of cis changes could involve the appearance or disappearance of a binding motif in the promoter of a target gene, whereas examples of trans changes can be the modifications in the interaction capabilities of a transcription factor. Both of these changes could be expected to be powerful drivers of evolutionary change. Most DNA-binding regulatory proteins recognize relatively short sequence motifs, so a limited number of nucleotide changes in a promoter region would be sufficient to include or exclude a particular gene from a regulon (13–17). Although modifications at the protein level can require more sophisticated changes, the modular nature of transcription regulators (18) means that a specific DNA binding domain could be hooked up to a variety of regulatory domains, and this would allow a cell to sample the evolutionary consequences of linking different upstream regulatory circuits to any given transcriptional unit.
Although it is relatively easy to visualize evolution testing out new regulatory circuitry for non-essential cellular processes, it is less clear how dramatic changes could occur in the regulation of central metabolic processes. In the case of the ribosomal protein regulon of the ascomycetes, the need to reconfigure the cis-acting sites to accommodate the DNA binding specificity of the Rap1 protein together with the need to maintain the connection to the environmental sensing circuitry puts severe constraints on how this exchange could be accomplished. Here we show that connection to the sensing circuitry was maintained by conserving the Ifh1-Fhl1 dimer in both the Rap1- and Tbf1-mediated regulons. However, the switch from linking the Ifh1-Fhl1 heterodimer to Rap1 rather than Tbf1 involved a dramatic reconfiguring of the protein-protein associations among the complexes. Rather than linking to the DNA-binding protein through the Fhl1 subunit as in the Tbf1-mediated circuit, a new interaction surface on Ifh1 allows this subunit to connect to Rap1 directly.
EXPERIMENTAL PROCEDURES
Strains, Media, and Plasmids
Cell growth, transformation, and DNA preparation were carried out using standard procedures (19, 20). Experiments were conducted in the SN148 strain background (21), and all C-terminally tagged strains were generated by using the plasmids pFA-Myc-HIS1, pFA-Myc-URA3, pFA-Myc-ARG4, pFA-HA-URA3, and pFA-HA-ARG4 as templates (22). Depending on the starting strain, promoter switches were constructed using the plasmid pFA-URA3-MET3p or pFA-HIS1-MET3p as a template (20) to create a strain with a full-length Tbf1 protein under the Met3p promoter. Briefly, suitable PCR products were amplified containing 100–120-base pair overhangs of the region just upstream and downstream of the termination codon or the starting codon of the gene being changed, and the PCR product was used to transform the C. albicans SN148 strain or a derivative. Transformants were selected on either −Ura, −His, −Arg, or double selective plates lacking two nutritional supplements (0.67% yeast nitrogen base, 2% glucose, amino acid dropout). Correct integrants were identified by diagnostic PCR and then grown until midlog phase either in non-selective liquid medium YPD (1% yeast extract, 2% peptone, 2% dextrose) in the C-terminal tagging experiments or in SD-Met+Cys+ or SD-Met−Cys− medium in the promoter switch experiments, and immunoblotting was done to test for actual expression of the tagged protein. All the strains thus generated are listed in Table 1.
TABLE 1.
Strain lists for C. albicans and S. cerevisiae
| Name of strain | Description | Parental strain | Ref./Source |
|---|---|---|---|
| C. albicans | |||
| SN148 | arg4/arg4 his1/his1 ura3/ura3 leu2/leu2 | SC 76 | 21 |
| CA-JDM001 | SN148 Ifh1-(FL)Myc-His1/IFH1;Fhl1-(FL)HA-Arg4/FHL1 | SN148 | This work |
| CA-JDM002 | SN148 Ifh1(1–700 aa)Myc-His1/IFH1;Fhl1-(FL)HA-Arg4/FHL1 | SN148 | This work |
| CA-JDM003 | SN148 Ifh1(1–600 aa)Myc-His1/Ifh1;Fhl1-(FL)HA-Arg4/FHL1 | SN148 | This work |
| CA-JDM004 | SN148 Ifh1(1–500 aa)Myc-His1/IFH1;Fhl1-(FL)HA-Arg4/FHL1 | SN148 | This work |
| CA-JDM005 | SN148 Ifh1(1–400 aa)Myc-His1/IFH1;Fhl1-(FL)HA-Arg4/FHL1 | SN148 | This work |
| CA-JDM006 | SN148 Fhl1-(FL)Myc-His1/FHL1 Ifh1-(FL)HA-Ura3/IFH1 | SN148 | This work |
| CA-JDM007 | SN148 Fhl1(1–900 aa)Myc-His1/FHL1;Ifh1-(FL)HA-Ura3/IFH1 | SN148 | This work |
| CA-JDM008 | SN148 Fhl1(1–700 aa)Myc-His1/FHL1;Ifh1-(FL)HA-Ura3/IFH1 | SN148 | This work |
| CA-JDM009 | SN148 Fhl1(1–160 aa)Myc-His1/FHL1;Ifh1-(FL)HA-Ura3/IFH1 | SN148 | This work |
| CA-JDM010 | SN148 Tbf1-(FL)Myc-Ura3/Tbf1-(FL)HA-Arg4 | SN148 | This work |
| CA-JDM011 | SN148 Tbf1(1–700 aa)Myc-His1/Tbf1-(FL)HA-Arg4 | SN148 | This work |
| CA-JDM012 | SN148 Tbf1(1–600 aa)Myc-His1/Tbf1-(FL)HA-Arg4 | SN148 | This work |
| CA-JDM013 | SN148 Tbf1(1–500 aa)Myc-His1/Tbf1-(FL)HA-Arg4 | SN148 | This work |
| CA-JDM014 | SN148 Tbf1(1–400 aa)Myc-His1/Tbf1-(FL)HA-Arg4 | SN148 | This work |
| CA-JDM015 | SN148 Tbf1(1–300 aa)Myc-His1/Tbf1-(FL)HA-Arg4 | SN148 | This work |
| CA-JDM016 | SN148 Tbf1-(FL)Myc-His1/TBF1 Ifh1-(FL)HA-Ura3/IFH1 | SN148 | This work |
| CA-JDM017 | SN148 Tbf1-(1–700 aa)Myc-His1/TBF1 Ifh1-(FL)HA-Ura3/IFH1 | SN148 | This work |
| CA-JDM018 | SN148 Tbf1-(1–600 aa)Myc-His1/TBF1 Ifh1-(FL)HA-Ura3/IFH1 | SN148 | This work |
| CA-JDM019 | SN148 Tbf1-(1–500 aa)Myc-His1/TBF1 Ifh1-(FL)HA-Ura3/IFH1 | SN148 | This work |
| CA-JDM020 | SN148 Tbf1-(1–400 aa)Myc-His1/TBF1 Ifh1-(FL)HA-Ura3/IFH1 | SN148 | This work |
| CA-JDM021 | SN148 Fhl1-(FL)Myc-His1/FHL1 Tbf1-(FL)HA-Arg4/Tbf1 | SN148 | This work |
| CA-JDM022 | SN148 Tbf1-(1–700 aa)Myc-His1/TBF1 Fhl1-(FL)HA-Arg4/Fhl1 | SN148 | This work |
| CA-JDM023 | SN148 Tbf1-(1–600 aa)Myc-His1/TBF1 Fhl1-(FL)HA-Arg4/Fhl1 | SN148 | This work |
| CA-JDM024 | SN148 Tbf1-(1–500 aa)Myc-His1/TBF1 Fhl1-(FL)HA-Arg4/Fhl1 | SN148 | This work |
| CA-JDM025 | SN148 His1-Met3p-Tbf1-(FL)Myc-Ura3/Tbf1-(FL)HA-Arg4 | SN148 | This work |
| CA-JDM026 | SN148 Tbf1(1–700 aa)Myc-His1/Ura3-Met3p-Tbf1-(FL)HA-Arg4 | SN148 | This work |
| CA-JDM027 | SN148 Tbf1(1–500 aa)Myc-His1/Ura3-Met3p-Tbf1-(FL)HA-Arg4 | SN148 | This work |
| BWP17 | ura3::imm434/ura3::imm434 his1::hisG/his1::hisG arg4::hisG arg4::hisG | 47 | |
| HLC 15 | BWP17 Fhl1-HA-His1, Tbf1-Myc-Arg4 | BWP17 | This work |
| S. cerevisiae | |||
| W303 a/α | MAT a/α ade2-1 leu2-3,112 ura3-1 his3-11,15 trp1-1 can1-100 ssd1-1 | 25 | |
| SC-JD 001 | W303 a/α Ifh1-(FL)FLAG-G418/IFH1 | W303 a/α | This work |
| SC-JD 002 | W303 a/α Ifh1-(FL)FLAG-G418/IFH1, Gal-HA-N-ter-Rap1-(FL)-His3/RAP1 | W303 a/α | This work |
| SC-JD 003 | W303 a/α Ifh1-(FL)FLAG-G418/IFH1, Gal-HA-N-ter-Rap1-(330–827)-His3/RAP1 | W303 a/α | This work |
| SC-JD 004 | W303 a/α Ifh1-(FL)FLAG-G418/IFH1, Gal-HA-N-ter-Rap1-(410–827)-His3/RAP1 | W303 a/α | This work |
| SC-JD 005 | W303 a/α Ifh1-(FL)FLAG-G418/IFH1, Gal-HA-N-ter-Rap1-(600–827)-His3/RAP1 | W303 a/α | This work |
| YZ154 | W303 a/α Ifh1-(FL)Myc-His3/IFH1 | W303 a/α | J. R. Warner |
| YZ155 | W303 a/α Ifh1-(1–800)Myc-His3/IFH1, Fhl1-(FL)HA-G418 | W303 a/α | J. R. Warner |
| YZ156 | W303 a/α Ifh1-(1–650)Myc-His3/IFH1, Fhl1-(FL)HA-G418 | W303 a/α | J. R. Warner |
| YZ157 | W303 a/α Ifh1-(1–500)Myc-His3/IFH1, Fhl1-(FL)HA-G418 | W303 a/α | J. R. Warner |
| YZ158 | W303 a/α Ifh1-(1–350)Myc-His3/IFH1, Fhl1-(FL)HA-G418 | W303 a/α | J. R. Warner |
| YZ159 | W303 a/α Ifh1-(1–200)Myc-His3/IFH1, Fhl1-(FL)HA-G418 | W303 a/α | J. R. Warner |
Yeast Two-hybrid Analysis
To map protein-protein interactions among domains of the ribosomal protein transcription pathway components, we used a S. cerevisiae two-hybrid system recently developed for the detection of protein-protein interactions in the cytoplasm. Briefly, this yeast two-hybrid system is based on the interaction of Ste11(MEKK) and Ste50 that is required for high osmolarity glycerol pathway activation and osmoadaptation, which is critical for the survival of yeast cells under hyperosmotic stress in the absence of the two-component osmosensing branch (23). The interaction of Ste11 and Ste50 through their respective SAM domains required to activate the high osmolarity glycerol pathway can be replaced by association of other protein-interacting modules (24). This behavior offers a unique potential to analyze bait-prey interactions by substituting them for the respective SAM domains and using the activation of the high osmolarity glycerol pathway as a reporter (23).
Plasmid pYL40 contains the fragment of STE11 encoding Ste11 lacking its SAM domain (amino acids (aa) 110–717) inserted at the SalI site of pGREG506 (22) and a HIS3 stuffer marker inserted at the SmaI site in front of the Ste11ΔSAM module. Similarly, pYL45 contains the fragment of STE50 encoding Ste50 without its SAM domain (aa 115–346) at the SalI site of pGREG503 (22) and a URA3 stuffer marker inserted at the SmaI site in front of the Ste50ΔSAM. All candidate ORFs or their fragments were PCR-amplified and cloned into the vector plasmids pYL40 and pYL45 at their SmaI sites through in vivo recombination in S. cerevisiae strains YCW1476 and YCW1477. All the primers used for the PCR amplification reactions contain gene-specific sequences and common sequences used for in vivo recombination in a layout as follows: 5′-ATTCTAGAGCGGCC GCACTAGTGGATCCCCCGGG-gene-specific sequence (starting with ATG)-3′ for the forward orientation and 5′-TCGATAAGCTTGATATCGAATTCCTGCAGCCCGGG-gene-specific sequence (delete the stop codon)-3′ for the reverse orientation. To query the bait-prey interaction, in vivo recombination positive clones (stuffer marker-negative with correct inserts) of the baits in one of the two yeast strains were crossed to the in vivo recombination positive clones of the preys in the other yeast strain, mating products were selected, and their ability to activate the high osmolarity glycerol pathway was measured by the ability to grow on hyperosmolarity medium (23).
Immunoblotting and Co-Immunoprecipitations
C. albicans Fhl1-HA, Ifh1-Myc, and the other double tagged strains or S. cerevisiae Ifh1-Myc tagged strains (Table 1) (21, 25) were grown to midlog phase in YPD/YP-Gal. Cells at a final A600 nm of 1.0–1.5 were harvested by centrifugation and lysed by bead beating in IP150 buffer (50 mm Tris-HCl (pH 7.4), 150 mm NaCl, 2 mm MgCl2, 0.1% Nonidet P-40) supplemented with Complete Mini protease inhibitor mixture tablet (Roche Applied Science) and 1 mm phenylmethylsulfonyl fluoride (PMSF). The lysates were then cleared by centrifugation, and protein concentration was estimated using the Bradford assay. One milligram of total protein was added to 40–50 μl of monoclonal mouse anti-Myc (9E10), anti-HA (12CA5) (Roche Applied Science), or anti-FLAG (Sigma)-conjugated beads; anti-Nhp2 rabbit polyclonal antibody coupled with protein A-agarose beads (Pierce); or anti-Rap1 antibody (a kind gift from J. Warner) and incubated at 4 °C with end-over-end mixing overnight. The next morning, beads were centrifuged at 2000 rpm at 4 °C, washed three times with IP150 buffer, boiled with SDS-PAGE loading buffer, and resolved by 4–20% gradient SDS-PAGE. Proteins were transferred onto a nitrocellulose membrane and analyzed by Western blotting using rabbit anti-Myc (1:1000) (Santa Cruz Biotechnology), anti-HA (1:2500) (Roche Applied Science), anti-FLAG (1:5000) (Sigma), anti-Nhp1, or anti-Rap1 polyclonal antibodies wherever applicable.
Bioinformatics Analysis
Multiple sequence alignments were performed with MAFFT (26) and viewed with Jalview (Version 2) (27). The protein sequences of the different yeast species were downloaded from the Fungal Orthogroups Repository hosted by the Broad Institute, Massachusetts Institute of Technology. Phylogenetic trees were drawn using the protein sequences with the help of the online programs available either at the MAFFT website, which uses the neighbor-joining method for building the tree and a map viewer known as Archaeopteryx (28, 29), or at the Phylogeny.fr website using their online programming for tree building using the “One Click” option for the default procedure (30, 31).
RESULTS
Ifh1 and Fhl1 Have an Evolutionarily Conserved Interaction
In the ascomycetes, Ifh1 and Fhl1 represent specialist transcription regulators that function in the control of ribosomal protein gene expression (5–9). Alignments of each protein among the fungi show that the family members contain only limited blocks of strong sequence similarity, although where investigated, they function in identical processes. All the Ifh1 proteins share a common FHB domain, which represents the only extended region of sequence similarity within the protein that was found across all the fungal species. However, there is also a region between amino acid residues 550 and 650 in the Ifh1 of S. cerevisiae that is conserved in closely related species of Saccharomycotina (Fig. 1, green rectangle) but is missing in the other fungi (Fig. 1). Similarly, multiple alignments of the Fhl1 protein sequences identify only two clear blocks of conserved similarity that are found throughout the fungi: the Forkhead-associated (FHA) domain and the Forkhead domain. The FHA domain represents the most highly conserved region of Fhl1, whereas the Forkhead domain shows comparatively less conservation, suggesting that the FHA domain may be involved in functions that have been most strongly conserved evolutionarily. In the yeast S. cerevisiae, the function of the FHA domain of Fhl1 is to direct the association of Fhl1 with Ifh1 by binding to the Ifh1 FHB domain (6–9, 32). We used two independent protein association assays to test whether similar Fhl1-Ifh1 binding occurred in the human pathogen C. albicans. The first method involved co-immunoprecipitation of tagged proteins and protein fragments, whereas the second involved a cytoplasmic yeast two-hybrid system (23). In both cases, the FHA domain of Fhl1 and the FHB region of Ifh1 were shown to be required for the interaction between these proteins in C. albicans.
FIGURE 1.
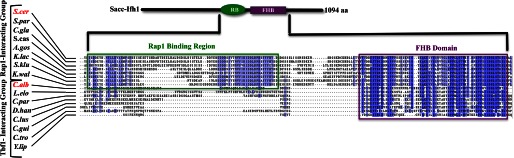
Alignment of Ifh1 protein sequences across the ascomycetes species highlighting the conserved and partially conserved regions. The FHB domain is the only extended stretch to show conservation across all species, whereas a smaller region is seen to be conserved across those Ifh1 proteins binding with Rap1; this region is missing in those species where Tbf1 is responsible for ribosomal protein transcriptional control in place of Rap1. Darker color indicates greater conservation. RB, Rap1 binding region; C.alb, C. albicans; S.cer, S. cerevisiae; S.par, Saccharomyces paradoxus; C.gla, Candida glabrata; S.cas, Saccharomyces castellii; A.gos, Ashbya gossypii; K.lac, Kluyveromyces lactis; K.wal, Kluyveromyces waltii; L.elo, Lodderomyces elongisporus; C.par, Candida parapsilosis; D.han, Debaryomyces hansenii; C.lus, Candida lusitaniae; C.gui, Candida guilliermondii; C.tro, Candida tropicalis; Y.lip, Yarrowia lipolytica; S.klu, Saccharomyces kluyveri.
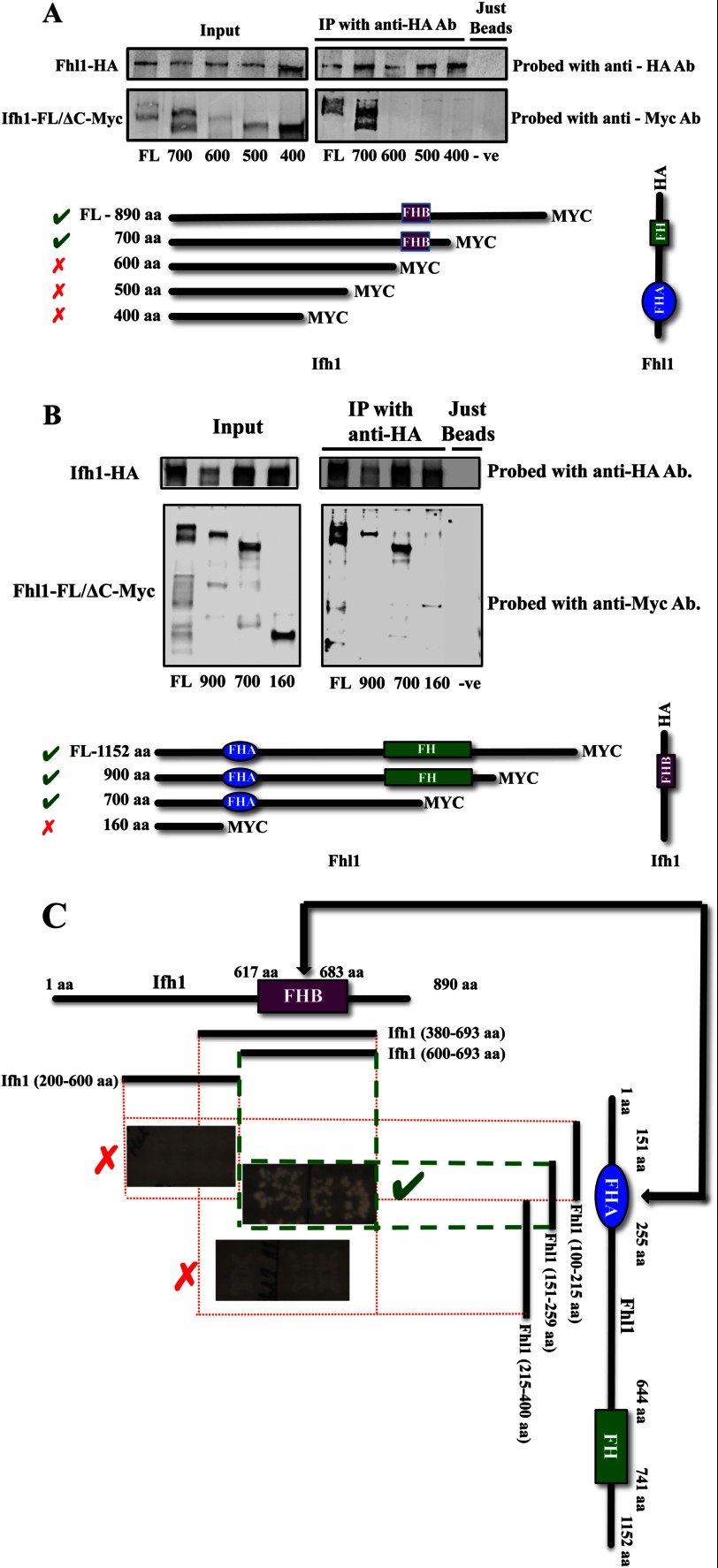
The in vivo association of Ifh1 and Fhl1 was investigated through co-immunoprecipitation. C-terminally Myc-tagged versions of full-length and truncated proteins along with the C-terminally HA-tagged versions of a potentially interacting protein were generated, and the interactions were monitored by co-immunoprecipitation. A series of strains was constructed in which different C-terminally deleted Myc-tagged Ifh1 constructs were expressed along with HA-tagged full-length Fhl1. The details of the tagged Ifh1 fragments and the tagged Fhl1 protein with which they were immunoprecipitated are given in Fig. 2A. The co-immunoprecipitation results show that Ifh1 is not pulled down with the full-length HA-tagged Fhl1 in the absence of an intact FHB domain (amino acids 617–683), implying that Ifh1 needs the FHB domain for binding Fhl1 (Fig. 2A). The reciprocal is also true; when a Myc-tagged fragment of Fhl1 is expressed without the FHA domain, it is not co-immunoprecipitated with full-length HA-tagged Ifh1. However, the full-length or smaller fragments of Fhl1 with an intact FHA domain can be pulled down with Ifh1 (Fig. 2B).
FIGURE 2.
Ifh1 and Fhl1 co-immunoprecipitate only if the FHB domain of Ifh1 and the FHA domain of Fhl1 are intact. A, co-immunoprecipitations from lysates of the different C. albicans strains with full-length (FL) and C-terminally deleted Myc-tagged versions of Ifh1 and the full-length Fhl1 C-terminally tagged with HA are shown (details of strains used are found in Table 1). B, co-immunoprecipitations from lysates of the different C. albicans strains with full-length (FL) and C-terminally deleted Myc-tagged versions of Fhl1 and the full-length Ifh1 C-terminally tagged with HA are shown (details of strains used are found in Table 1). C, the interaction between the FHB and FHA domains of Ifh1 and Fhl1 as observed in the yeast two-hybrid system. IP, immunoprecipitation; Ab, antibody; FH, Forkhead domain; −ve, negative.
To refine the in vivo data, a collection of protein fusion fragments was generated covering the entire length of the C. albicans Ifh1- and Fhl1-interacting domains and screened for their interactions using the two-hybrid assay. Different fragments tested along with whether they interacted or not are shown in Fig. 2C. Somewhat surprisingly, the full-length Ifh1 and the first 600-amino acid fragment of Fhl1 did not give a signal (data not shown), although smaller fragments of each protein generated a positive response. This could be because the larger proteins were able to fold to prevent interactions (33), but it also appears that the yeast two-hybrid system used here has a limitation on the maximum length of the fragments that can be tested. Sequence alignments of Fhl1 proteins showed that the FHA domain of the C. albicans protein extends from residues 150 to 255. When this region of Fhl1 (amino acids 151–259) was used as bait, it showed a clear interaction with Ifh1. In fact, all the fragments of Fhl1 containing the intact domain (with the exception of the first 600-aa fragment) tested were seen to interact with Ifh1. The interaction was abolished when this domain was even partially deleted. Similarly, multiple sequence alignments of Ifh1 identified the FHB domain in the C. albicans protein as extending from amino acid residues 617 to 683, and it was this region that was shown to interact with the FHA region of Fhl1. When this region was partially or completely deleted, no interaction was detected. Furthermore, a PCR-generated point mutant (G221W) was identified within the Fhl1 FHA domain that disrupted the interaction between Fhl1 and Ifh1, showing that specific residues in this interface are critical for the association (data not shown). Overall, the co-immunoprecipitation experiments and the S. cerevisiae two-hybrid system data support the model that the interaction between Ifh1 and Fhl1 in C. albicans occurs through the same domains as in S. cerevisiae, consistent with a conserved Ifh1-Fhl1 heteromer being a common feature of ribosomal expression control in the ascomycetes.
Interaction of Tbf1 with the Ifh1-Fhl1 Pair in C. albicans
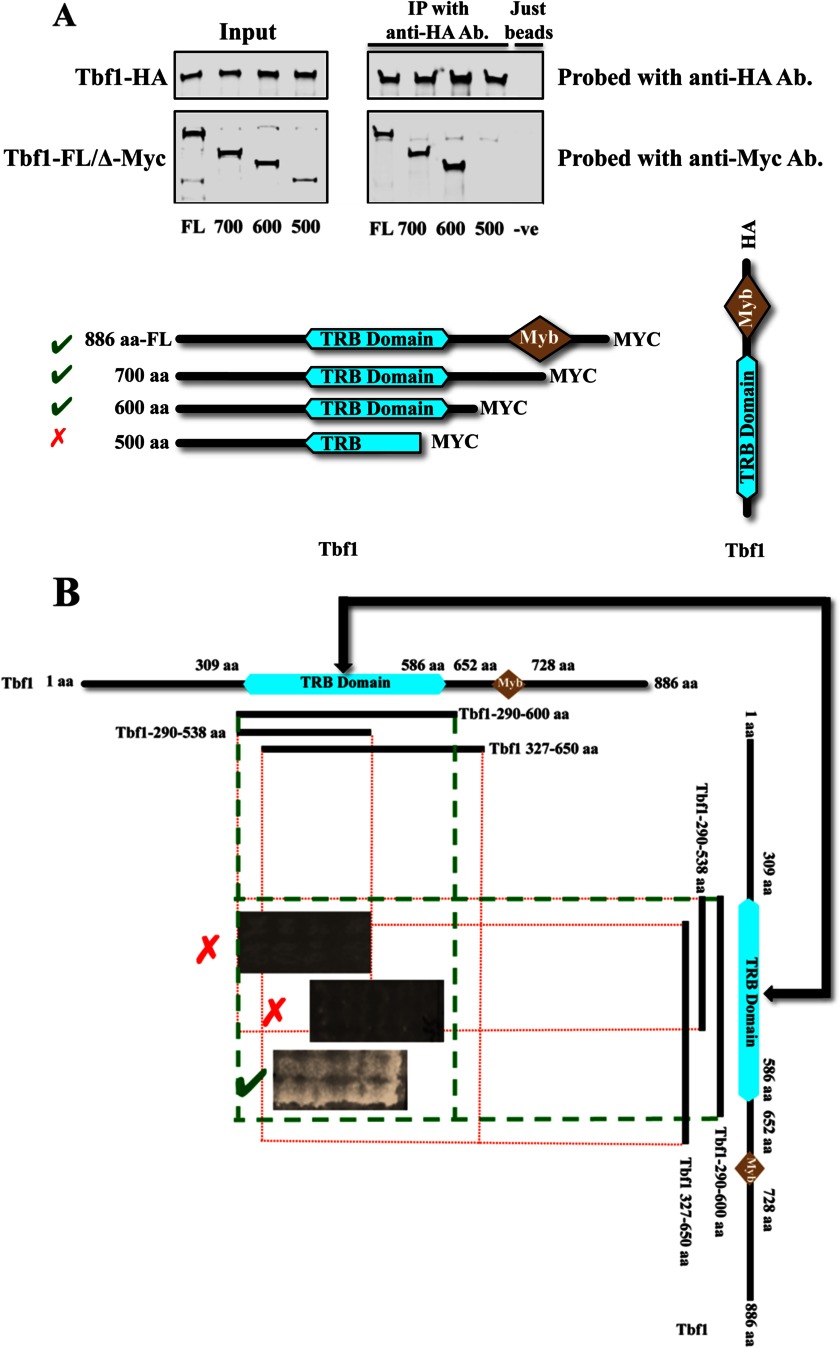
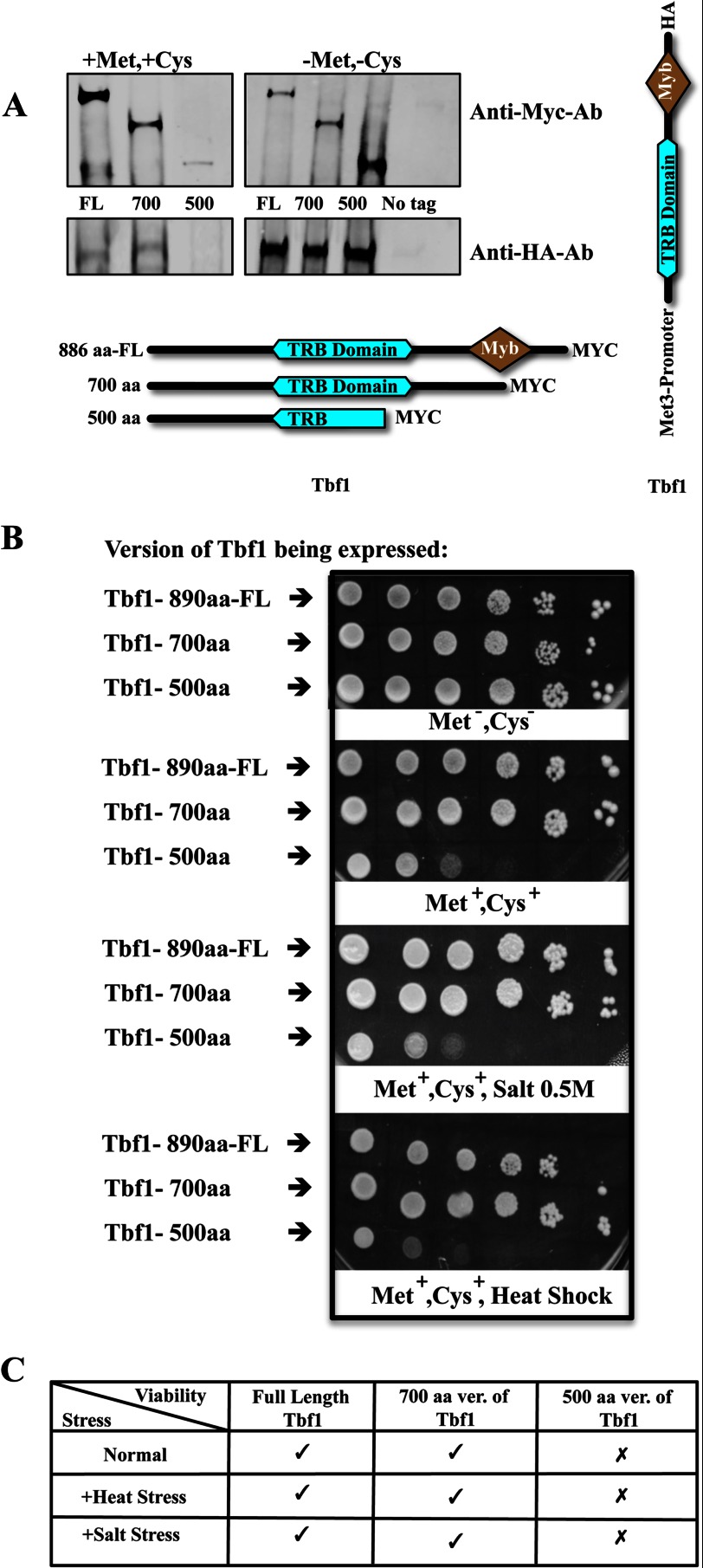
Our previous work (5) suggested that the Ifh1-Fhl1 pair interacts directly with Tbf1 in C. albicans in its function as a transcription factor controlling expression of ribosomal protein genes. Tbf1 in C. albicans is an 886-amino acid protein with two bioinformatically predicted domains, a telomere repeat binding (TRB) domain and a Myb domain (34, 35). Alignments of the Tbf1 protein sequences from various hemiascomycetes shows that in the C. albicans protein the TRB domain lies between aa residues 309 and 586, and the Myb domain is found between aa residues 652 and 728. ChIP-CHIP (chromatin immunoprecipitation followed by whole genome microarray) profiles of Tbf1 binding at RP promoters shows that along with a major peak, which coincides with the binding of Ifh1 and Fhl1, there is an extended shoulder of Tbf1 binding at most ribosomal protein promoters (5). Because the binding sites for Tbf1 are typically direct or inverted repeats of the binding motifs, it is possible that these shoulders result from binding of a dimerized Tbf1. To test for possible dimerization of Tbf1 in vivo, strains of C. albicans were constructed that express a C-terminally Myc-tagged full-length or deleted version of Tbf1 along with an HA-tagged full-length Tbf1. Co-immunoprecipitation experiments with anti-HA antibody followed by immunoblots probed with antibodies against the Myc and HA tags confirmed the dimerization of Tbf1 and suggested that Tbf1 subunits bind each other through their TRB domains (Fig. 3A). To confirm this observation, the intact and truncated TRB domain of Tbf1 was assayed for association with itself using the cytoplasmic S. cerevisiae two-hybrid system. The different fragments of Tbf1 tested are shown in Fig. 3B where it is seen that the assay detects interaction between constructs that both carry the intact 290–600-aa region.
FIGURE 3.
Tbf1 immunoprecipitates itself only if the TRB domain is intact. A, co-immunoprecipitations from lysates of the different C. albicans strains where one copy of Tbf1 is either full-length (FL) or C-terminally deleted with a Myc tag and the other copy of Tbf1 is full-length and C-terminally tagged with HA (details of strains used are shown in Table 1). B, the interaction of the TRB domain of Tbf1 with itself as observed in the yeast two-hybrid system. IP, immunoprecipitation; Ab, antibody; −ve, negative.
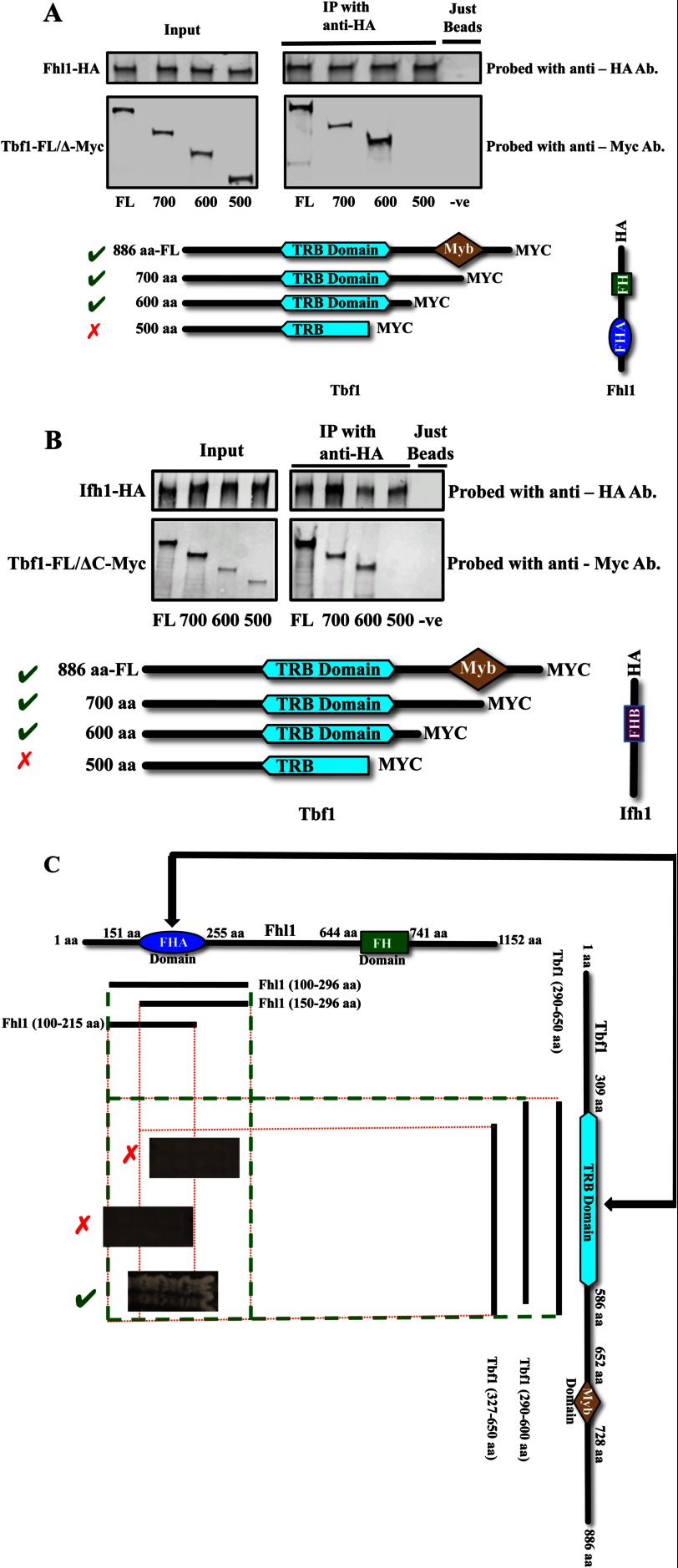
Next, to determine in vivo the interaction surfaces between Tbf1 and the Ifh1-Fhl1 heteromer, full-length or C-terminally deleted versions of Myc-tagged Tbf1 were expressed along with the full-length HA-tagged Fhl1 and co-immunoprecipitated. We observed that along with the full-length Tbf1 only those versions of Tbf1 possessing an intact TRB domain are pulled down with full-length Fhl1, implying that the TRB domain is necessary for the binding of Tbf1 with Fhl1 (Fig. 4A). When the full-length HA-tagged Ifh1 is expressed along with C-terminally deleted fragments of Myc-tagged Tbf1, both the 700- and the 600-aa fragments of Tbf1 are pulled down with Ifh1, whereas the smaller fragment is not, suggesting that the region of Tbf1 responsible for its interaction with Ifh1 is within the first 600 amino acids (Fig. 4B). This result is consistent with the TRB domain, which is found between amino acids 309 and 586, the region involved in the interaction. It is not possible from these two co-immunoprecipitation experiments to establish whether the observed interaction between either Ifh1 or Fhl1 and Tbf1 is direct; to ascertain this, we used the cytoplasmic yeast-two hybrid assay to examine the direct interaction between Tbf1 and Ifh1-Fhl1. Because Tbf1 also interacts with itself through the TRB domain, it is possible that a dimer of Tbf1 binds Fhl1 or Ifh1.
FIGURE 4.
Interactions among Fhl1, Ifh1, and Tbf1. A, co-immunoprecipitations from lysates of the different C. albicans strains with full-length (FL) and C-terminally deleted Myc-tagged versions of Tbf1 and a full-length Fhl1 C-terminally tagged with HA (details of strains used are shown in Table 1). B, co-immunoprecipitations from lysates of different C. albicans strains with full-length (FL) and C-terminally deleted Myc-tagged versions of Tbf1 and the full-length Ifh1 C-terminally tagged with HA (details of strains used are shown in Table 1). C, the interaction of the FHA domain of Fhl1 with TRB domain in Tbf1 as observed in the yeast two-hybrid system. IP, immunoprecipitation; Ab, antibody; FH, Forkhead domain; −ve, negative.
To identity the member of the Ifh1-Fhl1 heteromer interacting directly with Tbf1, fragments of Tbf1 were used in the cytoplasmic S. cerevisiae two-hybrid system to probe for interactions with the different components of the dimer. No direct interaction was observed between Tbf1 constructs and any of the fragments of Ifh1 tested (fragments of Ifh1 covering the whole protein have been used to test for potential interaction against fragments covering the whole of Tbf1) (data not shown). However, testing the interaction of Fhl1 with Tbf1 demonstrated that Fhl1 constructs containing the region between amino acids 100 and 296 (containing an intact FHA domain) interacted with the TRB domain of Tbf1 (Fig. 4C). These interactions were abolished if either of these domains was partially deleted. These results suggest that Fhl1, but not Ifh1, has a direct interaction with Tbf1 and that Fhl1 requires a region that includes the FHA domain for this association, whereas Tbf1 requires an intact TRB domain for interaction with Fhl1.
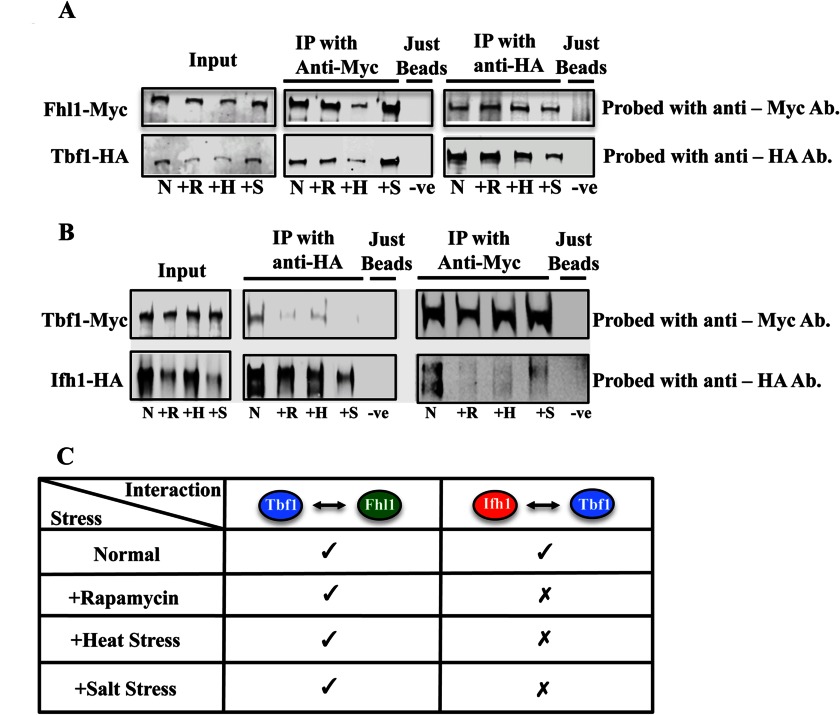
The interaction between Fhl1 and Tbf1 did not change in response to nutrient limitation or other stresses. When full-length tagged Tbf1 and Fhl1 were co-immunoprecipitated during rapamycin treatment and other stress conditions, we observed (Fig. 5A) that the interaction is not disrupted. This is in contrast to the Ifh1-Fhl1 association as the interaction between these proteins was observed to be reduced in response to rapamycin treatment and other similar kinds of stresses (see Fig. S13 of Ref. 5). Furthermore, it was observed that the Ifh1-Tbf1 interaction is subject to modification during nutritional and other kinds of stresses. When full-length tagged Tbf1 and Ifh1 were co-immunoprecipitated during rapamycin treatment and other stress conditions, we observed (Fig. 5B) that the interaction is disrupted, implying that the Tbf1-Ifh1 interaction is subject to nutritional conditions, osmotic stress, and heat. Overall, it appears that in C. albicans the binding of the Ifh1-Fhl1 heteromer with Tbf1 occurs through Fhl1 associating with the TRB domain of Tbf1 and that this association is not responsive to the environment. Because both the Ifh1-Fhl1 and Tbf1-Ifh1 associations are subject to environmental stresses, it is the Ifh1 protein that seems to be responsible for disassociating from the Tbf1-Fhl1-Ifh1 complex when the cells face difficult conditions and have to stop growth.
FIGURE 5.
Co-immunoprecipitations of full-length Fhl1 with Tbf1 and full-length Ifh1 with Tbf1 under different stress conditions. A, the interaction of Tbf1 and Fhl1 show no major differences under different stress conditions. B, the interaction between Ifh1 and Tbf1 is disrupted by different stress conditions. C, summary of the results seen in A and B. IP, immunoprecipitation; Ab, antibody; −ve, negative; N, normal conditions; +R, rapamycin treatment; +H, heat treatment; +S, salt treatment.
Disruption of the Tbf1 TRB Domain of Interaction Severely Compromises Cell Viability
We investigated whether specific domains defined by the interaction assays as being involved in the association of the transcription regulators were important for cellular function. We replaced the promoter of the full-length allele of Tbf1 with the inducible Met3p promoter in a strain where the other allele of Tbf1 had a C-terminal truncation (Fig. 6A). When the truncation extended into the TRB domain and the strain was incubated under repressing conditions to block expression of the full-length protein, cell growth was inhibited. This shutoff strain was also more sensitive to other conditions like heat and osmotic stress (Fig. 6B). However, when the truncation did not extend into the TRB domain, inhibition of the full-length protein did not compromise growth (Fig. 6B). The results shown in Fig. 6B have been summarized in Fig. 6C.
FIGURE 6.
A, immunoblot of different C. albicans strains with one copy of a Myc-tagged Tbf1, either full length (FL) or C-terminally deleted, and with the other full-length copy of Tbf1 tagged with HA and expressed under the Met3p promoter (details of strains used are in Table 1). The strains were grown in the presence or absence of methionine and cysteine. The presence of methionine and cysteine suppresses the expression of the HA-tagged full-length copy of Tbf1. B, the growth assay shows that in the absence of full-length Tbf1 the growth of the strains is severely compromised both in the absence and presence of stress-inducing factors. C shows the result of B in a tabular form. IP, immunoprecipitation; Ab, antibody; −ve, negative; ver., version.
Ifh1 Proteins That Link to Rap1 Contain a Distinct Domain
Although both Rap1 (in S. cerevisiae) and Tbf1 (in C. albicans) are capable of directing the Ifh1-Fhl1 heteromer to the promoters of the ribosomal protein genes, outside of the Myb domains, the Tbf1 and Rap1 proteins (36, 37) have very little similarity, and even these Myb domains are quite divergent. Therefore, we examined how the Ifh1-Fhl1 module interacted in S. cerevisiae with Rap1. Multiple sequence alignment analysis of the two branches of Ifh1 proteins (the Rap1-interacting group members and the Tbf1-interacting group members) showed that the Rap1-interacting proteins were generally larger and contained a conserved region that was missing in all the Tbf1-interacting group members (Fig. 1).
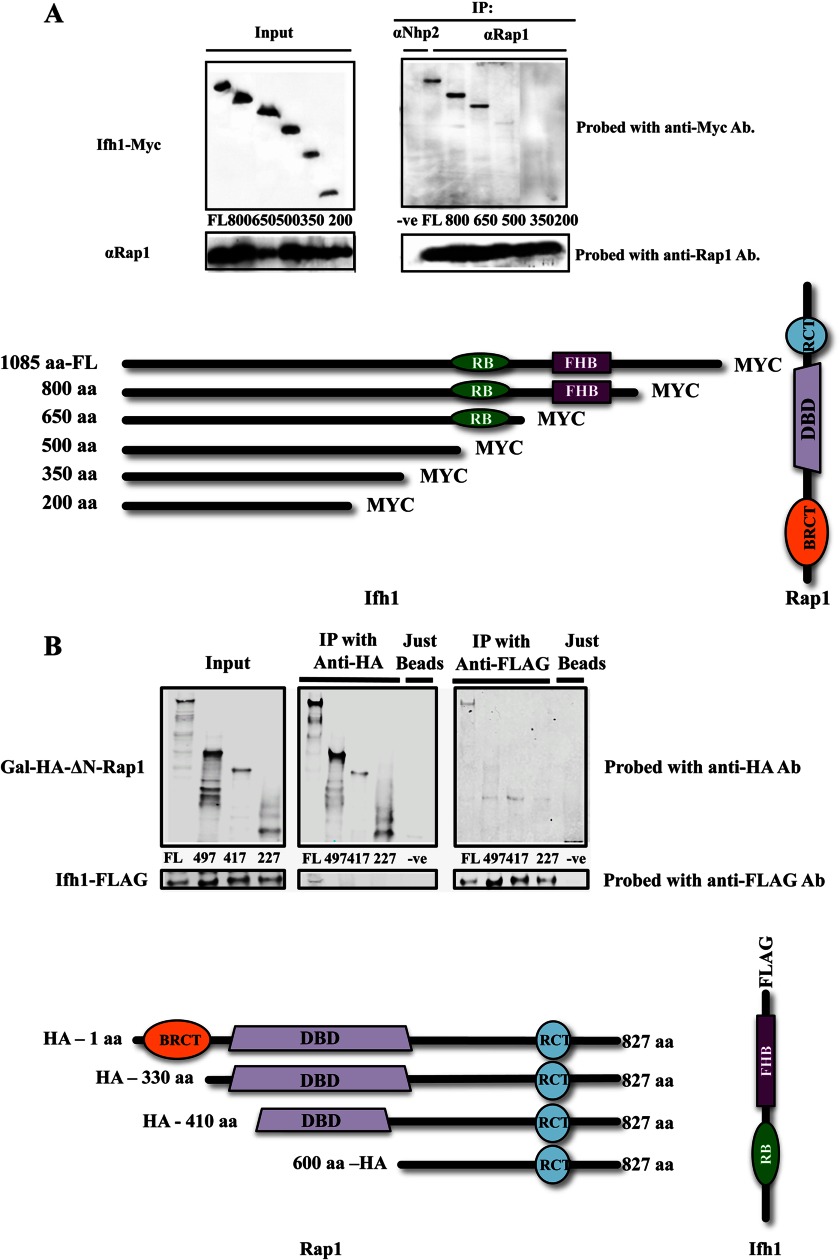
When C-terminally deleted Myc-tagged Ifh1 is expressed in S. cerevisiae and Rap1 is used as the bait for a co-immunoprecipitation, the smallest fragment of Ifh1 that is efficiently pulled down contains amino acids 1–650, whereas deletion to residue 500 blocks the interaction, establishing that the region in Ifh1 needed to bind Rap1 is within this stretch (Fig. 7A). The region that is uniquely conserved within those Ifh1 proteins that interact with Rap1 lies between amino acids 550 and 650, and thus, this domain appears to be required for efficient binding of Ifh1 to Rap1.
FIGURE 7.
A, co-immunoprecipitations from lysates of different S. cerevisiae strains with full-length (FL) and C-terminally deleted Myc-tagged versions of Ifh1 and the full-length Rap1 using anti-Rap1 antibody (details of strains used are shown in Table 1). B, co-immunoprecipitations from lysates of the different S. cerevisiae strains with full-length and N-terminally deleted HA-tagged versions of Rap1 where the full-length Ifh1 is C-terminally FLAG-tagged (details in Table 1). Ab, antibody; DBD, DNA binding domain; RCT, Rap1 C-terminal domain; RB, Rap1 binding region.
The region of Rap1 required for this interaction with Ifh1 was identified by constructing a series of strains expressing N-terminally deleted HA-tagged versions of Rap1 under the GAL1 promoter and expressing Ifh1 with the FLAG tag. When co-immunoprecipitations were performed with anti-FLAG and anti-HA antibodies followed by Western blots (Fig. 7B), only the full-length Rap1 co-immunoprecipitated Ifh1, and only full-length Ifh1 co-immunoprecipitated Rap1. All versions of Rap1 lacking the breast cancer susceptibility protein C terminus (BRCT) domain fail to co-immunoprecipitate Ifh1 or fail to be co-immunoprecipitated by Ifh1, suggesting that the BRCT domain of Rap1 is critical for the association with Ifh1.
Overall, in S. cerevisiae, Rap1 appears to interact with the Ifh1-Fhl1 heteromer through a domain of Ifh1 ∼100 amino acids long (Fig. 7A) that is missing in the Ifh1 family members that associate with Tbf1 (Fig. 1). Previous work has suggested that Fhl1 can bind an IFHL motif in the DNA (38) and that these IFHL sites are found in RP promoters adjacent to Rap1 binding sites. In addition, Ifh1 (but not Fhl1) is found to dissociate from RP promoters upon suppression of RP transcription by rapamycin treatment (6). These combined observations are consistent with a model wherein S. cerevisiae Ifl1 serves to bridge the two DNA-binding proteins, Rap1 and Fhl1, and that disruption of the Ifh1-Rap1 link serves to down-regulate RP gene expression. The way the Ifh1-Fhl1 pair interacts with Rap1 in S. cerevisiae is thus very different from the way the pair interacts with Tbf1 in C. albicans, demonstrating that these protein linkages have changed in concert with the genetic rewiring of the RP regulon from Tbf1 control to Rap1 control.
DISCUSSION
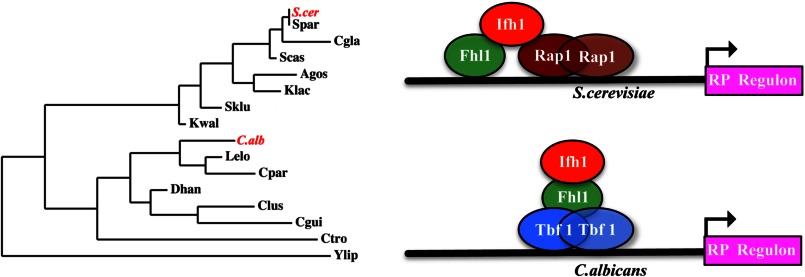
The human pathogenic fungus C. albicans and the bakers' yeast S. cerevisiae have very different ecological niches, but they have very similar responses to many environmental stresses, including that of starvation. In both organisms, poor growth conditions lead to a suppression of the entire regulon of genes encoding ribosomal proteins, whereas good conditions lead to rapid transcription of this group of genes (5, 39). This response is controlled by modulating gene expression in part through the control of a protein complex comprising Tbf1-Ifh1-Fhl1 in C. albicans and Rap1-Ifh1-Fhl1 in S. cerevisiae. The heterodimer Ifh1-Fhl1 of the complex responds to the same signal transduction pathways in both organisms, receiving the environmental and nutritional conditions through pathways such as those mediated by Tor and cAMP and directing whether the RP regulon is switched on or off (5, 10, 40). However, given the central importance of this circuit in the physiology of the fungi, it is remarkable that the DNA binding partner has changed from Tbf1 in C. albicans (3) to Rap1 in S. cerevisiae (6–9).
The process of rewiring this circuit has involved a fundamental change in the association of the regulatory proteins. Overall, the linking of the Ifh1 and Fhl1 proteins has remained the same in the different fungi with the FHB domain of Ifh1 binding to the FHA domain of Fhl1. However, the way this heterodimer connects to the DNA-binding specificity element appears quite different in the two classes of circuits. When the DNA-binding protein is Tbf1 as in C. albicans, the dimer is linked through the Fhl1 protein binding to the TRB domain of Tbf1. However, the shift to the Rap1-regulated circuit involved the transfer of the connection of the heterodimer with the DNA-binding protein to Ifh1 using a new domain specific to those Ifh1 proteins required to bind Rap1. In this configuration, Ifh1 binds to the BRCT domain of Rap1. This Rap1-mediated circuit arose in conjunction with the appearance of a new cis-acting DNA motif termed the IFHL element (9, 41). This suggests that in addition to reconfiguring the association among the proteins the new Rap1-mediated circuit involves a DNA binding capacity of the Ifh1-Fhl1 dimer that was not found in the Tbf1-mediated circuit. It is possible that this protein-DNA connection helps to stabilize the new association of the Ifh1-Fhl1 dimer with Rap1. This rewiring model is represented schematically in Fig. 8.
FIGURE 8.
Proposed model of restructuring of the protein-protein interactions within the RP regulon of C. albicans and S. cerevisiae alongside a scaled and unrooted phylogenetic tree whose branch lengths are proportional to the differences between pairs of neighboring species. C.alb, C. albicans; S.cer, S. cerevisiae; Spar, S. paradoxus; Cgla, C. glabrata; Scas, S. castellii; Agos, A. gossypii; Klac, K. lactis; Kwal, K. waltii; Lelo, L. elongisporus; Cpar, C. parapsilosis; Dhan, D. hansenii; Clus, C. lusitaniae; Cgui, C. guilliermondii; Ctro, C. tropicalis; Ylip, Y. lipolytica; Sklu, S. kluyveri.
A comparison of C. albicans and S. cerevisiae shows that transcription factor substitutions and reconfiguration occur in many other cellular pathways. For example, to control the expression of genes involved in glucose utilization, C. albicans uses Gal4 and Tye7 (42), whereas in S. cerevisiae, this circuit has been replaced by the transcription factors Gcr1 and Gcr2 (42, 43). In addition, in S. cerevisiae, the role of the Ino2/4 transcription factor is limited just to the control of lipid biosynthesis genes, whereas in C. albicans, it also regulates genes implicated in the β-oxidation process. Similarly, in S. cerevisiae, Gcn4 function is limited to regulation of amino acid biosynthesis genes, whereas it also includes control of genes for tRNA aminoacylation in C. albicans (44).
The broadening or narrowing of regulons such as those controlled by Gcn4 or Ino2/4 can occur through minor modifications in cis-acting DNA motifs that serve as the targets for DNA-binding transcription regulators (44, 45). The switching of the trans-acting transcription factor over an entire regulon is more of an evolutionary challenge, but in the case of control of glycolytic gene expression, the distinct carbohydrate metabolism in S. cerevisiae and C. albicans provides a framework that could direct the circuits in different directions (42). However, given the apparent similarity in the stress and nutritional responses of the ribosomal regulons in both S. cerevisiae and C. albicans, it appears that the pressure to replace the Tbf1 regulatory circuit with a Rap1-controlled process is not simply due to the requirements for distinct nutritional stress responses in the two species. Overall, there has been a steady shift in the ascomycetes from the use of Tbf1 as a DNA-binding regulator to the use of Rap1: first in the case of telomeres and later in the case of RP expression regulation (5, 44). Intriguingly, in S. cerevisiae, Tbf1 acts as a transcriptional regulator for small nucleolar RNA expression (46). This may represent the final component remaining in bakers' yeast of an evolutionarily ancient ribosome production regulon, still intact in C. albicans, consisting of the RP, rDNA, and small nucleolar RNA genes under the control of Tbf1. Overall, the switch in S. cerevisiae has been to put highly expressed genes such as the genes for glycolytic enzymes as well as those for ribosomal proteins under Rap1 regulation. Coordination of highly expressed genes may thus provide a selective advantage that can drive the rewiring process.
Currently, high throughput sequencing platforms are providing the genome sequences of a growing number of ascomycete fungi. Because many of these organisms are amenable to molecular manipulation, we are in a position to investigate the way these fungal genomes are expressed and regulated. Continuing investigations on how similar processes are differentially controlled should provide valuable insights into the way evolution rewires regulatory circuits to fit organisms to their ecological niches.
Acknowledgments
We thank Prof. Jonathan R Warner (Albert Einstein College of Medicine) for providing the YZ strains and anti-Rap1 antibody. The technical help of Dipayan Rudra in the co-immunoprecipitation of Fig. 7A and the technical help of Yu Zhao and Dipayan Rudra in the making of the YZ strains are gratefully acknowledged. We thank the members of the Whiteway laboratory, Biotechnology Research Institute, especially Cunle Wu, Andre Nantel, Doreen Harcus, Daniel Dignard, and members of the Biotechnology Research Institute Antibody Facility, especially Anne Marcil, Christine Gadoury, and Josée Ash, for technical assistance. We thank the members of the Whiteway laboratory, Concordia University, especially Nicholas Geoffrion, Hannah Regan, Tuana Mesquita, Faiza Tebbji, Julian Albert, and Pierre Cote, for critical reading of the manuscript. We especially thank Pierre Cote for help and suggestions about structuring the figures of the paper. We also thank Hugo Lavoie for strain HLC15. We thank Amandeep Glory and Catherine Bachewich (Biology Department, Concordia University) for help and constructive suggestions.
This work was supported by Canadian Institute for Health Research Grant MOP-42516 (to M. W.).
- Ifh1
- interacts with Forkhead 1
- RP
- ribosomal protein
- aa
- amino acid(s)
- Fhl1
- Forkhead-like 1
- Ca
- C. albicans
- Sc
- S. cerevisiae
- FHA
- Forkhead-associated
- FHB
- FHA binding
- SD
- synthetic dextrose
- SAM
- sterile α motif
- TRB
- telomere repeat binding
- BRCT
- breast cancer susceptibility protein C terminus.
REFERENCES
- 1. Warner J. R. (1999) The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24, 437–440 [DOI] [PubMed] [Google Scholar]
- 2. Lempiäinen H., Shore D. (2009) Growth control and ribosome biogenesis. Curr. Opin. Cell Biol. 21, 855–863 [DOI] [PubMed] [Google Scholar]
- 3. Hogues H., Lavoie H., Sellam A., Mangos M., Roemer T., Purisima E., Nantel A., Whiteway M. (2008) Transcription factor substitution during the evolution of fungal ribosome regulation. Mol. Cell 29, 552–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li B., Oestreich S., de Lange T. (2000) Identification of human Rap1: implications for telomere evolution. Cell 101, 471–483 [DOI] [PubMed] [Google Scholar]
- 5. Lavoie H., Hogues H., Mallick J., Sellam A., Nantel A., Whiteway M. (2010) Evolutionary tinkering with conserved components of a transcriptional regulatory network. PLoS Biol. 8, e1000329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rudra D., Zhao Y., Warner J. R. (2005) Central role of Ifh1p-Fhl1p interaction in the synthesis of yeast ribosomal proteins. EMBO J. 24, 533–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Martin D. E., Soulard A., Hall M. N. (2004) TOR regulates ribosomal protein gene expression via PKA and the Forkhead transcription factor FHL1. Cell 119, 969–979 [DOI] [PubMed] [Google Scholar]
- 8. Schawalder S. B., Kabani M., Howald I., Choudhury U., Werner M., Shore D. (2004) Growth-regulated recruitment of the essential yeast ribosomal protein gene activator Ifh1. Nature 432, 1058–1061 [DOI] [PubMed] [Google Scholar]
- 9. Wade J. T., Hall D. B., Struhl K. (2004) The transcription factor Ifh1 is a key regulator of yeast ribosomal protein genes. Nature 432, 1054–1058 [DOI] [PubMed] [Google Scholar]
- 10. Rudra D., Warner J. R. (2004) What better measure than ribosome synthesis? Genes Dev. 18, 2431–2436 [DOI] [PubMed] [Google Scholar]
- 11. Jorgensen P., Nishikawa J. L., Breitkreutz B. J., Tyers M. (2002) Systematic identification of pathways that couple cell growth and division in yeast. Science 297, 395–400 [DOI] [PubMed] [Google Scholar]
- 12. Powers T., Dilova I., Chen C. Y., Wedaman K. (2004) Yeast TOR signaling: a mechanism for metabolic regulation. Curr. Top. Microbiol. Immunol. 279, 39–51 [DOI] [PubMed] [Google Scholar]
- 13. Li H., Johnson A. D. (2010) Evolution of transcription networks–lessons from yeasts. Curr. Biol. 20, R746–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gasch A. P., Moses A. M., Chiang D. Y., Fraser H. B., Berardini M., Eisen M. B. (2004) Conservation and evolution of cis-regulatory systems in ascomycete fungi. PLoS Biol. 2, e398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Borneman A. R., Gianoulis T. A., Zhang Z. D., Yu H., Rozowsky J., Seringhaus M. R., Wang L. Y., Gerstein M., Snyder M. (2007) Divergence of transcription factor binding sites across related yeast species. Science 317, 815–819 [DOI] [PubMed] [Google Scholar]
- 16. Tompa M., Li N., Bailey T. L., Church G. M., De Moor B., Eskin E., Favorov A. V., Frith M. C., Fu Y., Kent W. J., Makeev V. J., Mironov A. A., Noble W. S., Pavesi G., Pesole G., Régnier M., Simonis N., Sinha S., Thijs G., van Helden J., Vandenbogaert M., Weng Z., Workman C., Ye C., Zhu Z. (2005) Assessing computational tools for the discovery of transcription factor binding sites. Nat. Biotechnol. 23, 137–144 [DOI] [PubMed] [Google Scholar]
- 17. Habib N., Wapinski I., Margalit H., Regev A., Friedman N. (2012) A functional selection model explains evolutionary robustness despite plasticity in regulatory networks. Mol. Syst. Biol. 8, 619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhu L., Huq E. (2011) Mapping functional domains of transcription factors. Methods Mol. Biol. 754, 167–184 [DOI] [PubMed] [Google Scholar]
- 19. Ausubel F. M. (1987) Current Protocols in Molecular Biology, John Wiley and Sons, Hoboken, NJ [Google Scholar]
- 20. Gola S., Martin R., Walther A., Dünkler A., Wendland J. (2003) New modules for PCR-based gene targeting in Candida albicans: rapid and efficient gene targeting using 100 bp of flanking homology region. Yeast 20, 1339–1347 [DOI] [PubMed] [Google Scholar]
- 21. Noble S. M., Johnson A. D. (2005) Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot. Cell 4, 298–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lavoie H., Sellam A., Askew C., Nantel A., Whiteway M. (2008) A toolbox for epitope-tagging and genome-wide location analysis in Candida albicans. BMC Genomics 9, 578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Côte P., Sulea T., Dignard D., Wu C., Whiteway M. (2011) Evolutionary reshaping of fungal mating pathway scaffold proteins. MBio 2, e00230–e00210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wu C., Jansen G., Zhang J., Thomas D. Y., Whiteway M. (2006) Adaptor protein Ste50p links the Ste11p MEKK to the HOG pathway through plasma membrane association. Genes Dev. 20, 734–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Thomas B. J., Rothstein R. (1989) Elevated recombination rates in transcriptionally active DNA. Cell 56, 619–630 [DOI] [PubMed] [Google Scholar]
- 26. Katoh K., Misawa K., Kuma K., Miyata T. (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Waterhouse A. M., Procter J. B., Martin D. M., Clamp M., Barton G. J. (2009) Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zmasek C. M., Eddy S. R. (2001) ATV: display and manipulation of annotated phylogenetic trees. Bioinformatics 17, 383–384 [DOI] [PubMed] [Google Scholar]
- 29. Han M. V., Zmasek C. M. (2009) phyloXML: XML for evolutionary biology and comparative genomics. BMC Bioinformatics 10, 356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., Chevenet F., Dufayard J. F., Guindon S., Lefort V., Lescot M., Claverie J. M., Gascuel O. (2008) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36, W465–W469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dereeper A., Audic S., Claverie J. M., Blanc G. (2010) BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol. Biol. 10, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rudra D., Mallick J., Zhao Y., Warner J. R. (2007) Potential interface between ribosomal protein production and pre-rRNA processing. Mol. Cell. Biol. 27, 4815–4824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brückner A., Polge C., Lentze N., Auerbach D., Schlattner U. (2009) Yeast two-hybrid, a powerful tool for systems biology. Int. J. Mol. Sci. 10, 2763–2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Brigati C., Kurtz S., Balderes D., Vidali G., Shore D. (1993) An essential yeast gene encoding a TTAGGG repeat-binding protein. Mol. Cell. Biol. 13, 1306–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bilaud T., Koering C. E., Binet-Brasselet E., Ancelin K., Pollice A., Gasser S. M., Gilson E. (1996) The telobox, a Myb-related telomeric DNA binding motif found in proteins from yeast, plants and human. Nucleic Acids Res. 24, 1294–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Konig P., Giraldo R., Chapman L., Rhodes D. (1996) The crystal structure of the DNA-binding domain of yeast RAP1 in complex with telomeric DNA. Cell 85, 125–136 [DOI] [PubMed] [Google Scholar]
- 37. Ribaud V., Ribeyre C., Damay P., Shore D. (2012) DNA-end capping by the budding yeast transcription factor and subtelomeric binding protein Tbf1. EMBO J. 31, 138–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhao Y., McIntosh K. B., Rudra D., Schawalder S., Shore D., Warner J. R. (2006) Fine-structure analysis of ribosomal protein gene transcription. Mol. Cell. Biol. 26, 4853–4862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jorgensen P., Rupes I., Sharom J. R., Schneper L., Broach J. R., Tyers M. (2004) A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 18, 2491–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Powers T., Walter P. (1999) Regulation of ribosome biogenesis by the rapamycin-sensitive TOR-signaling pathway in Saccharomyces cerevisiae. Mol. Biol. Cell 10, 987–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hall D. B., Wade J. T., Struhl K. (2006) An HMG protein, Hmo1, associates with promoters of many ribosomal protein genes and throughout the rRNA gene locus in Saccharomyces cerevisiae. Mol. Cell. Biol. 26, 3672–3679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Askew C., Sellam A., Epp E., Hogues H., Mullick A., Nantel A., Whiteway M. (2009) Transcriptional regulation of carbohydrate metabolism in the human pathogen Candida albicans. PLoS Pathog. 5, e1000612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Uemura H., Fraenkel D. G. (1990) gcr2, a new mutation affecting glycolytic gene expression in Saccharomyces cerevisiae. Mol. Cell. Biol. 10, 6389–6396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lavoie H., Hogues H., Whiteway M. (2009) Rearrangements of the transcriptional regulatory networks of metabolic pathways in fungi. Curr. Opin. Microbiol. 12, 655–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Martchenko M., Levitin A., Whiteway M. (2007) Transcriptional activation domains of the Candida albicans Gcn4p and Gal4p homologs. Eukaryot. Cell 6, 291–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Preti M., Ribeyre C., Pascali C., Bosio M. C., Cortelazzi B., Rougemont J., Guarnera E., Naef F., Shore D., Dieci G. (2010) The telomere-binding protein Tbf1 demarcates snoRNA gene promoters in Saccharomyces cerevisiae. Mol. Cell 38, 614–620 [DOI] [PubMed] [Google Scholar]
- 47. Sullivan B. A., Blower M. D., Karpen G. H. (2001) Determining centromere identity: cyclical stories and forking paths. Nat. Rev. Genet. 2, 584–596 [DOI] [PubMed] [Google Scholar]