Background: MLL5 protein regulates cell cycle progression.
Results: MLL5 regulates the expression of E2F1-target genes through an association with HCF-1.
Conclusion: MLL5 stimulates H3K4 trimethylation at E2F1 responsive promoters and cause transcriptional activation of E2F1 target genes to facilitate the G1 to S phase transition.
Significance: Our results reveal a novel molecular mechanism of MLL5 protein in the regulation of cell cycle progression.
Keywords: Cell Cycle, E2F Transcription Factor, Epigenetics, Gene Expression, Histone Methylation, H3K4, HCF-1, MLL5, Trithorax
Abstract
Trithorax group proteins methylate lysine 4 of histone 3 (H3K4) at active gene promoters. MLL5 protein, a member of the Trithorax protein family, has been implicated in the control of the cell cycle progression; however, the underlying molecular mechanism(s) have not been fully determined. In this study, we found that the MLL5 protein can associate with the cell cycle regulator “host cell factor” (HCF-1). The interaction between MLL5 and HCF-1 is mediated by the “HCF-1 binding motif” (HBM) of the MLL5 protein and the Kelch domain of the HCF-1 protein. Confocal microscopy showed that the MLL5 protein largely colocalized with HCF-1 in the nucleus. Knockdown of MLL5 resulted in reduced cell proliferation and cell cycle arrest in the G1 phase. Moreover, down-regulation of E2F1 target gene expression and decreased H3K4me3 levels at E2F1-responsive promoters were observed in MLL5 knockdown cells. Additionally, the core subunits, including ASH2L, RBBP5, and WDR5, that are necessary for effective H3K4 methyltransferase activities of the Trithorax protein complexes, were absent in the MLL5 complex, suggesting that a distinct mechanism may be used by MLL5 for exerting its H3K4 methyltransferase activity. Together, our findings demonstrate that MLL5 could associate with HCF-1 and then be recruited to E2F1-responsive promoters to stimulate H3K4 trimethylation and transcriptional activation, thereby facilitating the cell cycle G1 to S phase transition.
Introduction
Polycomb and Trithorx group proteins play opposite roles in the regulation of gene repression and activation by methylating lysine 27 (H3K27) or lysine 4 (H3K4), respectively, on histone H3 at the promoter regions of target genes (1). Two polycomb repressive complexes (PRC1 and PRC2) have been identified to date. The Ezh1/2 subunits in the PRC2 complex first catalyze the trimethylation of H3K27, then the Ring1A/B subunits of the PRC1 complex monoubiquitylate histone H2A on lysine 119 (H2AK119Ub1), resulting in strong transcriptional repression (2). In contrast, there are at least seven Trithorax group proteins in mammals, MLL,3 MLL2/ALR, MLL3, MLL4/WBP7, MLL5, SET1A, and SET1B, all of which contain a SET (Su(var)3–9, enhancer-of-zeste, Trithorax) domain that possess H3K4 methyltransferase activity (1).
It has been reported that the founding member of the Trithorax family, MLL, is located within the chromosome 11q23 region and is involved in chromosome translocations, partial tandem duplications, and amplifications in human leukemia (3). Studies on Mll gene knock-out mice showed that Mll plays a key role in both embryonic and adult hematopoiesis (4–6). Similarly, MLL5 is located within chromosome band 7q22, which is frequently deleted in human myeloid leukemia (7–9). Several recent studies on Mll5 gene knock-out mice have also revealed that Mll5 is an important regulator of hematopoietic stem cells (10–13). Thus, both MLL and MLL5 have been implicated in the regulation of hematopoiesis, indicating that a common molecular mechanism might be used by these two Trithorax proteins. It has been shown that MLL3 plays a vital role in adipogenesis (14, 15), whereas MLL4/WBP7 (also known as MLL2) is essential for mouse embryonic development (16), proper embryonic stem cell differentiation (17), and macrophage activation (18). The precise roles of MLL2/ALR, SET1A, and SET1B in embryonic development or hematopoiesis remain to be determined.
Like other members of the Trithorax family, the MLL5 protein contains a SET domain. However, H3K4 methyltransferase activity of the SET domain of the MLL5 protein had not been revealed until a recent study showed that the MLL5 protein exhibits its histone H3K4 methyltransferase activity only after O-GlcNAcylation at Thr-440 in the SET domain by O-GlcNAc transferase (OGT) (19). Additionally, several previous studies demonstrated that overexpression or down-regulation of the MLL5 protein resulted in cell cycle arrest at the G1/S and G2/M phases, and up-regulated p21 and de-phosphorylation of pRb may contribute, at least in part, to cell cycle arrest in MLL5 knockdown cells (20, 21). Nevertheless, whether additional mechanisms, especially the H3K4 methyltransferase activity of the MLL5 protein, are involved in MLL5-mediated cell cycle modulation is still unclear.
HCF-1 was originally discovered as a host cell factor for human herpes simplex virus infection, and it plays a crucial role in transcriptional regulation at specific cell-cycle phases (22). HCF-1-Pro protein undergoes a post-translationally proteolytic maturation process and results in two stably associated HCF-1-N and HCF-1-C fragments (23). Two recent reports showed that O-GlcNAcylation of HCF-1 by OGT was required for HCF-1 proteolytic maturation (24, 25). The HCF-1-N subunit promotes G1 phase progression, whereas the HCF-1-C subunit ensures proper mitosis and cytokinesis at the M phase. Moreover, it has been reported that HCF-1 and the associated SET-1/MLL histone H3K4 methyltransferase complex can be recruited at transcription factor E2Fs-responsive promoters to facilitate cell cycle S phase progression or induce E2F1-mediated DNA damage and apoptosis (26, 27).
In this study, we used affinity purification and a mass spectrum assay to search for MLL5-interacting proteins. We found that MLL5 associated with the cell cycle regulator HCF-1. Additionally, OGT, which has been reported to O-GlcNAcylate both MLL5 and HCF-1, also exists within the MLL5 complex. Common Trithorax scaffold proteins, such as ASH2L, RBBP5, and WDR5, are absent from the MLL5 complex, suggesting that the MLL5 protein may use a unique mechanism to exert its H3K4 methyltransferase activity. Knockdown of MLL5 caused impaired cell proliferation and cell cycle arrest at the G1/S phase. The transcriptional activation of E2F1 target genes and H3K4me3 levels at E2F1-responsive promoters were decreased significantly in MLL5 knockdown cells, suggesting that MLL5 protein could be recruited to E2F1 target promoters through an association with HCF-1. Collectively, our results reveal a novel molecular mechanism of MLL5 protein in the regulation of cell cycle progression at the G1/S phase.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
All cell cultures were maintained in a humidified atmosphere at 37 °C with 5% CO2. The HEK293T and HeLa cell lines were cultured in Dulbecco's modified Eagle's medium complemented with 10% fetal bovine serum (FBS, Hyclone), nonessential amino acids, β-mercaptoethanol, and penicillin/streptomycin antibiotics (Invitrogen). Cells were counted and seeded on 96-, 48-, 12-, and 6-well or 10-cm tissue culture plates (Corning). On the next day, they were either infected with specified lentiviruses or transfected with different plasmid DNAs using Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions.
Antibodies and Plasmids
Antibodies specific to the following proteins were purchased from the manufacturers as indicated: anti-FLAG M2 antibody (F1804, Sigma), anti-HA antibody (11583816001, Roche Applied Science), anti-V5 antibody (SC-271944, Santa Cruz), anti-β-actin antibody (A5316, Sigma), anti-MLL5 antibody (AP14173a, Abgent), anti-HCF-1 antibody (ab110896, Abacm), anti-trimethyl-H3K4 antibody (ab12209, Abcam), anti-WDR5 antibody (ab22512, Abcam), anti-ASH2L antibody (A300–112A, Bethyl Laboratories), anti-RBBP5 antibody (A300–109A, Bethyl Laboratories), anti-OGT antibody (SC-74547, Santa Cruz), anti-Cyclin A2 antibody (4656; Cell Signaling), anti-CDC2 antibody (9112, Cell Signaling), anti-CDC6 antibody (3387; Cell Signaling), anti-E2F1 antibody (SC-193; Santa Cruz), and anti-HCF1 antibody (A301–400A; Bethyl Laboratories).
Mammalian expression plasmid encoding the HCF-1 protein with V5-tag was a kind gift from Prof. Thomas M. Kristie, National Institutes of Health. Plasmids expressing MLL5 (tagged with 3× FLAG or HA) in mammalian cells were constructed by in-frame insertion of a human MLL5 cDNA into pCDEF3 expression vector.
Immunofluorescence and Confocal Microscopy
Cells were fixed for 15 min using 2% paraformaldehyde in PBS, permeabilized in PBS containing 0.25% Triton X-100 for 10 min. Following incubation in blocking solution (PBS-T containing 1% BSA), cells were stained with the polyclonal anti-HCF-1 antibody (ab110896, Abcam) and the monoclonal anti-FLAG (F1804, Sigma). Anti-mouse Alexa Fluor® 555 (A21422; Invitrogen) and anti-rabbit Alexa Fluor® 488 (A110081, Invitrogen) were used as secondary antibodies. Nuclei were stained with DAPI. Images were acquired using Leica DMRE microscope, HCXPL APO ×63/1.32–0.6 OIL CS objective, and Retiga Ex (Qimaging) camera.
Immunoprecipitation and MS of MLL5-containing Molecular Complexes
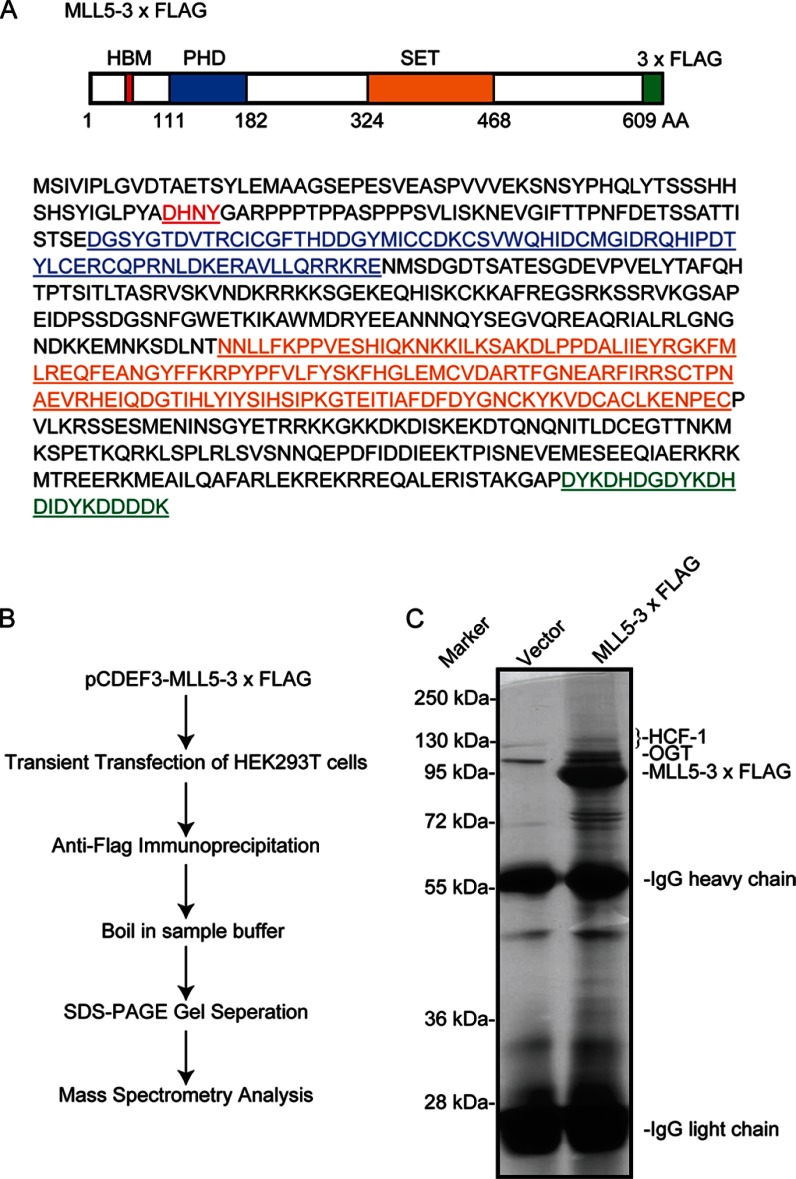
HEK293T cells were seeded to 24 10-cm tissue culture plates; on the next day, half of the plates were transfected with the pCDEF3-MLL5–3×FLAG expression vector, whereas the other half was mock-transfected. About 48 h post-transfection, cells were lysed with lysis buffer (0.05% Nonidet P-40, 20 mm HEPES, 20 mm KCl, 150 mm NaCl, 5 mm EDTA, 1 mm Na3VO4 and complete protease inhibitor mixture (04693132001, Roche Applied Science), pH 7.4, 3 × 107 cells/ml), and rotated at 4 °C for 30 min. Cell lysis supernatants were added to 100 μl of Sepharose beads covalently conjugated to FLAG-specific mAb (A2220, Sigma). Samples were incubated for 2 h with rotation at 4 °C. Beads were washed 3 times with wash buffer (0.05% Nonidet P-40, 20 mm HEPES, 20 mm KCl, 150 mm NaCl, 5 mm EDTA, 1 mm Na3VO4, pH 7.4). Bound proteins were eluted by boiling for 5 min in sample buffer, and the eluted samples were loaded on a 12% SDS-PAGE gel and separated. Gels were stained with silver staining. IgG heavy chain and light chain were excised out from the experiment and control lane; the left whole lane was digested with trypsinogen and analyzed by LC-MS/MS using an LTQ mass spectrometer.
Cell Lysis, Co-immunoprecipitation, and Western Blot Analysis
HEK293T cells were cultured on 10-cm tissue culture plates and co-transfected with the relevant plasmids as described above. Forty-eight hours post-transfection, cells were lysed with lysis buffer (0.05% Nonidet P-40, 20 mm HEPES, 20 mm KCl, 150 mm NaCl, 5 mm EDTA, 1 mm Na3VO4 and complete protease inhibitor mixture (04693132001, Roche Applied Science), pH 7.4, 3 × 107 cells/ml), and incubated on ice for 15 min. Cell lysates were centrifuged for 10 min at 1,3000 × g, at 4 °C. Supernatants were incubated with Protein G-Sepharose beads (P3296, Sigma) coupled to specific antibodies for 2 h with rotation at 4 °C. Beads were washed 3 times with wash buffer (0.05% Nonidet P-40, 20 mm HEPES, 20 mm KCl, 150 mm NaCl, 5 mm EDTA, 1 mm Na3VO4, pH 7.4). Bound proteins were eluted by boiling for 10 min in sample buffer, separated by SDS-PAGE, electrotransferred to PVDF membranes, and blocked for 1 h with 5% nonfat milk solution, followed by Western blotting with the appropriate antibodies and detection by enhanced chemiluminescence (ECL).
shRNA-mediated Knockdown of MLL5 or HCF-1
Specific knockdown of MLL5 or HCF-1 in cultured cells was induced by expression of corresponding shRNAs (shRNAs were cloned into the lentivirus vector pLKO.1 plasmid). In these experiments, MLL5 or HCF-1 are targeted by two different short hairpin-coding sequences (sh-MLL5–1063, 5′-ATC ATT GAA TAC AGA GGG AAG-3′; sh-MLL5–1248, 5′-GGT GAG GCA TGA AAT TCA AGA-3′; sh-HCF-1–889, 5′-GCT TGT CTC AAC CTG GAT ACC-3′; sh-HCF-1–2018, 5′-GCA GTG CTC TGA TTT CCA ATC-3′; sh-Scramble, 5′-TTC TCC GAA CGT GTC ACG TAC-3′), and their expression is driven by the ubiquitously expressed U6 promoter. pLKO.1 plasmids were first purified from bacterial cultures using a miniprep kit (Qiagen). Purified plasmids were used together with packaging plasmids (PSPAX2 and PMD2.G) to cotransfect HEK293T cells cultured in 10-cm dishes. Lentiviral stocks were used to transduce the HeLa cell line.
Chromatin Immunoprecipitation (ChIP) and Real-time PCR Quantification
HeLa cells or HEK293T cells were formaldehyde cross-linked, DNA were isolated and sonicated, and samples were immunoprecipitated, washed, and reverse cross-linked, the cells were lysed in 5 mm PIPES, pH 8.0, 85 mm KCl, and 0.5% Nonidet P-40, and the DNA was sonicated for 16 cycles of 10 s pulse at 25% amplitude using a Bioruptor (VCX500; Sonics & Materials, INC). ChIP DNA was detected by Gel-Red staining of PCR products after gel electrophoresis or by real-time PCR. Real-time PCR of ChIP DNAs was performed in triplicate using a SYBR Green quantitative PCR kit (DRR420A, Takara) and ABI 7900HT Fast Real-time PCR System (Applied Biosystems) under conditions standardized for each primer set (E2F1-Forward, 5′-GTG AGC ACA GCT GCT AGG GGA AG-3′, E2F1-Reverse, 5′-GAA GGC CTC CCA GCC AGG AG-3′; CYCLIN A-Forward, 5′-CTG GAT TAG CAT CTG ATA CCA GAA AGT GAC AC-3′, CYCLIN A-Reverse, 5′-TGA CTC AAT ATC CCT AGG GCA AAG AAA TAG-3′; CDC2-Forward, 5′-ATC TAG GTA ATT ATG CCT GGA GTT CTG ACG-3′, CDC2-Reverse, 5′-TGA GGT CAC AGA CAA ACA TGC TCA AGA C-3′; CDC6-Forward, 5′-TGT TAT TGC ATC GCT CAA TTG GTT AAG AAC-3′, CDC6-Reverse, 5′-GCT GAC ATT TGT TGT CTA CCT ACT ACG TGG-3′; U2C-Forward, 5′-TTT GCT CCC ACT GCC GTC-3′, U2C-Reverse, 5′-CTG AGT CTT TCG GTG CCC-3′). PCR quantification was done with Δ relative CT quantification, in which the values are calculated relative to input as follows: ΔCT = CT (input) − CT (sample); relative unit = 2ΔCT. Inputs correspond to 10% of total ChIP input DNA.
Cell Proliferation and Flow Cytometric Analysis
Bromodeoxyuridine (BrdU) incorporation and flow cytometric analysis were utilized to quantitate DNA-synthesizing cells in vitro. Cell proliferation was measured using an APC-BrdU Flow Kit (552598, BD Pharmingen). The BrdU assay was performed according to the manufacturer's protocol. Infected HeLa cells (1 × 106) were exposed to BrdU at a final concentration of 10 μm in DMEM for 1 h at 37 °C. After removing the labeling solution, cells were fixed and permeabilized in 100 μl of BD Bioscience Cytofix/Cytoperm buffer per well, incubated for 30 min on ice, finally washed 1 time with 1 ml of 1× BD Perm/Wash buffer and centrifuged at 300 × g for 5 min. The cells were incubated with 100 μl of BD Bioscience Cytoperm Plus buffer for 10 min on ice and refixed with 100 μl of BD Bioscience Cytofix/Cytoperm for 5 min on ice as the above fixation. Then cells were treated with 100 μl of diluted DNase (300 μg/ml) for 1 h at 37 °C to expose incorporated BrdU and followed by staining with anti-BrdU-APC for 20 min at room temperature and 20 μl of 7-aminoactinomycin D solution. Finally, the cells were resuspended and analyzed with a flow cytometer (BD LSRII). Data analysis was done using FlowJo 7.6 software.
RESULTS
Identification of MLL5-associated Proteins by Immunoprecipitation and Mass Spectrometry
Although MLL5 protein has been demonstrated to play critical roles in cell cycle regulation and hematopoiesis, the underlying molecular mechanisms have not been fully determined. To further explore the molecular mechanism of MLL5 in cell-cycle control, we sought to identify MLL5-interacting proteins using purification affinity combined with a mass spectrum assay. As described previously, the expression of a short N-terminal isoform of the MLL5 protein is more abundant than the long forms of MLL5 in human tissues (19). We therefore decided to use this short isoform of the MLL5 protein (1–609 amino acids) with both the PHD and SET domain in our research (Fig. 1A). Briefly, the expression vector for 3× FLAG-tagged MLL5 was transiently expressed in the HEK293T cell line, and extracts were prepared and immunoprecipitated with anti-FLAG antibody followed by SDS-PAGE gel separation and silver staining. The stained bands were excised from the gel and analyzed by mass spectrometry (Fig. 1B).
FIGURE 1.
Purification and identification of MLL5-associated proteins. A, schematic representation of 3× FLAG-tagged short isoform of MLL5 protein with a newly identified HBM motif (63–66 amino acids), a PHD (111–182 amino acids), and a SET domain (324–468 amino acids). B, the scheme for purification and identification of MLL5-associated proteins. C, silver staining analysis of MLL5-associated proteins using 3× FLAG-tagged short isoform of MLL5 protein as a bait. MLL5-associated proteins were purified by using anti-FLAG antibody from 2 × 107 MLL5 transfected HEK293T cells, and separated by SDS-PAGE and silver staining. The positions of molecular weight markers are indicated on the left, the overexpressed 3× FLAG-tagged MLL5 protein, the light and heavy chains of the anti-FLAG antibody, the bands of HCF-1 and OGT proteins are indicated on the right.
More than 30 putative MLL5-interacting proteins were identified by the mass spectrum analysis (Table 1 and Fig. 1C), suggesting that MLL5 may be involved in a wide range of biological processes. Among these putative MLL5-interacting proteins, HCF-1 and OGT attracted our attention. HCF-1 is a transcriptional coregulator, conserved among animal species, which is required for herpesvirus gene expression, cell cycle regulation, stem cell pluripotency, stress, and development (28–31). OGT is the only enzyme in the human genome that catalyzes the addition of O-GlcNac to many nuclear and cytoplasmic proteins in many fundamental cellular processes (32). Interestingly, a recent study demonstrated that the MLL5 protein can be GlcNAcylated by OGT, and possesses a histone H3K4 methyltransferase activity (19). Additionally, two recent studies showed that OGT could O-GlcNAcylate HCF-1 and induce HCF-1 proteolytic maturation in the cell-cycle regulation (24, 25). Thus, these results suggest that MLL5, HCF-1, and OGT may be associated in forming a protein complex and may play critical roles in diverse cellular processes.
TABLE 1.
List of MLL5-associated proteins identified by mass spectrum
| Gene symbol | Hits | Molecular mass | Activity |
|---|---|---|---|
| HCF-1 | 26 | 213 kDa | Nuclear coactivator |
| XRCC6 | 12 | 70 kDa | Ku70 |
| DHX9 | 10 | 14 1kDa | Putative RNA helicases |
| XRCC5 | 9 | 83 kDa | Ku80 |
| DDX21 | 9 | 87 kDa | Putative RNA helicases |
| DDX3X | 8 | 73 kDa | Putative RNA helicases |
| ILF2 | 7 | 43 kDa | Interleukin enhancer-binding factor |
| HNRNPU | 7 | 90 kDa | RNA-binding protein |
| SYNCRIP(HNRNPQ) | 7 | 70 kDa | RNA-binding protein |
| SSB Lupus La protein | 7 | 49 kDa | Lupus La protein |
| HNRNPR | 6 | 71 kDa | RNA-binding protein |
| IGF2BP1 | 6 | 63 kDa | IGF-II mRNA-binding protein |
| OGTa | 5 | 117 kDa | O-Linked N-acetylglucosamine (O-GlcNAc) transferase |
| YBX1 | 5 | 36 kDa | YB1 |
| DDX5 | 5 | 69 kDa | Putative RNA helicases |
| HNRNPA1 | 5 | 39 kDa | RNA-binding protein |
| ILF3 | 4 | 96 kDa | Interleukin enhancer-binding factor |
| FOXK1 | 4 | 75 kDa | Forkhead box protein K1 |
| PARP1 | 4 | 113 kDa | Ribosyltransferase |
| HNRNPM | 4 | 76 kDa | RNA-binding protein |
| KPNB1 | 4 | 97 kDa | NLS import receptor |
| PABPC1 | 4 | 71 kDa | Poly(A)-binding protein |
| CSDA | 4 | 40 kDa | DNA-binding protein A |
| NPM1 | 3 | 32 kDa | Assembly and/or transport of ribosome |
| TRIM28 | 3 | 89 kDa | E3 sumo; ligase |
| DDX17 | 3 | 80 kDa | Putative RNA helicases |
| U2AF2 | 3 | 54 kDa | A non-snRNP protein |
| HNRNPF | 3 | 46 kDa | RNA binding protein |
| DDX1 | 3 | 80 kDa | Putative RNA helicases |
| HNRNPA2B1 | 3 | 28 kDa | RNA-binding protein |
| HNRNPAB | 3 | 36 kDa | RNA-binding protein |
| HNRNPD | 3 | 38 kDa | RNA-binding protein |
| NSUN2 | 3 | 86 kDa | RNA methyltransferase |
| IGF2BP3 | 3 | 64 kDa | mRNA-binding protein |
| SNRNP200 | 3 | 245 kDa | Putative RNA helicases |
| G3BP1 | 3 | 52 kDa | DNA-unwinding enzymes |
| DNAJA1 | 3 | 45 kDa | DnaJ (Hsp40)homolog subfamily A member |
a HCF-1 and OGT were confirmed by co-immunoprecipitation assay in our study and are in boldface type.
The Kelch Domain of HCF-1 and the HBM Motif of MLL5 Mediate Their Interaction
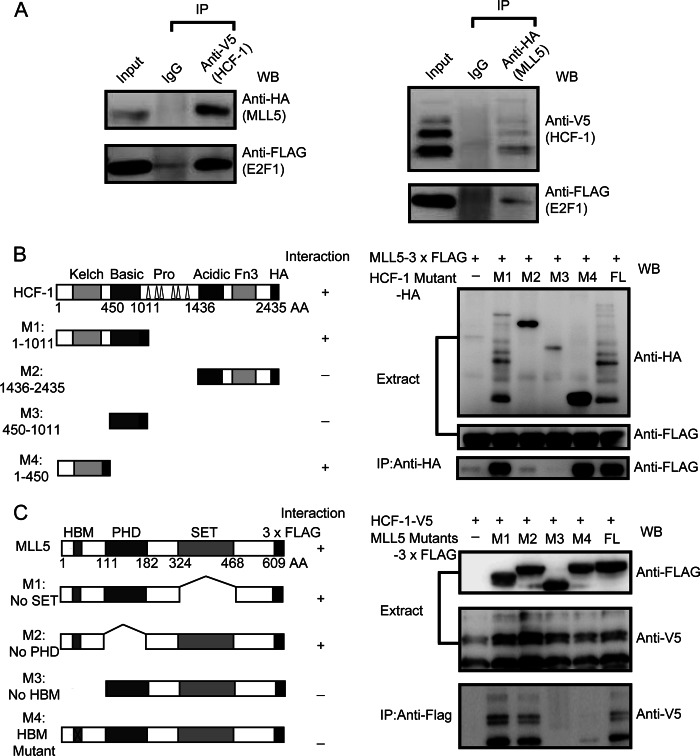
To further investigate the interaction between MLL5 and HCF-1, HEK293T cells were transiently co-transfected with plasmids encoding HA-tagged MLL5 protein and V5-tagged HCF-1 protein, and then subjected to immunoprecipitation and Western blotting analysis with either anti-HA or anti-V5 antibodies. As shown in Fig. 2A, HA-tagged MLL5 protein was able to immunoprecipitate with V5-tagged HCF-1, and V5-tagged HCF-1 was pulled down with HA-tagged MLL5 protein. Thus, the co-immunoprecipitation analysis confirmed the interaction between MLL5 and HCF-1, consistent with our previous mass spectrum results.
FIGURE 2.
Mapping of interacting domains of MLL5 and HCF-1 protein. A, shown is the association of MLL5, HCF-1, and E2F1 proteins. HEK293T cells were co-transfected with plasmids encoding HA-tagged MLL5, V5-tagged HCF-1, and FLAG-tagged E2F1. Left panel, V5-tagged HCF-1 protein was immunoprecipitated (IP) with anti-V5 antibody, and the presence of HA-tagged MLL5 and FLAG-tagged E2F1 protein were examined by Western blotting using anti-HA and anti-FLAG antibodies, respectively. Right panel, HA-tagged MLL5 protein was immunoprecipitated with anti-HA antibody, and the presence of V5-tagged HCF-1 and FLAG-tagged E2F1 proteins were examined by Western blotting using anti-V5 and anti-FLAG antibodies, respectively. B, mapping the MLL5 interacting domain in the HCF-1 protein. Left panel, a schematic representation of the domain structures of the full-length HCF-1 protein and truncated mutants. Right panel, HEK293T cells were co-transfected with plasmids encoding 3× FLAG-tagged MLL5 and HA-tagged full-length HCF-1 and truncated mutants; HA-tagged HCF-1 and truncated mutants were co-immunoprecipitated with anti-HA antibody, and the presence of 3× FLAG-tagged MLL5 protein was examined by Western blotting using anti-FLAG antibody. C, mapping the HCF-1 interacting domain in MLL5 protein. Left panel, a schematic representation of the domain structures of full-length MLL5 and truncated mutants. Right panel, HEK293T cells were co-transfected with plasmids encoding HA-tagged HCF-1 and 3× FLAG-tagged full-length MLL5 and truncated mutants, 3× FLAG-tagged MLL5 and truncated mutants were co-immunoprecipitated with anti-FLAG antibody, and the presence of HA-tagged HCF-1 protein were examined by Western blotting using anti-HA antibody.
We next sought to determine which regions of MLL5 and HCF-1 proteins were responsible for their interaction. HCF-1 exists as a heterodimer of the N- and C-terminal subunits derived from a precursor protein, which undergoes an unusual proteolytic maturation process (31). As shown in Fig. 2B, the HCF-1N subunit contains an N-terminal Kelch domain, which was first identified to bind to the HCF-1-binding motif (HBM) of herpesvirus protein VP16, followed by a basic domain; the HCF-1C contains an acidic domain and a C-terminal Fn3 domain. To identify the MLL5-interacting domains within HCF-1, we generated HA-tagged truncated versions of HCF-1 proteins and performed co-immunoprecipitations with FLAG-tagged MLL5. As shown in Fig. 2B, we found that the HCF-1N subunit was essential for binding of the MLL5 protein, whereas the HCF-1C subunit was dispensable for binding. Further analysis indicated that the Kelch domain of the HCF-1N subunit was crucial for binding the MLL5 protein (Fig. 2B).
We next decided to map the domain or motif of the MLL5 protein responsible for the binding to HCF-1. As mentioned above, the Kelch domain of HCF-1 is required for binding to the MLL5 protein. Moreover, it is known that the Kelch domain can recognize and bind to a conserved tetrapeptide motif (D/E)HXY, known as the HBM, and HBM motifs are found frequently in many known HCF-1-binding proteins, such as VP16 and OGT. Thus, we examined the protein sequence of MLL5 for possible HBMs. We found a previously unidentified HBM motif (PYADHNYGAR, HBM underlined) in the N terminus of the MLL5 protein (Figs. 1A and 2C). To test whether this HBM motif in MLL5 was responsible for interacting with HCF-1, we generated MLL5 mutant proteins lacking either the HBM motif or with a mutant HBM motif (PYADHNYGAR to PYAAAAAGAR, HBM underlined). As shown in Fig. 2C, the MLL5 proteins without the HBM motif and with the mutant HBM motif showed no ability to bind to HCF-1. In contrast, the MLL5 protein lacking either the PHD or SET domains retained their ability to bind to HCF-1. Thus, these results indicate that the newly identified HBM motif in the MLL5 protein was crucial for binding to HCF-1.
Partial Colocalization of MLL5 and HCF-1 Protein in the Nucleus
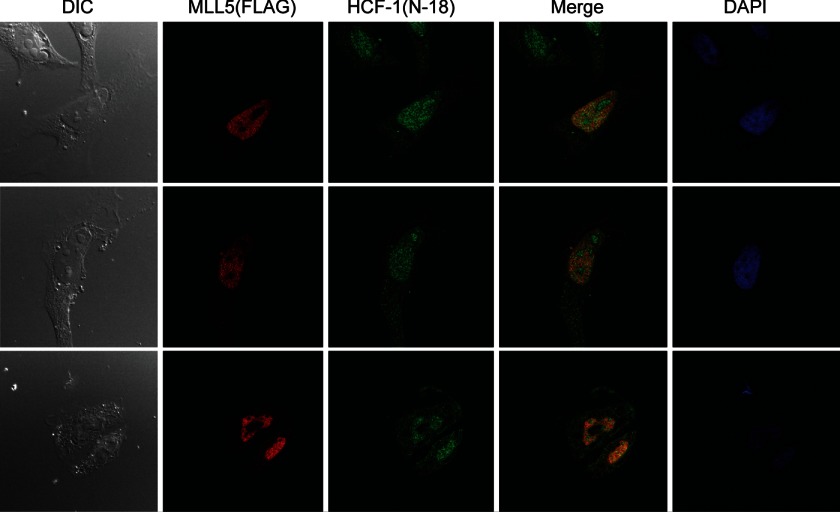
The experiments described above identified an interaction between MLL5 and HCF-1 proteins. We next sought to examine the subcellular localization of these two proteins and determine whether they could colocalize together within cells. Although several previous studies have shown that a short form of the MLL5 proteins (1–560 amino acids) was located mainly in the cell nucleus (20), and the HCF-1 has been detected primarily in the nucleus (30, 31, 33–39), whether they are able to colocalized within the cell nucleus has not been well investigated. To address this, HeLa cells were transiently co-transfected with plasmids encoding HCF-1 protein and FLAG-tagged MLL5 protein. Then 48 h after transfection, MLL5 and HCF-1 proteins were detected by immunofluorescent staining using a mouse anti-FLAG antibody (M2) and a rabbit anti-HCF-1 polyclonal antibody (N-18), respectively. As shown in Fig. 3, the MLL5 protein was detected almost exclusively in the cell nucleus, but largely excluded from the nucleoli. Also, HCF-1 was found predominantly in the nucleus, with a weaker signal also detected in the cytoplasm. Thus, these results show that MLL5 and HCF-1 proteins are largely colocalized in the cell nucleoplasm, consistent with the physical interaction between these two proteins in the co-immunoprecipitation assay.
FIGURE 3.
Partial colocalization of MLL5 and HCF-1 in the nucleus. HeLa cells were transiently co-transfected with plasmids encoding HCF-1 and 3× FLAG-tagged MLL5, and then fixed and subjected to indirect immunofluorescence using anti-HCF-1 polyclonal antibody (N-18) to detect HCF-1 and anti-FLAG monoclonal antibody to detect MLL5.
MLL5 Protein Does Not Associate with Core Component Proteins ASH2L, RBBP5, and WDR5
It has been demonstrated previously that MLL1/2, MLL3/4, and SET-1A/B complexes share common core components, including ASH2L, RBBP5, and WDR5, which are required for proper complex assembly and modulation of H3K4 methyltransferase activity (1). For example, the MLL1 protein is cleaved by Taspase-1 into two associated N-320 and C-180 fragments, and the C-180 fragment then interacts with ASH2L, RBBP5, and WDR5 proteins (40, 41). However, whether such common core components also associate with the MLL5 protein remained unclear; we thus sought to examine the possible association between MLL5 and those core component proteins by co-immunoprecipitation analysis.
The C-180 fragment of MLL was used as a positive control for the binding of these core component proteins in our experiment. HEK293T cells were transiently transfected with plasmids encoding 3× FLAG-tagged MLL5 protein or the C-180 fragment of the MLL protein, and the presence of endogenous ASH2L, RBBP5, and WDR5 in the MLL C-180 fragment or MLL5 precipitates was detected by using polyclonal antibodies against ASH2L, RBBP5, and WDR5, respectively (Fig. 4A). As shown in Fig. 4B, the expected co-immunoprecipitations were observed between the C-180 fragment of MLL and ASH2L, RBBP5, and WDR5. In contrast, none of these core components was detected in the MLL5 precipitates. Additionally, we found that OGT could associate with the MLL5 protein but not the MLL protein (Fig. 4B). Thus, along with a previous study (19), our results suggest that, unlike other members of the Trithorax protein family, the MLL5 protein may rely on the OGT-mediated O-GlcNAcylation rather than these common core component proteins for its H3K4 methyltransferase activity.
FIGURE 4.
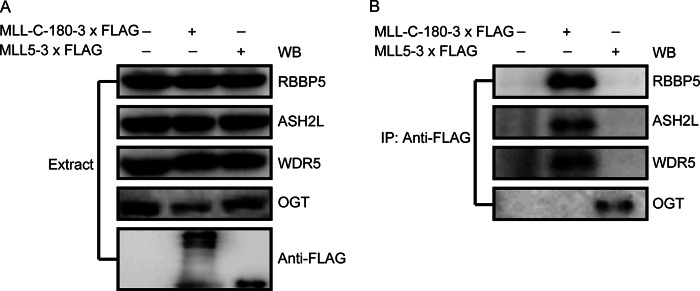
MLL5 protein does not associate with core component proteins ASH2L, RBBP5, and WDR5. A, HEK293T cells were transiently transfected with plasmids encoding 3× FLAG-tagged MLL5 protein or the C-180 fragment of the MLL protein. Expression of the MLL5 protein and MLL C-180 fragment were detected by using anti-FLAG antibody. Endogenous ASH2L, RBBP5, and WDR5 proteins were detected by Western blotting using polyclonal antibodies against ASH2L, RBBP5, and WDR5, respectively. B, the presence of ASH2L, RBBP5, WDR5, and OGT proteins were detected in the MLL C-180 fragment or MLL5 precipitates using polyclonal antibodies against ASH2L, RBBP5, WDR5, and OGT, respectively.
Knockdown of MLL5 Caused Cell Proliferation Defects, and the Levels of Both H3K4 Trimethylation at E2F1-responsive Promoters and the E2F1 Target Gene Expression Are Down-regulated in MLL5 Knockdown Cells
MLL5 has previously been implicated in cell-cycle regulation; overexpression or down-regulation of the MLL5 protein can induce cell-cycle arrest (20, 21). Interestingly, HCF-1 has also been identified as an important regulator of cell-cycle progression (42, 43). HCF-1 is a heterodimeric complex of two subunits: the HCF-1N subunit promotes passage through the G1/S phase, whereas the HCF-1C subunit controls the proper exit from mitosis in the M phase (31). Thus, our finding of a physical interaction between MLL5 and HCF-1 prompted us to further investigate the biological significance of their interaction in cell-cycle progression.
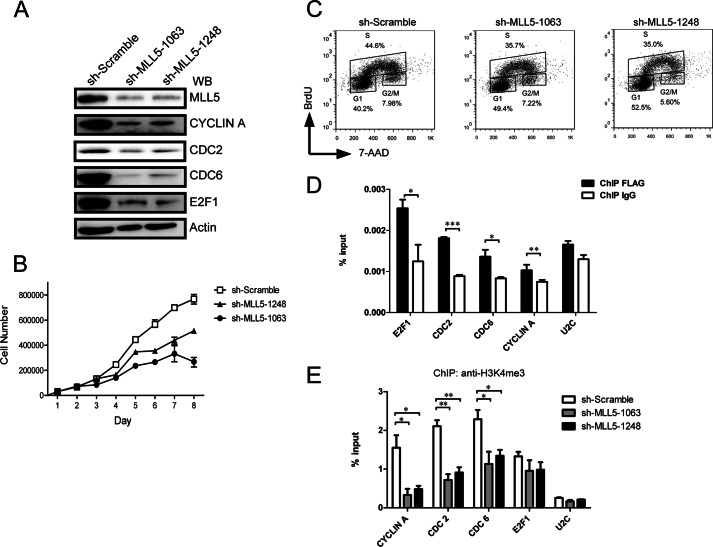
We first used a lentivirus-based short hairpin RNA (shRNA) approach to knockdown MLL5 expression in HeLa cells. Two different shRNAs targeted to mRNA sequences starting at 1063 and 1248 bp downstream of the translation start site were used; both could knockdown MLL5 protein effectively (Fig. 5A). We then detected cell growth in MLL5 knockdown cells, and observed a dramatic decrease in cell proliferation in MLL5 knockdown cells (Fig. 5B), consistent with previous results (21). Moreover, a BrdU incorporation assay was used to monitor cell proliferation in MLL5 knockdown cells, and we found a dramatic decrease in the percentage of S-phase cells in MLL5 knockdown cells (Fig. 5C), suggesting a delay in the G1 to S phase transition may occur in MLL5 knockdown cells.
FIGURE 5.
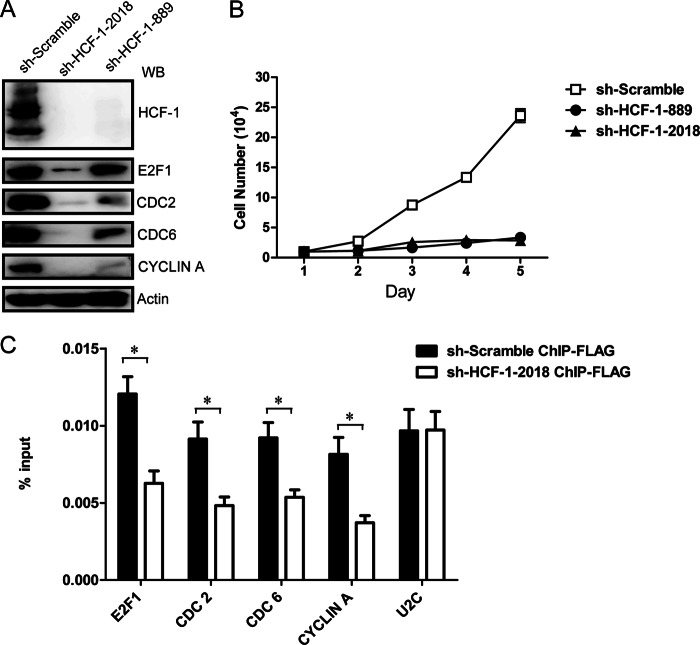
Knockdown of MLL5 caused cell proliferation defects, and the levels of both H3K4 trimethylation at E2F1-responsive promoters and the E2F1 target gene expression are down-regulated in MLL5 knockdown cells. A, knockdown of MLL5 protein in HeLa cells leads to down-regulation of E2F1 target genes. Two different lentivirus-based shRNAs target to an mRNA sequence starting 1063 and 1248 bp downstream of the translation start site were used to knockdown MLL5 expression in HeLa cells. Whole cell lysates were harvested at day 6 post-infection. The expression of endogenous E2F1, CYCLIN A, CDC2, and CDC6 were detected by polyclonal antibodies. Actin was used as a loading control. B, defective cell proliferation in MLL5 knockdown cells. 3 × 104 cells were seeded in 48-well plates at day 1, at each indicated time point, triplicates of cells from each group were harvested and counted in a hemocytometer. C, BrdU incorporation assay in MLL5 knockdown cells. BrdU was added to the culture medium for 1 h, and cells were collected, fixed, and then stained with anti-BrdU antibody conjugated with allophycocyanine, and 7-aminoactinomycin D (7-AAD) was used to stain genomic DNA. D, MLL5 localizes to E2F1-responsive promoters. HEK293T cells were transiently transfected with plasmids encoding 3× FLAG-tagged MLL5, and the cells were harvested 48 h later. Quantitation of ChIP analysis of transfected HEK293T cells with an anti-FLAG antibody by triplicate real-time PCR of the indicated promoters is shown. E, the H3K4 trimethylation levels at E2F1 target promoters were decreased significantly in MLL5 knockdown cells. Quantitation of H3K4 trimethylation ChIP analysis of HeLa cells infected with lentivirus-based scramble control or shRNA by triplicate real-time PCR of the indicated promoters is shown. Two-tailed unpaired Student's t tests were performed, *, p < 0.05; **, p < 0.01; ***, p < 0.001.
The G1 to S phase transition is a key regulatory point in the cell cycle, and E2Fs transcription factors coordinate the expression of a number of genes involved in cell-cycle progression, particularly those involved in progression through G1 and into the S phase of the cell cycle (44). At late G1 phase, E2F1 disassociates from phosphorylated retinoblastoma (RB) proteins and binds to promoters of target genes to induce the activation of transcription of various genes required for S phase entry (45). Deregulated expression of E2F1 target genes has been shown to induce inappropriate S phase entry (46). It has been shown previously that HCF-1 can physically interact with the E2F1 protein, and is involved in E2F1-mediated transcriptional activation in the late G1 phase (26). Interestingly, in our experiment, we found that MLL5 could be co-immunoprecipitated with HCF-1 and E2F1 (Fig. 2A), indicating that MLL5 may associate with E2F1 through binding to HCF-1. This observation prompted us to hypothesize that the MLL5 protein could be recruited by HCF-1 to E2F1-responsive promoters, and then stimulate H3K4 trimethylation at these promoters and facilitate the transactivation of E2F1 target genes during the G1/S phase.
To test this, ChIP assays were performed using an anti-FLAG antibody in HEK293T cells transiently transfected with 3× FLAG-tagged MLL5. As shown in Fig. 5D, an enrichment of MLL5 protein was detected at the promoters of E2F1 target genes, including CYCLIN A (47), CDC2 (48), CDC6 (49–51), and E2F1 itself (44). These results suggest that MLL5 localizes to the promoters of E2F1 target genes. Moreover, we also found that the expression levels of these E2F1 target genes were significantly down-regulated in MLL5 knockdown cells (Fig. 5A). We further examined the H3K4 trimethylation levels at these E2F1-responsive promoters in both scramble control cells and MLL5 knockdown cells by a ChIP assay with anti-H3K4me3 antibody followed by real-time PCR analysis. As shown in Fig. 5E, the H3K4 trimethylation levels at the CYCLIN A, CDC2, and CDC6 promoters were reduced significantly in MLL5 knockdown cells. However, H3K4 trimethylation levels at the E2F1 promoter were only mildly decreased in MLL5 knockdown cells, suggesting that other H3K4 methyltransferases may play redundant roles in histone H3K4 trimethylation at the E2F1 promoters. Thus, our results suggest that MLL5 localizes to the promoters of E2F1 target genes and induces H3K4 trimethylation and gene transcriptional activation.
HCF-1 Is Required for MLL5 Recruitment to E2F1-responsive Promoters
We next examined whether the recruitment of MLL5 to E2F1-responsive promoters is dependent on HCF-1. To address this, we used a lentivirus-based shRNA approach to knockdown HCF-1 expression in HeLa cells. Two different shRNAs targeted to mRNA sequences starting at 889 and 2018 bp downstream of the translation start site were used; both could effectively knockdown the HCF-1 expression (Fig. 6A). Consistent with previous reports (42, 43), loss of HCF-1 causes cell proliferation defects (Fig. 6B). Moreover, the expression levels of CYCLIN A, CDC2, CDC6, and E2F1 proteins were also significantly down-regulated in HCF-1 knockdown cells (Fig. 6A).
FIGURE 6.
HCF-1 is required for MLL5 recruitment to E2F1-responsive promoters. A, knockdown of HCF-1 in HeLa cells leads to down-regulation of E2F1 target genes. Two different lentivirus-based shRNAs target to an mRNA sequence starting 889 and 2018 bp downstream of the translation start site were used to knockdown HCF-1 expression in HeLa cells. Whole cell lysates were harvested at day 6 post-infection. The expression of endogenous HCF-1, E2F1, CYCLIN A, CDC2, and CDC6 were detected by polyclonal antibodies. Actin was used as a loading control. WB, Western blot. B, defective cell proliferation in HCF-1 knockdown cells. 1 × 104 HeLa cells were seeded in 48-well plates at day 1, at each indicated time point, triplicates of cells from each group were harvested and counted in a hemocytometer. C, binding of the MLL5 protein to E2F1 target promoters were markedly reduced in HCF-1 knockdown cells. 3× FLAG-tagged MLL5 were transiently transfected in HEK293T cell lines that have been infected with lentivirus-based HCF-1 shRNA or scramble control, and the cells were harvested 72 h later. ChIP analyses were performed with an anti-FLAG antibody in HEK293T previously infected with lentivirus-based scramble control or HCF-1 shRNA. Two-tailed unpaired Student's t tests were performed, * p < 0.05.
Next, HEK293T cell lines stably expressing lentivirus-based HCF-1 shRNA or scramble control were transiently transfected with 3× FLAG-tagged MLL5 protein. The association of MLL5 with the E2F1-responsive promoters were then detected by a ChIP assay with anti-FLAG antibody followed by real-time PCR analysis. As shown in Fig. 6C, knockdown of HCF-1 markedly reduces the binding of MLL5 to the promoters of E2F1 target genes, suggesting that HCF-1 is required for MLL5 recruitment to these E2F1-responsive promoters.
Taken together, our results demonstrate that the MLL5 protein could be recruited to E2F1-responsive promoters through association with HCF-1, and could stimulate H3K4 trimethylation at these promoters and then facilitate transcriptional activation of E2F1 target genes during the G1 to S phase transition.
DISCUSSION
In the current study, we revealed a new molecular mechanism to explain how the MLL5 protein regulates cell cycle progression. We demonstrated that the MLL5 protein physically associated with the cell cycle regulator HCF-1 and the O-GlcNAc transferase OGT, and then was recruited to E2F1-responsive promoters to stimulate H3K4 trimethylation at the promoters and cause transcriptional activation of E2F1 target genes to facilitate the G1 to S phase transition. Thus, our results provide new insights into the molecular mechanism of MLL5 action in the cell-cycle transition from G1 to S phase (Fig. 7).
FIGURE 7.
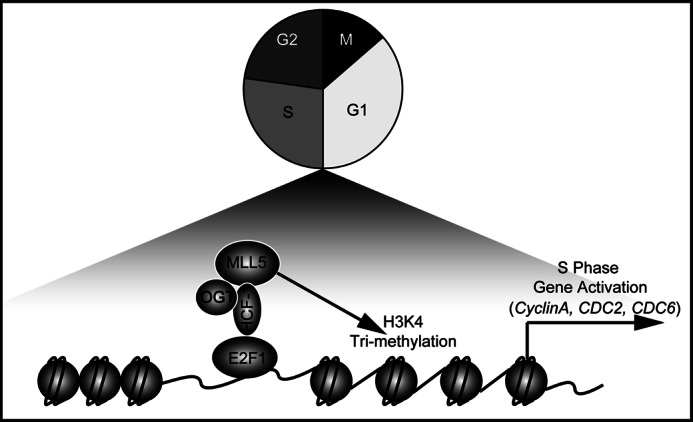
Model for MLL5-mediated cell cycle control. MLL5 form a complex with HCF-1 and OGT, and then was recruited to E2F1 responsive promoters (CYCLIN A, CDC2, and CDC6) to stimulate H3K4 trimethylation and cause transcriptional activation of E2F1 target genes to facilitate the G1 to S phase transition. See text for details.
The HCF-1 protein was discovered originally as a host cell factor for human herpes simplex virus infection, and later studies indicated that HCF-1 plays a key role in cell-cycle progression, partially through its interactions with various members of the E2F transcription factor family to induce cell cycle-specific transcription (22, 26). In this study, we further showed that MLL5 could associate with HCF-1 within the nucleus, and a previously unknown HBM motif in the MLL5 protein mediated the interaction with the Kelch domain of the HCF-1 protein. Interestingly, we found that the O-GlcNAc transferase OGT protein can also be co-immunoprecipitated with MLL5 and HCF-1. It has previously been shown that OGT-mediated GlcNAcylation at Thr-440 in the MLL5 SET domain is essential for its H3K4 methyltransferase activity (19). Moreover, OGT-directed GlcNAcylation of HCF-1 is necessary for proteolytic maturation of HCF-1 (24, 25). Thus, these results suggest that MLL5 and OGT/HCF-1 may form a protein complex to modulate cell-cycle progression. Interestingly, several lines of evidence suggest that HCF-1/OGT complexes are not only limited to cell cycle regulation, but are also implicated in a wide range of biological processes, including gluconeogenesis (52) and leukemia transformation (53). Thus it is of particular interest in the future to study whether MLL5 also contributes to these HCF-1/OGT-involved biological processes.
Although defective cell-cycle progression has been observed in MLL5 knockdown cells, the roles of HCF-1 and E2F1 in this process have not been well investigated previously. The E2F family of transcription factors plays a pivotal role in coordinating the transcriptional activation of genes important for the G1 to S phase cell-cycle transition. In late G1 phase, the E2F1 protein disassociates from phosphorylated RB protein and then transactivates E2F1 target genes, facilitating the G1 to S phase transition. In a previous study, Cheng et al. (21) found up-regulation of the dephosphorylation of the RB protein in MLL5 knockdown cells, suggesting that the RB-E2F1 pathway may play a role in MLL5 knockdown-induced cell-cycle arrest. In this study, we further provide new evidence showing that the MLL5 protein can be recruited to E2F1-responsive promoters through association with HCF-1, and stimulate the transcription of E2F1 target genes, thereby ensuring proper execution of the G1 to S phase transition.
H3K4 trimethylation at gene promoters has been implicated in transcriptional activation, and Trithorax group proteins are responsible for H3K4 methylation (1). Interestingly, the H3K4 methyltransferase activity of the MLL5 protein had not been verified until a recent study by Fujiki et al. (19) showed that the H3K4 methyltransferase activity of MLL5 can be evoked by OGT-mediated GlcNAcylation at Thr-440 in the MLL5 SET domain. Consistent with this, we found that levels of H3K4 trimethylation at E2F1 target promoters were decreased significantly in MLL5 knockdown cells, suggesting that the MLL5 protein may possess H3K4 methyltransferase activity at E2F1 target gene promoters and stimulate gene transcription. Interestingly, MLL complexes could also be recruited to E2F1-responsive promoters by HCF-1 to stimulate H3K4 trimethylation and transcriptive expression during the G1 to S phase cell cycle transition (26). Thus, our results, along with the previous observations (26), suggest that MLL and MLL5 may use a similar molecular pathway to promote E2F1 target gene expression during cell-cycle progression.
A core protein scaffold composed of WDR5, RBBP5, and ASH2L subunits is essential for the H3K4 methyltransferase activities of the Trithorax protein complexes (1, 54–56). However, we were unable to find these core subunits in MLL5 protein complexes using either mass spectrum or co-immunoprecipitation assays, suggesting that these core subunits may be dispensable for the H3K4 methyltransferase activity of the MLL5 protein. Moreover, the MLL5 protein is structurally different from other members of the Trithorax family, as evidenced by the observation that the SET domain of the MLL5 protein is located in the N-terminal region, rather than at the C terminus of the protein, as well as the fact that a post-SET motif is present in other Trithorax family proteins, whereas it is absent from the MLL5 protein (7). Thus, these results suggest that, unlike the other Trithorax family proteins, MLL5 may use a distinct mechanism to exert its H3K4 methyltransferase activity.
Although MLL5 was originally identified as a novel Trithorax family member and a potential tumor suppressor in human myeloid leukemia, its biological function in vivo had not been revealed until recently (7, 13). Several recent studies on Mll5 knock-out mice, including one of ours, showed that Mll5 plays an important role in hematopoiesis (10–12). Inactivation of Mll5 in hematopoietic stem cells (HSCs) causes a severe impairment in long-term hematopoietic reconstitution under competitive conditions. Intriguingly, Mll-deficient HSCs also exhibit impaired long-term reconstitution ability (5, 6). Because one of the cellular bases for hematopoietic reconstitution is the cell cycling of transplanted HSCs in the recipients, it seems reasonable to hypothesize that the impaired reconstitution ability of Mll- or Mll5-deficient HSC might result, at least in part, from the altered cell cycle progression of the transplanted Mll- or Mll5-deficient HSC. Furthermore, it is of interest to determine whether other members of the Trithorax family also regulate cell cycle progression or HSCs self-renewal.
In summary, our study provides new insights into the molecular mechanisms of MLL5 in cell-cycle regulation. MLL5 and MLL proteins may share a common molecular mechanism to control cell-cycle progression; it will, therefore, be of interest to further investigate the roles of other Trithorax family members, particularly in cell-cycle progression and the regulation of HSCs, in the future. Moreover, increasing evidence shows that the HCF-1/OGT complex is involved in a wide range of biological processes. Therefore, future studies should determine whether MLL5 is involved in these HCF-1/OGT complex-related biological processes.
Acknowledgments
We thank Prof. Thomas M. Kristie for the gift of HCF-1 cDNA. We gratefully acknowledge the assistance of the Core Facility for Flow Cytometry, Institut Pasteur of Shanghai, and Core Facility for Proteomics in the Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences.
This work was supported by National Natural Science Foundation of China Grants 30971672, 81090410, and 81270618, National Basic Research Program of China Grants 2010CB945600 and 2011CB966300, and grants from the 100 Talent Program of the Chinese Academy of Sciences (to Y. Z.), Shanghai Pujiang Program Grant 11PJ1401800 (to Q. L.), Research Fund for the Doctoral Program from Ministry of Education of China Grant 20093704120004, and Award Research Funds for Young Scholars from Shandong Province Grant BS2010SW031 (to J. Y.).
- MLL
- mixed lineage leukemia
- HCF
- host cell factor
- HBM
- HCF-1-binding motif
- RB
- retinoblastoma
- HSC
- hematopoietic stem cell
- OGT
- O-GlcNAc transferase.
REFERENCES
- 1. Schuettengruber B., Martinez A. M., Iovino N., Cavalli G. (2012) Trithorax group proteins. Switching genes on and keeping them active. Nat. Rev. Mol. Cell Biol. 12, 799–814 [DOI] [PubMed] [Google Scholar]
- 2. Margueron R., Reinberg D. (2011) The Polycomb complex PRC2 and its mark in life. Nature 469, 343–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hess J. L. (2004) MLL. A histone methyltransferase disrupted in leukemia. Trends Mol. Med. 10, 500–507 [DOI] [PubMed] [Google Scholar]
- 4. Yagi H., Deguchi K., Aono A., Tani Y., Kishimoto T., Komori T. (1998) Growth disturbance in fetal liver hematopoiesis of Mll-mutant mice. Blood 92, 108–117 [PubMed] [Google Scholar]
- 5. McMahon K. A., Hiew S. Y., Hadjur S., Veiga-Fernandes H., Menzel U., Price A. J., Kioussis D., Williams O., Brady H. J. (2007) Mll has a critical role in fetal and adult hematopoietic stem cell self-renewal. Cell Stem Cell 1, 338–345 [DOI] [PubMed] [Google Scholar]
- 6. Jude C. D., Climer L., Xu D., Artinger E., Fisher J. K., Ernst P. (2007) Unique and independent roles for MLL in adult hematopoietic stem cells and progenitors. Cell Stem Cell 1, 324–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Emerling B. M., Bonifas J., Kratz C. P., Donovan S., Taylor B. R., Green E. D., Le Beau M. M., Shannon K. M. (2002) MLL5, a homolog of Drosophila trithorax located within a segment of chromosome band 7q22 implicated in myeloid leukemia. Oncogene 21, 4849–4854 [DOI] [PubMed] [Google Scholar]
- 8. Fischer K., Fröhling S., Scherer S. W., McAllister Brown J., Scholl C., Stilgenbauer S., Tsui L. C., Lichter P., Döhner H. (1997) Molecular cytogenetic delineation of deletions and translocations involving chromosome band 7q22 in myeloid leukemias. Blood 89, 2036–2041 [PubMed] [Google Scholar]
- 9. Kratz C. P., Emerling B. M., Donovan S., Laig-Webster M., Taylor B. R., Thompson P., Jensen S., Banerjee A., Bonifas J., Makalowski W., Green E. D., Le Beau M. M., Shannon K. M. (2001) Candidate gene isolation and comparative analysis of a commonly deleted segment of 7q22 implicated in myeloid malignancies. Genomics 77, 171–180 [DOI] [PubMed] [Google Scholar]
- 10. Heuser M., Yap D. B., Leung M., de Algara T. R., Tafech A., McKinney S., Dixon J., Thresher R., Colledge B., Carlton M., Humphries R. K., Aparicio S. A. (2009) Loss of MLL5 results in pleiotropic hematopoietic defects, reduced neutrophil immune function, and extreme sensitivity to DNA demethylation. Blood 113, 1432–1443 [DOI] [PubMed] [Google Scholar]
- 11. Madan V., Madan B., Brykczynska U., Zilbermann F., Hogeveen K., Döhner K., Döhner H., Weber O., Blum C., Rodewald H. R., Sassone-Corsi P., Peters A. H., Fehling H. J. (2009) Impaired function of primitive hematopoietic cells in mice lacking the Mixed-Lineage-Leukemia homolog MLL5. Blood 113, 1444–1454 [DOI] [PubMed] [Google Scholar]
- 12. Zhang Y., Wong J., Klinger M., Tran M. T., Shannon K. M., Killeen N. (2009) MLL5 contributes to hematopoietic stem cell fitness and homeostasis. Blood 113, 1455–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu H., Westergard T. D., Hsieh J. J. (2009) MLL5 governs hematopoiesis. A step closer. Blood 113, 1395–1396 [DOI] [PubMed] [Google Scholar]
- 14. Lee J., Saha P. K., Yang Q. H., Lee S., Park J. Y., Suh Y., Lee S. K., Chan L., Roeder R. G., Lee J. W. (2008) Targeted inactivation of MLL3 histone H3-Lys-4 methyltransferase activity in the mouse reveals vital roles for MLL3 in adipogenesis. Proc. Natl. Acad. Sci. U.S.A. 105, 19229–19234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee S., Lee D. K., Dou Y., Lee J., Lee B., Kwak E., Kong Y. Y., Lee S. K., Roeder R. G., Lee J. W. (2006) Coactivator as a target gene specificity determinant for histone H3 lysine 4 methyltransferases. Proc. Natl. Acad. Sci. U.S.A. 103, 15392–15397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Glaser S., Schaft J., Lubitz S., Vintersten K., van der Hoeven F., Tufteland K. R., Aasland R., Anastassiadis K., Ang S. L., Stewart A. F. (2006) Multiple epigenetic maintenance factors implicated by the loss of Mll2 in mouse development. Development 133, 1423–1432 [DOI] [PubMed] [Google Scholar]
- 17. Lubitz S., Glaser S., Schaft J., Stewart A. F., Anastassiadis K. (2007) Increased apoptosis and skewed differentiation in mouse embryonic stem cells lacking the histone methyltransferase mll2. Mol. Biol. Cell 18, 2356–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Austenaa L., Barozzi I., Chronowska A., Termanini A., Ostuni R., Prosperini E., Stewart A. F., Testa G., Natoli G. (2012) The histone methyltransferase Wbp7 controls macrophage function through GPI glycolipid anchor synthesis. Immunity 36, 572–585 [DOI] [PubMed] [Google Scholar]
- 19. Fujiki R., Chikanishi T., Hashiba W., Ito H., Takada I., Roeder R. G., Kitagawa H., Kato S. (2009) GlcNAcylation of a histone methyltransferase in retinoic-acid-induced granulopoiesis. Nature 459, 455–459 [DOI] [PubMed] [Google Scholar]
- 20. Deng L. W., Chiu I., Strominger J. L. (2004) MLL 5 protein forms intranuclear foci, and overexpression inhibits cell cycle progression. Proc. Natl. Acad. Sci. U.S.A. 101, 757–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cheng F., Liu J., Zhou S. H., Wang X. N., Chew J. F., Deng L. W. (2008) RNA interference against mixed lineage leukemia 5 resulted in cell cycle arrest. Int. J. Biochem. Cell Biol. 40, 2472–2481 [DOI] [PubMed] [Google Scholar]
- 22. Zargar Z., Tyagi S. (2012) Role of host cell factor-1 in cell cycle regulation. Transcription 3, 187–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wilson A. C., LaMarco K., Peterson M. G., Herr W. (1993) The VP16 accessory protein HCF is a family of polypeptides processed from a large precursor protein. Cell 74, 115–125 [DOI] [PubMed] [Google Scholar]
- 24. Capotosti F., Guernier S., Lammers F., Waridel P., Cai Y., Jin J., Conaway J. W., Conaway R. C., Herr W. (2011) O-GlcNAc transferase catalyzes site-specific proteolysis of HCF-1. Cell 144, 376–388 [DOI] [PubMed] [Google Scholar]
- 25. Daou S., Mashtalir N., Hammond-Martel I., Pak H., Yu H., Sui G., Vogel J. L., Kristie T. M., Affar el B. (2011) Crosstalk between O-GlcNAcylation and proteolytic cleavage regulates the host cell factor-1 maturation pathway. Proc. Natl. Acad. Sci. U.S.A. 108, 2747–2752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tyagi S., Chabes A. L., Wysocka J., Herr W. (2007) E2F activation of S phase promoters via association with HCF-1 and the MLL family of histone H3K4 methyltransferases. Mol. Cell 27, 107–119 [DOI] [PubMed] [Google Scholar]
- 27. Tyagi S., Herr W. (2009) E2F1 mediates DNA damage and apoptosis through HCF-1 and the MLL family of histone methyltransferases. EMBO J. 28, 3185–3195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dejosez M., Levine S. S., Frampton G. M., Whyte W. A., Stratton S. A., Barton M. C., Gunaratne P. H., Young R. A., Zwaka T. P. (2010) Ronin/Hcf-1 binds to a hyperconserved enhancer element and regulates genes involved in the growth of embryonic stem cells. Genes Dev. 24, 1479–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dejosez M., Krumenacker J. S., Zitur L. J., Passeri M., Chu L. F., Songyang Z., Thomson J. A., Zwaka T. P. (2008) Ronin is essential for embryogenesis and the pluripotency of mouse embryonic stem cells. Cell, 133, 1162–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Peng H., Nogueira M. L., Vogel J. L., Kristie T. M. (2010) Transcriptional coactivator HCF-1 couples the histone chaperone Asf1b to HSV-1 DNA replication components. Proc. Natl. Acad. Sci. U.S.A. 107, 2461–2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Julien E., Herr W. (2003) Proteolytic processing is necessary to separate and ensure proper cell growth and cytokinesis functions of HCF-1. EMBO J. 22, 2360–2369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hanover J. A., Krause M. W., Love D. C. (2012) Bittersweet memories. Linking metabolism to epigenetics through O-GlcNAcylation. Nat. Rev. Mol. Cell Biol. 13, 312–321 [DOI] [PubMed] [Google Scholar]
- 33. Kristie T. M., Pomerantz J. L., Twomey T. C., Parent S. A., Sharp P. A. (1995) The cellular C1 factor of the herpes simplex virus enhancer complex is a family of polypeptides. J. Biol. Chem. 270, 4387–4394 [DOI] [PubMed] [Google Scholar]
- 34. La Boissière S., Hughes T., O'Hare P. (1999) HCF-dependent nuclear import of VP16. EMBO J. 18, 480–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wilson A. C., Boutros M., Johnson K. M., Herr W. (2000) HCF-1 amino- and carboxyl-terminal subunit association through two separate sets of interaction modules. Involvement of fibronectin type 3 repeats. Mol. Cell. Biol. 20, 6721–6730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Julien E., Herr W. (2004) A switch in mitotic histone H4 lysine 20 methylation status is linked to M phase defects upon loss of HCF-1. Mol Cell 14, 713–725 [DOI] [PubMed] [Google Scholar]
- 37. Narayanan A., Nogueira M. L., Ruyechan W. T., Kristie T. M. (2005) Combinatorial transcription of herpes simplex virus and varicella zoster virus immediate early genes is strictly determined by the cellular coactivator HCF-1. J. Biol. Chem. 280, 1369–1375 [DOI] [PubMed] [Google Scholar]
- 38. Misaghi S., Ottosen S., Izrael-Tomasevic A., Arnott D., Lamkanfi M., Lee J., Liu J., O'Rourke K., Dixit V. M., Wilson A. C. (2009) Association of C-terminal ubiquitin hydrolase BRCA1-associated protein 1 with cell cycle regulator host cell factor 1. Mol. Cell. Biol. 29, 2181–2192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mangone M., Myers M. P., Herr W. (2010) Role of the HCF-1 basic region in sustaining cell proliferation. PLoS One 5, e9020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hsieh J. J., Ernst P., Erdjument-Bromage H., Tempst P., Korsmeyer S. J. (2003) Proteolytic cleavage of MLL generates a complex of N- and C-terminal fragments that confers protein stability and subnuclear localization. Mol. Cell. Biol. 23, 186–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hsieh J. J., Cheng E. H., Korsmeyer S. J. (2003) Taspase1. A threonine aspartase required for cleavage of MLL and proper HOX gene expression. Cell 115, 293–303 [DOI] [PubMed] [Google Scholar]
- 42. Goto H., Motomura S., Wilson A. C., Freiman R. N., Nakabeppu Y., Fukushima K., Fujishima M., Herr W., Nishimoto T. (1997) A single-point mutation in HCF causes temperature-sensitive cell-cycle arrest and disrupts VP16 function. Genes Dev. 11, 726–737 [DOI] [PubMed] [Google Scholar]
- 43. Wilson A. C., Freiman R. N., Goto H., Nishimoto T., Herr W. (1997) VP16 targets an amino-terminal domain of HCF involved in cell cycle progression. Mol. Cell. Biol. 17, 6139–6146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Trimarchi J. M., Lees J. A. (2002) Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell Biol. 3, 11–20 [DOI] [PubMed] [Google Scholar]
- 45. Blais A., Dynlacht B. D. (2004) Hitting their targets. An emerging picture of E2F and cell cycle control. Curr. Opin. Genet. Dev. 14, 527–532 [DOI] [PubMed] [Google Scholar]
- 46. Johnson D. G., Schneider-Broussard R. (1998) Role of E2F in cell cycle control and cancer. Front. Biosci. 3, d447–448 [DOI] [PubMed] [Google Scholar]
- 47. Woo R. A., Poon R. Y. (2003) Cyclin-dependent kinases and S phase control in mammalian cells. Cell Cycle 2, 316–324 [PubMed] [Google Scholar]
- 48. Aleem E., Kiyokawa H., Kaldis P. (2005) Cdc2-cyclin E complexes regulate the G1/S phase transition. Nat. Cell Biol. 7, 831–836 [DOI] [PubMed] [Google Scholar]
- 49. Hateboer G., Kiyokawa H., Kaldis P. (1008) Cell cycle-regulated expression of mammalian CDC6 is dependent on E2F. Mol. Cell. Biol. 18, 6679–6697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ohtani K., Tsujimoto A., Ikeda M., Nakamura M. (1998) Regulation of cell growth-dependent expression of mammalian CDC6 gene by the cell cycle transcription factor E2F. Oncogene 17, 1777–1785 [DOI] [PubMed] [Google Scholar]
- 51. Yan Z., DeGregori J., Shohet R., Leone G., Stillman B., Nevins J. R., Williams R. S. (1998) Cdc6 is regulated by E2F and is essential for DNA replication in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 95, 3603–3608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ruan H. B., Han X., Li M. D., Singh J. P., Qian K., Azarhoush S., Zhao L., Bennett A. M., Samuel V. T., Wu J., Yates J. R., 3rd, Yang X. (2012) O-GlcNAc transferase/host cell factor C1 complex regulates gluconeogenesis by modulating PGC-1α stability. Cell Metab. 16, 226–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Dey A., Seshasayee D., Noubade R., French D. M., Liu J., Chaurushiya M. S., Kirkpatrick D. S., Pham V. C., Lill J. R., Bakalarski C. E., Wu J., Phu L., Katavolos P., LaFave L. M., Abdel-Wahab O., Modrusan Z., Seshagiri S., Dong K., Lin Z., Balazs M., Suriben R., Newton K., Hymowitz S., Garcia-Manero G., Martin F., Levine R. L., Dixit V. M. (2012) Loss of the tumor suppressor BAP1 causes myeloid transformation. Science 337, 1541–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dou Y., Milne T. A., Ruthenburg A. J., Lee S., Lee J. W., Verdine G. L., Allis C. D., Roeder R. G. (2006) Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat. Struct. Mol. Biol. 13, 713–719 [DOI] [PubMed] [Google Scholar]
- 55. Cao F., Chen Y., Cierpicki T., Liu Y., Basrur V., Lei M., Dou Y. (2010) An Ash2L/RbBP5 heterodimer stimulates the MLL1 methyltransferase activity through coordinated substrate interactions with the MLL1 SET domain. PLoS One 5, e14102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cho Y. W., Hong T., Hong S., Guo H., Yu H., Kim D., Guszczynski T., Dressler G. R., Copeland T. D., Kalkum M., Ge K. (2007) PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J. Biol. Chem. 282, 20395–20406 [DOI] [PMC free article] [PubMed] [Google Scholar]