Background: Hedgehog signaling plays important roles during intestinal development.
Results: Mesenchymal Gli2 activation, but not Gli3 removal, rescued Hedgehog pathway defects. Screen identified small leucine-rich glycoproteins (SLRPs) downstream of Hedgehog in the gut.
Conclusion: Hedgehog activity is mediated by Gli2 and involves SLRP regulation in the developing intestine.
Significance: This study uncovers novel genetic and molecular mechanisms underlying Hedgehog function in intestinal development.
Keywords: Development, Embryo, Hedgehog, Intestine, Signaling, Gli, Mesenchyme
Abstract
Hedgehog (Hh) signaling is involved in multiple aspects of embryonic gut development, including mesenchymal growth and smooth muscle differentiation. The Gli family transcription factors is thought to collectively mediate Hh signaling in mammals. However, the function of different Gli proteins in gut development remains uncharacterized. Here, we genetically dissect the contribution of Gli transcriptional activation and de-repression in intestinal growth and patterning. We find that removal of the Gli3 repressor is dispensable for intestinal development and does not play a major role in Hh-controlled gut development. However, Gli2 activation is able to fully rescue the Smoothened (Smo)-null intestinal phenotype, suggesting that the Gli2 transcription factor is the main effector for Hh signaling in the intestine. To understand further the molecular mechanism underlying Hh/Gli function in the developing gut, we identify a subset of small leucine-rich glycoproteins (SLRPs) that may function downstream of Hh signaling in the mesenchyme. We show that osteoglycin, a SLRP, inhibits Hh-induced differentiation toward the smooth muscle lineage in C3H10T1/2 pluripotent mesenchymal cells. Taken together, our study reveals, for the first time, the distinct roles of Gli proteins in intestine development and suggests SLRPs as novel regulators of smooth muscle cell differentiation.
Introduction
Hedgehog (Hh)3 signaling has emerged as one of the critical pathways regulating the development, homeostasis, and tumorigenesis of the gastrointestinal tract (1–8). Hh ligands, including Sonic hedgehog (Shh) and Indian hedgehog (Ihh), are produced by the endodermal gut epithelium and are believed to signal to the adjacent mesenchyme to regulate tissue growth and patterning (4, 5, 9). In a recent study, we genetically removed both Shh and Ihh ligands from the early embryonic gut endoderm and found that Hh signaling is essential for gut mesenchymal development (5). Specifically, we found that Hh proteins are the primary factor responsible for maintenance and expansion of early mesenchymal progenitor populations (5). In addition, our results suggested that Hh signaling is important for later differentiation, including specification of the smooth muscle lineage from primitive mesenchyme, a prominent event during intestinal mesenchymal development (5). Despite its importance, the genetic and cellular mechanisms underlying Hh control of intestinal development remain poorly understood.
In vertebrates, Hh signaling is mediated by the Gli family of transcription factors, Gli1, Gli2, and Gli3. Hh ligands bind to its transmembrane receptor Patched1 (Ptch1), relieving inhibition of another transmembrane protein, Smoothened (Smo), which in turn activates downstream intracellular signaling events to regulate Gli activity (10–12). Of the Gli proteins, Gli2 and Gli3 are the primary signal responders, with Gli2 mainly acting as a transcriptional activator and Gli3 predominantly acting as a repressor (13, 14). Gli1, a direct transcriptional target of Hh signaling, is dispensable for mouse embryonic development (15). Therefore, the balance between the activities of Gli2/3 transcriptional activator and repressor dictates Hh responses that are both tissue-specific and developmental stage-specific. During limb and ureter development, Shh acts mainly by opposing Gli3 repression (16, 17). During neural tube and skeletal development, integrated regulation of Gli2 activation and Gli3 de-repression has both overlapping and distinct functions (18–21). Surprisingly, the Gli regulatory mechanism and the relative contribution of Gli proteins to Hh function in the intestine have yet to be examined.
In this study we demonstrate the direct genetic requirement of Hh responsiveness in the gut mesenchyme, and found that removal of Gli3 repression had no effect on Hh-mediated mesenchymal growth and differentiation. However, activation of Gli2 fully rescued Smo-null defects in the intestine, suggesting that Gli2, but not Gli3, is the major effector for Hh signaling during intestine development. Furthermore, our study suggests that a subset of SLRPs may act downstream of Hh signaling in the mesenchyme to regulate smooth muscle differentiation in the developing intestine.
EXPERIMENTAL PROCEDURES
Mouse Strains
Nkx3.2Cre, SmoC, Gli3C, R26SmoM2, and R26mT/mG mice have been described before (22–26). To generate the R26Gli2 allele, the cDNA fragment encoding Gli2 fused with a C-terminal FLAG tag was inserted into the shuttling vector pBigT before being cloned into pROSA-PAS to produce the final targeting construct. ES cell targeting and blastocyst injection were performed by University of Massachusetts Medical School Transgenic Core to generate chimeric animals. Nkx3.2Cre;R26Gli2, Nkx3.2Cre;R26SmoM2, and Nkx3.2Cre;R26mT/mG mice were generated by crossing Nkx3.2Cre to R26Gli2;R26SmoM2 or R26mT/mG mice. Nkx3.2Cre;SmoC/C and Nkx3.2Cre;Gli3C/C mice were obtained by crossing Nkx3.2Cre;SmoC/+ and Nkx3.2Cre;Gli3C/+ to SmoC/C or Gli3C/C mice. To generate Nkx3.2Cre;SmoC/C;Gli3C/C and Nkx3.2Cre;SmoC/C; R26Gli2 embryos, Nkx3.2Cre;SmoC/+;Gli3C/+ or Nkx3.2Cre;moC/+ mice were crossed to SmoC/C;Gli3C/C and SmoC/C;R26Gli2 mice. All mouse experiments were performed according to the guidelines of IACUC at University of Massachusetts Medical School.
Tissue Collection and Histology
Upon euthanasia, embryonic intestine was dissected and fixed in 4% paraformaldehyde for 6 h. For paraffin sections, tissues were dehydrated, embedded in paraffin blocks, and cut at a thickness of 6 μm. For frozen sections, tissues were dehydrated in 30% sucrose and embedded in OCT, and sections were cut at a thickness of 12 μm. For RNA analysis, tissues were flash frozen in liquid nitrogen. Tissue sections (6 μm) were stained with hematoxylin and eosin (H&E) using standard reagents and protocols.
Immunohistochemistry, Immunofluorescence, and Immunoblotting
For immunohistochemistry, high temperature antigen retrieval was conducted on paraffin sections in sodium citrate buffer (pH 6.0) for 30 min. Sections were blocked in a buffer containing 5% BSA and 0.1% Triton X-100 in PBS and incubated overnight at 4 °C in primary antibodies diluted in blocking buffer. Primary antibodies used were: Ki67 (Abcam), cleaved caspase 3 (Cell Signaling), α-smooth muscle actin (SMA; Sigma), β-tubulin (Covance), β-catenin (Santa Cruz Biotechnology), and CD44 (eBioscience). Signal was detected with biotinylated secondary antibodies in the Vectastain ABC kit (Vector Laboratories).
For immunofluorescence, tissue frozen sections or C3H10T1/2 cells were washed with PBS, fixed with 4% paraformaldehyde for 5 min, incubated in blocking buffer containing 5% sheep serum, 1% FBS, and 0.1% Triton X-100 for 1 h, and incubated overnight at 4 °C with SMA antibody (1:800) diluted in blocking buffer. Secondary antibodies were Alexa Fluor- conjugated antibodies (Invitrogen) at a 1:500 dilution in blocking buffer. Slides were mounted in mounting media containing DAPI. For immunoblotting, primary antibodies were: V5 (ThermoFisher), FLAG (Sigma), and β-actin (Sigma). HRP-conjugated secondary antibodies were obtained from Jackson Laboratories.
C3H10T1/2 Cell Culture, Lentiviral Infection, and Cell Differentiation Assay
C3H10T1/2 cells were obtained from ATCC and maintained in DMEM with 10% FBS at 37 °C in 5% CO2. Cells were cultured at 30–50% confluence to prevent differentiation. For lentiviral infection, a C-terminally V5-tagged osteoglycin (OGN) cDNA fragment was inserted into the pGIPZ based lentiviral expression vector. The OGN lentiviral construct was transfected along with the packing plasmids into growing HEK293T cells, and viral supernatants were collected 48 h later. C3H10T1/2 cells were infected with OGN lentivirus in the presence of Polybrene and selected with puromycin for 4 days to establish the OGN-expressing cell line. For smooth muscle cell differentiation, cells were cultured in growth media for 24 h before switching to differentiation medium (DMEM with 0.5% FBS) for 12 h, followed by addition of Shh conditioned media (1:100) in culture for 36 h. Smooth muscle differentiation was assayed by immunostaining or qPCR analysis of SMA expression.
Luciferase Reporter Analysis
C3H10T1/2 cells with or without OGN overexpression were transfected with the luciferase reporter construct, GliBS-Luc (gift of Dr. H. Kondoh, Osaka University) and the Renilla luciferase expression vector. The cells were either treated with Shh conditioned medium or co-transfected with a Gli2 expression construct. Luciferase assays were conducted 72 h after transfection using the dual-luciferase reporter kit (Promega). Assays were conducted in triplicate.
Quantitative RT-PCR
cDNA synthesis was conducted using an Invitrogen Superscript II kit. Primers used for quantitative RT-PCR were: mouse β-actin (forward, TGACAGGATGCAGAAGGAGA; reverse, CTGGAAGGTGGACAGTGAGG), Ptch1 (forward, AACAAAAATTCAACCAAACCTC; reverse, TGTCTTCATTCCAGTTGATGTG), HIP1 (forward, CTATTGGGCCTCACGACCAC; reverse, TTCCAGAAACACCCTGGCTG), Myocd (forward, AAGGTCCATTCCAACTGCTC; reverse, CCATCTCTACTGCTGTCATCC), SMA (forward, GAACACGGCATCATCACCAAC; reverse, CTCCAGAGTCCAGCACAATACC), Axin2 (forward, TGACTCTCCTTCCAGATCCCA; reverse, TGCCCACACTAGGCTGACA), and Lgr5 (forward, CGGGACCTTGAAGATTTCCT; reverse, GATTCGGATCAGCCAGCTAC). All qPCR assays were conducted in triplicates, and standard deviation was used to calculate error bars.
Affymetrix Gene Chip Analysis
Intestinal tissues were dissected from E13.5 wild type and Nkx3.2Cre;R26SmoM2 embryos. RNA was isolated, labeled, and hybridized to mouse GeneST1.0 chips (Affymetrix) according to Affymetrix protocols. Three independent biological samples were used for chip analysis. Statistical analyses were performed using R, a system for statistical computation and graphics. Genes with p value <0.05 and -fold change >1.5 were considered potential targets for further investigation.
RESULTS
Genetic Removal of Smo from the Developing Intestinal Mesenchyme
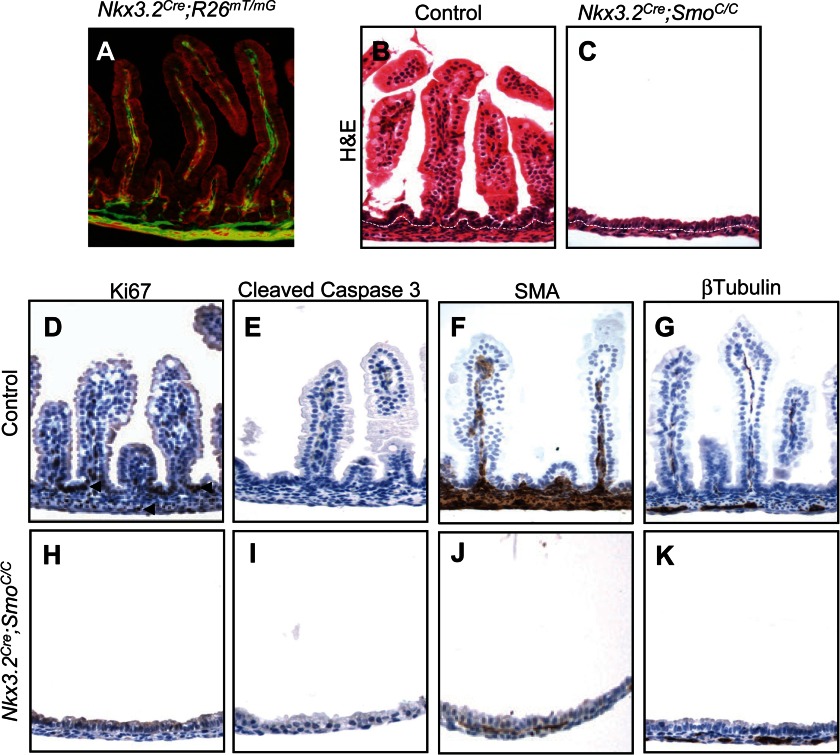
Our previous analysis of mouse embryos lacking both Shh and Ihh from early gut endoderm provided strong evidence that endoderm-derived Hh proteins control adjacent mesenchymal development (5). The majority of evidence from the literature also supports a paracrine mode of Hh signaling from the epithelium to the mesenchyme (4, 6, 7, 9); however, direct genetic evidence for such a requirement within the gut mesenchyme is still lacking. Furthermore, the extremely small size of the intestine in the endodermal Shh/Ihh mutants restricted our previous analysis mainly to fetal stomach mesenchyme (5). Therefore, we set out to examine further the role of Hh signaling in intestinal development and determine whether cell-autonomous Hh reception is genetically required in the mesenchyme. We performed targeted deletion of Smo from the intestinal mesenchymal compartment. Smo, a cell surface transmembrane protein, is essential for all Hh signal transduction in recipient cells (27). We crossed a conditional Smo allele, SmoC (23), to a recently established Nkx3.2Cre line (22), which initiates Cre expression in the gut mesenchyme at E9.5. When Nkx3.2Cre was crossed to a dual-fluorescent Rosa26 reporter allele, R26mT/mG (26), widespread Cre-mediated recombination was observed exclusively in the intestinal mesenchyme of Nkx3.2Cre;R26mT/mG mice (Fig. 1A). Nkx3.2Cre;SmoC/C mutant embryos survived gestation but died immediately after birth, therefore allowing us to analyze the late stage development of embryonic intestine. Intestinal mesenchymal proliferation and expansion in the Nkx3.2Cre;SmoC/C embryos were dramatically affected (Fig. 1, B and C), producing a phenotype similar to that seen in the Shh/Ihh double mutant (5). At E18.5, significantly fewer mesenchymal cells remained between the thin layers of mesentery and intestinal epithelium (Fig. 1,B and C) in the Smo mutant embryos. In agreement with our previous report for a primary mitogenic role of Hh signaling on mesenchymal populations (5), mesenchymal proliferation was absent, although apoptosis was not increased in the Smo-mutated intestinal mesenchyme, as assayed by Ki67 and cleaved caspase 3 staining, respectively (Fig. 1, D, E, H, and I).
FIGURE 1.
Mesenchymal removal of Smo in the developing intestine. A, Nkx3.2Cre-mediated mesenchymal recombination in the intestine. Fluorescence image of the Nkx3.2Cre;R26mT/mG intestine shows widespread Cre-mediated recombination and GFP expression exclusively in the intestinal mesenchymal compartment. B and C, H&E staining of the intestine of control and Nkx3.2Cre;SmoC/C embryos at E18.5. White dashed lines delineate the boundary of gut epithelium and mesenchyme. D–K, defects in mesenchymal growth and patterning in Nkx3.2Cre;SmoC/C embryos at E18.5. Shown is immunohistochemical staining of Ki67 (D and H), cleaved caspase 3 (E and I), SMA (F and J), and β-tubulin (G and K) in the intestine of control and Nkx3.2Cre;SmoC/C mutant embryos.
Smo deletion also dramatically affected mesenchymal differentiation, including smooth muscle differentiation. Smooth muscle cells, detected by the expression of SMA, are specified from local intestinal mesenchyme (28). Earlier studies suggested that Hh signaling may have an inhibitory effect on this process (29, 30). However, our analysis of fetal intestine harboring deletion of endodermal Shh/Ihh suggests that Hh signaling is required for smooth muscle specification in the developing gut (5). Consistent with this, we found that in E18.5 Nkx3.2Cre;SmoC/C embryos, the number of SMA-expressing or Desmin-expressing intestinal smooth muscle cells was drastically decreased (Fig. 1, F and J, and data not shown). Further, the β-tubulin-expressing enteric neuron population was also decreased in the Smo mutant embryos (Fig. 1, G and K). Together, these findings demonstrate the genetic requirement of Hh responsiveness in the mesenchyme to maintain growth and promote differentiation in the fetal intestine.
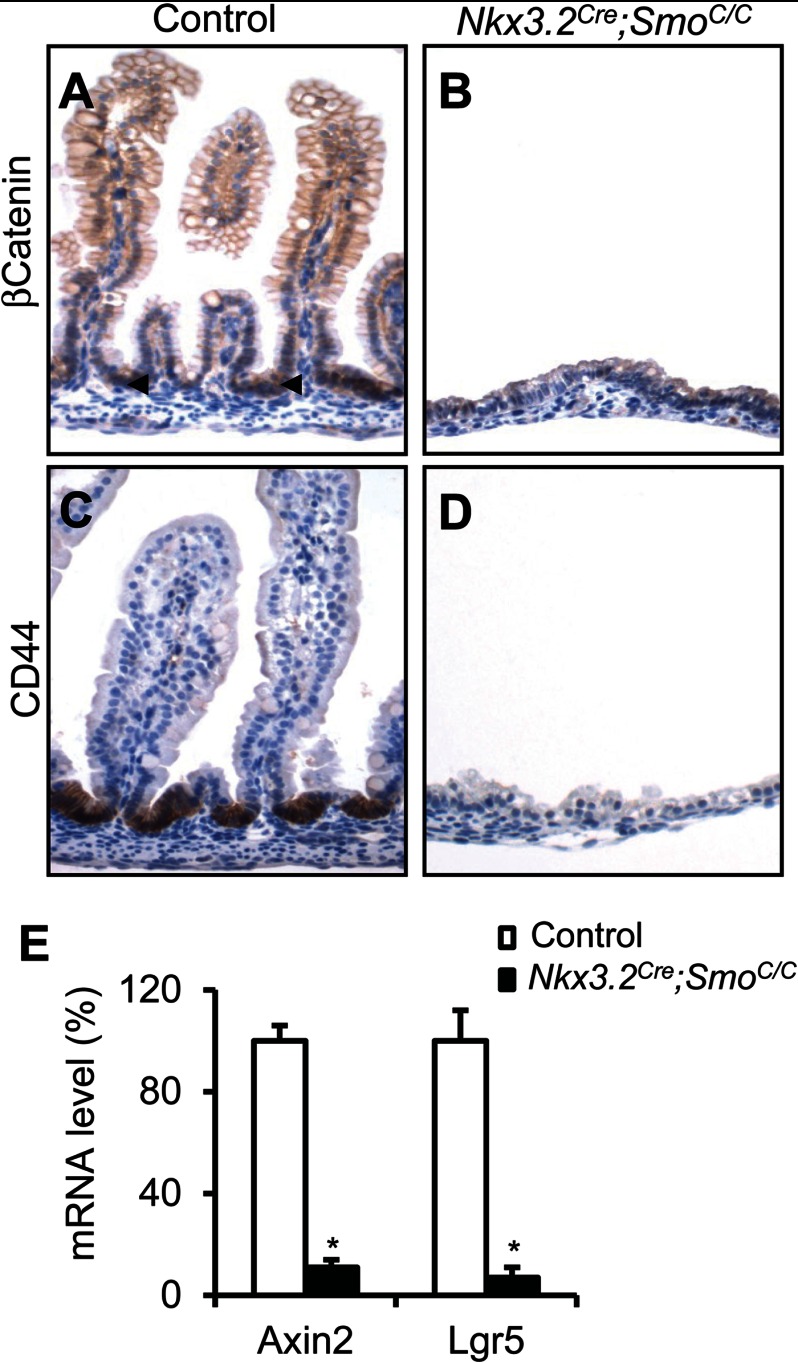
Hh signaling mediates the reciprocal interaction between intestinal epithelium and mesenchyme (2, 6, 7, 31), although its exact role in regulation of epithelial differentiation is not fully understood. In the Villin-HIP1 and VillinCre;IhhC/C mutant mice, partial inhibition of Hh signaling leads to Wnt pathway activation and intestinal epithelial overproliferation in newborn animals (2, 31). Therefore, we further analyzed the intestinal epithelial development in Nkx3.2Cre;SmoC/C mutant embryos. We found that both epithelial proliferation and differentiation were severely affected by mesenchymal Smo deletion. The intestinal epithelium of mutant embryos at E18.5 was not proliferative (Fig. 1, D and H), as assayed by Ki67 staining, and lacked differentiation of the Goblet and enteroendocrine cell lineages (data not shown). Smo mesenchymal deletion also blocked epithelial activation of Wnt/β-catenin signaling, measured by β-catenin nuclear staining and the expression of the Wnt target gene CD44 (Fig. 2, A–D), as well as qPCR analysis of the Wnt target genes, Axin2 and Lgr5 (Fig. 2E). Although the perinatal death of mutant embryos prevented postnatal analysis, these results highlight the importance of Hh signaling in mediating the interactions between the epithelial and mesenchymal compartments, and underline the fact that these two tissue layers are highly interdependent during intestinal development.
FIGURE 2.
Epithelial Wnt activity in embryonic intestine with mesenchymal Smo deletion. A–D, immunohistochemical staining of β-catenin and CD44 in the intestine of control and Nkx3.2Cre;SmoC/C embryos at E18.5. E, qPCR analysis of mRNA levels of Axin2 and Lgr5 in control and Nkx3.2Cre;SmoC/C intestine. *, p < 0.01; error bars indicate mean ± S.D.
Gli3 Is Dispensable for Intestinal Mesenchymal Development
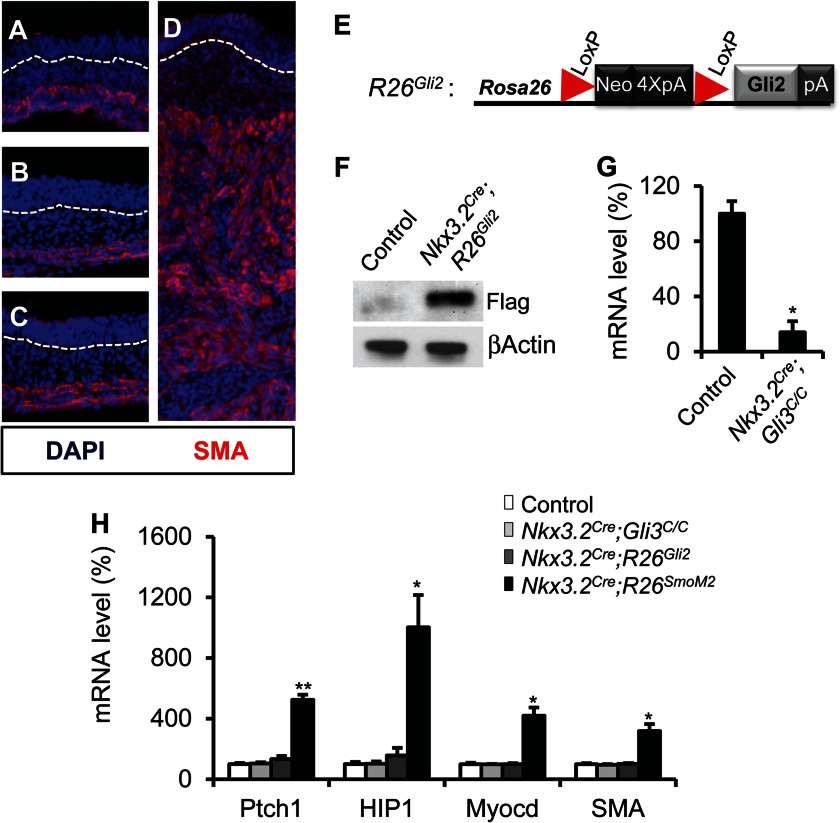
Removal of Gli3 transcriptional repression plays a key role in mediating Hh signaling in different developmental contexts (14, 16, 17, 19). Although a modest glandular hyperplasia has been reported in the Gli3 mutated embryonic stomach (32), the role of Gli3 in intestinal development and contribution to Hh function remain unknown. We generated mice in which Gli3 was deleted in the intestinal mesenchyme using the Nkx3.2Cre and Gli3C allele (24). Nkx3.2Cre;Gli3C/C mice were born at the expected Mendelian frequency and did not display a gross phenotype compared with the littermate controls (data not shown). At the embryonic stage, Nkx3.2Cre;Gli3C/C mutant embryos exhibited normal fetal intestinal epithelial and mesenchymal development (Fig. 3, B and G, and 4B). This suggests that Gli3 is not required for intestinal development.
FIGURE 3.
Removal of Gli3 and ectopic expression of Gli2 in the intestinal mesenchyme. A–D, immunofluorescence staining of SMA in the developing intestine of control (A), Nkx3.2Cre;Gli3C/C (B), Nkx3.2Cre;R26Gli2 (C), and Nkx3.2Cre;R26SmoM2 (D) embryos at E13.5. White dashed lines delineate the boundary of epithelium and mesenchyme. Note the normal thickness and SMA expression pattern in the Nkx3.2Cre;Gli3C/C and Nkx3.2Cre;R26Gli2 embryos compared with control embryos. E, schematic representation of the Rosa26 conditional R26Gli2 allele. F, immunoblot analysis of Gli2-FLAG protein expression in control and Nkx3.2Cre;R26Gli2 embryos. G, qPCR analysis of Gli3 mRNA levels in control and Nkx3.2Cre;Gli3C/C gut. H, relative mRNA levels of Ptch1, HIP1, Myocd and SMA in control, Nkx3.2Cre;Gli3C/C, Nkx3.2Cre;R26Gli2, and Nkx3.2Cre;R26SmoM2 intestine at E13.5. *, p < 0.05; **, p < 0.01; error bars indicate mean ± S.D.
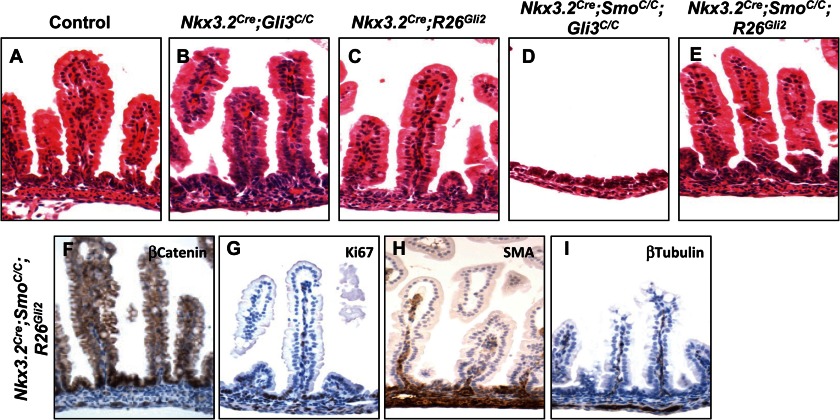
Despite the lack of phenotype after Gli3 deletion in mouse ureter mesenchyme, Gli3 removal completely rescues Smo mutant phenotype (17). Therefore, we further investigated whether Gli3 de-repression contributes to intestinal development in the context of Hh pathway inactivation. We generated embryos with both Smo and Gli3 deficiency in the intestinal mesenchyme. Surprisingly, we found that elimination of Gli3 in the Nkx3.2Cre;SmoC/C background failed to restore the Smo-null defects in the intestine (Fig. 4D), suggesting that removal of Gli3 repression does not play a significant role in mediating Hh signaling in the developing intestinal mesenchyme.
FIGURE 4.
Gli2 activation, but not Gli3 de-repression, mediates Hh function in the developing gut. A–E, histology of the control, Nkx3.2Cre;Gli3C/C, Nkx3.2Cre;R26Gli2, Nkx3.2Cre;SmoC/C;Gli3C/C, and Nkx3.2Cre;SmoC/C;R26Gli2 intestine at E18.5. F–I, epithelial and mesenchymal development in the Nkx3.2Cre;SmoC/C;R26Gli2 intestine at E18.5. Immunohistochemical staining of β-catenin (F), Ki67 (G), SMA (H), and β-tubulin (I) in the Nkx3.2Cre;SmoC/C;R26Gli2 embryos is shown.
Gli2 Activation Rescues Intestinal Development in the Smo-null Embryos
Previous studies report that both Gli2 and Gli3 are expressed in the developing gut mesenchyme (4). Given that Gli3 removal failed to restore intestinal development in the Smo-null embryos, we hypothesized that Gli2-mediated transcriptional activation is the predominant mediator of Hh signaling in the gut. To test this idea, we generated a Rosa26 knock-in allele of Gli2, R26Gli2, which allowed Cre-dependent expression of a FLAG-tagged form of Gli2 from the ubiquitously expressed Rosa26 locus (Fig. 3, E and F).
We generated Nkx3.2Cre;R26Gli2 embryos by crossing Nkx3.2Cre to the R26Gli2 mice. Interestingly, we found that similar to Gli3 removal, Gli2 activation alone by R26Gli2 did not significantly affect embryonic intestinal development (Figs. 3 and 4). At E18.5, intestinal mesenchymal and epithelial development appeared phenotypically normal in the Nkx3.2Cre;R26Gli2 mutant embryos (Fig. 4, A and C). This is drastically different from the phenotype we observed in the Nkx3.2Cre;R26SmoM2 mutant embryos (Ref. 5 and Fig. 3D). R26SmoM2 is a Rosa26 conditional knock-in allele of SmoM2, a constitutively active form of Smo (25). Nkx3.2Cre;R26SmoM2 mutant embryos died at approximately E14.5 (5). SmoM2 activation dramatically expanded the intestinal mesenchymal compartment and induced the expression of the smooth muscle lineage markers, myocardin (Myocd) and SMA at E13.5, as assayed by immunostaining and qPCR (Fig. 3, D and H). We further examined Hh pathway activity in Gli3-deleted, Gli2- or SmoM2-expressing embryonic intestines and found that expression of the Hh target genes, such as HIP1 and Ptch1, was markedly increased in the Nkx3.2Cre;R26SmoM2 mutant intestine at E13.5, but was largely unaffected in the Nkx3.2Cre;R26Gli2 or Nkx3.2Cre;Gli3C/C embryos (Fig. 3H), which likely accounts for the lack of Hh gain-of-function phenotype in Gli3 or R26Gli2 mutant embryos. However, it is interesting to note that in transiently transfected NIH3T3 cells, Gli2 induced stronger downstream transcriptional activation than SmoM2 (data not shown), suggesting that additional posttranslational regulatory mechanisms may modulate Gli2 activity or stability in vivo.
To examine whether Gli2 activation contributes to Hh function in the intestine, we generated Nkx3.2Cre;SmoC/C;R26Gli2 embryos and found that Gli2 expression from the Rosa26 locus completely rescued the Smo-null intestinal phenotypes. Histological analyses of E18.5 embryos revealed that intestinal development in the Nkx3.2Cre;SmoC/C;R26Gli2 embryos is normal (Fig. 4E). We observed the rescue of mesenchymal growth, smooth muscle development, and intestinal epithelial differentiation (Fig. 4, F–I). Together, these results strongly suggest that Gli2, but not Gli3, is the major effector for Hh signaling during embryonic intestinal development.
Hh-Gli Regulates Expression of the Small Leucine-rich Glycoproteins
We performed expression profiling using Affymatrix chips in the wild-type and Hh gain-of-function (Nkx3.2Cre;R26SmoM2) E13.5 mutant gut to investigate the Hh-Gli downstream mechanism in developing gut. Our analysis identified 150 significantly up-regulated genes in Nkx3.2Cre;R26SmoM2 embryos (supplemental Table S1), including Hip1, Ptch1, Ptch2, and Gli1, which are known Hh target genes involved in the feedback regulation of pathway signal transduction (Fig. 5A and supplemental Table S1). Consistent with the idea that the Hh pathway positively regulates smooth muscle cell differentiation, we found that the genes associated with smooth muscle lineage specification, such as Myocd and SMA, were up-regulated as well (Fig. 5A and supplemental Table S1).
FIGURE 5.
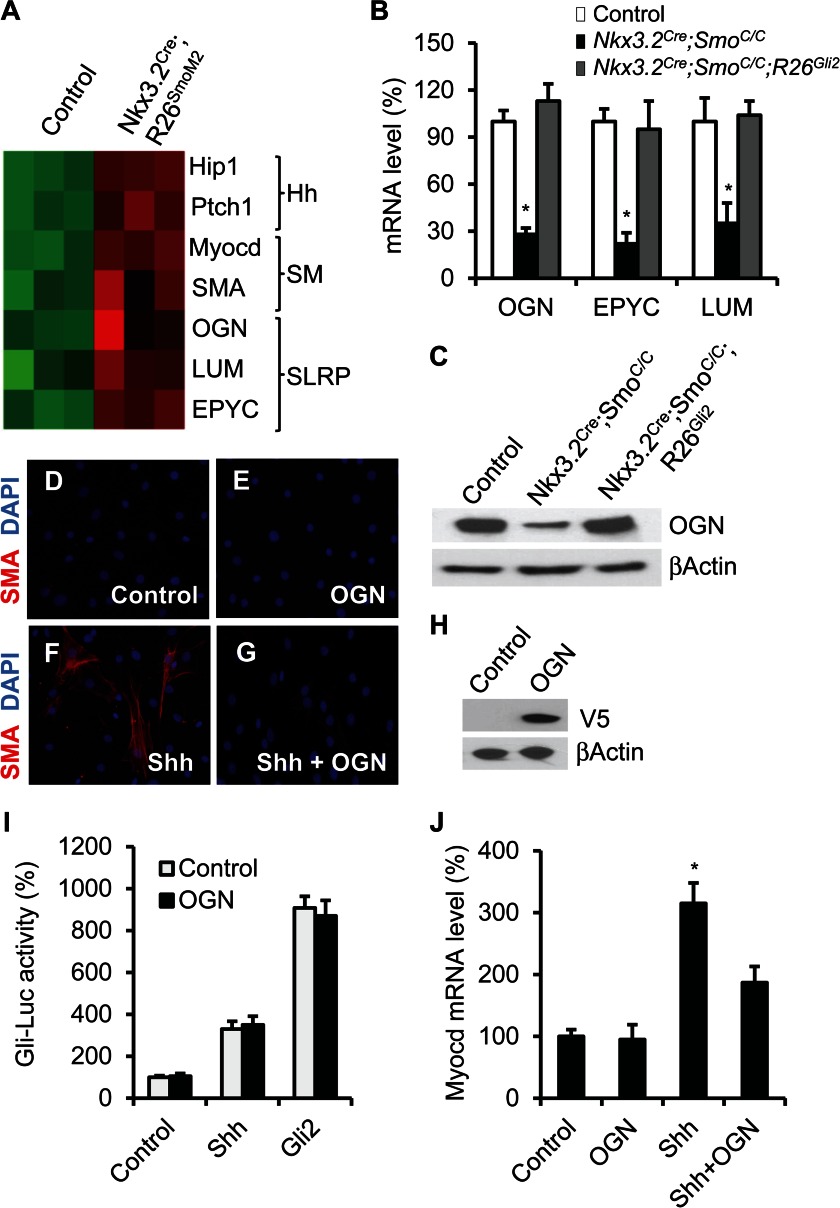
SLRPs regulate smooth muscle cell differentiation. A, heat map illustrating mRNA expression of selected genes in control and SmoM2-expressing developing gut. SM, smooth muscle differentiation. B, qPCR analysis of mRNA expression of OGN, EPYC, and LUM in control, Nkx3.2Cre;SmoC/C, and Nkx3.2Cre;SmoC/C;R26Gli2 intestine at E18.5. C, immunoblot analysis of OGN expression in control, Nkx3.2Cre;SmoC/C, and Nkx3.2Cre;SmoC/C;R26Gli2 intestine. D–G, immunofluorescence staining of SMA in the control and OGN-overexpressing C3H10T1/2 cells with or without Shh treatment. H, immunoblot analysis of expression of V5-tagged OGN proteins in C3H10T1/2 cells. I, Gli-luciferase reporter activity in the control and OGN-expressing C3H10T1/2 cells with Shh treatment or Gli2 overexpression. J, qPCR analysis of Myocd mRNA level in the control and OGN-overexpressing C3H10T1/2 cells with or without Shh treatment. *, p < 0.05, error bars indicate mean ± S.D.
Interestingly, we found that several SLRP glycoproteins, including epiphycan (EPYC), osteoglycin (OGN), and lumican (LUM), were also among the top genes up-regulated in the mutant gut (Fig. 5A and supplemental Table S1). EPYC, OGN, and LUM belong to the SLRP family of proteins, which are characterized by the presence of the N-terminal cysteine-rich clusters (33). SLRPs have been reported to regulate cell growth and differentiation (33), although it is not known whether they are involved in intestinal development. To further characterize the potential Hh regulation of SLRP, we performed qPCR and immunoblot analyses of the expression of OGN, EPYC, and LUM in the intestine of wild type, Smo-deficient (Nkx3.2Cre;SmoC/C) and Gli2-rescue (Nkx3.2Cre;SmoC/C;R26Gli2) embryos. Consistent with our transcriptional profiling data, we found that mRNA levels of OGN, EPYC, and LUM, and protein expression of OGN, were down-regulated in the Smo-null embryos, but were restored by Gli2 expression (Fig. 5, B and C). Together, these data suggest that these SLRPs may act downstream of Hh signaling in the developing intestinal mesenchyme.
OGN Regulates Smooth Muscle Cell Differentiation of C3H10T1/2 Cells in Vitro
To examine the involvement of SLRPs in smooth muscle differentiation, we utilized the in vitro C3H10T1/2 cell differentiation model. C3H10T1/2 is a mouse embryonic pluripotent mesenchymal cell line that differentiates into SMA-positive smooth muscle cells in response to treatment with Hh proteins and has been used as a surrogate system to study the Hh-induced differentiation program (8).
To further examine the effect of SLRPs on C3H10T1/2 cell differentiation, we used lentiviral infection to generate a C3H10T1/2 cell line that stably expresses C-terminally V5-tagged OGN proteins (Fig. 5H). We found that OGN expression alone was not sufficient to induce cell differentiation toward smooth muscle lineage (Fig. 5, D and E) and did not affect Gli-dependent luciferase reporter activity in the C3H10T1/2 cells with or without Shh treatment or Gli2 expression (Fig. 5I), suggesting that the proteoglycan is unlikely directly involved in Hh signal transduction. However, we found that OGN ectopic expression was able to inhibit Shh-mediated promotion of smooth muscle cell differentiation. Shh-induced induction of smooth muscle cell markers SMA and Myocd was decreased in OGN-expressing C3H10T1/2 cells, as assayed by immune-fluorescence staining or mRNA qPCR (Fig. 5, F, G, and J). Together, these data suggest that SLRPs are novel regulators of intestinal mesenchymal development and may function downstream of Hh signaling in a negative feedback fashion to modulate smooth muscle cell differentiation.
DISCUSSION
The current study investigated the genetic function of the Gli transcriptional effectors of Hh signaling in intestinal development and identified a novel function of SLRPs in smooth muscle cell differentiation. Recent studies from our laboratory and others established that Shh and Ihh proteins produced by the endodermal epithelium play a key role in intestinal development by controlling mesenchymal growth and patterning (1, 4, 5, 8, 9). However, the mechanisms through which Hh ligands carry out their diverse activities in the intestine are not well understood, specifically which Gli effectors mediate Hh function. Here, we show the first genetic analysis for the role of different Gli proteins during intestinal development and present strong evidence that the major Hh function in the intestinal mesenchyme is achieved through the Gli2 transcriptional activator.
Removal of Gli3-mediated transcriptional repression plays many critical roles in embryonic development (13, 14, 16, 19, 20). In the developing ureter, Hh signaling functions in a similar fashion as it does in the gastrointestinal system. Hh ligands are secreted by the epithelium and signal to the adjacent ureter mesenchyme (17, 30), and a recent report showed that removal of Gli3 repression is able to fully rescue the Smo-null mesenchymal phenotypes (17). Therefore, it is somewhat surprising that our genetic analysis revealed that Gli3 activity is dispensable for Hh function in the intestine, although both Gli2 and Gli3 are expressed in the mesenchyme (4). Our results highlight the context-dependent manner of Gli regulation in Hh-controlled developmental processes.
The molecular mechanism underlying gut mesenchymal development remains poorly characterized. Our transcriptional profiling and functional analysis suggest that a subset of SLRPs may act downstream of Hh signaling and is involved in regulating smooth muscle cell differentiation. Hh signaling has been reported to regulate expression of certain extracellular matrix proteins, such as collagens (34), but it is the first time that Hh signaling is linked to SLRPs. The SLRP family consists of 17 member proteins that are largely characterized by the presence of N-terminal cysteine-rich clusters (33), and the exact role of SLRPs in vivo is poorly understood. It has been reported that they may regulate cell proliferation and differentiation in certain developmental contexts (33), and genetic mutations in decorin (DCN) and LUM have been implicated in inherited ocular disorders such as congenital stromal dystrophy and high myopia in the cornea (35, 36). Our study suggests that SLRPs are novel regulators for smooth muscle differentiation. In Hh-mediated intestinal development, it appears that OGN acts in a negative feedback manner to modulate smooth muscle cell differentiation. Several SLRPs, including DCN and biglycan, have been reported to regulate different signaling pathways, such as insulin growth factor, epidermal growth factor, and transforming growth factor/bone morphogenetic protein (33). It is interesting to speculate that SLRPs may function to integrate critical signaling pathways in the embryonic gut mesenchyme. Clearly, future genetic studies are needed to establish the function of SLRPs in intestinal development.
Acknowledgments
We thank Annie Pang for technical assistance and members of the Mao laboratory for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Training Grant T32 CA130807 (to J. L. C.). This work was also supported by American Cancer Society Grant 20376-RSG-11-040-01-DDC, the Charles H. Hood Foundation and Worcester Biomedical Research Foundation (to J. M.), and National Natural Science Foundation of China Grant 81141068 (to H. H.).

This article contains supplemental Table S1.
- Hh
- Hedgehog
- En
- embryonic day n
- EPYC
- epiphycan
- Ihh
- Indian Hedgehog
- LUM
- lumican
- Myocd
- myocardin
- OGN
- osteoglycin
- Ptch
- Patched
- qPCR
- quantitative PCR
- Shh
- Sonic Hedgehog
- SLRP
- small leucine-rich glycoprotein
- SMA
- α-smooth muscle actin
- Smo
- Smoothened.
REFERENCES
- 1. van den Brink G. R. (2007) Hedgehog signaling in development and homeostasis of the gastrointestinal tract. Physiol. Rev. 87, 1343–1375 [DOI] [PubMed] [Google Scholar]
- 2. Kosinski C., Stange D. E., Xu C., Chan A. S., Ho C., Yuen S. T., Mifflin R. C., Powell D. W., Clevers H., Leung S. Y., Chen X. (2010) Indian hedgehog regulates intestinal stem cell fate through epithelial-mesenchymal interactions during development. Gastroenterology 139, 893–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yauch R. L., Gould S. E., Scales S. J., Tang T., Tian H., Ahn C. P., Marshall D., Fu L., Januario T., Kallop D., Nannini-Pepe M., Kotkow K., Marsters J. C., Rubin L. L., de Sauvage F. J. (2008) A paracrine requirement for hedgehog signalling in cancer. Nature 455, 406–410 [DOI] [PubMed] [Google Scholar]
- 4. Kolterud A., Grosse A. S., Zacharias W. J., Walton K. D., Kretovich K. E., Madison B. B., Waghray M., Ferris J. E., Hu C., Merchant J. L., Dlugosz A. A., Kottmann A. H., Gumucio D. L. (2009) Paracrine hedgehog signaling in stomach and intestine: new roles for hedgehog in gastrointestinal patterning. Gastroenterology 137, 618–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mao J., Kim B. M., Rajurkar M., Shivdasani R. A., McMahon A. P. (2010) Hedgehog signaling controls mesenchymal growth in the developing mammalian digestive tract. Development 137, 1721–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zacharias W. J., Li X., Madison B. B., Kretovich K., Kao J. Y., Merchant J. L., Gumucio D. L. (2010) Hedgehog is an anti-inflammatory epithelial signal for the intestinal lamina propria. Gastroenterology 138, 2368–2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. van Dop W. A., Heijmans J., Büller N. V., Snoek S. A., Rosekrans S. L., Wassenberg E. A., van den Bergh Weerman M. A., Lanske B., Clarke A. R., Winton D. J., Wijgerde M., Offerhaus G. J., Hommes D. W., Hardwick J. C., de Jonge W. J., Biemond I., van den Brink G. R. (2010) Loss of Indian Hedgehog activates multiple aspects of a wound healing response in the mouse intestine. Gastroenterology 139, 1665–1676 [DOI] [PubMed] [Google Scholar]
- 8. Zacharias W. J., Madison B. B., Kretovich K. E., Walton K. D., Richards N., Udager A. M., Li X., Gumucio D. L. (2011) Hedgehog signaling controls homeostasis of adult intestinal smooth muscle. Dev. Biol. 355, 152–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ramalho-Santos M., Melton D. A., McMahon A. P. (2000) Hedgehog signals regulate multiple aspects of gastrointestinal development. Development 127, 2763–2772 [DOI] [PubMed] [Google Scholar]
- 10. McMahon A. P., Ingham P. W., Tabin C. J. (2003) Developmental roles and clinical significance of hedgehog signaling. Curr. Top. Dev. Biol. 53, 1–114 [DOI] [PubMed] [Google Scholar]
- 11. Lum L., Beachy P. A. (2004) The Hedgehog response network: sensors, switches, and routers. Science 304, 1755–1759 [DOI] [PubMed] [Google Scholar]
- 12. Hooper J. E., Scott M. P. (2005) Communicating with Hedgehogs. Nat. Rev. Mol. Cell Biol. 6, 306–317 [DOI] [PubMed] [Google Scholar]
- 13. Ruiz i Altaba A. (2011) Hedgehog signaling and the Gli code in stem cells, cancer, and metastases. Sci. Signal 4, pt9. [DOI] [PubMed] [Google Scholar]
- 14. Hui C. C., Angers S. (2011) Gli proteins in development and disease. Annu. Rev. Cell Dev. Biol. 27, 23.21–23.25 [DOI] [PubMed] [Google Scholar]
- 15. Bai C. B., Auerbach W., Lee J. S., Stephen D., Joyner A. L. (2002) Gli2, but not Gli1, is required for initial Shh signaling and ectopic activation of the Shh pathway. Development 129, 4753–4761 [DOI] [PubMed] [Google Scholar]
- 16. Litingtung Y., Dahn R. D., Li Y., Fallon J. F., Chiang C. (2002) Shh and Gli3 are dispensable for limb skeleton formation but regulate digit number and identity. Nature 418, 979–983 [DOI] [PubMed] [Google Scholar]
- 17. Cain J. E., Islam E., Haxho F., Blake J., Rosenblum N. D. (2011) GLI3 repressor controls functional development of the mouse ureter. J. Clin. Invest. 121, 1199–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ding Q., Motoyama J., Gasca S., Mo R., Sasaki H., Rossant J., Hui C. C. (1998) Diminished Sonic hedgehog signaling and lack of floor plate differentiation in Gli2 mutant mice. Development 125, 2533–2543 [DOI] [PubMed] [Google Scholar]
- 19. Hilton M. J., Tu X., Cook J., Hu H., Long F. (2005) Ihh controls cartilage development by antagonizing Gli3, but requires additional effectors to regulate osteoblast and vascular development. Development 132, 4339–4351 [DOI] [PubMed] [Google Scholar]
- 20. Litingtung Y., Chiang C. (2000) Specification of ventral neuron types is mediated by an antagonistic interaction between Shh and Gli3. Nat. Neurosci. 3, 979–985 [DOI] [PubMed] [Google Scholar]
- 21. Joeng K. S., Long F. (2009) The Gli2 transcriptional activator is a crucial effector for Ihh signaling in osteoblast development and cartilage vascularization. Development 136, 4177–4185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Verzi M. P., Stanfel M. N., Moses K. A., Kim B. M., Zhang Y., Schwartz R. J., Shivdasani R. A., Zimmer W. E. (2009) Role of the homeodomain transcription factor Bapx1 in mouse distal stomach development. Gastroenterology 136, 1701–1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Long F., Chung U. I., Ohba S., McMahon J., Kronenberg H. M., McMahon A. P. (2004) Ihh signaling is directly required for the osteoblast lineage in the endochondral skeleton. Development 131, 1309–1318 [DOI] [PubMed] [Google Scholar]
- 24. Blaess S., Stephen D., Joyner A. L. (2008) Gli3 coordinates three-dimensional patterning and growth of the tectum and cerebellum by integrating Shh and Fgf8 signaling. Development 135, 2093–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mao J., Ligon K. L., Rakhlin E. Y., Thayer S. P., Bronson R. T., Rowitch D., McMahon A. P. (2006) A novel somatic mouse model to survey tumorigenic potential applied to the Hedgehog pathway. Cancer Res. 66, 10171–10178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Muzumdar M. D., Tasic B., Miyamichi K., Li L., Luo L. (2007) A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605 [DOI] [PubMed] [Google Scholar]
- 27. Zhang X. M., Ramalho-Santos M., McMahon A. P. (2001) Smoothened mutants reveal redundant roles for Shh and Ihh signaling including regulation of L/R asymmetry by the mouse node. Cell 105, 781–792 [PubMed] [Google Scholar]
- 28. McHugh K. M. (1995) Molecular analysis of smooth muscle development in the mouse. Dev. Dyn. 204, 278–290 [DOI] [PubMed] [Google Scholar]
- 29. Sukegawa A., Narita T., Kameda T., Saitoh K., Nohno T., Iba H., Yasugi S., Fukuda K. (2000) The concentric structure of the developing gut is regulated by Sonic hedgehog derived from endodermal epithelium. Development 127, 1971–1980 [DOI] [PubMed] [Google Scholar]
- 30. Yu J., Carroll T. J., McMahon A. P. (2002) Sonic hedgehog regulates proliferation and differentiation of mesenchymal cells in the mouse metanephric kidney. Development 129, 5301–5312 [DOI] [PubMed] [Google Scholar]
- 31. Madison B. B., Braunstein K., Kuizon E., Portman K., Qiao X. T., Gumucio D. L. (2005) Epithelial hedgehog signals pattern the intestinal crypt-villus axis. Development 132, 279–289 [DOI] [PubMed] [Google Scholar]
- 32. Kim J. H., Huang Z., Mo R. (2005) Gli3 -null mice display glandular overgrowth of the developing stomach. Dev. Dyn. 234, 984–991 [DOI] [PubMed] [Google Scholar]
- 33. Schaefer L., Iozzo R. V. (2008) Biological functions of the small leucine-rich proteoglycans: from genetics to signal transduction. J. Biol. Chem. 283, 21305–21309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ormestad M., Astorga J., Landgren H., Wang T., Johansson B. R., Miura N., Carlsson P. (2006) Foxf1 and Foxf2 control murine gut development by limiting mesenchymal Wnt signaling and promoting extracellular matrix production. Development 133, 833–843 [DOI] [PubMed] [Google Scholar]
- 35. Bredrup C., Knappskog P. M., Majewski J., Rødahl E., Boman H. (2005) Congenital stromal dystrophy of the cornea caused by a mutation in the decorin gene. Invest. Ophthalmol. Vis. Sci. 46, 420–426 [DOI] [PubMed] [Google Scholar]
- 36. Majava M., Bishop P. N., Hägg P., Scott P. G., Rice A., Inglehearn C., Hammond C. J., Spector T. D., Ala-Kokko L., Männikkö M. (2007) Novel mutations in the small leucine-rich repeat protein/proteoglycan (SLRP) genes in high myopia. Hum. Mutat. 28, 336–344 [DOI] [PubMed] [Google Scholar]