Background: Parkinson disease protein α-synuclein has sequence similarity with apolipoproteins.
Results: α-Synuclein can generate nanoparticles from phospholipid membranes and fatty acids by taking up a broken helical structure.
Conclusion: These nanoparticles have shapes and dimensions reminiscent of apolipoprotein nanodiscs.
Significance: The data suggest apolipoprotein-like roles for α-synuclein as a lipid- or fatty acid-carrying protein.
Keywords: α-Synuclein, Electron Microscopy (EM), Electron Paramagnetic Resonance (EPR), Membrane Proteins, Neurodegenerative Diseases, Nanoparticles
Abstract
α-Synuclein (αS) is a membrane-binding protein with sequence similarity to apolipoproteins and other lipid-carrying proteins, which are capable of forming lipid-containing nanoparticles, sometimes referred to as “discs.” Previously, it has been unclear whether αS also possesses this property. Using cryo-electron microscopy and light scattering, we found that αS can remodel phosphatidylglycerol vesicles into nanoparticles whose shape (ellipsoidal) and dimensions (in the 7–10-nm range) resemble those formed by apolipoproteins. The molar ratio of αS to lipid in nanoparticles is ∼1:20, and αS is oligomeric (including trimers and tetramers). Similar nanoparticles form when αS is added to vesicles of mitochondrial lipids. This observation suggests a mechanism for the previously reported disruption of mitochondrial membranes by αS. Circular dichroism and four-pulse double electron electron resonance experiments revealed that in nanoparticles αS assumes a broken helical conformation distinct from the extended helical conformation adopted when αS is bound to intact vesicles or membrane tubules. We also observed αS-dependent tubule and nanoparticle formation in the presence of oleic acid, implying that αS can interact with fatty acids and lipids in a similar manner. αS-related nanoparticles might play a role in lipid and fatty acid transport functions previously attributed to this protein.
Introduction
α-Synuclein (αS)4 is an abundant protein in the brain that has been implicated in the pathogenesis of Parkinson disease (1–7). Although its physiological functions are not fully understood, various membrane-related roles have been proposed as follows: in neuronal plasticity; stabilization of synaptic vesicles; modulation of neurotransmitters; regulation of vesicular trafficking; and brain metabolism (8–10). Membrane interaction has also been proposed to play a significant role in the pathogenesis of Parkinson disease, as αS can disrupt cellular membranes. Damage to mitochondria is a commonly found feature of αS expression (11–15), but disruption of other cellular membranes such as Golgi (16, 17), lysosomes (18), endoplasmic reticulum, and nucleus (19) has also been reported.
The membrane interaction of αS is mediated by seven 11-amino acid repeats present in the first ∼90 amino acids. Remarkably, this region can take up a variety of structures upon membrane interaction. Most commonly observed is an extended helical structure (20–25), but broken helical structures (23, 26–29) or much shorter helical structures (30) have also been reported. It is clear that the structure of αS can be quite variable and depends on the particular lipids involved (23, 25). An important determinant for αS structure is whether or not vesicles stay intact during binding (23, 31). Vesicle-bound αS typically has an extended helical structure, whereas conditions that disrupt vesicles can lead to rather different structures (23). We previously reported that αS can induce membrane curvature and that it can convert large phospholipid vesicles into bilayer tubes and cylindrical micelles (25, 31). In addition, small vesicles and small nonvesicular structures can form at higher protein-to-lipid ratios. We refer to the latter structures as nanoparticles because their dimensions, albeit variable, are of the order of 10 nm. Tubulation was also independently observed using a supported bilayer system (32), and vesiculation was found in the case of mitochondrial membranes (12). Induction of membrane curvature occurs at physiological concentrations using brain lipid extracts (33) and is thought to affect SNARE-mediated vesicle fusion (34).
Whereas tubulation is mediated by an extended helical structure of αS (25), much less is known about the nanoparticles. Their dimensions are reminiscent of those seen in lipoprotein particles formed by apolipoproteins, perilipins, TIP47, and adipophilin (35, 36). Apolipoproteins are involved in lipid transport, whereas perilipins, TIP47, and adipophilin bind to lipid droplets and control triglyceride metabolism (37, 38). Their ability to complex with and carry lipids is thought to be of functional relevance (36, 38, 39). These lipoprotein particles are often referred to as discs, but recent evidence indicates that their structures are ellipsoidal in solution (40–43). A range of such structures is likely to exist depending on the exact conditions, including protein-to-lipid ratio. αS shares sequence similarity with apolipoproteins, perilipins, TIP47, and adipophilin in the sense that all of these proteins contain 11-amino acid repeat regions. There is some indication that αS plays a role in the transport and trafficking of lipids. αS has been shown to accumulate on triglyceride-rich lipid droplets (44, 45) and to be associated with microsomes. αS is associated with neuromelanin, lipid droplets that are present in neurons of substantia nigra (46). It is not known, however, whether αS can form lipoprotein particles similar to those of other 11-amino acid repeat containing proteins.
αS is also thought to participate in fatty acid transport as well as metabolism (47–51), and fatty acids induce αS oligomerization (51–53). However, the nature of these oligomers and specifics of the interactions of lipids and fatty acids with αS remain to be determined.
To ascertain the properties of the αS-lipid nanoparticles and assess their similarity to previously reported apolipoprotein-like proteolipid particles, we investigated their structure using circular dichroism (CD), cryo-electron microscopy (cryo-EM), fluorescence spectroscopy, electron paramagnetic resonance (EPR) spectroscopy, and light scattering. We find that they have an overall ellipsoidal architecture analogous to that reported for apolipoproteins. In these complexes, αS forms helical oligomers and assumes a shorter broken helical structure, indicating that in nanoparticles its interaction with lipid is distinctively different from its interaction with liposomes. We observe the formation of nanoparticles from a variety of negatively charged membranes, including liposomes that mimic mitochondrial membranes. In addition, we find that αS can form small oligomeric complexes in the presence of fatty acids. These data strongly suggest that αS has the ability to function as an apolipoprotein-like, intracellular lipid-carrying protein. It might be important to control this property in vivo to avoid disruption of cellular membranes.
EXPERIMENTAL PROCEDURES
Purification of Wild-type and Mutant α-Synuclein
The human wild-type and mutant α-synuclein were expressed in Escherichia coli BL21 (DE3) pLysS cells and generated as reported earlier (54). Briefly, cells were lysed by boiling, followed by acid precipitation. Supernatant was passed through anion exchange columns and eluted with a 0–1.0 m NaCl gradient.
Preparation of Phospholipid Vesicles
The following synthetic lipids were used to prepare different membrane compositions: 1-palmitoyl-2-oleoyl-sn-glycero-3-[phospho-RAC-(1-glycerol)] (POPG), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine, 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-l-serine (POPS), l-α-phosphatidylinositol (liver, bovine), cardiolipin (heart, bovine), cholesterol, and sphingomyelin (brain). All lipids were purchased from Avanti Polar Lipids Inc. (Alabaster, AL). Large nonextruded vesicles were prepared by vortexing the dried lipid film in the required buffer.
Purification of Lipoprotein Particles
α-Synuclein was incubated with POPG-containing vesicles at a protein-to-lipid molar ratio of 1:10 for an hour. The mixture was then subjected to ultracentrifugation for 10 min at 120,000 × g. The supernatant was passed through a Superdex 200 size exclusion column at a flow rate of 0.5 ml/min. Fractions were collected and used for further analyses.
Fluorescence Resonance Energy Transfer (FRET)
A single cysteine mutant of αS (position 131) was conjugated to fluorescent dye Alexa488 or Alexa546 using the thiol-reactive Alexa Fluor 488 C5 maleimide and Alexa Fluor 546 C5 maleimide (Invitrogen). Unconjugated dye was separated using a PD10 desalting column (GE Healthcare). FRET was monitored by recording the spectra from 500 to 650 nm on a Jasco fluorometer (FP-6500), using excitation at 488 nm. The FRET measurements of the monomer were from a freshly prepared sample that was filtered through a 30-kDa cutoff filter (Millipore).
Cross-linking and SDS-PAGE
Purified nanoparticles and αS were subjected to cross-linking using 0.1% glutaraldehyde (Electron Microscopy Sciences) followed by SDS-PAGE. 12% NuPAGE Bistris gels (Invitrogen) were used with NuPAGE MES running buffer for separation. Visualization was done by staining the gels with SimplyBlue safe stain (Invitrogen) for 1 h followed by destaining with ultra pure water for 12 h.
Estimation of Protein-to-Lipid Ratio in Nanoparticles
The protein-to-lipid ratio of gel filtration-purified nanoparticles was estimated by simultaneously labeling protein and lipid. αS was labeled at position 131 with N,N′-dimethyl-N-(iodoacetyl)-N′-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)ethylenediamine (IANBD) (Invitrogen) as described previously (25). The protein concentration could then be estimated by measuring the absorbance at 478 nm and using the extinction coefficient (18,492 m−1) of IANBD (after correction for labeling efficiency). Lipid amounts were estimated in an analogous manner. An extinction coefficient of 58,600 m−1 was used for 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) (rhodamine-DOPE) to calculate lipid concentration. We used 1% rhodamine-DOPE for both POPG and cardiolipin/POPC 20:80% nanoparticles. After collecting different gel filtration fractions, we measured the absorbance at 478 and 570 nm using a UV-visible spectrophotometer and then calculated the protein (CP) and lipid (CL) concentrations, respectively. Finally, the protein-to-lipid molar ratio in different gel filtration fractions was obtained by obtaining CP/CL.
Circular Dichroism
The CD spectra were obtained using a Jasco J-810 spectropolarimeter with a 1-mm quartz cell at room temperature. The parameters used were as follows: scan rate of 50 nm/min, bandwidth of 1 nm, time response of 0.1 nm, and step resolution of 0.5 nm. Protein concentration was determined using the extinction coefficient of protein at 280 nm based on the number of tyrosine molecules in the protein. Appropriate blanks were collected under similar conditions and were subtracted to obtain the final spectra. A 10 mm sodium phosphate (pH 7.4) buffer was used for all CD studies.
Dynamic Light Scattering
Dynamic light scattering measurements of lipid vesicles and samples obtained after gel filtration were performed in a quartz micro-cuvette with a 1-cm path length and a 10-μl volume at room temperature using a Wyatt DynaPro Nanostar Dynamic Light Scattering instrument (Wyatt Technology, Santa Barbara, CA). The data were produced by a regularization fit and analyzed using Dynamics 7.0 software. Percent mass was used as the y axis value. The calculated radii were sphere-equivalent radii obtained using the Rayleigh sphere model.
Electron Microscopy
To prepare negatively stained specimens, carbon-coated Formvar films mounted on copper grids (EMS) were floated on 10-μl droplets of sample for 5 min, and the excess liquid was blotted off before staining with 1% uranyl acetate. A JEOL 1400 transmission electron microscope operated at 100 kV was used for imaging.
Cryo-electron Microscopy/Image Analysis
Samples were applied to holey carbon grids (Quantifoil) and vitrified in a Vitrobot cryo-station (FEI). The humidity was carefully controlled to avoid drying-related deformation of membranes. Specimens were observed with a CM200-FEG (FEI), and images were recorded on film (SO163, Eastman Kodak Co.). Digitization was performed on a SCAI scanner (Carl Zeiss) at final sampling rates of 1.84 or 0.966 Å/pixel. 628 particles were selected for further analysis. K-means-based reference-free classification and subsequent averaging were performed using SPIDER (55) and EMAN (56). For detailed classification, principal component analysis was also used.
Spin Labeling of Cysteine Mutants and Continuous Wave-EPR
αS single or double cysteine mutants were incubated overnight at 4 °C with a 5-fold molar excess of the 1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl-methane-thiosulfonate spin label (R1). PD10 columns (GE Healthcare) were used to remove excess spin label. The spin labeled αS is referred to as R1-labeled protein.
Continuous wave-EPR spectra were recorded from vesicle- or oleic acid-bound αS at a protein-to-lipid/fatty acid molar ratio of 1:100 in 20 mm Hepes (pH 7.4), 100 mm NaCl buffer. Unbound protein was removed from vesicle- or oleic acid-bound αS by pelleting in an ultracentrifuge for 1 h at 120,000 × g. Spectra were recorded using a Bruker EMX X-band continuous wave-EPR spectrometer with a dielectric resonator at 1.59-milliwatt incident microwave power, using a field modulation of 1.5 G. As reported previously (22), sharp lines from background labeling were subtracted using software provided by Christian Altenbach (UCLA). Subsequently, all spectra were amplitude-normalized.
Pulsed EPR and Distance Analysis
Samples were prepared at a protein-to-lipid ratio of 1:10. For all experiments, 10–20% fully spin-labeled protein, containing two spin labels per protein, was mixed with unlabeled wild-type protein and incubated for 5 min before adding it to the lipid vesicles. To perform four-pulse double electron electron resonance (DEER) measurements on nanoparticles formed from POPG, the mixture of αS and POPG vesicles was incubated for 1 h and then ultracentrifuged for 10 min at 120,000 × g to separate the tubes from unbound protein and lipoprotein particles. Purification of lipoprotein particles was performed by passing through a Superdex 200 size exclusion column at a flow rate of 0.5 ml/min. Fractions were collected and used for analysis. In the case of oleic acid, the nanoparticles could not be purified to concentrations required for pulsed EPR measurements using the abovementioned methodology. Therefore, we carried out DEER experiments with samples at different protein-to-lipid ratios where nanoparticles (1:5) and tubes (1:40) are strongly favored. In both cases unbound protein was removed by ultracentrifugation for 1 h at 200,000 × g. The pellet consisting of nanoparticles or tubes was used for DEER experiments. Four-pulse DEER experiments were performed using a Bruker Elexsys E580 X-band pulse EPR spectrometer fitted with a 3-mm split ring (MS-3) resonator, a continuous-flow cryostat (CF935, Oxford Instruments), and a temperature controller (ITC503S, Oxford Instruments). All samples, in 15% sucrose, were flash-frozen, and data were acquired at 78 K. The data were fitted using Tikhonov regularization (57) as implemented in DEERAnalysis2011 packages (58). The background contribution from nonspecific interaction was subtracted, using a three-dimensional model for nanoparticle-bound αS. Tikhonov regularization was used with regularization parameters of 100 or less obtained from the L-curve analysis to fit distances.
RESULTS
α-Synuclein Forms Small Lipoprotein Particles
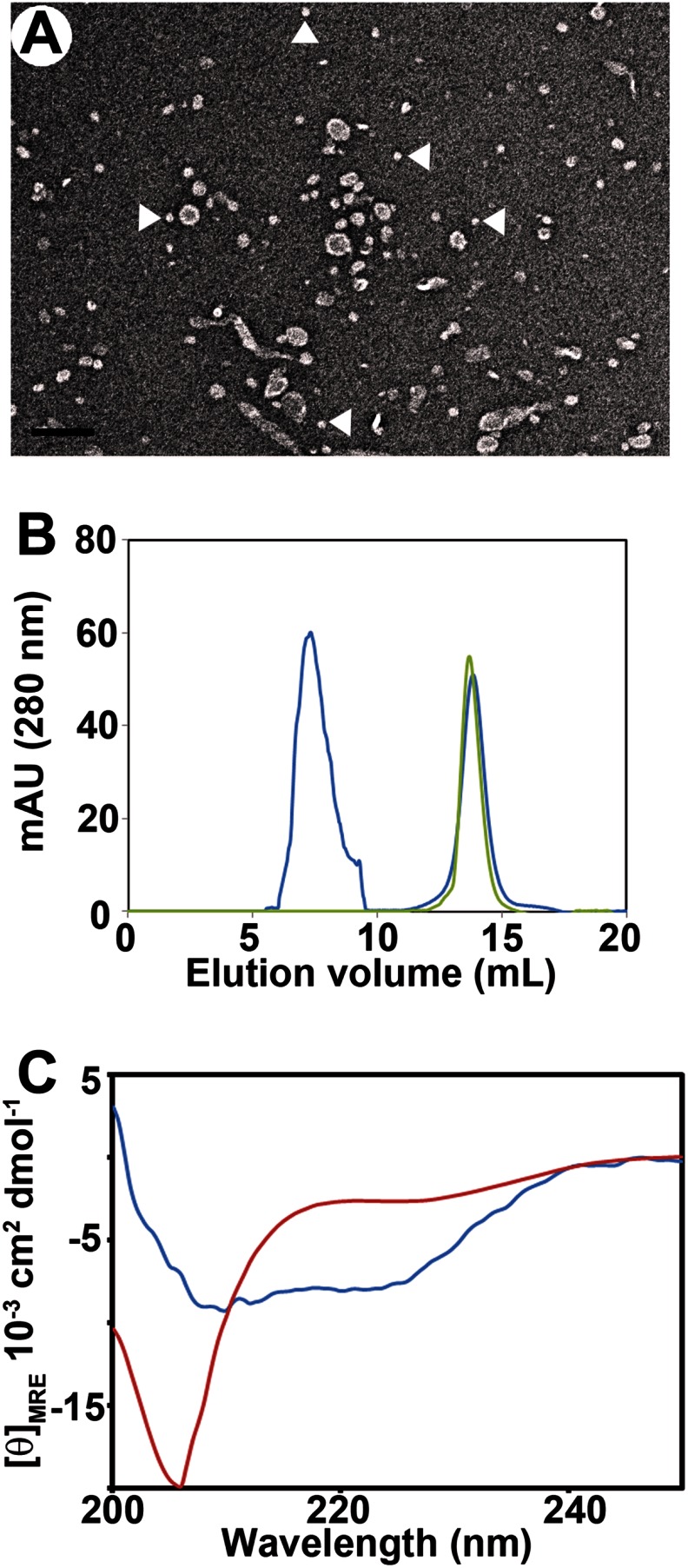
To investigate the nanoparticles, we first devised a protocol to separate them from small vesicles and tubes that also form when αS is incubated with large phosphatidylglycerol-containing vesicles (25, 31). High speed ultracentrifugation was used to pellet the long tubular structures. The supernatant, which contained a mixture of small tubes, vesicles, and nanoparticles (Fig. 1A), was then further fractionated by gel filtration. Two main elution peaks were observed (Fig. 1B). The latter peak eluted at the same position as free αS (∼12.5 ml) and had the characteristic random coil CD spectrum (Fig. 1C, red spectrum). In contrast, all fractions from the first peak were α-helical, according to CD (Fig. 1C, blue spectrum).
FIGURE 1.
Purification of lipoprotein nanoparticles formed by α-synuclein. A, supernatant obtained after ultracentrifugation of wild-type αS incubated with POPG vesicles at a protein-to-lipid molar ratio of 1:10. White arrowheads indicate nanoparticles. Scale bar, 100 nm. B, separation of smaller nanoparticles by gel filtration after incubating αS with POPG vesicles. Green trace represents protein alone, and blue trace shows the supernatant obtained as described for A. C, circular dichroism spectra of αS alone (red curve) and fraction 8–8.5 ml (blue curve) of gel filtration after incubating wild-type αS with POPG vesicles. mAU, milliabsorbance unit.
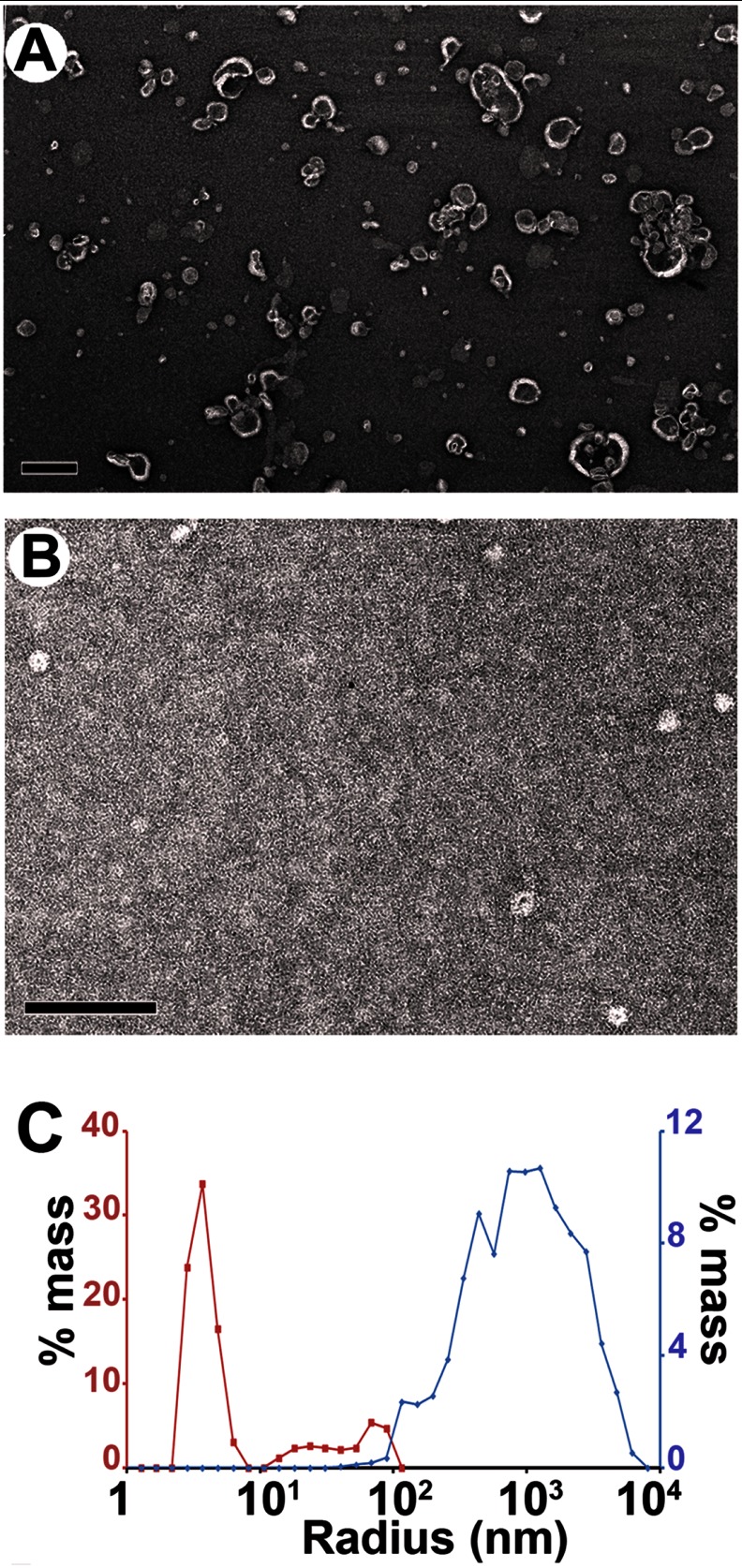
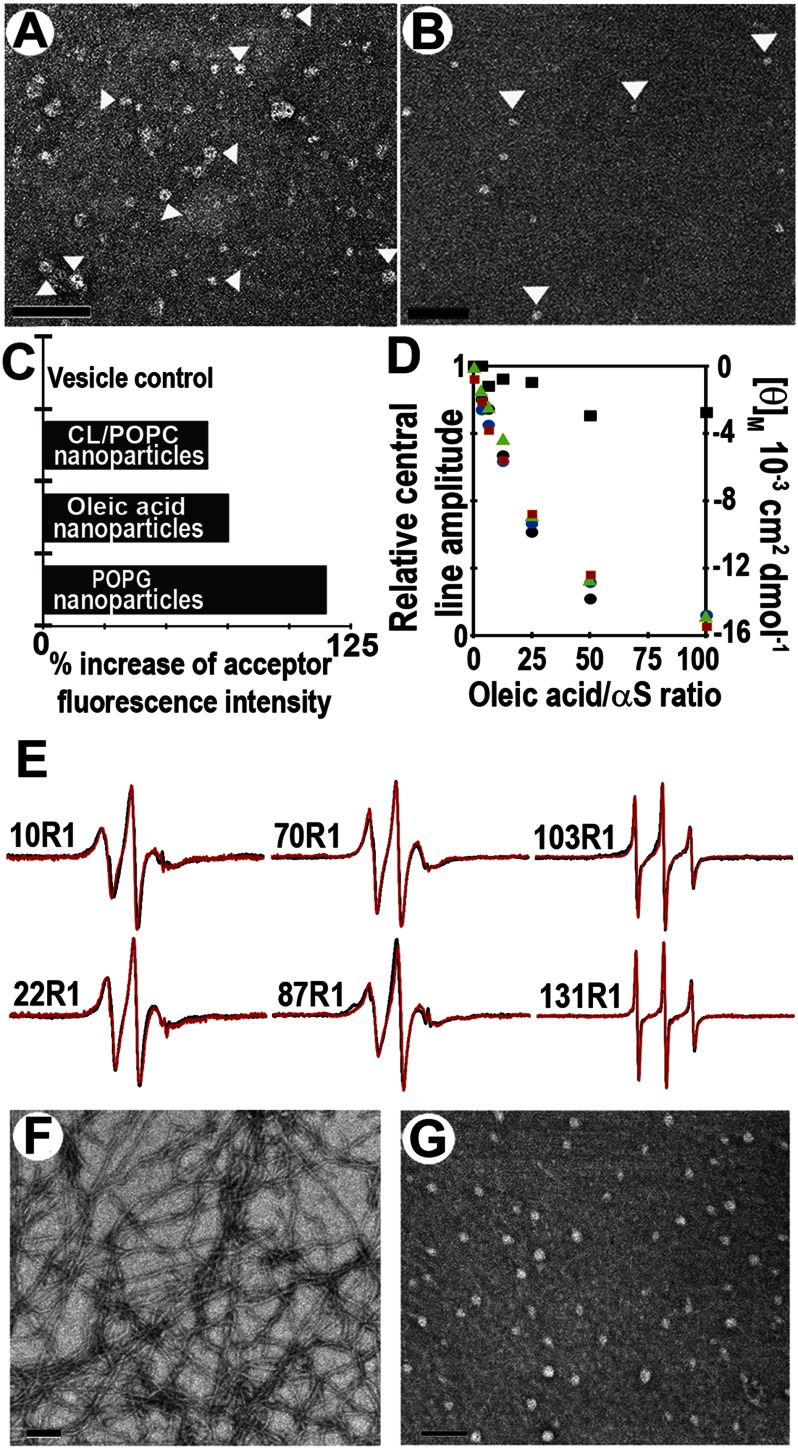
Negative stain EM indicated that this peak contained two main species. Whereas the earlier fractions consisted mainly of small vesicles (Fig. 2A) with some short tubular structures, the latter portion of this peak was mainly composed of small roundish structures, i.e. the nanoparticles (Fig. 2B). To further characterize them in solution, we performed dynamic light scattering experiments. As illustrated in Fig. 2C, a hydrodynamic radius peak was observed at ∼7 nm. This is too small to represent vesicles, which are generally at least 10 nm in radius (59).
FIGURE 2.
α-Synuclein forms proteolipid nanoparticles. A, negative stain electron micrograph of gel filtration fraction (6–8 ml) of supernatant obtained after high speed ultracentrifugation; see Fig. 1B. Scale bar, 100 nm. B, negative stain electron micrograph of purified (fraction 8–8.5 ml) ∼10–17-nm sized particles formed when large POPG vesicles (600 μm) were incubated with αS (60 μm). Scale bar, 100 nm. C, analysis by dynamic light scattering showed that particles of ∼7 nm average hydrodynamic radius are formed in the presence of αS (8–8.5 ml) of gel filtration (red trace) as compared with POPG vesicles without protein (blue trace).
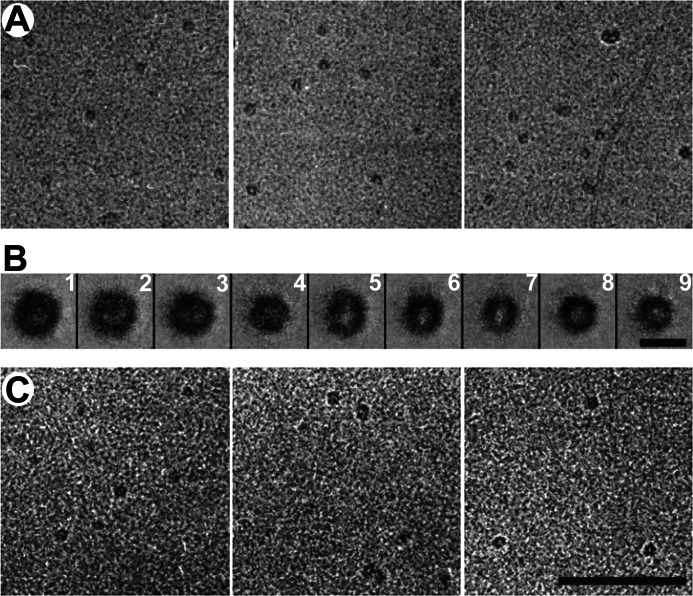
The nanoparticles were then examined by cryo-EM (Fig. 3A). A sizeable set of images (n = 628) was subjected to classification and class-averaging procedures. The number of distinct classes was not sharply defined, but classes that gave consistent features in the averages were a robust feature of the analysis, regardless of how many classes were specified (Fig. 3B). The majority of class averages show round particles with low density central regions, which vary in diameter from 10 nm (classes 1 and 2) to about 8.5 nm (classes 4–6) to about 6.5 nm (classes 7–9). Some of the particles are oval rather than round (class 5) with an axial ratio of about 1.2, and in some cases, the peripheral zone of higher density intrudes into the central region (class 2). Overall, the data suggest a certain degree of intrinsic heterogeneity as the smallest round particles cannot represent an alternative view (a potential source of image variability) of the largest round particles. All of the class averages present higher peripheral density, suggesting that αS is wrapping around the lipids in a ring-like manner. Such an architecture has previously been suggested for early discoidal high density lipoprotein particles formed by apolipoprotein A-1 (apoA-1) (43, 60).
FIGURE 3.
CryoEM images of αS-induced nanoparticles at a protein-to-lipid ratio of 1:5 from MLVs (A) and SUVs (C) are shown. B, class averages of 628 selected nanoparticles induced by αS-MLVs. The size of these particles varies from 7 to 10 nm. Scale bar, A and C, 100 nm; B, 10 nm.
Small unilamellar vesicles (SUVs) composed of POPG are commonly used for studying the interactions of αS with membranes (23, 27, 61–63). We have previously shown that αS can also disrupt such vesicles (23), but the morphology of the resulting structures has not been described in detail. As shown in Fig. 3C, incubation of αS with SUVs also resulted in the formation of small lipoprotein particles. Their size and shape were indistinguishable from those obtained from large vesicles (Fig. 3A) (25). Again, αS-containing nanoparticles could be purified by gel filtration, eluting at the same position and with an α-helical signature, as judged by CD (data not shown). The main difference encountered when using SUV was the absence of the long tubular structures, sometimes up to microns in length that were produced when large vesicles were used (25, 31). Thus, it appears that the much fewer lipids available in a single SUV are insufficient for the formation of micrometer long tubes, which require a more substantial amount of lipid. In fact, assuming average sizes of SUV and large vesicles of 25 and 1000 nm, the large vesicles contain about ∼3000-fold more lipid. This ratio in lipid numbers approximately correlates with the different tubule lengths, which are on the order of 100 nm and 100 μm, respectively.
αS Is Oligomeric on Nanoparticles
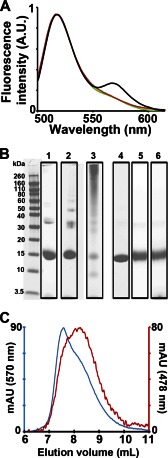
To test for oligomerization of αS, we performed FRET and cross-linking experiments. For FRET experiments, αS was labeled with Alexa488 as donor and Alexa546 as acceptor. The respective derivatives were then mixed, and the fluorescence emission spectra were recorded for different preparations. A FRET signal between 550 and 600 nm was observed from αS nanoparticles that were isolated by gel filtration (Fig. 4A, black trace). This signal was absent for monomeric derivatives in the absence of lipids (Fig. 4A, red trace). Binding of the fluorescently labeled derivatives to SUVs under conditions that prevent membrane remodeling and keep vesicles intact (23) also did not result in enhanced FRET compared with monomer (Fig. 4A, green).
FIGURE 4.
Nanoparticles contain multiple αS molecules. A, increase in fluorescence resonance energy transfer (FRET) on nanoparticles formed from POPG vesicles (black) was measured by mixing them with αS labeled with Alexa488 and Alexa546. The POPG nanoparticles were purified using the aforementioned gel filtration protocol before testing for FRET. No FRET enhancement compared with monomer (red) was observed when αS was added to POPS/POPC 3/7 SUVs (green) under conditions known to keep the vesicles intact (23). Increasing the concentration of monomeric protein 10-fold higher than nanoparticles did not lead to an increase of emission intensity at 570 nm. A protein-to-lipid molar ratio of 1:10 was used. B, SDS-PAGE with or without cross-linking was used to ascertain the number of αS-molecules per nanoparticle. Lane 1, noncross-linked nanoparticles from POPG; lane 2, cross-linked nanoparticles after 30 s of incubation with 0.1% glutaraldehyde; lane 3, cross-linked nanoparticles after 1 min of incubation with 0.1% glutaraldehyde; lane 4, noncross-linked monomeric αS control; lane 5, cross-linked monomeric αS control after 1 min of incubation with 0.1% glutaraldehyde; lane 6, αS cross-linked with 0.1% glutaraldehyde for 1 min in the presence of POPS/POPC 3/7 SUVs. C, gel filtration trace of the early nanoparticle-containing fractions. The red trace represents the absorbance at 478 nm of IANBD-labeled αS, and the blue trace represents the absorbance at 570 nm of POPG containing 1% rhodamine-DOPE. A CP/CL (see “Experimental Procedures”) of ∼1:20 was obtained for the nanoparticle-containing fraction (8–8.5 ml) indicating the presence of ∼1 αS molecule per 20 molecules of lipid. AU, arbitrary units of fluorescence intensity; mAU, milliabsorbance units.
To further characterize the oligomeric state of αS on nanoparticles, we performed SDS-PAGE (Fig. 4B). Using glutaraldehyde cross-linking, dimeric, trimeric, and tetrameric bands were well resolved for nanoparticles (Fig. 4B, lane 2) after 30 s of cross-linking. At longer times (1 min) of cross-linking (Fig. 4B, lane 3), the cross-linking became more pronounced but the bands were also more smeared. Importantly, even the longer cross-linking times did not show appreciable cross-linking of monomeric αS (Fig. 4B, lane 5) or vesicle-bound αS (lane 6). In the absence of cross-linker, the trimer and tetramer bands of the nanoparticles weakened (Fig. 4B, lane 1), but a dimer band remained. Collectively, these data indicate that αS forms oligomers on nanoparticles, but not on un-remodeled, intact vesicles.
The simultaneous use of fluorescently labeled proteins and lipids allowed us to provide an estimate of the protein-to-lipid ratio in nanoparticles. For these experiments, we used gel filtration to purify nanoparticles, and from the absorbances of the labeled proteins and lipids we were able to estimate the respective concentrations and obtain protein-to-lipid ratios of ∼1:20–25 (Fig. 4C).
αS in Lipoprotein Nanoparticles Has a Different Structure than on Membrane Tubes or Vesicles
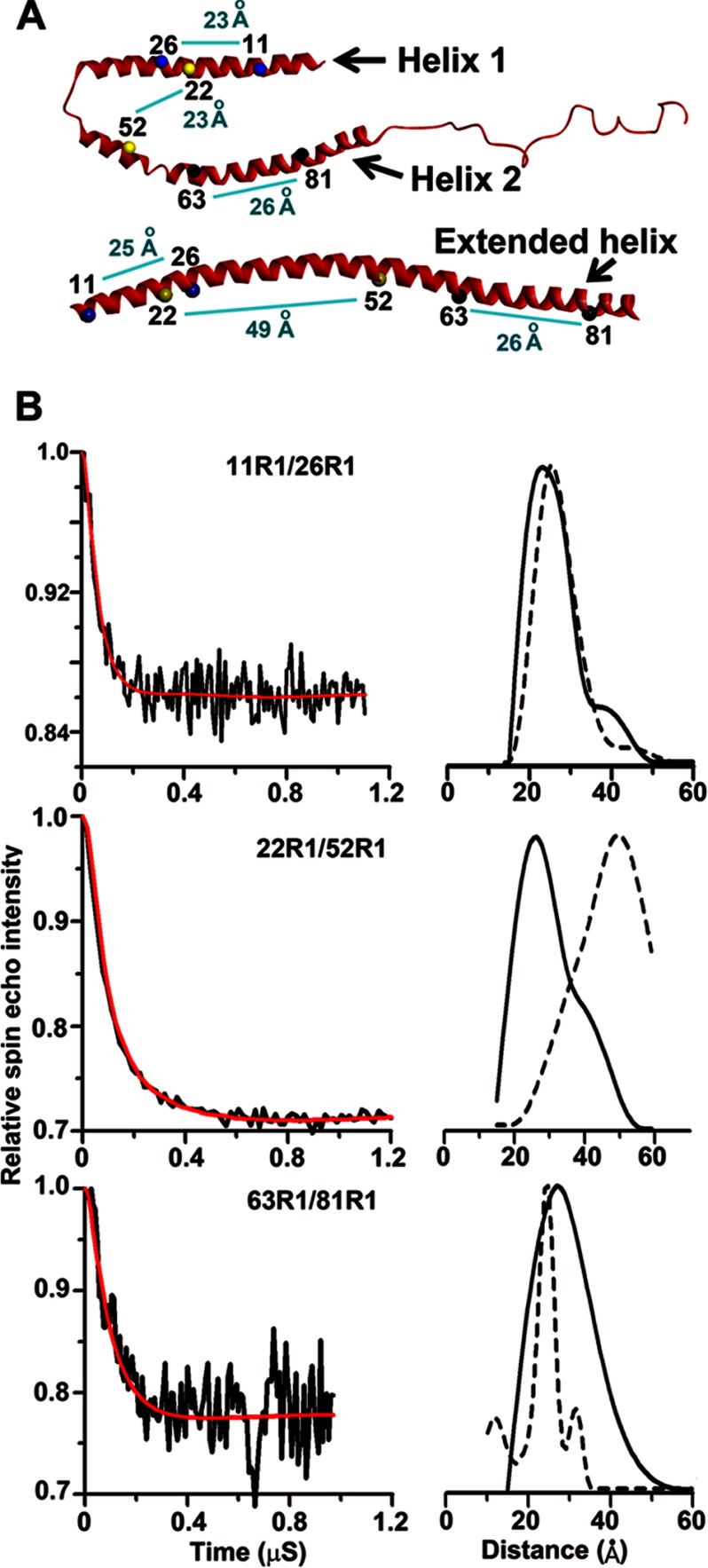
αS takes up an extended helical conformation when bound to vesicles (20–24) or tubes (25). To investigate its structure in nanoparticles, we measured intramolecular distances using four-pulse DEER experiments. Nanoparticles were generated from either large vesicles or SUVs and purified using the aforementioned gel filtration protocol. Three double mutants were used with spin labels (R1) either at positions 11 and 26 (11R1/26R1), at positions 22 and 52 (22R1/52R1), or at positions 63 and 81 (63R1/81R1) (Fig. 5A). Residues 11 and 26 are expected to be in the same helix regardless of whether a single extended helix or a two-segment helix is formed (23, 29). Indeed, with the 11R1/26R1 derivative, the nanoparticles gave rise to distance distributions (Fig. 5B, top panel) rather similar to those obtained in prior studies on vesicular and tubular structures (23, 25). In contrast, the distance separating residues 22 and 52 is strongly dependent on the structure into which αS is incorporated (23, 28, 29). With the 22R1/52R1 derivative, very different spacings were observed (Fig. 5B, middle panel). Unlike the 46–50 Å observed on tubes and vesicles, the DEER data from nanoparticles yielded distances of ∼27 Å (Fig. 5B). These data are incompatible with an extended helical structure, but they are compatible with a two-segment helical structure. 63R1/81R1 yielded a distance compatible with a helix within this region, but the distance distribution was broader than that obtained in our previous study with SUVs (Fig. 5B, bottom panel). This could indicate the presence of multiple conformations of αS, perhaps including some less ordered structures in this region, as suggested previously (30). Moreover, similar distances were obtained for nanoparticles regardless of whether they were formed from SUVs or large vesicles (data not shown). Taking these observations together with their similar appearance in cryo-EM, it is likely that nanoparticles from both sources are closely related.
FIGURE 5.
Spin-label intramolecular distances from four-pulse DEER experiment. A, numbers mark the positions of double mutants in a bent helix (SDS-bound structure of αS top) versus extended helix (SUV-bound structure of αS bottom). The light blue line shows the connectivity between the two double mutants used in this study. The distances shown are from previous NMR studies of the SDS-bound structure of αS (top) and from 4-pulse DEER experiments of the SUV-bound structure of αS (bottom). B, dipolar evolution for αS 11R1–26R1 (top left), 22R1–52R1 (middle left), and 63R1–81R1 (bottom left) for nanoparticles purified after incubating αS with POPG SUVs is shown. The black trace denotes background-corrected experimental data. The red curve depicts fit using L-curve Tikhonov regularizations, and the right panels show the resulting intramolecular distance distributions in solid black. Because of significantly better signal-to-noise ratios, shorter scans were used to carry out Tikhonov regularization. An enhanced signal-to-noise ratio compensated the increased uncertainty arising from shortened time scan. To compare with distances obtained for same R1 pairs on tubes, data obtained from our previous study for respective R1 pairs on micellar tubes have been indicated by broken lines.
α-Synuclein Forms Nanoparticles with Physiologically Relevant Lipids and Fatty Acids
Next we investigated whether αS forms nanoparticles in the presence of more physiologically relevant lipids. It has been reported that αS can interact with mitochondrial membranes and that overexpression of αS can disrupt mitochondrial membrane integrity (11–14, 64). Damage to mitochondrial membrane has been attributed to the direct membrane interaction of αS and specifically to cardiolipin, which is present in both outer and inner mitochondrial membranes (15). To investigate how αS might interact with mitochondrial membranes and potentially disrupt their integrity, we incubated αS for 1 h with multilamellar vesicles with a lipid composition commonly used to mimic mitochondrial membranes. After ultracentrifugation, the supernatant was visualized by EM (Fig. 6, A and B). Indeed, αS formed nanoparticles similar to those observed with POPG vesicles. When subjected to gel filtration, the nanoparticles again eluted after the void volume peak. By monitoring the absorbance of fluorescently labeled αS and lipid of these isolated nanoparticles, we estimated the protein-to-lipid ratio to be on the order of ∼1:20–25 for these nanoparticles (data not shown). FRET data further confirmed the presence of αS oligomers in nanoparticles formed from cardiolipin/POPC vesicles (Fig. 6C). Unfortunately, the lower yields under these conditions (compared with POPG) prevented us from obtaining high quality four-pulse DEER data. It should be noted that negatively charged membranes are required as we could not generate nanoparticles from the neutral phosphatidylcholine-containing vesicles (data not shown).
FIGURE 6.
α-Synuclein forms nanoparticles from vesicles mimicking mitochondrial membranes. A, αS with vesicles mimicking mitochondrial membrane (POPC/1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine/POPS/l-α-phosphatidylinositol/cardiolipin/sphingomyelin/cholesterol = 39:34:1:5:18:1:2%). Scale bar, 100 nm. B, αS with vesicles composed of cardiolipin/POPC (20:80%). Arrowheads point to some of the nanoparticles. The P/L ratio was 1:10. Scale bar, 100 nm. C, nanoparticles formed from POPG, oleic acid, and cardiolipin/POPC vesicles contain multiple αS molecules. Percent increase in acceptor fluorescence (FRET) intensity on nanoparticles formed by αS labeled with Alexa488 and Alexa546 is shown. Fluorescence intensity of αS labeled with Alexa488 and Alexa546 in the absence of lipid was used as control to calculate percent increase of acceptor fluorescence intensity at 570 nm. No FRET enhancement was seen with POPS/POPC SUV vesicle control, which was intact under conditions previously shown (23). D, αS helicity increases with increasing amounts of oleic acid. Mean residue molar ellipticity (θM, right y axis) at 222 nm (red square) is plotted against the indicated oleic acid/αS molar ratios. Ordering of amino acids at position 10 (green triangle), 22 (black circle), 70 (blue circle), and 131 (black square) was monitored by monitoring the change in central line amplitude (left y axis) as a function of increasing oleic acid concentration using continuous wave EPR. E, continuous wave EPR spectra of αS in the presence of oleic acid or POPG. Residues in the N-terminal membrane binding region tested for their ability to bind to oleic acid (red spectra) showed comparable structuring to that reported with POPG (black spectra) vesicles. C-terminal residues (103R1 and 131R1) largely remained unstructured with oleic acid or POPG vesicles. F, tubes formed after incubating αS with oleic acids at a protein-to-fatty acid molar ratio of 1:20. Scale bar, 100 nm. G, nanoparticles formed upon incubating αS with oleic acids at a molar ratio of 1:5. Scale bar, 100 nm.
The interaction of αS with free fatty acids is thought to be of physiological relevance, and it is known that αS oligomerizes upon addition of free fatty acids (45, 48, 50, 52, 65) possibly causing the formation of helical conformations related to those described above (66). To test whether αS has a similar means of interacting with fatty acids as with lipids, we incubated the protein with oleic acid under various αS-to-oleic acid ratios. According to CD, increasing amounts of oleic acid give rise to enhanced helicity (Fig. 6D, red squares). Next, we used continuous wave EPR to test whether fatty acid binding, like membrane binding, is mediated by the N-terminal region of the molecule. Toward this end, EPR spectra of spin-labeled αS derivatives were recorded as a function of oleic acid concentration. When spin labels were introduced in the N-terminal region of the molecule (10R1, 22R1, and 70R1), the interaction with oleic acid caused a progressive decrease in the amplitude of the EPR spectra (amplitudes are plotted in Fig. 6D) that corresponded to the structuring observed by CD (Fig. 6D). In contrast, only minor changes in spectral amplitude were obtained for the C-terminal labeling site (131R1) (Fig. 6D). The EPR spectra of the fatty acid-bound forms (Fig. 6E, red traces) show that sites in the N-terminal membrane-binding region (10R1, 22R1, 70R1, and 87R1) have intermediate mobility consistent with an ordered structure. In contrast, sites in the C terminus (103R1 and 131R1) exhibit a high mobility that is characteristic for a disordered structure. Thus, similar regions are involved in fatty acid and membrane binding. For comparison, we also recorded the EPR spectra of POPG-bound αS derivatives under the equivalent protein-to-lipid ratios (Fig. 6E, black traces). Qualitatively similar spectra were observed further supporting potential similarities between fatty acid and lipid binding.
Next, we used electron microscopy to test whether similar morphologies could be observed in the case of lipid and fatty acid-bound αS. Indeed, at a protein-to-fatty acid molar ratio of 1:20 to 1:40, we observed narrow tubular structures that have dimensions similar to those seen in cylindrical micelles from POPG (Fig. 6F) (25). At a higher protein content (e.g. a ratio of 1:5), we observed small round structures reminiscent of those observed with phosphatidylglycerol or mitochondrial lipids (Fig. 6G). The fatty acid-containing nanoparticles (∼20–30 nm in diameter by negative stain EM) were stable during gel filtration, and FRET measurements of the purified nanoparticles again indicated the presence of αS oligomers (Fig. 6C).
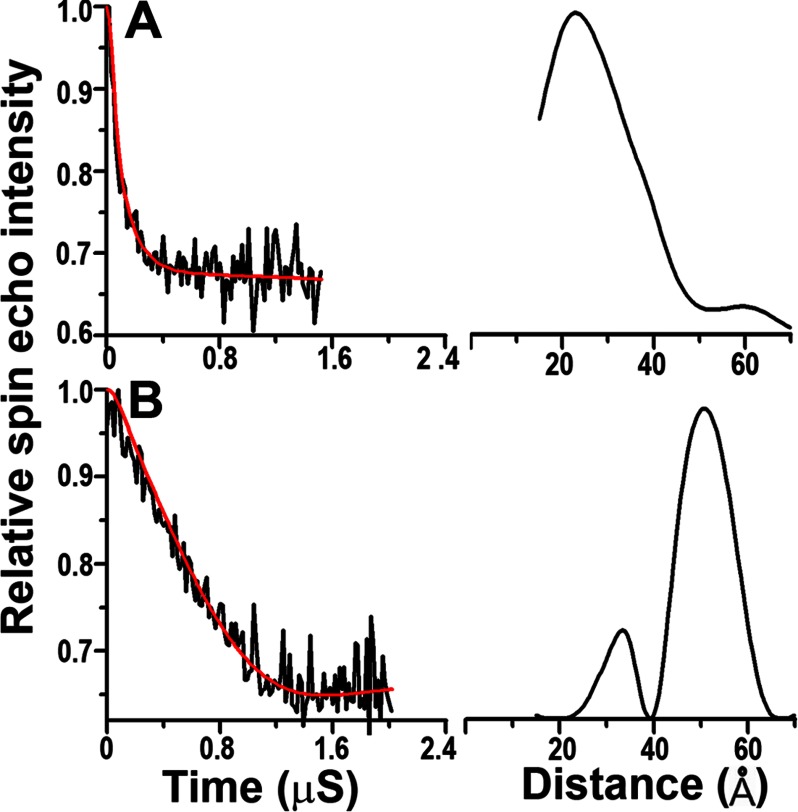
To test for further structural similarities between POPG and oleic acid-bound αS, we used four-pulse DEER experiments at different αS to fatty acid ratios. The 22R1/52R1 derivative was used, because it is a sensitive indicator for extended or broken helical structures (Fig. 5A). Under conditions that favor the formation of cylindrical micelles (protein-to-oleic acid ratio of 1:40), we mainly observed long distances (Fig. 7) consistent with an extended helical structure (Fig. 5A). In contrast, larger protein-to-fatty acid ratios (1:5), which favor the formation of nanoparticles, gave short distances that are incompatible with an extended helical structure (Fig. 7). These data further illustrate the structural similarities between lipid and fatty acid-bound αS.
FIGURE 7.
Intramolecular distance measurements of oleic acid-bound αS using four-pulse DEER. Measurements were carried out using the 22R1–52R1 derivative at 1:5 (A) and 1:40 (B) protein to fatty acid ratios. The dipolar evolution of αS 22R1–52R1 is shown on the left. The black traces denote background-corrected experimental data. The red curves depict a fit using Tikhonov regularization, and the right panels show the resulting intramolecular distance distributions in black.
DISCUSSION
Based on the sequence similarity between apolipoproteins, TIP47, perilipins, and αS, it has long been suspected that αS might be capable of forming lipoprotein particles. However, in the presence of neutral lipids commonly used with apolipoproteins, such structures could not be generated (67). The key difference in this study is the use of negatively charged lipids or fatty acids. Otherwise, our conditions, including the protein-to-lipid ratios, were comparable with those used in earlier studies with apolipoproteins, TIP47, and perilipin lipoprotein particles (35, 36, 43).
The αS-containing nanoparticles share a number of similarities with those described for the aforementioned proteins. First, their sizes as determined by both EM and light scattering are closely similar. Second, multiple apolipoprotein molecules wrap their helices around the lipid core to form apolipoprotein-lipid particles. Our FRET and SDS-PAGE data are consistent with αS being oligomeric on nanoparticles as well. The largest oligomer we can clearly resolve on gels is a tetramer (Fig. 4B). Based on our estimate of a protein-to-lipid ratio of 1:20 to 1:25, such a tetramer would contain ∼80–100 lipids, a number that is close to that obtained for apolipoprotein nanoparticles or discs (43). Based on the average size of a lipid, a bilayer made from 80–100 lipids would be about 5–6 nm in diameter. If αS molecules were to be wrapped around such a lipid core, the nanoparticle would take up a size similar to that observed by cryo-EM studies. In fact, the class average images for the αS-lipid particles (Fig. 3B) are consistent with a model in which multiple helical αS wrap around a central lipid-filled core. Third, the class averages have the appearance of discoid structures, but, as explained above, these particles are not likely to be perfectly flat; rather, the cryo-EM data suggest that these “discs” have ellipsoidal shapes. This feature is in agreement with recent studies on apolipoproteins that showed by other approaches that the apolipoprotein/lipid particles are ellipsoidal in solution (40, 42, 43). A fourth similarity between apolipoproteins and αS is the ability to clear the light scattering of large vesicles (Fig. 2C) (31, 68) by converting them into smaller structures, including tubes, small vesicles, and lipoprotein particles. We previously reported that apolipoproteins are also capable of forming tubes (31). Whether tubes act as precursors of nanoparticles remains to be tested.
Considering that αS is thought to play a role in lipid transport and metabolism in vivo (47, 69–75), its apolipoprotein-like lipid binding and carrying abilities are likely to be of functional relevance. Its preference for negatively charged membranes might be important in this context. The extracellular localization of apolipoproteins exposes them to more neutral lipids, although the intracellular localization of αS leads to it encountering more negatively charged phospholipids. In fact, under some conditions αS is exposed to very high concentrations (∼20%) of negatively charged cardiolipin in the inner mitochondrial membrane. An increased accumulation of αS at the inner mitochondrial membrane is linked to mitochondrial dysfunction in Parkinson disease (15, 76). Our present data indicate that this disruption could be due, at least in part, to the ability of αS to remove and carry negatively charged lipids. As in prior studies on apolipoproteins (43, 77, 78), the protein-to-lipid ratios in the present experiments were high. Although the exact concentrations in the brain are not known, αS has been estimated to be present at concentrations as high as hundreds of micromolars (79–81). The overall concentration of lipid membranes in a typical cell has been estimated to be in the low millimolar range (82). Thus, the ratios used may not be too different from intracellular conditions.
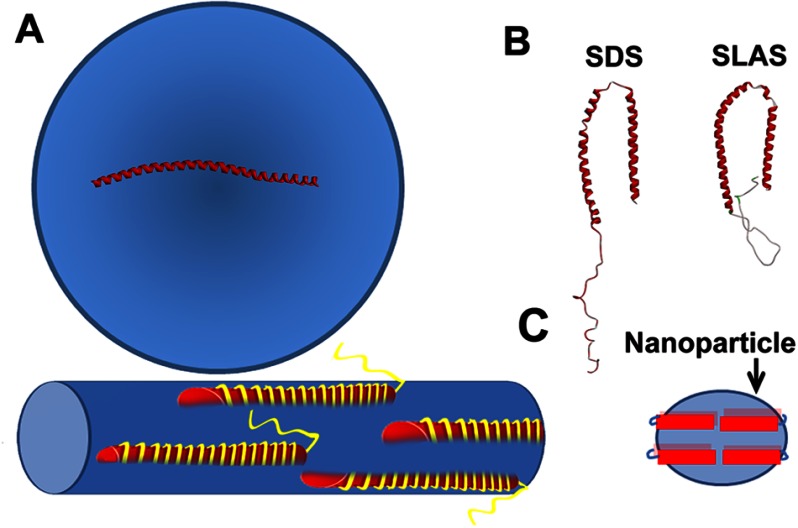
The structural characterization of membrane-bound αS has led to some controversy because many different structures have been invoked. Here, we find that these seemingly contradictory results can be reconciled if one considers the ability of αS to remodel membranes. When αS binds to intact vesicles, it takes up an extended helical structure (Fig. 8A). A related extended helical structure is also present on tubular assemblies (Fig. 8A) (25). In contrast, αS does not take up an extended helical structure on nanoparticles (Fig. 8C). Our EPR data (Fig. 5) are consistent with a broken helical structure in which helix 2 may not be fully ordered. Thus, αS structure strongly depends on the nature of the lipid assembly in which it is incorporated. This plasticity also applies to its association with detergent micelles where slightly different two-helix structures have been reported when αS interacts with SDS (26, 29) and SLAS (28) micelles (Fig. 8B). Although the energy difference between the various αS structures is probably not very large (83), the protein appears to have a general tendency to take up an extended helical structure in the presence of larger lipid assemblies, such as vesicles and tubes. These assemblies are large enough to accommodate an extended helical structure, some 14 nm in length. In contrast, the diameters of typical detergent micelles and nanoparticles are too small to accommodate the extended helix. The present data also have an important practical consideration. When studying the interaction of αS with liposomes, it is essential to verify that no membrane remodeling occurred. The formation of small lipid particles could otherwise lead to the erroneous conclusion that αS takes up a broken helical structure on liposomes.
FIGURE 8.
Summary of α-synuclein structure on different lipid assemblies. A, αS forms an extended helical structure on SUVs and tubes; B, with SDS and SLAS, it forms bent a helical structure. C, αS does not take up a single extended helix on αS-lipid nanoparticles. Red cylinders indicate the shorter helical structure as compared with an extended helix. Multiple αS molecules could be bound on nanoparticles similar to that proposed for apolipoprotein particles (see text).
Several in vitro and in vivo studies have indicated that αS oligomerizes upon exposure to fatty acids (51, 66, 84, 85). The present data provide further insight by finding that αS can form fatty acid-containing complexes that are structurally similar to those formed with lipids. In addition to cylindrical micelles, αS can form small nanoparticles with fatty acids. The DEER distances indicate that αS forms an extended helical structure on the former, but a broken helical structure on the latter. These structural features are analogous to those formed on the corresponding lipid-bound equivalents.
Collectively, our data illustrate the ability of αS to bind to a range of amphiphilic molecules via related mechanisms. This property mandates that extra precautions are needed when handling αS. During the course of our experiments, we found that when αS is run through columns previously exposed to lipid, it can pick up lipid molecules and form complexes in which it has a helical conformation, similar to those described here (data not shown). Accordingly, to verify that columns are free of amphiphilic molecules that could interact with αS in this way, we routinely pre-run our columns with monomeric αS.
αS is widely perceived as an intrinsically disordered monomeric protein, a concept that has been well supported by a number of studies (86–88). However, two recent studies presented evidence that αS might also be able to form small oligomers (trimers and tetramers) with an α-helical conformation (89, 90). The dimensions of these oligomers are similar to those of the nanoparticles generated here. Our data further show that nanoparticles contain helical oligomers (including trimers and tetramers), and these can be generated with both lipids as well as fatty acids. The promiscuous interactions of αS with various amphiphilic molecules raise the possibility that the previously reported oligomers might have been stabilized by amphiphilic molecules. The structural relationship between those oligomers and the nanoparticles studied here remains to be investigated.
Acknowledgment
We thank Sowmya Bekshe Lokappa for help with acquiring dynamic light scattering data.
This work was supported, in whole or in part, by National Institutes of Health Grant 2 R01 M063915 (to R. L.) and by the Intramural Research Program of the NIAMS.
- αS
- α-synuclein
- POPG
- 1-palmitoyl-2-oleoyl-sn-glycero-3-[phospho-RAC-(1-glycerol)]
- POPC
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- POPS
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-l-serine
- SUV
- small unilamellar vesicle
- DEER
- double electron electron resonance
- IANBD
- N,N′-dimethyl-N-(iodoacetyl)-N′-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)ethylenediamine
- Bistris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- rhodamine-DOPE
- 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl).
REFERENCES
- 1. Ibáñez P., Bonnet A. M., Débarges B., Lohmann E., Tison F., Pollak P., Agid Y., Dürr A., Brice A. (2004) Causal relation between α-synuclein gene duplication and familial Parkinson's disease. Lancet 364, 1169–1171 [DOI] [PubMed] [Google Scholar]
- 2. Krüger R., Kuhn W., Müller T., Woitalla D., Graeber M., Kösel S., Przuntek H., Epplen J. T., Schöls L., Riess O. (1998) Ala30Pro mutation in the gene encoding α-synuclein in Parkinson's disease. Nat. Genet. 18, 106–108 [DOI] [PubMed] [Google Scholar]
- 3. Lee M. K., Stirling W., Xu Y., Xu X., Qui D., Mandir A. S., Dawson T. M., Copeland N. G., Jenkins N. A., Price D. L. (2002) Human α-synuclein-harboring familial Parkinson's disease-linked Ala-53 → Thr mutation causes neurodegenerative disease with α-synuclein aggregation in transgenic mice. Proc. Natl. Acad. Sci. U.S.A. 99, 8968–8973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Norris E. H., Giasson B. I., Lee V. M. (2004) α-Synuclein: normal function and role in neurodegenerative diseases. Curr. Top. Dev. Biol. 60, 17–54 [DOI] [PubMed] [Google Scholar]
- 5. Polymeropoulos M. H., Lavedan C., Leroy E., Ide S. E., Dehejia A., Dutra A., Pike B., Root H., Rubenstein J., Boyer R., Stenroos E. S., Chandrasekharappa S., Athanassiadou A., Papapetropoulos T., Johnson W. G., Lazzarini A. M., Duvoisin R. C., Di Iorio G., Golbe L. I., Nussbaum R. L. (1997) Mutation in the α-synuclein gene identified in families with Parkinson's disease. Science 276, 2045–2047 [DOI] [PubMed] [Google Scholar]
- 6. Singleton A. B., Farrer M., Johnson J., Singleton A., Hague S., Kachergus J., Hulihan M., Peuralinna T., Dutra A., Nussbaum R., Lincoln S., Crawley A., Hanson M., Maraganore D., Adler C., Cookson M. R., Muenter M., Baptista M., Miller D., Blancato J., Hardy J., Gwinn-Hardy K. (2003) α-Synuclein locus triplication causes Parkinson's disease. Science 302, 841. [DOI] [PubMed] [Google Scholar]
- 7. Tofaris G. K., Spillantini M. G. (2007) Physiological and pathological properties of α-synuclein. Cell. Mol. Life Sci. 64, 2194–2201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Beyer K. (2007) Mechanistic aspects of Parkinson's disease: α-synuclein and the biomembrane. Cell Biochem. Biophys. 47, 285–299 [DOI] [PubMed] [Google Scholar]
- 9. Bisaglia M., Mammi S., Bubacco L. (2009) Structural insights on physiological functions and pathological effects of α-synuclein. FASEB J. 23, 329–340 [DOI] [PubMed] [Google Scholar]
- 10. Auluck P. K., Caraveo G., Lindquist S. (2010) α-Synuclein: membrane interactions and toxicity in Parkinson's disease. Annu. Rev. Cell Dev. Biol. 26, 211–233 [DOI] [PubMed] [Google Scholar]
- 11. Martin L. J., Pan Y., Price A. C., Sterling W., Copeland N. G., Jenkins N. A., Price D. L., Lee M. K. (2006) Parkinson's disease α-synuclein transgenic mice develop neuronal mitochondrial degeneration and cell death. J. Neurosci. 26, 41–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nakamura K., Nemani V. M., Azarbal F., Skibinski G., Levy J. M., Egami K., Munishkina L., Zhang J., Gardner B., Wakabayashi J., Sesaki H., Cheng Y., Finkbeiner S., Nussbaum R. L., Masliah E., Edwards R. H. (2011) Direct membrane association drives mitochondrial fission by the Parkinson disease-associated protein α-synuclein. J. Biol. Chem. 286, 20710–20726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Song D. D., Shults C. W., Sisk A., Rockenstein E., Masliah E. (2004) Enhanced substantia nigra mitochondrial pathology in human α-synuclein transgenic mice after treatment with MPTP. Exp. Neurol. 186, 158–172 [DOI] [PubMed] [Google Scholar]
- 14. Xie W., Chung K. K. (2012) α-synuclein impairs normal dynamics of mitochondria in cell and animal models of Parkinson's disease. J. Neurochem. 122, 404–414 [DOI] [PubMed] [Google Scholar]
- 15. Nakamura K. (2013) Neurotherapeutics, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fujita Y., Ohama E., Takatama M., Al-Sarraj S., Okamoto K. (2006) Fragmentation of Golgi apparatus of nigral neurons with α-synuclein-positive inclusions in patients with Parkinson's disease. Acta Neuropathol. 112, 261–265 [DOI] [PubMed] [Google Scholar]
- 17. Gosavi N., Lee H. J., Lee J. S., Patel S., Lee S. J. (2002) Golgi fragmentation occurs in the cells with prefibrillar α-synuclein aggregates and precedes the formation of fibrillar inclusion. J. Biol. Chem. 277, 48984–48992 [DOI] [PubMed] [Google Scholar]
- 18. Meredith G. E., Totterdell S., Petroske E., Santa Cruz K., Callison R. C., Jr., Lau Y. S. (2002) Lysosomal malfunction accompanies α-synuclein aggregation in a progressive mouse model of Parkinson's disease. Brain Res. 956, 156–165 [DOI] [PubMed] [Google Scholar]
- 19. Stichel C. C., Zhu X. R., Bader V., Linnartz B., Schmidt S., Lübbert H. (2007) Mono- and double-mutant mouse models of Parkinson's disease display severe mitochondrial damage. Hum. Mol. Genet. 16, 2377–2393 [DOI] [PubMed] [Google Scholar]
- 20. Ferreon A. C., Gambin Y., Lemke E. A., Deniz A. A. (2009) Interplay of α-synuclein binding and conformational switching probed by single-molecule fluorescence. Proc. Natl. Acad. Sci. U.S.A. 106, 5645–5650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Georgieva E. R., Ramlall T. F., Borbat P. P., Freed J. H., Eliezer D. (2008) Membrane-bound α-synuclein forms an extended helix: long-distance pulsed ESR measurements using vesicles, bicelles, and rodlike micelles. J. Am. Chem. Soc. 130, 12856–12857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jao C. C., Der-Sarkissian A., Chen J., Langen R. (2004) Structure of membrane-bound α-synuclein studied by site-directed spin labeling. Proc. Natl. Acad. Sci. U.S.A. 101, 8331–8336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jao C. C., Hegde B. G., Chen J., Haworth I. S., Langen R. (2008) Structure of membrane-bound α-synuclein from site-directed spin labeling and computational refinement. Proc. Natl. Acad. Sci. U.S.A. 105, 19666–19671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Trexler A. J., Rhoades E. (2009) α-Synuclein binds large unilamellar vesicles as an extended helix. Biochemistry 48, 2304–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mizuno N., Varkey J., Kegulian N. C., Hegde B. G., Cheng N., Langen R., Steven A. C. (2012) Remodeling of lipid vesicles into cylindrical micelles by α-synuclein in an extended α-helical conformation. J. Biol. Chem. 287, 29301–29311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Borbat P., Ramlall T. F., Freed J. H., Eliezer D. (2006) Inter-helix distances in lysophospholipid micelle-bound α-synuclein from pulsed ESR measurements. J. Am. Chem. Soc. 128, 10004–10005 [DOI] [PubMed] [Google Scholar]
- 27. Drescher M., van Rooijen B. D., Veldhuis G., Subramaniam V., Huber M. (2010) A stable lipid-induced aggregate of α-synuclein. J. Am. Chem. Soc. 132, 4080–4082 [DOI] [PubMed] [Google Scholar]
- 28. Rao J. N., Jao C. C., Hegde B. G., Langen R., Ulmer T. S. (2010) A combinatorial NMR and EPR approach for evaluating the structural ensemble of partially folded proteins. J. Am. Chem. Soc. 132, 8657–8668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ulmer T. S., Bax A., Cole N. B., Nussbaum R. L. (2005) Structure and dynamics of micelle-bound human α-synuclein. J. Biol. Chem. 280, 9595–9603 [DOI] [PubMed] [Google Scholar]
- 30. Bodner C. R., Dobson C. M., Bax A. (2009) Multiple tight phospholipid-binding modes of α-synuclein revealed by solution NMR spectroscopy. J. Mol. Biol. 390, 775–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Varkey J., Isas J. M., Mizuno N., Jensen M. B., Bhatia V. K., Jao C. C., Petrlova J., Voss J. C., Stamou D. G., Steven A. C., Langen R. (2010) Membrane curvature induction and tubulation are common features of synucleins and apolipoproteins. J. Biol. Chem. 285, 32486–32493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pandey A. P., Haque F., Rochet J. C., Hovis J. S. (2011) α-Synuclein-induced tubule formation in lipid bilayers. J. Phys. Chem. B 115, 5886–5893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Westphal C. H., Chandra S. S. (2013) Monomeric synucleins generate membrane curvature. J. Biol. Chem. 288, 1829–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dewitt D. C., Rhoades E. (2013) α-synuclein can inhibit SNARE-mediated vesicle fusion through direct interactions with lipid bilayers. Biochemistry 52, 2385–2387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Silva R. A., Huang R., Morris J., Fang J., Gracheva E. O., Ren G., Kontush A., Jerome W. G., Rye K. A., Davidson W. S. (2008) Structure of apolipoprotein A-I in spherical high density lipoproteins of different sizes. Proc. Natl. Acad. Sci. U.S.A. 105, 12176–12181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bulankina A. V., Deggerich A., Wenzel D., Mutenda K., Wittmann J. G., Rudolph M. G., Burger K. N., Höning S. (2009) TIP47 functions in the biogenesis of lipid droplets. J. Cell Biol. 185, 641–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Brasaemle D. L. (2007) Thematic review series: adipocyte biology. The perilipin family of structural lipid droplet proteins: stabilization of lipid droplets and control of lipolysis. J. Lipid Res. 48, 2547–2559 [DOI] [PubMed] [Google Scholar]
- 38. Segrest J. P., Jones M. K., De Loof H., Brouillette C. G., Venkatachalapathi Y. V., Anantharamaiah G. M. (1992) The amphipathic helix in the exchangeable apolipoproteins: a review of secondary structure and function. J. Lipid Res. 33, 141–166 [PubMed] [Google Scholar]
- 39. Davidson W. S., Thompson T. B. (2007) The structure of apolipoprotein A-I in high density lipoproteins. J. Biol. Chem. 282, 22249–22253 [DOI] [PubMed] [Google Scholar]
- 40. Peters-Libeu C. A., Newhouse Y., Hall S. C., Witkowska H. E., Weisgraber K. H. (2007) Apolipoprotein E*dipalmitoylphosphatidylcholine particles are ellipsoidal in solution. J. Lipid Res. 48, 1035–1044 [DOI] [PubMed] [Google Scholar]
- 41. Catte A., Patterson J. C., Jones M. K., Jerome W. G., Bashtovyy D., Su Z., Gu F., Chen J., Aliste M. P., Harvey S. C., Li L., Weinstein G., Segrest J. P. (2006) Novel changes in discoidal high density lipoprotein morphology: a molecular dynamics study. Biophys. J. 90, 4345–4360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jones M. K., Gu F., Catte A., Li L., Segrest J. P. (2011) “Sticky” and “promiscuous”, the yin and yang of apolipoprotein A-I termini in discoidal high-density lipoproteins: a combined computational-experimental approach. Biochemistry 50, 2249–2263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gu F., Jones M. K., Chen J., Patterson J. C., Catte A., Jerome W. G., Li L., Segrest J. P. (2010) Structures of discoidal high density lipoproteins: a combined computational-experimental approach. J. Biol. Chem. 285, 4652–4665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cole N. B., Murphy D. D., Grider T., Rueter S., Brasaemle D., Nussbaum R. L. (2002) Lipid droplet binding and oligomerization properties of the Parkinson's disease protein α-synuclein. J. Biol. Chem. 277, 6344–6352 [DOI] [PubMed] [Google Scholar]
- 45. Outeiro T. F., Lindquist S. (2003) Yeast cells provide insight into α-synuclein biology and pathobiology. Science 302, 1772–1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Halliday G. M., Ophof A., Broe M., Jensen P. H., Kettle E., Fedorow H., Cartwright M. I., Griffiths F. M., Shepherd C. E., Double K. L. (2005) α-Synuclein redistributes to neuromelanin lipid in the substantia nigra early in Parkinson's disease. Brain 128, 2654–2664 [DOI] [PubMed] [Google Scholar]
- 47. Golovko M. Y., Barceló-Coblijn G., Castagnet P. I., Austin S., Combs C. K., Murphy E. J. (2009) The role of α-synuclein in brain lipid metabolism: a downstream impact on brain inflammatory response. Mol. Cell. Biochem. 326, 55–66 [DOI] [PubMed] [Google Scholar]
- 48. Golovko M. Y., Faergeman N. J., Cole N. B., Castagnet P. I., Nussbaum R. L., Murphy E. J. (2005) α-Synuclein gene deletion decreases brain palmitate uptake and alters the palmitate metabolism in the absence of α-synuclein palmitate binding. Biochemistry 44, 8251–8259 [DOI] [PubMed] [Google Scholar]
- 49. Assayag K., Yakunin E., Loeb V., Selkoe D. J., Sharon R. (2007) Polyunsaturated fatty acids induce α-synuclein-related pathogenic changes in neuronal cells. Am. J. Pathol. 171, 2000–2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Castagnet P. I., Golovko M. Y., Barceló-Coblijn G. C., Nussbaum R. L., Murphy E. J. (2005) Fatty acid incorporation is decreased in astrocytes cultured from α-synuclein gene-ablated mice. J. Neurochem. 94, 839–849 [DOI] [PubMed] [Google Scholar]
- 51. Sharon R., Goldberg M. S., Bar-Josef I., Betensky R. A., Shen J., Selkoe D. J. (2001) α-Synuclein occurs in lipid-rich high molecular weight complexes, binds fatty acids, and shows homology to the fatty acid-binding proteins. Proc. Natl. Acad. Sci. U.S.A. 98, 9110–9115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ben Gedalya T., Loeb V., Israeli E., Altschuler Y., Selkoe D. J., Sharon R. (2009) α-Synuclein and polyunsaturated fatty acids promote clathrin-mediated endocytosis and synaptic vesicle recycling. Traffic 10, 218–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lücke C., Gantz D. L., Klimtchuk E., Hamilton J. A. (2006) Interactions between fatty acids and α-synuclein. J. Lipid Res. 47, 1714–1724 [DOI] [PubMed] [Google Scholar]
- 54. Der-Sarkissian A., Jao C. C., Chen J., Langen R. (2003) Structural organization of α-synuclein fibrils studied by site-directed spin labeling. J. Biol. Chem. 278, 37530–37535 [DOI] [PubMed] [Google Scholar]
- 55. Shaikh T. R., Gao H., Baxter W. T., Asturias F. J., Boisset N., Leith A., Frank J. (2008) SPIDER image processing for single-particle reconstruction of biological macromolecules from electron micrographs. Nat. Protoc. 3, 1941–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ludtke S. J. (2010) 3-D structures of macromolecules using single-particle analysis in EMAN. Methods Mol. Biol. 673, 157–173 [DOI] [PubMed] [Google Scholar]
- 57. Chiang Y. W., Borbat P. P., Freed J. H. (2005) The determination of pair distance distributions by pulsed ESR using Tikhonov regularization. J. Magn. Reson. 172, 279–295 [DOI] [PubMed] [Google Scholar]
- 58. Jeschke G., Chechik V., Ionita P., Godt A., Zimmermann H., Banham J., Timmel C. R., Hilger D., Jung H. (2006) DeerAnalysis2006 - A Comprehensive Software Package for Analyzing Pulsed ELDOR Data. Appl. Magn. Res. 30, 473–498 [Google Scholar]
- 59. Huang C., Mason J. T. (1978) Geometric packing constraints in egg phosphatidylcholine vesicles. Proc. Natl. Acad. Sci. U.S.A. 75, 308–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mishra V. K., Palgunachari M. N., Datta G., Phillips M. C., Lund-Katz S., Adeyeye S. O., Segrest J. P., Anantharamaiah G. M. (1998) Studies of synthetic peptides of human apolipoprotein A-I containing tandem amphipathic α-helixes. Biochemistry 37, 10313–10324 [DOI] [PubMed] [Google Scholar]
- 61. Wang G. F., Li C., Pielak G. J. (2010) 19F NMR studies of α-synuclein-membrane interactions. Protein Sci. 19, 1686–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Drescher M., Godschalk F., Veldhuis G., van Rooijen B. D., Subramaniam V., Huber M. (2008) Spin-label EPR on α-synuclein reveals differences in the membrane binding affinity of the two antiparallel helices. ChemBioChem 9, 2411–2416 [DOI] [PubMed] [Google Scholar]
- 63. Drescher M., Veldhuis G., van Rooijen B. D., Milikisyants S., Subramaniam V., Huber M. (2008) Antiparallel arrangement of the helices of vesicle-bound α-synuclein. J. Am. Chem. Soc. 130, 7796–7797 [DOI] [PubMed] [Google Scholar]
- 64. Nakamura K., Nemani V. M., Wallender E. K., Kaehlcke K., Ott M., Edwards R. H. (2008) Optical reporters for the conformation of α-synuclein reveal a specific interaction with mitochondria. J. Neurosci. 28, 12305–12317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sharon R., Bar-Joseph I., Mirick G. E., Serhan C. N., Selkoe D. J. (2003) Altered fatty acid composition of dopaminergic neurons expressing α-synuclein and human brains with α-synucleinopathies. J. Biol. Chem. 278, 49874–49881 [DOI] [PubMed] [Google Scholar]
- 66. De Franceschi G., Frare E., Pivato M., Relini A., Penco A., Greggio E., Bubacco L., Fontana A., de Laureto P. P. (2011) Structural and morphological characterization of aggregated species of α-synuclein induced by docosahexaenoic acid. J. Biol. Chem. 286, 22262–22274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Davidson W. S., Jonas A., Clayton D. F., George J. M. (1998) Stabilization of α-synuclein secondary structure upon binding to synthetic membranes. J. Biol. Chem. 273, 9443–9449 [DOI] [PubMed] [Google Scholar]
- 68. Mishra V. K., Palgunachari M. N., Lund-Katz S., Phillips M. C., Segrest J. P., Anantharamaiah G. M. (1995) Effect of the arrangement of tandem repeating units of class A amphipathic α-helixes on lipid interaction. J. Biol. Chem. 270, 1602–1611 [DOI] [PubMed] [Google Scholar]
- 69. Lee S. J., Jeon H., Kandror K. V. (2008) α-Synuclein is localized in a subpopulation of rat synaptic vesicles. Acta Neurobiol. Exp. 68, 509–515 [DOI] [PubMed] [Google Scholar]
- 70. Abeliovich A., Schmitz Y., Fariñas I., Choi-Lundberg D., Ho W. H., Castillo P. E., Shinsky N., Verdugo J. M., Armanini M., Ryan A., Hynes M., Phillips H., Sulzer D., Rosenthal A. (2000) Mice lacking α-synuclein display functional deficits in the nigrostriatal dopamine system. Neuron 25, 239–252 [DOI] [PubMed] [Google Scholar]
- 71. Cabin D. E., Shimazu K., Murphy D., Cole N. B., Gottschalk W., McIlwain K. L., Orrison B., Chen A., Ellis C. E., Paylor R., Lu B., Nussbaum R. L. (2002) Synaptic vesicle depletion correlates with attenuated synaptic responses to prolonged repetitive stimulation in mice lacking α-synuclein. J. Neurosci. 22, 8797–8807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Murphy D. D., Rueter S. M., Trojanowski J. Q., Lee V. M. (2000) Synucleins are developmentally expressed, and α-synuclein regulates the size of the presynaptic vesicular pool in primary hippocampal neurons. J. Neurosci. 20, 3214–3220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Nemani V. M., Lu W., Berge V., Nakamura K., Onoa B., Lee M. K., Chaudhry F. A., Nicoll R. A., Edwards R. H. (2010) Increased expression of α-synuclein reduces neurotransmitter release by inhibiting synaptic vesicle reclustering after endocytosis. Neuron 65, 66–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Willingham S., Outeiro T. F., DeVit M. J., Lindquist S. L., Muchowski P. J. (2003) Yeast genes that enhance the toxicity of a mutant huntingtin fragment or α-synuclein. Science 302, 1769–1772 [DOI] [PubMed] [Google Scholar]
- 75. Withers G. S., George J. M., Banker G. A., Clayton D. F. (1997) Delayed localization of synelfin (synuclein, NACP) to presynaptic terminals in cultured rat hippocampal neurons. Brain Res. Dev. Brain Res. 99, 87–94 [DOI] [PubMed] [Google Scholar]
- 76. Devi L., Raghavendran V., Prabhu B. M., Avadhani N. G., Anandatheerthavarada H. K. (2008) Mitochondrial import and accumulation of α-synuclein impair complex I in human dopaminergic neuronal cultures and Parkinson disease brain. J. Biol. Chem. 283, 9089–9100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Newhouse Y., Peters-Libeu C., Weisgraber K. H. (2005) Crystallization and preliminary x-ray diffraction analysis of apolipoprotein E-containing lipoprotein particles. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 61, 981–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Li L., Chen J., Mishra V. K., Kurtz J. A., Cao D., Klon A. E., Harvey S. C., Anantharamaiah G. M., Segrest J. P. (2004) Double belt structure of discoidal high density lipoproteins: molecular basis for size heterogeneity. J. Mol. Biol. 343, 1293–1311 [DOI] [PubMed] [Google Scholar]
- 79. Uversky V. N., Li J., Bower K., Fink A. L. (2002) Synergistic effects of pesticides and metals on the fibrillation of α-synuclein: implications for Parkinson's disease. Neurotoxicology 23, 527–536 [DOI] [PubMed] [Google Scholar]
- 80. Uversky V. N., Li J., Fink A. L. (2001) Metal-triggered structural transformations, aggregation, and fibrillation of human α-synuclein. A possible molecular NK between Parkinson's disease and heavy metal exposure. J. Biol. Chem. 276, 44284–44296 [DOI] [PubMed] [Google Scholar]
- 81. Iwai A., Masliah E., Yoshimoto M., Ge N., Flanagan L., de Silva H. A., Kittel A., Saitoh T. (1995) The precursor protein of non-Aβ component of Alzheimer's disease amyloid is a presynaptic protein of the central nervous system. Neuron 14, 467–475 [DOI] [PubMed] [Google Scholar]
- 82. Tertov V. V., Orekhov A. N., Rudchenko S. A., Mukhin D. N., Smirnov V. N., Molotkovsky YuG., Bergelson L. D. (1985) Determination of total intracellular lipid content by flow cytofluorometry. Biochem. Biophys. Res. Commun. 128, 1196–1202 [DOI] [PubMed] [Google Scholar]
- 83. Lokappa S. B., Ulmer T. S. (2011) α-Synuclein populates both elongated and broken helix states on small unilamellar vesicles. J. Biol. Chem. 286, 21450–21457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Perrin R. J., Woods W. S., Clayton D. F., George J. M. (2001) Exposure to long chain polyunsaturated fatty acids triggers rapid multimerization of synucleins. J. Biol. Chem. 276, 41958–41962 [DOI] [PubMed] [Google Scholar]
- 85. Sharon R., Bar-Joseph I., Frosch M. P., Walsh D. M., Hamilton J. A., Selkoe D. J. (2003) The formation of highly soluble oligomers of α-synuclein is regulated by fatty acids and enhanced in Parkinson's disease. Neuron 37, 583–595 [DOI] [PubMed] [Google Scholar]
- 86. Fauvet B., Fares M. B., Samuel F., Dikiy I., Tandon A., Eliezer D., Lashuel H. A. (2012) Characterization of semisynthetic and naturally Nα-acetylated α-synuclein in vitro and in intact cells: implications for aggregation and cellular properties of α-synuclein. J. Biol. Chem. 287, 28243–28262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Fauvet B., Mbefo M. K., Fares M. B., Desobry C., Michael S., Ardah M. T., Tsika E., Coune P., Prudent M., Lion N., Eliezer D., Moore D. J., Schneider B., Aebischer P., El-Agnaf O. M., Masliah E., Lashuel H. A. (2012) α-Synuclein in central nervous system and from erythrocytes, mammalian cells, and Escherichia coli exists predominantly as disordered monomer. J. Biol. Chem. 287, 15345–15364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Weinreb P. H., Zhen W., Poon A. W., Conway K. A., Lansbury P. T., Jr. (1996) NACP, a protein implicated in Alzheimer's disease and learning, is natively unfolded. Biochemistry 35, 13709–13715 [DOI] [PubMed] [Google Scholar]
- 89. Bartels T., Choi J. G., Selkoe D. J. (2011) α-Synuclein occurs physiologically as a helically folded tetramer that resists aggregation. Nature 477, 107–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Wang W., Perovic I., Chittuluru J., Kaganovich A., Nguyen L. T., Liao J., Auclair J. R., Johnson D., Landeru A., Simorellis A. K., Ju S., Cookson M. R., Asturias F. J., Agar J. N., Webb B. N., Kang C., Ringe D., Petsko G. A., Pochapsky T. C., Hoang Q. Q. (2011) A soluble α-synuclein construct forms a dynamic tetramer. Proc. Natl. Acad. Sci. U.S.A. 108, 17797–17802 [DOI] [PMC free article] [PubMed] [Google Scholar]