Background: The subunit-specific roles of the CCT subunits in the chaperonin, TRiC, have not been elucidated.
Results: When expressed in E. coli, CCT4 and CCT5 form TRiC-like homo-oligomeric rings.
Conclusion: TRiC does not require all eight CCT subunits to form functional rings.
Significance: The unexpected formation of homo-oligomeric CCT rings provides clues into the assembly of TRiC as a complex.
Keywords: Protein Assembly, Protein Expression, Protein Folding, Protein Self-assembly, Protein-Protein Interactions, CCT, TRiC
Abstract
Chaperonins are a family of chaperones that encapsulate their substrates and assist their folding in an ATP-dependent manner. The ubiquitous eukaryotic chaperonin, TCP-1 ring complex (TRiC), is a hetero-oligomeric complex composed of two rings, each formed from eight different CCT (chaperonin containing TCP-1) subunits. Each CCT subunit may have distinct substrate recognition and ATP hydrolysis properties. We have expressed each human CCT subunit individually in Escherichia coli to investigate whether they form chaperonin-like double ring complexes. CCT4 and CCT5, but not the other six CCT subunits, formed high molecular weight complexes within the E. coli cells that sedimented about 20S in sucrose gradients. When CCT4 and CCT5 were purified, they were both organized as two back-to-back rings of eight subunits each, as seen by negative stain and cryo-electron microscopy. This morphology is consistent with that of the hetero-oligomeric double-ring TRiC purified from bovine testes and HeLa cells. Both CCT4 and CCT5 homo-oligomers hydrolyzed ATP at a rate similar to human TRiC and were active as assayed by luciferase refolding and human γD-crystallin aggregation suppression and refolding. Thus, both CCT4 and CCT5 homo-oligomers have the property of forming 8-fold double rings absent the other subunits, and these complexes carry out chaperonin reactions without other partner subunits.
Introduction
In the cell, molecular chaperones guide misfolded and nascent proteins to their native states or shield misfolded proteins from detrimental aggregation (1, 2). There are two main types of chaperones that bind to newly synthesized proteins and influence their folding in an ATP-dependent manner: the heat shock protein (Hsp) 70 family and the chaperonin or Hsp60 family (3). Hsp70 chaperones bind to proteins immediately after exiting the ribosome and hinder the proteins from forming improper interactions in the cell cytoplasm (3). Substrate folding by chaperonins occurs in a cavity within the chaperonin where the substrate is isolated, in part or in whole, away from the environment of the cell (4, 5).
Chaperonins are composed of two back-to-back rings with seven to nine subunits each (2). Each subunit is divided into three domains: equatorial, which contains the site of ATP binding and subunit-subunit contacts; intermediate, which acts as a hinge region; and apical, which recognizes and binds substrates (6). Chaperonins are classified as group I (found in prokaryotes and in the mitochondria and chloroplasts of eukaryotes) or group II (found in the archaeal and eukaryotic cytosol) (3). The archaeal group II chaperonins consist of rings that have one to three different subunits (7). Due to their decreased complexity compared with the eukaryotic chaperonins, most of the group II chaperonin research has focused on archaeal chaperonins such as Methanococcus maripaludis chaperonin (Mm-Cpn).2 The mechanism of lid closure and substrate release has been elucidated, and high resolution structures of Mm-Cpn are available (8–11).
The eukaryotic group II chaperonin TRiC, TCP-1 (tailless complex polypeptide-1) ring complex, consists of two identical rings, each with eight different CCT (chaperonin containing TCP-1) subunits (12). Through a variety of structural, functional, and cell biology methods, interactions between TRiC and its main substrates, actin and tubulin, have been well characterized (13–19). However, TRiC binding is not limited to actin and tubulin; TRiC binds 9–15% of newly synthesized proteins in [35S]methionine pulse-labeled baby hamster kidney cells (4). Recent research has focused on the arrangement of the eight CCT subunits in TRiC, the binding and hydrolysis of ATP in TRiC, and the recognition of substrates by specific CCT subunits of TRiC.
The arrangement of CCT subunits in TRiC has been a source of controversy (12, 20–22). However, recently, a novel method has established a consistent arrangement for bovine and yeast TRiC (23, 24). This does not explicitly exclude the existence of other CCT subunit arrangements. With the eight CCT subunits expressed from seven different genes, the assembly of TRiC must be regulated to ensure one of each subunit per mature ring (25). In fact, TRiC could contain a different arrangement and ratio of CCT subunits in different tissues or in different stages of embryonic development. Furthermore, there is evidence that TRiC variants containing specific subunits may have different roles (26) and that the CCT subunits may have additional functions in the cell independent of TRiC chaperonin function (27).
It has recently been found that the different CCT subunits of TRiC bind ATP with different affinities (28). For the TRiC chaperonin to close, every subunit does not need to bind ATP, unlike the ATP-binding mechanism in GroEL/ES (2), where every GroEL subunit has to bind an ATP for closure. Only four of the CCT subunits (CCT1, CCT2, CCT4, and CCT5) seemed to bind ATP at physiological concentrations, representing high ATP-affinity subunits (28). Introducing ATP binding-deficient and ATP hydrolysis-deficient mutations into the other subunits (CCT3, CCT6, CCT7, and CCT8) in yeast did not affect yeast growth (28). Combining this information with the recent consistent arrangement of CCT subunits around TRiC (where the high ATP-affinity subunits are located together on one half of the ring) suggests that the high ATP-affinity subunits regulate an asymmetrical power stroke that drives ATP hydrolysis (23, 28).
The apical substrate-recognition domain exhibits the largest divergence of sequence among the CCT subunits, suggesting that this heterogeneity among CCT subunits evolved to recognize and refold a variety of substrates in the eukaryotic cytosol (1, 29, 30). Although only a limited number of substrates have been investigated, binding of non-native state substrates to TRiC may not involve all CCT subunits (13, 14, 30, 31). Many substrates appear to bind across the ring, thus contacting subunits on either side of the ring (32, 33). The apical domains of CCT1 and CCT4 have been implicated in TRiC binding to a construct made up of exon one of the huntingtin protein (34, 35), whereas CCT1 and CCT7 were shown to bind von Hippel-Lindau tumor suppressor protein (pVHL) (30). Although it seems that not all CCT subunits bind a substrate, it is uncertain whether only those specific CCT subunits can bind that particular substrate.
Eukaryotic TRiC has been purified from yeast (22, 23, 36) and from bovine (18, 31, 37) and mouse (20, 32, 38) testes, and more recently from HeLa cells (39). The potential of TRiC as a target of therapeutic agent will benefit from access to human TRiC (39). However, purification of human TRiC from HeLa cells is expensive (39), and recombinant co-expression of all eight CCT subunits has resulted in very low yields (40). To understand how individual human CCT subunits function (both in terms of ATP binding and hydrolysis, and substrate recognition and folding), we have successfully expressed single subunits in Escherichia coli. When we purified two of the CCT proteins, CCT4 and CCT5, to our surprise, they were organized into chaperonin-like homo-oligomeric rings that exhibited chaperonin activities.
EXPERIMENTAL PROCEDURES
CCT Subunit Expression
The pET21b vector was modified (pET21b*) to include a TEV protease cleavage site between the end of the inserted gene and the C-terminal His6 tag. The human CCT genes were synthesized by Genescript (Piscataway, NJ) and inserted into the pET21b* vector using the following restriction sites per gene: CCT1, NdeI and NheI; CCT2, NdeI and BamHI; CCT3, SacII and BamHI; CCT4, NdeI and BamHI; CCT5, NdeI and BamHI; CCT6, NdeI and BamHI; CCT7, SacII and BamHI; CCT8, SacII and BamHI. Each plasmid was confirmed by sequencing (Genewiz). Recombinant single CCT subunits were prepared by plasmid transformation into E. coli BL21 (DE3) RIL cells. The cells were grown in Super Broth to A600 of 5.0 at 37 °C and then shifted to 18 °C and induced with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside. After an overnight induction, cultures were pelleted by centrifugation for 15 min. The cells were resuspended in CCT-A (20 mm HEPES/KOH, pH 7.4, 300 mm NaCl, 10 mm MgCl2, 10% glycerol, 1 mm DTT, 1 mm ATP) with addition of one EDTA-free cOmplete protease inhibitor (Roche Applied Science) per liter of culture.
CCT Subunit Purification
After the addition of 1 mm DTT, 5 mm MgCl2, and 5 μg/ml DNase, the cells were lysed via French Press at a pressure of 16,000 pounds per square inch. The lysate was centrifuged at 20,000 × g for 45 min. The supernatant was removed by pipetting, 0.45-μm filtered, and passed over a nickel-nitrilotriacetic acid column (Pierce). After loading, the column was first washed with 100% CCT-A, then the CCT single subunit was eluted off the column in a linear gradient from 10 to 100% CCT-B (CCT-A but with 250 mm imidazole). The fractions containing the CCT single subunit were combined and concentrated using Vivaspin ultraconcentrators (Satorius Stedim). The protein was diluted with CCT-A down to 25 mm imidazole. After the addition of TEV protease, the CCT single subunit was incubated overnight at 4 °C with gentle rocking.
The His tag-cleaved CCT single subunit was 0.45-μm filtered and applied again to the nickel-nitrilotriacetic acid column, to which it no longer bound. The flowthrough fractions containing the CCT single subunit were combined, further concentrated, and passed over a Superose 6 10/300 GL size exclusion column (GE Healthcare). CCT4 and CCT5 single subunits eluted by CCT-SEC (CCT-A but with 5% glycerol and no ATP) around 12–14.5 ml off the size exclusion column, consistent with that of a 1-MDa complex. These fractions were pooled and concentrated, and the protein concentration was measured using the BCA assay (Pierce) with BSA as the standard. The purified CCT subunit band was cut out, trypsin-digested, and LC-MS/MS analysis was run on a Qstar mass spectrometer by Biopolymer and Proteomics Core Facility at the Koch Institute (Cambridge, MA). Peptides were identified by searching for hits in the Mascot database. N-terminal sequencing was conducted by Tufts University Core Facility (Boston, MA).
Human TRiC and Mm-Cpn Purification
The human TRiC control sample was purified as described at length by Knee et al. (39). Mm-Cpn was purified as described by Knee et al. with the slight variation that the protein was grown up in Super Broth (41).
Sucrose Gradient Sedimentation
Isokinetic 5–40% sucrose (in CCT-SEC buffer) gradients were prepared via the gradient master (BioComp Instruments) and ultracentrifuged at 4 °C using a SW50 rotor for 18 h at 28,000 rpm (Beckman). Twenty-four fractions were collected using a gradient fractionator (BioComp Instruments).
SDS-PAGE and Immunoblotting
Proteins were separated by SDS-PAGE (14% or 10% gel) at 165 V for 1 h after boiling in reducing buffer (60 mm Tris, pH 6.8, 2% SDS, 5% β-mercaptoethanol, 10% glycerol, bromphenol blue for color) for 5 min. The gels were stained with Coomassie Blue or Krypton (Pierce). Transfer was conducted for 1.5 h at 300 mA in transfer buffer (10% methanol, 25 mm Tris, 192 mm glycine) onto 0.45-μm polyvinylidene difluoride (PVDF) membranes (Millipore). The primary antibodies used for CCT1–8 were from Santa Cruz Biotechnology: CCT1, sc-53454; CCT2, sc-28556; CCT3, sc-33145; CCT4, sc-48865; CCT5, sc-13886; CCT6, sc-100958; CCT7, sc-130441; and CCT8, sc-13891. The secondary antibodies were alkaline phosphatase-conjugated (Millipore), and the membranes were visualized using the alkaline phosphatase-conjugate substrate kit (Bio-Rad).
Electron Microscopy
Copper grids with Formvar carbon coating (400 mesh; Ted Pella) were glow-discharged for 20 s, and 5 μl of purified chaperonin was placed on the grids for 5 min. Excess sample on the grids was blotted off using filter paper, and the grids were floated onto a drop of filtered 1.5% uranyl acetate (Sigma-Aldrich) for 45 s. Grids were visualized under a JEOL 1200 SX transmission electron microscope, and digital micrographs were taken using an AMT 16000S camera system.
Cryo-electron Microscopy
For the apo state, 0.5 mg/ml CCT4 or CCT5 was used for cryo-EM sample preparation. For the closed state, 0.35 mg/ml CCT5 was incubated for 30 min at 37 °C with 5 mm Al(NO3)3, 30 mm NaF, and 1 mm ATP. A volume of 2.5 μl was applied onto a plasma-cleaned grid (R1.2/1.3; Quantifoil Micro Tools) and plunge-frozen into liquid ethane operated automatically by Vitrobot Mark III device (FEI). Images for both states were taken at 71,361× detector magnification on a JEM 2200FS microscope (JEOL) with Omega in-column energy filter (energy slit, 20 eV) and recorded on a Gatan 4K×4K CCD with a dose of 20 e/A2. A total of 54 (apo state CCT5) and 160 (ATP-AlFx state CCT5) CCD frames were taken with defocuses ranging from 2 to 3.5 μm.
A total of 5,000 particles (apo) or 6,307 particles (ATP-AlFx state) were boxed out semiautomatically by e2boxer.py (EMAN2) (42). The reference-free two-dimensional class averages were generated by using e2refine2d.py (EMAN2) to group 5,000 particles in each dataset into 100 classes. For three-dimensional reconstruction, all of the 6,307 particles in ATP-AlFx state were fit using the automatic CTF fitting program fitctf.py (EMAN), and then manually examined and adjusted using the EMAN program ctfit.py. A C8 symmetry-imposed multiple model refinement approach was used initially to separate conformationally heterogeneous particles into different groups using multirefine as described previously (43, 44). The initial model was generated from the dataset by startcsym.py (EMAN). Three initial models for refinement were generated by adding different random noise at a level of σ = 0.15 to the initial model, and the dataset is divided into three subclasses. After iterative refinements, the refined models converged, and one refined model was associated with 2,974 (∼47%) particles. This subset of particle images was reprocessed from scratch without imposing any symmetry.
An initial asymmetric model was generated for this subset of particle images and no symmetry was applied during the refinement process. When the refined map converged, a Gaussian low pass filter with cut-off frequency of 0.04 Å−1 (25 Å) was applied to interpret the low resolution features of the map. A rotational correlation plot was used to assess the symmetry of the map, and a strong 8-fold symmetry was observed.
After C8 symmetry was observed in the map, it was imposed during another round of three-dimensional refinement (EMAN) providing a C8-symmetrized initial model. After refinement, the final resolution was measured to be 22 Å (0.143 criterion (45)). To further validate the map, the dataset of 2,974 particles was divided into two independent sub-datasets, with 1,487 particles each. Two phase-randomized symmetry-free models were used as initial models for these two datasets. The phase-randomized models were generated as follows. The symmetry-free refined model was subject to a phase randomization process by using a low pass phase randomization filter in EMAN2 to randomize the Fourier phases below 33 Å. Fourier shell correlation (FSC) between these two template models demonstrated the expected zero mean correlation beyond 33 Å resolution. The choice of 33 Å was based on an expectation that the final resolution would be better than this. The two independent datasets were then refined independently from these two starting models. This “gold standard” resolution (46) was measured as 22 Å based on the Fourier shell correlation = 0.143 cut-off criterion (45).
Thermal Denaturation by CD
The secondary structure of the chaperonins was assayed at 100 μg/ml protein in filtered and degassed 10 mm Tris, 20 mm KCl. For protein in the closed state, 1 mm ATPγS was added to the buffer. The temperature was raised from 25 to 100 °C in 5 °C steps and equilibrated for 5 min at each temperature. Far-UV CD spectra from 260 to 195 nm were obtained for each chaperonin, and the buffer using an AVIV model 202 CD spectrophotometer at each temperature. The buffer was subtracted at each temperature, and the signal at 227 nm was selected for thermal denaturation analysis. Transition midpoints were determined using a two-state unfolding fit in Prism (GraphPad).
ATP Hydrolysis Assay
The ATP hydrolysis assays were performed as described by Reissmann et al. (47). Briefly, 250 nm chaperonin was incubated for 5 min at 30 °C in 1.25× reaction mix. At time zero, ATP was added to a final concentration of 2 mm with [α-32P]ATP (PerkinElmer Life Sciences) at a concentration of 0.002 μCi/μl, and the reaction proceeded at 30 °C. At each indicated point, 2 μl of the sample was taken out of the reaction and spotted onto a PEI-cellulose TLC plate (Macherey-Nagel). The plates were run using a mobile phase of 1 m LiCl and 2 m formic acid, air-dried, and exposed to a phosphor screen. After 24 h, the screen was scanned by a Typhoon imager (GE Healthcare), and the amount of [α-32P]ADP was quantified using ImageJ.
Luciferase and Human γD-Crystallin (HγD-Crys) Refolding Assays
The luciferase refolding assay was performed as described by Knee et al. (39). The HγD-Crys aggregation suppression assay is described in detail by Acosta-Sampson and King (48) and Knee et al. (41) and was modified in this study by use of a decreased chaperonin concentration of 145 nm.
RESULTS
Expression and Purification of CCT Subunits
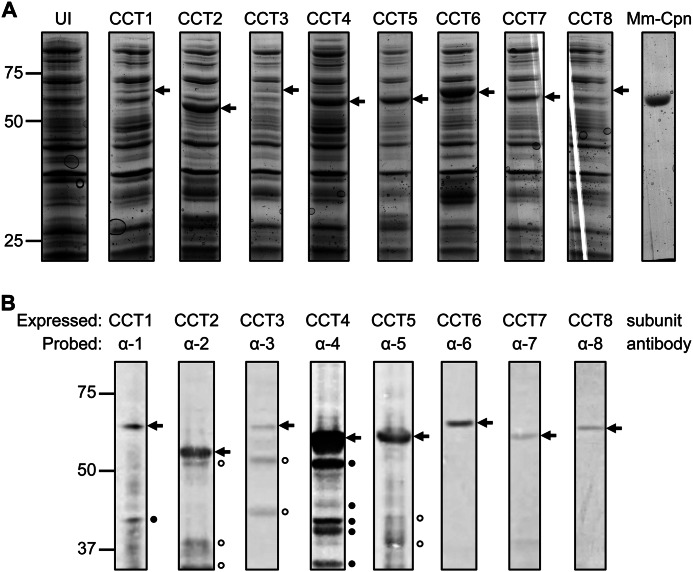
The CCT subunits were successfully cloned into a modified pET21b vector that included a TEV protease site before the C-terminal His6 tag. Due to the variations in DNA sequences of the CCT subunits, two of four different restriction enzymes were used to insert each CCT subunit DNA sequence into the vector. Of four different E. coli expression lines (BL21 (DE3) Gold, BL21 (DE3) pLysS, Rosetta (DE3) pLysS, BL21 (DE3) RIL), BL21 (DE3) RIL was found to express full-length CCT subunit protein to the highest level. The CCT subunit sequences were nonoptimized human sequences and the BL21 (DE3) RIL cell line is enhanced for expressing human sequences in E. coli. Some of the CCT subunits accumulated at much higher levels than other CCT subunits as seen by the cell lysates electrophoresed through 10% SDS-PAGE and Coomassie-stained (Fig. 1A). The expression levels were verified by immunoblots of the cell lysate proteins probed by each of the respective CCT antibodies (Fig. 1B).
FIGURE 1.
Expression of human CCT subunits in BL21 (DE3) RIL E. coli cells. A, cells expressing each of the eight subunits (CCT1–8), an Mm-Cpn control (Mm-Cpn), and an uninduced control (UI) were electrophoresed through 10% SDS-PAGE and stained with Coomassie Blue. Arrows indicate the major overexpressed band in each lane that has an induced plasmid. B, the same samples in A were separated by 10% SDS-PAGE, transferred, and probed with each of the eight CCT antibodies. We saw no cross-reaction of each of the CCT antibodies with any other CCT subunit. Arrows mark the antigenic band. Antibodies to CCT2, CCT3, CCT5, and CCT8 were polyclonal, whereas antibodies to CCT1, CCT4, CCT6, and CCT7 were monoclonal. Filled circles designate bands that are CCT subunits whereas open circles designate bands that may not be CCT subunits.
For a number of the subunits, lower molecular species were clearly visible in the immunoblots. Four of these antibodies are monoclonal, CCT1, CCT4, CCT6, and CCT7, indicating that the lower molecular weight species are CCT fragments produced by degradation. These fragment levels were not sensitive to time of incubation of the lysates, suggesting that proteolysis was happening within the expressing cells. Variation of temperature of cell growth, conditions of induction, and treatment of the lysed cells, did not have a significant effect on the differences in expression among the eight subunits. A 53-kDa fragment was present in the CCT4 expression. Mass spectrometry and N-terminal sequencing identified the fragment as lacking the first 60 amino acids of CCT4. We considered that fragment might be the result of late translation initiation, but mutations of the suspected methionine did not significantly decrease the level of the fragment (data not shown). Therefore, the fragment might be the result of a specific protease acting within the cell.
In our attempts to express subunits without a His tag, we had difficulty separating the CCT subunits from the endogenous GroEL/S complexes. Although we cannot rule out differences in transcription or translation, we suspect that the differences in subunit accumulation may reflect whether or not the translated CCT chains are able to utilize the E. coli chaperone/chaperonin apparatus to assist their folding and assembly.
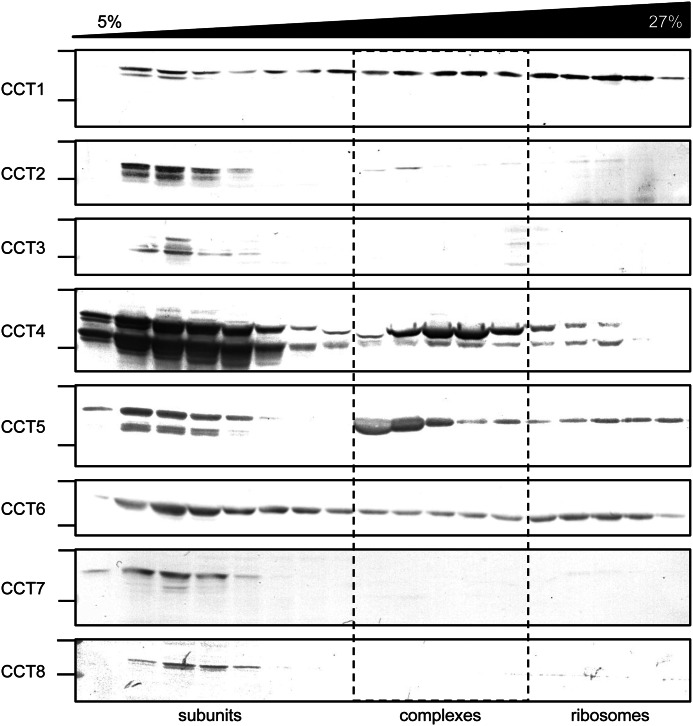
To test whether the expressed CCT subunits formed higher order complexes, lysates of cells expressing the CCT subunits were fractionated on sucrose gradients. Use of the lysates rather than purified proteins allowed us to verify that these complexes are forming within the E. coli cells and that that the C-terminal His tag did not impede subunit assembly. The sedimentation of the CCT subunits in the sucrose gradients was compared via immunoblots due to the low expression of some of the subunits and the abundance of E. coli proteins present in the lysate (Fig. 2). Whereas CCT4 and CCT5 formed higher order complexes of similar sedimentation to human TRiC and Mm-Cpn, the other CCT subunits did not. CCT2 was the only other CCT subunit possibly forming very low levels of a ∼20S complex; however, we did not observe rings in these samples by electron microscopy (EM). CCT1 and CCT6 subunits were found throughout the sucrose gradients; these may represent aggregated states or association with ribosome subunits. Mass spectrometry and EM of purified CCT1 showed that CCT1 associated with ribosomes, consistent with its position in the sucrose gradients (data not shown). The rest of the subunits, CCT2, CCT3, CCT7, and CCT8, were recovered as slowly sedimenting species.
FIGURE 2.
Sucrose ultracentrifugation gradients of CCT subunits. BL21 (DE3) RIL cells expressing each of the eight subunits (CCT1–8) were lysed, fractionated through sucrose gradients, separated by 10% SDS-PAGE, transferred, and probed with their respective antibodies. Fractions from the top (5%) through two-thirds (27%) of the sucrose gradients are shown. For each CCT subunit, on the left, the immunoblot region between 75 kDa (top line) and 50 kDa (bottom line) is shown. Some CCT subunits were sedimenting as soluble subunits, others as complexes, and some were sticking to ribosomes.
CCT4 and CCT5 were chosen for further purification due to their assembled state and high expression level. The purification of the CCT subunits followed a standard His6-tagged protein purification. The cells were lysed with a French Press; this was found to be most effective in maximizing CCT subunit protein yield, compared with sonication or chemical lysis. Supernatant/pellet separation of the lysed species did show that a fraction of the CCT protein ended up in the pellet. We did not investigate the nature of these chains but believe that they resided in inclusion bodies.
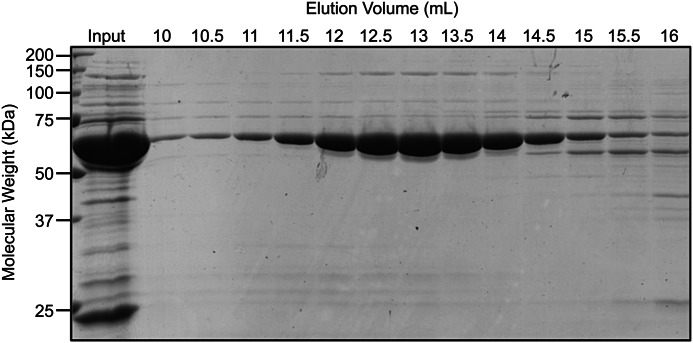
The lysates were passed over a nickel-nitrilotriacetic acid column and eluted off with a gradient of imidazole concentrations from 25 to 250 mm. The protein was concentrated, diluted to a lower imidazole concentration (25 mm) and incubated with TEV protease overnight. The TEV-cleaved CCT subunit protein was passed again over the nickel-nitrilotriacetic acid column to which it no longer bound. The resulting protein fractions were concentrated and passed over a size exclusion column. The major protein peak eluted between 12 and 14.5 ml (CCT5 shown in Fig. 3). This elution was consistent with a 1-MDa complex and corresponding to the elution volume of both human TRiC and Mm-Cpn (39, 41). The symmetry of the distribution and absence of a trailing edge indicate that the complexes were not dissociating into subunits under the conditions of the fractionation. Sucrose gradients on purified protein confirmed the existence of a complex with no dissociated monomers (data not shown).
FIGURE 3.
CCT5 purified by size exclusion chromatography as a 1-MDa complex. The input (Input) and various elution volumes (10–16 ml) were electrophoresed through 14% SDS-PAGE and stained with Coomassie Blue. CCT5 appeared as a band ∼60 kDa in size eluted in volumes of 12–14.5 ml consistent with a 1-MDa complex.
The final yield was ∼2 mg/liter of lysate for CCT4 and 5 mg/liter of lysate for CCT5. Interestingly, CCT5 was concentrated up to 10 mg/ml without issues whereas CCT4 tended to precipitate above 1.5 mg/ml. The purified CCT4 and CCT5 subunits were verified by immunoblots and mass spectrometry (data not shown). From the mass spectrometry, there was no detectable GroEL/ES in the preparations of purified His tag-cleaved CCT single subunits complexes.
Structural Characterization of the CCT4 and CCT5 Homo-oligomers
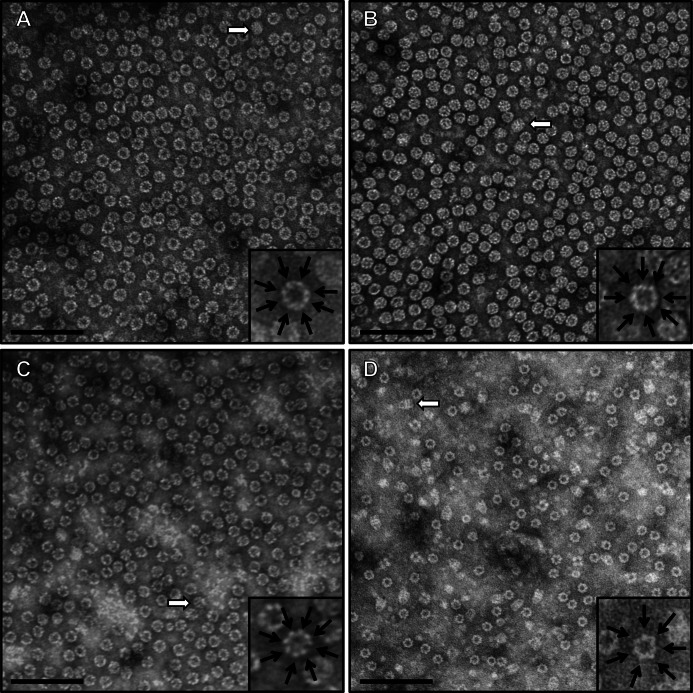
When viewed by negative stain EM, both CCT4 and CCT5 homo-oligomers formed rings (Fig. 4, A and B). These rings were ∼160 Å wide and 180 Å tall, consistent with those of other group II chaperonins, such as human TRiC and Mm-Cpn (39, 41). The rings seen for CCT4 and CCT5 homo-oligomers were similar to those of human TRiC (Fig. 4C) but distinctly different from those of GroEL/ES from E. coli (Fig. 4D). The difference was seen in not only the top views of the rings (eight in CCT4/CCT5/human TRiC and seven in GroEL, shown as insets), but also in the side views (as shown with open arrows). The CCT4 homo-oligomer structure looked hollow in the center, but CCT5 seemed to contain extra density. This extra density was present throughout the three steps of purification and persisted with or without the ATP presence (data not shown).
FIGURE 4.
Negative stain transmission electron microscopy of purified CCT4 and CCT5 homo-oligomers showed morphology similar to human TRiC and distinct from GroEL/ES. The morphology of CCT4 (A) and CCT5 (B) was consistent with that of group II chaperonins. The complexes were ∼160 Å in diameter and ∼180 Å in height and shown here at ×200,000 magnification. The morphology of human TRiC (C) and GroEL/ES (D) is shown as a control to the CCT4/CCT5 morphology. These GroEL/ES complexes can be distinguished due to their subunit per ring differences (seven for GroEL, eight for TRiC; shown as insets) and unique side view morphology (shown by open arrows). Scale bars, 100 nm.
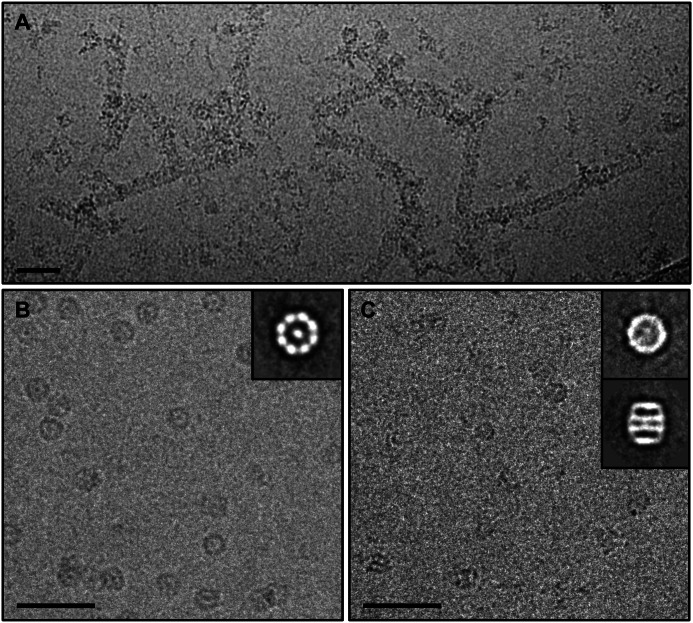
Additionally, we observed end-to-end homo-oligomer polymers in EM for CCT4 but not CCT5 homo-oligomers. Trent et al. reported similar filaments formed by the chaperonin of the archaea Sulfolobus shibatae and postulated cytoskeletal or regulatory roles for such polymers (49). Whereas these were present at low level in negative stain EM, they were common when CCT4 was viewed in cryo-EM (Fig. 5A). The presence of these polymers impeded structural study of CCT4 by cryo-EM.
FIGURE 5.
Raw cryo-EM images of CCT5 homo-oligomers and two-dimensional class averages indicated two rings of eight subunits per ring. A, raw cryo-EM image of CCT4 homo-oligomers end-on-end polymers. B, raw cryo-EM image of the apo/open state CCT5 homo-oligomer with an inset of the two-dimensional classification in the top view, showing eight subunits per ring. C, raw cryo-EM image of the ATP-AlFx/closed state CCT5 homo-oligomer with an inset of the two-dimensional classification of the top and side view, showing that the CCT5 chaperonin complex consisted of two back-to-back rings with eight subunits per ring. Scale bars, 50 nm.
To further understand the quaternary structure of CCT5 homo-oligomers, we performed cryo-EM of this complex in both the apo and ATP-AlFx states. In the apo state (Fig. 5B), a reference-free two-dimensional class average approach was taken to demonstrate that the top view class average (inset) displayed eight density blobs without imposing any assumption on the symmetry in the analysis. The apo state resulted in preferred orientation of end-on views, which has been encountered in the cryo-EM studies of TRiC or Mm-Cpn in their apo states (11, 44). When incubated with ATP-AlFx, similar features were also seen in the raw images and two orthogonal views of two-dimensional class averages of CCT5 (Fig. 5C and insets). However, the density became more continuous from the end-on view of the two-dimensional class average, which was also observed with TRiC/CCT or Mm-Cpn in ATP-AlFx states compared with their apo states (11, 44). This suggests that CCT5 homo-oligomer is capable of hydrolyzing ATP and closing the complex. To carry out the three-dimensional reconstruction of CCT5 homo-oligomers, we used the ATP-AlFx condition because it allowed us to obtain sufficient number of particle images in different orientations needed for a three-dimensional structure determination.
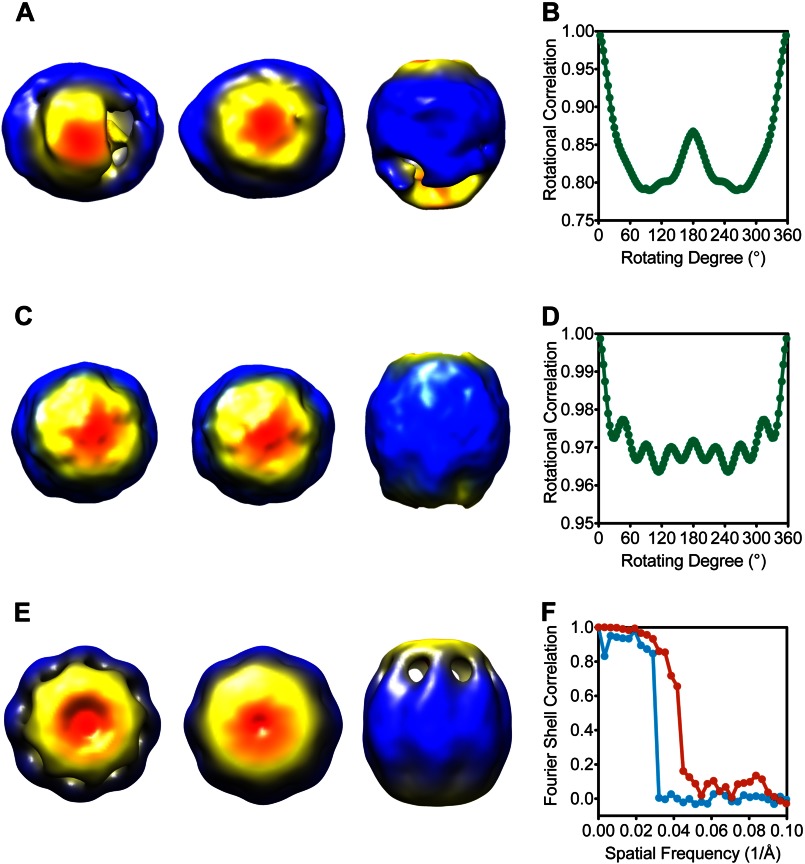
In the image reconstruction of CCT5 particle images, we noted significant conformational heterogeneity, not unusual for reconstructions of group II chaperonins. However ∼47% of particles could be sorted out computationally to have homologous conformation. This data subset was reprocessed from scratch with a symmetry-free initial model (Fig. 6, A and B). A symmetry-free reconstruction of this subset of particle images (Fig. 6C) clearly showed that the CCT5 complex had similar quaternary structure as TRiC or Mm-Cpn (10, 12, 38).
FIGURE 6.
Cryo-EM reconstructions of CCT5 homo-oligomers suggested TRiC-like structures. A, symmetry-free initial template of CCT5 homo-oligomers in the ATP-AlFx state is shown from three different views (top, bottom, and side). B, rotational correlation analysis of the symmetry-free reconstructed map along the central axis from 0° to 360° shows only C2 symmetry. C, three different views show the symmetry-free three-dimensional reconstruction map of ATP-AlFx state of CCT5 homo-oligomers. D, rotational correlation analysis of the symmetry-free reconstructed map along the central axis from 0° to 360° shows eight peaks with ∼45° spacing, suggesting 8-fold symmetry of the reconstruction. E, three-dimensional reconstruction of CCT5 homo-oligomers in the ATP-AlFx/closed state with C8 symmetry imposed shows a TRiC-like structure in three views. F, resolution measured at 0.143 cutoff in the Fourier shell correlation between the two initial models with random phase was ∼33 Å (blue) whereas between the two C8 symmetry imposed final maps was ∼22 Å (red). The maps shown in A, C, and E are radially colored.
A rotational correlation analysis was carried out for the symmetry-free reconstructed map and eight peaks were observed with ∼45° spacing when the structure was rotated along the central axis from 0° to 360°, indicating the presence of 8-fold symmetry in the complex along the central axis (Fig. 6D). With C8 symmetry imposed, the reconstructed map (Fig. 6E) further improved to 22 Å resolution based on phase randomized resolution test with two independent datasets (Fig. 6F). Interestingly, the CCT5 complex had a more elongated conformation along the symmetry axis compared with TRiC and the two rings were not exactly identical (i.e. lack of 2-fold symmetry). One possibility is that the heterogeneous subunits of TRiC have stronger intra-ring interactions that could be conducive to a more compact closed state.
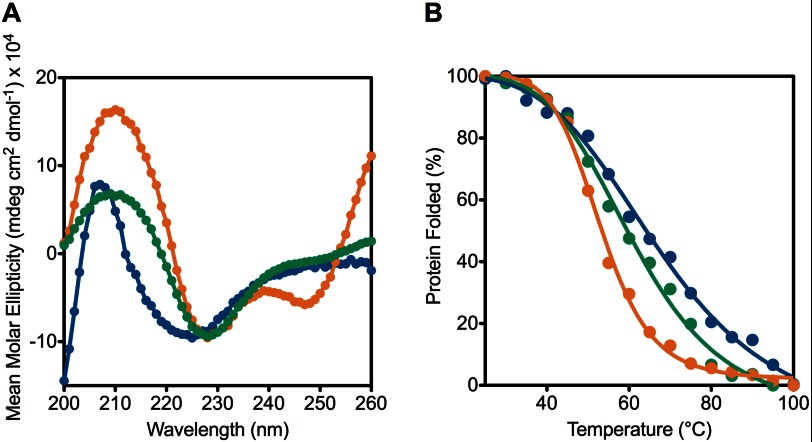
To investigate this further, we performed thermal denaturation studies by circular dichroism (CD) on human TRiC, CCT4, and CCT5. All three chaperonins had similar far UV CD scans with human TRiC having a minimum at 225 nm whereas CCT4 and CCT5 had minima at 228 nm (Fig. 7A). Occasionally, CCT4 and CCT5 had an unusual minimum at 247 nm, attributed to ATP or ADP self-association (50). This signal was reduced during purification of CCT5; however, CCT4 samples retained this signal, but the stoichiometry of the nucleotide was <1% of the protein chains (50).
FIGURE 7.
Human TRiC is more stable than CCT4 and CCT5 homo-oligomers by thermal denaturation using CD. A, CD scans of CCT4 (orange), CCT5 (green), and human TRiC (blue) are shown from 260 to 200 nm. CCT4 and CCT5 had minima at 228 nm whereas human TRiC had a minimum at 225 nm. B, CD signal at 226 nm was monitored while CCT4 (orange), CCT5 (green), and human TRiC (blue) were thermally denatured from 25 to 100 °C. The denaturation midpoint of CCT4 was 53 °C and CCT5 was 60 °C whereas that of human TRiC was 68 °C, suggesting that CCT4 and CCT5 complexes were less stable than that of human TRiC. CCT4 had the most cooperative transition, followed by CCT5 and then human TRiC consistent with the hetero-oligomeric wild type nature of human TRiC. Adding ATP slightly decreased the denaturation midpoint of the chaperonins, primarily due to the increase in cooperativity.
Both CCT4 and CCT5 melted at lower temperature than human TRiC (53 °C for CCT4; 60 °C for CCT5; 68 °C for human TRiC) (Fig. 7B). This suggested that subunit-subunit interactions within TRiC stabilized its secondary structure more than the structures of CCT5 and CCT4. The transition was much more cooperative for CCT4 and CCT5 than for human TRiC. Whereas no aggregation was visible upon heating with any of these chaperonins, the denaturation process was not reversible, and rings were not observed by EM after sample denaturation.
When ATP was added in the form of ATPγS, we saw a very small decrease in melting temperature (2 °C for CCT5 and 5 °C for human TRiC and CCT4), primarily attributed to increased cooperativity of melting (data not shown). Although addition of ATP should not change the actual secondary structure, and therefore melting temperature, our results are consistent with the loss of flexibility in the apical domains of the closed structure, resulting in a more symmetric, uniform structure.
Functional Characterization of the CCT4 and CCT5 Homo-oligomers
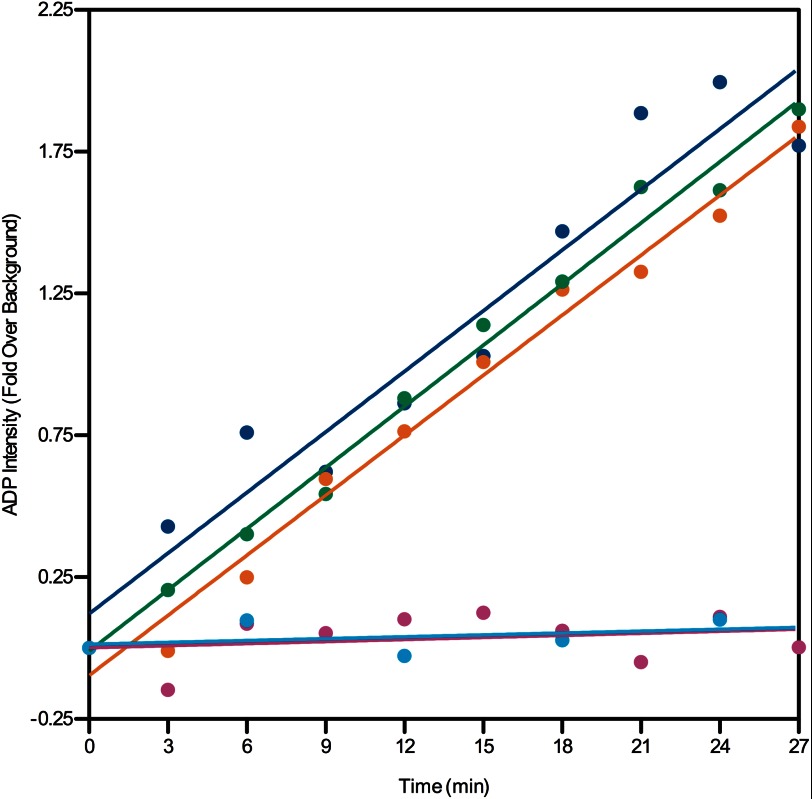
The hydrolysis of ATP is an important functional characteristic of the chaperonins. The ATP hydrolysis properties of CCT4 and CCT5 were assayed using radioactive ATP. Surprisingly, CCT4 and CCT5 hydrolyzed ATP at a rate comparable with human TRiC (Fig. 8).
FIGURE 8.
CCT4 and CCT5 homo-oligomers hydrolyze ATP at a similar rate to human TRiC. The generation of [α-32P]ADP was quantified over time for 250 nm CCT4 (orange), CCT5 (green), human TRiC (blue), and BSA (magenta), and a water control (cyan). CCT4 and CCT5 show very similar ATP hydrolysis properties as human TRiC.
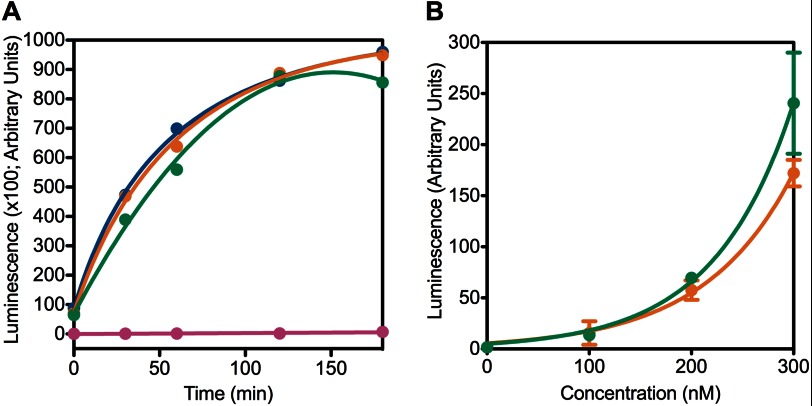
CCT4 and CCT5 homo-oligomers were assayed for refolding of luciferase (51), which we previously used to test the substrate refolding activity of human TRiC (39). In the experiment, unfolded luciferase was diluted into buffer with chaperonin (51). Addition of luciferin and subsequent monitoring of luminescence production assayed the presence of refolded luciferase in the mixture. At a concentration of 400 nm, human TRiC, and CCT4, and CCT5 homo-oligomers refolded luciferase to about the same level, leveling off after 2 h (Fig. 9A). When the concentration of chaperonin was varied, CCT4 and CCT5 homo-oligomers showed higher activity at higher concentrations, as evidenced by higher luciferase activity (Fig. 9B). Whereas the range of chaperonin concentrations (measured as a 16-mer) varied from 0 to 300 nm, luciferase concentration was constant at ∼10 nm. The luciferase refolding activity of CCT4 and CCT5 homo-oligomers over this range indicated that the folding could be attributed directly to these two chaperonins and not to any buffer component.
FIGURE 9.
CCT4 and CCT5 homo-oligomers were active in refolding luciferase. A, CCT4 (orange), CCT5 (green), and human TRiC (blue) at 400 nm were active in refolding luciferase compared with the BSA (magenta) control for >2 h at room temperature. B, when the chaperonin concentration was varied, CCT4 (orange) and CCT5 (green) homo-oligomers are more active in refolding luciferase with increasing concentration. The luciferase concentration was constant at 10 nm. For this experiment, n = 3, and the error bars shown are S.E.
Whereas luciferase is a model substrate for the chaperonins, a more stringent human substrate is HγD-Crys (39, 41). HγD-Crys is found in the lens of the eye, and its folding and unfolding have been studied extensively (52, 53). Whereas some chaperones, such as the major lens chaperone, α-crystallin, can only suppress HγD-Crys aggregation, group II chaperonins have been shown to actively suppress and refold HγD-Crys molecules in vitro (39, 41, 48, 54). Knee et al. found that this suppression and refolding ability was strictly ATP-dependent (41). In this assay, when unfolded HγD-Crys was diluted from high concentration of guanidinium hydrochloride into buffer at concentrations of 50 μg/ml, partially folded intermediates partitioned between productive refolding and off-pathway aggregation. This aggregation was monitored by sample turbidity (A350).
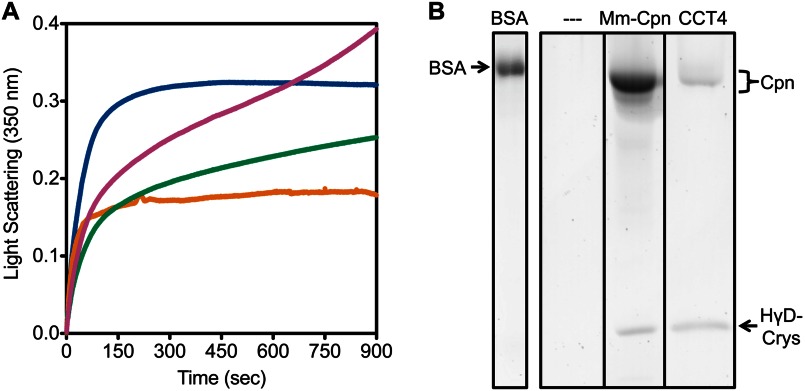
Under the conditions of this assay, containing residual 0.55 m guanidinium hydrochloride, both CCT4 and CCT5 homo-oligomers exhibited slow polymerization by themselves (data not shown). Therefore, the concentration of the chaperonin was decreased (16-fold) to 145 nm, compared with the 2.3 μm used in previous studies, but the concentration of HγD-Crys was unchanged (41). When CCT4 or CCT5 homo-oligomers were added to the reaction, aggregation of partially folded HγD-Crys was significantly suppressed (Fig. 10A). Whereas turbidity in the HγD-Crys aggregation suppression by CCT4 homo-oligomer reached a plateau, HγD-Crys aggregation suppression by CCT5 homo-oligomer showed continuing increase in turbidity. We attributed this to CCT5 homo-oligomer polymerization. Previous studies showed that Mm-Cpn and human TRiC suppress HγD-Crys aggregation by 60–80% (39, 41). At the significantly reduced concentrations used in this study, both CCT4 and CCT5 homo-oligomers still suppressed HγD-Crys aggregation by ∼50%. When CCT5 homo-oligomer was assayed without ATP, there was less HγD-Crys aggregation suppression, and the CCT5 homo-oligomer polymerization was even more distinct. The initial curve of HγD-Crys aggregation suppression by CCT5 homo-oligomer without ATP was consistent with that seen for HγD-Crys aggregation suppression by Mm-Cpn without ATP (41). At the conclusion of the assay, the samples were filtered to remove large aggregates and electrophoresed through 14% SDS-PAGE. A 20-kDa band consistent with HγD-Crys was seen in the sample with CCT4 and CCT5 homo-oligomer, but not in the HγD-Crys alone or BSA control samples, indicating that a fraction of the partially folded HγD-Crys was refolded to native-like state specifically by the chaperonins (Fig. 10B; only CCT4 and controls are shown for clarity). The activity of human TRiC to refold HγD-Crys is reported in Knee et al. and is consistent in levels seen here with CCT4 and CCT5 (39).
FIGURE 10.
CCT4 and CCT5 homo-oligomers suppressed aggregation of partially folded HγD-Crys and promoted HγD-Crys native-like state refolding. A, aggregation of HγD-Crys (blue) was suppressed by the addition of human CCT4 (orange) or CCT5 (green) by ∼50% after 15 min at 37 °C. CCT5 tended to self-polymerize showing a higher turbidity. Without ATP (magenta), HγD-Crys aggregation suppression by CCT5 was decreased, and the CCT5 polymerization was seen more clearly. Curves shown are representative; the assay was repeated three to five times for each chaperonin. B, after filtering, the samples of HγD-Crys with or without chaperonins were separated by 14% SDS-PAGE and stained by Krypton. Without chaperonin (- - -) and with BSA, no HγD-Crys band was present, but with Mm-Cpn and CCT4, HγD-Crys was seen, indicating that it was refolded to native-like state.
DISCUSSION
To our surprise, CCT4 and CCT5 subunits purified out of E. coli formed homo-oligomeric chaperonin-like complexes. These novel complexes not only possessed morphology consistent with human TRiC, but were also active in refolding two different substrates. Because these homo-oligomeric complexes lack many of the wild type subunit-subunit interactions and are less stable than the complete endogenous complex, they may not have the structural integrity of the complete complexes. However, they are clearly active double 8-fold barrels.
The differential expression levels of CCT subunits have been observed in fibroblasts and mouse tissues (25, 55), but may be present in many other tissues and cell types. The expression differences we saw for the CCT subunits in E. coli cells may reflect differential folding efficiency or stability of the CCT subunits in these cells. The expression of CCT4 and CCT5 may have been robust in part because the CCT subunit proteins folded successfully and were assembled into rings inside the cells and were therefore resistant to degradation. Cheng et al. showed that the folding and assembly of Hsp60 after import into mitochondria depended on the existence of preassembled Hsp60 complexes (56). That result implied that folding of Hsp60 depends on Hsp60 chaperonin function. The human CCT subunits may also require chaperone or chaperonin assistance in their folding, at least within the E. coli cytoplasm.
Most of the CCT5 particles contained density within the chaperonin. Although there are minor contaminating bands seen by SDS-PAGE, no one impurity could account for the density seen within most of the particles. One hypothesis is that many different newly synthesized E. coli proteins may be recognized and bound by the CCT5 homo-oligomers. Another explanation for that density is that CCT5 chains synthesized within the E. coli cells or damaged during the purification are recognized by the CCT5 homo-oligomeric chaperonin and bound. Further cryo-EM and mass spectroscopy studies may distinguish between these hypotheses.
The ATP hydrolysis properties of CCT4 and CCT5 were surprising because recently Reissmann et al. revealed that CCT4 and CCT5 have higher affinity for ATP than the other CCT subunits (28). The authors postulated that CCT4 and CCT5, which in the latest consistent TRiC arrangement are on one side of the ring (23), drive an asymmetrical power stroke of ATP hydrolysis that pushes the folding cycle. For CCT4 and CCT5 homo-oligomers to have ATP hydrolysis properties similar to those of human TRiC may mean that in these complexes, each subunit binds ATP as in GroEL/ES or that the identical subunits take turns hydrolyzing ATP as seen with the ClpX protease rings (2, 57). The ATP hydrolysis properties of the homo-oligomers should be further explored to better understand the mechanism involved in folding cycle in these chaperonins.
In response to the model that specific CCT subunits recognize particular substrates, we were interested to see whether one CCT subunit homo-oligomer but not the other could recognize and refold our tested substrates. However, both CCT4 and CCT5 homo-oligomers recognized and refolded both luciferase and HγD-Crys. To study accurately the specificity or redundancy of the CCT subunits, we plan to study substrates that are proposed to only interact with some of the subunits such as actin, tubulin, huntingtin, and pVHL (14, 30, 34).
All reported structures of TRiC purified from tissues describe rings of eight different subunits. It had been assumed that all eight were obligatory for assembly. Machida et al. have co-expressed all eight subunits in baby hamster kidney cells and showed that they formed a TRiC-like complex (40). Although they showed that the CCT subunits were in equal stoichiometry, the complexes they observed could have contained some CCT4 or CCT5 homo-oligomers.
The results reported here raise the question of what prevents homo-oligomers from forming in cells. There is no evidence for regulation at the level of transcription or translation that would prevent this. Recently, work with the V0 ring of the V-ATPase showed that the specific arrangement of subunits evolved due to mutations in interfaces between subunits, rather than evolution of subunit function (58). In light of that work, it may be that CCT4 and CCT5 retained the ability to form homo-oligomer contacts but the rest of the CCT subunits did not. Therefore, TRiC may only be regulated at the level of assembly, as in bacteriophage, where the interactions of soluble subunits with growing complexes drive the specificity of association (59). More recently, Zhang et al. showed that the chaperonin-like Biedel Bradet syndrome subunits help assemble the final Biedel Bradet syndrome complex by sequential addition of each subunit (60). This needs to be explored for TRiC through direct in vitro dissociation and reassembly experiments.
In summary, we have successfully purified CCT homo-oligomer subunit complexes from E. coli. These subunits have TRiC-like morphology and are active in refolding two substrates. This novel system will be employed to further study the subunit specificity and redundancy of the CCT subunits within TRiC and to further provide insight into the assembly of TRiC in the cell.
Acknowledgments
We thank Jeannie Chew and Kelly Knee for initiating this project and Meme Tran and Rebecca Dillard for excellent technical assistance. Ioannis Papayannopoulos and Richard Schiavoni at the Koch Institute Biopolymers and Proteomics Core Facility, Debby Pheasant and the MIT Biophysical Instrumentation Facility for the Study of Complex Macromolecular Systems (NSF-007031), and Michael Berne at the Tufts University Core Facility are gratefully acknowledged.
This work was supported, in whole or in part, by National Institutes of Health Common Fund Grants PN2EY016525, R01EY015834, and P41GM103832.
The cryo-electron microscope maps have been deposited in the EMDataBank, http://www.emdatabank.org (accession numbers EMD-5640 and EMD-5641).
- Mm-Cpn
- M. maripaludis chaperonin
- ATPγS
- adenosine 5′-O-(3-thiotriphosphate)
- CCT
- chaperonin containing TCP-1
- HγD-Crys
- human γD-crystallin
- TCP-1
- tailless complex polypeptide-1
- TEV
- tobacco etch virus
- TRiC
- TCP-1 ring complex
- pVHL
- von Hippel-Lindau tumor suppressor protein
- ATP-AlFx
- adenosine triphosphate-aluminum fluoride.
REFERENCES
- 1. Frydman J. (2001) Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu. Rev. Biochem. 70, 603–647 [DOI] [PubMed] [Google Scholar]
- 2. Horwich A. L., Fenton W. A., Chapman E., Farr G. W. (2007) Two families of chaperonin: physiology and mechanism. Annu. Rev. Cell Dev. Biol. 23, 115–145 [DOI] [PubMed] [Google Scholar]
- 3. Hartl F. U., Bracher A., Hayer-Hartl M. (2011) Molecular chaperones in protein folding and proteostasis. Nature 475, 324–332 [DOI] [PubMed] [Google Scholar]
- 4. Thulasiraman V., Yang C. F., Frydman J. (1999) In vivo newly translated polypeptides are sequestered in a protected folding environment. EMBO J. 18, 85–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tang Y. C., Chang H. C., Roeben A., Wischnewski D., Wischnewski N., Kerner M. J., Hartl F. U., Hayer-Hartl M. (2006) Structural features of the GroEL-GroES nano-cage required for rapid folding of encapsulated protein. Cell 125, 903–914 [DOI] [PubMed] [Google Scholar]
- 6. Braig K., Otwinowski Z., Hegde R., Boisvert D. C., Joachimiak A., Horwich A. L., Sigler P. B. (1994) The crystal structure of the bacterial chaperonin GroEL at 2.8 Å. Nature 371, 578–586 [DOI] [PubMed] [Google Scholar]
- 7. Bigotti M. G., Clarke A. R. (2008) Chaperonins: the hunt for the group II mechanism. Arch. Biochem. Biophys. 474, 331–339 [DOI] [PubMed] [Google Scholar]
- 8. Douglas N. R., Reissmann S., Zhang J., Chen B., Jakana J., Kumar R., Chiu W., Frydman J. (2011) Dual action of ATP hydrolysis couples lid closure to substrate release into the group II chaperonin chamber. Cell 144, 240–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pereira J. H., Ralston C. Y., Douglas N. R., Kumar R., Lopez T., McAndrew R. P., Knee K. M., King J. A., Frydman J., Adams P. D. (2012) Mechanism of nucleotide sensing in group II chaperonins. EMBO J. 31, 731–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pereira J. H., Ralston C. Y., Douglas N. R., Meyer D., Knee K. M., Goulet D. R., King J. A., Frydman J., Adams P. D. (2010) Crystal structures of a group II chaperonin reveal the open and closed states associated with the protein folding cycle. J. Biol. Chem. 285, 27958–27966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang J., Baker M. L., Schröder G. F., Douglas N. R., Reissmann S., Jakana J., Dougherty M., Fu C. J., Levitt M., Ludtke S. J., Frydman J., Chiu W. (2010) Mechanism of folding chamber closure in a group II chaperonin. Nature 463, 379–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cong Y., Baker M. L., Jakana J., Woolford D., Miller E. J., Reissmann S., Kumar R. N., Redding-Johanson A. M., Batth T. S., Mukhopadhyay A., Ludtke S. J., Frydman J., Chiu W. (2010) 4.0-Å resolution cryo-EM structure of the mammalian chaperonin TRiC/CCT reveals its unique subunit arrangement. Proc. Natl. Acad. Sci. U.S.A. 107, 4967–4972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hynes G. M., Willison K. R. (2000) Individual subunits of the eukaryotic cytosolic chaperonin mediate interactions with binding sites located on subdomains of β-actin. J. Biol. Chem. 275, 18985–18994 [DOI] [PubMed] [Google Scholar]
- 14. Llorca O., Martín-Benito J., Gómez-Puertas P., Ritco-Vonsovici M., Willison K. R., Carrascosa J. L., Valpuesta J. M. (2001) Analysis of the interaction between the eukaryotic chaperonin CCT and its substrates actin and tubulin. J. Struct. Biol. 135, 205–218 [DOI] [PubMed] [Google Scholar]
- 15. Muñoz I. G., Yébenes H., Zhou M., Mesa P., Serna M., Park A. Y., Bragado-Nilsson E., Beloso A., de Cárcer G., Malumbres M., Robinson C. V., Valpuesta J. M., Montoya G. (2011) Crystal structure of the open conformation of the mammalian chaperonin CCT in complex with tubulin. Nat. Struct. Mol. Biol. 18, 14–19 [DOI] [PubMed] [Google Scholar]
- 16. Neirynck K., Waterschoot D., Vandekerckhove J., Ampe C., Rommelaere H. (2006) Actin interacts with CCT via discrete binding sites: a binding transition-release model for CCT-mediated actin folding. J. Mol. Biol. 355, 124–138 [DOI] [PubMed] [Google Scholar]
- 17. Yaffe M. B., Farr G. W., Miklos D., Horwich A. L., Sternlicht M. L., Sternlicht H. (1992) TCP1 complex is a molecular chaperone in tubulin biogenesis. Nature 358, 245–248 [DOI] [PubMed] [Google Scholar]
- 18. Frydman J., Nimmesgern E., Erdjument-Bromage H., Wall J. S., Tempst P., Hartl F. U. (1992) Function in protein folding of TRiC, a cytosolic ring complex containing TCP-1 and structurally related subunits. EMBO J. 11, 4767–4778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lewis V. A., Hynes G. M., Zheng D., Saibil H., Willison K. (1992) T-complex polypeptide-1 is a subunit of a heteromeric particle in the eukaryotic cytosol. Nature 358, 249–252 [DOI] [PubMed] [Google Scholar]
- 20. Liou A. K., Willison K. R. (1997) Elucidation of the subunit orientation in CCT (chaperonin containing TCP1) from the subunit composition of CCT micro-complexes. EMBO J. 16, 4311–4316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Martín-Benito J., Grantham J., Boskovic J., Brackley K. I., Carrascosa J. L., Willison K. R., Valpuesta J. M. (2007) The inter-ring arrangement of the cytosolic chaperonin CCT. EMBO Rep. 8, 252–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dekker C., Roe S. M., McCormack E. A., Beuron F., Pearl L. H., Willison K. R. (2011) The crystal structure of yeast CCT reveals intrinsic asymmetry of eukaryotic cytosolic chaperonins. EMBO J. 30, 3078–3090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Leitner A., Joachimiak L. A., Bracher A., Mönkemeyer L., Walzthoeni T., Chen B., Pechmann S., Holmes S., Cong Y., Ma B., Ludtke S., Chiu W., Hartl F. U., Aebersold R., Frydman J. (2012) The molecular architecture of the eukaryotic chaperonin TRiC/CCT. Structure 20, 814–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kalisman N., Adams C. M., Levitt M. (2012) Subunit order of eukaryotic TRiC/CCT chaperonin by cross-linking, mass spectrometry, and combinatorial homology modeling. Proc. Natl. Acad. Sci. U.S.A. 109, 2884–2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kubota H., Yokota S., Yanagi H., Yura T. (1999) Structures and co-regulated expression of the genes encoding mouse cytosolic chaperonin CCT subunits. Eur. J. Biochem. 262, 492–500 [DOI] [PubMed] [Google Scholar]
- 26. Roobol A., Holmes F. E., Hayes N. V., Baines A. J., Carden M. J. (1995) Cytoplasmic chaperonin complexes enter neurites developing in vitro and differ in subunit composition within single cells. J. Cell Sci. 108, 1477–1488 [DOI] [PubMed] [Google Scholar]
- 27. Roobol A., Carden M. J. (1999) Subunits of the eukaryotic cytosolic chaperonin CCT do not always behave as components of a uniform hetero-oligomeric particle. Eur. J. Cell Biol. 78, 21–32 [DOI] [PubMed] [Google Scholar]
- 28. Reissmann S., Joachimiak L. A., Chen B., Meyer A. S., Nguyen A., Frydman J. (2012) A gradient of ATP affinities generates an asymmetric power stroke driving the chaperonin TRiC/CCT folding cycle. Cell Rep. 2, 866–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kim S., Willison K. R., Horwich A. L. (1994) Cytosolic chaperonin subunits have a conserved ATPase domain but diverged polypeptide-binding domains. Trends Biochem. Sci. 19, 543–548 [DOI] [PubMed] [Google Scholar]
- 30. Spiess C., Miller E. J., McClellan A. J., Frydman J. (2006) Identification of the TRiC/CCT substrate binding sites uncovers the function of subunit diversity in eukaryotic chaperonins. Mol. Cell 24, 25–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Feldman D. E., Spiess C., Howard D. E., Frydman J. (2003) Tumorigenic mutations in VHL disrupt folding in vivo by interfering with chaperonin binding. Mol. Cell 12, 1213–1224 [DOI] [PubMed] [Google Scholar]
- 32. Llorca O., Martín-Benito J., Ritco-Vonsovici M., Grantham J., Hynes G. M., Willison K. R., Carrascosa J. L., Valpuesta J. M. (2000) Eukaryotic chaperonin CCT stabilizes actin and tubulin folding intermediates in open quasi-native conformations. EMBO J. 19, 5971–5979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Martín-Benito J., Bertrand S., Hu T., Ludtke P. J., McLaughlin J. N., Willardson B. M., Carrascosa J. L., Valpuesta J. M. (2004) Structure of the complex between the cytosolic chaperonin CCT and phosducin-like protein. Proc. Natl. Acad. Sci. U.S.A. 101, 17410–17415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tam S., Geller R., Spiess C., Frydman J. (2006) The chaperonin TRiC controls polyglutamine aggregation and toxicity through subunit-specific interactions. Nat. Cell Biol. 8, 1155–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sontag E. M., Joachimiak L. A., Tan Z., Tomlinson A., Housman D. E., Glabe C. G., Potkin S. G., Frydman J., Thompson L. M. (2013) Exogenous delivery of chaperonin subunit fragment ApiCCT1 modulates mutant Huntingtin cellular phenotypes. Proc. Natl. Acad. Sci. U.S.A. 110, 3077–3082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pappenberger G., McCormack E. A., Willison K. R. (2006) Quantitative actin folding reactions using yeast CCT purified via an internal tag in the CCT3/γ subunit. J. Mol. Biol. 360, 484–496 [DOI] [PubMed] [Google Scholar]
- 37. Ferreyra R. G., Frydman J. (2000) Purification of the cytosolic chaperonin TRiC from bovine testis. Methods Mol. Biol. 140, 153–160 [DOI] [PubMed] [Google Scholar]
- 38. Llorca O., McCormack E. A., Hynes G., Grantham J., Cordell J., Carrascosa J. L., Willison K. R., Fernandez J. J., Valpuesta J. M. (1999) Eukaryotic type II chaperonin CCT interacts with actin through specific subunits. Nature 402, 693–696 [DOI] [PubMed] [Google Scholar]
- 39. Knee K. M., Sergeeva O. A., King J. A. (2013) Human TRiC complex purified from HeLa cells contains all eight CCT subunits and is active in vitro. Cell Stress Chaperones 18, 137–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Machida K., Masutani M., Kobayashi T., Mikami S., Nishino Y., Miyazawa A., Imataka H. (2012) Reconstitution of the human chaperonin CCT by co-expression of the eight distinct subunits in mammalian cells. Protein Expr. Purif. 82, 61–69 [DOI] [PubMed] [Google Scholar]
- 41. Knee K. M., Goulet D. R., Zhang J., Chen B., Chiu W., King J. A. (2011) The group II chaperonin Mm-Cpn binds and refolds human γD crystallin. Protein Sci. 20, 30–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tang G., Peng L., Baldwin P. R., Mann D. S., Jiang W., Rees I., Ludtke S. J. (2007) EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 [DOI] [PubMed] [Google Scholar]
- 43. Chen D. H., Song J. L., Chuang D. T., Chiu W., Ludtke S. J. (2006) An expanded conformation of single-ring GroEL-GroES complex encapsulates an 86 kDa substrate. Structure 14, 1711–1722 [DOI] [PubMed] [Google Scholar]
- 44. Cong Y., Schröder G. F., Meyer A. S., Jakana J., Ma B., Dougherty M. T., Schmid M. F., Reissmann S., Levitt M., Ludtke S. L., Frydman J., Chiu W. (2012) Symmetry-free cryo-EM structures of the chaperonin TRiC along its ATPase-driven conformational cycle. EMBO J. 31, 720–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rosenthal P. B., Henderson R. (2003) Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 333, 721–745 [DOI] [PubMed] [Google Scholar]
- 46. Scheres S. H., Chen S. (2012) Prevention of overfitting in cryo-EM structure determination. Nat. Methods 9, 853–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Reissmann S., Parnot C., Booth C. R., Chiu W., Frydman J. (2007) Essential function of the built-in lid in the allosteric regulation of eukaryotic and archaeal chaperonins. Nat. Struct. Mol. Biol. 14, 432–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Acosta-Sampson L., King J. (2010) Partially folded aggregation intermediates of human γD-, γC-, and γS-crystallin are recognized and bound by human αB-crystallin chaperone. J. Mol. Biol. 401, 134–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Trent J. D., Kagawa H. K., Yaoi T., Olle E., Zaluzec N. J. (1997) Chaperonin filaments: the archaeal cytoskeleton? Proc. Natl. Acad. Sci. U.S.A. 94, 5383–5388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Heyn M. P., Bretz R. (1975) The self-association of ATP: thermodynamics and geometry. Biophys. Chem. 3, 35–45 [DOI] [PubMed] [Google Scholar]
- 51. Thulasiraman V., Ferreyra R. G., Frydman J. (2000) Folding assays: assessing the native conformation of proteins. Methods Mol. Biol. 140, 169–177 [DOI] [PubMed] [Google Scholar]
- 52. Flaugh S. L., Kosinski-Collins M. S., King J. (2005) Contributions of hydrophobic domain interface interactions to the folding and stability of human γD-crystallin. Protein Sci. 14, 569–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kosinski-Collins M. S., King J. (2003) In vitro unfolding, refolding, and polymerization of human γD crystallin, a protein involved in cataract formation. Protein Sci. 12, 480–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Moreau K. L., King J. A. (2012) Cataract-causing defect of a mutant γ-crystallin proceeds through an aggregation pathway which bypasses recognition by the α-crystallin chaperone. PLoS ONE 7, e37256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Satish L., Lo N., Gallo P. H., Johnson S., Haberman S., Kathju S. (2011) Chaperonin containing T-complex polypeptide (CCT) subunit expression in oral mucosal wounds and fibroblasts. Cell Stress Chaperones 16, 675–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cheng M. Y., Hartl F. U., Horwich A. L. (1990) The mitochondrial chaperonin hsp60 is required for its own assembly. Nature 348, 455–458 [DOI] [PubMed] [Google Scholar]
- 57. Glynn S. E., Martin A., Nager A. R., Baker T. A., Sauer R. T. (2009) Structures of asymmetric ClpX hexamers reveal nucleotide-dependent motions in a AAA+ protein-unfolding machine. Cell 139, 744–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Finnigan G. C., Hanson-Smith V., Stevens T. H., Thornton J. W. (2012) Evolution of increased complexity in a molecular machine. Nature 481, 360–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kikuchi Y., King J. (1975) Assembly of the tail of bacteriophage T4. J. Mol. Biol. 99, 695–716 [DOI] [PubMed] [Google Scholar]
- 60. Zhang Q., Yu D., Seo S., Stone E. M., Sheffield V. C. (2012) Intrinsic protein-protein interaction-mediated and chaperonin-assisted sequential assembly of a stable Bardet-Biedl syndrome protein complex, the BBSome. J. Biol. Chem. 287, 20625–20635 [DOI] [PMC free article] [PubMed] [Google Scholar]