Background: Some iron-dependent enzymes acquire their cofactor from iron chaperones, such as PCBP1.
Results: PCBP2, and PCBP3 interact with iron and ferritin in yeast and human cells.
Conclusion: All PCBP family members may function as iron chaperones.
Significance: The PCBP family members are multifunctional adaptors, mediating interactions between iron or nucleic acids and proteins that act on these molecules.
Keywords: Iron, Metals, Molecular Chaperone, Ribonuclear Protein (RNP), Transport Metals, Yeast Metabolism, hnRNP
Abstract
The mechanisms through which iron-dependent enzymes receive their metal cofactors are largely unknown. Poly r(C)-binding protein 1 (PCBP1) is an iron chaperone for ferritin; both PCBP1 and its paralog PCBP2 are required for iron delivery to the prolyl hydroxylase that regulates HIF1. Here we show that PCBP2 is also an iron chaperone for ferritin. Co-expression of PCBP2 and human ferritins in yeast activated the iron deficiency response and increased iron deposition into ferritin. Depletion of PCBP2 in Huh7 cells diminished iron incorporation into ferritin. Both PCBP1 and PCBP2 were co-immunoprecipitated with ferritin in HEK293 cells, and expression of both PCBPs was required for ferritin complex formation in cells. PCBP1 and -2 exhibited high affinity binding to ferritin in vitro. Mammalian genomes encode 4 PCBPs, including the minimally expressed PCBPs 3 and 4. Expression of PCBP3 and -4 in yeast activated the iron deficiency response, but only PCBP3 exhibited strong interactions with ferritin. Expression of PCBP1 and ferritin in an iron-sensitive, ccc1 yeast strain intensified the toxic effects of iron, whereas expression of PCBP4 protected the cells from iron toxicity. Thus, PCBP1 and -2 form a complex for iron delivery to ferritin, and all PCBPs may share iron chaperone activity.
Introduction
Transition metal ions function as essential cofactors for hundreds of cellular proteins in archaea, bacteria, and eukaryotes. Most metalloproteins contain iron and/or zinc, whereas a smaller number contain copper, manganese, cobalt, nickel, and molybdenum (1). The simplest model to account for the incorporation of metals into proteins posits a stochastic process, in which pools of free metal ions interact with nascent polypeptides through Brownian motion. If the correct metal ion finds its cognate ligand binding site, the metal binds, and the protein folds into its active form. In this model, the interaction of a metal ion with a non-cognate binding site is weak and transient.
Several observations make clear, however, that the incorporation of metals onto proteins is a more complex process that cannot be described using this simplest of models. First, the protein binding sites that coordinate divalent metal ions can be structurally very similar and, at least in vitro, accommodate a variety of metal ions with high affinity (1). Incorporation of the non-native metal ion typically inactivates a metalloprotein. Yet in vivo, metalloproteins are typically found with only their correct metal ligand. Second, the concentration of “free” metal ions within cells can be extremely low. For example, kinetic measurements indicate that essentially all cytosolic zinc and copper ions are found tightly associated with cytosolic binding proteins (2). Thus, free metal ions are not available to activate enzymes. Third, iron and copper ions are redox-active and potentially toxic to cells. These ions occupy multiple valence states under physiological conditions. Reduced iron and copper can, in the presence of oxygen, trigger the formation of reactive oxygen species that can damage many cellular components. Yet cells are capable of maintaining these metal ions in a usable form that also minimizes their toxicity in an aerobic environment.
Although the mechanisms through which most metalloproteins acquire their cognate metal ions are unknown, some proteins are activated via metallochaperones; that is, proteins that bind specific metal ions and deliver them to target proteins via a metal-mediated, protein-protein interaction (3). In a genetic screen conducted in budding yeast, human poly (C)-binding protein 1 (PCBP1)7 was identified as an iron chaperone that delivers iron to human ferritin, a cellular iron storage protein (4). Mammalian ferritin is a 24-subunit heteropolymer composed of H and L peptides that form a hollow sphere into which iron is deposited (5). Ferrous iron binds to ferritin via carboxylate side chains that line the pores formed between adjacent subunits on the 3-fold axis of symmetry.
Oxidation of the Fe(II) to Fe(III) and deposition of Fe(III) into the mineral core occurs in the interior of the sphere at ferroxidase active sites located on H-subunits. Mitochondrial and nuclear forms of ferritin have also been recently described (6). Functional assays in yeast indicate that PCBP1 can facilitate the incorporation of iron into ferritin through a direct protein-protein interaction. Complementary studies in human cells demonstrate that depletion of PCBP1 impairs the incorporation of iron into ferritin. Purified recombinant PCBP1 binds Fe(II) with low micromolar affinity in a three Fe:1 polypeptide ratio and can donate iron to ferritin in vitro. Subsequent studies have identified additional targets for iron delivery through PCBP1; they are the prolyl hydroxylases (PHDs) and asparagyl hydroxylase (FIH1) that modify the α subunit of hypoxia-inducible factor (7).
The PHDs are part of a large family of Fe(II)- and 2-oxoglutarate-dependent dioxygenases (8–10). Enzymes of this class coordinate a single ferrous ion deep in the active site via two histidines and a single acidic amino acid residue. A recent study has shown that PCBP1 is required for the incorporation of iron into PHD2 in cultured human cells, especially when cells are made transiently iron deficient with iron chelators (7). Depletion of PCBP1 in cells using siRNA leads to loss of iron incorporation into PHD2 and loss of PHD activity. PCBP1 directly interacts with PHD2 in cells, and a purified PCBP1-Fe(II) complex could activate PHD in vitro. PCBP1 is also required to maintain the activity of FIH1 in cells, and an iron-dependent interaction between PCBP1 and FIH1 was demonstrated in vivo.
PCBP1 (also called heterogeneous nuclear ribonucleoprotein E1 or α-CP1) is a multifunctional protein. It acts as a sequence-specific RNA- and DNA-binding protein to regulate the translation or stability of a number of cellular and viral RNA targets containing C-rich sequences (11–13). PCBP1 can modulate gene expression at the transcriptional level and can physically interact with and alter the fate of other cellular proteins. PCBP1 is a member of a family of four homologous proteins that includes PCBP2, PCBP3, and PCBP4. Although PCBP1 is an intronless gene, the other PCBP family members undergo alternative splicing that results in greater diversity in the expression of PCBPs in mammals. Each member of this gene family contains three heterogeneous nuclear ribonucleoprotein K-homology (KH) domains. These are ancient, conserved RNA binding domains that mediate sequence-specific interactions with single-stranded RNA and DNA. Although the KH domains are highly conserved between PCBP family members, sequences outside these domains are much less conserved. PCBP1 exhibits the greatest sequence similarity (81%) to PCBP2. Both are typically expressed at high levels in cells, and they can be found together in association with RNA and protein targets. Both PCBP1 and PCBP2 can function independently in nucleic acid binding and exhibit specificity for different protein targets. Depletion of PCBP2 in mammalian cells was also associated with defects in the incorporation of iron into PHDs (7). PCBP3 and PCBP4 exhibit much more limited levels and patterns of expression, and little is known about their cellular functions, although both have been demonstrated to bind poly-C RNA. Overexpression of splice variants of PCBP4 (also called MCG10) can induce apoptosis and cell cycle arrest in cultured cells (14, 15). PCBP3 has been shown to interact with C-rich promoter sequences of the μ-opioid receptor gene to influence transcription (16, 17).
Here we demonstrate that PCBP2 also functions as an iron chaperone for ferritin in yeast and mammalian cells and that both PCBP1 and PCBP2 are required to form a stable complex with ferritin. PCBP3 and PCBP4 also exhibit evidence of iron chaperone activity when exogenously expressed in yeast, and PCBP3 can directly interact with ferritin in cells.
EXPERIMENTAL PROCEDURES
Yeast Strains, Plasmids, and Media
Saccharomyces cerevisiae W303a (Mata ade2-1 trp1-1 can1-100 leu2-3,112 his3-1,15 ura3-1) was used to construct ferritin H and L/FRE1-HIS3 reporter strains as described (4). Ferritin H- and L-expressing strains in the YPH499 background were previously described. Deletion of CCC1 in the YPH499 strains with and without H and L ferritin was performed by PCR-mediated gene replacement. The KanMx cassette was amplified from genomic DNA isolated from the YLR220W haploid deletion mutant strain, the PCR product was transformed into the ferritin strains, and G418-resistant clones that also exhibited characteristic iron sensitivity were selected. Strains were tested for correct genome integration by PCR. Open reading frames corresponding to full-length PCBP1, PCBP2, PCBP4, and FMR1 and the PCBP3.2 coding sequence were purchased from Open Biosystems and subcloned into the yeast expression vector pYX212. To construct a full-length PCBP3, sequences corresponding to amino acids 201–240 were amplified from rat PCBP3 and subcloned into pPCBP3.2 by in vivo recombination in yeast. PCBPs were subcloned into pYES2 with 2×FLAG, fused in-frame at the amino terminus. All plasmids were sequenced to confirm correct inserts. Yeast complete synthetic (SC) medium and defined-iron medium were prepared as described (18).
Human Cell Culture
Stable cell lines expressing doxycycline-inducible, FLAG-tagged versions of PCBPs were constructed in the Flp-In T-Rex 293 cell line (Invitrogen) according to the manufacturer's instructions. FLAG-tagged PCBPs in the pYES2 expression vectors were PCR-amplified and subcloned into pcDNA5/FRT/TO and confirmed by sequencing. The resulting plasmids and pOG44 were co-transfected into the Flp-In T-Rex 293, cell line and hygromycin-resistant cells were selected. Stable cell lines and Huh7 cells were maintained in high glucose Dulbecco's modified Eagle's medium containing 5% or 10% fetal bovine serum, penicillin G 50 units/ml, and streptomycin 50 μg/ml.
Yeast Assays
For FRE1-HIS3 reporter assays, congenic W303a strains expressing no ferritin, H ferritin, L ferritin, or H and L ferritin and containing the FRE1-HIS3 reporter were transformed with pYX212 or corresponding pPCBPs, and were transformants plated in serial 10-fold dilutions on defined-iron medium without uracil or without uracil and histidine containing 1 mm ferrozine, 10 μm copper sulfate, and 250 μm ferrous ammonium sulfate. Plates were incubated for 3 days at 30 °C. For iron toxicity plate assays, the ccc1Δ strains expressing no ferritin or H and L ferritin were transformed with pYX212 or pPCBPs, and transformants were plated in serial dilutions on SC medium without uracil, supplemented with no extra iron or 5 or 6 mm ferrous ammonium sulfate. Plates were incubated at 30 °C for 3 or 4 days. For [55Fe]ferritin iron loading assays in yeast, YPH499 expressing H and L ferritin was transformed with pYX212 or the corresponding pPCBPs. Transformants were grown from very low density to A600 of 0.5 in SC medium supplemented with 10 μm 55Fe2Cl3 at 2 μCi/mg. Cells were washed, lysed, and analyzed by native gel electrophoresis and phosphorimaging or Western blotting as previously described (4). Images were obtained on Typhoon Trio and analyzed using ImageQuant (GE Biosciences). For immunoprecipitations in yeast, W303a was co-transformed with pYX212 or pPCBP1 and pYES2-FLAG-PCBP2, pYES2-FLAG-PCBP3 or pYES2-FLAG-PCBP4 and pGEV (19). Transformants were grown in SC medium, and expression of FLAG-PCBPs was induced with 1 μm β-estradiol for 4 h. Yeast cellular proteins were extracted by glass bead lysis with 50 mm Tris-HCl, pH 7.4, 40 mm KCl, 1 mm DTT, and complete protease inhibitor (Roche Applied Science). Protein extracts were clarified by centrifugation, and 1 mg of extract was subjected to immunoprecipitation using M2 anti-FLAG antibody (Sigma) on protein G Dynabeads (Invitrogen). Samples were eluted in 2× lithium dodecyl sulfate buffer and subjected to Western blotting using anti-FLAG (1:10,000) and anti-PCBP1 (1:10,000). Anti-mouse and anti-chicken secondary antibodies (LiCor) were conjugated with infrared dyes (680 and 800 nm, respectively) and used at 1:10,000. Fluorescent images were collected using the Odyssey system (LiCor).
Real-time PCR
Yeast transformed with pPCBP plasmids were grown in SC medium from very low density to mid-log phase and harvested at A600 of 0.7. RNA was extracted, and real-time PCR was performed as described (20). Primers to amplify FIT2 were 5′-TTTGACAAACGGTTCAGGTTCA-3′ and 5′-TGATTCGACGGCTTGAGTGA-3′ and for FRE2 were 5′-GACGTCCATCTTGAGCGCTAT-3′ and 5′-GTCTTTGCAGGTGATGCTCTTG-3′. Quantitative real-time PCR on human cell RNA was performed as described (7) using the double-stranded DNA dye SYBR Green (Applied Biosystems) on an ABI 7500 system according to the manufacturer's protocols.
Electrophoretic Mobility Shift Assays
A yeast pep4Δ strain was transformed with pYES2 and pYES2 FLAG-PCBP1, FLAG-PCBP2, FLAG-PCBP3, FLAG-PCBP3.2, and FLAG-PCBP4. Transformants were grown on SC raffinose medium and induced with 0.2% galactose for 4 h before harvesting. Cells were subjected to glass bead lysis in binding buffer (50 mm Tris, pH 7.4, 150 mm NaCl) plus 1 mm DTT and protease inhibitor tablet (EDTA-free), and lysates were analyzed by Western blotting with anti-FLAG antibody. For oligonucleotide binding, the poly-C probe (5′-CCCCACCCCTCTTCCCCCCACCCC-3′) (21) was end-labeled with [γ-32P]ATP using T4 polynucleotide kinase. Assays contained 1 μg of yeast lysate and poly-C probe (100 nm) in binding buffer with or without unlabeled poly-C probe (10 μm) as specific competitor or 10 μm nonspecific competitor from the CMV 3′ UTR (5′-CGCAAATGGGCGGTAGGCGTG-3′). For determination of binding constants, labeled probed was added at 0.15 to 50 nm. Binding reactions were separated on 6% polyacrylamide DNA gels (Invitrogen) and analyzed by phosphorimaging. Binding constants were calculated by nonlinear regression analysis using Prism5.0 (Graphpad).
Human Cell Assays
PCBP1 and PCBP2 were depleted in Huh7 cells or HEK cell lines by transfection of siRNA (Invitrogen), and 55Fe loading into ferritin measured as described (4). Ferritin levels in Huh7 cells were measured by enzyme-linked immunosorbent assay (Bio-Quant) as described. For immunoprecipitations, PCBP and ferritin expression were induced in stable cell lines by overnight treatment with 1 μg/ml doxycycline and 100 μg/ml ferric ammonium citrate. Cells were harvested and lysed in 0.1% Igepal-CA 630, 100 mm Tris-HCl, pH 8.0, 150 mm NaCl, 5% glycerol, 1 mm DTT, EDTA-free protease inhibitor. Cleared lysates were subjected to immunoprecipitation with anti-ferritin antibody (Sigma) and protein G-Sepharose. Immune complexes and whole cell extracts were analyzed by Western blotting using anti-FLAG (1:10,000), anti-ferritin (1:5000), anti-PCBP1 (1:10,000), anti-PCBP2 (Novus, 1:500), or anti-tubulin (1:5,000) and detected using horseradish peroxidase-conjugated secondary antibodies.
Isothermal Titration Calorimetry
Recombinant PCBP1 and PCBP2 were purified and subjected to isothermal titration calorimetry (4). PCBP1 (440 μm) and PCBP2 (300 μm) were each incubated anaerobically with 3 mol eq of Fe(II) for ∼45 min before loading into the isothermal titration calorimetry (ITC) syringe. Apoferritin from equine spleen (Sigma) was argon-purged and brought to a final concentration of 3 μm before loading into the ITC cell. Both the protein and Fe(II) solutions were prepared in buffer containing 50 mm Tris, 150 mm NaCl, pH 7.6. ITC was conducted at 25 °C on a MicroCal VP-ITC instrument (MicroCal Inc., Northampton, MA), maintaining a constant stirring speed of 416 rpm. Titrates of Fe(II) alone, Fe(II)-PCBP1, or Fe(II)-PCBP2 were individually injected into ferritin solutions in 10-μl aliquots, giving rise to exothermic profiles in each case. A total of 29 injections were performed with a spacing of 300 s (600s for PCBP1-ferritin) between injections to allow for the detected temperature signal to return to the base line before every new injection. Origin 7.0 software package supplied by MicroCal was used to analyze and fit the raw isotherm. All data were best fit using a two-binding site model.
RESULTS
Delivery of Iron to Ferritin via PCBP2 in Yeast
We tested whether PCBP2 could act as an iron chaperone for human ferritin in yeast. To accomplish this, we used yeast strains that convert from histidine auxotrophy to histidine prototrophy when cytosolic iron is diverted into exogenously expressed human ferritin (4). Fungi, such as S. cerevisiae, do not express ferritins. We constructed yeast strains in which human H ferritin, human L ferritin, or both are stably integrated into the genome in a single copy under the control of the strong constitutive PGK1 promoter. These strains express ferritins and assemble the peptides into multimeric complexes similar to those of native human ferritins, except that they contain only small amounts of iron. These strains were further modified to express an iron-dependent reporter. This reporter construct contains the HIS3 coding sequence under the control of the iron-regulated FRE1 promoter. After integration into the genome, these strains exhibit histidine auxotrophy (FRE1-HIS3 off) under iron-sufficient conditions but switch to histidine prototrophy (FRE1-HIS3 on) under iron-deficient conditions or when cytosolic iron is diverted into ferritin.
Previously, we demonstrated that expression of PCBP1 could activate the FRE1-HIS3 reporter, especially when cells also expressed H and L ferritin (4). To test whether PCBP2 could independently act as an iron chaperone for ferritin, we transformed the ferritin reporter strains with plasmids containing PCBP1, PCBP2, or the empty vector and plated them in serial dilutions on medium with and without histidine (Fig. 1A). Expression of ferritins with the empty vector did not activate the reporter, and the strains did not grow on medium lacking histidine (−Ura-His, right panel). In contrast, expression of PCBP1 or PCBP2 in a strain without ferritin or a strain expressing only L ferritin weakly activated the reporter and resulted in a small amount of growth on the plates lacking histidine. Expression of PCBP1 or PCBP2 in strains expressing H ferritin or H and L ferritin more strongly activated the reporter and resulted in a larger amount of growth on the plates lacking histidine. Only H ferritin manifests the ferroxidase activity that is required for incorporation of Fe(II) into the mineral core as Fe(III); thus, requirement of H ferritin for full activation of the reporter is consistent with the incorporation of cytosolic iron into the mineral core. PCBP1 and PCBP2 exhibited a similar requirement for the co-expression of H ferritin for full activation of the reporter.
FIGURE 1.
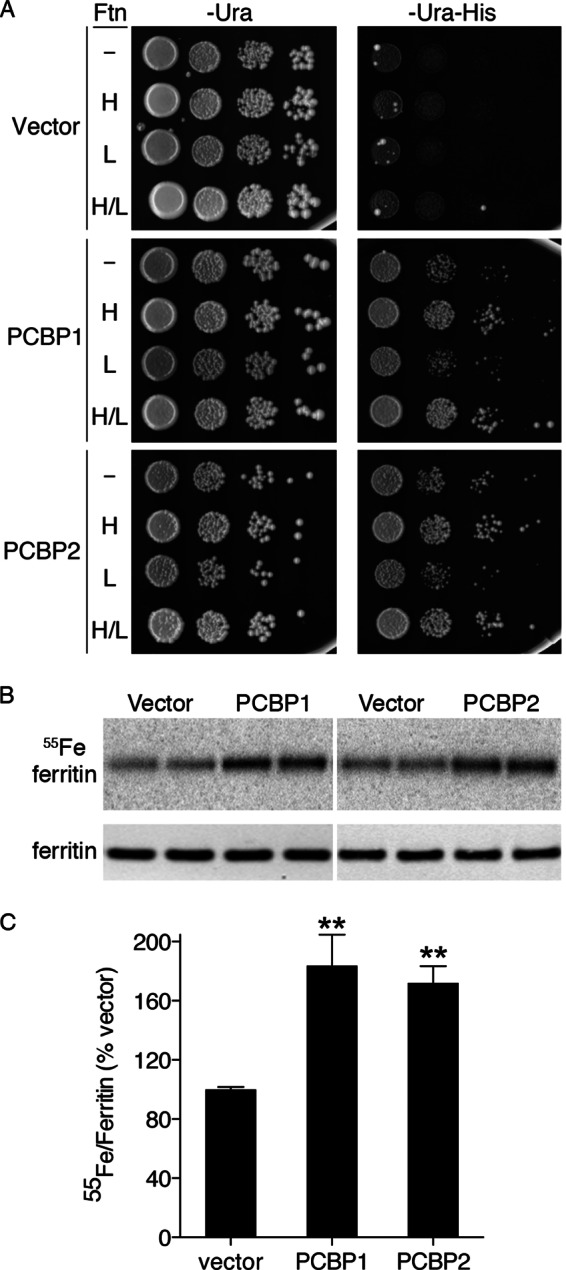
PCBP2-mediated delivery of iron to ferritin in yeast. A, shown is activation of iron-dependent reporter in yeast expressing PCBP1 and PCBP2 with H ferritin. Congenic yeast strains (W303a background) containing the FeRE/HIS3 reporter and either no ferritin (Ftn, −), human H ferritin (H), human L ferritin (L), or both H and L ferritin (H/L) were transformed with pYX212 (Vector), pPCBP1 (PCBP1), or pPCBP2 (PCBP2). Transformants were plated in serial 10-fold dilutions on defined iron medium containing 1 mm ferrozine and 250 μm iron but lacking either uracil (−Ura) or lacking uracil and histidine (−Ura-His). Plates were incubated at 30 °C for 3 days. Activation of the FeRE/HIS3 reporter is indicated by growth on −Ura-His medium. B and C, enhanced incorporation of 55Fe into ferritin in yeast expressing PCBP1 and PCBP2 is shown. A yeast strain expressing human H and L ferritin was transformed with pYX212, pPCBP1, or pPCBP2. Transformants were grown in SC medium supplemented with 10 μm 55FeCl3. Cell lysates were subjected to native PAGE followed by phosphorimaging (55Fe ferritin, upper panel in B) or denaturing PAGE followed by Western blotting with anti-ferritin antibody (ferritin, lower panel in B). Duplicate samples from each culture were loaded. Ferritin was the only radiolabeled species detected by phosphorimaging. C, shown is quantitation of data in B. 55[Fe]Ferritin/ferritin protein ratios were expressed as percentage of vector control. Experiments were replicated three or four times. Error bars depict S.E. ** indicates p < 0.002.
We directly tested whether co-expression of PCBP2 with H and L ferritin could affect the amount of iron incorporated into ferritin in vivo. Yeast expressing H and L ferritin were transformed with plasmids containing PCBP1, PCBP2, or the empty vector. These transformants were grown in medium containing 55FeCl3, and the ferritin in lysates was examined by native gel electrophoresis and autoradiography (Fig. 1, B and C). Under these conditions, [55Fe]ferritin is the only species detectable by autoradiography. Strains expressing PCBP1 or PCBP2 exhibited a 2-fold increase in the amount of iron loaded into ferritin, and this increase was not due to changes in the level of ferritin protein in the cell (Fig. 1B, lower panel). These data indicated that, when expressed in yeast, PCBP2 was similar to PCBP1 in its capacity to enhance iron loading into ferritin.
Loss of Ferritin Iron Loading in Mammalian Cells Lacking PCBP2
Previously, we demonstrated that depletion of PCBP1 in Huh7 cells using siRNA resulted in a loss of iron loading into ferritin (4). We tested whether Huh7 cells depleted of PCBP2 exhibited similar defects in ferritin iron loading by transfecting cells with siRNAs directed against PCBP1, PCBP2, and PCBP1 and -2 or a non-targeting control siRNA. Cells were then incubated with 55FeCl3, and the amount of iron incorporated into endogenously expressed ferritin was measured by non-denaturing electrophoresis and autoradiography (Fig. 2A). Depletion of PCBP1, PCBP2, or both PCBP1 and PCBP2 together led to similar reductions in iron incorporation. Again, the differences in ferritin iron loading were not due to changes in the levels of total ferritin protein (Fig. 2B). We confirmed that siRNA treatments resulted in depletion of PCBP mRNAs by real-time PCR (Fig. 2C). These data suggested that both PCBP1 and PCBP2 were independently required for efficient delivery of iron to ferritin in Huh7 cells.
FIGURE 2.
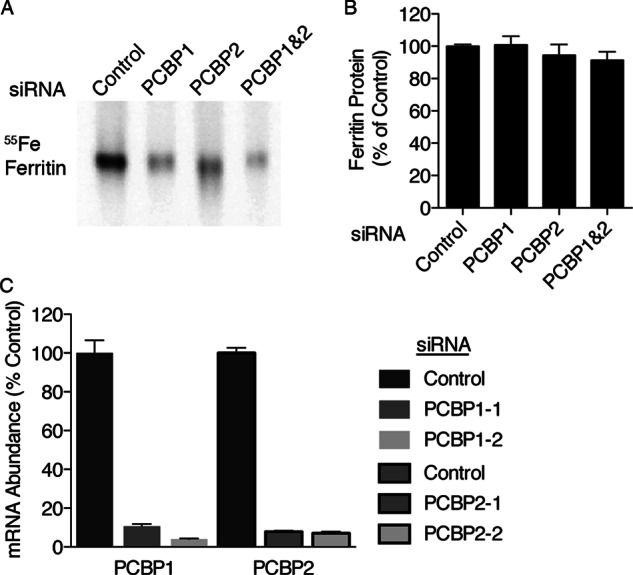
PCBP2-mediated delivery of iron to ferritin in mammalian cells. A, decreased ferritin iron loading in Huh7 cells depleted of PCBP1 and PCBP2 is shown. Huh7 cells were transfected with non-targeting control, PCBP1, PCBP2, or PCBP1 and -2 siRNAs. Cells were labeled overnight with 55FeCl3, and lysates were subjected to native-PAGE and phosphorimaging. Experiments were replicated three times. B, total ferritin protein in Huh7 cells lacking PCBP1 and PCBP2 is shown. Ferritin protein was detected in lysates from A using ELISA. C, efficiency of independent siRNAs in depleting PCBP1 and PCBP2 is shown. Huh7 cells transfected with the indicated siRNAs were analyzed by real-time PCR. Samples were analyzed in triplicate and expressed as a % of control. Error bars indicate S.E.
Binding of Ferritin to Both PCBP1 and PCBP2 in Mammalian Cells
Metallochaperones are proposed to function through metal-facilitated, direct protein-protein interactions. PCBP1 was demonstrated to physically interact with ferritin in vivo when both were exogenously expressed in yeast cells (4). To examine whether PCBP1 or PCBP2 could interact with ferritin in mammalian cells, we constructed cells lines that stably expressed epitope-tagged versions of PCBP1 and PCBP2 under the control of a tetracycline-regulable promoter (Fig. 3A). These cell lines offered two advantages. First, both cell lines expressed their respective FLAG-tagged PCBP at a level similar to the endogenously expressed protein (Fig. 3A, right panels). Because high levels of overexpression can lead to non-native protein-protein interactions, these lines avoided this type of artifact. Second, the use of epitope-tagged PCBPs allowed for quantitative comparisons of PCBP interactions with ferritin by co-immunoprecipitation (Fig. 3B). Cell lines were treated with doxycycline to induce PCBP expression and with FeCl3 to induce ferritin expression. Lysates were subjected to immunoprecipitation with or without anti-ferritin antibodies, and immune complexes were analyzed by Western blotting. FLAG-PCBP1 was readily detected in anti-ferritin immune complexes but not in control immunoprecipitates. Although FLAG-PCBP2 was initially not detected in anti-ferritin immunoprecipitates using lower amounts of cell extract (Fig. 3B, left panel), it was readily detected when larger quantities of lysate were subjected to immunoprecipitation (Fig. 3B, right panel). Again, no FLAG-PCBP2 was detected in control immunoprecipitates. Although this requirement for more lysate might suggest that the affinity of the ferritin interaction is lower for PCBP2 than for PCBP1, FLAG-PCBP2 was also expressed at lower levels than FLAG-PCBP1 in these cell lines (Fig. 3C).
FIGURE 3.
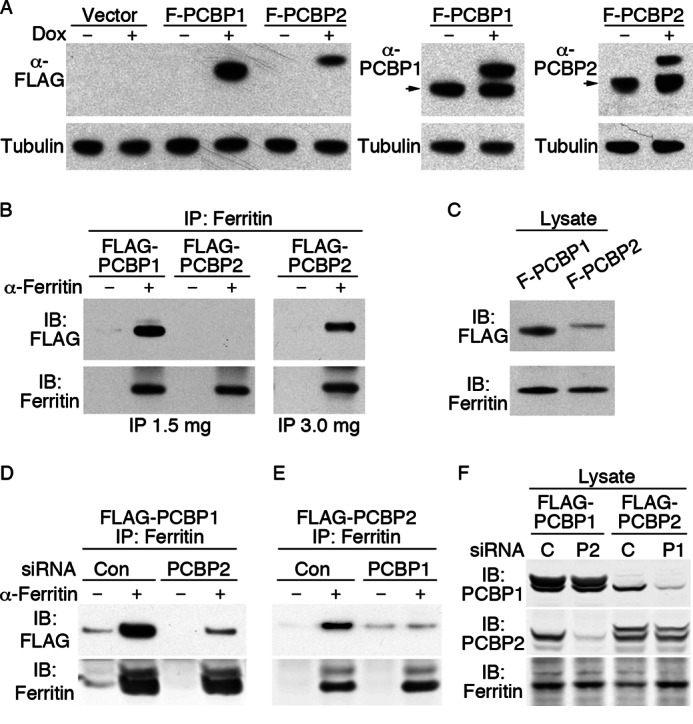
Co-immunoprecipitation of epitope-tagged PCBP1 and PCBP2 with ferritin. A, shown is construction of cell lines inducibly expressing FLAG-PCBP1 and FLAG-PCBP2 at levels similar to endogenous PCBP1 and -2. T-Rex 293 cells containing integrated copies of empty vector, FLAG-PCBP1, and FLAG-PCBP2 under the control of a tetracycline-regulable promoter were untreated or treated with 1 μg/ml doxycycline for 36 h. Lysates were subjected to Western blotting with anti-FLAG antibody (left panel), anti-PCBP1 (center panel), and anti-PCBP2 (right panel) and anti-tubulin. The arrow indicates migration of endogenous PCBP. B, shown is detection of FLAG-PCBP1 and FLAG-PCBP2 in immunoprecipitates (IP) of ferritin. Cell lines were treated as in A with the addition of 100 μg/ml ferric citrate for 18 h. Lysates were subjected to immunoprecipitation with anti-ferritin antibody or protein A beads alone, and immune complexes were analyzed by Western blotting with anti-FLAG or anti-ferritin antibodies, as indicated. Co-immunoprecipitation of FLAG-PCBP2 was only detected when 3 mg of lysate was used. C, detection of FLAG-PCBP1 (F-PCBP1), FLAG-PCBP2, and ferritin in whole cell lysates is shown. Whole cell lysates used in B were subjected to Western blot analysis as indicated. D, shown is the requirement of PCBP2 in PCBP-ferritin complexes. FLAG-PCBP1 cells were transfected with non-targeting control (con) and PCBP2 siRNAs, then treated with ferric citrate and doxycycline. Lysates were subjected to immunoprecipitation with or without anti-ferritin antibodies, and immune complexes analyzed by Western blotting as in B. E, shown is the requirement of PCBP1 in PCBP-ferritin complexes. FLAG-PCBP2 cells were treated with control and PCBP1 siRNAs and analyzed as in D. F, shown is detection of PCBP1 and PCBP2 in whole cell lysates from D and E. IB denotes the antibody used for immunoblotting.
Requirement for Both PCBP1 and PCBP2 in Ferritin Complex Formation
Our data indicated that PCBP1 and PCBP2 were both independently required for efficient ferritin-iron loading in human cells. To test this mechanistically, we quantitatively examined the formation of PCBP-ferritin complexes when one of the PCBP components was depleted by siRNA. We treated the FLAG-PCBP1 cell line with non-targeting siRNA or siRNA against PCBP2. When ferritin was immunoprecipitated, cells lacking PCBP2 exhibited greatly reduced amounts of FLAG-PCBP1 in the anti-ferritin immune complexes (Fig. 3D) even though similar amounts of ferritin were precipitated and levels of FLAG-PCBP1 were similar in both lysates (Fig. 3F). In a reciprocal experiment, FLAG-PCBP2 cells were depleted of PCBP1, and ferritin was immunoprecipitated. Again, the amount of FLAG-PCBP2 that co-precipitated with ferritin was greatly reduced in cells lacking PCBP1 (Fig. 3E), even though FLAG-PCBP2 was present at the same level in both lysates (Fig. 3F). These data suggest that both PCBP1 and PCBP2 are required to form a stable complex with ferritin.
High Affinity Binding of Fe-PCBP1 and Fe-PCBP2 to Ferritin in Vitro
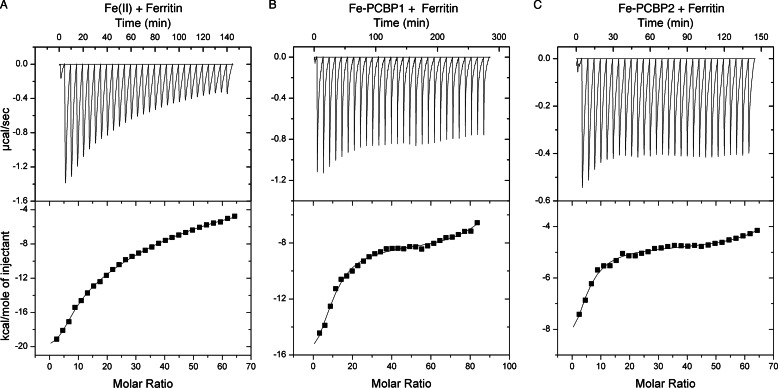
To further characterize the interactions of PCBP1 and PCBP2 with ferritin, we used ITC to directly measure the binding of individual PCBPs with ferritin. ITC is a quantitative method of analyzing the thermodynamic properties of a ligand binding reaction. By measuring the heat evolved in a binding reaction, the binding constants, stoichiometries, entropies, and enthalpies may be determined. Both PCBP1 and PCBP2 bind ferrous iron with low micromolar affinity, with PCBP1 binding in a 3:1 stoichiometric ratio and PCBP2 exhibiting a higher stoichiometry (4).8 Titration of Fe(II) into solutions of equine spleen apoferritin under anaerobic conditions produced negative peaks in the raw thermogram, indicating iron binding to ferritin in an exothermic manner (Fig. 4A, upper panel). Integration of the area within the titration peaks of the raw thermogram yielded the processed spectrum (Fig. 4A, lower panel). Non-linear regression analysis indicated that the data best fit a two-site binding model and that iron bound to ferritin with a Kd1 of 5.6 μm in a 10:1 stoichiometry at the higher affinity sites (Table 1). These data were similar to those previously published (22). Next, purified PCBP1 or PCBP2 was anaerobically loaded with Fe(II) in a 3:1 ratio and titrated into apoferritin (Fig. 4, B and C). Again, negative peaks in the raw thermograms indicated Fe-PCBP binding to ferritin in an exothermic reaction. Analysis of the integrated thermograms, however, indicated that Fe-PCBP1 and Fe-PCBP2 bound to ferritin with affinities that were 30- and 20-fold higher, respectively, than the affinity of Fe(II) alone. Multiple molar equivalents of the Fe-PCBPs bound to ferritin polymers, consistent with the known structure of ferritin, which contains eight iron binding pores formed at the 3-fold axes of symmetry. PCBPs did not show significant interactions with ferritin in the absence of Fe(II) (data not shown), indicating that iron facilitated the protein-protein interaction, possibly through the formation of PCBP-Fe-ferritin intermediates. These in vitro interactions of Fe-PCBP1 and Fe-PCBP2 with ferritin are consistent with the function of both PCBP1 and PCBP2 as iron chaperones in vivo.
FIGURE 4.
Binding of Fe-PCBP1 and Fe-PCBP2 to ferritin in vitro. ITC of Fe(II) (A), Fe(II)-PCBP1 (B), and Fe(II)-PCBP2 (C) into ferritin is shown. Anerobically prepared Fe(II) and Fe(II)-PCBP in 3:1 ratios were titrated into equine spleen ferritin. Raw isotherm of heat released per injection (top) and calculated total heat evolved per injection (bottom) is shown.
TABLE 1.
Binding affinities and stoichiometries of Fe-PCBP1 and Fe-PCBP2 for ferritin N-stoichiometry
| Sample | N1 | Kd1 μm | N2 | Kd2 μm |
|---|---|---|---|---|
| Fe(II) + ferritin | 10 ± 1 | 5.6 ± 3.0 | 66 ± 12 | 48 ± 46 |
| Fe-PCBP1 + ferritin | 9.5 ± 0.9 | 0.2 ± 0.1 | 95 ± 12 | 5.3 ± 5.0 |
| Fe-PCBP2 + ferritin | 4.0 ± 1.2 | 0.3 ± 0.1 | 74 ± 9 | 2.1 ± 0.6 |
Low or Absent Expression of Paralogs of PCBP1 and PCBP2 in Cultured Human Cells
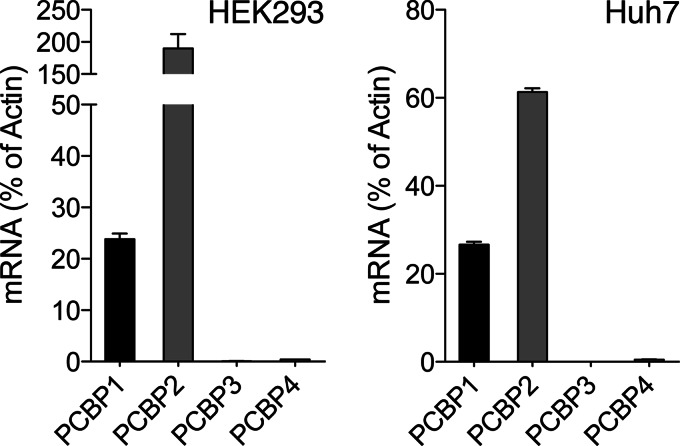
PCBP1 and PCBP2 are part of a multigene family that includes PCBP3, PCBP4, and the more distantly related heterogeneous nuclear ribonucleoprotein K (12). All members contain a similar domain structure that includes three highly conserved KH domains. Two of the KH domains are near the amino terminus and are separated from the third, carboxyl-terminal KH domain, by a region of low homology. PCBP3 is 66% identical to PCBP1, and PCBP4 is 40% identical to PCBP1. Although PCBP1 and PCBP2 are abundantly expressed in all human tissues, PCBP3 and PCBP4 are expressed in many fewer tissues and at much lower levels. We measured the expression levels of PCBPs in HEK293 and Huh7 cells using real-time PCR and found that, as previously observed, transcripts for PCBP1 and PCBP2 were very abundant, whereas transcripts for PCBP3 and PCBP4 were detectable but present at exceedingly low levels (Fig. 5). Thus we were unable to study endogenous PCBP3 or PCBP4 in cultured cells.
FIGURE 5.
Very low expression of PCBP3 and PCBP4 in cultured cells. Hek293 (left) and Huh7 (right) cells were harvested and analyzed by real-time PCR using primers for PCBP1, PCBP2, PCBP3, PCBP4, and actin. Transcript abundance is expressed as a % of actin mRNA. All five transcripts were detected. Error bars indicate S.E.
Differential Activation of the Iron Deficiency Response in Yeast Expressing PCBP3 and PCBP4
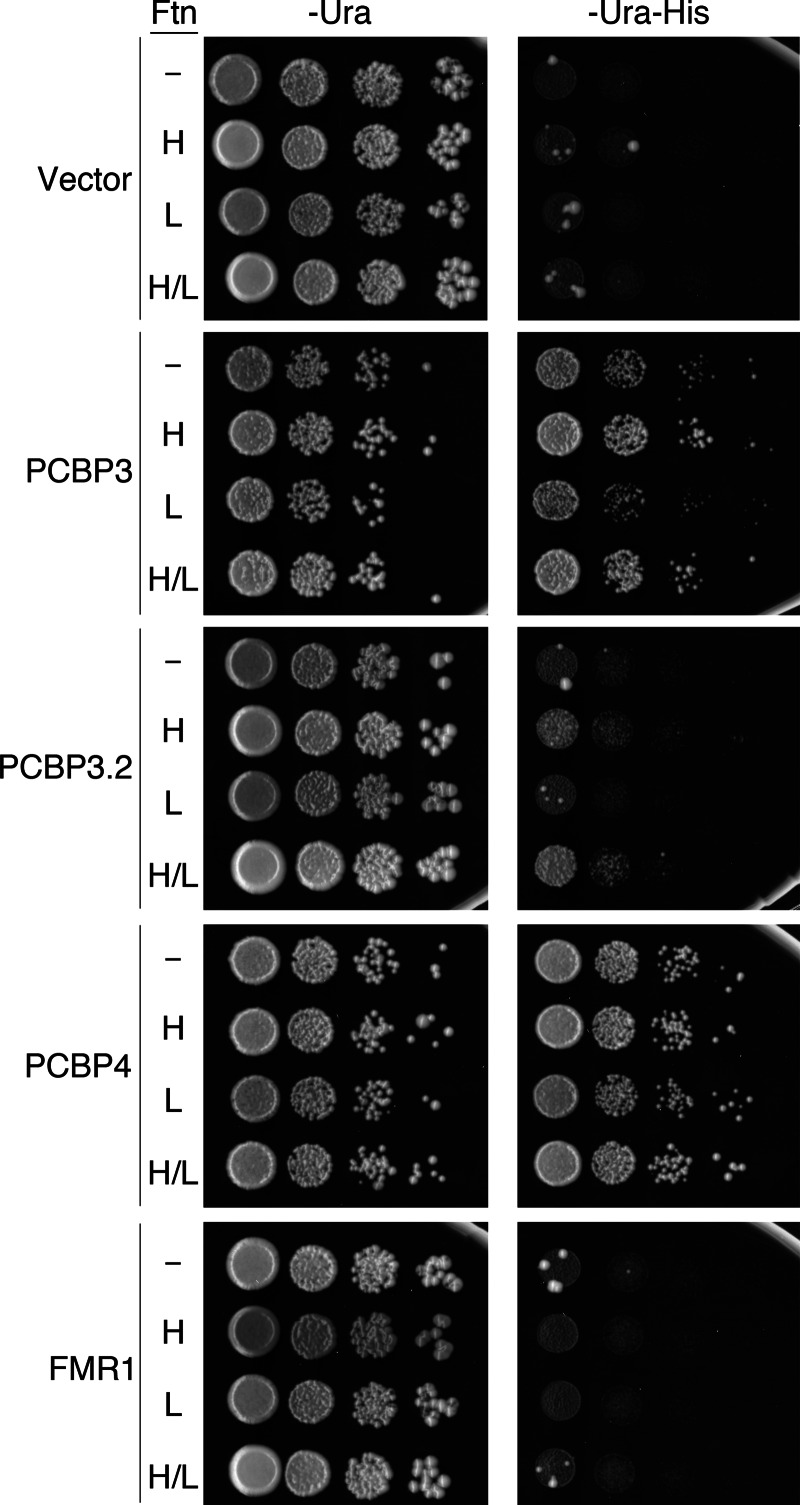
To examine the function of PCBP3 and PCBP4 and to compare their functions to PCBPs 1 and 2, we subcloned the open reading frames of PCBP3 and PCBP4 into yeast expression vectors and transformed the resulting plasmids into the ferritin reporter strains as in Fig. 1A, upper panel. We also examined a naturally occurring splice variant of PCBP3, called PCBP3.2, that lacks a 26-amino acid sequence located between the second and third KH domains. As a control, we examined an additional KH domain protein from outside the PCBP family. FMR1 (fragile X mental retardation 1) encodes an RNA-binding protein with two tandem KH domains (23); it is not known to play a role in metal metabolism. These transformants were plated on medium with and without histidine as in Fig. 1A (Fig. 6). Similar to PCBP1 and -2, strains expressing PCBP3 and either no ferritin or L ferritin exhibited slow growth on medium without histidine, indicating weak activation of the FRE1-HIS3 reporter. In contrast, strains expressing PCBP3 with H ferritin or H and L ferritin grew well on medium without histidine, indicating strong activation of the reporter. These data indicated that PCBP3 activated the iron deficiency response in yeast and that PCBP3 genetically interacted with ferritin. A similar examination of PCBP3.2 demonstrated only a very slight increase in activation of the reporter when compared with the vector-transformed strains, although growth in the H ferritin-expressing strains was slightly greater than in the strains without H ferritin. These data suggested that the effects of PCBP3.2 on yeast iron were much smaller than those of the other PCBPs tested.
FIGURE 6.
Activation of iron-dependent reporter in yeast expressing H ferritin and PCBP3 or PCBP4. Yeast strains (W303a background) expressing human ferritins were transformed with pYX212 (Vector), pPCBP3, pPCBP3.2, pPCBP4, and pFMR1. Transformants were plated on defined-iron medium lacking uracil (−Ura) or lacking uracil and histidine (−Ura−His) as in Fig. 1A. Rare, fast-growing colonies on a background of slow-growing colonies represent spontaneously occurring suppressor mutants. Ftn, ferritin; H, human H ferritin; L, human L ferritin; H/L, both H and L ferritin.
Yeast expressing PCBP4 exhibited a different pattern of growth in the reporter strains than the other PCBPs. Growth was rapid, and the FRE1-HIS3 reporter was strongly activated when PCBP4 was expressed. Furthermore, similar growth was observed in strains with and without H ferritin, suggesting a lack of genetic interaction with ferritin. Yeast strains expressing FMR1 exhibited essentially no growth above vector control levels on medium without histidine and no genetic interaction with ferritin. These data indicated that expression of KH domains alone was not sufficient to activate the iron deficiency response or interact with ferritin. Activation of the FRE1-HIS3 reporter in each strain was dependent on Aft1, the major iron-dependent transcription factor of yeast, as no transformants activated the reporter when AFT1 was also deleted (data not shown).
RNA Binding Activity of Epitope-tagged PCBPs Expressed in Yeast
To confirm that all of the PCBPs expressed in yeast were expressed and folded in an active form, we constructed plasmids containing FLAG-epitope-tagged versions of PCBPs under the control of the galactose-inducible promoter, transformed the resulting plasmids into yeast, and examined the proteins by Western blotting and oligonucleotide binding activity (Fig. 7). All of the FLAG-tagged strains were expressed at similar levels in yeast (Fig. 7A). Binding activity to a 32P-labeled poly-C oligonucleotide probe was measured using an electrophoretic mobility-shift assay coupled with phosphorimaging (Fig. 7, B and C). Vector-transformed yeast lysates exhibited no specific protein- poly-C complexes. Lysates from yeast expressing PCBPs exhibited a single PCBP-poly-C complex that was 1) absent when a 100-fold excess of unlabeled poly-C probe was added as a specific competitor (lanes marked S) but 2) present when a 100-fold excess of control probe was added as a nonspecific competitor (lanes marked N). Each of the PCBPs expressed in yeast exhibited high affinity binding to poly-C oligonucleotide. Titration of increasing amounts of probe into the yeast lysates was performed, and the individual binding affinities of the protein-DNA complexes was calculated (Table 2). All of the PCBPs exhibited low nanomolar binding affinities similar to those previously published, with PCBP4 exhibiting a slightly lower affinity than the other PCBPs (21).
FIGURE 7.
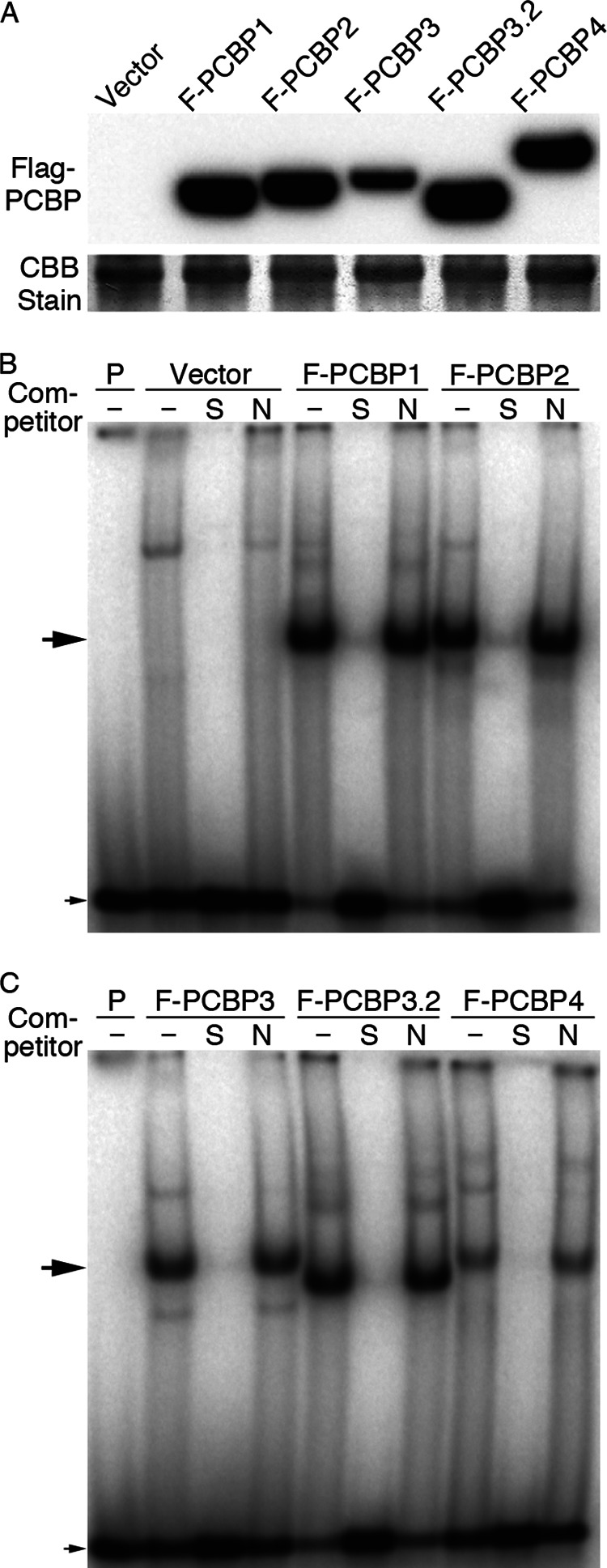
FLAG-tagged PCBPs expressed in yeast bind oligonucleotides. A, shown is expression of FLAG-PCBPs in yeast. A pep4Δ strain was transformed with pYES2 and pYES2 containing amino-terminal 2×FLAG-tagged PCBP1, PCBP2, PCBP3, PCBP3.2, and PCBP4. Transformants were grown on SC raffinose medium and induced with 0.2% galactose for 4 h before harvesting. Cell lysates were analyzed by Western blotting with anti-FLAG antibody. A major band from the Coomassie-stained gel is shown as a loading control. B and C, shown is specific binding of FLAG-PCBPs to a poly-C oligonucleotide probe. 32P-Labeled poly-C oligonucleotide probe was mixed with buffer alone (P) or yeast lysates from A and separated by PAGE. Unlabeled poly-C oligonucleotide and mutated oligonucleotide at 10-fold molar excesses were added as specific competitor (S) and nonspecific competitor (N), respectively. The large arrow corresponds to specific PCBP-probe complex; the small arrow corresponds to free probe.
TABLE 2.
Binding affinities of PCBPs expressed in yeast for poly-C oligonucleotide
CI, confidence interval. r2, goodness of fit for non-linear regression.
| PCBP1 | PCBP2 | PCBP3 | PCBP3.2 | PCBP4 | |
|---|---|---|---|---|---|
| Kd ± 95% CI (nm) | 3.4 ± 0.9 | 3.7 ± 0.6 | 3.1 ± 1.0 | 2.5 ± 0.2 | 12.4 ± 1.5 |
| r2 | 0.91 | 0.97 | 0.89 | 0.99 | 0.98 |
Activation of Aft1-mediated Transcription by Expression of PCBPs and Ferritin
Activation of the FRE1-HIS3 reporter in strains expressing PCBPs and H ferritin suggested that PCBPs could generally activate the iron deficiency response in yeast. To more quantitatively measure the activation of Aft1 and to confirm the requirement of ferritin co-expression, we directly measured the capacity of PCBP and ferritin co-expression to activate transcription of other Aft1-dependent genes (Fig. 8). FRE2 and FIT2 are strongly transcribed in an Aft1-dependent manner in cells exposed to iron deficiency (24). We transformed the wild-type parent strain (YPH499) and the congenic H and L ferritin strain with plasmids for PCBP1, PCBP2, PCBP3, PCBP4, and FMR1 and the empty vector. Transformants were grown to mid-log phase in iron-replete, SC medium, and individual mRNA levels were measured using real-time PCR. In the YPH499 strain background, grown in standard SC medium, expression of FRE2 and FIT2 was relatively low and did not increase when PCBPs were expressed in the absence of ferritin. Expression of H and L ferritin without PCBPs did not activate the transcription of FRE2 (Fig. 8A) but did activate transcription of FIT2 (Fig. 8B), a gene that is very highly induced by small amounts of iron deficiency. In both cases, co-expression of ferritin with PCBP1, PCBP2, PCBP3, or PCBP4, but not FMR1, further activated the transcription of FRE2 and FIT2. These data confirmed that each of the PCBPs could activate the iron deficiency response in yeast and suggested that ferritin was also required.
FIGURE 8.
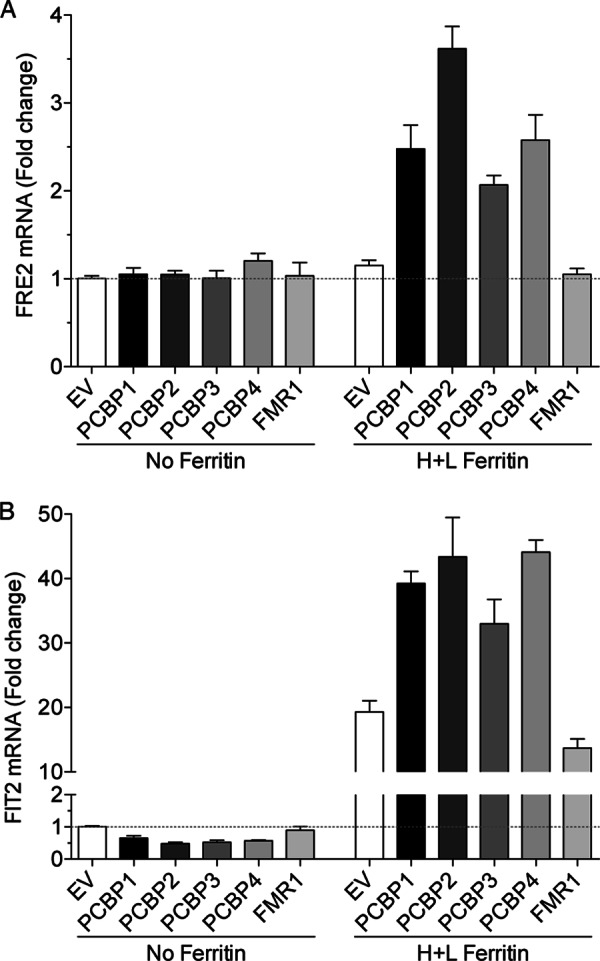
Activation of Aft1-dependent transcription in cells expressing both ferritin and PCBPs. A yeast strain expressing human H and L ferritin and the congenic parent strain (YPH499 background) was transformed with pYX212, pPCBP1, pPCBP2, pPCBP3, pPCBP4, or pFMR1. Transformants were grown in SC−Ura medium to mid-log phase, and cells were collected an analyzed by real-time PCR for Aft1 target genes FRE2 (A) and FIT2 (B). Transcript levels were expressed as –fold change over pYX212-transformed cells (EV) without ferritin. Experiments were replicated three times. Error bars indicate S.E.
PCBP3-mediated Ferritin Iron Loading in Yeast
We directly measured the capacity of PCBP3 and PCBP4 to augment iron loading into ferritin in yeast using the 55FeCl3 labeling assay. Yeast expressing H and L ferritin were transformed with PCBPs and FMR1, and the transformants were labeled with 55FeCl3. Lysates were analyzed by native gel electrophoresis and phosphorimaging or by Western blotting (Fig. 9). When compared with vector-transformed yeast, strains expressing PCBP3 exhibited a 1.8-fold increase in ferritin iron accumulation. Expression of PCBP3.2 or PCBP4 was also associated with increased ferritin iron accumulation but to a lesser degree than that of PCBP3. Expression of FMR1 had no effect on ferritin iron accumulation. These data were consistent with a role for PCBP3 in the delivery of iron to ferritin and a weaker role for PCBP3.2 and PCBP4.
FIGURE 9.
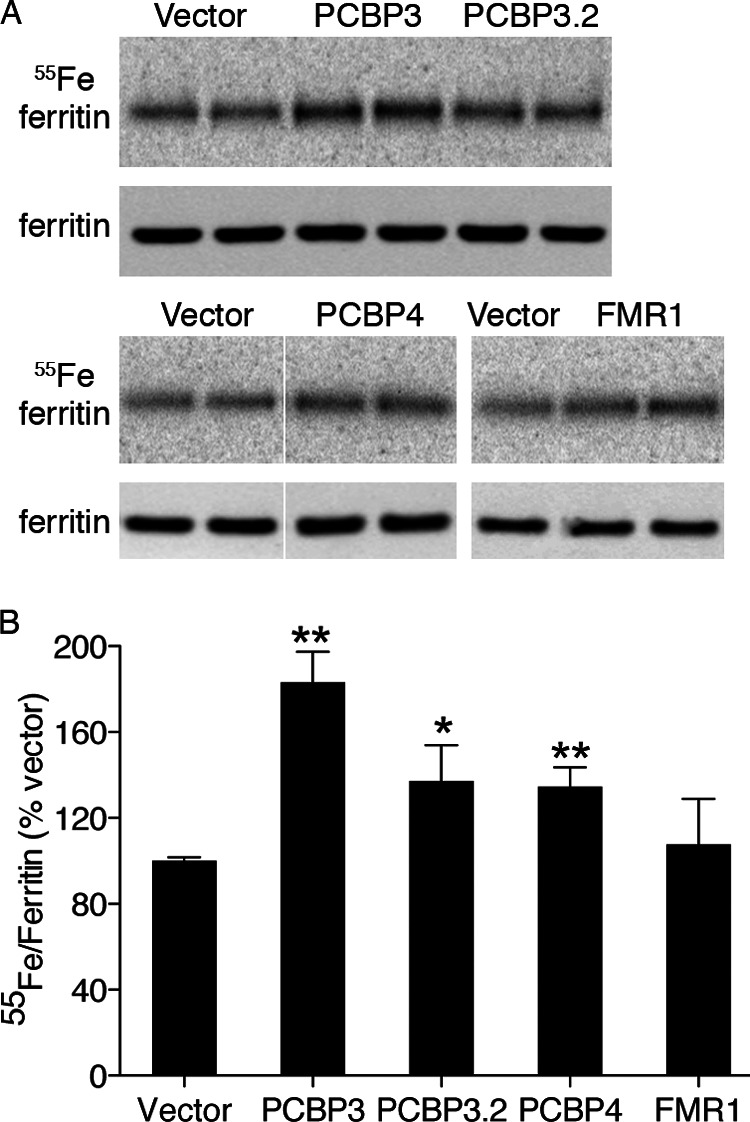
Enhanced incorporation of 55Fe into ferritin in yeast expressing PCBP3 and PCBP4. A and B, a yeast strain expressing human H and L ferritin was transformed with pYX212, pPCBP3, pPCBP3.2, or pPCBP4. Transformants were grown in medium supplemented with 55FeCl3 and analyzed in duplicate as in Fig. 1, B and C. Experiments were replicated three times. Error bars indicate S.E. ** indicates p < 0.002, * indicates p < 0.05 when compared with vector.
PCBP4- but Not PCBP1-mediated Reversal of Iron Toxicity
Data from Fig. 6 suggested that PCBP4-mediated activation of the FRE1-HIS3 reporter was ferritin-independent. In contrast, data from Fig. 8 suggested that PCBP4 was similar to other PCBPs in its requirement for ferritin to activate Aft1-dependent transcription, and Fig. 9 suggested that PCBP4 weakly augmented ferritin iron loading. To clarify the activity of PCBP4, we compared the capacities of PCBP4 and PCBP1 to affect the iron sensitivity of a strain lacking CCC1. CCC1 encodes a polytopic integral membrane protein that facilitates the transfer of iron from the cytosol to the lumen of the vacuole in yeast (25). The vacuole is the major site of iron storage; strains without Ccc1 are unable to store excess iron in the vacuole and are sensitive to the toxic effects of iron. We tested whether PCBP1 or PCBP4, with or without ferritin, could mitigate the toxic effects of iron in the ccc1Δ strain (Fig. 10). The ccc1Δ strain grew well on standard medium (no added iron) but exhibited slow (5 mm) or almost no growth (6 mm) on media with high concentrations of added iron. Expression of H and L ferritin in this strain did not change this sensitivity to high concentrations of iron. Surprisingly, expression of PCBP1 in the ccc1Δ strain resulted in very slow (5 mm) or no growth (6 mm), suggesting it enhanced, rather than mitigated, the toxic effects of iron. Co-expression of PCBP1 and ferritin slightly aggravated this effect. In contrast, expression of PCBP4 in the ccc1Δ strain resulted in more rapid growth on high concentrations of iron, suggesting PCBP4 mitigated the toxic effects of iron. Co-expression of ferritin with PCBP4 was similar to expression of PCBP4 alone. These data indicated that PCBP1 and PCBP4 exhibited qualitatively different activities when expressed in yeast, with PCBP1 increasing the toxicity of iron and PCBP4 protecting against iron toxicity. Although these genetic data do not reveal a biochemical mechanism for the differences between PCBP1 and PCBP4, these data clearly indicate that PCBP1 and PCBP4 function differently in yeast. A possible interpretation of these results is that PCBP1, perhaps by delivering iron to ferritin, maintains cytosolic iron in a bioavailable pool that is potentially toxic. PCBP4 may act to bind and sequester iron, thus making it less toxic.
FIGURE 10.
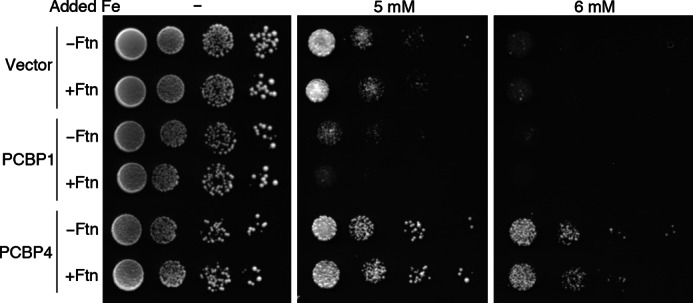
PCBP1 enhancement and PCBP4 mitigation of iron toxicity in a ccc1Δ yeast strain. Yeast strains expressing no ferritin or human H and L ferritin (Ftn) and deleted for CCC1 were constructed and transformed with pYX212, pPCBP1, and pPCBP4. Transformants were plated in serial 10-fold dilutions on SC−Ura medium supplemented with the indicated amounts of ferrous ammonium sulfate. Plates were incubated at 30 °C for 3 days.
Binding of PCBP3, PCBP3.2, and PCBP4 to Ferritin in Human Cells
Although we could not study the functions of endogenously expressed PCBP3 and PCBP4 in cultured human cell lines, we could construct cell lines that inducibly expressed PCBP3 and PCBP4 at levels similar to those of PCBP1 and PCBP2. Cell lines expressing FLAG epitope-tagged PCBP3, PCBP3.2, and PCBP4 were constructed and used to measure the physical interaction of PCBP3 and PCBP4 with ferritin. Both FLAG-PCBP3 and FLAG-PCBP3.2 were detected in immune complexes precipitated with anti-ferritin antibodies but not in precipitates without anti-ferritin antibody (Fig. 11A). In contrast, FLAG-PCBP4 was not detected in immunoprecipitates, although all three PCBPs were expressed at similar levels (Fig. 11B). These data indicated that PCBP3 formed a complex with ferritin and could potentially deliver iron to ferritin in cells. Conversely, these data suggested that PCBP4 did not form a stable complex with ferritin and was unlikely to act as a ferritin iron chaperone.
FIGURE 11.
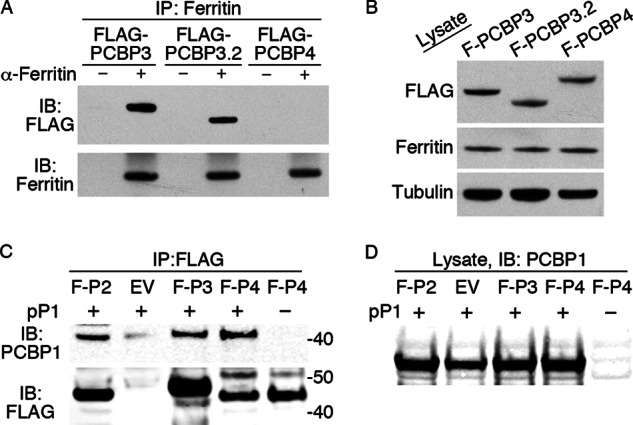
Co-immunoprecipitation of PCBP3 and PCBP3.2 with ferritin in human cells. A, ferritin binding to PCBP3 and PCBP3.2 is shown. T-Rex 293 cells containing stably integrated copies of FLAG-PCBP3, FLAG-PCBP3.2, and FLAG-PCBP4 were treated with doxycycline and iron as in Fig. 3 and subjected to immunoprecipitation with anti-ferritin antibody. Immune complexes were analyzed by Western blotting with anti-FLAG (IB:FLAG) and anti-ferritin (IB:Ferritin) antibodies. B, PCBPs were expressed at similar levels. Whole cell lysates from A were subjected to Western blot analysis as indicated. C, co-immunoprecipitation of PCBP1 with PCBP2, -3, and -4 is shown. Wild-type yeast were transformed with pPCBP1 (pP1) or the corresponding empty vector and amino-terminal FLAG-tagged PCBP2 (F-P2), PCBP3 (F-P3), PCBP4 (F-P4), or the empty pYES2 vector (EV) and pGEV. Transformants were grown on SC−Ura−Leu−His medium to early log phase, then FLAG-PCBP expression was induced with β-estradiol for 4 h. Lysates were subjected to immunoprecipitation with anti-FLAG antibody, and immune complexes were analyzed by Western blotting (IB). D, shown is equal expression of PCBP1 in all transformants. Whole cell lysates from E were subjected to Western blotting with anti-PCBP1 antibody.
Binding of PCBP2, PCBP3, and PCBP4 to PCBP1 in Yeast
Previously, we demonstrated that PCBP2 could be detected in immunoprecipitates of PCBP1 (7). To determine whether PCBP3 and PCBP4 could also interact with PCBP1, we expressed PCBP1 and FLAG-tagged versions of PCBP2, PCBP3, and PCBP4 in yeast cells and examined the co-precipitation of PCBP1 in anti-FLAG immunoprecipitates (Fig. 11C). PCBP1 was detected in immune complexes from each of the FLAG-PCBP-transformed strains but not in yeast transformed with the empty vector that expressed no FLAG-PCBP and not in yeast that were not transformed with the pPCBP1 plasmid. All yeast transformed with pPCBP1 expressed PCBP1 at similar levels (Fig. 11D). Thus, PCBP3 and PCBP4 appeared to be similar to PCBP2 in their capacity to interact and form a complex with PCBP1.
DISCUSSION
PCBP2 Is an Iron Chaperone for Ferritin
Recent studies have only begun to illuminate the roles of PCBP family members in the delivery of iron to target proteins in the cell. Previously, PCBP1 was shown to act as an iron chaperone for ferritin in both yeast and mammalian cells. Here we have presented evidence that PCBP2 is similar to PCBP1 in its capacity to 1) activate the iron deficiency response, 2) genetically interact with H ferritin, but not L ferritin, and 3) enhance the incorporation of iron into ferritin when both PCBP2 and ferritin are heterologously expressed in yeast. Similar to PCBP1, PCBP2 was required for efficient delivery of iron to ferritin in human cells, as depletion of PCBP2 by siRNA in Huh7 cells led to impaired incorporation of iron into ferritin. Both PCBP1 and PCBP2 were co-immunoprecipitated with ferritin in iron-treated HEK293 cells, and expression of both PCBP1 and PCBP2 was required for the formation of a stable PCBP-ferritin complex in cells. These studies suggest that, rather than working independently as iron chaperones, PCBP1 and PCBP2 form a hetero-oligomeric complex with ferritin for iron delivery. These studies also highlight the utility of heterologous expression in budding yeast for the functional analysis of mammalian iron proteins.
ITC studies confirmed that iron-loaded PCBP1 and PCBP2 bound to ferritin in vitro in multiple mol equivalents with high nanomolar affinity, far higher that the binding affinity of Fe(II) alone to ferritin and far higher than the affinity of apoPCBP for ferritin. The higher binding affinity of Fe-PCBP for ferritin (compared with iron alone) offers a thermodynamic explanation for the observation that the iron chaperones increase the efficiency of iron loading into ferritin. The finding that multiple molar equivalents of Fe-PCBP can bind to ferritin mechanistically supports a model in which iron is transferred through a direct protein-protein interaction that occurs at the pores formed between ferritin subunits. Fe-PCBP1 exhibited a stoichiometry of binding of 9.5 ± 0.9 Fe-PCBP1 per ferritin oligomer. Iron enters the ferritin core through pores formed along the 3-fold axis of symmetry, of which there are 8 in the ferritin polymer. Thus the ITC data support a model in which PCBP1 interacts with ferritin at the pores where iron enters.
The studies presented here and previously published data on the capacity of PCBP1 and PCBP2 to deliver iron to PHDs suggest that the two chaperones also function cooperatively to deliver iron to PHD. Defects in the delivery of iron to PHDs are observed in mammalian cells only when levels of PCBP1 or PCBP2 are severely depleted. This observation suggests the unaffected PCBP paralog in siRNA studies cannot rescue the activities of the depleted paralog even when present in abundance. This was demonstrated directly; when purified, recombinant PCBP1 was shown to restore activity to PHDs in lysates from cells lacking PCBP1 but not from cells lacking PCBP2 (7). Both PCBP1 and PCBP2 participate independently in other cellular processes and seem to function as adaptor proteins that mediate the interactions of single-stranded RNA and DNA with other protein targets. Some of these interactions may be affected by iron binding, as exogenous iron chelators can enhance the interactions of PCBP2 with dicer and increase the efficiency of microRNA processing (26).
PCBP3 and PCBP4 Also Have Iron Chaperone Activity
The data presented here indicated that in yeast PCBP3 behaved identically to PCBP1 and PCBP2. Data from human cells indicated that exogenously expressed PCBP3 co-immunoprecipitated with ferritin, and preliminary data suggest that purified recombinant PCBP3 also binds iron in vitro (data not shown). Thus, PCBP3 could potentially function as an iron chaperone in human cells. mRNA transcripts for PCBP3 have been detected at very low levels in many tissues (27). Exogenously expressed PCBP3 can bind to the promoter and repress the expression of the μ-opioid receptor gene in mouse neuronal cells, and it has been shown to bind a poly-C oligonucleotide (16, 17), but beyond these observations little is known about the functions of PCBP3 in mammals. We observed that the protein derived from the alternatively spliced PCBP3.2 had reduced activity in yeast, although it co-immunoprecipitated with ferritin in human cells and bound to poly-C oligonucleotide. These observations suggest that the alternatively spliced forms of PCBP paralogs may be functionally different and that the sequences between KH domains two and three have a role in iron binding or delivery. Further studies will be required to determine whether PCBP3.2 differs in its capacity to bind iron or whether an analogous mutation in PCBP1 or PCBP2 has functional consequences.
PCBP4 behaved differently from PCBP1, PCBP2, and PCBP3 in our yeast assays. The strong activation of the iron deficiency response suggests that PCBP4 expression lowers the pool of cytosolic iron sensed by Aft1, likely due to iron binding and sequestration by PCBP4. The mitigation of the toxic effects of iron in the ccc1Δ strain expressing PCBP4 supports the concept that PCBP4 can bind and sequester iron and that it is functionally different from the other PCBPs. Our data suggest that PCBP4 does not efficiently deliver iron to ferritin. First, evidence for a genetic interaction was variable. In the plate assay of Aft1 activation, performed in the W303a background, no interaction with ferritin was apparent. In contrast, direct measurements of Aft1 target mRNAs, performed in the YPH499 background, indicated activation was ferritin-dependent. The variable genetic interaction with ferritin and the relatively lower ferritin iron loading activity suggests that although PCBP4 may bind iron, it may not efficiently deliver it to ferritin. This reduced interaction with ferritin is supported by the observation in human cells that PCBP4-ferritin interactions were not detectable when PCBP4 was exogenously expressed at moderate levels. PCBP4 may have a preference for delivering iron to target enzymes other than ferritin, or it could make iron available to other PCBPs, as it is found complexed with PCBP1 when co-expressed in yeast cells. PCBP4 is expressed at significantly elevated levels in neuronal tissues and in embryonic stem cells, suggesting a role in neuronal development (27). Whether this role involves a specialized function in iron delivery remains to be determined, but brain tissue has a marked sensitivity to perturbations in iron homeostasis and may require slightly different systems for iron uptake and delivery (28).
Acknowledgment
We thank David Eide for generously sharing plasmids.
This work was supported in part by the Intramural Research Program of the NIDDK, National Institutes of Health.
S. Leidgens, K. Z. Bullough, H. Shi, F. Li, M. Shakoury-Elizeh, T. Yabe, P. Subramanian, E. Hsu, N. Natarajan, A. Nandal, T. L. Stemmler, and C. C. Philpott, manuscript in preparation.
- PCBP1
- poly (C)-binding protein 1
- PHD
- prolyl hydroxylase
- KH
- K-homology
- hnRNP
- heterogeneous nuclear ribonucleoprotein
- SC
- synthetic complete medium
- ITC
- isothermal titration calorimetry
- FMR1
- fragile X mental retardation 1.
REFERENCES
- 1. Waldron K. J., Rutherford J. C., Ford D., Robinson N. J. (2009) Metalloproteins and metal sensing. Nature 460, 823–830 [DOI] [PubMed] [Google Scholar]
- 2. Outten C. E., O'Halloran T. V. (2001) Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science 292, 2488–2492 [DOI] [PubMed] [Google Scholar]
- 3. Rosenzweig A. C. (2002) Metallochaperones. Bind and deliver. Chem. Biol. 9, 673–677 [DOI] [PubMed] [Google Scholar]
- 4. Shi H., Bencze K. Z., Stemmler T. L., Philpott C. C. (2008) A cytosolic iron chaperone that delivers iron to ferritin. Science 320, 1207–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hintze K. J., Theil E. C. (2006) Cellular regulation and molecular interactions of the ferritins. Cell. Mol. Life Sci. 63, 591–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Arosio P., Levi S. (2010) Cytosolic and mitochondrial ferritins in the regulation of cellular iron homeostasis and oxidative damage. Biochim. Biophys. Acta 1800, 783–792 [DOI] [PubMed] [Google Scholar]
- 7. Nandal A., Ruiz J. C., Subramanian P., Ghimire-Rijal S., Sinnamon R. A., Stemmler T. L., Bruick R. K., Philpott C. C. (2011) Activation of the HIF prolyl hydroxylase by the iron chaperones PCBP1 and PCBP2. Cell Metab. 14, 647–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kaelin W. G., Jr., Ratcliffe P. J. (2008) Oxygen sensing by metazoans. The central role of the HIF hydroxylase pathway. Mol. Cell 30, 393–402 [DOI] [PubMed] [Google Scholar]
- 9. Loenarz C., Schofield C. J. (2008) Expanding chemical biology of 2-oxoglutarate oxygenases. Nat. Chem. Biol. 4, 152–156 [DOI] [PubMed] [Google Scholar]
- 10. Ozer A., Bruick R. K. (2007) Non-heme dioxygenases. Cellular sensors and regulators jelly rolled into one? Nat. Chem. Biol. 3, 144–153 [DOI] [PubMed] [Google Scholar]
- 11. Chaudhury A., Chander P., Howe P. H. (2010) Heterogeneous nuclear ribonucleoproteins (hnRNPs) in cellular processes. Focus on hnRNP E1 multifunctional regulatory roles. RNA 16, 1449–1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Makeyev A. V., Liebhaber S. A. (2002) The poly(C)-binding proteins. A multiplicity of functions and a search for mechanisms. RNA 8, 265–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ostareck-Lederer A., Ostareck D. H. (2004) Control of mRNA translation and stability in haematopoietic cells. The function of hnRNPs K and E1/E2. Biol. Cell 96, 407–411 [DOI] [PubMed] [Google Scholar]
- 14. Scoumanne A., Cho S. J., Zhang J., Chen X. (2011) The cyclin-dependent kinase inhibitor p21 is regulated by RNA-binding protein PCBP4 via mRNA stability. Nucleic Acids Res. 39, 213–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhu J., Chen X. (2000) MCG10, a novel p53 target gene that encodes a KH domain RNA-binding protein, is capable of inducing apoptosis and cell cycle arrest in G2-M. Mol. Cell. Biol. 20, 5602–5618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Choi H. S., Kim C. S., Hwang C. K., Song K. Y., Law P. Y., Wei L. N., Loh H. H. (2007) Novel function of the poly(C)-binding protein αCP3 as a transcriptional repressor of the μ-opioid receptor gene. FASEB J. 21, 3963–3973 [DOI] [PubMed] [Google Scholar]
- 17. Kang D. H., Song K. Y., Choi H. S., Law P. Y., Wei L. N., Loh H. H. (2012) Novel dual binding function of a poly(C)-binding protein 3, transcriptional factor which binds the double-strand and single-stranded DNA sequence. Gene 501, 33–38 [DOI] [PubMed] [Google Scholar]
- 18. Sherman F. (1991) in Guide to Yeast Genetics and Molecular Biology (Guthrie C., Fink G., eds) pp. 3–20, Academic Press, New York [Google Scholar]
- 19. Gao C. Y., Pinkham J. L. (2000) Tightly regulated, β-estradiol dose-dependent expression system for yeast. Biotechniques 29, 1226–1231 [DOI] [PubMed] [Google Scholar]
- 20. Protchenko O., Shakoury-Elizeh M., Keane P., Storey J., Androphy R., Philpott C. C. (2008) Role of PUG1 in inducible porphyrin and heme transport in Saccharomyces cerevisiae. Eukaryot. Cell 7, 859–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chkheidze A. N., Lyakhov D. L., Makeyev A. V., Morales J., Kong J., Liebhaber S. A. (1999) Assembly of the α-globin mRNA stability complex reflects binary interaction between the pyrimidine-rich 3′-untranslated region determinant and poly(C)-binding protein αCP. Mol. Cell. Biol. 19, 4572–4581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bou-Abdallah F., Arosio P., Santambrogio P., Yang X., Janus-Chandler C., Chasteen N. D. (2002) Ferrous ion binding to recombinant human H-chain ferritin. An isothermal titration calorimetry study. Biochemistry 41, 11184–11191 [DOI] [PubMed] [Google Scholar]
- 23. Valverde R., Edwards L., Regan L. (2008) Structure and function of KH domains. FEBS J. 275, 2712–2726 [DOI] [PubMed] [Google Scholar]
- 24. Shakoury-Elizeh M., Tiedeman J., Rashford J., Ferea T., Demeter J., Garcia E., Rolfes R., Brown P. O., Botstein D., Philpott C. C. (2004) Transcriptional Remodeling in response to iron deprivation in Saccharomyces cerevisiae. Mol. Biol. Cell 15, 1233–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li L., Chen O. S., McVey Ward D., Kaplan J. (2001) CCC1 is a transporter that mediates vacuolar iron storage in yeast. J. Biol. Chem. 276, 29515–29519 [DOI] [PubMed] [Google Scholar]
- 26. Li Y., Lin L., Li Z., Ye X., Xiong K., Aryal B., Xu Z., Paroo Z., Liu Q., He C., Jin P. (2012) Iron homeostasis regulates the activity of the microRNA pathway through poly(C)-binding protein 2. Cell Metab. 15, 895–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Su A. I., Wiltshire T., Batalov S., Lapp H., Ching K. A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Cooke M. P., Walker J. R., Hogenesch J. B. (2004) A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. U.S.A. 101, 6062–6067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rouault T. A., Cooperman S. (2006) Brain iron metabolism. Semin. Pediatr. Neurol. 13, 142–148 [DOI] [PubMed] [Google Scholar]