Background: Heterochromatin condenses in the middle of rod cell nuclei during retina maturation.
Results: The level of linker histone H1c increases during retina maturation. Rod photoreceptors in triple H1 knock-out mice have less compact chromatin.
Conclusion: H1c is a key architectural factor for chromatin condensation in the rod photoreceptor.
Significance: Histone H1c expression may be genetically modified to promote rod photoreceptor maturation and retina integrity.
Keywords: Development, Heterochromatin, Histones, Photoreceptors, Retina, Histone H1c, Retina Development, Rod Photoreceptor
Abstract
Mature rod photoreceptor cells contain very small nuclei with tightly condensed heterochromatin. We observed that during mouse rod maturation, the nucleosomal repeat length increases from 190 bp at postnatal day 1 to 206 bp in the adult retina. At the same time, the total level of linker histone H1 increased reaching the ratio of 1.3 molecules of total H1 per nucleosome, mostly via a dramatic increase in H1c. Genetic elimination of the histone H1c gene is functionally compensated by other histone variants. However, retinas in H1c/H1e/H10 triple knock-outs have photoreceptors with bigger nuclei, decreased heterochromatin area, and notable morphological changes suggesting that the process of chromatin condensation and rod cell structural integrity are partly impaired. In triple knock-outs, nuclear chromatin exposed several epigenetic histone modification marks masked in the wild type chromatin. Dramatic changes in exposure of a repressive chromatin mark, H3K9me2, indicate that during development linker histone plays a role in establishing the facultative heterochromatin territory and architecture in the nucleus. During retina development, the H1c gene and its promoter acquired epigenetic patterns typical of rod-specific genes. Our data suggest that histone H1c gene expression is developmentally up-regulated to promote facultative heterochromatin in mature rod photoreceptors.
Introduction
During mammalian development, cells committed to differentiate into a specific tissue withdraw from the cell cycle and start a process of tissue maturation that involves up-regulation of genes specific for the differentiated tissue and down-regulation of genes specific for progenitor cells (1, 2). This developmentally regulated process culminates in formation of compact blocks of heterochromatin inside the nuclei and spatial segregation of heterochromatin and euchromatin domains (3). Chromatin condensation prevents certain transcription factors from activating genes in the compact chromatin (4). Recent studies suggest that each tissue type has its own unique program that orchestrates tissue maturation, with specific sets of chromatin architectural and remodeling proteins, histone modifications, and micro-RNA (5–12).
The mammalian retina is a part of the CNS and is derived from the optic cup, an outgrowth of forebrain neuroepithelium formed early in CNS development. Six major neuronal and one glial cell types are generated from the multipotential retinal progenitors in a stereotypic temporal sequence. In mice the terminal differentiation of retinal cells spans from embryonic day 12 to approximately postnatal day 7. Around 80% of the cells in adult mouse retina are photoreceptors, and rod photoreceptors outnumber cone photoreceptors more than 30-fold (13). Mouse rods have compact heterochromatin and its nuclear architecture may be particularly adapted to nocturnal vision (14). This heterochromatin is accumulated in the middle of the small rod cell nucleus (D ∼4–5 μm), and spans over 70% of the nuclear diameter, pushing euchromatin out to a peripheral ring (14, 15). Spinocerebellar ataxia type 7 (SCA7),4 a neurodegenerative disease caused by polyglutamine expansion in ataxin 7, a component of the chromatin-remodeling complex, also causes decondensation of rod photoreceptor chromatin and leads to visual impairment and blindness (16).
Although heterochromatin reorganization and chromatin condensation in terminally differentiated cells are conserved features of vertebrate tissues, its exact mechanism and factors mediating chromatin condensation are largely unknown. One of the most basic elements of chromatin structure that varies among cell types is the average distance between nucleosomes or the nucleosomal repeat length. Among various eukaryotic cells and tissues, this length varies from the shortest 155-bp nucleosomal repeat length in fission yeast (17) to the longest ∼240-bp nucleosomal repeat length in echinoderm sperm (18). Within the mammalian CNS, chromatin in different neuronal cells has a wide range of nucleosomal repeat length, from 160 bp in cortical neurons to 201 bp in cortical glia and 215 bp in cerebellar granule cells (19–22). Previous studies showed that accumulation of developmentally regulated architectural proteins such as linker histone H5 (23) in chicken blood cells and HP1 in differentiating muscle cells (24) are involved in developmentally regulated chromatin reorganization. For other maturing blood cells, such as mouse lymphocytes and erythroblasts, our previous studies suggested that specific sets of histone modifications, in particular histone H3K9 dimethylation (H3K9me2) and histone H3 and H4 acetylation, rather than any specific protein, orchestrate chromatin condensation and spatial rearrangement in the nuclei of the mature cells (7, 25). Remarkably, H3K9me2 has been shown in other studies to be associated with HP1 (26) and linker histones (27, 28) suggesting that this histone modification is “read” by architectural proteins that induce chromatin structural transitions.
In this study we show that developing rod photoreceptor cells undergo a dramatic increase in the nucleosomal repeat length from 190 bp in postnatal day 1 (PN1) to 206 bp in adult retina. Consistent with this, the amount of linker histone H1 is increased during rod maturation. Both biochemical and genetic experiments indicate that H1c is a key linker histone variant regulating tight condensation of chromatin in rod photoreceptors and helping maintain structural integrity of these cells.
EXPERIMENTAL PROCEDURES
Mice
All animal breeding and experiments were conducted in accordance with NIH guidelines and approved by the Animal Care and Use Committee of the Pennsylvania State University School of Medicine and Georgia Tech Institutional Animal Care and Use Committee. C57BL/6j and rd1/rd1-Pde6b-RD1 mice (29) were purchased from The Jackson Laboratory (Bar Harbor, ME). Single (H1c), double (H1c/H1e), and triple (H1c/H1e/H10) linker histone knock-out mice have been extensively characterized previously (30–34). These mutations are in a C57BL/6j background. For electron microscopy study two H1c/H1e/H10 KO (knock-out) mice (4 eyes) and one H1c/H1e KO mouse (1 eye) were used. For the immunohistochemistry study three H1c/H1e/H10 KO mice (6 eyes) and two H1c/H1e KO mice (4 eyes) were used.
Antibodies and Reagents
Chemicals were purchased from Fisher Scientific (Pittsburgh, PA), unless otherwise noted. Anti-H3K4me2 (07–030), anti-HP1α (05–689), anti-HP1γ (07–332), and anti-H3K27me3 (07–449) were from Millipore (Billerica, MA), anti-H3K9me2 (ab1220), anti-H1 (ab1938), and anti-H2AZ (ab4174) were from Abcam (Cambridge, MA), anti-HP1β (MAB3448) was from Chemicon.
Immunohistochemistry
Methods were as previously described (35). Antigen retrieval was achieved by boiling samples for 5 min in 10 mm citrate buffer, pH 6.0. Sections were labeled with primary antibodies and secondary antibodies conjugated with Alexa Fluor 488 or 594 (Invitrogen/Molecular Probes). Digital images were recorded using an Olympus FV1000 confocal microscope.
Electron Microscopy and Semi-thin Sections
Whole eyes were dissected from mice and fixed in 2.5% glutaraldehyde plus 2% paraformaldehyde in 0.08 m sodium cacodylate buffer, pH 7.3, containing 3% sucrose and 0.025% CaCl2 for 1 h at 4 °C. Eyes were opened through the iris and the lens was removed. Intact eye cups were fixed overnight in the same fixative by rotating at 4 °C. Eye cups were cut in half and then into peripheral, middle, and central regions of retina and stored in 0.08 m sodium cacodylate buffer containing 3% sucrose and 0.025% CaCl2. The tissues were postfixed in 1% OsO4 and 0.08 m sodium cacodylate buffer containing 3% sucrose and 0.025% CaCl2 for 2 h, dehydrated, and embedded in EMbed 812. Thin sections were stained with 2% uranyl acetate, followed by 0.4% lead citrate, and viewed with a Jeol 1400 electron microscope. Semi-thin sections were stained by a modified method (36) without fuchsin counterstaining. Hematoxylin and eosin for H&E staining were from Harleco/EMD (Gibbstown, NJ).
SDS-PAGE and Western Blotting
Isolated nuclei were dissolved in SDS-containing loading buffer and electrophoresis was carried out in 15% polyacrylamide SDS-containing gels (37). Proteins were transferred to Immuno-Blot PVDF membrane (Bio-Rad) and detected with primary and secondary HRP-conjugated antibodies as described (7). For semiquantitative analysis of relative protein levels in nuclear samples, the Coomassie-stained gels or autoradiographs after ECL (Thermo Scientific, Rockford, IL) detection were scanned and digitized and the intensity of protein bands was quantitated using OptiQuant version 03.00 (Packard Instrument Co, Meriden, CT) or ImageJ (NIH) software packages.
Cell and Nuclei Isolation from Retina
These were carried out according to Ref. 7. Eyes were collected on ice from animals at different developmental stages and dissected to isolate retinas. 10–20 retinas were collected in 1 ml of cold PBS and triturated by pipetting up and down with a pipetman with a 1-ml tip. The suspension of cells was briefly spun down at 500 × g for 3 min. For nuclei isolation cells were resuspended in 0.5% IGEPAL CA-630 (Sigma) in RSB (10 mm NaCl, 3 mm MgCl2, 10 mm HEPES, pH 7.5) plus 1 mm PMSF and a protease inhibitor mixture. Cells were vortexed several times during a 20-min incubation on ice and then centrifuged 7 min at 3500 × g. Nuclei were resuspended in RSB plus 1 mm PMSF and Halt Protease Inhibitors Mixture.
Nuclease Digestion
Micrococcal nuclease (Nuclease S7, Roche applied Science) was used for digestion of isolated retina nuclei as described (7, 19, 20, 34). Nuclease at a concentration of 2.5 units/ml was added to an aliquot of nuclei preparation containing 1 mg/ml of DNA in RSB, 0.5 m PMSF, 1 mm CaCl2. The reaction was carried out at 37 °C for various time intervals and terminated by adding EDTA to a final concentration of 10 mm. The resulting chromatin was treated with 1% SDS and 50 μg/ml of Proteinase K for 1 h at 55 °C. DNA was purified by 2× phenol-chloroform extraction and ethanol precipitation and subjected to electrophoresis in 1% agarose in Tris acetate/EDTA buffer with constant buffer recirculation using a 1-kb DNA ladder (Invitrogen) as size marker. After ethidium bromide staining, gels were recorded digitally and band detection and DNA length assignment (on basis of the positions of marker bands) was used for data analysis and graphing; nucleosome repeat length = length of DNA fragment/number of nucleosomes in fragment. Accuracy of the calculated distance between nucleosomes or nucleosome repeat length is increased with an increase of the number of nucleosomes in the DNA fragment.
Histone Isolation
Histone isolation was carried out using a H2SO4 acid extraction method for preparation of total histone (31).
Histone Preparation and Reverse-phase High-performance Liquid Chromatography (HPLC) Analysis
Retinas were dissected and histones were extracted as previously described (31). Histones were dissolved in ddH2O and HPLC was performed using System Gold from Beckman Coulter with a 168NM Detector on a Vydac HPLC C18 column, or Äkta purifier UPC 10 as described (7, 38, 39).
Statistical Analysis
Student's t test for two samples assuming equal variances was performed with Excel Data Analysis (Microsoft Excel 2007) or GraphPad Prism4 software.
RESULTS
Heterochromatin Accumulation in the Rod Nuclei during Mouse Retina Development
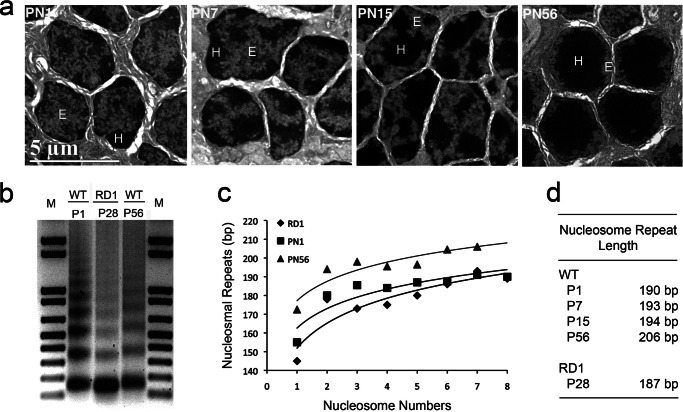
Nuclei of mouse adult rod photoreceptors contain abundant heterochromatin. To visualize mouse rod photoreceptor maturation and heterochromatin formation during postnatal development we used electron microscopy of ultrathin cell sections (Fig. 1a). At postnatal day 1 (PN1), electron dense heterochromatin were seen as several foci located mostly at the nuclear periphery and there was no sharp border between the dark heterochromatin and less electron dense euchromatin. During rod maturation the amount of heterochromatin started to increase around PN7 as the rod outer segments developed. Heterochromatin also began to relocate toward the center of the rod nuclei and the heterochromatic foci started to fuse together at PN15, so that in adult eyes (PN56) heterochromatin occupied most of the nuclear volume pushing euchromatin out to the nuclear periphery (Fig. 1a). The accumulation of heterochromatin in rod nuclei has been well described at the light microscopic level (14). Consistent with previous reports (14, 15), we measured the total nuclear and heterochromatin diameters to be 4.47 μm (S.D. = 0.27 μm) and 3.46 μm, respectively, so that heterochromatin occupies ∼46% of the volume of the mature rod nuclei. The ratio between the area taken up by heterochromatin and the nuclear area is 0.6 for PN56; whereas at PN1 the nuclear diameter is bigger, 6.33 μm, but the nuclear/heterochromatin area ratio is only 0.18 (Fig. 1a).
FIGURE 1.
Mouse rod photoreceptor development is accompanied by heterochromatin accumulation and increasing nucleosomal repeat length. a, electron microscopy of mouse rod photoreceptor nuclei at PN1, 7, 15, and 56. Scale bar is 5 μm. E, euchromatin; H, heterochromatin area inside the nucleus. b–d, nucleosomal spacing is increased during development. b, agarose gel electrophoresis of DNA isolated from nuclei of wild type mouse retina at PN1 (lane P1) and PN56 (lane P56) and RD1 mutant retina (lane RD1). Isolated nuclei were treated with 2.5 units/ml of micrococcal nuclease for 6 min. Lanes M show DNA molecular size markers. Chromatin from PN56 mouse retina migrates slower than chromatin from the early stage of development. c, median sizes of the micrococcal nuclease digests for wild type retina at PN1 (squares) and PN56 (triangles) and for RD1 retina (diamonds), were calculated from densitometry of the gel, divided by the number of nucleosomes in DNA fragments (b) and plotted against nucleosome number. Accuracy of the calculated nucleosome repeat length is increased with an increase of the number of nucleosomes in the DNA fragment. d, nucleosomal repeat length for mouse wild type retina at different developmental stages in comparison to RD1 mutant retina (S.D. 2 bp).
Chromatin Condensation in Rods Is Accompanied by Increase of the Nucleosome Repeat Length
For several mature cell types, such as chicken erythrocytes or mouse cerebellar granule cells, accumulation of heterochromatin during maturation is coincident with an increase in nucleosomal repeat length (22, 23, 40). To analyze nucleosomal repeat length in rod photoreceptors we performed micrococcal nuclease digestion on isolated retinal nuclei from different developmental stages (Fig. 1, b–d). Another published method to plot the data (34) and to calculate nucleosome repeat length gave the same results (data not shown). By measuring the MNase-digested DNA fragment length divided by the nucleosome number we calculated that the size of the nucleosome repeat increased from 190 bp at PN1 retina to 193 bp at PN7 (by which stage essentially all cells have become post-mitotic) to 206 bp at PN56 (adult) (Fig. 1, c and d).
To determine whether this increase was specific to rod photoreceptors, we used samples from RD1 mutant retinas as control. After PN28, retinas of these mutant mice lack the outer nuclear layer of rods and cones and are composed of mostly bipolar, amacrine, and other cells from the inner nuclear and ganglion cell layers (29). The size of the nucleosomal repeat length in RD1 retina was 187 bp (Fig. 1, b–d), which was even less than the nucleosomal repeat length for PN1 retina and indicates that the 206-bp nucleosomal repeat length is characteristic of rod photoreceptors but not the cell of the inner nuclear and ganglion cell layers. Thus, we observed that a significant increase in nucleosomal repeat length accompanies the global chromatin condensation in mouse rod photoreceptors, indicating a dramatic change in nucleosome positioning during development.
Specific Enrichment of Linker Histone Subtype H1c in Rod Photoreceptors Is Associated with Chromatin Condensation
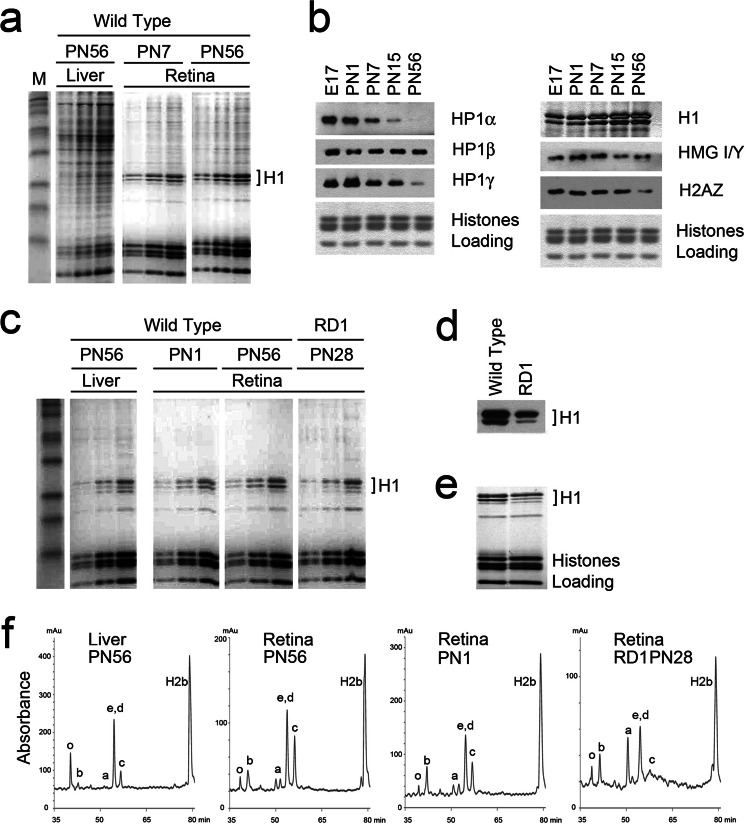
The progressive chromatin condensation during terminal rod differentiation raised the question of which developmentally regulated factors are responsible for these changes. To compare changes in nuclear protein composition during retina development, we first isolated total proteins from nuclei of mouse adult retinas and compared them with nuclear proteins from mouse adult liver by Laemmli PAGE showing prominent core and linker histones (Fig. 2a). When compared with mouse liver, retina nuclei appear to contain much less nonhistone and linker histone protein proteins.
FIGURE 2.
Nuclear protein composition in differentiating mouse retina. a, total protein from nuclei analyzed by SDS-PAGE and stained with Coomassie R-250. Each sample was loaded on the gel in 1, 2, and 4 times the amount. b, Western blots of nuclear samples isolated from mouse retina during development and probed with antibodies against HP1α, β, γ, H1, H2AZ, and HMG I/Y. Coomassie staining of core histones was used as loading control. c, SDS-PAGE of acid-extracted histones from wild type liver PN56, wild type retina at PN1 and PN56, and mutant RD1 retina. Each sample was loaded on the gel in 1, 2, and 4 times the amounts. d, Western blot of histone proteins from wild type retina (PN56) and RD1 mutant (RD1) probed with antibody against H1 histone. e, Coomasie staining of the same samples shows equal loading of core histones but a different composition of linker histones H1. f, reverse-phase HPLC chromatography of isolated histones eluted with a linear gradient of acetonitrile (7, 34, 35) shows that the amount of linker histone H1c is increased during development. The abscissa represents the elution time and the ordinate represents absorbance at 214 nm. The identity of the histone subtypes in each peak is indicated.
We next compared the levels of linker histones and several other heterochromatin architectural proteins at different developmental stages using Western blotting (Fig. 2b). We found that heterochromatin proteins 1 (HP1) α and γ were dramatically decreased during rod maturation and almost disappeared in adult retina, consistent with previous findings for blood cell maturation (7, 41, 42). Also, there was notably less HP1β in adult tissues than in embryos (Fig. 2b, left panels). Levels of two other chromatin architectural proteins, H2AZ and HMG I/Y, associated with heterochromatin in mature trophoblasts (43) and senescent fibroblasts (44), respectively, also decreased during retina maturation (Fig. 2b, bottom right panels). The amount of linker histones gradually increased during development (Fig. 2b, upper right panel). Thus, among the heterochromatin proteins tested, only the linker histone H1 level increased during retina maturation.
Comparison of acid-extracted histones in PN1 with adult retina (PN56) showed an increased amount of total H1 in PN56, and comparison of wild-type retina with RD1 or liver showed an increased total amount of H1 in the wild type retina, as well as a different composition of H1 subtypes (Fig. 2, c–e). To compare linker histone levels in retina with previously published data for liver, we examined stoichiometry of H1 per nucleosome in these 2 types of tissues by quantitative densitometry of the Coomassie staining. We used data from Ref. 40 that liver nuclei have 0.7 H1 per nucleosome as a reference, and calculated that mouse adult retina has 1.3 molecules of total H1 per nucleosome.
To analyze which particular subtype(s) of linker histone H1 are specifically expressed during mouse retina differentiation we performed HPLC fractionation of purified histones (31) isolated from retinas of PN1 and PN56 animals, mutant RD1 retina, and wild type liver (Fig. 2f). HPLC separation profiles show that whereas adult liver nuclei have mostly H1e, H1d, and H10 subtypes (Fig. 2f, see also in Ref. 30), in retinal chromatin the linker histone subtype H1c is the one most increased during development. When normalized to H2b, H1c was increased by ∼1.8 times between PN1 and PN56 (Table 1). We used HPLC data to calculate the combined amount of all histones H1 per nucleosome in PN56 retina (Table 1) and the resulting 1.25-fold increase is in good agreement with a 1.3-fold increase shown by quantitative gel densitometry. Histone H10 is underrepresented in all retina samples (PN1, PN56, and RD1) when compared with liver, whereas subtypes H1a and H1b were enriched in all retina samples, especially in RD1 (Fig. 2f).
TABLE 1.
Normalized amount of linker histones in retina
| Linker histone | Genotype/stage |
Ratio P56/P1 | ||
|---|---|---|---|---|
| H1c KO(P56) | PN1 | PN56 | ||
| H2b | 1 | 1 | 1 | |
| H1c | 0 | 0.21 | 0.37 | 1.76 |
| H1d/e | 0.73 | 0.42 | 0.55 | 1.31 |
| H1a | 0.06 | 0.13 | 0.11 | 0.85 |
| H1b | 0.08 | 0.2 | 0.11 | 0.55 |
| H1o | 0.12 | 0.06 | 0.11 | 1.83 |
| Total H1 | 0.99 | 1.02 | 1.25 | 1.23 |
Thus, the subtype composition changes of linker histones during rod development, in parallel with nucleosomal repeat length increase, strikingly resemble the process first observed with maturing chicken erythrocytes (45). However, in contrast to chicken erythrocytes, there is a pronounced increase in H1c, rather than H10 (the mouse homolog of H5), suggesting that the ability to condense chromatin during cell differentiation is not unique for the H10/H5 histone subtype.
Reduction of the H1 Level in H1c Knock-out Retina Is Compensated by Other Linker Histones
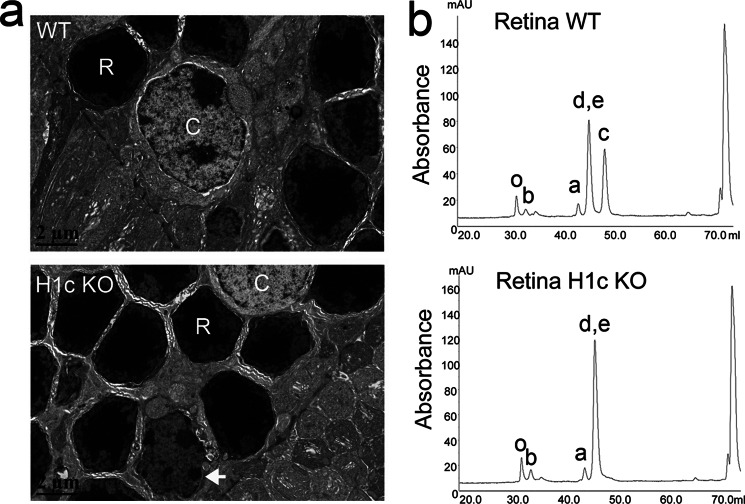
Because we found that the level of linker histone H1c correlates with chromatin condensation during rod development, and this process is accompanied by a dramatic increase in nucleosomal repeat length, we asked if rod development is affected in H1c knock-out mice. We first looked at Hematoxylin and Eosin (H&E) general staining of cryosections and paraffin sections of eyes from H1c KO mice and found no obvious anatomical or cellular abnormalities in the eyes of these mice (data not shown). Electron microscopy of ultrathin cell sections shows a rare event of poor nuclear condensation in occasional rod nuclei of H1c KO mice (white arrow in Fig. 3a), but overall retinal ultrastructure was not perturbed in knock-out animals.
FIGURE 3.
Linker histone level in H1c knock-out retina is compensated by other linker histones. a, electron micrograph of the outer nuclear layer in wild type (top) and H1c KO (bottom) mice retina. Note the difference in the nuclear structure of rod (R) and cone (C) photoreceptors. b, reverse-phase HPLC chromatography of isolated histones from wild type and H1c KO mice retina. The abscissa represents the linear gradient of acetonitrile elution time and the ordinate represents absorbence at 214 nm. The identity of the histone subtypes in each peak is indicated.
To analyze the amount of remaining linker histones in retina of H1c KO mice we compared HPLC profiles of histones in wild type and H1c KO adult retina. As shown in Fig. 3b, in addition to the absence of H1c there was an increase in the H1d/e peak in H1c KO retinas suggesting compensation of H1c by H1d or H1e. Nevertheless, the total level of linker histone in H1c KO is reduced to the level of linker histones at PN1 (Table 1).
Retinal Tissue Morphology in H1 Triple and Double Knock-out Mice
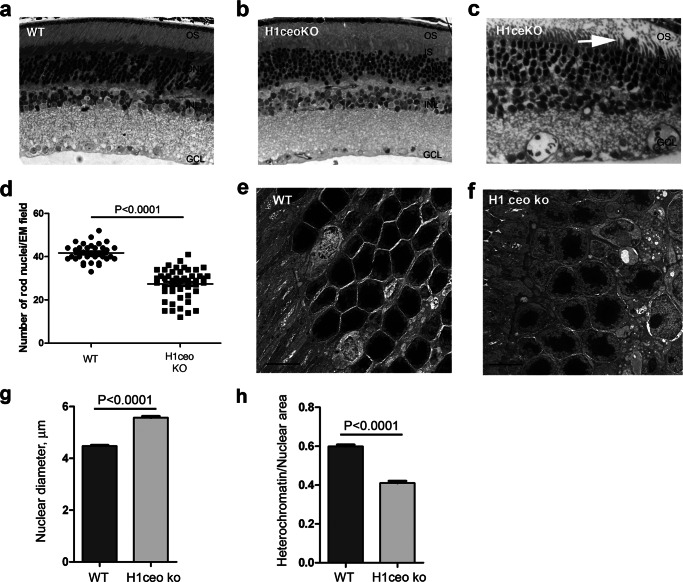
Triple H1-null mice lacking H1c, H1e, and H10 subtypes of linker histone (H1c/H1e/H10 KO) had a reduced H1 content in thymus, liver, and spleen, a moderate (less than 5 bp) decrease in nucleosomal repeat length, but no changes in nuclear size (30, 32). Because linker histone H1c is compensated by other linker subtypes in retina of single knock-out H1c mice, most probably by H1e, we asked if rod development and chromatin condensation would be affected in rod photoreceptor nuclei of double H1c/H1e and H1c/H1e/H10 triple KO mice. As shown in Fig. 4, a and b, the inner retinal structure appeared normal, but the outer nuclear layer contained more loosely packed rod photoreceptor nuclei in triple KO retina. The same was true for double KO retina (Fig. 4c). We used electron microscopy to quantify the number of rod photoreceptor nuclei in wild type and triple H1c/H1e/H10 KO mouse retina and showed that the outer nuclei layer of mutant retina has 30% fewer rod photoreceptor nuclei per unit area than retina of wild type mouse (Fig. 4d). This analysis does not distinguish whether the rod cell packing abnormalities are accumulated during development or through degeneration.
FIGURE 4.
Changes in the retina outer nuclear layer structure in H1 triple null mice. a–c, semi-thin sections of central mouse retina from wild type (a), H1c/H1e/H10 KO (b), and H1c/H1e KO (c). d, outer nuclei layer of triple H1c/H1e/H10 KO mouse retinas have fewer rod photoreceptor nuclei than retinas of wild type mice. Nuclei of rod photoreceptors were counted on EM sections from the middle portion of retina in wild type mice (34 EM fields) and triple KO mice (48 EM fields). e and f, electron micrographs of the outer nuclear layer in wild type (e) and H1c/H1e/H10 KO mice retina (f). Bar represents 5 μm. g and h, around 250 rod photoreceptor nuclei from the middle part of the 2 retinas for each genotype were measured and analyzed on the electron micrographs. Average maximum diameter (g) is larger in H1 triple null retina (p < 0.0001, Student's t test, two-sample assuming equal variances), but the ratio between heterochromatin area/nuclei area (h) is greater in wild type (WT) retinas (p < 0.0001, Student's t test, two-sample assuming equal variances).
When we compared the retinal ultrastructure by electron microscopy (Fig. 4, e and f) of wild type and H1c/H1e/H10-triple KO mice, we found that mutant photoreceptors had less compact heterochromatin. Although the average maximum diameter of the rod photoreceptor nuclei was increased in the H1c/H1e/H10 KO retina (Fig. 4g) the fraction of the nuclear area occupied by heterochromatin is decreased (Fig. 4h). This suggests that the process of chromatin condensation is partly impaired in mutant retina.
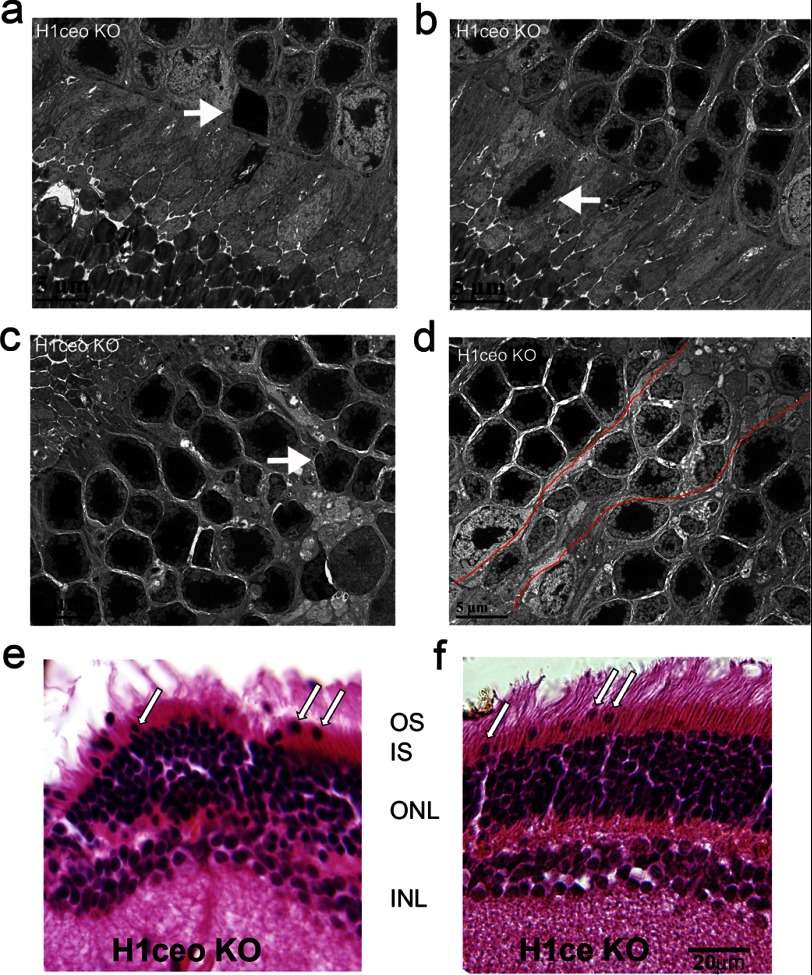
Additionally we found several abnormalities in the nuclear morphology of double and triple KO retinas (Figs. 4c and 5). Some of the rod nuclei were displaced out of the outer nuclear layer beyond the outer limiting membrane. These abnormalities were more pronounced in retina periphery, than in central and middle regions of H1c/H1e/H10 and H1c/H1e KO retinas (Figs. 4c and 5, b, e, and f, arrows).
FIGURE 5.
Specific changes in chromatin structure in the nuclei of rod photoreceptors in triple H1c/H1e/H10 KO and double H1c/H1e KO mice. a–d, nuclei of rod photoreceptors in H1c/H1e/H10 KO mice show structural abnormalities such as cell death (a, arrow), mislocation to the layer of rod outer segments (b, arrow), folding of nuclear membrane (c, arrow), and patches of nuclei of the rod with poorly condensed chromatin inside the outer nuclear layer (ONL) (d, nuclei inside red contour). e and f, H&E staining of retina from H1c/H1e/H10 KO (e) and H1c/H1e KO (f) with several mislocated from the ONL to OS rod photoreceptor nuclei (arrows).
Several rod photoreceptors had very electron dense nuclear material (Fig. 5a, arrow) characteristic of apoptotic cells and other nuclei showed an abnormal folded structure (Fig. 5c, arrow, and data not shown for H1c/H1e KO). Apoptotic events in H1c/H1e/H10 KO were rare; we found only 8 apoptotic cells per more than 1,000 scored. Nevertheless, because these were not observed at all in the wild type and single H1c knock-out retinas it suggests that further H1 knockouts could lead to more extensive apoptotic effects. The levels of chromatin condensation were variably altered in the H1c/H1e/H10 KO mouse; there were patches of nuclei with poor chromatin condensation adjacent to apparently normal rod nuclei with highly condensed heterochromatin (Fig. 5d, rod nuclei inside red lines). The level of chromatin condensation varied in double KO retina too; whereas some rod nuclei appeared normal others had very low chromatin condensation (data not shown). These phenotypic defects in rod photoreceptors of KO mice could be because of a generalized cellular pathology, but specific changes inside the rod nuclei suggest that the level of linker histone helps to maintain not only heterochromatin compaction inside the rod nuclei, but proper retina structure as well.
Nuclei of Rod Photoreceptors in H1c/H1e/H10 Knock-out Mice Have More Accessible Chromatin
Nuclei of mature mouse rod photoreceptors have a unique “inverted” heterochromatin architecture (14) in which combinations of histone modifications define distinct concentric heterochromatin and euchromatin zones (15). When we probed wild type and H1c/H1e/H10 KO retinas with antibodies against several histone modifications, we found that H1c/H1e/H10 KO retinas showed much brighter and more extensive immunofluorescence staining than wild type retina under identical labeling and image acquisition conditions (Fig. 6). We first stained retina cryosections with anti-H3K4me2, a known marker for euchromatin, and with anti-H3K9me2, a marker associated with facultative heterochromatin (Fig. 6a) and measured fluorescence intensity profiles across the equators of several nuclei for each genotype (Fig. 6b). In the wild type specimen, both antibodies weakly stained the nuclear periphery outside of the Hoechst-positive chromocenter. In the knock-out retina, not only did both marks show a higher intensity of immunofluorescence, but the H3K9me2 signal became dramatically stronger in the apocentric zone at the periphery of pericentric heterochromatin. Although stronger, the fluorescence shown by anti-H3K4me2 remained confined to the euchromatic region of the nucleus.
FIGURE 6.
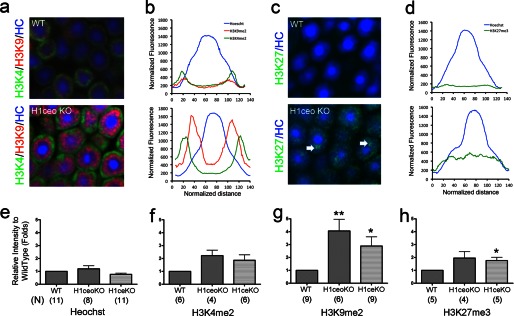
Nuclei of rod photoreceptors of H1 triple null mice with deleted H1c,- e, and -o linker histone genes acquired less compact chromatin conformation and exposed epigenetic histone modification marks masked in wild type nuclei. a, distribution of histone modifications in the nuclei of rod photoreceptor in wild type (top) and H1c/H1e/H10 KO (bottom) mice. Sections of retina tissue were stained with: anti-H3K9me2 (red) and anti-H3K4me2 (green). All sections were counterstained with Hoechst 33258 (blue). b and d, averaged and normalized intensity profiles for fluorescence of each specific antibody or Hoechst staining across the nuclear centers of rod photoreceptor nuclei in wild type (top) or H1c/H1e/H10 KO (bottom) retinas. c, distribution of histone modifications in the nuclei of rod photoreceptor wild type (top) and in H1c/H1e/H10 KO (bottom) mice. Sections of retina tissue were stained with: anti-H3K27me3 (green), and counterstained with Hoechst 33358 (blue). e–h, histograms show comparison of the average peak fluorescence intensity for immunofluorescence staining in wild type and H1c/H1e/H10 KO. Maximum fluorescence intensity for 20 or more nuclei profiles per slide for 4 or more samples of each genotype was measured and normalized to wild type samples for Hoechst 33258 (e), H3K4me2 (f), H3K9me2 (g), and H2K27me3 (h) staining. N, number of samples measured for each experimental condition. g, p value: *, <0.05; **, <0.02. h, p value: *, <0.05.
H3K27me3 is generally considered a mark of transcriptionally silenced chromatin. In wild type rod nuclei it shows weak punctuate staining at the apocentric zone around the Hoechst 33258-positive chromocenter (Fig. 6, c and d, upper panel, and Ref. 15). In the H1 knock-out retina, anti-H3K27 showed increased immunofluorescence and was redistributed all over the nuclear volume (Fig. 6, c and d, lower panel). On cryosections of H1c/H1e/H10 KO female mice (but not control female mice), we were able to observe Barr body staining (inactive X chromosome), a well known feature of facultative heterochromatin marked by H3K27me3 (Fig. 5c, arrows, lower panel).
We measured peak heights on fluorescence intensity profiles for more than 20 nuclei per slide for several samples of retinal cryosections from wild type and H1c/H1e/H10 KO (Fig. 6, e–h). Although Hoechst staining intensity did not significantly change in knock-out mice (Fig. 6e), all epigenetic marks showed much greater staining in mutant retinas (Fig. 6, f–h). The most dramatic change was the 3–4-fold increase in fluorescence intensity for H3K9me2 (p < 0.02) (Fig. 6g). Consistently, we observed similar phenotypes and accessibility of chromatin in H1c/H1e KO mice showing that H1e and H1c are two linker histone subtypes essential for chromatin condensation (Fig. 6, e–h).
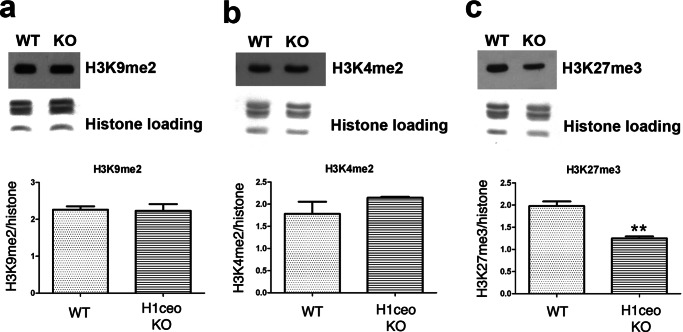
To distinguish whether increases in fluorescence intensity reflect differences in accessibility rather than differences in abundance of these histone modifications we performed Western blots (Fig. 7) and demonstrated that amounts of H3K9me2 (Fig. 7a) and H3K4me2 (Fig. 7b) histone modifications do not significantly change in the triple H1c/H1e/H10 KO retina in comparison with the wild type retina, and the H3K27me3 (Fig. 7c) modification was significantly decreased in triple knock-out retina. This supports our suggestion, that rod photoreceptor nuclei have more accessible chromatin.
FIGURE 7.
Amounts of H3K9me2 (a), H3K4me2 (b), and H3K27me3 (c) histone modifications do not increase in the triple H1c/H1e/H10 KO retina in comparison with wild type retina. Upper panels show Western blots of total protein samples isolated from the retina of triple H1c/H1e/H10 KO and wild type mice. Coomassie staining of the core histones was used as loading control. Bottom panels show quantification of Western blots (n = 3). Intensity of bands on blots for histone modifications were normalized on intensity for the staining of core histones. p value: **, <0.02.
Taken together, experiments with H1c/H1e/H10 and H1c/H1e KO mice demonstrated that without a sufficient amount of linker histones in the rod photoreceptor nuclei, chromatin acquired a less compact, more open conformation and more accessible epigenetic markers. Additionally, dramatic changes in the H3K9me2 profiles and fluorescence intensity in the nuclei of mutant retina suggested that developmental accumulation of linker histone led to a tighter condensation of facultative heterochromatin and, potentially, decreased access to DNA-binding transcriptional factors.
Epigenetic and Transcriptional Changes at the Linker Histones Loci in Mouse Genome during Retina Maturation
In our previous work, we have described stage- and gene-specific signatures for two histone modifications, H3K4me2 and H3K27me3 that accompany mouse retina tissue maturation in vivo (12). Using the data set from this work we examined chromatin modification patterns during retina development for six H1 genes. As ubiquitously expressed genes, linker histone genes all have a noticeable level of H3K4me2 accumulation around the TSS and no H3K27me3 accumulation at any developmental stage (Fig. 8a, blue boxes). When we compared developmental changes in H3K4me2 occupancy in the area around TSS (±2kb) we noticed that, for H10 (H1f0), H1b (Hist1h1b), and H1d (Hist1h1d) this histone modification was constantly present during development. For H1a (Hist1h1a) and H1e (Hist1h1e) there was a moderate increase, and the amount of H3K4me2 around the TSS of H1c (Hist1h1c) was increased dramatically (Fig. 8b). Such developmentally regulated epigenetic changes specific for the H1c gene are strikingly similar to that seen for other rod photoreceptor-specific genes (12). We then compared H3K4me2 occupancy around the TSS for all linker histone subtypes between retinas of the wild type and RD1 mutant animals. H3K4me2 occupancy was not changed for H1b (Hist1h1b) and H1d (Hist1h1d) (Fig. 8c). H3K4me2 occupancy of TSS of other H1 histone genes was reduced in RD1 mutant retina, with the largest changes seen for H1c (Hist1h1c) and H1e (Hist1h1e) (Fig. 8c). This is consistent with the role of H1e (Hist1h1e) in compensating for H1c (Hist1h1c) in chromatin of H1c KO rods (Fig. 3b).
FIGURE 8.

Epigenetic changes in linker histones loci in mouse genome during retina maturation. a, histone modification patterns for H3K4me2 and H3K27me3 around six mouse linker histone genes at four different stages during wild type retina development at E17.5, PN1, PN7, PN15, and mutant retina RD1 at PN28. ChIP-Seq with anti-GFP antibody was used as a negative control. Normalized sequenced tags from ChIP-Seq samples were mapped to the mouse genome (12). TSS for each gene is marked by a blue box. Note that only histone H1c has a special genomic region (marked with red box) that accumulates the H3K4me2 mark later in the retina development. b, comparison of changes of the H3K4me2 occupancy (normalized amount of reads) around TSS for six linker histone genes during retina development. Radial distance represents occupancy. Each developmental stage is marked by a color. c, comparison of H3K4me2 occupancy around TSS for six linker histone genes in wild type retina and RD1 mutant retina. d, detailed map of H3K4me2 accumulation at the H1c gene locus at PN15. The H1c transcript marked in black, the CRX binding site (CRXb) in maroon, and the retina-specific transcript BG293685 from region upstream of H1c gene in green, the arrows show the direction of transcription. Locations of primer pairs that were used to study transcription from the H1c locus are shown above the map in red. e, comparison of the transcription level by quantitative RT-PCR at different developmental stages for the H1c gene and the upstream region that shows specific epigenetic regulation; normalization done by using Gapdh quantitative RT-PCR.
The Hist1h1c gene is highly transcribed in adult rod photoreceptors (46). Among all linker histone subtype genes only H1c (Hist1h1c) has a binding site at the promoter area for CRX, a retinal specific transcriptional factor (47) (Fig. 8d, maroon box). Additionally, we noticed another very interesting feature of the epigenetic landscape in the area between 5.3 and 8.5 kb upstream of the H1c (Hist1h1c) gene TSS (Fig. 8a, red box). Accumulation of H3K4me2 at this site was highly developmentally regulated, reaches a maximum at PN15, and showed a dramatic decrease in RD1 retina. Two possible explanations for this observation are that this DNA sequence harbors a rod-specific enhancer or a transcriptional site for noncoding RNA that may play a role in the enhanced level of H1c expression seen in mature rod photoreceptors. A search through the University of Southern California EST database resulted in a ∼600 bp transcript BG293685 starting from 8.5 kb upstream of H1c (Hist1h1c) TSS with transcription going in the same direction as the H1c (Hist1h1c) gene. This transcript is from an adult retina EST library (48) suggesting that a regulatory noncoding RNA is transcribed from this locus in mature mouse retina.
To test transcription from the regions upstream of the H1c (Hist1h1c) gene and to compare it with gene transcription, we performed RT-PCR on RNA samples from mouse retina at 3 developmental stages: PN1, PN15, and adult. Three pairs of primers (Pr1, Pr2, and Pr3) were designed for the upstream putative regulatory region of the H1c (Hist1h1c) gene (8.5 kb upstream of H1c TSS) and one pair of primers corresponded to the H1c (Hist1h1c) gene (Fig. 8d). Only primer pair 2 for the upstream region gave a positive signal with retina cDNA. Transcription from the upstream region, as well as from the H1c gene showed a 1.5–2-fold increase at developmental stages later than PN1 (Fig. 8e).
Taken together, the developmental epigenetic changes and genome organization of the H1c (Hist1h1c) gene locus support our biochemical finding of tissue-specific up-regulated H1c expression through retina maturation and suggests that a new gene-specific regulatory mechanism controls the expression of the subtype of the linker histone in rod photoreceptors.
DISCUSSION
Maturation of vertebrate tissues is associated with accumulation of heterochromatin in the nuclei of terminal differentiated cells that withdraw from the cell cycle. During rod photoreceptor development, highly condensed heterochromatin accumulates in the middle of the small nuclei (14). These changes in nuclear morphology correlate with terminal rod differentiation starting from birth and are completed in mice at around 3 weeks of age (49). We found that heterochromatin condensation in rod cell nuclei involves gradual increases in the nucleosomal repeat length and the amount of linker histone per nucleosome. The bulk of the increase in nucleosomal repeat length occurs after PN7, indicating that it is due to structural changes within the existing rod cell population as essentially all retinal cells have become post-mitotic by this age (50). Correlation of nucleosomal repeat length and the histone H1/nucleosome ratio in rods is in good agreement with the regression analysis shown in a recent review (51) and puts rod photoreceptors among the cells containing the most compact chromatin, like chicken erythrocytes (40) and cerebral cortex glia (21) all of which have a H1/nucleosome ratio >1 and nucleosomal repeat length >200 bp. The structure of rod cell chromatin is similar to that of cerebellar granular cells with nucleosomal repeat length ∼208 bp (20) but strikingly different from that of cerebral cortex neurons (19, 21), which have a H1/nucleosome ratio <0.5 and nucleosomal repeat length <170.
Despite the fact that linker histone subtypes act as redundant proteins in knock-out mice (30, 33), new data show that each tissue is characterized by a specific set of expressed linker histone subtypes (27, 52, 53). The globular central domain of the linker histone is highly conserved among the subtypes, but the N- and C-terminal domains are divergent with the C-terminal domain showing especially strong variations in its length and charge. H1a and -c subtypes have relatively short C-terminal sequences, H1b and -e have the longest sequences, whereas H10 and -d have C-tails of intermediate length. H10, -a, -c, and -d were found in euchromatin and H1b and -e in the heterochromatin of the neuroblastoma cell line (54). Additionally it was shown that histone H1a and -c act as weak chromatin condensers, H1d as an intermediate, and H10, -e, and -b as strong condensers in vitro (55).
Several previous studies have proposed that H10, the closest mammalian homologue of avian histone H5, plays a major role in chromatin condensation in cultured cells (56, 57) and in terminally differentiated cells that undergo global heterochromatization (58, 59). In contrast to H10, H1c has been implicated in organizing more open chromatin structures (57, 60). However, up to now there has been no direct evidence for H10 being responsible for chromatin condensation in terminally differentiated cells. Furthermore, a FRAP study with GFP transgenes showed that both H1c and H10 have similar chromatin retention properties inside the cell nuclei (61). Knock-out of H10 revealed no measurable structural or morphological changes in chromatin (33). In our study we found a surprisingly low amount of H10 and a marked increase in H1c suggesting that during retinal maturation in vivo, H1c rather than H10 participates in global heterochromatin condensation. Our experiments with linker knock-out mice showed that a single knock-out of H1c does not affect the rod cell phenotype and that other linker histone(s), H1e and/or H1d, compensate for this loss. Based on the preferential expression in rods and the specific changes in H3K4me2 around the TSS, as well as the phenotype of the double H1c/H1e KO (Figs. 4c, 5f, 6, e–h, and 8, b and c), we suggest that compensation is primarily due to the subtype H1e.
We found that decreases in the amount of linker histones in triple KO retina led to an increase of the diameter of the rod photoreceptor nuclei, whereas the fraction of the nuclear area occupied by heterochromatin is decreased (Fig. 4, g and h) in parallel to the decrease in heterochromatin electron density and increased in exposure of histone modifications typical of facultative heterochromatin. This suggests that the process of facultative heterochromatin condensation is partly impaired in mutant retina. An interesting observation is that the total heterochromatin diameter is not decreased as much as the nuclear diameter is increased, implying that it is the heterochromatin density that decreased rather than its volume, perhaps by extending some of the chromatin loops from the heterochromatin to euchromatin compartment. This is consistent with reduced electron density of heterochromatin in the triple KO retina in EM (Fig. 4f).
We have shown previously that in terminally differentiated blood cells, such as lymphocytes and erythrocytes, compact chromatin forms layers in which 3 different zones could be seen (3, 25). Regions composed of juxtaposed chromosome centromeres are often located in the center of the heterochromatic foci in the nucleus. Constitutive pericentromeric heterochromatin occupies chromosomal positions around centromeres and has a distinctive pattern of post-translational histone modifications with abundant H3K9me3. The third, apocentric, zone forms in the process of terminal cell differentiation, concomitant with large-scale gene repression. This zone is composed of facultative heterochromatin marked by H3K9me2 (3, 25). Although in mouse and chicken blood cells heterochromatin is mostly located at the nuclear periphery, in mouse rod photoreceptors it is accumulated as a single massive structure in the middle of the rod nucleus occupying ∼45% of its volume. Nuclei of mouse rod photoreceptors have all 3 of these zones and an additional zone of the H3K27me3 chromatin, organized concentrically around the central core (12, 14, 15). We found that the decreased amount of linker histones in the H1c/H1e/H10 triple KO retina led to a dramatically increased exposure of H3K9me2 chromatin (Fig. 6). This suggests that the developmentally regulated accumulation of linker histone caused specific hypercondensation of facultative heterochromatin marked by this modification and might contribute to forming the concentric heterochromatin architecture in the nucleus. A specific association of linker histone with H3K9me2 was recently reported for human cells from all three embryonic germ layers (27). Recent studies have also shown that linker histone H1 organizes H3K9me2 in the heterochromatin of Drosophila by direct interaction with HMTase Su(var)3–9 (28, 62). It remains to be determined if linker histone levels control H3K9me2 positions in the mouse genome.
In addition to changes in linker histone levels and subtype composition during development, it was shown in vivo that the Cdk2 kinase could extensively phosphorylate linker histones during cell cycle progression. In general, H1 phosphorylation results in a more open chromatin structure (63), H1 eviction from promoter regions that leads to transcription initiation (64) and hormonal gene activation (65, 66). Linker histone is dephosphorylated during induced differentiation of mouse erythroleukemia cells, a model system for erythroid differentiation, whereas expression of histone H1 with phosphorylation-mimicking mutations effectively blocked the differentiation (67). It will be interesting to determine whether dephosphorylation of H1c or other linker histones plays a role during development of rod photoreceptors when heterochromatin compaction occurs.
Previous studies have documented large-scale chromatin decondensation in the neurodegenerative disease SCA7 (16). One recent study demonstrated that the amount of H1c was decreased in rods of a SCA7 (polyglutamine expansion in ataxin 7, subunit of the SAGA coactivator) mouse model. In this disease, neurodegeneration is accompanied by chromatin decondensation in rod nuclei suggesting a contribution of H1c depletion to the SCA7 nuclear phenotype (68). However, in our mouse experiments we found no effect of the single H1c knock-out and a less severe disruption of heterochromatin in the triple H1c/H1e/H10 knock-out than in SCA7 retina. Cumulatively, current evidence suggests that whereas H1c is involved in chromatin condensation, its function is redundant. In the disease systems, it must be synergistic with other linker histones and, likely, other architectural factors such as histone acetylation (16).
Rod photoreceptors are one of the few types of cell with nuclei in which chromatin folds into distinct 30 nm fibers in situ (15). In situ, this chromatin structure was also observed only in sea urchin and chicken erythrocytes (69). The chromatin structure from mouse retina, in contrast to cells from most other organisms, can be easily dissected by genetic means. That makes mouse retina a powerful genetic system to study chromatin higher-order transitions associated with cell differentiation and disease and the role of linker histone expression and histone modifications in formation of condensed chromatin fibers.
Acknowledgments
We thank J. M. Flanagan and members of his lab for help with HPLC experiments and R. Myers for help with electron microscopy.
This work was supported, in whole or in part, by National Institutes of Health Grants EY013865 (to C. J. B.), CA079057 (to A. I. S.), and GM085261 (to Y. F.), a grant from the Macula Vision Research Foundation (to C. J. B.), and National Science Foundation Grant MCB-1021681 (to S. A. G.).
- SCA7
- spinocerebellar ataxia type 7
- HP
- heterochromatin protein
- PN
- postnatal
- TSS
- transcriptional start site.
REFERENCES
- 1. Zhang S. S., Xu X., Liu M. G., Zhao H., Soares M. B., Barnstable C. J., Fu X. Y. (2006) A biphasic pattern of gene expression during mouse retina development. BMC Dev. Biol. 6, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Welch J. J., Watts J. A., Vakoc C. R., Yao Y., Wang H., Hardison R. C., Blobel G. A., Chodosh L. A., Weiss M. J. (2004) Global regulation of erythroid gene expression by transcription factor GATA-1. Blood 104, 3136–3147 [DOI] [PubMed] [Google Scholar]
- 3. Grigoryev S. A., Bulynko Y. A., Popova E. Y. (2006) The end adjusts the means. Heterochromatin remodelling during terminal cell differentiation. Chromosome Res. 14, 53–69 [DOI] [PubMed] [Google Scholar]
- 4. Rawlings J. S., Gatzka M., Thomas P. G., Ihle J. N. (2011) Chromatin condensation via the condensin II complex is required for peripheral T-cell quiescence. EMBO J. 30, 263–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zaret K. S., Watts J., Xu J., Wandzioch E., Smale S. T., Sekiya T. (2008) Pioneer factors, genetic competence, and inductive signaling. Programming liver and pancreas progenitors from the endoderm. Cold Spring Harbor Symp. Quant. Biol. 73, 119–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kioussis D., Ellmeier W. (2002) Chromatin and CD4, CD8A and CD8B gene expression during thymic differentiation. Nat. Rev. Immunol. 2, 909–919 [DOI] [PubMed] [Google Scholar]
- 7. Popova E. Y., Krauss S. W., Short S. A., Lee G., Villalobos J., Etzell J., Koury M. J., Ney P. A., Chasis J. A., Grigoryev S. A. (2009) Chromatin condensation in terminally differentiating mouse erythroblasts does not involve special architectural proteins but depends on histone deacetylation. Chromosome Res. 17, 47–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Popova E. Y., Claxton D. F., Lukasova E., Bird P. I., Grigoryev S. A. (2006) Epigenetic heterochromatin markers distinguish terminally differentiated leukocytes from incompletely differentiated leukemia cells in human blood. Exp. Hematol. 34, 453–462 [DOI] [PubMed] [Google Scholar]
- 9. McKinsey T. A., Zhang C. L., Olson E. N. (2002) Signaling chromatin to make muscle. Curr. Opin. Cell Biol. 14, 763–772 [DOI] [PubMed] [Google Scholar]
- 10. Asp P., Blum R., Vethantham V., Parisi F., Micsinai M., Cheng J., Bowman C., Kluger Y., Dynlacht B. D. (2011) Genome-wide remodeling of the epigenetic landscape during myogenic differentiation. Proc. Natl. Acad. Sci. U.S.A. 108, E149–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang L., Flygare J., Wong P., Lim B., Lodish H. F. (2011) miR-191 regulates mouse erythroblast enucleation by down-regulating Riok3 and Mxi1. Genes Dev. 25, 119–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Popova E. Y., Xu X., DeWan A. T., Salzberg A. C., Berg A., Hoh J., Zhang S. S., Barnstable C. J. (2012) Stage and gene specific signatures defined by histones H3K4me2 and H3K27me3 accompany mammalian retina maturation in vivo. PLoS One 7, e46867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jeon C. J., Strettoi E., Masland R. H. (1998) The major cell populations of the mouse retina. J. Neurosci. 18, 8936–8946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Solovei I., Kreysing M., Lanctôt C., Kösem S., Peichl L., Cremer T., Guck J., Joffe B. (2009) Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell 137, 356–368 [DOI] [PubMed] [Google Scholar]
- 15. Kizilyaprak C., Spehner D., Devys D., Schultz P. (2010) In vivo chromatin organization of mouse rod photoreceptors correlates with histone modifications. PLoS One 5, e11039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Helmlinger D., Hardy S., Abou-Sleymane G., Eberlin A., Bowman A. B., Gansmüller A., Picaud S., Zoghbi H. Y., Trottier Y., Tora L., Devys D. (2006) Glutamine-expanded ataxin-7 alters TFTC/STAGA recruitment and chromatin structure leading to photoreceptor dysfunction. PLoS Biol. 4, e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lantermann A. B., Straub T., Strålfors A., Yuan G. C., Ekwall K., Korber P. (2010) Schizosaccharomyces pombe genome-wide nucleosome mapping reveals positioning mechanisms distinct from those of Saccharomyces cerevisiae. Nat. Struct. Mol. Biol. 17, 251–257 [DOI] [PubMed] [Google Scholar]
- 18. Athey B. D., Smith M. F., Rankert D. A., Williams S. P., Langmore J. P. (1990) The diameters of frozen-hydrated chromatin fibers increase with DNA linker length. Evidence in support of variable diameter models for chromatin. J. Cell Biol. 111, 795–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Thomas J. O., Thompson R. J. (1977) Variation in chromatin structure in two cell types from the same tissue. A short DNA repeat length in cerebral cortex neurons. Cell 10, 633–640 [DOI] [PubMed] [Google Scholar]
- 20. Jaeger A. W., Kuenzle C. C. (1982) The chromatin repeat length of brain cortex and cerebellar neurons changes concomitant with terminal differentiation. EMBO J. 1, 811–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pearson E. C., Bates D. L., Prospero T. D., Thomas J. O. (1984) Neuronal nuclei and glial nuclei from mammalian cerebral cortex. Nucleosome repeat lengths, DNA contents and H1 contents. Eur. J. Biochem. 144, 353–360 [DOI] [PubMed] [Google Scholar]
- 22. Berkowitz E. M., Sanborn A. C., Vaughan D. W. (1983) Chromatin structure in neuronal and neuroglial cell nuclei as a function of age. J. Neurochem. 41, 516–523 [DOI] [PubMed] [Google Scholar]
- 23. Weintraub H. (1978) The nucleosome repeat length increases during erythropoiesis in the chick. Nucleic Acids Res. 5, 1179–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Agarwal N., Hardt T., Brero A., Nowak D., Rothbauer U., Becker A., Leonhardt H., Cardoso M. C. (2007) MeCP2 interacts with HP1 and modulates its heterochromatin association during myogenic differentiation. Nucleic Acids Res. 35, 5402–5408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Grigoryev S. A., Nikitina T., Pehrson J. R., Singh P. B., Woodcock C. L. (2004) Dynamic relocation of epigenetic chromatin markers reveals an active role of constitutive heterochromatin in the transition from proliferation to quiescence. J. Cell Sci. 117, 6153–6162 [DOI] [PubMed] [Google Scholar]
- 26. Jacobs S. A., Khorasanizadeh S. (2002) Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science 295, 2080–2083 [DOI] [PubMed] [Google Scholar]
- 27. Li J. Y., Patterson M., Mikkola H. K., Lowry W. E., Kurdistani S. K. (2012) Dynamic distribution of linker histone H1.5 in cellular differentiation. PLoS Genet. 8, e1002879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lu X., Wontakal S. N., Emelyanov A. V., Morcillo P., Konev A. Y., Fyodorov D. V., Skoultchi A. I. (2009) Linker histone H1 is essential for Drosophila development, the establishment of pericentric heterochromatin, and a normal polytene chromosome structure. Genes Dev. 23, 452–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chang B., Hawes N. L., Hurd R. E., Davisson M. T., Nusinowitz S., Heckenlively J. R. (2002) Retinal degeneration mutants in the mouse. Vision Res. 42, 517–525 [DOI] [PubMed] [Google Scholar]
- 30. Fan Y., Sirotkin A., Russell R. G., Ayala J., Skoultchi A. I. (2001) Individual somatic H1 subtypes are dispensable for mouse development even in mice lacking the H1(0) replacement subtype. Mol. Cell Biol. 21, 7933–7943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fan Y., Skoultchi A. I. (2004) Genetic analysis of H1 linker histone subtypes and their functions in mice. Methods Enzymol. 377, 85–107 [DOI] [PubMed] [Google Scholar]
- 32. Fan Y., Nikitina T., Zhao J., Fleury T. J., Bhattacharyya R., Bouhassira E. E., Stein A., Woodcock C. L., Skoultchi A. I. (2005) Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell 123, 1199–1212 [DOI] [PubMed] [Google Scholar]
- 33. Sirotkin A. M., Edelmann W., Cheng G., Klein-Szanto A., Kucherlapati R., Skoultchi A. I. (1995) Mice develop normally without the H1(0) linker histone. Proc. Natl. Acad. Sci. U.S.A. 92, 6434–6438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fan Y., Nikitina T., Morin-Kensicki E. M., Zhao J., Magnuson T. R., Woodcock C. L., Skoultchi A. I. (2003) H1 linker histones are essential for mouse development and affect nucleosome spacing in vivo. Mol. Cell Biol. 23, 4559–4572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang S. S., Wei J., Qin H., Zhang L., Xie B., Hui P., Deisseroth A., Barnstable C. J., Fu X. Y. (2004) STAT3-mediated signaling in the determination of rod photoreceptor cell fate in mouse retina. Invest. Ophthalmol. Vis. Sci. 45, 2407–2412 [DOI] [PubMed] [Google Scholar]
- 36. Jenö L., Géza L. (1975) A simple differential staining method for semi-thin sections of ossifying cartilage and bone tissues embedded in epoxy resin. Mikroskopie 31, 1–4 [PubMed] [Google Scholar]
- 37. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 38. Zhang Y., Cooke M., Panjwani S., Cao K., Krauth B., Ho P. Y., Medrzycki M., Berhe D. T., Pan C., McDevitt T. C., Fan Y. (2012) Histone h1 depletion impairs embryonic stem cell differentiation. PLoS Genet. 8, e1002691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Medrzycki M., Zhang Y., Cao K., Fan Y. (2012) Expression analysis of mammalian linker-histone subtypes. J. Vis. Exp. 19, 3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bates D. L., Thomas J. O. (1981) Histones H1 and H5. One or two molecules per nucleosome? Nucleic Acids Res. 9, 5883–5894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Istomina N. E., Shushanov S. S., Springhetti E. M., Karpov V. L., Krasheninnikov I. A., Stevens K., Zaret K. S., Singh P. B., Grigoryev S. A. (2003) Insulation of the chicken β-globin chromosomal domain from a chromatin-condensing protein, MENT. Mol. Cell Biol. 23, 6455–6468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gilbert N., Boyle S., Sutherland H., de Las Heras J., Allan J., Jenuwein T., Bickmore W. A. (2003) Formation of facultative heterochromatin in the absence of HP1. EMBO J. 22, 5540–5550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fan J. Y., Rangasamy D., Luger K., Tremethick D. J. (2004) H2A.Z alters the nucleosome surface to promote HP1α-mediated chromatin fiber folding. Mol. Cell 16, 655–661 [DOI] [PubMed] [Google Scholar]
- 44. Narita M., Narita M., Krizhanovsky V., Nuñez S., Chicas A., Hearn S. A., Myers M. P., Lowe S. W. (2006) A novel role for high-mobility group a proteins in cellular senescence and heterochromatin formation. Cell 126, 503–514 [DOI] [PubMed] [Google Scholar]
- 45. Weintraub H. (1984) Histone-H1-dependent chromatin superstructures and the suppression of gene activity. Cell 38, 17–27 [DOI] [PubMed] [Google Scholar]
- 46. Siegert S., Cabuy E., Scherf B. G., Kohler H., Panda S., Le Y. Z., Fehling H. J., Gaidatzis D., Stadler M. B., Roska B. (2012) Transcriptional code and disease map for adult retinal cell types. Nat. Neurosci. 15, 487–495 [DOI] [PubMed] [Google Scholar]
- 47. Corbo J. C., Lawrence K. A., Karlstetter M., Myers C. A., Abdelaziz M., Dirkes W., Weigelt K., Seifert M., Benes V., Fritsche L. G., Weber B. H., Langmann T. (2010) CRX ChIP-seq reveals the cis-regulatory architecture of mouse photoreceptors. Genome Res. 20, 1512–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Blackshaw S., Harpavat S., Trimarchi J., Cai L., Huang H., Kuo W. P., Weber G., Lee K., Fraioli R. E., Cho S. H., Yung R., Asch E., Ohno-Machado L., Wong W. H., Cepko C. L. (2004) Genomic analysis of mouse retinal development. PLoS Biol. 2, E247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Morrow E. M., Furukawa T., Cepko C. L. (1998) Vertebrate photoreceptor cell development and disease. Trends Cell Biol. 8, 353–358 [DOI] [PubMed] [Google Scholar]
- 50. Young R. W. (1985) Cell differentiation in the retina of the mouse. Anat. Rec. 212, 199–205 [DOI] [PubMed] [Google Scholar]
- 51. Woodcock C. L., Skoultchi A. I., Fan Y. (2006) Role of linker histone in chromatin structure and function. H1 stoichiometry and nucleosome repeat length. Chromosome Res. 14, 17–25 [DOI] [PubMed] [Google Scholar]
- 52. Happel N., Doenecke D. (2009) Histone H1 and its isoforms. Contribution to chromatin structure and function. Gene 431, 1–12 [DOI] [PubMed] [Google Scholar]
- 53. Kamieniarz K., Izzo A., Dundr M., Tropberger P., Ozretic L., Kirfel J., Scheer E., Tropel P., Wisniewski J. R., Tora L., Viville S., Buettner R., Schneider R. (2012) A dual role of linker histone H1.4 Lys-34 acetylation in transcriptional activation. Genes Dev. 26, 797–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Th'ng J. P., Sung R., Ye M., Hendzel M. J. (2005) H1 family histones in the nucleus. Control of binding and localization by the C-terminal domain. J. Biol. Chem. 280, 27809–27814 [DOI] [PubMed] [Google Scholar]
- 55. Clausell J., Happel N., Hale T. K., Doenecke D., Beato M. (2009) Histone H1 subtypes differentially modulate chromatin condensation without preventing ATP-dependent remodeling by SWI/SNF or NURF. PLoS One 4, e0007243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gunjan A., Alexander B. T., Sittman D. B., Brown D. T. (1999) Effects of H1 histone variant overexpression on chromatin structure. J. Biol. Chem. 274, 37950–37956 [DOI] [PubMed] [Google Scholar]
- 57. Brown D. T., Alexander B. T., Sittman D. B. (1996) Differential effect of H1 variant overexpression on cell cycle progression and gene expression. Nucleic Acids Res. 24, 486–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sekeri-Pataryas K. E., Sourlingas T. G. (2007) The differentiation-associated linker histone, H1.0, during the in vitro aging and senescence of human diploid fibroblasts. Ann. N.Y. Acad. Sci. 1100, 361–367 [DOI] [PubMed] [Google Scholar]
- 59. Izzo A., Kamieniarz K., Schneider R. (2008) The histone H1 family. Specific members, specific functions? Biol. Chem. 389, 333–343 [DOI] [PubMed] [Google Scholar]
- 60. Huang H. C., Cole R. D. (1984) The distribution of H1 histone is nonuniform in chromatin and correlates with different degrees of condensation. J. Biol. Chem. 259, 14237–14242 [PubMed] [Google Scholar]
- 61. Misteli T., Gunjan A., Hock R., Bustin M., Brown D. T. (2000) Dynamic binding of histone H1 to chromatin in living cells. Nature 408, 877–881 [DOI] [PubMed] [Google Scholar]
- 62. Lu X., Wontakal S. N., Kavi H., Kim B. J., Guzzardo P. M., Emelyanov A. V., Xu N., Hannon G. J., Zavadil J., Fyodorov D. V., Skoultchi A. I. (2013) Drosophila H1 regulates the genetic activity of heterochromatin by recruitment of Su(var)3–9. Science 340, 78–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Contreras A., Hale T. K., Stenoien D. L., Rosen J. M., Mancini M. A., Herrera R. E. (2003) The dynamic mobility of histone H1 is regulated by cyclin/CDK phosphorylation. Mol. Cell Biol. 23, 8626–8636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Koop R., Di Croce L., Beato M. (2003) Histone H1 enhances synergistic activation of the MMTV promoter in chromatin. EMBO J. 22, 588–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Bhattacharjee R. N., Banks G. C., Trotter K. W., Lee H. L., Archer T. K. (2001) Histone H1 phosphorylation by Cdk2 selectively modulates mouse mammary tumor virus transcription through chromatin remodeling. Mol. Cell. Biol. 21, 5417–5425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Vicent G. P., Nacht A. S., Font-Mateu J., Castellano G., Gaveglia L., Ballaré C., Beato M. (2011) Four enzymes cooperate to displace histone H1 during the first minute of hormonal gene activation. Genes Dev. 25, 845–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Yellajoshyula D., Brown D. T. (2006) Global modulation of chromatin dynamics mediated by dephosphorylation of linker histone H1 is necessary for erythroid differentiation. Proc. Natl. Acad. Sci. U.S.A. 103, 18568–18573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kizilyaprak C., Spehner D., Devys D., Schultz P. (2011) The linker histone H1C contributes to the SCA7 nuclear phenotype. Nucleus 2, 444–454 [DOI] [PubMed] [Google Scholar]
- 69. Grigoryev S. A., Woodcock C. L. (2012) Chromatin organization. The 30-nm fiber. Exp. Cell Res. 318, 1448–1455 [DOI] [PubMed] [Google Scholar]