Abstract
Multi-species conserved non-coding elements occur in the vertebrate genome and are clustered in the vicinity of developmentally regulated genes. Many are known to act as cis-regulators of transcription and may reside at long distances from the genes they regulate. However, the relationship of conserved sequence to encoded regulatory information and indeed, the mechanism by which these contribute to long-range transcriptional regulation is not well understood. The ZRS, a highly conserved cis-regulator, is a paradigm for such long-range gene regulation. The ZRS acts over approximately 1 Mb to control spatio-temporal expression of Shh in the limb bud and mutations within it result in a number of limb abnormalities, including polydactyly, tibial hypoplasia and syndactyly. We describe the activity of this developmental regulator and discuss a number of mechanisms by which regulatory mutations in this enhancer function to cause congenital abnormalities.
Keywords: Shh, long-range regulation, limb development, preaxial polydactyly
1. Introduction
The vast majority of the human genome does not encode protein and within the genome are large gene-free tracts called gene deserts. These are of increasing interest to developmental biologists and in contradiction to the use of the phrase ‘gene desert’, these are not necessarily regions devoid of function but rather may contain a large amount of transcriptional regulatory information. In fact, a subset of gene deserts, most often those associated with highly regulated genes controlling developmental processes, are distinguished by composition. Highly conserved sequence elements that are found in multiple vertebrate species are scattered within this subset of gene deserts [1], and these were initially considered candidates as gene enhancers. With the recent publication of the ENCODE project, we are gaining insights into the function of this intergenic DNA (for an overview see [2]). One role of ENCODE was to assign genetic and epigenetic signatures to enhancers and other long-range gene regulators to identify common features to help understand their regulatory activity, and to highlight the potential genomic positions of novel regulatory elements. However, despite these genome wide approaches, the analysis of single developmental genes as paradigms for long-range control is of continuing value in attempts to understand long-range regulatory activity. One such example of long-range control is that of the Shh gene [3–6]. This gene is expressed in a large number of developing tissues and as a result is under the influence of a number of enhancers, many of which have been identified. Thus, the genomic landscape associated with Shh represents a good model for investigating the level of control required by a typical developmental gene. Here, we discuss the role of Shh during limb development and its participation in a spectrum of limb abnormalities that are associated with the alteration of its long-range regulatory activity.
2. Structure of the Shh gene
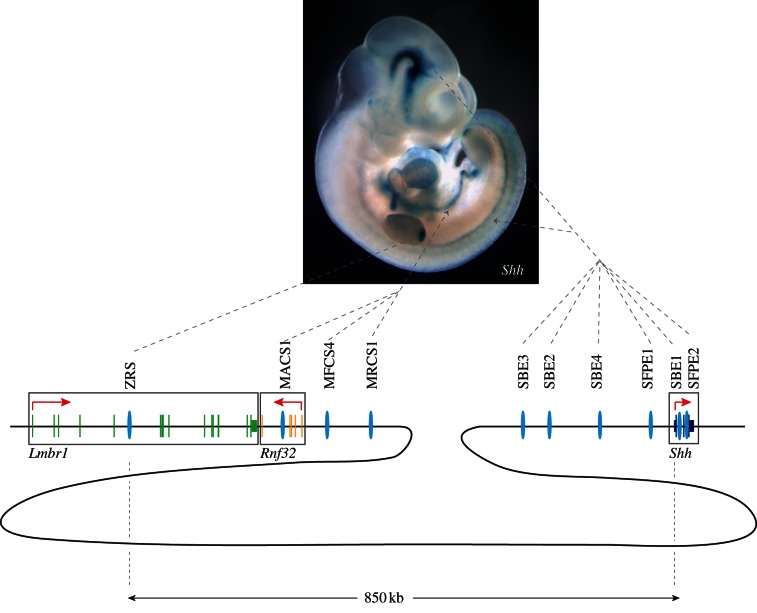
The 5′ end of the Shh gene is juxtaposed to a long gene desert of about 750 kb in the mouse from which follows a gene-dense region of the genome (figure 1). Cis-regulators that contribute to the complex temporal and spatial pattern of Shh are located within the Shh gene, throughout the gene desert [3,4] and interestingly, spilling over into the next two nearest genes (called Rnf32 and Lmbr1). In all, the known cis-regulators extend over 800 kb of DNA in mouse and approximately 1 Mb in human [5,6]. The position of six neural enhancers start inside the Shh gene and extend to over 400 kb at the 5′ end, whereas three epithelial enhancers that direct expression to the pharyngeal, lung and gut endodermal linings reside further 5′, and cluster near the Rnf32 gene. The furthest regulator, called the ZRS (also called MFCS1 [7]), lies inside intron 5 of the Lmbr1 gene.
Figure 1.
Complex long-range regulators control Shh expression during development. The mouse Shh gene is depicted (not to scale) showing the coding region and the relative location of the known Shh enhancers (blue ovals). Brain (SBE1–4) and floorplate enhancers (SFPE1,2) reside inside the gene and extend to about 400 kb. Those enhancers that regulate expression in the epithelial linings are at the extreme end of the gene desert extending into the Rnf32 gene at the beginning of a gene-dense region. The limb-specific enhancer, the ZRS, is inside intron 5 of the Lmbr1 gene which is the next gene over. The coding regions for Shh, Rnf32 and Lmbr1 are boxed in and the direction of transcription is shown by the red arrow. The complex expression of Shh in the mouse embryo at E10.5 is shown, and an example of where each group of enhancers function is shown by the dashed arrows.
Our interest in Shh gene regulation stems from work investigating the genetic basis of limb abnormalities and particularly those that affect the pattern of skeletal elements that compose our hands and feet. We showed the association of a condition called preaxial polydactyly with mutations in the highly conserved ZRS element [5,8]. The ZRS regulates expression of Shh in the developing limb bud, within the mesenchyme at the posterior margin. SHH acts as a morphogen and is secreted from a region of the posterior mesenchyme, called the zone of polarizing activity, patterning the digits in a posterior to anterior direction [5,8,9]. Thus, the focused expression pattern is crucial for regulating the number of digits and for specifying digit identity. If this precise regulation is altered then the patterning of the limb is compromised.
3. Evolution of Shh expression in the limb
The ZRS is conserved in a number of vertebrate species including the fishes [5,7]. For example, comparison of the mouse sequence with that of the coelacanth (Latimeria chalumnae) is striking and shows approximately 75 per cent identity (unpublished data). The ZRS is also found in the Chondrichthyes (sharks and rays) genome [10], and Shh is expressed in the posterior domain of the developing shark fin. In both of these classes of fish, the relative genomic location of the ZRS is the same as in mammals, with ZRS sitting inside the Lmbr1 gene. In an analysis to establish the activity of the fish enhancer, the conserved ZRS sequence was isolated from the Fugu (pufferfish) genome and attached to a reporter gene for analysis in transgenic mouse embryos. The fish regulator was capable of directing specific expression to the appropriate site at the posterior margin of the developing limb bud [5]. The deep conservation of the ZRS for over 400 Myr of evolution suggests that exact sequence is important for function and that the set of factors that bind and regulate the ZRS are similar in fishes as well as mammals. The skeletal elements that compose the bones of the hands and feet, the carpals/tarsals, metacarpals/metatarsals and phalanges, appeared to arise from a series of transformations that eventually resulted in a novel structure, the handplate. Within this evolutionary time frame, the ZRS initially regulated expression of Shh in the fin and subsequently, this regulatory network was co-opted in the tetrapods for a function to specify digits in the autopod.
4. Regulatory mutations cause congenital abnormalities
Shh is a model for genes that are regulated by long-distance enhancers and from a genetic perspective, understanding mutational mechanisms that disrupt this genomic configuration will aid in understanding both normal regulation and processes that underlie congenital abnormalities. Chromosomal translocations that disrupt the regulatory information or cause position affects are well known for SHH [11], as well as for many other genes [12]. These SHH chromosomal lesions usually result in holoprosencephaly; however, we recently reported a patient with additional malformations displaying severe syndactyly (fusion of the digits) of the hands and feet. An intrachromosomal inversion was identified which interrupts the normal regulatory landscape of the SHH gene [13]. However, the limb defects were not owing to the loss of SHH regulatory information, but rather the influence of newly juxtaposed enhancers. One of the enhancers adopted in this chromosomal rearrangement displayed limb-specific activity in mouse transgenics experiments. This enhancer directs expression broadly throughout the limb bud and additionally maintains expression to late stages of limb development. Further, a mouse transgenic model was developed in which this enhancer was used to drive Shh expression and the novel spatio-temporal pattern of Shh expression directed by this enhancer caused syndactyly in mouse embryos and is, therefore, most likely responsible for the severe syndactyly found in this patient.
Of further interest is the large number of regulatory mutations that directly affect the activity of the ZRS and result in a spectrum of skeletal limb abnormalities [14]. Perhaps contrary to expectations, the mutations that reside in this regulatory element do not just lower or raise the expression levels of the SHH/Shh gene but effect spatial and temporal changes in limb expression, and these changes underlie the abnormal phenotype. In addition, the ZRS mutations show an interesting genotype–phenotype correlation leading to distinct clinical classifications of the limb malformations; although, these display an overlapping spectrum of digital abnormalities. These are preaxial polydactyly type 2 (PPD2 which includes isolated triphalangial thumb), triphalangial thumb-polysyndactyly syndrome (TPTPS), syndactyly type IV (SD4) and Werner mesomelic syndrome (WMS). As these defects stem from ZRS mutations, Wieczorek et al [15] suggested that these limb defects be collectively referred to as ‘ZRS-associated syndromes’.
Initially, only point mutations were identified in the ZRS in patients with preaxial polydactyly [8,15–23], however, recent developments have added to the mutation spectrum. A 13 bp insertion was found in a Swedish family with limb defects ([24]; figure 2a). In addition, a number of reports, particularly one that reports a large study of Chinese families [25], showed the occurrence of small intrachromosomal duplications that affect ZRS copy number [15,26]. Two tandem copies of the ZRS give rise to a spectrum of limb defects with the most severe being polydactyly associated with complete syndactyly (TPTPS and SD4). In a recent review, Klopocki & Mundlos [27] suggest that the Laurin–Sandrow syndrome should be included in the spectrum of defects associated with ZRS duplications associated with severe syndactyly. These patients have additional digits that more closely resemble mirror image duplications with one or two thumbs in the middle of the digital array. Some of these patients display long bone involvement with ulnar and fibular duplications.
Figure 2.
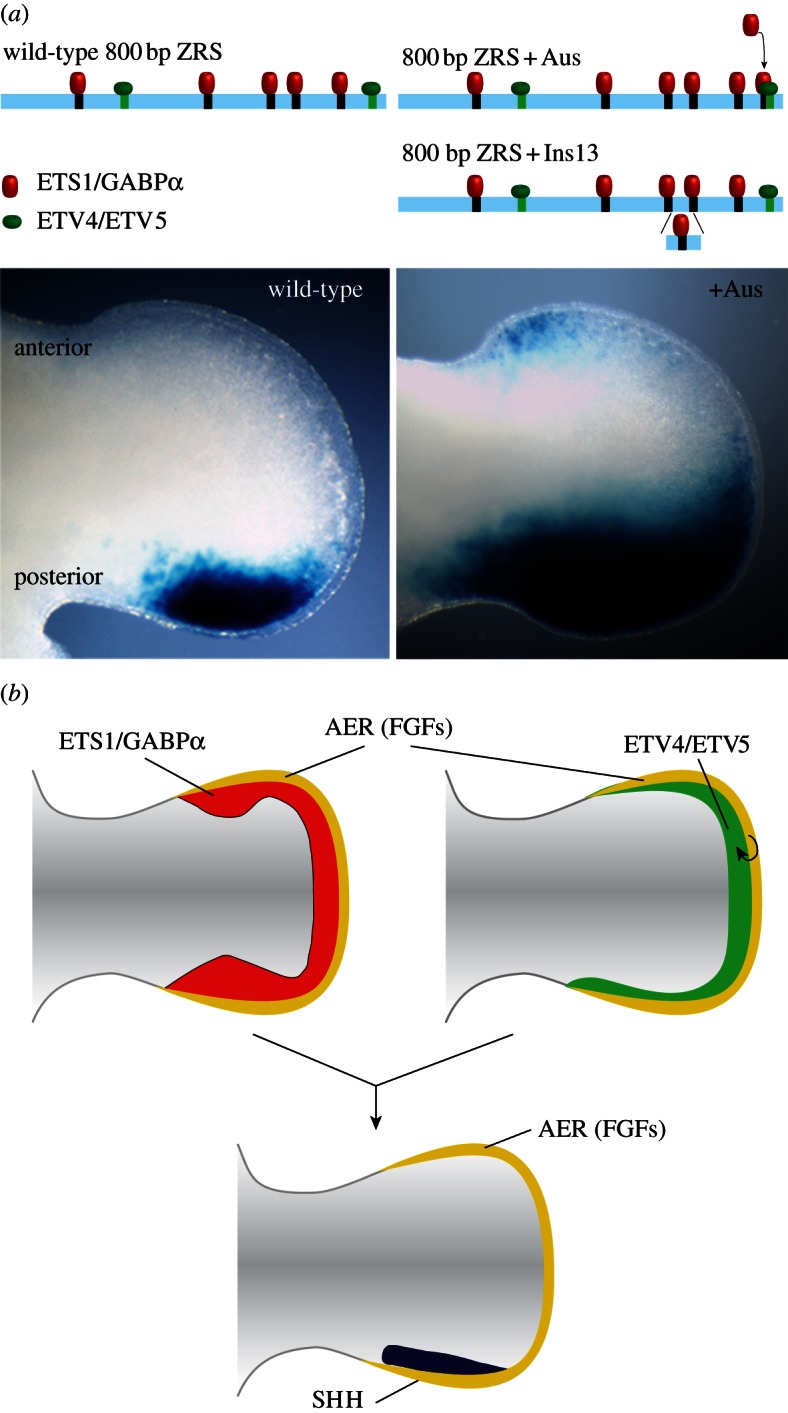
ETS transcription factors act to set the boundary of Shh expression; too many binding sites cause polydactyly. (a) The wild-type (left-hand column) and mutant (right-hand column) ZRS is depicted with the binding sites for ETS1/GABPα (black rectangles) and ETV4/ETV5 (green rectangles). The mutant ZRS has an additional site either created by a point mutation (+AUS) or added within an insertion of 13 bp (+Ins13). The expression pattern of the wild-type and the +Aus mutant in the limb bud is shown below (transgenic embryos expressing LacZ under the influence of wild-type or mutant ZRS). The expression patterns of the four ETS transcription factors are depicted in (b), showing the overlapping patterns for the activating ETS1/GABPα factors and the repressing ETV4/ETV5 factors which together function to restrict the spatial expression of Shh. The Etv4/5 genes are under the influence of the apical ectodermal ridge (AER) through fibroblast growth factor production.
5. Mechanisms that underlie the limb abnormalities
It has been known for well over 40 years, since the classic experiments of Saunders [28], that digit duplications are induced by transplanting mesenchyme from the posterior margin of the limb bud to the anterior margin during the early stages of development. The inducing tissue was defined as originating in the zone of polarizing activity (ZPA) of the limb bud and later the protein responsible for this activity was identified as SHH. We and others [29–31] showed that in mouse models for preaxial polydactyly Shh is mis-expressed and, in addition to the normal posterior pattern of expression, is found along the anterior margin; thus recreating by genetic mutation those earlier transplantation experiments. Identification of the point mutations gave a molecular basis for the mis-expression. These mutations provided a clear example of the capacity of regulatory mutations to cause human disease, and in addition showed the disease potential of a single base pair change embedded in a million bases of DNA. However, the mechanism for driving the ectopic expression was not clear, and the genetic evidence suggests that there are multiple means for modifying the expression of a gene.
The greater than 20 different point mutations associated with limb defects are scattered over the whole approximately 770 bp of the conserved ZRS sequence and are always located at a highly conserved nucleotide. As most evidence suggests, an enhancer functions as a DNA platform for protein interactions and hence, it is expected that the ZRS mutations would affect the binding of a number of different proteins, with perhaps each mutation influencing the binding of a different transcription factor. A point mutation can effect DNA/protein interactions either by changing the affinity of a binding site or by altering the specificity of the site so a different factor binds. Whatever the molecular input from each mutation, these produce a common outcome which is the ectopic expression of Shh.
In an attempt to examine possible mechanisms, it was shown that two of the ZRS mutations (called the AUS and the AC mutations) result in the change in binding specificity ([32]; figure 2a). These mutations create additional sites that supplement five consensus binding sites that already reside in the wild-type ZRS. The sites, which share the sequence AGGAAG/AT, bind members of the ETS family of transcription factors. Previously, it was shown by ChIPseq that the two ETS factors, ETS1 and GABPα, are often found together and when associated with this consensus are usually present in putative enhancers located at a distance from the nearest gene [33]. Accordingly, using ChIP-chip ETS1 and GABPα were shown to bind to the ZRS in the limb bud [32]. To establish the role of these two ETS factors, the five binding sites were systematically disrupted in expression constructs in mouse transgenics [32]. The sites play a role in establishing the expression boundary in the posterior of the limb, and disruption of these sites (particularly sites 1 and 3) resulted in a narrower expression domain. When the AUS mutation was added to the ZRS creating an additional binding site, the boundary of expression expanded, but in addition, appreciable ectopic expression was detected [32]. Interestingly, the 13 bp insertion mutation in the Swedish family resulting in polydactyly (figure 2a) mentioned above [24] contains an AGGAAGT sequence and may effect ectopic expression by a similar mechanism.
Both ETS1 and GABPα are expressed throughout the distal mesenchyme of the limb bud (figure 2b). In addition, two other closely related ETS transcription factors, ETV4 and ETV5, also bind to two sites within the ZRS. These factors are also expressed throughout the distal mesenchyme of the limb and act to restrict expression of the SHH gene to the posterior margin [34,35]. Disruption of both of the ETV4/5-binding sites together cause expression in the ectopic domain [32], so the transgenic data is consistent with genetic data that shows ETV4/5 are involved in repressing ectopic expression and restricting Shh to the anterior boundary. Therefore, the two sets of ETS factors play a role in which the overall outcome is to define the spatial pattern of Shh expression (figure 2b). The broad expression of the Ets1 and Gabpα genes in the limb, together with the ChIP-qPCR data that shows binding of ETS1 at the ZRS in the anterior half of the limb [32], are consistent with the notion that the addition of one extra binding site is sufficient to override the inhibition of Shh at the ectopic site.
Analysis of the point mutation responsible for the polydactylous mouse DZ [23] defined another mechanism by which a novel binding site causes an abnormal limb phenotype. This point change creates a high-affinity binding site for HnRNP U. Previous data shows that the ZRS must move into the vicinity of the Shh promoter to activate transcription [36]. Binding of HnRNP U was postulated to mediate chromosomal looping required for interaction between the ZRS and the promoter region of the Shh gene. As in the anterior limb domain the Shh gene is primed but inactive, the binding of HnRNP U is sufficient to direct the looping and cause the ectopic expression of Shh.
Perhaps a different molecular mechanism is at play in a condition which has been called WMS [15]. Patients from three unrelated families have mutations at the same position in the ZRS (position 404), and these are either a G > C or G > A change [8,15]. Each patient presents with a similar and very severe limb phenotype. These patients have polydactyly but also exhibit a short-limb dwarfism that results from hypoplasia of the tibia (severe shortening of the long bone in the legs); transgenic analysis of the G > C change showed an extreme pattern of ectopic expression [8] suggesting that these mutations result in production of a high concentration of SHH perhaps disrupting development of the tibia in addition to the polydactyly. It is not clear how the mutation affects ZRS activity; however, the fact that different nucleotide changes occur in the same position suggest that WMS may result from loss of binding of a particular factor. Several factors other than ETV4/5, such as GLI3 and ALX4 [37], are known to repress Shh expression in the anterior of the limb and the disrupted binding of one of these or another, as yet unknown, repressor may give the severe phenotype seen.
Triphalangial thumb and the extra digits that constitute PPD2 spectrum result from mis-expression of the SHH gene. However, the question remains as to how SHH is involved in tibial hypoplasia and syndactyly. As stated above, the tibial hypoplasia may be due to high concentrations of SHH in the ectopic domain. Syndactyly, however, is more difficult to explain. Recent analysis that led to mis-expression of Shh in the distal mesenchyme of the limb at later stages of limb bud development (at stages when wild-type expression has turned-off) led to syndactyly in the mouse [13]. In these analyses, the interdigital tissue that normally undergoes programmed cell death persisted. It seems likely that the ZRS duplications that result in the severe forms of TPTPS and SD4 may be affecting both the spatial and temporal pattern of SHH expression. Taken together, these examples suggest several different mechanisms that can alter long-range regulation of the SHH gene.
6. How does the ZRS operate from such a long distance?
It is not clear how a regulator that resides at a long distance activates the promoter of the target gene or whether there are different classes of enhancers separated by the ability to act over very long genomic distances. In attempting to establish some of the rules that regulate the chromatin dynamics of the large Shh regulatory region, Amano et al. [36] examined the ZRS (MFCS1) and its interactions with its target gene. The extreme distance between regulator and gene makes this an excellent system for measuring intranuclear distances. By measuring physical distances by 3D–FISH (3 dimensional fluorescent in situ hybridization) in the ZPA region of the limb bud, it was demonstrated that the Shh gene and the ZRS showed a significant decrease in their intranuclear distance; whereas, an intermediate sequence in the gene desert did not; data that is indicative of looping. By analysing just those cells actively transcribing Shh, the percentage of cells with a close association of ZRS and the Shh gene increased dramatically. A physical interaction was further confirmed by 3C (chromosome conformation capture). A striking observation from these studies was that only a proportion of ZPA cells were productively transcribing the Shh gene at any one time. However, in the notochord and neural tissue, where expression is associated with enhancers acting at a closer range, the expression of Shh is strong and long-lasting. This led to the suggestion that a different mode of action occurs between short-range and long-range enhancers and may provide a means for regulating levels in different developing tissues.
Another observation from this study is that the ZRS appears primed, but inactive, at the anterior margin of the limb bud. We postulate that this poised state is crucial to the ectopic expression that leads to the limb abnormalities and that the regulatory mutations are sufficient to drive the ZRS from this state into an active configuration. A contributing factor to this poised state may be the location of the ZRS inside an actively transcribed gene (the Lmbr1 gene). The activity of a number of putative enhancers have been associated with transcription though the enhancer sequence producing small non-coding RNAs, so-called enhancer RNAs [36,38]. Hence, transcription through the ZRS may play a role in this priming process. Further studies are required to understand this relationship, and because a number of developmental enhancers reside inside neighbouring active genes, this may be a common mechanism for regulating genes during development.
7. Conclusions
Long-range regulation of genes is an important process during development. Each regulator has its own unique signature encoded by the sequence resulting in a distinct set of DNA-binding factors. These factors will direct the long-range activity to seek out the appropriate promoter within hundreds of thousands of base pairs, and as a consequence provide the spatial and temporal regulatory information for transcription. By studying individual models for long-range gene expression, we will be able to ask specific questions about transcriptional specificity unique to developmental tissue and stage, and begin to understand what happens when mutations cause the inbuilt precision of the system to break down.
Reference
- 1.Ovcharenko I, Loots GG, Nobrega MA, Hardison RC, Miller W, Stubbs L. 2005. Evolution and functional classification of vertebrate gene deserts. Genome Res. 15, 137–145 10.1101/gr.3015505 (doi:10.1101/gr.3015505) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 10.1038/nature11247 (doi:10.1038/nature11247) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jeong Y, Epstein DJ. 2003. Distinct regulators of Shh transcription in the floor plate and notochord indicate separate origins for these tissues in the mouse node. Development 130, 3891–3902 10.1242/dev.00590 (doi:10.1242/dev.00590) [DOI] [PubMed] [Google Scholar]
- 4.Jeong Y, El Jaick K, Roessler E, Muenke M, Epstein DJ. 2006. A functional screen for sonic hedgehog regulatory elements across a 1 Mb interval identifies long-range ventral forebrain enhancers. Development 133, 761–772 10.1242/dev.02239 (doi:10.1242/dev.02239) [DOI] [PubMed] [Google Scholar]
- 5.Lettice LA, et al. 2003. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum. Mol. Genet. 12, 1725–1735 10.1093/hmg/ddg180 (doi:10.1093/hmg/ddg180) [DOI] [PubMed] [Google Scholar]
- 6.Sagai T, Amano T, Tamura M, Mizushina Y, Sumiyama K, Shiroishi T. 2009. A cluster of three long-range enhancers directs regional Shh expression in the epithelial linings. Development 136, 1665–1674 10.1242/dev.032714 (doi:10.1242/dev.032714) [DOI] [PubMed] [Google Scholar]
- 7.Sagai T, Hosoya M, Mizushina Y, Tamura M, Shiroishi T. 2005. Elimination of a long-range cis-regulatory module causes complete loss of limb-specific Shh expression and truncation of the mouse limb. Development 132, 797–803 10.1242/dev.01613 (doi:10.1242/dev.01613) [DOI] [PubMed] [Google Scholar]
- 8.Lettice LA, Hill AE, Devenney PS, Hill RE. 2008. Point mutations in a distant sonic hedgehog cis-regulator generate a variable regulatory output responsible for preaxial polydactyly. Hum. Mol. Genet. 17, 978–985 10.1093/hmg/ddm370 (doi:10.1093/hmg/ddm370) [DOI] [PubMed] [Google Scholar]
- 9.Hill RE. 2007. How to make a zone of polarizing activity: insights into limb development via the abnormality preaxial polydactyly. Dev. Growth Differ. 49, 439–448 10.1111/j.1440-169X.2007.00943.x (doi:10.1111/j.1440-169X.2007.00943.x) [DOI] [PubMed] [Google Scholar]
- 10.Dahn RD, Davis MC, Pappano WN, Shubin NH. 2007. Sonic hedgehog function in chondrichthyan fins and the evolution of appendage patterning. Nature 445, 311–314 10.1038/nature05436 (doi:10.1038/nature05436) [DOI] [PubMed] [Google Scholar]
- 11.Belloni E, et al. 1996. Identification of Sonic hedgehog as a candidate gene responsible for holoprosencephaly. Nat. Genet. 14, 353–356 10.1038/ng1196-353 (doi:10.1038/ng1196-353) [DOI] [PubMed] [Google Scholar]
- 12.Kleinjan DJ, van Heyningen V. 1998. Position effect in human genetic disease. Hum. Mol. Genet. 7, 1611–1618 10.1093/hmg/7.10.1611 (doi:10.1093/hmg/7.10.1611) [DOI] [PubMed] [Google Scholar]
- 13.Lettice LA, et al. 2011. Enhancer-adoption as a mechanism of human developmental disease. Hum. Mutat. 32, 1492–1499 10.1002/humu.21615 (doi:10.1002/humu.21615) [DOI] [PubMed] [Google Scholar]
- 14.Anderson E, Peluso S, Lettice LA, Hill RE. 2012. Human limb abnormalities caused by disruption of hedgehog signaling. Trends Genet. 28, 364–373 10.1016/j.tig.2012.03.012 (doi:10.1016/j.tig.2012.03.012) [DOI] [PubMed] [Google Scholar]
- 15.Wieczorek D, et al. 2010. A specific mutation in the distant sonic hedgehog (SHH) cis-regulator (ZRS) causes Werner mesomelic syndrome (WMS) while complete ZRS duplications underlie Haas type polysyndactyly and preaxial polydactyly (PPD) with or without triphalangeal thumb. Hum. Mutat. 31, 81–89 10.1002/humu.21142 (doi:10.1002/humu.21142) [DOI] [PubMed] [Google Scholar]
- 16.Al-Qattan MM, Al Abdulkareem I, Al Haidan Y, Al Balwi M. 2012. A novel mutation in the SHH long-range regulator (ZRS) is associated with preaxial polydactyly, triphalangeal thumb, and severe radial ray deficiency. Am. J. Med. Genet. A 158A, 2610–2615 10.1002/ajmg.a.35584 (doi:10.1002/ajmg.a.35584) [DOI] [PubMed] [Google Scholar]
- 17.Albuisson J, Isidor B, Giraud M, Pichon O, Marsaud T, David A, Le Caignec C, Bezieau S. 2011. Identification of two novel mutations in Shh long-range regulator associated with familial pre-axial polydactyly. Clin. Genet. 79, 371–377 10.1111/j.1399-0004.2010.01465.x (doi:10.1111/j.1399-0004.2010.01465.x) [DOI] [PubMed] [Google Scholar]
- 18.Farooq M, et al. 2010. Preaxial polydactyly/triphalangeal thumb is associated with changed transcription factor-binding affinity in a family with a novel point mutation in the long-range cis-regulatory element ZRS. Eur. J. Hum. Genet. 18, 733–736 10.1038/ejhg.2009.225 (doi:10.1038/ejhg.2009.225) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Furniss D, Lettice LA, Taylor IB, Critchley PS, Giele H, Hill RE, Wilkie AO. 2008. A variant in the sonic hedgehog regulatory sequence (ZRS) is associated with triphalangeal thumb and deregulates expression in the developing limb. Hum. Mol. Genet. 17, 2417–2423 10.1093/hmg/ddn141 (doi:10.1093/hmg/ddn141) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gurnett CA, Bowcock AM, Dietz FR, Morcuende JA, Murray JC, Dobbs MB. 2007. Two novel point mutations in the long-range SHH enhancer in three families with triphalangeal thumb and preaxial polydactyly. Am. J. Med. Genet. A 143, 27–32 10.1002/ajmg.a.31563 (doi:10.1002/ajmg.a.31563) [DOI] [PubMed] [Google Scholar]
- 21.Semerci CN, Demirkan F, Ozdemir M, Biskin E, Akin B, Bagci H, Akarsu NA. 2009. Homozygous feature of isolated triphalangeal thumb-preaxial polydactyly linked to 7q36: no phenotypic difference between homozygotes and heterozygotes. Clin. Genet. 76, 85–90 10.1111/j.1399-0004.2009.01192.x (doi:10.1111/j.1399-0004.2009.01192.x) [DOI] [PubMed] [Google Scholar]
- 22.Vandermeer JE, Afzal M, Alyas S, Haque S, Ahituv N, Malik S. 2012. A novel ZRS mutation in a Balochi tribal family with triphalangeal thumb, pre-axial polydactyly, post-axial polydactyly, and syndactyly. Am. J. Med. Genet. A 158A, 2031–2035 10.1002/ajmg.a.35473 (doi:10.1002/ajmg.a.35473) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao J, Ding J, Li Y, Ren K, Sha J, Zhu M, Gao X. 2009. HnRNP U mediates the long-range regulation of Shh expression during limb development. Hum. Mol. Genet. 18, 3090–3097 10.1093/hmg/ddp250 (doi:10.1093/hmg/ddp250) [DOI] [PubMed] [Google Scholar]
- 24.Laurell T, et al. 2012. A novel 13 base pair insertion in the sonic hedgehog ZRS limb enhancer (ZRS/LMBR1) causes preaxial polydactyly with triphalangeal thumb. Hum. Mutat. 33, 1063–1066 10.1002/humu.22097 (doi:10.1002/humu.22097) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun M, et al. 2008. Triphalangeal thumb-polysyndactyly syndrome and syndactyly type IV are caused by genomic duplications involving the long-range, limb-specific SHH enhancer. J. Med. Genet. 45 589–595 10.1136/jmg.2008.057646 (doi:10.1136/jmg.2008.057646) [DOI] [PubMed] [Google Scholar]
- 26.Klopocki E, Ott CE, Benatar N, Ullmann R, Mundlos S, Lehmann K. 2008. A microduplication of the long range SHH limb regulator (ZRS) is associated with triphalangeal thumb-polysyndactyly syndrome. J. Med. Genet. 45, 370–375 10.1136/jmg.2007.055699 (doi:10.1136/jmg.2007.055699) [DOI] [PubMed] [Google Scholar]
- 27.Klopocki E, Mundlos S. 2011. Copy-number variations, noncoding sequences, and human phenotypes. Annu. Rev. Genomics Hum. Genet. 12, 53–72 10.1146/annurev-genom-082410-101404 (doi:10.1146/annurev-genom-082410-101404) [DOI] [PubMed] [Google Scholar]
- 28.Saunders JW, Gasseling MT. 1968. Ectodermal–mesenchymal interactions in the origin of limb symmetry. In Epithelial–mesenchymal interactions (eds Fleischmeyer R, Billingham RE.), pp. 78–97 Baltimore, MD: Williams & Wilkins [Google Scholar]
- 29.Blanc I, Bach A, Robert B. 2002. Unusual pattern of Sonic hedgehog expression in the polydactylous mouse mutant Hemimelic extra-toes. Int. J. Dev. Biol. 46, 969–974 [PubMed] [Google Scholar]
- 30.Sharpe J, Lettice L, Hecksher-Sorensen J, Fox M, Hill R, Krumlauf R. 1999. Identification of sonic hedgehog as a candidate gene responsible for the polydactylous mouse mutant Sasquatch. Curr. Biol. 9, 97–100 10.1016/S0960-9822(99)80022-0 (doi:10.1016/S0960-9822(99)80022-0) [DOI] [PubMed] [Google Scholar]
- 31.Masuya H, Sagai T, Wakana S, Moriwaki K, Shiroishi T. 1995. A duplicated zone of polarizing activity in polydactylous mouse mutants. Genes Dev. 9, 1645–1653 10.1101/gad.9.13.1645 (doi:10.1101/gad.9.13.1645) [DOI] [PubMed] [Google Scholar]
- 32.Lettice LA, et al. 2012. Opposing functions of the ETS factor family define Shh spatial expression in limb buds and underlie polydactyly. Dev. Cell 22, 459–467 10.1016/j.devcel.2011.12.010 (doi:10.1016/j.devcel.2011.12.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hollenhorst PC, Shah AA, Hopkins C, Graves BJ. 2007. Genome-wide analyses reveal properties of redundant and specific promoter occupancy within the ETS gene family. Genes Dev. 21, 1882–1894 10.1101/gad.1561707 (doi:10.1101/gad.1561707) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mao J, McGlinn E, Huang P, Tabin CJ, McMahon AP. 2009. Fgf-dependent Etv4/5 activity is required for posterior restriction of sonic hedgehog and promoting outgrowth of the vertebrate limb. Dev. Cell 16, 600–606 10.1016/j.devcel.2009.02.005 (doi:10.1016/j.devcel.2009.02.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Z, Verheyden JM, Hassell JA, Sun X. 2009. FGF-regulated Etv genes are essential for repressing Shh expression in mouse limb buds. Dev. Cell 16, 607–613 10.1016/j.devcel.2009.02.008 (doi:10.1016/j.devcel.2009.02.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Amano T, Sagai T, Tanabe H, Mizushina Y, Nakazawa H, Shiroishi T. 2009. Chromosomal dynamics at the Shh locus: limb bud-specific differential regulation of competence and active transcription. Dev. Cell 16, 47–57 10.1016/j.devcel.2008.11.011 (doi:10.1016/j.devcel.2008.11.011) [DOI] [PubMed] [Google Scholar]
- 37.Qu S, Niswender KD, Ji Q, van der MR, Keeney D, Magnuson MA, Wisdom R. 1997. Polydactyly and ectopic ZPA formation in Alx-4 mutant mice. Development 124, 3999–4008 [DOI] [PubMed] [Google Scholar]
- 38.Kim TK, et al. 2010. Widespread transcription at neuronal activity-regulated enhancers. Nature 465, 182–187 10.1038/nature09033 (doi:10.1038/nature09033) [DOI] [PMC free article] [PubMed] [Google Scholar]