Abstract
Vertebrate genes are characterized by the presence of cis-regulatory elements located at great distances from the genes they control. Alterations of these elements have been implicated in human diseases and evolution, yet little is known about how these elements interact with their surrounding sequences. A recent survey of the mouse genome with a regulatory sensor showed that the regulatory activities of these elements are not organized in a gene-centric manner, but instead are broadly distributed along chromosomes, forming large regulatory landscapes with distinct tissue-specific activities. A large genome-wide collection of expression data from this regulatory sensor revealed some basic principles of this complex genome regulatory architecture, including a substantial interplay between enhancers and other types of activities to modulate gene expression. We discuss the implications of these findings for our understanding of non-coding transcription, and of the possible consequences of structural genomic variations in disease and evolution.
Keywords: gene regulation, regulatory landscapes and remote enhancers, repressors and latent regulatory activity, specificity of gene–enhancer regulatory interactions
1. Introduction
An essential part of the control of gene expression is achieved at the transcriptional level. This level of regulation integrates the contribution of multiple types of cis-acting genomic elements. Beyond the promoter region, which is in close proximity to the transcriptional start site, the importance of elements located at much farther distances is increasingly being recognized [1–3]. Growing numbers of human genetic conditions have been found to result from mutations, deletions or other alterations of regulatory elements, mostly enhancers, that can be located more than 1 Mb away from the gene they regulate [4–11]. Genome-wide association studies, which analyse the genetic basis of phenotypic variation, have also frequently identified genomic intervals devoid of protein-coding genes as causal regions, further suggesting that variations or changes in gene regulatory elements can have profound physiological effects [12].
For these reasons, defining the regulatory sequences that control gene activity is becoming a key biomedical issue. Recent technological advances are providing an increasing repertoire of methods to identify regulatory elements, particularly enhancers. As illustrated recently by the ENCODE project, chromatin immuno-precipitation (ChIP) for different transcription factors (TFs), chromatin-associated marks and transcription-associated protein complexes enables the cataloguing of sequences with characteristics of regulatory elements [13]. Strikingly, these efforts revealed that in a given cell, a very large number of elements can regulate a given gene. These findings were reinforced by data from chromatin conformation capture (3C) analyses, which showed that promoters and enhancers are engaged in complex interlaced interactions [14,15], indicating that interactions may extensively reshape the individual activities of gene regulatory elements in a non-additive manner. It is therefore important to complement the approaches that deconstruct the genome into its most basic constitutive elements (promoters, enhancers, silencers, insulators), with more holistic approaches that address how the activities of large arrays of cis-regulatory elements are integrated, to ultimately give rise to gene-specific programmes.
Here, we provide a brief review on the evidence that emphasizes the role of remote cis-regulatory elements in gene expression and disease. We describe the different methods that can be used to identify and characterize these elements, particularly enhancers. We present and discuss different findings, which demonstrate that the activity of enhancers is not purely determined by their sequence but can also be highly dependent on the surrounding genomic regions. We summarize our own observations obtained with genome regulatory organization mapping with integrated transposons (GROMIT), a strategy, which we developed to investigate the regulatory activities present within the genome [16]. GROMIT revealed the widespread presence of tissue-specific regulatory activities throughout the genome: these activities are distributed along large intervals, forming broad regulatory landscapes, which extend far away from genes. Comparison of the activities displayed within these regulatory landscapes with those determined for isolated single enhancer elements, demonstrated that a substantial part of these latter, potential activities may be silenced or repressed in one way or another. We discuss what these findings tell us about the nature of the genome's regulatory architecture, as well as their implications for human disease and for the evolution of gene expression.
2. Cis-regulatory elements, disease and phenotypic variation
The importance of regulatory effects in disease and more generally in shaping vertebrate phenotypic variation has almost become a paradigm in modern genomics. Early on, the observation that identical or overlapping clinical symptoms could be caused either by mutations in a gene or by chromosomal rearrangements with breakpoints hundreds to thousands of kilobases away from these genes, strongly suggested the presence of remote influences in controlling gene activities. Initially poorly characterized, these influences were collectively defined under the catch-all notion of ‘position effects’ (reviewed in Kleinjan & van Heyningen [17]). It is now known that several of these conditions result from the disruption of a remote regulatory element [7,8,18,19], sometimes present more than 1 Mb away from the gene it is controlling. New technologies, such as array-comparative genome hybridization and next-generation sequencing, enable mapping of genomic structural variations in patients and in normal populations with greater resolution (reviewed in Alkan et al. [20]). This facilitates the identification and analysis of cases with potential ‘position effects’, including hitherto difficult-to-detect genomic changes. Indeed, thanks to these developments, microduplications that encompass regulatory elements have been revealed as a substantial cause of human developmental malformations [21–25].
Beyond Mendelian diseases, genomic and genetic variations affecting regulatory elements also contribute more broadly to phenotypic diversity: a large fraction of variants associated with disease susceptibility and small-effect quantitative traits resides in intergenic regions [12,26,27]. Observations that intra- and inter-species phenotypic variation is also commonly due to changes in regulatory regions [28–31] further suggest that modulation of gene function through regulatory innovation or modulation can be a driving force of evolutionary changes.
Because of this important role of regulatory elements in health, disease and evolution, extensive efforts have been applied to identify such elements, and to understand how they contribute to gene expression.
3. Identifying regulatory elements
Broadly, the regulatory elements described to date can be grouped into (proximal) promoters, enhancers, repressors and insulators [32] (figure 1a). Our ability to detect such elements, either computationally or experimentally, has improved markedly over the past decade or so, thanks to the discovery of a number of characteristic features (recently reviewed by Hardison & Taylor [33]). For example, the availability of whole genome sequences has enabled the rapid identification of evolutionarily conserved non-coding sequences, which frequently have regulatory functions [34,35], although some elements, undergoing more rapid turnover, are not amenable to such screens [13,36]. Cis-regulatory elements are bound by clustered TFs [37], and TF occupancy is linked to diverse chromatin features that can be ascertained: cis-regulatory elements often overlap nucleosome-depleted regions, and are characterized by distinct histone composition, histone modifications and by binding of specific proteins, such as transcription cofactors and chromatin remodelling proteins (figure 1a). Chromatin profiling has therefore become an efficient method to allow genome-wide identification of such sites [33]. Nucleosome-depleted regions can be detected by nuclease hypersensitivity [38] or FAIRE [39], whereas regions associated with specific histones, histone marks or proteins can be identified by ChIP with appropriate antibodies. Such methods, together with the decreasing cost of sequencing, have recently led to detailed inventories of regulatory elements [13,40–42]. It is noteworthy that the presence of specific marks such as H3K27ac on distant enhancer elements can also help distinguish elements that are active, from elements that may only be in a poised state, thus providing ways to use biochemical profiles to infer biological activities [43,44]. In addition, the development of methods, such as 3C and its derivatives (4C, 5C and HiC), which detect chromosomal regions that physically interact with promoters, has also been useful to identify regulatory elements, particularly distant ones [45,46].
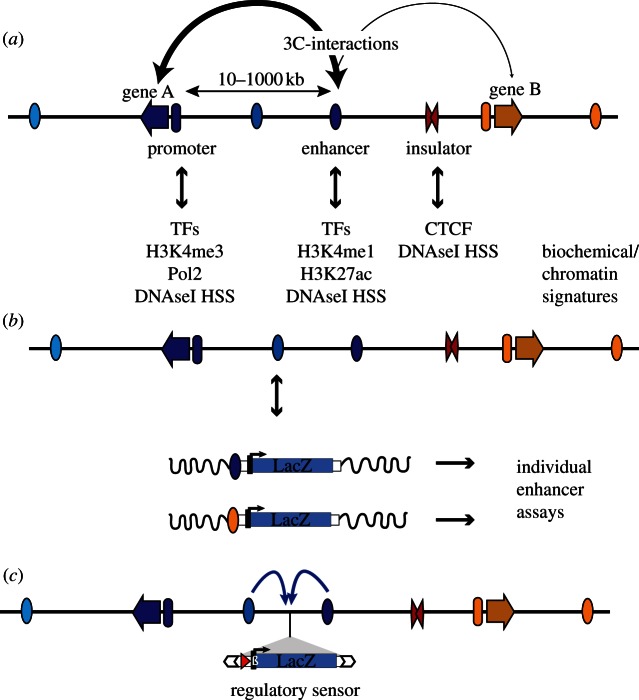
Figure 1.
Regulatory elements and how to detect them. (a) Multiple regulatory elements are involved in the tissue-specific transcriptional regulation of genes (arrows). These elements include promoters (ovals preceding genes), enhancers (ovals, with tissue-specific activity indicated by different colours) and insulators (red double triangle). Enhancers can be located at great distance from their target gene. Different regulatory elements are marked by distinct chromatin signatures and binding of proteins, which can be used to identify active elements in accessible tissues using biochemical methods. It is generally accepted that activation is achieved through direct interactions between enhancers and target promoters, which can be detected by chromosome conformation capture (3C) techniques (the stroke of the connecting arrows represents the frequency/strength of the interactions). TFs, transcription factors; DNaseI HSS, DNaseI hypersensitive site. (b) The activity of individual regulatory modules can be tested in in vivo reporter assays. This is usually achieved by cloning the element upstream of a reporter gene, by measuring the expression of this transgene integrated randomly into the genome (indicated by wavy black line). (c) In comparison, GROMIT uses a transposon (white arrows), which carries regulatory sensor (LacZ gene, driven by minimal promoter) as cargo. Through random transposition, the reporter gene can be distributed throughout the genome, showing integrated regulatory input from the multiple modules (indicated by arrows) acting on that position.
However, it should be emphasized that these approaches exploit indirect properties of enhancers and do not assess them in an operational manner (i.e. whether an enhancer actually contributes to gene expression). Indeed, estimating the proportion of TF binding events or chromatin marks that are truly functional is therefore an important on-going debate in the field, as is the definition of ‘biological function’ [47–49].
In vivo reporter assays provide a more functional approach to test individual elements, by assessing their ability to drive gene expression. A frequently used reporter assay consists of cloning a putative enhancer fragment upstream of a reporter gene driven by a promoter. The promoter used is often a small neutral promoter region, with minimal or no activity by itself, but that responds accurately to the input of the adjacent enhancer. Accordingly, the activity of the enhancer is revealed by the expression pattern of the reporter gene (figure 1b). The recent development of massively parallel reporter assays [50,51] offers ways to test the activities of thousands of elements simultaneously, and to dissect individual enhancers by testing the influence of thousands of random mutations on their activity. These high-throughput approaches are opening important avenues, but are currently largely restricted to cell lines. They can be applied to in vivo conditions, with some limits: for example, in mice, hydrodynamic tail vein injection of DNA constructs results in episomal uptake of fragments, but primarily in the liver [50]. Thus, despite these new technological developments, getting detailed information about enhancer activity across multiple tissues, developmental stages, in both the proper physiologic and epigenetic contexts, may still be better achieved with enhancer-reporter gene transgenes integrated in one-cell embryos. Importantly, new improved vectors and integration systems (transposons, lentiviruses, integrases [52–55]) may facilitate and improve the efficiency of in vivo integrative transgenesis. Already, systematic in vivo transgenic reporter assays have led to collections of enhancer activities that provide direct and important clues about the nature of tissue-specific regulatory elements [56,57].
These transgenic assays have revealed that a great part of the transcriptional activity of genes can be attributed to the action of autonomous regulatory modules, each in charge of a subset of the overall expression pattern. Their action is often described as additive, and adjacent modules, active in different tissues, do not interfere with each other [58]. However, in essence, these assays are reductionist, and test relatively short pieces of DNA sequences in isolation, which are usually randomly integrated into the genome, and therefore out of their natural genomic context. In several instances, enhancer elements at the endogenous locus do not recapitulate the activity that they showed when tested in enhancer assays [59–61]. The observed differences have been put down to non-additive activity of regulatory elements in their native region, or position effects owing to the genomic context where the element is inserted.
It may be worth underlining that such ‘context-dependent’ functions have also been reported by other studies, and probably do not simply reflect technical artefacts of transgenic assays. For example, sequence variation explains only a very small fraction of the differential TF occupancy found in human samples [62], suggesting that distant elements or ‘epigenetic’ factors could contribute to the activity of the same element (see Voss et al. [63]). Detailed studies of some endogenous loci, including their functional dissection through deletions and inversions, highlight the importance of regulatory interactions, either through physical interactions as a chromatin hub or regulatory archipelagos [64,65], or through more complex types of ‘regulatory priming’ of an enhancer by another element [66]. Consequently, it is imperative not only to catalogue regulatory elements, but also to establish how regulation is achieved mechanistically. Importantly, in most transgenic assays, the genomic element of interest is cloned just next to the reporter promoter, whereas the biological activity of remote enhancers results not only from their recruitment of TFs, but also from their capacity to interact with appropriate target gene(s) in the appropriate tissue or cell type. Some of these aspects of enhancer function can be addressed by testing their activity in the context of yeast or bacterial artificial chromosomes (BACs) [10,67]. Owing to their large size, BACs are generally assumed to represent endogenous regulatory landscapes more accurately, although large, complex landscapes may still not be fully covered within an individual clone.
4. From enhancers to target genes
A vital step to comprehend how gene regulation is achieved mechanistically lies in understanding how the interactions between genes and surrounding regulatory elements are controlled. However, the assignment of target genes to regulatory elements can be ambiguous, because regulatory regions can extend over hundreds of kilobases, and the gene most proximal to an enhancer is not necessarily its target [10,11,68,69]. One approach to predict target genes has been to search for co-occurrence of marks specific for active enhancers and promoters, thereby establishing enhancer–promoter units [42]. Alternatively, current views support the idea that most enhancers are engaged in direct physical interactions with their target gene promoters; these interactions can be detected by 3C or its more high-throughput derivatives (4C, 5C, HiC) [46]. These methods allow the identification of regions physically interacting with promoters [15], or—if combined with ChIP for specific proteins (ChIA–PET)—the detections of interactions between regions bound by those proteins [14,70]. Interestingly though, interactions between distal elements and promoters are not exclusive: instead, promoters as well as enhancers are frequently engaged in multiple interactions [14,15]. Whether these interactions are functionally relevant (e.g. to achieve co-regulation of genes), or simply a consequence of other properties remains unclear. Physical proximity of co-regulated genes located on different chromosomes has also been observed [71], but these situations most probably represent co-localization to discrete subnuclear domains (transcription factories [72]) optimized for coordinated regulation, rather than trans-regulation through elements on another chromosome. The small number of documented cases where genes are controlled by regulatory elements localized on a different chromosome [73–75] most probably represent exceptional—and sometimes debated—systems. Furthermore, overall interchromosomal interactions have been reported to be rather indiscriminate, with no evidence for their being organized by few, specific regions, and the frequency of interactions correlates strongly with the average distance of a given locus from the edge of a chromosome territory [76,77]. Together, with the paucity of interchromosomal interactions whose frequency reaches the levels of known distant promoter–enhancer pairs [15], this argues that for most genes, the elements that contribute to their expression will be found in cis, yet at distances that could frequently be above hundreds of kilobases. Further evidence for the overwhelming importance of cis-regulation comes from genetic crosses between different mouse strains, which has indicated that potentially more than 90 per cent of gene expression differences can be attributed—either partially or completely—to variants acting in cis [78].
In this context, different systems are known to contribute to the specificity of these often distant enhancer–promoter cis interactions, either through direct tethering in favour of target genes [79], or through insulator sequences that block the ectopic activation of other neighbouring genes [80]. Intriguingly, at the same time, multiple observations suggest that enhancers can act also promiscuously, resulting in activation of neighbouring but biologically irrelevant genes. Such collateral activity has been documented for Lnp, adjacent to the Hoxd cluster [10], Nme4 associated with the α-globin locus [81] and Igβ in the pituitary [82].
These caveats in our current understanding stem in part from the different approaches used to identify individual elements and to assert their function with frequently indirect readouts. This highlights the need for complementary and alternative approaches that may allow assessing regulatory activity in vivo, and in the endogenous context to add to the accumulated molecular information with more functional insights.
5. Charting regulatory landscapes with GROMIT
To bridge this gap in our understanding, we have recently developed a novel method, GROMIT, that enables us to chart the organization and distribution of regulatory activities along chromosomes, providing an integrated and non-gene-centric view of cis-regulatory activity [16]. The basic principle of GROMIT is to distribute a regulatory sensor throughout the mouse genome, by harnessing the properties of the Sleeping Beauty transposon [83,84]. The regulatory sensor consists of a LacZ reporter gene, driven by a 50 bp long fragment of the human β-globin promoter (figure 1c). This short promoter is essentially neutral: it has no activity by itself, but is very sensitive to endogenous regulatory information, without perturbing endogenous gene expression [16]. Therefore, this system makes it possible to determine the regulatory input acting on a given genomic position, where the transposon in inserted. Because the sensor is incorporated into the genome, it measures the integrated regulatory activity of all elements (activating and repressing) acting on that position. This distinguishes GROMIT from reporter assays, which test individual elements in isolation and at random positions. Importantly, the transposon carrying this sensor gene can be remobilized efficiently in vivo, in a cut-and-paste manner, to generate animals with new insertions. The insertion sites can be precisely mapped, and in contrast to other systems show no integration bias towards particular regions or genomic hallmarks [16]. This property allows the production of a very large number of insertions, and the study of the expression patterns at those positions (figure 2a). These insertions collectively establish a map of how regulatory influences that control gene expression are distributed.
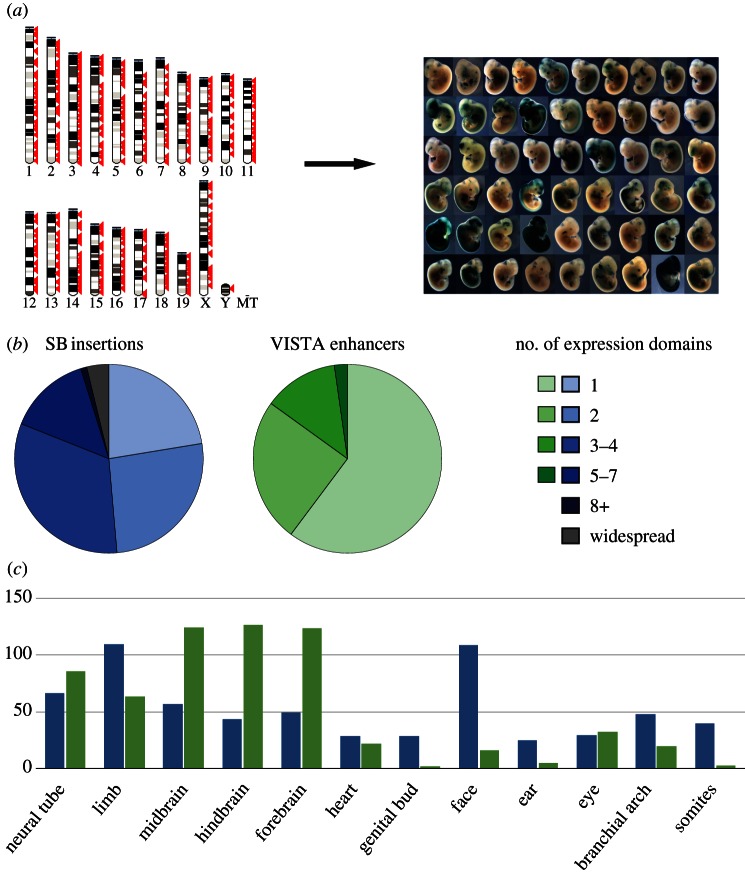
Figure 2.
Regulatory activity captured by GROMIT and comparison with enhancer activity. (a) We generated a collection of random, genome-wide insertion sites (left, each red arrow representing an insertion site), which we tested one by one for LacZ expression at E11.5 of embryonic development, revealing pervasive, tissue-specific expression throughout the genome (right, a representative set of obtained expression patterns). A complete list of all insertions and the associated expression patterns is available in the TRACER database (tracerdatabase.embl.de). (b) Comparison of the activity of GROMIT insertions (SB insertions) and the activities documented in the VISTA enhancer browser (VISTA enhancers) [56]. The pie charts show how frequently insertions or enhancers displayed activity in a given number of expression domains (indicated by colours). For the analysis, we removed all insertions from the GROMIT dataset that were clearly part of the same regulatory landscapes (showing similar expression patterns and less than 200 kb apart), and only took into consideration VISTA enhancers from early screens (IDs between 1 and 1290), to avoid bias introduced by tissue-specific p300-bound regions [85]. (c) Distribution of tissue-specific activities between GROMIT insertions (blue bars) and VISTA enhancers (green bars). Tissues are indicated along the x-axis, and the y-axis shows the number of observed insertions/enhancers with activity in a given tissue.
6. Widespread distribution of tissue-specific inputs
Initial analysis of β-galactosidase stainings of more than 150 insertions, collected at stage E11.5 of embryonic development, revealed that almost 60 per cent of tested locations showed expression [16], regardless of their position relative to genes. An expanded dataset confirmed this initial observation (figure 2a). The vast majority of insertions showed restricted, tissue-specific expression patterns (figure 2a), with fewer than 5 per cent showing widespread expression. This propensity towards tissue-specific expression from most genomic positions highlighted that regulatory activities are distributed along chromosomes, and not centred towards the vicinity of specific regions such as gene promoters. Importantly, the observed patterns frequently shared striking similarity with the activities of neighbouring enhancers that had been characterized previously or with flanking genes or other insertions. Collectively, these results indicated that GROMIT captures biologically relevant regulatory activities, including those acting far from genes.
First, we took advantage of the VISTA enhancer browser [56] to compare the autonomous activities of individual enhancer elements defined by in vivo enhancer assays with the patterns observed at endogenous genomic positions using GROMIT. On a global level, the comparison showed that the expression patterns captured by our regulatory sensor were overall more complex, with more than 75 per cent of insertions showing activity in more than one tissue, whereas in the reporter assay, more than a half of the enhancers showed reproducible expression in only a single tissue (figure 2b). This broader specificity suggested that—in line with our expectations—the GROMIT sensor generally captured the overlapping activity of more than one enhancer at a given position. These findings were confirmed by case studies of individual loci (two examples shown in figure 3, others in [16,65]).
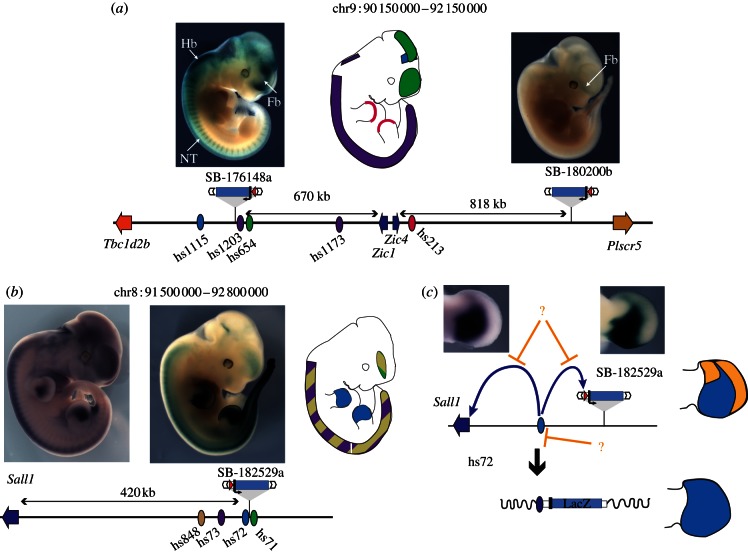
Figure 3.
Comparing the activities captured by GROMIT with individual enhancer activity and expression of endogenous genes. (a) Extent of the Zic1–Zic4 locus. The insertion SB-176148a, approximately 670 kb centromeric of Zic1 and Zic4 captures most of the expression domains predicted by the individual enhancers identified at this locus, In contrast, insertion SB-180200b, located almost 820 kb telomeric of Zic4, shows a different and weak forebrain expression. (b,c) Differences between enhancer activity and gene expression at the Sall1 locus. (b) Insertion SB-182529a, located 420 kb away from the Sall1 gene shows a similar expression pattern to the endogenous gene. These tissue-specific expressions overlap with the activities assigned to a series of enhancers within the region. (c) Interestingly, the expression patterns of Sall1 (left) and of the regulatory sensor (right) only partially capture the activities displayed by the enhancer hs72 when tested in isolation. Whereas the tested enhancer drove expression throughout the whole limb bud (bottom drawing, enhancer activity shown in blue), limb expression within the genomic context is restricted to a proximal posterior domain (top drawing, with orange shapes showing the regions where enhancer activity is restricted). Whether this suppression of enhancer activity is achieved by repression of the enhancer itself (indicated by lower, repressive orange arrow), or by disabling the interaction between the enhancer and its potential target promoters (indicated by upper, repressive orange arrows), remains unclear. For all panels, the locus is represented schematically at the bottom, with the position of genes (arrows), enhancers (ovals) and insertions indicated. Enhancers are shown as ovals, their tissue-specific activity schematically depicted on the embryo outline, using the same colour code. Pictures of the LacZ staining patterns for GROMIT insertions SB-182529a and the RNA in situ hybridization pattern for Sall1 are adapted with permission from Ruf et al. [16].
The relative distribution of tissue-specific activities obtained by GROMIT insertions and VISTA enhancers was quite different (figure 2c). For example, the set of enhancers analysed by VISTA was strongly biased to drive expression in fore-, mid- and hindbrain, whereas only 4 per cent of these enhancers were active in the face. The expression of the regulatory sensor did not show a similar preference for neural tissues, but exposed a frequent regulatory potential for expression in facial tissues (18% of all insertions, counting insertions clustered within 200 kb as one). Possibly, these differences may be purely of technical nature: the VISTA dataset was compiled using an Hsp68 promoter fragment [56], whereas GROMIT uses the β-globin minimal promoter [16], and although enhancers, by their classical definition, should not show promoter preference, some exceptions have been described [86]. Thus, promoter bias might add to the different distribution of tissue-specific activities. But, these discrepancies could also reflect biological phenomena. For example, the VISTA enhancers used for the analysis were picked from sequences with a high degree of evolutionary conservation. It is known that the degree of sequence conservation can be quite variable for enhancers active in different tissues [36], and therefore the different distributions may mirror the different evolutionary constraints associated with regulatory elements active in different tissues. The observed discrepancies could also arise from tissue-specific differences in the regulatory architecture: if the distribution of particular enhancers differs (e.g. if brain enhancers cluster together), or if their range of action is dissimilar (e.g. brain enhancers have a shorter range of action), then a similar shift would be observed. Further studies will be needed to investigate these questions, but this comparison shows the need of combining diverse approaches to get a more complete picture of gene regulatory mechanisms.
7. From enhancers to regulatory landscapes
Frequently, adjacent insertions (or an insertion-endogenous gene pair) shared extensive similarities in their expression patterns, with several cases where insertions were several hundred kilobases apart (examples in figure 3 and in [16]). The widespread activities and large intervals exposed by these observations are reminiscent of the previously described ‘regulatory landscapes’ (genomic domains where otherwise unrelated genes shared expression specificities) [10,11], and ‘genomic regulatory blocks’ (regions of conserved synteny between genes and non-coding elements among evolutionary distant relatives) [87]. The data obtained by GROMIT imply that their presence is a pervasive feature, and is not restricted to few loci around key developmental regulatory genes. It is yet unclear, if one or few enhancers with particular long-range properties define large regulatory landscapes, or if the conjugated action of multiple co-interacting regulatory modules dispersed across a large interval determines these co-expression territories, as suggested by recent work on the Lnp–Hoxd interval [65].
Naturally, similar gene expression patterns by themselves cannot be considered proof that a given enhancer is regulating reporter gene expression, or that the same enhancers are regulating the endogenous and the reporter gene. Accordingly, it will be exciting to subject tissues with insertions obtained with GROMIT to other assays, such as chromosome conformation capture, to determine whether the insertion sites physically interact with their putative enhancers, and whether the interaction profile of the endogenous gene and the reporter gene is similar, given their often near-identical expression pattern. Similarly, if the TFs binding a given enhancer are known, ChIP and ChIA–PET will provide interesting avenues to detect whether these proteins mediate direct physical interactions and influence the genomic range and distribution of enhancer activities.
8. From regulatory landscapes to gene expression
As mentioned earlier, the expression patterns displayed by the regulatory sensor often overlap with the expression domains of a neighbouring gene, and in line with this, the expression domains captured by GROMIT were generally a composite of the individual activities of the multiple enhancers that surround it (figure 3).
Yet, at a given genomic position, the reporter gene frequently showed only a subset of the expression domains of the endogenous gene (figure 3), as well as sometimes differences with an immediately adjacent enhancer. As mentioned earlier, these discrepancies can be, in part, technical (promoter bias, sensitivity of in situ probes, etc.). However, adjacent insertions sometimes revealed different subsets of the expression pattern of the flanking endogenous gene, showing that all domains could be captured by our promoter, and the causes for these differences are more intricate [16]. This implies that the regulatory elements that control gene expression may have distinct ranges of action, thereby defining different regulatory landscapes, and ultimately resulting in differential gene expression at different positions within a locus. The observation that GROMIT insertions can report activity of an enhancer across hundreds of kilobases underlines that GROMIT is not ideal for precise identification of enhancer location. However, it enables us to define the range of action of enhancers, the extent of large regulatory landscapes, as well as their boundaries. The distance separating two adjacent insertions showing different expression patterns can sometimes be as short as a few tens of kilobases [16], hinting to the position of possible regulatory insulator elements, which can hardly be identified by direct means.
In addition, detailed side-by-side comparison of reporter and endogenous gene expression, with the enhancer activities obtained by transgenic assays, showed that a substantial subset of autonomous enhancer activities were absent at the intact locus, and accordingly failed to activate the endogenous target gene or a nearby sensor. How this is achieved remains unclear (figure 3c). It is possible that enhancer activity per se is inhibited in certain tissues, for example by direct repressors of enhancer activity, possibly by establishing a repressive chromatin structure, which prevents binding of TFs. Alternatively, the ability of an enhancer to activate a gene or a sensor at a distance may be restricted by the chromosomal conformation of the locus or the interaction profile of that enhancer. Regardless of the mechanistic cause, however, these results imply that long-range regulatory activity is not only prevalent throughout the genome, but that the expression domains determined by enhancers are fine tuned locally by their interplay with other factors involved in gene regulation, resulting in a ‘latent’ potential that can be revealed in another genomic context.
Intriguingly, with few insertions within large gene-deserts, we observed expression of the reporter sensor, which was completely at odds with the activity of flanking endogenous protein-coding genes, defining regulatory landscapes that do not seem to include target genes. It is possible that these regulatory activities control the expression of un-annotated or non-coding genes, such as microRNAs (miRNAs) or long non-coding RNAs (lncRNAs) or even act in trans, towards a different chromosome or at extremely long distance. However, it could also suggest that gene-poor regions may be inhabited by a plethora of elements with tissue-specific regulatory potential, without attributed target genes and only latent biological functions.
9. Outlining the regulatory map of the genome
Taken together, the operational scan of regulatory potential provided by GROMIT, and our prior knowledge of transcriptional regulation revealed some novel principles of the global regulatory architecture of the mouse genome (figure 4a). First, we observed the pervasive presence of extended regulatory landscapes. Within these landscapes, related expression patterns were observed at multiple positions by both our regulatory sensor and the endogenous genes. A comparison with known enhancers revealed that these expression patterns are often the integrated output of multiple regulatory elements. Importantly, an endogenous gene may be associated with overlapping yet distinct landscapes, each one with different tissue specificities and covering different intervals. The subdivision of a single genomic locus into different regulatory landscapes may arise from the relative positions and properties of the regulatory elements that lie within it. In addition to the range of action of individual enhancers, structural constraints and higher-order three-dimensional organization of the genome, including lamin-associated domains [90,91], CTCF-loops [92] or topologically associated domains [93,94] may also influence the formation of these landscapes. Thus, the extended regulatory landscapes observed with GROMIT are probably the functional consequence of chromatin and conformational structures, as well as of the spatial range of enhancers and of their extensive interactions, as described for the Hoxd regulatory archipelago [65].
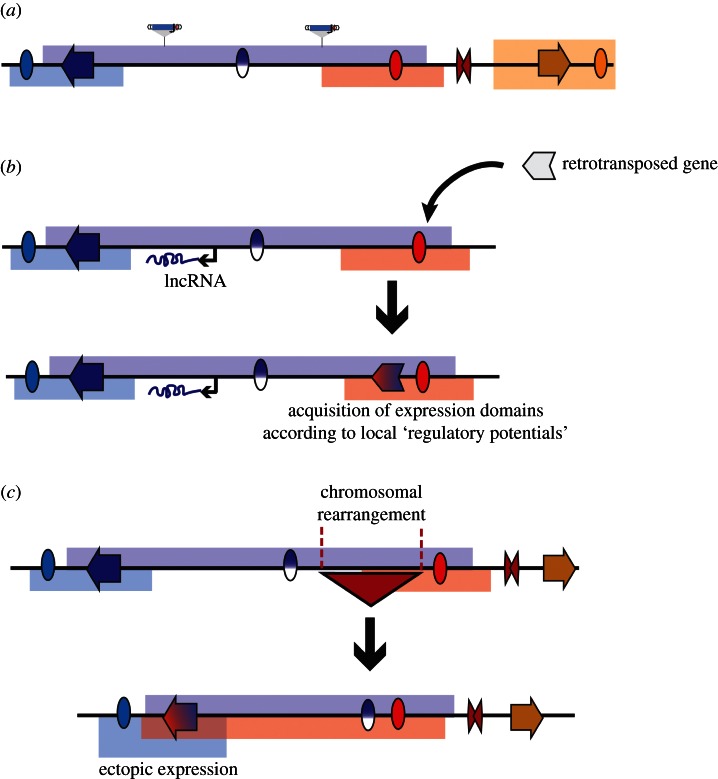
Figure 4.
An integrated view of regulatory landscapes and their implications. (a) The activity of enhancers (ovals) is distributed throughout extended regions (regulatory landscapes, indicated by bars in the same colour as the enhancer), rather than only activating specific promoters. Activities of different enhancers can overlap, giving rise to complex expression patterns. Some enhancer activities are masked within their endogenous environment, giving rise to latent activities (indicated by split blue-white oval). Distinct regulatory landscapes can be separated by insulator-like elements (red double triangle). Because regulatory activities are not targeted towards genes, this results in the widespread presence of tissue-specific expression potential, captured with GROMIT insertions (indicated by schematic of transposon). (b) The pervasive presence of regulatory activities within the regulatory landscapes can contribute to transcription of non-coding genes, such as long non-coding RNAs (lncRNAs, indicated by black arrow), giving rise to their tissue-specific expression patterns (shown by transcript in the same colour as regulatory landscape). Novel genes can easily acquire a specific expression, for example by retrotransposition into an existing regulatory landscape. Thus, a retrogene with no activity by itself (white arrow) will gain expression as part of the regulatory landscape (red–blue arrow). (c) Chromosomal rearrangements can also lead to novel expression patterns. As demonstrated by a schematic deletion of the region overlapping the red triangle, rearrangements can results in alterations of the regulatory landscape, thereby putting genes within the range of action of remote regulatory elements. Such ‘position effects’ can contribute to disease [17,88] and to the evolution of gene expression [89].
A future challenge will remain to precisely relate our operational view of regulatory elements and transcriptional output, with the underlying mechanics of gene regulation, and define the causal and functional relationships between regulatory and structural domains. To obtain such an understanding will require a multi-pronged approach, identifying single elements and their intrinsic activity, generating high-resolution, high-density data from systems such as GROMIT to determine the integrated regulatory output, as well as from biochemical assays to map epigenetic modifications and chromosomal interactions. Such detailed studies will also identify what factors contribute to limit the range of action of enhancers, and what causes the local differences in their activity.
10. Further implications: from regulatory maps to gene expression and phenotype
GROMIT exposed the fact that enhancer activities are not exclusively and selectively targeted to gene promoters, but distributed throughout the genome. This suggests that similar to our regulatory sensor, any type of cryptic promoter may acquire intergenic non-coding transcripts, the existence of which has been widely documented [95]. In the light of our findings, the tissue-specific expression of non-coding transcripts could therefore be a trait that can arise simply from their genomic location, and is not automatically a strong indication of functionality. Consequently, the overlap of expression between lncRNAs and their flanking protein-coding genes may not necessarily indicate functional regulation in cis of the latter by the first [96,97], but simply their common location within the same regulatory landscape (figure 4b). This is not to say that all non-coding RNAs are simply transcriptional noise. But the widespread and promiscuous distribution of tissue-specific regulatory activities revealed by GROMIT indicate that expression specificity may not be sufficient indication of functionality, and that additional experimental evidence is necessary to determine the biological relevance of non-coding transcripts [98].
On the other hand, the prevalence of tissue-specific regulatory activities within large intergenic domains also constitutes opportunities for evolutionary tinkering: they provide activities that can facilitate the emergence of functional lncRNAs, and explain how retrotransposed genes [99], and evolutionarily young genes such as orphan genes [100] or protogenes [101] can obtain specific expression domains (figure 4b).
Importantly, the local modulation of intrinsic regulatory activities implies that structural changes in the genome, such as deletions, duplications and inversion, could alter this interplay, leading to alterations in the regulatory landscape, through specific gain or loss of expression patterns (i.e. the masking or unmasking of regulatory potential). Whereas the juxtaposition of existing genes in a novel regulatory environment could result in acquisition of new expression patterns (figure 4c). This phenomenon could be the cause of several pathological conditions, where genomic rearrangements ‘move’ genes in new regulatory environments, exposing them to novel regulatory influences, either by ‘adopting’ regulatory influences normally acting on a different gene [10,88,102], or by unmasking ‘latent’ regulatory activities. The hereditary mixed polyposis syndrome caused by a duplication upstream of the GREMLIN gene [103] and pre-axial polydactyly caused by duplication of the limb-enhancer region of SHH [24,25] may illustrate this latter phenomenon well. More generally, such a phenomenon provides a new framework for understanding the phenotypic impact of the widespread structural variation found in the human population [104]. Thus, phenotypes may not only arise directly from the deletion or duplication of regulatory elements, but in part also from shifting the boundaries of existing regulatory landscapes, leading to modulation of gene expression in the vicinity of structural variants, up to few megabases away. Importantly, this is in line with experimental evidence of altered gene expression as a consequence of genomic rearrangements, both in humans and mice.
Thus, ultimately, an integrated map of the regulatory organization of the genome, including the position of enhancers, but also their range of action, their interactions, the location of regulatory boundaries and latent regulatory potentials will be instrumental to translate individual genomic sequence information into phenotypic predictions.
Acknowledgements
We thank the members of the laboratory for helpful discussions and for contributing to the development of the GROMIT system and dataset, as well as the anonymous reviewers for helpful comments. F.S.'s research is supported by grants from the European Commission-FP7 (Health 223210/CISSTEM), Human Frontier Science Programme (grant no. RGY0081/2008-C) and the German Research Foundation (DFG-SP1331/3-1). O.S. is supported by a fellowship awarded by the Louis-Jeantet Foundation.
References
- 1.Visel A, Rubin EM, Rubin EM, Pennacchio LA. 2009. Genomic views of distant-acting enhancers. Nature 461, 199–205 10.1038/nature08451 (doi:10.1038/nature08451) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Williamson I, Hill RE, Bickmore WA. 2011. Enhancers: from developmental genetics to the genetics of common human disease. Dev. Cell 21, 17–19 10.1016/j.devcel.2011.06.008 (doi:10.1016/j.devcel.2011.06.008) [DOI] [PubMed] [Google Scholar]
- 3.Bulger M, Groudine M. 2010. Enhancers: the abundance and function of regulatory sequences beyond promoters. Dev. Biol. 339, 250–257 10.1016/j.ydbio.2009.11.035 (doi:10.1016/j.ydbio.2009.11.035) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lettice LA, et al. 2003. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum. Mol. Genet. 12, 1725–1735 10.1093/hmg/ddg180 (doi:10.1093/hmg/ddg180) [DOI] [PubMed] [Google Scholar]
- 5.Antonellis A. 2005. Deletion of long-range sequences at Sox10 compromises developmental expression in a mouse model of Waardenburg-Shah (WS4) syndrome. Hum. Mol. Genet. 15, 259–271 10.1093/hmg/ddi442 (doi:10.1093/hmg/ddi442) [DOI] [PubMed] [Google Scholar]
- 6.Ghiasvand NM, Rudolph DD, Mashayekhi M, Brzezinski JA, Goldman D, Glaser T. 2011. Deletion of a remote enhancer near ATOH7 disrupts retinal neurogenesis, causing NCRNA disease. Nat. Neurosci. 14, 578–586 10.1038/nn.2798 (doi:10.1038/nn.2798) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.D'haene B, et al. 2009. Disease-causing 7.4 kb cis-regulatory deletion disrupting conserved non-coding sequences and their interaction with the FOXL2 promotor: implications for mutation screening. PLoS Genet. 5, e1000522. 10.1371/journal.pgen.1000522 (doi:10.1371/journal.pgen.1000522) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loots GG, Kneissel M, Keller H, Baptist M, Chang J, Collette NM, Ovcharenko D, Plajzer-Frick I, Rubin EM. 2005. Genomic deletion of a long-range bone enhancer misregulates sclerostin in Van Buchem disease. Genome Res. 15, 928–935 10.1101/gr.3437105 (doi:10.1101/gr.3437105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sabherwal N, et al. 2007. Long-range conserved non-coding SHOX sequences regulate expression in developing chicken limb and are associated with short stature phenotypes in human patients. Hum. Mol. Genet. 16, 210–222 10.1093/hmg/ddl470 (doi:10.1093/hmg/ddl470) [DOI] [PubMed] [Google Scholar]
- 10.Spitz F, Gonzalez F, Duboule D. 2003. A global control region defines a chromosomal regulatory landscape containing the HoxD cluster. Cell 113, 405–417 10.1016/S0092-8674(03)00310-6 (doi:10.1016/S0092-8674(03)00310-6) [DOI] [PubMed] [Google Scholar]
- 11.Zuniga A, et al. 2004. Mouse limb deformity mutations disrupt a global control region within the large regulatory landscape required for Gremlin expression. Genes Dev. 18, 1553–1564 10.1101/gad.299904 (doi:10.1101/gad.299904) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sakabe NJ, Savic D, Nobrega MA. 2012. Transcriptional enhancers in development and disease. Genome Biol. 13, 238. 10.1186/gb-2012-13-1-238 (doi:10.1186/gb-2012-13-1-238) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Consortium TEP. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 10.1038/nature11247 (doi:10.1038/nature11247) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li G, et al. 2012. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell 148, 84–98 10.1016/j.cell.2011.12.014 (doi:10.1016/j.cell.2011.12.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanyal A, Lajoie BR, Jain G, Dekker J. 2012. The long-range interaction landscape of gene promoters. Nature 489, 109–113 10.1038/nature11279 (doi:10.1038/nature11279) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ruf S, Symmons O, Uslu VV, Dolle D, Hot C, Ettwiller L, Spitz F. 2011. Large-scale analysis of the regulatory architecture of the mouse genome with a transposon-associated sensor. Nat. Genet. 43, 379–386 10.1038/ng.790 (doi:10.1038/ng.790) [DOI] [PubMed] [Google Scholar]
- 17.Kleinjan DA, van Heyningen V. 2005. Long-range control of gene expression: emerging mechanisms and disruption in disease. Am. J. Hum. Genet. 76, 8–32 10.1086/426833 (doi:10.1086/426833) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benko S, et al. 2009. Highly conserved non-coding elements on either side of SOX9 associated with Pierre Robin sequence. Nat. Genet. 41, 359–364 10.1038/ng.329 (doi:10.1038/ng.329) [DOI] [PubMed] [Google Scholar]
- 19.Birnbaum RY, et al. 2012. Coding exons function as tissue-specific enhancers of nearby genes. Genome Res. 22, 1059–1068 10.1101/gr.133546.111 (doi:10.1101/gr.133546.111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alkan C, Coe BP, Eichler EE. 2011. Genome structural variation discovery and genotyping. Nat. Rev. Genet. 12, 363–376 10.1038/nrg2958 (doi:10.1038/nrg2958) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dathe K, et al. 2009. Duplications involving a conserved regulatory element downstream of BMP2 are associated with brachydactyly type A2. Am. J. Hum. Genet. 84, 483–492 10.1016/j.ajhg.2009.03.001 (doi:10.1016/j.ajhg.2009.03.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurth I, et al. 2009. Duplications of noncoding elements 5′ of SOX9 are associated with brachydactyly-anonychia. Nat. Genet. 41, 862–863 10.1038/ng0809-862 (doi:10.1038/ng0809-862) [DOI] [PubMed] [Google Scholar]
- 23.de Mollerat XJ, et al. 2003. A genomic rearrangement resulting in a tandem duplication is associated with split hand–split foot malformation 3 (SHFM3) at 10q24. Hum. Mol. Genet. 12, 1959–1971 [DOI] [PubMed] [Google Scholar]
- 24.Klopocki E, Ott C, Benatar N, Ullmann R, Mundlos S, Lehmann K. 2008. A microduplication of the long range SHH limb regulator (ZRS) is associated with triphalangeal thumb-polysyndactyly syndrome. J. Med. Genet. 45, 370–375 10.1136/jmg.2007.055699 (doi:10.1136/jmg.2007.055699) [DOI] [PubMed] [Google Scholar]
- 25.Sun M, et al. 2008. Triphalangeal thumb-polysyndactyly syndrome and syndactyly type IV are caused by genomic duplications involving the long-range, limb-specific SHH enhancer. J. Med. Genet. 45, 589–595 10.1136/jmg.2008.057646 (doi:10.1136/jmg.2008.057646) [DOI] [PubMed] [Google Scholar]
- 26.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. 2009. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl Acad. Sci. USA 106, 9362–9367 10.1073/pnas.0903103106 (doi:10.1073/pnas.0903103106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keane TM, et al. 2011. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 477, 289–294 10.1038/nature10413 (doi:10.1038/nature10413) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chan YF, et al. 2010. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science 327, 302–305 10.1126/science.1182213 (doi:10.1126/science.1182213) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McLean CY, et al. 2011. Human-specific loss of regulatory DNA and the evolution of human-specific traits. Nature 471, 216–219 10.1038/nature09774 (doi:10.1038/nature09774) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prud'homme B, Gompel N, Rokas A, Kassner VA, Williams TM, Yeh S-D, True JR, Carroll SB. 2006. Repeated morphological evolution through cis-regulatory changes in a pleiotropic gene. Nature 440, 1050–1053 10.1038/nature04597 (doi:10.1038/nature04597) [DOI] [PubMed] [Google Scholar]
- 31.Jones FC, et al. 2012. The genomic basis of adaptive evolution in threespine sticklebacks. Nature 484, 55–61 10.1038/nature10944 (doi:10.1038/nature10944) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bulger M, Groudine M. 2011. Functional and mechanistic diversity of distal transcription enhancers. Cell 144, 327–339 10.1016/j.cell.2011.01.024 (doi:10.1016/j.cell.2011.01.024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hardison RC, Taylor J. 2012. Genomic approaches towards finding cis-regulatory modules in animals. Nat. Rev. Genet. 13, 469–483 10.1038/nrg3242 (doi:10.1038/nrg3242) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Woolfe A, Elgar G. 2008. Organization of conserved elements near key developmental regulators in vertebrate genomes. Adv. Genet. 61, 307–338 10.1016/S0065-2660(07)00012-0 (doi:10.1016/S0065-2660(07)00012-0) [DOI] [PubMed] [Google Scholar]
- 35.Pennacchio LA, et al. 2006. In vivo enhancer analysis of human conserved non-coding sequences. Nature 444, 499–502 10.1038/nature05295 (doi:10.1038/nature05295) [DOI] [PubMed] [Google Scholar]
- 36.Blow MJ, et al. 2010. ChIP-Seq identification of weakly conserved heart enhancers. Nat. Genet. 42, 806–810 10.1038/ng.650 (doi:10.1038/ng.650) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kadonaga JT. 2004. Regulation of RNA polymerase II transcription by sequence-specific DNA binding factors. Cell 116, 247–257 10.1016/S0092-8674(03)01078-X (doi:10.1016/S0092-8674(03)01078-X) [DOI] [PubMed] [Google Scholar]
- 38.Gross DS, Garrard WT. 1988. Nuclease hypersensitive sites in chromatin. Annu. Rev. Biochem. 57, 159–197 10.1146/annurev.bi.57.070188.001111 (doi:10.1146/annurev.bi.57.070188.001111) [DOI] [PubMed] [Google Scholar]
- 39.Giresi PG, Kim J, McDaniell RM, Iyer VR, Lieb JD. 2007. FAIRE (formaldehyde-assisted isolation of regulatory elements) isolates active regulatory elements from human chromatin. Genome Res. 17, 877–885 10.1101/gr.5533506 (doi:10.1101/gr.5533506) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heintzman ND, et al. 2009. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459, 108–112 10.1038/nature07829 (doi:10.1038/nature07829) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ernst J, et al. 2011. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473, 43–49 10.1038/nature09906 (doi:10.1038/nature09906) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shen Y, et al. 2012. A map of the cis-regulatory sequences in the mouse genome. Nature 488, 116–120 10.1038/nature11243 (doi:10.1038/nature11243) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. 2011. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470, 279–283 10.1038/nature09692 (doi:10.1038/nature09692) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Creyghton MP, et al. 2010. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl Acad. Sci. USA 107, 21 931–21 936 10.1073/pnas.1016071107 (doi:10.1073/pnas.1016071107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dekker J. 2008. Gene regulation in the third dimension. Science 319, 1793–1794 10.1126/science.1152850 (doi:10.1126/science.1152850) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Wit E, de Laat W. 2012. A decade of 3C technologies: insights into nuclear organization. Genes Dev. 26, 11–24 10.1101/gad.179804.111 (doi:10.1101/gad.179804.111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li X-Y, et al. 2008. Transcription factors bind thousands of active and inactive regions in the Drosophila blastoderm. PLoS Biol. 6, e27. 10.1371/journal.pbio.0060027 (doi:10.1371/journal.pbio.0060027) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Biggin MD. 2010. MyoD, a lesson in widespread DNA binding. Dev. Cell 18, 505–506 10.1016/j.devcel.2010.04.004 (doi:10.1016/j.devcel.2010.04.004) [DOI] [PubMed] [Google Scholar]
- 49.Spitz F, Furlong EEM. 2012. Transcription factors: from enhancer binding to developmental control. Nat. Rev. Genet. 13, 613–626 10.1038/nrg3207 (doi:10.1038/nrg3207) [DOI] [PubMed] [Google Scholar]
- 50.Patwardhan RP, et al. 2012. Massively parallel functional dissection of mammalian enhancers in vivo. Nat. Biotechnol. 30, 265–270 10.1038/nbt.2136 (doi:10.1038/nbt.2136) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Melnikov A, et al. 2012. Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nat. Biotechnol. 30, 271–277 10.1038/nbt.2137 (doi:10.1038/nbt.2137) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kawakami K. 2005. Transposon tools and methods in zebrafish. Dev. Dyn. 234, 244–254 10.1002/dvdy.20516 (doi:10.1002/dvdy.20516) [DOI] [PubMed] [Google Scholar]
- 53.Belteki G, Gertsenstein M, Ow DW, Nagy A. 2003. Site-specific cassette exchange and germline transmission with mouse ES cells expressing phiC31 integrase. Nat. Biotechnol. 21, 321–324 10.1038/nbt787 (doi:10.1038/nbt787) [DOI] [PubMed] [Google Scholar]
- 54.Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D. 2002. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science 295, 868–872 10.1126/science.1067081 (doi:10.1126/science.1067081) [DOI] [PubMed] [Google Scholar]
- 55.Friedli M, et al. 2010. A systematic enhancer screen using lentivector transgenesis identifies conserved and non-conserved functional elements at the Olig1 and Olig2 locus. PLoS ONE 5, e15741. 10.1371/journal.pone.0015741 (doi:10.1371/journal.pone.0015741) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Visel A, Minovitsky S, Dubchak I, Pennacchio LA. 2007. VISTA Enhancer Browser–a database of tissue-specific human enhancers. Nucleic Acids Res. 35, D88–D92 10.1093/nar/gkl822 (doi:10.1093/nar/gkl822) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li Q, Ritter D, Yang N, Dong Z, Li H, Chuang JH, Guo S. 2010. A systematic approach to identify functional motifs within vertebrate developmental enhancers. Dev. Biol. 337, 484–495 10.1016/j.ydbio.2009.10.019 (doi:10.1016/j.ydbio.2009.10.019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Visel A, Akiyama JA, Shoukry M, Afzal V, Rubin EM, Pennacchio LA. 2009. Functional autonomy of distant-acting human enhancers. Genomics 93, 509–513 10.1016/j.ygeno.2009.02.002 (doi:10.1016/j.ygeno.2009.02.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dunipace L, Ozdemir A, Stathopoulos A. 2011. Complex interactions between cis-regulatory modules in native conformation are critical for Drosophila snail expression. Development 138, 4075–4084 10.1242/dev.069146 (doi:10.1242/dev.069146) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Summerbell D, Ashby PR, Coutelle O, Cox D, Yee S, Rigby PW. 2000. The expression of Myf5 in the developing mouse embryo is controlled by discrete and dispersed enhancers specific for particular populations of skeletal muscle precursors. Development 127, 3745–3757 [DOI] [PubMed] [Google Scholar]
- 61.Prazak L, Fujioka M, Gergen JP. 2010. Non-additive interactions involving two distinct elements mediate sloppy-paired regulation by pair-rule transcription factors. Dev. Biol. 344, 1048–1059 10.1016/j.ydbio.2010.04.026 (doi:10.1016/j.ydbio.2010.04.026) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kasowski M, et al. 2010. Variation in transcription factor binding among humans. Science 328, 232–235 10.1126/science.1183621 (doi:10.1126/science.1183621) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Voss AK, Vanyai HK, Collin C, Dixon MP, McLennan TJ, Sheikh BN, Scambler P, Thomas T. 2012. MOZ regulates the Tbx1 locus, and Moz mutation partially phenocopies DiGeorge syndrome. Dev. Cell 23, 652–663 10.1016/j.devcel.2012.07.010 (doi:10.1016/j.devcel.2012.07.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Patrinos GP, de Krom M, de Boer E, Langeveld A, Imam AMA, Strouboulis J, de Laat W, Grosveld FG. 2004. Multiple interactions between regulatory regions are required to stabilize an active chromatin hub. Genes Dev. 18, 1495–1509 10.1101/gad.289704 (doi:10.1101/gad.289704) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Montavon T, Soshnikova N, Mascrez B, Joye E, Thevenet L, Splinter E, de Laat W, Spitz F, Duboule D. 2011. A regulatory archipelago controls Hox genes transcription in digits. Cell 147, 1132–1145 10.1016/j.cell.2011.10.023 (doi:10.1016/j.cell.2011.10.023) [DOI] [PubMed] [Google Scholar]
- 66.Leddin M, et al. 2011. Two distinct auto-regulatory loops operate at the PU.1 locus in B cells and myeloid cells. Blood 117, 2827–2838 10.1182/blood-2010-08-302976 (doi:10.1182/blood-2010-08-302976) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schedl A, Ross A, Lee M, Engelkamp D, Rashbass P, van Heyningen V, Hastie ND. 1996. Influence of PAX6 gene dosage on development: overexpression causes severe eye abnormalities. Cell 86, 71–82 10.1016/S0092-8674(00)80078-1 (doi:10.1016/S0092-8674(00)80078-1) [DOI] [PubMed] [Google Scholar]
- 68.Lettice LA, et al. 2002. Disruption of a long-range cis-acting regulator for Shh causes preaxial polydactyly. Proc. Natl Acad. Sci. USA 99, 7548–7553 10.1073/pnas.112212199 (doi:10.1073/pnas.112212199) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kleinjan DA, Seawright A, Mella S, Carr CB, Tyas DA, Simpson TI, Mason JO, Price DJ, van Heyningen V. 2006. Long-range downstream enhancers are essential for Pax6 expression. Dev. Biol. 299, 563–581 10.1016/j.ydbio.2006.08.060 (doi:10.1016/j.ydbio.2006.08.060) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fullwood MJ, et al. 2009. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature 462, 58–64 10.1038/nature08497 (doi:10.1038/nature08497) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schoenfelder S, et al. 2010. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat. Genet. 42, 53–61 10.1038/ng.496 (doi:10.1038/ng.496) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Edelman LB, Fraser P. 2012. Transcription factories: genetic programming in three dimensions. Curr. Opin. Genet. Dev. 22, 110–114 10.1016/j.gde.2012.01.010 (doi:10.1016/j.gde.2012.01.010) [DOI] [PubMed] [Google Scholar]
- 73.Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R. 2006. Interchromosomal interactions and olfactory receptor choice. Cell 126, 403–413 10.1016/j.cell.2006.06.035 (doi:10.1016/j.cell.2006.06.035) [DOI] [PubMed] [Google Scholar]
- 74.Noordermeer D, et al. 2011. Variegated gene expression caused by cell-specific long-range DNA interactions. Nat. Cell Biol. 13, 944–951 10.1038/ncb2278 (doi:10.1038/ncb2278) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA. 2005. Interchromosomal associations between alternatively expressed loci. Nature 435, 637–645 10.1038/nature03574 (doi:10.1038/nature03574) [DOI] [PubMed] [Google Scholar]
- 76.Yaffe E, Tanay A. 2011. Probabilistic modeling of Hi-C contact maps eliminates systematic biases to characterize global chromosomal architecture. Nat. Genet. 43, 1059–1065 10.1038/ng.947 (doi:10.1038/ng.947) [DOI] [PubMed] [Google Scholar]
- 77.Kalhor R, Tjong H, Jayathilaka N, Alber F, Chen L. 2012. Genome architectures revealed by tethered chromosome conformation capture and population-based modeling. Nat. Biotechnol. 30, 90–98 10.1038/nbt.2057 (doi:10.1038/nbt.2057) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gonçalves A, Leigh-Brown S, Thybert D, Stefflova K, Turro E, Flicek P, Brazma A, Odom DT, Marioni JC. 2012. Extensive compensatory cis-trans regulation in the evolution of mouse gene expression. Genome Res. 22, 2376–2384 10.1101/gr.142281.112 (doi:10.1101/gr.142281.112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Calhoun VC, Stathopoulos A, Levine M. 2002. Promoter-proximal tethering elements regulate enhancer-promoter specificity in the Drosophila Antennapedia complex. Proc. Natl Acad. Sci. USA 99, 9243–9247 10.1073/pnas.142291299 (doi:10.1073/pnas.142291299) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yang J, Corces VG. 2011. Chromatin insulators: a role in nuclear organization and gene expression. Adv. Cancer Res. 110, 43–76 10.1016/B978-0-12-386469-7.00003-7 (doi:10.1016/B978-0-12-386469-7.00003-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lower KM, et al. 2009. Adventitious changes in long-range gene expression caused by polymorphic structural variation and promoter competition. Proc. Natl Acad. Sci. USA 106, 21 771–21 776 10.1073/pnas.0909331106 (doi:10.1073/pnas.0909331106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cajiao I, Zhang A, Yoo EJ, Cooke NE, Liebhaber SA. 2004. Bystander gene activation by a locus control region. EMBO J. 23, 3854–3863 10.1038/sj.emboj.7600365 (doi:10.1038/sj.emboj.7600365) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ivics Z, Hackett PB, Plasterk RH, Izsvák Z. 1997. Molecular reconstruction of sleeping beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell 91, 501–510 10.1016/S0092-8674(00)80436-5 (doi:10.1016/S0092-8674(00)80436-5) [DOI] [PubMed] [Google Scholar]
- 84.Keng VW, et al. 2005. Region-specific saturation germline mutagenesis in mice using the sleeping beauty transposon system. Nat. Methods 2, 763–769 10.1038/nmeth795 (doi:10.1038/nmeth795) [DOI] [PubMed] [Google Scholar]
- 85.Visel A, et al. 2009. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457, 854–858 10.1038/nature07730 (doi:10.1038/nature07730) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Butler JE, Kadonaga JT. 2001. Enhancer-promoter specificity mediated by DPE or TATA core promoter motifs. Genes Dev. 15, 2515–2519 10.1101/gad.924301 (doi:10.1101/gad.924301) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kikuta H, et al. 2007. Genomic regulatory blocks encompass multiple neighbouring genes and maintain synteny in vertebrates. Genome Res. 17, 545–555 10.1101/gr.6086307 (doi:10.1101/gr.6086307) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lettice LA, et al. 2011. Enhancer-adoption as a mechanism of human developmental disease. Hum. Mutat. 32, 1492–1499 10.1002/humu.21615 (doi:10.1002/humu.21615) [DOI] [PubMed] [Google Scholar]
- 89.Cande JD, Chopra VS, Levine M. 2009. Evolving enhancer-promoter interactions within the tinman complex of the flour beetle, Tribolium castaneum. Development 136, 3153–3160 10.1242/dev.038034 (doi:10.1242/dev.038034) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Guelen L, et al. 2008. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453, 948–951 10.1038/nature06947 (doi:10.1038/nature06947) [DOI] [PubMed] [Google Scholar]
- 91.Peric-Hupkes D, et al. 2010. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol. Cell 38, 603–613 10.1016/j.molcel.2010.03.016 (doi:10.1016/j.molcel.2010.03.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Handoko L, et al. 2011. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat. Genet. 43, 630–638 10.1038/ng.857 (doi:10.1038/ng.857) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 10.1038/nature11082 (doi:10.1038/nature11082) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nora EP, et al. 2012. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485, 381–385 10.1038/nature11049 (doi:10.1038/nature11049) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kapranov P, Willingham AT, Gingeras TR. 2007. Genome-wide transcription and the implications for genomic organization. Nat. Rev. Genet. 8, 413–423 10.1038/nrg2083 (doi:10.1038/nrg2083) [DOI] [PubMed] [Google Scholar]
- 96.Ravasi T, et al. 2006. Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Res. 16, 11–19 10.1101/gr.4200206 (doi:10.1101/gr.4200206) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dinger ME, et al. 2008. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 18, 1433–1445 10.1101/gr.078378.108 (doi:10.1101/gr.078378.108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ponting CP, Oliver PL, Reik W. 2009. Evolution and functions of long noncoding RNAs. Cell 136, 629–641 10.1016/j.cell.2009.02.006 (doi:10.1016/j.cell.2009.02.006) [DOI] [PubMed] [Google Scholar]
- 99.Marques AC, Dupanloup I, Vinckenbosch N, Reymond A, Kaessmann H. 2005. Emergence of young human genes after a burst of retroposition in primates. PLoS Biol. 3, e357. 10.1371/journal.pbio.0030357 (doi:10.1371/journal.pbio.0030357) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tautz D, Domazet-Lošo T. 2011. The evolutionary origin of orphan genes. Nat. Rev. Genet. 12, 692–702 10.1038/nrg3053 (doi:10.1038/nrg3053) [DOI] [PubMed] [Google Scholar]
- 101.Carvunis A-R, et al. 2012. Proto-genes and de novo gene birth. Nature 487, 370–374 10.1038/nature11184 (doi:10.1038/nature11184) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kokubu C, et al. 2003. Undulated short-tail deletion mutation in the mouse ablates Pax1 and leads to ectopic activation of neighboring Nkx2-2 in domains that normally express Pax1. Genetics 165, 299–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jaeger E, et al. 2012. Hereditary mixed polyposis syndrome is caused by a 40 kb upstream duplication that leads to increased and ectopic expression of the BMP antagonist GREM1. Nat. Genet. 44, 699–703 10.1038/ng.2263 (doi:10.1038/ng.2263) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Feuk L, Carson AR, Scherer SW. 2006. Structural variation in the human genome. Nat. Rev. Genet. 7, 85–97 10.1038/nrg1767 (doi:10.1038/nrg1767) [DOI] [PubMed] [Google Scholar]