Abstract
Transcriptional enhancers are a major class of functional element embedded in the vast non-coding portion of the human genome. Acting over large genomic distances, enhancers play critical roles in the tissue and cell type-specific regulation of genes, and there is mounting evidence that they contribute to the aetiology of many human diseases. Methods for genome-wide mapping of enhancer regions are now available, but the functional architecture contained within human enhancer elements remains unclear. Here, we review recent approaches aimed at understanding the functional anatomy of individual enhancer elements, using systematic qualitative and quantitative assessments of mammalian enhancer variants in cultured cells and in vivo. These studies provide direct insight into common architectural characteristics of enhancers including the presence of multiple transcription factor-binding sites and the mixture of both transcriptionally activating and repressing domains within the same enhancer. Despite such progress in understanding the functional composition of enhancers, the inherent complexities of enhancer anatomy continue to limit our ability to predict the impact of sequence changes on in vivo enhancer function. While providing an initial glimpse into the mutability of mammalian enhancers, these observations highlight the continued need for experimental enhancer assessment as genome sequencing becomes routine in the clinic.
Keywords: enhancer, gene regulation, mutation, mouse genomics
1. Introduction
Transcriptional enhancers are non-coding regulatory sequences, important for the temporal and spatial in vivo expression of genes [1]. They can be located tens to hundreds of thousands of base pairs away from their target genes and function through chromatin remodelling and DNA looping to activate transcription of their target genes' promoters [2,3]. Recent evidence suggests the existence of hundreds of thousands of enhancers distributed throughout our genome [4,5]. Furthermore, a majority of polymorphisms associated with human diseases through genome-wide association studies do not fall within protein-encoding sequence, nor are they in substantial linkage disequilibrium with protein-encoding sequences [1,6,7]. In conjunction with examples of individual enhancers implicated in human diseases, as outlined below, this raises the possibility that sequence changes in regulatory elements, particularly enhancers, contribute to a wide spectrum of human phenotypes.
Despite their proposed important roles in development and disease, major unanswered questions remain about enhancers. Unlike coding sequence, which has clearly defined and standardized structures, little is known about the sequence architecture present within enhancers. This lack of structural insight has thus far hampered efforts to predict enhancers computationally using DNA sequence alone [8], although recent computational methods using additional information such as transcription-factor binding data and collections of experimentally verified enhancers allow for substantially improved prediction of tissue-specific enhancers [9]. Furthermore, this lack of understanding about enhancer structure makes it difficult to assess the functional consequences of sequence changes within enhancers. Moving forward, as whole-genome sequencing becomes standard in human disease studies, sequence variants in enhancers will be identified with regularity, and the ability to quickly distinguish between functionally neutral, deleterious and possibly advantageous mutations will be of paramount importance. An understanding of enhancer architecture is needed to help predict the functional consequences of enhancer sequence variants in a way that is analogous to using in silico methods to predict the functional consequences of non-sense and missense protein-coding mutations.
Here, we review the role of enhancers in human disease and recent studies that have begun to illuminate the functional architecture present in mammalian enhancers. We also describe current experimental methods of assessing the impact of sequence changes on enhancer function. These studies suggest that mammalian enhancer architecture is highly heterogeneous, supporting the need for additional experimental characterization of such elements. Despite the variability, a few common characteristics of enhancer architecture are emerging. Mammalian enhancers often exhibit a high density of transcription-factor binding sites (TFBSs), a high degree of functional redundancy and a mixture of both transcriptionally activating and repressing elements.
2. Enhancers in human disease
The first human disease-associated enhancer mutations were identified in β-thalassemia patients who harboured unexplained deletions of non-coding sequence within the β-globin locus. Upon further study, it was recognized that these deletions removed important non-coding DNA that regulated β-globin expression, thereby linking enhancer loss to human disease [10,11]. Additional subsequent studies have identified alterations, including both internal mutations and full deletions, of enhancer elements that contribute to a variety of rare developmental disorders. These include limb malformations such as preaxial polydactyly [12], the bone morphology disorder van Buchem disease [13–15], the intestinal disorder Hirschsprung disease [16] and the eye malformation disorder aniridia [17]. A second set of examples of human disease in which enhancers likely play a role include disease-associated balanced translocations that disrupt the sequence contiguity between non-coding sequences and nearby genes [18]. These changes in genome structure, typically referred to as ‘position effects,’ have long been thought to cause disease by separating genes from the distant-acting regulatory elements required for their normal expression.
Evidence for a potential role of enhancers in more common human diseases has emerged from the observation that a significant fraction of disease-associated loci identified through genome-wide association studies contain no linked gene-coding sequence variants [1,6,7]. Furthermore, such non-coding disease-associated variants are highly enriched in putative enhancers [6,19]. Recently, there have been several reports of common and rare non-coding variants that alter gene expression and are associated with common human phenotypes. For example, large deletions or duplications of nearby non-coding sequences that change the expression levels of IRGM [20,21] and VIPR2 [22] have been associated with Crohn's disease and schizophrenia, respectively. Single nucleotide variants in a non-coding locus at 9p21 near the CDKN2A/B genes have been proposed to affect the risk for cardiovascular disease by changing the regulatory function of enhancers present in this interval [23–26]. Furthermore, prostate cancer-associated variants located in a 17q24.3 gene desert have been shown to alter the function of an enhancer regulating the expression of SOX9 [27,28].
Because most human disease studies have initially focused on functionally characterizing the effects of protein-coding variants, the examples listed above likely represent only a small subset of a potentially much larger pool of enhancer variants that contribute to both rare and common diseases. Human disease studies are currently poised to shift from whole-exome sequencing and genome-wide single nucleotide polymorphism genotyping to whole-genome sequencing as sequencing costs continue to decrease. This shift in technology will mean the discovery of a deluge of novel non-coding sequence variants. The first challenge in characterizing such variants will be identifying whether or not novel variants fall within functional non-coding elements, such as enhancers. Effective experimental methods for the genome-wide identification of enhancers have been developed [29], and the ENCODE project [4] and many others are currently using such methods to systematically identify where enhancers are located in the human genome. The second, currently more difficult, challenge will be determining whether or not a newly discovered enhancer variant is likely to be pathogenic. In contrast to enhancers, protein-coding sequences have clearly defined and well-understood structures. This has allowed for the development of computational programs that can quickly assess the likelihood that a coding variant is pathogenic [30,31], which has greatly facilitated human disease studies that use exome sequencing [32]. However, in silico tools are not currently available for assessing enhancer variants, and a better understanding of enhancer architecture is needed for such tools to be developed. What, then, is the current understanding of the architecture present in enhancers, and what experimental methods can be used to assess enhancer variation and facilitate the development of computational tools for predicting the pathogenic effects of enhancer variants?
3. Architectural studies of enhancers
Studies of enhancer architecture have largely focused on characterizing enhancers identified in invertebrate organisms, particularly the sea urchin Endo16 and CyIIIa enhancers [33], and the Drosophila even-skipped (eve) stripe 2 enhancer [34]. Architectural studies of these enhancers have identified numerous regulatory modules contained within each enhancer, and these modules are typically composed of one or a few TFBSs. These modules are often capable of carrying out specialized functions, generally independent of the other modules contained within the enhancer. These invertebrate examples have led to the so-called billboard, or information display, hypothesis of enhancer architecture: enhancers act as a collection of independent TFBS modules rather than as a cooperative unit [35]. Under this model, an enhancer is made up of several, often functionally redundant, modules, some of which activate transcription, some of which repress transcription and some of which amplify these other signals. The overall regulatory output of an enhancer is, therefore, produced by the net sum of all the independent elements contained within, and the order of the modules should have little effect on enhancer function [35]. As a consequence of functional redundancy and the lack of constraint on internal spatial organization, enhancers conforming to this model are predicted to be buffered against the effects of many mutations.
In contrast to the enhancer architectures described in invertebrates, one of the most well-characterized mammalian enhancers, the human interferon-β 1 (IFNB1) enhancer, shows very limited modularity and a strong dependence upon proper spatial organization [36]. This enhancer contains several distinct regulatory domains [37,38], but the domains are highly interdependent. Individually mutating any of the domains, or altering the spacing between them, is sufficient to significantly decrease or eliminate enhancer activity [36–38]. This locus has led to the ‘enhanceosome’ model of enhancer architecture: enhancers contain TFBSs that recruit proteins that act in a highly cooperative manner [35,36]. Proper spatial organization of these proteins, determined by the relative placement of their binding sites, is required for this synergistic activity and, thus, for proper enhancer function. As a consequence of these spatial constraints, enhancers conforming to this model are predicted to display little functional redundancy and be highly susceptible to inactivating mutations. Structural studies of transcription factor binding to the IFNB1 enhancer have experimentally demonstrated several aspects of this model and offer a mechanistic explanation for the susceptibility of this enhancer to inactivating mutations [39,40]. These studies have shown that a variety of transcription factors simultaneously interact with this enhancer and, collectively, make physical contact with nearly every nucleotide within the highly conserved core domain, providing additional support for the recruitment of an enhanceosome to this site.
The billboard and the enhanceosome models, both shaped by evidence derived from a relatively small set of prototypic examples, are useful approaches to explain general characteristics of enhancers, but evidence available for many other enhancers suggests that they merely represent the extreme ends of a spectrum of architectural diversity [34]. Supporting this, studies in Drosophila indicate that enhancers often fall somewhere on a continuum between complete modularity, where the spatial relationship between domains is unimportant, and total spatial constraint [41,42]. This raises the possibility that, likewise, observations at the well-studied human IFNB1 enhancer may not be useful as a generalized model of mammalian enhancers. Indeed, several recent in vivo studies examining the architecture found in mammalian enhancer sequences showcase the high degree of architectural diversity present in mammalian enhancers. We have divided these studies into those that examine qualitative versus quantitative effects of sequence variation on enhancer function to highlight the differences and trade-offs between the two types of experimental approaches.
(a). Qualitative in vivo assays of enhancer variation
Transgenic mouse reporter assays are one of the most widely used qualitative measures of mammalian in vivo enhancer activity. For these experiments, allelic variants of enhancers are linked to a reporter gene (for example, LacZ) and then individually delivered into mouse zygotes through pronuclear injection [43]. The resulting transgenic embryos or animals can then be scored visually for changes in reporter gene expression patterns. The strength of these in vivo experiments lies in their usefulness in assessing enhancer activity in whole organs or other structures found throughout the body of the organism, allowing for the identification of changes in both the intensity and the spatial pattern of gene expression resulting from enhancer mutations.
Like the IFNB1 enhancer discussed above, recent transgenic studies of several mouse enhancers are consistent with some enhancers having a low degree of functional redundancy and a high degree of domain interdependence. For these loci, modest enhancer variation can have dramatic effects on enhancer function. For example, dissection of two independent enhancers near the Gata4 gene [44,45] and one enhancer near Gjd3 [46] have demonstrated that mutating a single TFBS can be sufficient to abolish the enhancer's activity. Indeed, these enhancers contain several necessary TFBSs, and individually mutating any one of a handful of these sites appears to be sufficient to abolish activity, indicating a potentially high susceptibility of these enhancers to inactivating sequence changes.
By contrast, some mammalian enhancers appear to display the more modular, functionally redundant architecture common to invertebrate enhancers. Supporting this are two elegant studies that have recently examined the architecture present in the ZRS, the distant-acting enhancer that regulates limb expression of Sonic Hedgehog (SHH) during embryonic development [47,48]. Single base-pair changes in this highly conserved enhancer lead to preaxial polydactyly (extra digits occurring on the thumb side of the hand) and other limb abnormalities in humans [49], mice [49], cats [48] and chickens [50]. In vivo characterization of both naturally occurring variants and artificial variants affecting ZRS TFBSs have identified important domains throughout the 800 bp long ZRS that contribute to its activity [47,48]. Decreasing or eliminating normal ZRS activity requires rather severe mutations, such as the simultaneous removal of at least two TFBSs, indicating that the numerous TFBSs in this enhancer exhibit a high degree of functional redundancy. How, then, do single point mutations confer the polydactyly phenotype? Interestingly, the ZRS, like many other enhancers, contains a mixture of activating and repressing functional domains [47]. It is this balance between activation and repression that is responsible for the discrete activity of the ZRS, which primarily drives SHH expression only in the posterior portion of both the fore- and hindlimbs. When this balance is tipped further towards activation, as in the case of at least two of the preaxial polydactyly mutations that have been shown to create additional activating TFBSs, the spatial activity of the ZRS can expand into the anterior portion of the limbs. This, in turn causes ectopic anterior expression of SHH and, thereby, polydactyly. Although less common than loss-of-function mutations, this locus highlights that gain-of-function mutations in enhancers, which can be caused either by the creation of additional activating TFBSs or the disruption of repressing TFBSs, are important to consider in human disease studies.
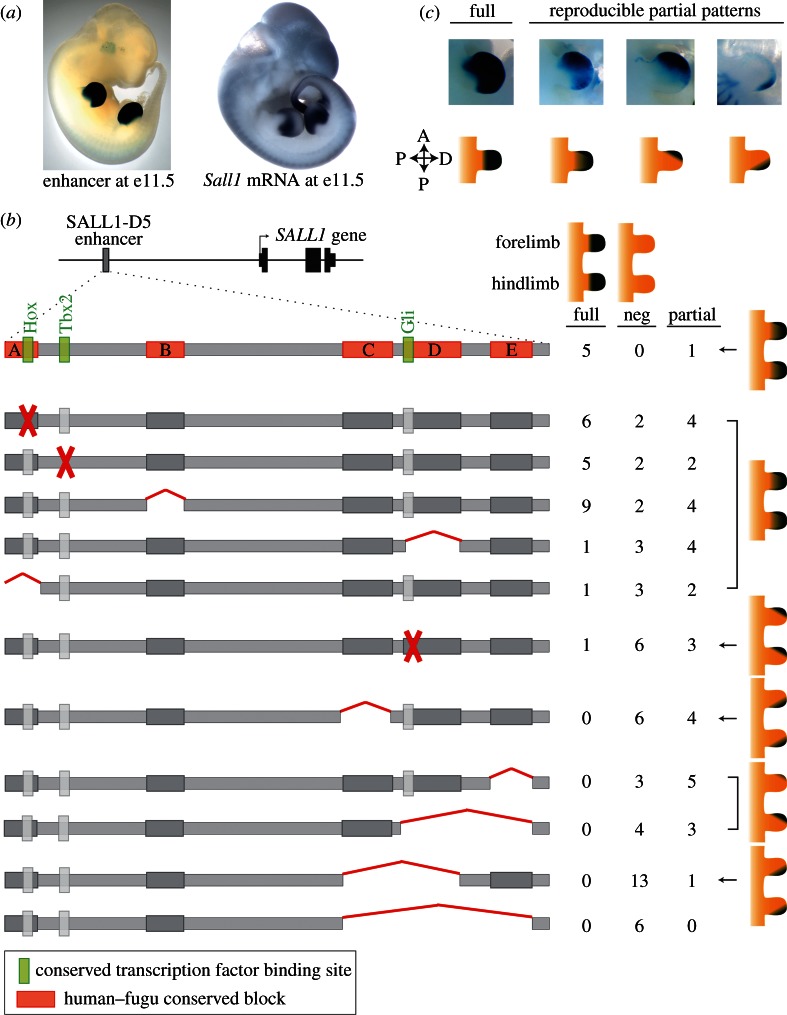
In addition to the ZRS, we provide here another, previously unpublished, example of a mammalian limb enhancer that displays characteristics of functional modularity and redundancy. This enhancer, SALL1-D5, is located approximately 500 kb upstream of the human SALL1 gene and was originally identified based on its extreme sequence conservation in most vertebrates [51]. When fused to a minimal promoter and LacZ reporter transgene, SALL1-D5 drives highly reproducible reporter gene expression throughout mouse embryonic limbs at embryonic day (e) 11.5 (figure 1a) in a pattern that recapitulates the expression pattern of Sall1 mRNA in the limb (figure 1a; [52]). Together, these observations suggest that SALL1-D5 is a regulator of SALL1 expression during vertebrate limb development. To better understand which sequences within SALL1-D5 are important for its enhancer function, we constructed a series of alleles containing substitutions that disrupt predicted binding sites for transcription factors active in limb development: Hox, Tbx2 and Gli (figure 1b). To help define the minimum sequence necessary for enhancer activity and to further elucidate the regulatory architecture present within SALL1-D5, we also constructed alleles containing deletions of sequences within the enhancer that are highly conserved between human and Fugu (figure 1b). These alleles were fused to a minimal promoter and LacZ reporter gene, and their effect on reporter gene expression in limb was tested at mouse embryonic day 11.5.
Figure 1.
Mutations to the ultraconserved SALL1 enhancer alter the spatial gene expression pattern. (a) Enhancer SALL1-D5 drives robust LacZ expression throughout the fore- and hindlimbs in embryonic day 11.5 (e11.5) mouse embryos in a pattern that is similar to limb expression of Sall1. (b) The SALL1-D5 enhancer is approximately 500 kb upstream of the transcriptional start site of human SALL1. To identify which sequences within the highly conserved 179 bp core of SALL1-D5 (grey bar) are crucial for limb expression, we engineered a series of substitution or deletion alleles, diagrammed here, and tested their effect on LacZ expression in e11.5 transgenic mouse embryos. To the right of each allele, the number of transgenic e11.5 embryos showing full, partial and no (neg) reporter gene expression in the limbs are indicated. The cartoons on the far right show the partial gene expression pattern observed for each allele. (c) Representative pictures of mouse e11.5 forelimbs showing full SALL1-D5 activity compared with partial activity patterns resulting from SALL1-D5 mutations. Partial patterns ranged from an overall reduction of SALL1-D5 activity throughout the limb to the restriction of activity to one side of the limb (anterior or posterior). A↔P: anterior-to-posterior axis, P↔D: proximal-to-distal axis.
Alterations to SALL1-D5 led to a variety of reproducible LacZ expression patterns in the developing limbs (figure 1b,c). Where wild-type SALL1-D5 drove strong LacZ expression throughout both the fore- and hindlimbs, some alleles resulted in an expression pattern that, while still present throughout the developing limb buds, was fainter, consistent with overall decreased enhancer activity. Substitutions abolishing the predicted Hox and Tbx2 binding sites, along with the deletion of three subregions highly conserved in vertebrates (A, B and D in figure 1b), had very modest effects on the enhancer activity of SALL1-D5 (figure 1b). Most of the embryos with these alleles displayed either full or mildly diluted enhancer activity throughout the limb buds. Other alleles resulted in LacZ patterns that were restricted to either the anterior or posterior portions of the limbs. Three alleles—a substitution to a predicted Gli-binding site, deletion of conserved site E and deletion of conserved sites D and E together—resulted in reporter gene expression that was restricted to the anterior portion of the limb buds. The deletion of conserved element C led to LacZ expression that was restricted to the posterior side of the limb bud. Only a substantial deletion of 62 bp, encompassing conserved elements C through E, completely abolished SALL1-D5 activity.
Taken together, these results are consistent with the presence of independent domains within the SALL1-D5 enhancer that are each responsible for only a portion of the enhancer's full spatial activity. It appears that sequences within conserved site C are responsible for gene expression in the anterior regions of the fore- and hindlimbs. Sequences within conserved site E, and to a lesser extent the putative Gli-binding site, appear responsible for enhancing gene expression in the posterior portions of the limb buds. Despite the observed modularity, the SALL1-D5 enhancer does not display a purely ‘billboard’ architecture. Some deletion alleles, particularly those missing conserved site C, lead to an increase in the number of transgenic embryos that have no reporter gene expression. These results suggest that there may be some limited cooperation between functional sites within the enhancer, and the loss of conserved site C may partly disrupt the activities of other functional elements. Like many of the Drosophila enhancers discussed previously, mammalian enhancers likely display characteristics of both the modular and the highly interdependent architectural models of enhancers.
(b). High-throughput quantitative assays of enhancer variation
Qualitative transgenic assessments of enhancers are powerful tools to detect spatial changes or the complete loss of enhancer activity resulting from mutation, but these methods have very limited ability to detect more modest alterations to enhancer intensity, primarily owing to copy number and position effect differences in transgenic animals. In addition, despite their elegance, transgenic experiments suffer from limitations in throughput, in part owing to their relatively high cost. Instead, modest quantitative effects of enhancer mutations have been studied predominantly using in vitro reporter assays, whereby allelic variants of an enhancer are coupled to a luciferase reporter gene and transfected into cells [53–55]. The resulting reporter gene intensity can then be measured quantitatively with a luminometer, allowing for the detection of modest changes to gene expression. Two recent studies have reported major advances in the high-throughput quantitative assessment of enhancers in cultured cells [56] and in vivo in the context of a mouse organ [57].
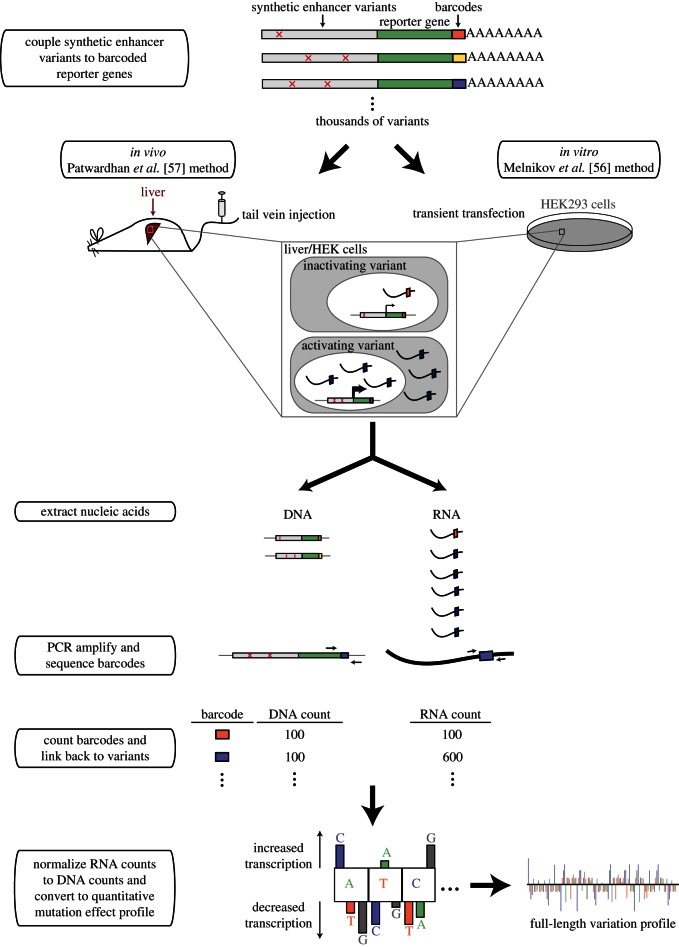
Both methods use technological advances in DNA synthesis and high-throughput DNA sequencing to parallelize enhancer–reporter assays, resulting in the ability to test many enhancer variants in a single experiment. As outlined in figure 2, allelic enhancer variants are synthesized de novo, and each allele is then coupled to a minimal promoter and a reporter gene containing a unique DNA sequence, or barcode, in its 3′-untranslated region. This barcoding allows for the testing of multiple variants at once because sequencing of these unique sites can be used to distinguish between the transcripts associated with different enhancer alleles. The linked enhancer–reporter constructs are then delivered to cells, where the reporter genes are transcribed according to the instructions contained within the enhancer sequences. RNA is harvested from the cells and reverse transcribed, and the barcodes within the transcripts are PCR amplified and sequenced using high-throughput sequencing. To control for the copy number of each enhancer–reporter construct, DNA is also collected, and the barcodes contained within are amplified and sequenced in parallel. The number of RNA sequence reads for each barcode is normalized by its number of DNA sequence reads, and this ratio is used as a measure of reporter gene expression. Comparing the reporter gene expression of each enhancer variant relative to the wild-type enhancer yields a quantitative mutation effect profile that shows which mutations increase and which decrease transcription.
Figure 2.
High-throughput methods to quantitatively assess enhancer variation. In both methods discussed, enhancer variants and unique barcodes are synthesized and linked to reporter genes. Enhancer–reporter constructs are then delivered in vivo to liver cells through mouse tail vein injection (Patwardhan et al. [57]) or by transient transfection to human HEK293T cells (Melnikov et al. [56]). RNA and DNA are collected, and the barcodes contained in each nucleic acid fraction are PCR amplified and sequenced. The number of RNA sequence reads for each barcode is normalized by dividing by its number of DNA reads to create a measure of reporter gene expression. Comparing reporter gene expression from enhancer variants to the wild-type enhancer results in a quantitative mutation effect profile.
The primary differences between the two methods are the cells used and how the enhancer–reporter constructs are delivered to these different cell types. While Melnikov et al. [56] used standard in vitro transient transfection of plasmid DNA into human HEK293T cells, Patwardhan et al. [57] used a method for in vivo transfection via mouse tail vein injection. For this method, purified plasmid DNA is dissolved in a large volume of saline and quickly injected into the tail vein of a mouse [58]. The large injection volume and fast delivery causes the DNA solution to flow into internal organs, particularly the liver, where the DNA is taken up into cells.
Unlike the qualitative transgenic assays described in the previous section, these methods are less amenable to studying spatial changes in reporter gene expression. Performed purely in vitro, the Melnikov et al. [56] method is incompatible with studying spatial patterns of gene expression, which requires the presence of organs or other body structures. The Patwardhan et al. [57] method could, in principle, be coupled with fine-scale liver dissection prior to RNA and DNA sequencing to assess spatial changes in reporter gene activity, but this remains to be demonstrated. The real strength of these methods is the ability to simultaneously test the effects of thousands of mutations on the intensity of enhancer activity, which can be used for mapping enhancer architecture with base-pair resolution. To this end, Patwardhan et al. [57] studied the effect of several thousand base-pair substitutions on three mammalian liver enhancers, and Melnikov et al. [56] tested the effects of every possible single base-pair substitution, a variety of longer consecutive substitutions, and small insertions on one mammalian enhancer: the human IFNB1 enhancer described above.
For IFNB1, Melnikov et al. [56] found that nucleotide substitutions or insertions to the core domains of the enhancer were largely functionally deleterious, replicating the previous lower-throughput mutagenesis studies and the structural analysis of this enhancer [36,37,39,40]. This study also demonstrated the utility of empirical data for enhancer engineering. Using their experimental findings, Melnikov and co-workers were able to make predictions of how to mutagenize the IFNB1 enhancer to alter its activity and successfully demonstrated an increase in its inducible activity.
In contrast to the IFNB1 example, the liver enhancers studied by Patwardhan et al. [57] were more functionally resistant to mutagenesis. Although individual substitutions to many of the bases within these enhancers resulted in altered activity, and most of these activity-affecting substitutions (approx. 70% of bases) resulted in a decrease of reporter gene expression, the vast majority of substitutions had quantitatively modest effects on expression. Only 3 per cent of polymorphisms altered enhancer activity more than twofold, suggesting that these enhancers are largely robust to single base alterations. These results are consistent with a large degree of functional redundancy contained within these loci, similar to the SHH ZRS and the SALL1-D5 enhancers discussed above. Also similar to the ZRS, this study observed a few instances where mutating a single site to all three alternative base sequences resulted in increased enhancer activity, consistent with these sites acting as part of negative regulatory elements.
The divergent findings of these two recent high-resolution studies emphasize the vast potential diversity of enhancer architectures present in mammals. On one end of the spectrum lies the IFNB1 enhancer, conforming to the enhanceosome model with its high sensitivity to sequence variation. On the other end of the spectrum lie the tested liver enhancers, conforming more closely to the billboard model with their large amount of functional redundancy and robustness to mutation. Combined with the findings from the in vivo qualitative assessments of enhancer architecture, these studies clearly show that mammalian enhancer architecture is highly heterogeneous.
4. Conclusions
Collectively, analyses of mammalian enhancers have shown that they can display a wide range of architectures, but several universal characteristics of these sites have begun to emerge. First, the mammalian enhancers studied to date all contain a collection of putative or experimentally validated TFBSs, and these sites typically play functional roles within the enhancers. Second, enhancers often contain both activating and repressing domains, and it is likely that this interplay between transcriptional activation and repression accounts for the very specific spatial and temporal gene expression patterns produced by enhancers. The primary source of heterogeneity in enhancers is the degree to which they display functional redundancy and, relatedly, how important their internal spatial organization is for proper function. Many mammalian enhancers are robust to sequence alterations, consistent with a high degree of functional redundancy. By contrast, others, such as the canonical IFNB1 enhancer, are highly susceptible to inactivation, consistent with a high degree of domain synergism and a low degree of redundancy. Functional enhancer redundancy for the genes regulated by such enhancers may, instead, be established by the presence of multiple independent enhancers with overlapping activities. This is particularly likely for Gata4, for which at least four separate enhancers have thus far been identified, including two with overlapping spatio-temporal activities [45,59,60].
The finding of a high degree of functional redundancy within mammalian enhancers has posed an interesting conundrum: if mammalian enhancers can exhibit a large degree of internal functional redundancy, why do many of them also exhibit strong evolutionary sequence conservation? For example, the SHH ZRS and SALL1-D5 enhancers discussed above exhibit functional redundancy but are also highly conserved across vertebrate evolution. If mutating or deleting a functional domain has apparently little effect on the enhancer's function, what selective forces are acting to maintain these apparently unnecessary or redundant sequences? Is proper gene expression so important that alterations of even minimal effect are strongly selected out of populations? Or could these sites instead be conserved because they are active in regulating gene expression at a different developmental time point than the ones examined? Does this mean that enhancer architecture can be different depending upon the spatio-temporal context of the enhancer within a developing embryo or organism? Clearly, additional studies are needed, including the functional dissection of enhancers under a variety of conditions.
The number of enhancers that have been architecturally assessed by any type of in vivo or high-throughput in vitro method remains very small, and the few that have been studied hint at a rather high level of heterogeneity in enhancer architecture. We have highlighted common characteristics of enhancer anatomy, but we are still far from being able to make de novo predictions regarding the effects of enhancer variants using sequence data alone. If enhancers do, in fact, have a high degree of architectural heterogeneity, then the universal rules required for such predictions may not exist or may be highly tissue-specific. Therefore, experimental assessments will continue to be necessary to characterize the pathogenicity of enhancer sequence variants.
The methods described above will enable substantial progress towards a deeper understanding of enhancer architecture, but they also have a few limitations. Qualitative in vivo assessments can provide detailed spatial information about enhancers, but they are prohibitively expensive to use for assessing more than a handful of variants. High-throughput quantitative assays can be used for testing a multitude of variants, but they are limited to in vitro cellular systems or a very small number of in vivo organs (e.g. liver for tail vein assays). High-throughput in vivo assays that work in a wider variety of cell types could potentially be developed by exploring the viral-based DNA delivery methods used for gene therapy. Continued characterization of architectural elements within enhancers and the development of better assays for such characterization will thus be a major focus of functional genomics moving forward. As human disease studies transition from whole-exome to whole-genome sequencing, the need for rapid experimental and computational methods to assess regulatory sequence variants will soon become acute.
5. Material and methods
(a). Plasmid construction and transgenic enhancer assay
Mutations were made in the SALL1-D5 enhancer using Quikchange II XL site-directed mutagenesis kit (Stratagene). The electronic supplementary material, table S1 shows the primers used to make the eleven different variants tested. The plasmids were transformed using One Shot Top10 chemically competent cells (Invitrogen) and extracted with the QIAprep Miniprep kit (Qiagen). Sanger sequencing was used to verify that each of the constructs contained the expected mutation. Transgenic enhancer assays were carried out as described previously [61] with one modification: embryos were harvested and stained at 11.5 days post-conception. Embryos were tested for transgenesis as previously described [61].
Acknowledgements
All animal protocols were approved by the Lawrence Berkeley National Laboratory Animal Welfare and Research Committee.
We thank Nadav Ahituv and Marianna Ivanov for their work in characterizing the SALL1-D5 enhancer. A.V. and L.A.P. were supported by National Institute of Neurological Disorders and Stroke grant no. R01NS062859A and by National Human Genome Research Institute grants nos. R01HG003988 and U54HG006997. A.V. was supported by NIDCR grant no. U01-DE020060. D.E.D. was supported by the National Heart Lung and Blood Institute grant no. 5T32HL098057 (to Children's Hospital Oakland Research Institute). Research was conducted at the E.O. Lawrence Berkeley National Laboratory and performed under Department of Energy Contract DE-AC02-05CH11231, University of California. All animal work was reviewed and approved by the LBNL Animal Welfare and Research Committee.
References
- 1.Visel A, Rubin EM, Pennacchio LA. 2009. Genomic views of distant-acting enhancers. Nature 461, 199–205 10.1038/nature08451 (doi:10.1038/nature08451) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ptashne M. 1986. Gene regulation by proteins acting nearby and at a distance. Nature 322, 697–701 10.1038/322697a0 (doi:10.1038/322697a0) [DOI] [PubMed] [Google Scholar]
- 3.Li Q, Barkess G, Qian H. 2006. Chromatin looping and the probability of transcription. Trends Genet. 22, 197–202 10.1016/j.tig.2006.02.004 (doi:10.1016/j.tig.2006.02.004) [DOI] [PubMed] [Google Scholar]
- 4.ENCODE Project Consortium Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 10.1038/nature11247 (doi:10.1038/nature11247) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.May D, et al. 2012. Large-scale discovery of enhancers from human heart tissue. Nat. Genet. 44, 89–93 10.1038/ng.1006 (doi:10.1038/ng.1006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maurano MT, et al. 2012. Systematic localization of common disease-associated variation in regulatory DNA. Science 337, 1190–1195 10.1126/science.1222794 (doi:10.1126/science.1222794) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.1000 Genomes Project Consortium 2010. A map of human genome variation from population-scale sequencing. Nature 467, 1061–1073 10.1038/nature09534 (doi:10.1038/nature09534) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Su J, Teichmann SA, Down TA. 2010. Assessing computational methods of cis-regulatory module prediction. PLoS Comput. Biol. 6, e1001020. 10.1371/journal.pcbi.1001020 (doi:10.1371/journal.pcbi.1001020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zinzen RP, Girardot C, Gagneur J, Braun M, Furlong EEM. 2009. Combinatorial binding predicts spatio-temporal cis-regulatory activity. Nature 462, 65–70 10.1038/nature08531 (doi:10.1038/nature08531) [DOI] [PubMed] [Google Scholar]
- 10.Kioussis D, Vanin E, deLange T, Flavell RA, Grosveld FG. 1983. β-globin gene inactivation by DNA translocation in gamma beta-thalassaemia. Nature 306, 662–666 10.1038/306662a0 (doi:10.1038/306662a0) [DOI] [PubMed] [Google Scholar]
- 11.Semenza GL, Delgrosso K, Poncz M, Malladi P, Schwartz E, Surrey S. 1984. The silent carrier allele: beta thalassemia without a mutation in the beta-globin gene or its immediate flanking regions. Cell 39, 123–128 10.1016/0092-8674(84)90197-1 (doi:10.1016/0092-8674(84)90197-1) [DOI] [PubMed] [Google Scholar]
- 12.Lettice LA, et al. 2002. Disruption of a long-range cis-acting regulator for Shh causes preaxial polydactyly. Proc. Natl Acad. Sci. USA 99, 7548–7553 10.1073/pnas.112212199 (doi:10.1073/pnas.112212199) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Balemans W, et al. 2002. Identification of a 52 kb deletion downstream of the SOST gene in patients with van Buchem disease. J. Med. Genet. 39, 91–97 10.1136/jmg.39.2.91 (doi:10.1136/jmg.39.2.91) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Staehling-Hampton K, et al. 2002. A 52 kb deletion in the SOST-MEOX1 intergenic region on 17q12-q21 is associated with van Buchem disease in the Dutch population. Am. J. Med. Genet. 110, 144–152 10.1002/ajmg.10401 (doi:10.1002/ajmg.10401) [DOI] [PubMed] [Google Scholar]
- 15.Collette NM, Genetos DC, Economides AN, Xie L, Shahnazari M, Yao W, Lane NE, Harland RM, Loots GG. 2012. Targeted deletion of Sost distal enhancer increases bone formation and bone mass. Proc. Natl Acad. Sci. USA 109, 14 092–14 097 10.1073/pnas.1207188109 (doi:10.1073/pnas.1207188109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Emison ES, et al. 2005. A common sex-dependent mutation in a RET enhancer underlies Hirschsprung disease risk. Nature 434, 857–863 10.1038/nature03467 (doi:10.1038/nature03467) [DOI] [PubMed] [Google Scholar]
- 17.Lauderdale JD, Wilensky JS, Oliver ER, Walton DS, Glaser T. 2000. 3′ deletions cause aniridia by preventing PAX6 gene expression. Proc. Natl Acad. Sci. USA 97, 13 755–13 759 10.1073/pnas.240398797 (doi:10.1073/pnas.240398797) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kleinjan DA, Lettice LA. 2008. Long-range gene control and genetic disease. Adv. Genet. 61, 339–388 10.1016/S0065-2660(07)00013-2 (doi:10.1016/S0065-2660(07)00013-2) [DOI] [PubMed] [Google Scholar]
- 19.Ernst J, et al. 2011. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473, 43–49 10.1038/nature09906 (doi:10.1038/nature09906) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCarroll SA, et al. 2008. Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn's disease. Nat. Genet. 40, 1107–1112 10.1038/ng.215 (doi:10.1038/ng.215) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prescott NJ, et al. 2010. Independent and population-specific association of risk variants at the IRGM locus with Crohn's disease. Hum. Mol. Genet. 19, 1828–1839 10.1093/hmg/ddq041 (doi:10.1093/hmg/ddq041) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vacic V, et al. 2011. Duplications of the neuropeptide receptor gene VIPR2 confer significant risk for schizophrenia. Nature 471, 499–503 10.1038/nature09884 (doi:10.1038/nature09884) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Helgadottir A, et al. 2007. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science 316, 1491–1493 10.1126/science.1142842 (doi:10.1126/science.1142842) [DOI] [PubMed] [Google Scholar]
- 24.McPherson R, et al. 2007. A common allele on chromosome 9 associated with coronary heart disease. Science 316, 1488–1491 10.1126/science.1142447 (doi:10.1126/science.1142447) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Visel A, et al. 2010. Targeted deletion of the 9p21 non-coding coronary artery disease risk interval in mice. Nature 464, 409–412 10.1038/nature08801 (doi:10.1038/nature08801) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harismendy O, et al. 2011. 9p21 DNA variants associated with coronary artery disease impair interferon-gamma signalling response. Nature 470, 264–268 10.1038/nature09753 (doi:10.1038/nature09753) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gudmundsson J, et al. 2007. Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nat. Genet. 39, 977–983 10.1038/ng2062 (doi:10.1038/ng2062) [DOI] [PubMed] [Google Scholar]
- 28.Zhang X, Cowper-Sal Lari R, Bailey SD, Moore JH, Lupien M. 2012. Integrative functional genomics identifies an enhancer looping to the SOX9 gene disrupted by the 17q24.3 prostate cancer risk locus. Genome Res. 22, 1437–1446 10.1101/gr.135665.111 (doi:10.1101/gr.135665.111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Visel A, et al. 2009. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457, 854–858 10.1038/nature07730 (doi:10.1038/nature07730) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. 2010. A method and server for predicting damaging missense mutations. Nat Methods 7, 248–249 10.1038/nmeth0410-248 (doi:10.1038/nmeth0410-248) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar P, Henikoff S, Ng PC. 2009. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 4, 1073–1081 10.1038/nprot.2009.86 (doi:10.1038/nprot.2009.86) [DOI] [PubMed] [Google Scholar]
- 32.Bamshad MJ, Ng SB, Bigham AW, Tabor HK, Emond MJ, Nickerson DA, Shendure J. 2011. Exome sequencing as a tool for Mendelian disease gene discovery. Nat. Rev. Genet. 12, 745–755 10.1038/nrg3031 (doi:10.1038/nrg3031) [DOI] [PubMed] [Google Scholar]
- 33.Davidson EH. 1999. A view from the genome: spatial control of transcription in sea urchin development. Curr. Opin. Genet. Dev. 9, 530–541 10.1016/S0959-437X(99)00013-1 (doi:10.1016/S0959-437X(99)00013-1) [DOI] [PubMed] [Google Scholar]
- 34.Borok MJ, Tran DA, Ho MCW, Drewell RA. 2010. Dissecting the regulatory switches of development: lessons from enhancer evolution in Drosophila. Development 137, 5–13 10.1242/dev.036160 (doi:10.1242/dev.036160) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arnosti DN, Kulkarni MM. 2005. Transcriptional enhancers: intelligent enhanceosomes or flexible billboards? J. Cell. Biochem. 94, 890–898 10.1002/jcb.20352 (doi:10.1002/jcb.20352) [DOI] [PubMed] [Google Scholar]
- 36.Thanos D, Maniatis T. 1995. Virus induction of human IFN β gene expression requires the assembly of an enhanceosome. Cell 83, 1091–1100 10.1016/0092-8674(95)90136-1 (doi:10.1016/0092-8674(95)90136-1) [DOI] [PubMed] [Google Scholar]
- 37.Goodbourn S, Maniatis T. 1988. Overlapping positive and negative regulatory domains of the human beta-interferon gene. Proc. Natl Acad. Sci. USA 85, 1447–1451 10.1073/pnas.85.5.1447 (doi:10.1073/pnas.85.5.1447) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Du W, Maniatis T. 1992. An ATF/CREB binding site is required for virus induction of the human interferon beta gene. Proc. Natl Acad. Sci. USA 89, 2150–2154 10.1073/pnas.89.6.2150 (doi:10.1073/pnas.89.6.2150) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Panne D, Maniatis T, Harrison SC. 2007. An atomic model of the interferon-beta enhanceosome. Cell 129, 1111–1123 10.1016/j.cell.2007.05.019 (doi:10.1016/j.cell.2007.05.019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Panne D, Maniatis T, Harrison SC. 2004. Crystal structure of ATF-2/c-Jun and IRF-3 bound to the interferon-beta enhancer. EMBO J. 23, 4384–4393 10.1038/sj.emboj.7600453 (doi:10.1038/sj.emboj.7600453) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swanson CI, Evans NC, Barolo S. 2010. Structural rules and complex regulatory circuitry constrain expression of a Notch- and EGFR-regulated eye enhancer. Dev. Cell 18, 359–370 10.1016/j.devcel.2009.12.026 (doi:10.1016/j.devcel.2009.12.026) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu F, Posakony JW. 2012. Role of architecture in the function and specificity of two notch-regulated transcriptional enhancer modules. PLoS Genet. 8, e1002796. 10.1371/journal.pgen.1002796 (doi:10.1371/journal.pgen.1002796) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nagy A, Gertsenstein M, Vintersten K, Behringer R. 2003. Manipulating the mouse embryo, 3rd edn Cold Spring Harbor, NY: CSHL Press [Google Scholar]
- 44.Rojas A, De Val S, Heidt AB, Xu S-M, Bristow J, Black BL. 2005. Gata4 expression in lateral mesoderm is downstream of BMP4 and is activated directly by Forkhead and GATA transcription factors through a distal enhancer element. Development 132, 3405–3417 10.1242/dev.01913 (doi:10.1242/dev.01913) [DOI] [PubMed] [Google Scholar]
- 45.Rojas A, Schachterle W, Xu S-M, Black BL. 2009. An endoderm-specific transcriptional enhancer from the mouse Gata4 gene requires GATA and homeodomain protein-binding sites for function in vivo. Dev. Dyn. 238, 2588–2598 10.1002/dvdy.22091 (doi:10.1002/dvdy.22091) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Munshi NV, McAnally J, Bezprozvannaya S, Berry JM, Richardson JA, Hill JA, Olson EN. 2009. Cx30.2 enhancer analysis identifies Gata4 as a novel regulator of atrioventricular delay. Development 136, 2665–2674 10.1242/dev.038562 (doi:10.1242/dev.038562) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lettice LA, et al. 2012. Opposing functions of the ETS factor family define Shh spatial expression in limb buds and underlie polydactyly. Dev. Cell 22, 459–467 10.1016/j.devcel.2011.12.010 (doi:10.1016/j.devcel.2011.12.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lettice LA, Hill AE, Devenney PS, Hill RE. 2008. Point mutations in a distant sonic hedgehog cis-regulator generate a variable regulatory output responsible for preaxial polydactyly. Hum. Mol. Genet. 17, 978–985 10.1093/hmg/ddm370 (doi:10.1093/hmg/ddm370) [DOI] [PubMed] [Google Scholar]
- 49.Lettice LA, et al. 2003. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum. Mol. Genet. 12, 1725–1735 10.1093/hmg/ddg180 (doi:10.1093/hmg/ddg180) [DOI] [PubMed] [Google Scholar]
- 50.Maas SA, Suzuki T, Fallon JF. 2011. Identification of spontaneous mutations within the long-range limb-specific Sonic hedgehog enhancer (ZRS) that alter Sonic hedgehog expression in the chicken limb mutants oligozeugodactyly and silkie breed. Dev. Dyn. 240, 1212–1222 10.1002/dvdy.22634 (doi:10.1002/dvdy.22634) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pennacchio LA, et al. 2006. In vivo enhancer analysis of human conserved non-coding sequences. Nature 444, 499–502 10.1038/nature05295 (doi:10.1038/nature05295) [DOI] [PubMed] [Google Scholar]
- 52.Nishinakamura R, et al. 2001. Murine homolog of SALL1 is essential for ureteric bud invasion in kidney development. Development 128, 3105–3115 [DOI] [PubMed] [Google Scholar]
- 53.Rosenthal N. 1987. Identification of regulatory elements of cloned genes with functional assays. Methods Enzymol. 152, 704–720 10.1016/0076-6879(87)52075-4 (doi:10.1016/0076-6879(87)52075-4) [DOI] [PubMed] [Google Scholar]
- 54.Naylor LH. 1999. Reporter gene technology: the future looks bright. Biochem. Pharmacol. 58, 749–757 10.1016/S0006-2952(99)00096-9 (doi:10.1016/S0006-2952(99)00096-9) [DOI] [PubMed] [Google Scholar]
- 55.Schenborn E, Groskreutz D. 1999. Reporter gene vectors and assays. Mol. Biotechnol. 13, 29–44 10.1385/MB:13:1:29 (doi:10.1385/MB:13:1:29) [DOI] [PubMed] [Google Scholar]
- 56.Melnikov A, et al. 2012. Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nat. Biotechnol. 30, 271–277 10.1038/nbt.2137 (doi:10.1038/nbt.2137) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Patwardhan RP, et al. 2012. Massively parallel functional dissection of mammalian enhancers in vivo. Nat. Biotechnol. 30, 265–270 10.1038/nbt.2136 (doi:10.1038/nbt.2136) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu F, Song Y, Liu D. 1999. Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Ther. 6, 1258–1266 10.1038/sj.gt.3300947 (doi:10.1038/sj.gt.3300947) [DOI] [PubMed] [Google Scholar]
- 59.Schachterle W, Rojas A, Xu S-M, Black BL. 2012. ETS-dependent regulation of a distal Gata4 cardiac enhancer. Dev. Biol. 361, 439–449 10.1016/j.ydbio.2011.10.023 (doi:10.1016/j.ydbio.2011.10.023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rojas A, Schachterle W, Xu S-M, Martín F, Black BL. 2010. Direct transcriptional regulation of Gata4 during early endoderm specification is controlled by FoxA2 binding to an intronic enhancer. Dev. Biol. 346, 346–355 10.1016/j.ydbio.2010.07.032 (doi:10.1016/j.ydbio.2010.07.032) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Poulin F, Nobrega MA, Plajzer-Frick I, Holt A, Afzal V, Rubin EM, Pennacchio LA. 2005. In vivo characterization of a vertebrate ultraconserved enhancer. Genomics 85, 774–781 10.1016/j.ygeno.2005.03.003 (doi:10.1016/j.ygeno.2005.03.003) [DOI] [PubMed] [Google Scholar]