Abstract
Developmental gene expression programmes are coordinated by the specialized distal cis-regulatory elements called enhancers, which integrate lineage- and signalling-dependent inputs to guide morphogenesis. In previous work, we characterized the genome-wide repertoire of active enhancers in human neural crest cells (hNCC), an embryonic cell population with critical roles in craniofacial development. We showed that in hNCC, co-occupancy of a master regulator TFAP2A with nuclear receptors NR2F1 and NR2F2 correlates with the presence of permissive enhancer chromatin states. Here, we take advantage of pre-existing human genetic variation to further explore potential cooperation between TFAP2A and NR2F1/F2. We demonstrate that isolated single nucleotide polymorphisms affecting NR2F1/F2-binding sites within hNCC enhancers can alter TFAP2A occupancy and overall chromatin features at the same enhancer allele. We propose that a similar strategy can be used to elucidate other cooperative relationships between transcription factors involved in developmental transitions. Using the neural crest and its major contribution to human craniofacial phenotypes as a paradigm, we discuss how genetic variation might modulate the molecular properties and activity of enhancers, and ultimately impact human phenotypic diversity.
Keywords: neural crest, craniofacial, enhancer, single nucleotide polymorphism, chromatin, cooperativity
1. Developmental enhancers as platforms for cooperative transcription factor recruitment and integration of signalling cues
In the course of embryonic development multiple cell types, tissues and organs are progressively generated from a single cell, the zygote. These different cell types share nearly identical genetic information, yet each possesses unique properties that are driven by expression of a characteristic set of genes. It is now widely accepted that enhancers, a group of distal cis-regulatory elements, are crucial in the establishment of cell-type- and developmental stage-specific gene expression patterns [1–6]. According to their classical definition, enhancers are compact (approx. 200–500 bp) DNA sequences containing clusters of transcription factor (TF)-binding motifs that control expression of target promoters over long distances and in an orientation-independent manner [2,7]. The spatio-temporal activity of developmental enhancers is largely determined by the combinatorial binding of TFs to their cognate motifs [5,8–10]. The large number of TFs encoded by mammalian genomes, their frequent tissue specificity and their responsiveness to signalling ensures a rich repertoire of context-dependent TF combinations. TFs representing both lineage-specific regulators as well as sequence-dependent effectors of signal transduction pathways commonly converge at enhancer elements to activate transcription, thus integrating intrinsic and extrinsic environmental signalling cues [1,11–13]. Such integration allows for exquisite spatial and temporal control of gene expression during development.
To gain access to DNA, TFs have to compete with nucleosomes that occlude TF-binding motifs and block protein–DNA interactions [14–16]. The DNA-binding affinity of an individual TF is typically much lower than that of a histone octamer–DNA complex, hence cooperative binding of multiple factors is thought to play a major role in overcoming the nucleosomal barrier in TF recruitment [17–19]. Such cooperativity may depend on direct physical association between TFs, or may occur in the absence of direct interaction by simply promoting nucleosomal eviction [5,18]. In some instances recruitment of TFs to enhancer elements is sequential, with the so-called ‘pioneer’ factors able to first gain access to the nucleosomal DNA, either in isolation or through interaction with chromatin remodellers and histone chaperones [20–27]. Such pioneer factors can subsequently facilitate binding of other TFs and the assembly of coactivator complexes [20,22–24]. Coactivator recruitment allows an additional layer of regulatory integration, as TF cooperativity provides multiple binding surfaces for general coactivators or for an increasing repertoire of coactivators with distinct chromatin remodelling and modifying activities [2,28,29]. Cumulatively, this enables unique TF combinations to synergistically establish permissive chromatin states at enhancer elements and promote long-range communication with promoters [2,30,31]. Despite the cell-type specificity of these TF combinations, transcriptionally permissive chromatin states share many conserved features regardless of cellular context. For example, active enhancers display common epigenomic profiles such as nucleosomal depletion at TF-occupied sites flanked by regions enriched for nucleosomes marked by certain histone modifications, including H3K4me1 and H3K27ac (reviewed in [2,32]). In recent years, these features have been exploited to annotate cis-regulatory repertoires through genome-wide mapping of DNase hypersensitive sites, histone modifications and general coactivator occupancy, revealing the staggering preponderance and developmental dynamics of regulatory elements in the human genome [33–36].
2. Epigenomic landscapes of human neural crest
Human embryonic stem cells (hESC) and their in vitro differentiation models combined with epigenomic mapping offer an opportunity to uncover regulatory elements used in transient cell states that arise during early human development which have previously been largely inaccessible for study. One transient cell type of particular developmental and medical relevance is the neural crest (NC), a vertebrate-specific cell population specified early in embryogenesis at the neural plate border territory separating neuroectoderm from the epidermis [37,38]. After specification, NC progenitors undergo an epithelial-to-mesenchymal transition, delaminate from the dorsal neural tube, migrate throughout the body and acquire an extraordinarily broad differentiation potential, giving rise to elements of the craniofacial skeleton, the middle ear, the peripheral nervous system, pigment cells, and certain cardiac structures [37,38]. Cranial neural crest cells originating from the cephalic region of the neural tube produce a large variety of mesenchymal cell types including bone and cartilage, which elsewhere in the body are formed solely by mesodermal derivatives [39,40]. Thus, neural crest cells (NCC) are truly unique, as they not only migrate over unparalleled distances, but also effectively broaden their developmental potential upon specification, thus traversing traditionally delineated germ layer fate restrictions. In addition, aberrant NC development is associated with a broad variety of congenital malformations often including deafness and complex craniofacial defects, seen in a large number of congenital disorders known as neurocristopathies as well as in more common non-syndromic manifestations such as cleft lip and palate [41,42].
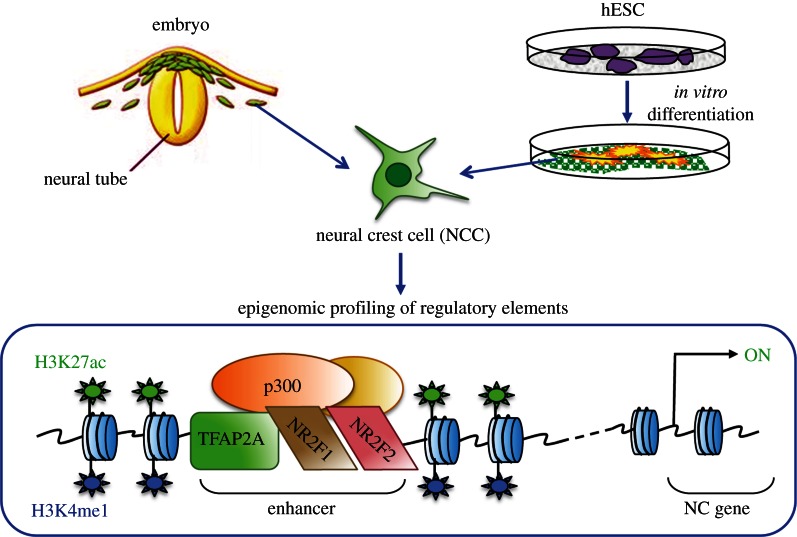
To study gene regulatory mechanisms underlying development of this unique cell type, our laboratory has developed a hESC-based in vitro model that recapitulates specification, migration and maturation of the cranial neural crest [43]. Recently, through epigenomic profiling we demonstrated that differentiation of hESC to human neural crest (hNCC) is accompanied by dramatic changes in enhancer chromatin landscapes ([44]; figure 1). Over 4000 promoter-distal genomic elements were marked by an active chromatin enhancer signature (defined by the presence of coactivator p300 flanked by nucleosomes modified by H3K27ac and H3K4me1) in the hNCC, and these putative enhancers showed strong association with genomic regions implicated in craniofacial development and disease [44]. Moreover, analysis of sequence motifs enriched at the annotated hNCC enhancers predicted major TFs that bind at these elements. We subsequently validated novel predictions coming out of our epigenomic profiling through in vitro and in vivo follow-up studies. For example, we demonstrated that nuclear receptors NR2F1 and NR2F2 (aka COUP-TF1 and COUP-TF2; [45]) are novel regulators of NC gene expression and craniofacial morphogenesis, which bind at a subset of NC enhancers along with a NC master regulator TFAP2A [46]. Importantly, genomic regions with simultaneous co-occupancy of TFAP2A and NR2F1/2 are associated with permissive chromatin states, characterized by high levels of p300 and H3K27ac [44]. These results suggest that, as has been reported in other systems [12,13], cooperative function of lineage specifiers (e.g. TFAP2A) and signalling effectors (e.g. NR2F1/F2) converges at active enhancer elements (figure 1). Here, we present new data that more directly illustrate cooperation between TFAP2A and NR2F1/F2 in NCCs, and discuss implications of these observations for studies of craniofacial variation.
Figure 1.
Graphical summary of major findings arising from the epigenomic characterization of hNCC. hNCC derived from hESC were used for genome-wide profiling of histone modifications, TF and coactivator occupancy. Simultaneous presence of a master NC regulator TFAP2A and of nuclear receptors NR2F1/F2 at enhancers is associated with permissive chromatin states characterized by high levels of H2K27ac and p300.
3. Human genetic variation as a tool to investigate molecular mechanisms of enhancer function
As discussed above, cooperativity between TFs in DNA binding, coactivator recruitment and establishment of permissive chromatin states are important features of enhancer-mediated gene regulation. Unfortunately, analyses of such synergies are often confounded by the fact that major developmental TFs commonly regulate each other's expression and are essential for maintenance of the cell fate of interest, making loss-of-function studies difficult to interpret. Synergistic function of TFAP2A and NR2F1/F2 in NCCs, for example, is strongly suggested by their physical co-association and the observation that genomic regions co-bound by TFAP2A and NR2F1/2 are characterized by elevated levels of histone marks associated with active enhancers compared with regions bound by either factor alone [44]. Though compelling, these observations are nonetheless only correlative. To complicate the matter, a more direct examination of potential cooperativity between TFAP2A and NR2F1/F2 is precluded by the observation that these factors control each other's expression, and depletion of either TF has a profound effect on the NC gene expression program [44], demonstrating the difficulty in dissecting direct versus indirect effects in knockdown studies.
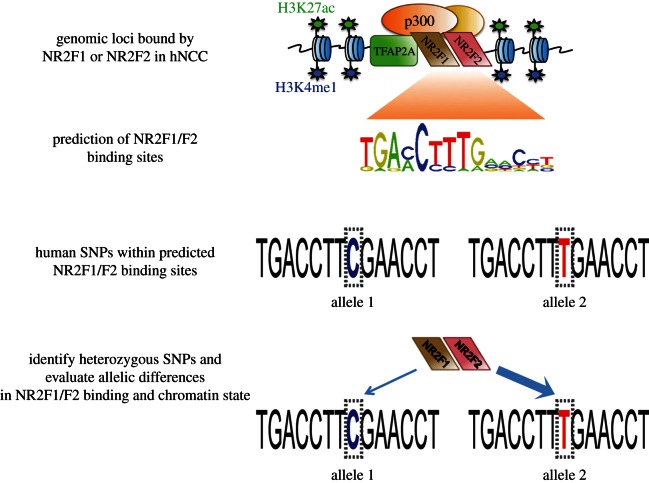
These caveats motivated us to explore alternative methods for studying TF cooperativity in a developmentally dynamic system. To this end we decided to take advantage of the natural genetic variation that occurs in the human genome (figure 2). We hypothesized that if two TFs cooperate in binding to DNA and the establishment of permissive chromatin states, then a single nucleotide polymorphism (SNP) affecting DNA-binding affinity of one factor should in turn affect occupancy of the other factor, as well as chromatin state at the co-bound enhancer (figure 2). To test this hypothesis we analysed NCC enhancers bound by NR2F1/F2, as reported in Rada-Iglesias et al. [44]. We then identified sequence variants falling within NR2F1/F2 recognition motifs at these enhancers, focusing on SNPs characterized by high heterozygosity in the human population. We selected 19 such common SNPs and genotyped them in a human H9 hESC line, revealing nine SNPs heterozygous in this genetic background. Using these heterozygous variants, we could directly compare biases in occupancy of TFs and histone modifications between two enhancer alleles within the same cell population (figure 2).
Figure 2.
Strategy for using human genetic variation as a tool to investigate binding cooperativity between NR2F1/F2 and TFAP2A at hNCC enhancers. Location of NR2F1/F2 binding motif within distal genomic sites bound by NR2F1 or NR2F2 in hNCC (n = 2748, reported in [44]) was predicted using the ‘MISP:Motif-based interval screener with PSSM’ tool from Cistrome (http://cistrome.org/ap/), which resulted in 4712 predicted NR2F1/F2 sites. For site predictions, the NR2F1/F2 motif IDs used were MA0017, M00155, UP00009 with a cut-off of 100 for the motifs screen. Next, all human SNPs (based on dbSNP build 130) overlapping the predicted NR2F1/F2 binding sites were identified (n = 373). Then, 19 SNPs of high heterozygosity within human population were selected and genotyped in H9 hESC, yielding nine heterozygous SNPs. hNCC were derived from H9 hESC, and ChIP and quantitative genotyping were combined in order to evaluate allelic differences in TF binding and active enhancer chromatin marks at these heterozygous loci.
4. Single nucleotide polymorphisms can affect cooperative binding of transcription factors and chromatin states at enhancers
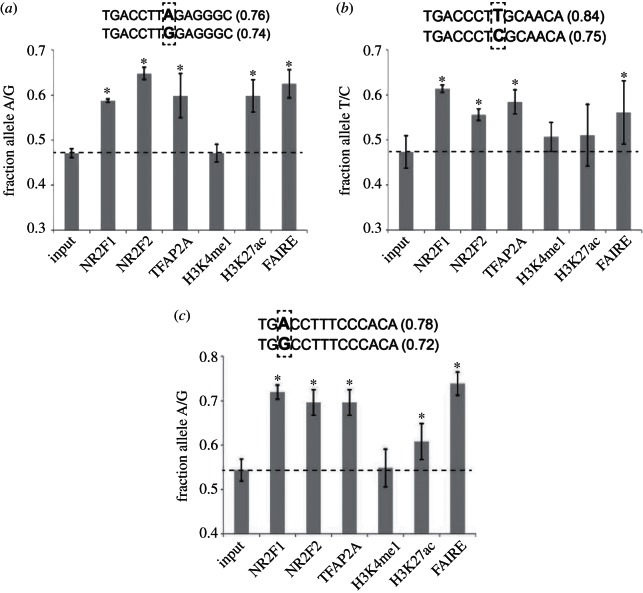
We differentiated H9 hESC to hNCC and performed chromatin immuno-precipitation (ChIP) analyses with NR2F1, NR2F2, TFAP2A, H3K27ac and H3K4me1 antibodies, followed by quantitative genotyping of enhancers harbouring aforementioned heterozygous SNPs in NR2F1/F2-binding motifs. In addition, we also analysed nucleosomal depletion at these regions using a FAIRE assay. For three of the nine SNPs we detected modest, but significant and reproducible allelic differences in binding of NR2F1 and NR2F2 (see figure 3, electronic supplementary material, figure S1). In each case stronger binding was associated with the allelic variant that more closely matched the recognition consensus of these nuclear receptors (note that NR2F1 and NR2F2 heterodimerize and share the same DNA binding motif [45,47]). Importantly, quantitative genotyping of TFAP2A ChIP DNA demonstrated that binding of TFAP2A is consistently increased at the same allele that shows preferential NR2F1/F2 enrichment, even though in each case both alleles harbour equivalent TFAP2A binding motifs, located within 100 bp of the investigated SNPs. These data suggests that analysed genetic variants indirectly affect TFAP2A occupancy via binding cooperativity with NR2F1/F2. Albeit we cannot formally exclude a possibility that elevated TFAP2A signal in ChIP assays results from the increased interaction with NR2F1/F2 (leading in turn in the enhanced formaldehyde cross-linkability) rather than a direct increase in TFAP2A DNA binding levels, the latter interpretation is supported by observation that the enhancer alleles displaying preferential TF binding were characterized by the stronger nucleosomal depletion in FAIRE assay (figure 3). Moreover, enrichment of H3K27ac, a chromatin mark closely correlated with enhancer activity [34,48], was also commonly increased at the enhancer alleles showing preferential TF binding, although the differences were not always statistically significant. In contrast, we did not detect allelic biases in H3K4me1 at any of the analysed enhancers. This result is consistent with our earlier observations that H3K4me1 enrichment often precedes enhancer activation and is not dependent on the presence of TFAP2A and/or NR2F1/F2 [44].
Figure 3.
Allelic differences in TF binding and chromatin states demonstrate cooperative binding of NR2F1/F2 and TFAP2A. Quantitative genotyping of total genomic input DNA, ChIP DNAs obtained for NR2F1, NR2F2, TFAP2A, H3K4me1 and H3K27ac and FAIRE DNA was performed for three heterozygous SNPs: (a) rs6759359, (b) rs2911258 and (c) rs1672997. For each of these SNPs the two alleles are presented within the predicted NR2F1/F2 binding sites, with the number in brackets denoting the similarity score with respect to the NR2F1/F2 consensus binding motif. For quantitative genotyping, genomic regions spanning the different SNPs were amplified by PCR, followed by purification and Sanger sequencing (see the electronic supplementary material, table S1). For each SNP and DNA sample, the height of each of the two alleles in the sequencing chromatograms was estimated using ImageJ and allelic frequencies (x-axis) were calculated dividing the height of the allele representing a better match to the NR2F1/F2 consensus motif by the sum of the height of the two alleles. PCRs and subsequent sequencing reactions were performed for each sample at least in triplicates, with the error bars representing standard deviations. Two-tailed t-test was used to determine whether allelic frequencies were significantly different between ChIP or FAIRE DNAs and total genomic input DNA, with the asterisks denoting a p-value < 0.05. Overall similar results (see the electronic supplementary material, figure S1) were obtained when independent biological replicates of the ChIP and FAIRE DNA samples were used to evaluate allelic differences by an alternative quantitative genotyping method (Pyrosequencing, Quiagen). Binding of all TFs, enrichment of H3K4me1 and H3K27ac and low-nucleosomal density as measured by FAIRE were confirmed at genomic regions spanning the three investigated SNPs by qPCR (data not shown). hNCC differentiation, ChIP and FAIRE were performed as previously described [44].
Taken together, our results further support a model in which cooperative function of TFAP2A and NR2F1/F2 (with likely input from other TFs) promotes establishment or maintenance of permissive chromatin states at NC enhancers. Analogous experimental strategies can be applied to investigate the functional relationships between major transcriptional regulators in other cell types and developmental processes, particularly in cases when loss of function studies are confounded by the caveats discussed above. The relationships between human genetic variation, TF binding, allele-specific chromatin states and gene expression are becoming an area of intense scientific interest, and several forays to characterize such relationships in a genome-wide manner have already been made [49–54]. For example, genome-wide occupancy measurements of 24 different TFs in a human lymphoblastoid cell line found that as much as 5 per cent of TF-binding sites show an allelic imbalance in occupancy. Importantly, the factors binding within these regions often exhibited a coordinated reaction to functional variants, and only approximately 12 per cent of allele-specific enrichments could be explained by sequence variants falling directly within binding motif for a given factor, suggesting cooperative effects at these regions [49]. Furthermore, systematic computational approaches such as those used and experimentally validated for NF- κB have taken advantage of this functional genomic covariance to accurately predict co-associated TFs [50]. Moreover, when coupled to high-throughput sequencing methods, analysis of allelic imbalances in chromatin signatures may provide a powerful, unbiased way to identify ‘strong’ and ‘weak’ functional enhancer variants, particularly in a heterozygous setting where even subtle allelic biases can be reliably quantified and measured. Several recent studies support the role of non-coding genetic diversity as a major driver of individual- and allele-specific chromatin states and uncover association of such variants with modulation in gene expression [51–54].
5. Molecular and phenotypic impact of genetic variation at enhancers
As discussed above, a single SNP can affect not only the binding of the TF whose motif is altered, but also binding of additional TFs that are recruited in cis to the same enhancer. Given the cooperative principles governing enhancer activity [2,5,9], a single SNP, therefore, has the potential to alter the overall enhancer state, as reflected by changes in TF binding, nucleosomal occupancy and histone modifications typical of active enhancers. We will hereafter refer to genetic variants that measurably affect enhancer states as enhancer-SNPs (eSNPs). eSNPs represent only a small subset of genetic variation within enhancers, because these polymorphisms not only have to affect DNA recognition of a TF critical for enhancer activity, but this occupancy change must not be compensated for by other factors or additional binding sites for the same TF [3]. Even among eSNPs, those that dramatically alter enhancer activity are most likely rare. Instead, more subtle allelic biases in enhancer states are probably much more common. An outstanding question remains as to what extent such moderate changes in enhancer states elicit changes in level or timing of gene expression. Again, the answer will vary depending on the specific enhancer–promoter pair, on the degree of redundancy with other enhancers acting on the same target and on the environmental conditions, which have been shown to reveal essentiality for seemingly redundant enhancers [3,55,56]. Nonetheless, evidence emerges that cis-regulatory variation does result in transcriptional changes and that even subtle differences in gene expression level and spatio-temporal control can have important consequences for phenotypes [57–64]. In one example of particular relevance, a single SNP within a craniofacial-associated IRF6 enhancer element disrupting TFAP2A binding has already been shown to confer elevated risk for non-syndromic cleft lip [65].
Comparison of the developmental basis of body-pattern evolution in animal models shows that morphological variation is largely a product of quantitative and spatio-temporal changes in deployment of conserved regulatory networks [66,67]. Some variation in gene expression may arise from somatic mosaicism, environmental perturbation, epigenetic influences or simple biological stochasticity, but a significant proportion of phenotypic diversity is encoded within heritable genetic information. Direct genome sequencing, linkage analysis and genome-wide association studies reveal that much of this transmissible information responsible for trait modulation is buried within non-coding regions of the genome. This can be rationalized as a consequence of genetic pleiotropy, because mutations disrupting function within a coding region of an important developmental gene may confer widespread detrimental effects throughout a developing organism. As mentioned above, regulatory elements such as enhancers are thought instead to act in a highly tissue- and stage-specific manner, commonly in the context of other redundant or partially redundant regulatory elements. Variation arising within these non-coding regulatory regions is, therefore, less likely to be deleterious to the organism as a whole, rendering enhancers more accommodating of functional polymorphism on a population-wide level because they permit small changes in transcriptional regulation to arise without a massive reshuffling of developmental patterning networks.
In addition to their impact on intraspecies variation, eSNPs can also play an important role in speciation and evolutionary adaptation to changing environments. Since the discovery that coding regions of the genome remain largely conserved across species, it has long been postulated that evolutionary divergence mostly arises from quantitative (and spatio-temporal) rather than qualitative changes to gene function [68]. Recent advances in sequencing technologies have substantiated this prediction that changes within regulatory elements are a major source of evolutionary divergence, even between closely related species [69–71]. Importantly, there is now theory and evidence to argue that the same expression perturbations driving intraspecies variation may ultimately be responsible for speciation and fixed interspecies divergence [72].
6. Genetic variation at neural crest regulatory elements: implications for human craniofacial diversity
One of the most interesting examples of intraspecies variation is the human face, a single feature that best distinguishes an individual while also connecting each of us to our broader ethnic and familial ancestry. Although craniofacial morphology is known to be highly heritable, genetic factors that underlie normal variation in human face and skull shape remain poorly understood [73]. Craniofacial structures originate largely from the cranial neural crest, a highly developmentally plastic cell population, which arises in the anterior part of the nascent neural tube and forms the majority of bone, cartilage and connective tissue of the head and face. Interestingly, in contrast to endochondral bone formation in which bone develops from a cartilage template, recent research suggests that craniofacial bone and cartilage (which undergo largely intramembranous ossification) may represent independent tissue modules and, although they all derive from the NC, are controlled by different genes and form separate condensations [71,74–78]. This modularity, combined with developmental robustness imparted by the crest's plasticity, means that specific traits can be adjusted in a fairly autonomous manner while maintaining integration with surrounding structures. A quick glance at the spectrum of shapes and sizes of facial features throughout the human population demonstrates this potential for quantitative phenotype modulation and reflects a multifactorial genetic contribution. Facial morphology, therefore, provides a ripe and tractable model for investigating the link between genotype, molecular phenotype and trait modulation in a complex human developmental context.
We anticipate that enhancers acting within the NCCs and their derivatives during gestational development are major drivers of facial phenotypic diversity. In support of early developmental and NC-driven origins of facial variation, avian xenotransplantation studies show that the NC contains autonomous morphogenetic information that can coordinate with surrounding tissues to drive species-specific (and probably individual-specific) facial morphology [79]. Indeed, manipulation of conserved signalling pathway effectors such as BMP4 in specific regions of avian crest-originated facial prominences is able to stimulate local proliferation domains that parallel evolutionary differences between species of Darwin finches and chick [80]. This suggests that heterochronic and heterotonic changes within the early crest itself are sufficient to influence facial traits and underlines the necessity for establishing models of human neural crest development to better understand the regulatory control of facial morphogenesis.
Our studies of the human neural crest epigenome represent initial attempts to characterize a cis-regulatory repertoire relevant for early steps in the formation of the human face [44]. Although we identified thousands of putative enhancers, we probably did not capture the full complexity of craniofacial cis-regulation, and therefore later stages of craniofacial development must also be considered in subsequent functional genomic analyses. In experiments described here, we took advantage of natural genetic variation to provide further support for cooperative function of TFAP2A and NR2F1/F2 at NC enhancers and supplied a proof-of-principle demonstrating that eSNPs exists within cranial neural crest enhancers. The future challenge will be not only to systematically identify such eSNPs in the human population, but to link them with diversity of specific aspects of human craniofacial morphologies.
Acknowledgements
This work was supported by NIH RO1 GM095555 and CIRM RB3-05100 grants for J.W. and Siebel Scholarship for A.R-I.
References
- 1.Buecker C, Wysocka J. 2012. Enhancers as information integration hubs in development: lessons from genomics. Trends Genet. 28, 276–284 10.1016/j.tig.2012.02.008 (doi:10.1016/j.tig.2012.02.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bulger M, Groudine M. 2011. Functional and mechanistic diversity of distal transcription enhancers. Cell 144, 327–339 10.1016/j.cell.2011.01.024 (doi:10.1016/j.cell.2011.01.024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lagha M, Bothma JP, Levine M. 2012. Mechanisms of transcriptional precision in animal development. Trends Genet. 28, 409–416 10.1016/j.tig.2012.03.006 (doi:10.1016/j.tig.2012.03.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ong CT, Corces VG. 2012. Enhancers: emerging roles in cell fate specification. EMBO Rep. 13, 423–430 10.1038/embor.2012.52 (doi:10.1038/embor.2012.52) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spitz F, Furlong EE. 2012. Transcription factors: from enhancer binding to developmental control. Nat. Rev. Genet. 13, 613–626 10.1038/nrg3207 (doi:10.1038/nrg3207) [DOI] [PubMed] [Google Scholar]
- 6.Levine M. 2010. Transcriptional enhancers in animal development and evolution. Curr. Biol. 20, R754–R763 10.1016/j.cub.2010.06.070 (doi:10.1016/j.cub.2010.06.070) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Banerji J, Olson L, Schaffner W. 1983. A lymphocyte-specific cellular enhancer is located downstream of the joining region in immunoglobulin heavy chain genes. Cell 33, 729–740 10.1016/0092-8674(83)90015-6 (doi:10.1016/0092-8674(83)90015-6) [DOI] [PubMed] [Google Scholar]
- 8.Sandmann T, Girardot C, Brehme M, Tongprasit W, Stolc V, Furlong EE. 2007. A core transcriptional network for early mesoderm development in Drosophila melanogaster. Genes Dev. 21, 436–449 10.1101/gad.1509007 (doi:10.1101/gad.1509007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yanez-Cuna JO, Kvon EZ, Stark A. 2012. Deciphering the transcriptional cis-regulatory code. Trends Genet. 29, 11–22 10.1016/j.tig.2012.09.007 (doi:10.1016/j.tig.2012.09.007) [DOI] [PubMed] [Google Scholar]
- 10.Zinzen RP, Girardot C, Gagneur J, Braun M, Furlong EE. 2009. Combinatorial binding predicts spatio-temporal cis-regulatory activity. Nature 462, 65–70 10.1038/nature08531 (doi:10.1038/nature08531) [DOI] [PubMed] [Google Scholar]
- 11.Halfon MS, Carmena A, Gisselbrecht S, Sackerson CM, Jimenez F, Baylies MK, Michelson AM. 2000. Ras pathway specificity is determined by the integration of multiple signal-activated and tissue-restricted transcription factors. Cell 103, 63–74 10.1016/S0092-8674(00)00105-7 (doi:10.1016/S0092-8674(00)00105-7) [DOI] [PubMed] [Google Scholar]
- 12.Mullen AC, et al. 2011. Master transcription factors determine cell-type-specific responses to TGF-β signaling. Cell 147, 565–576 10.1016/j.cell.2011.08.050 (doi:10.1016/j.cell.2011.08.050) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trompouki E, et al. 2011. Lineage regulators direct BMP and Wnt pathways to cell-specific programs during differentiation and regeneration. Cell 147, 577–589 10.1016/j.cell.2011.09.044 (doi:10.1016/j.cell.2011.09.044) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li B, Carey M, Workman JL. 2007. The role of chromatin during transcription. Cell 128, 707–719 10.1016/j.cell.2007.01.015 (doi:10.1016/j.cell.2007.01.015) [DOI] [PubMed] [Google Scholar]
- 15.Lorch Y, LaPointe JW, Kornberg RD. 1992. Initiation on chromatin templates in a yeast RNA polymerase II transcription system. Genes Dev. 6, 2282–2287 10.1101/gad.6.12a.2282 (doi:10.1101/gad.6.12a.2282) [DOI] [PubMed] [Google Scholar]
- 16.Workman JL, Taylor IC, Kingston RE. 1991. Activation domains of stably bound GAL4 derivatives alleviate repression of promoters by nucleosomes. Cell 64, 533–544 10.1016/0092-8674(91)90237-S (doi:10.1016/0092-8674(91)90237-S) [DOI] [PubMed] [Google Scholar]
- 17.Miller JA, Widom J. 2003. Collaborative competition mechanism for gene activation in vivo. Mol. Cell. Biol. 23, 1623–1632 10.1128/MCB.23.5.1623-1632.2003 (doi:10.1128/MCB.23.5.1623-1632.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mirny LA. 2010. Nucleosome-mediated cooperativity between transcription factors. Proc. Natl Acad. Sci. USA 107, 22 534–22 539 10.1073/pnas.0913805107 (doi:10.1073/pnas.0913805107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Voss TC, et al. 2011. Dynamic exchange at regulatory elements during chromatin remodeling underlies assisted loading mechanism. Cell 146, 544–554 10.1016/j.cell.2011.07.006 (doi:10.1016/j.cell.2011.07.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zaret KS, Carroll JS. 2011. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 25, 2227–2241 10.1101/gad.176826.111 (doi:10.1101/gad.176826.111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Biddie SC, et al. 2011. Transcription factor AP1 potentiates chromatin accessibility and glucocorticoid receptor binding. Mol. Cell 43, 145–155 10.1016/j.molcel.2011.06.016 (doi:10.1016/j.molcel.2011.06.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de la Serna IL, Ohkawa Y, Berkes CA, Bergstrom DA, Dacwag CS, Tapscott SJ, Imbalzano AN. 2005. MyoD targets chromatin remodeling complexes to the myogenin locus prior to forming a stable DNA-bound complex. Mol. Cell. Biol. 25, 3997–4009 10.1128/MCB.25.10.3997-4009.2005 (doi:10.1128/MCB.25.10.3997-4009.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, Carroll JS, Liu XS, Brown M. 2008. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell 132, 958–970 10.1016/j.cell.2008.01.018 (doi:10.1016/j.cell.2008.01.018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liber D, Domaschenz R, Holmqvist PH, Mazzarella L, Georgiou A, Leleu M, Fisher AG, Labosky PA, Dillon N. 2010. Epigenetic priming of a pre-B cell-specific enhancer through binding of Sox2 and Foxd3 at the ESC stage. Cell Stem Cell 7, 114–126 10.1016/j.stem.2010.05.020 (doi:10.1016/j.stem.2010.05.020) [DOI] [PubMed] [Google Scholar]
- 25.Xu J, Pope SD, Jazirehi AR, Attema JL, Papathanasiou P, Watts JA, Zaret KS, Weissman IL, Smale ST. 2007. Pioneer factor interactions and unmethylated CpG dinucleotides mark silent tissue-specific enhancers in embryonic stem cells. Proc. Natl Acad. Sci. USA 104, 12 377–12 382 10.1073/pnas.0704579104 (doi:10.1073/pnas.0704579104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gualdi R, Bossard P, Zheng M, Hamada Y, Coleman JR, Zaret KS. 1996. Hepatic specification of the gut endoderm in vitro: cell signaling and transcriptional control. Genes Dev. 10, 1670–1682 10.1101/gad.10.13.1670 (doi:10.1101/gad.10.13.1670) [DOI] [PubMed] [Google Scholar]
- 27.Bossard P, Zaret KS. 1998. GATA transcription factors as potentiators of gut endoderm differentiation. Development 125, 4909–4917 [DOI] [PubMed] [Google Scholar]
- 28.Merika M, Thanos D. 2001. Enhanceosomes. Curr. Opin. Genet. Dev. 11, 205–208 10.1016/S0959-437X(00)00180-5 (doi:10.1016/S0959-437X(00)00180-5) [DOI] [PubMed] [Google Scholar]
- 29.Wu WH, Hampsey M. 1999. Transcription: common cofactors and cooperative recruitment. Curr. Biol. 9, R606–R609 10.1016/S0960-9822(99)80385-6 (doi:10.1016/S0960-9822(99)80385-6) [DOI] [PubMed] [Google Scholar]
- 30.Krivega I, Dean A. 2012. Enhancer and promoter interactions—long distance calls. Curr. Opin. Genet. Dev. 22, 79–85 10.1016/j.gde.2011.11.001 (doi:10.1016/j.gde.2011.11.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kagey MH, et al. 2010. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467, 430–435 10.1038/nature09380 (doi:10.1038/nature09380) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zentner GE, Scacheri PC. 2012. The chromatin fingerprint of gene enhancer elements. J. Biol. Chem. 287, 30 888–30 896 10.1074/jbc.R111.296491 (doi:10.1074/jbc.R111.296491) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heintzman ND, et al. 2009. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459, 108–112 10.1038/nature07829 (doi:10.1038/nature07829) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. 2011. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470, 279–283 10.1038/nature09692 (doi:10.1038/nature09692) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Visel A, et al. 2009. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457, 854–858 10.1038/nature07730 (doi:10.1038/nature07730) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thurman RE, et al. 2012. The accessible chromatin landscape of the human genome. Nature 489, 75–82 10.1038/nature11232 (doi:10.1038/nature11232) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gammill LS, Bronner-Fraser M. 2003. Neural crest specification: migrating into genomics. Nat. Rev. Neurosci. 4, 795–805 10.1038/nrn1219 (doi:10.1038/nrn1219) [DOI] [PubMed] [Google Scholar]
- 38.Sauka-Spengler T, Bronner-Fraser M. 2008. A gene regulatory network orchestrates neural crest formation. Nat. Rev. Mol. Cell Biol. 9, 557–568 10.1038/nrm2428 (doi:10.1038/nrm2428) [DOI] [PubMed] [Google Scholar]
- 39.Cordero DR, Brugmann S, Chu Y, Bajpai R, Jame M, Helms JA. 2010. Cranial neural crest cells on the move: Their roles in craniofacial development. Am. J. Med. Genet. A 155A, 270–279 10.1002/ajmg.a.33702 (doi:10.1002/ajmg.a.33702) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Minoux M, Rijli FM. 2010. Molecular mechanisms of cranial neural crest cell migration and patterning in craniofacial development. Development 137, 2605–2621 10.1242/dev.040048 (doi:10.1242/dev.040048) [DOI] [PubMed] [Google Scholar]
- 41.Birnbaum S, et al. 2009. Key susceptibility locus for nonsyndromic cleft lip with or without cleft palate on chromosome 8q24. Nat. Genet. 41, 473–477 10.1038/ng.333 (doi:10.1038/ng.333) [DOI] [PubMed] [Google Scholar]
- 42.Passos-Bueno MR, Ornelas CC, Fanganiello RD. 2009. Syndromes of the first and second pharyngeal arches: a review. Am. J. Med. Genet. A 149A, 1853–1859 10.1002/ajmg.a.32950 (doi:10.1002/ajmg.a.32950) [DOI] [PubMed] [Google Scholar]
- 43.Bajpai R, et al. 2010. CHD7 cooperates with PBAF to control multipotent neural crest formation. Nature 463, 958–962 10.1038/nature08733 (doi:10.1038/nature08733) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rada-Iglesias A, Bajpai R, Prescott S, Brugmann SA, Swigut T, Wysocka J. 2012. Epigenomic annotation of enhancers predicts transcriptional regulators of human neural crest. Cell Stem Cell 11, 633–648 10.1016/j.stem.2012.07.006 (doi:10.1016/j.stem.2012.07.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin FJ, Qin J, Tang K, Tsai SY, Tsai MJ. 2011. Coup d'Etat: an orphan takes control. Endocr. Rev. 32, 404–421 10.1210/er.2010-0021 (doi:10.1210/er.2010-0021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Croze N, Maczkowiak F, Monsoro-Burq AH. 2011. Reiterative AP2a activity controls sequential steps in the neural crest gene regulatory network. Proc Natl Acad Sci USA 108, 155–160 10.1073/pnas.1010740107 (doi:10.1073/pnas.1010740107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsai SY, Tsai MJ. 1997. Chick ovalbumin upstream promoter-transcription factors (COUP-TFs): coming of age. Endocr. Rev. 18, 229–240 10.1210/er.18.2.229 (doi:10.1210/er.18.2.229) [DOI] [PubMed] [Google Scholar]
- 48.Creyghton MP, et al. 2010. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl Acad. Sci. USA 107, 21 931–21 936 10.1073/pnas.1016071107 (doi:10.1073/pnas.1016071107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reddy TE, et al. 2012. Effects of sequence variation on differential allelic transcription factor occupancy and gene expression. Genome Res. 22, 860–869 10.1101/gr.131201.111 (doi:10.1101/gr.131201.111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Karczewski KJ, Tatonetti NP, Landt SG, Yang X, Slifer T, Altman RB, Snyder M. 2011. Cooperative transcription factor associations discovered using regulatory variation. Proc. Natl Acad. Sci. USA 108, 13 353–13 358 10.1073/pnas.1103105108 (doi:10.1073/pnas.1103105108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maurano MT, et al. 2012. Systematic localization of common disease-associated variation in regulatory DNA. Science 337, 1190–1195 10.1126/science.1222794 (doi:10.1126/science.1222794) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Degner JF, et al. 2012. DNase I sensitivity QTLs are a major determinant of human expression variation. Nature 482, 390–394 10.1038/nature10808 (doi:10.1038/nature10808) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McDaniell R, et al. 2010. Heritable individual-specific and allele-specific chromatin signatures in humans. Science 328, 235–239 10.1126/science.1184655 (doi:10.1126/science.1184655) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gaulton KJ, et al. 2010. A map of open chromatin in human pancreatic islets. Nat. Genet. 42, 255–259 10.1038/ng.530 (doi:10.1038/ng.530) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Frankel N, Davis GK, Vargas D, Wang S, Payre F, Stern DL. 2010. Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature 466, 490–493 10.1038/nature09158 (doi:10.1038/nature09158) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hong JW, Hendrix DA, Levine MS. 2008. Shadow enhancers as a source of evolutionary novelty. Science 321, 1314. 10.1126/science.1160631 (doi:10.1126/science.1160631) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lettice LA, et al. 2003. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum. Mol. Genet. 12, 1725–1735 10.1093/hmg/ddg180 (doi:10.1093/hmg/ddg180) [DOI] [PubMed] [Google Scholar]
- 58.Kleinjan DA, van Heyningen V. 2005. Long-range control of gene expression: emerging mechanisms and disruption in disease. Am. J. Hum. Genet. 76, 8–32 10.1086/426833 (doi:10.1086/426833) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stitzel ML, et al. 2010. Global epigenomic analysis of primary human pancreatic islets provides insights into type 2 diabetes susceptibility loci. Cell Metab. 12, 443–455 10.1016/j.cmet.2010.09.012 (doi:10.1016/j.cmet.2010.09.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pittman AM, et al. 2010. Allelic variation at the 8q23.3 colorectal cancer risk locus functions as a cis-acting regulator of EIF3H. PLoS Genet. 6 10.1371/journal.pgen.1001126 (doi:10.1371/journal.pgen.1001126) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ahmadiyeh N, et al. 2010. 8q24 prostate, breast, and colon cancer risk loci show tissue-specific long-range interaction with MYC. Proc. Natl Acad. Sci. USA 107, 9742–9746 10.1073/pnas.0910668107 (doi:10.1073/pnas.0910668107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sotelo J, et al. 2010. Long-range enhancers on 8q24 regulate c-Myc. Proc. Natl Acad. Sci. USA 107, 3001–3005 10.1073/pnas.0906067107 (doi:10.1073/pnas.0906067107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wasserman NF, Aneas I, Nobrega MA. 2010. An 8q24 gene desert variant associated with prostate cancer risk confers differential in vivo activity to a MYC enhancer. Genome Res. 20, 1191–1197 10.1101/gr.105361.110 (doi:10.1101/gr.105361.110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sakabe NJ, Savic D, Nobrega MA. 2012. Transcriptional enhancers in development and disease. Genome Biol. 13, 238. 10.1186/gb-2012-13-1-238 (doi:10.1186/gb-2012-13-1-238) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rahimov F, et al. 2008. Disruption of an AP-2α binding site in an IRF6 enhancer is associated with cleft lip. Nat. Genet. 40, 1341–1347 10.1038/ng.242 (doi:10.1038/ng.242) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Peter IS, Davidson EH. 2011. Evolution of gene regulatory networks controlling body plan development. Cell 144, 970–985 10.1016/j.cell.2011.02.017 (doi:10.1016/j.cell.2011.02.017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gellon G, McGinnis W. 1998. Shaping animal body plans in development and evolution by modulation of Hox expression patterns. Bioessays 20, 116–125 (doi:10.1002/(SICI)1521-1878(199802)20:2<116::AID-BIES4>3.0.CO;2-R) [DOI] [PubMed] [Google Scholar]
- 68.King MC, Wilson AC. 1975. Evolution at two levels in humans and chimpanzees. Science 188, 107–116 10.1126/science.1090005 (doi:10.1126/science.1090005) [DOI] [PubMed] [Google Scholar]
- 69.Wray GA, Hahn MW, Abouheif E, Balhoff JP, Pizer M, Rockman MV, Romano LA. 2003. The evolution of transcriptional regulation in eukaryotes. Mol. Biol. Evol. 20, 1377–1419 10.1093/molbev/msg140 (doi:10.1093/molbev/msg140) [DOI] [PubMed] [Google Scholar]
- 70.Carroll SB. 2008. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134, 25–36 10.1016/j.cell.2008.06.030 (doi:10.1016/j.cell.2008.06.030) [DOI] [PubMed] [Google Scholar]
- 71.Stark A, et al. 2007. Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. Nature 450, 219–232 10.1038/nature06340 (doi:10.1038/nature06340) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wittkopp PJ, et al. 2009. Intraspecific polymorphism to interspecific divergence: genetics of pigmentation in Drosophila. Science 326, 540–544 10.1126/science.1176980 (doi:10.1126/science.1176980) [DOI] [PubMed] [Google Scholar]
- 73.Johannsdottir B, Thorarinsson F, Thordarson A, Magnusson TE. 2005. Heritability of craniofacial characteristics between parents and offspring estimated from lateral cephalograms. Am. J. Orthod. Dentofacial Orthop. 127, 200–207 10.1016/j.ajodo.2004.07.033 (doi:10.1016/j.ajodo.2004.07.033) [DOI] [PubMed] [Google Scholar]
- 74.Hall BK, Miyake T. 1992. The membranous skeleton: the role of cell condensations in vertebrate skeletogenesis. Anat. Embryol. (Berl) 186, 107–124 [DOI] [PubMed] [Google Scholar]
- 75.Lengele B, Schowing J, Dhem A. 1996. Chondroid tissue in the early facial morphogenesis of the chick embryo. Anat. Embryol. (Berl) 193, 505–513 [DOI] [PubMed] [Google Scholar]
- 76.Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, Behringer RR, de Crombrugghe B. 2002. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell 108, 17–29 10.1016/S0092-8674(01)00622-5 (doi:10.1016/S0092-8674(01)00622-5) [DOI] [PubMed] [Google Scholar]
- 77.Yoshida CA, et al. 2004. Runx2 and Runx3 are essential for chondrocyte maturation, and Runx2 regulates limb growth through induction of Indian hedgehog. Genes Dev. 18, 952–963 10.1101/gad.1174704 (doi:10.1101/gad.1174704) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mallarino R, Grant PR, Grant BR, Herrel A, Kuo WP, Abzhanov A. 2011. Two developmental modules establish 3D beak-shape variation in Darwin's finches. Proc. Natl Acad. Sci. USA 108, 4057–4062 10.1073/pnas.1011480108 (doi:10.1073/pnas.1011480108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schneider RA, Helms JA. 2003. The cellular and molecular origins of beak morphology. Science 299, 565–568 10.1126/science.1077827 (doi:10.1126/science.1077827) [DOI] [PubMed] [Google Scholar]
- 80.Abzhanov A, Protas M, Grant BR, Grant PR, Tabin CJ. 2004. Bmp4 and morphological variation of beaks in Darwin's finches. Science 305, 1462–1465 10.1126/science.1098095 (doi:10.1126/science.1098095) [DOI] [PubMed] [Google Scholar]