Abstract
We have combined the circular chromosome conformation capture protocol with high-throughput, genome-wide sequence analysis to characterize the cis-acting regulatory network at a single locus. In contrast to methods which identify large interacting regions (10–1000 kb), the 4C approach provides a comprehensive, high-resolution analysis of a specific locus with the aim of defining, in detail, the cis-regulatory elements controlling a single gene or gene cluster. Using the human α-globin locus as a model, we detected all known local and long-range interactions with this gene cluster. In addition, we identified two interactions with genes located 300 kb (NME4) and 625 kb (FAM173a) from the α-globin cluster.
Keywords: α-globin locus, cis-regulatory elements, chromatin conformation capture
1. Introduction
The past few years have seen the development of techniques that allow us to detect physical interactions between key chromosomal elements that regulate gene expression. Such analyses are based on chromosome conformation capture (3C), designed to identify and quantitate novel ligation products between any two specific DNA sequences (in chromatin) that become closely juxtaposed in the nucleus in vivo [1–3]. By quantifying ligation products in large populations of cells, the relative frequency of such physical interactions can be inferred. Using 3C, it has been shown that cis-acting elements located up to 1 Mb apart on a chromosome may interact (forming a chromosomal loop) when genes are switched on [4] or off [5,6].
More recently, related techniques have been developed to analyse many intra- and inter-chromosomal interactions simultaneously, in an unbiased manner rather than focusing on pre-selected pairs of sequences [7–12]. Such methods ([8,9,11–15]; referred to as circularized 3C, or circular chromosome conformation capture (4C)) are based on the assumption that, as a result of cross-linking, cutting and ligating regions of interacting chromatin, circular molecules containing various combinations of associated segments of DNA will be formed (figure 1). Using inverse PCR primers located near the ends of a chosen fragment of interest (the ‘bait’), the amplified 4C library should capture all sequences (within a population of cells) that interact with the bait. An important aim of this approach is to characterize in detail the entire cis-regulatory network controlling a single gene.
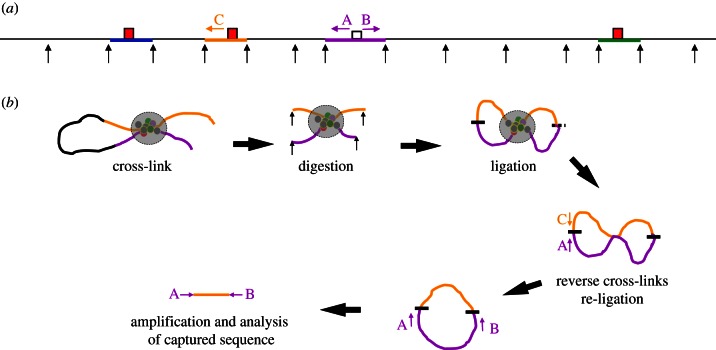
Figure 1.
Outline of 4C methodology. (a) Shows a hypothetical genomic locus in a linear configuration, black arrows represent the digestion sites for a frequently cutting restriction endonuclease. Four restriction fragments are highlighted in colour with the capture point (bait) to be analysed in purple. Three interacting cis-elements (red boxes) are shown contained within fragments coloured in blue, yellow and green. The positions of primer sequences used in 4C analysis (A and B) and in 3C analysis (A and C, for example) are shown as arrows. (b) The locus is shown with the yellow cis-element interacting with the bait by looping out the intervening sequence. The proteins associated with the two fragments (coloured circles) are shown after formaldehyde cross-linking (large grey circle) to fix the structure. The chromatin is digested after fixation (black arrows). Next, ligation produces permanent links between the two interacting fragments. The two fragments can ligate (shown as a short black line) in two ways; at both ends to produce a circle or at one end (shown as a short-dashed black line) to produce a linear molecule. These DNA/protein structures are then de-cross-linked, and the DNA isolated. Linear molecules can be converted to a circle by a second round of ligation (discussed in the main text). An interaction (for example, between the purple and yellow fragments) can be assayed using conventional 3C using primers A and C. All of the fragments interacting with the purple capture fragment (bait) can be amplified by inverse PCR using the primers A and B.
In the work presented here, we have used a slightly modified 4C protocol with sufficient resolution to identify individual elements interacting with a chosen cis-element, and used this to analyse the human α-globin locus[16,17] that has previously been analysed using conventional 3C (from multiple fixed positions) in both erythroid (expressing) and non-erythroid (silent) cells [18,19]. All previously known interactions with the α-globin regulatory elements and interactions with two additional non-globin genes (NME4 and FAM173a) were identified.
2. Results
(a). Generating a library of chromatin interactions using a circular chromosome conformation capture protocol
The methodology is based on the established 3C assay (fig. 1 provided in reference Gondor et al. [20] and in §4). In brief, living cells were treated with formaldehyde to cross-link protein and DNA [21,22]. Cells were then lysed to allow restriction enzyme digestion of the cross-linked chromatin. The bulk of chromatin was cut into small fragments, while cross-linked, interacting protein/DNA complexes remained physically associated with each other (figure 1). These non-random interactions were covalently linked by ligation of the DNA fragments (while still in the context of chromatin) in a large volume to maximize the difference between infrequent ligation of random individual fragments and the more frequent ligation of closely associated fragments that interact in chromatin. Finally, cross-links were reversed and DNA was isolated.
In the standard 3C protocol, the enrichment for an interaction between two specific fragments (relative to the random background level of interactions) is assayed using quantitative PCR with a primer in each fragment (see primers A and C in figure 1 and [2,3]). However, interacting molecules can be circularized (figure 1). By designing appropriate PCR primers for the chosen bait, a library containing all of the fragments that were interacting at the time of the initial cross-linking can be made by inverse PCR (primers A and B in figure 1; [9]).
A frequently cutting restriction enzyme (DpnII, ^GATC) that efficiently digests cross-linked chromatin was used to cut the cross-linked chromatin. Its average fragment size is 433 bp, improving resolution compared with using 6-cutter enzymes [13–15]. Circles containing relatively short fragments can be efficiently amplified in subsequent PCR reactions (see below). Furthermore, the large 4 bp over-hang (5′-3′) remaining after DpnII digestion provides a very efficient substrate for ligation.
A second ligation step that facilitates the probability of re-circularization after isolation of DNA was found to improve the detection of previously characterized cis-interactions involving the α-globin promoters (see the electronic supplementary material, figure S1).
(b). Analysis of an amplified circular chromosome conformation capture library on tiled genomic microarrays using a promoter as the bait
Using this protocol, 4C libraries were generated from primary erythroid cells (eight replicates), primary T cells (two replicates) and Epstein–Barr virus (EBV)-transformed B lymphocytes (four replicates). These libraries were amplified using inverse PCR primers designed to capture all sequences interacting with the chosen bait (figure 1).
Primers for the α-globin gene promoters were designed within an 899 bp DpnII fragment (chr16:162 450–163 349 and chr16:166 254–167 153) containing the promoters of the duplicated HBA genes (α1 and α2, figure 2a). Labelled DNA was hybridized to tiled microarrays representing the human α-globin genes, their known regulatory elements and 500 kb of flanking DNA [23]. Enrichment was measured relative to labelled, sonicated input DNA. In both non-erythroid cell types (T-lymphocyte and EBV-transformed B lymphocyte), positively enriched signals on the microarray were only found in regions over and immediately adjacent to the bait (figure 2b). No additional distant, long-range interactions were seen. As a positive control, we analysed the non-erythroid 4C libraries using inverse PCR primers within a previously characterized constitutive CTCF-bound region in the same genomic interval. As for other CTCF-bound regions [7,24], this bait was shown to participate in looping events with other CTCF-bound regions (see the electronic supplementary material, figure S2).
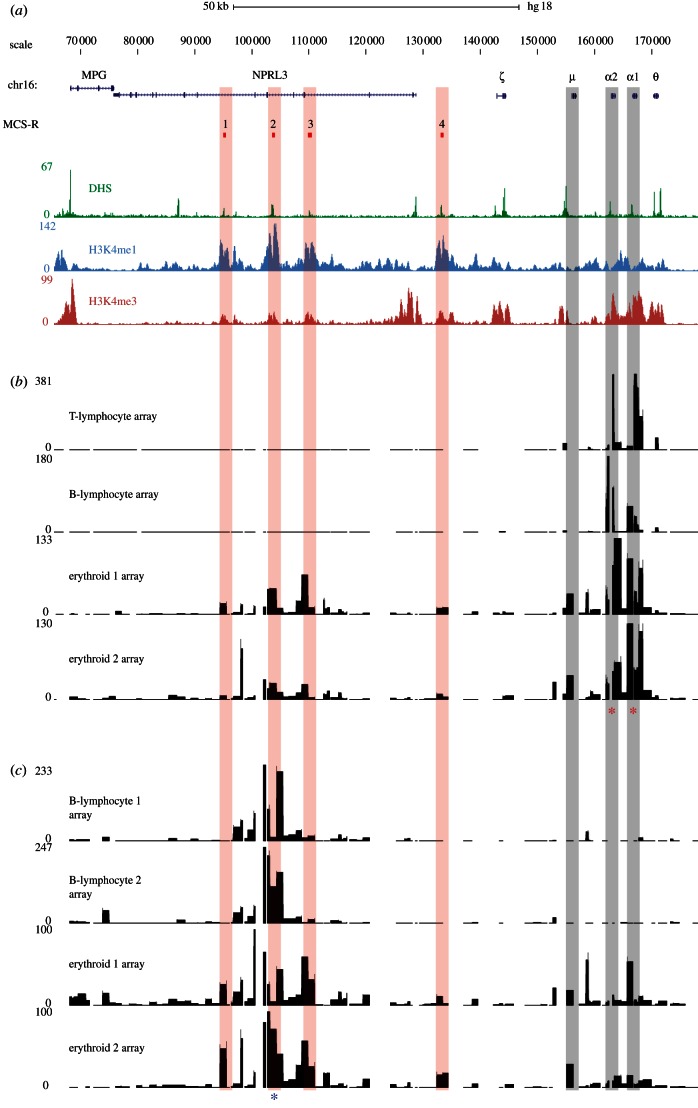
Figure 2.
Identification of cis-regulatory elements on tiled genomic microarrays using α-globin as bait. (a) A 100 kb region of human chromosome 16 from 70–170 kb of genome build HG18. RefSeq genes are shown as blue lines for gene extent, and blue ticks for exons. The MCS-R track shows the multi-species conserved sequences associated with the α-globin regulatory elements (MCS-R1–4), and their alignments to microarray features are indicated with vertical red bars. Below this are plots of DNase-seq (green) and chromatin ChIP-seq (H3K4me1 in light blue, H3K4me3 in red) data for the K562 cell line from the ENCODE project on genome build HG18. Position of HBM gene (μ) and HBA genes (α1 and α2) are indicated by vertical grey bars. (b) Inverse PCR amplification from the α-globin promoters (indicated by red asterisks) of 4C libraries derived from T lymphocytes, B lymphocytes and two biological replicates from primary erythroid cells. Samples were hybridized to tiled genomic PCR microarrays [23] using sonicated genomic DNA as an input to correct for probe hybridization efficiency. The signal on the y-axis is normalized for the total signal on the array. Genomic distance is shown, to scale with annotation, on the x-axis and fold enrichment relative to input on the y-axis. The duplicated α-globin genes have almost identical DNA sequences and, therefore, the primers simultaneously assay interactions with each promoter. (c) Inverse PCR amplification from the α-globin regulatory element MCS-R2 (HS-40; indicated by a blue asterisk) of 4C libraries derived from two B lymphocyte and two erythroid biological replicates. Samples were hybridized to the same tiled genomic PCR microarrays (above) using sonicated genomic DNA as an input. Note that the level of signal over the gene promoters is lower than that of the reverse experiment performed from the gene promoters. This highlights a technical limitation of the 4C assay. The cis-elements (MCS-R1 to R4) themselves have a relatively low GC content (54%) equal to the locus average, whereas the promoters of the HBA genes are large CpG islands with a very high GC content (73%). Although the PCR system used in this protocol is optimized for GC-rich templates, the promoters of the globin genes are difficult to amplify even using standard, non-inverse PCR. Furthermore, they are contained within relatively large DpnII fragments (899 bp). Together, these features are likely to make inverse PCR amplification relatively inefficient for these loci. This idea is supported by the stronger enrichment seen for the promoter of HBM which, while having a high GC content (72%), lies within a smaller (368 bp) DpnII fragment. Because the 4C assay relies on an inverse PCR step, large and very GC-rich fragments may be under-represented in the data produced by this assay.
In erythroid cells, the α-globin promoters can be seen to interact with their flanking sequences to a much greater degree with new, specific and strong interactions (figure 2b). The most proximal strong interaction is with the promoter of a neighbouring gene HBM (µ globin), a globin-related gene expressed only in erythroid cells [25]. The lack of such a signal in the non-erythroid tissues demonstrates that this is not a random interaction and is not due to cross-hybridization with the amplified HBA promoter. This comparison between chromatin derived from expressing and non-expressing cells shows that when both the HBM and HBA genes are active, their promoters frequently interact, which has been reported for other active promoters [26–28].
The main distal interaction occurs over the body of the C16orf35 gene containing the α-globin regulatory elements (multiple conserved sequence (MCS)-R1 to R3). In erythroid samples, the signal is maximal over this area with distinct peaks of interaction seen over each element clearly identifying MCS-R1 (hypersensitive site, HS48); MCS-R2 (HS-40) and MCS-R3 (HS-33). A smaller peak can also be seen over a fourth conserved erythroid cis-element, MCS-R4 (HS-10), located between the C16orf35 gene and the HBA genes. Therefore, using this 4C protocol with the α-globin promoter as bait, we identified all known, non-random, tissue-specific interactions between the promoters and their upstream regulatory elements previously established by 3C analysis [18].
(c). Analysis of an amplified circular chromosome conformation capture library on tiled genomic microarrays using a distal regulatory element as the bait
To further validate these interactions, we reversed the direction of the capture by designing primers in a region (identified above) that interacts with the HBA genes. We, therefore, amplified the same 4C libraries with inverse primers designed within a 1205 bp DpnII fragment containing the MCS-R2 cis-element (chr16:102 658–103 864) and analysed this material on tiled microarrays. Figure 2c compares two erythroid and two non-erythroid (EBV-transformed B lymphocyte) samples. As with the promoter capture, there is little evidence of any long-range interactions in the B-lymphocyte cultures, in which the MCS-R2 element and HBA genes are inactive. By contrast in erythroid cells, strong peaks of interaction are seen over two neighbouring cis-elements, MCS-R1 and MCS-R3, with a smaller peak over the more distal MCR-R4 element. Signal is also seen over the genomic area containing the HBA genes, and a strong peak is seen over the promoter of HBM.
The overlap and reciprocity of the interactions identified by 4C using the gene promoters as bait compared with those identified using MCS-R2 as bait, supports the previously proposed model in which these two sequences interact specifically in erythroid cells [19]. Furthermore, not only do the promoters of the HBA genes interact with all four characterized cis-elements (MCS-R1-4), but when using just one of these elements (MCS-R2) as bait, we identify the α-globin promoters and also capture the other regulatory elements (MCR-R1, 3 and 4) strongly suggesting that all of these elements come together (possibly simultaneously) forming an active chromatin hub, as previously suggested for the β-globin gene cluster [29].
(d). Analysis of circular chromosome conformation capture libraries by high-throughput sequencing using paired-end reads
High-throughput sequencing (HTS) has revolutionized genome-wide analysis, and in 4C experiments has an advantage over microarray experiments by confirming the specificity of amplification and allowing the exclusion of mis-primed amplimers from downstream analysis (see below). A feature of the Illumina platform is the ability to sequence both ends of a DNA fragment and link the forward and reverse reads. This paired-end HTS (peHTS) protocol produces 50 bp of sequence from either end of a single DNA fragment.
Initially in our analysis, each end of the fragment was considered as a single read and mapped to a specific DpnII fragment. At this stage, the relationship between the two paired-ends was masked from the standard mapping tools. Once the individual ends had been mapped, the paired-end information (linking one single copy read to another) was then used to unequivocally score interactions with one end in the α-promoter and the other in the ligated, interacting fragment (see figure 3 and electronic supplementary material, figure S3a). Mis-primed fragments that lacked the expected sequence associated with the bait fragment could be excluded. Fragments that lie adjacent to each other in the genome (possibly resulting from non-digestion of chromatin or ligation simply based on their proximity) were also removed from the analysis.
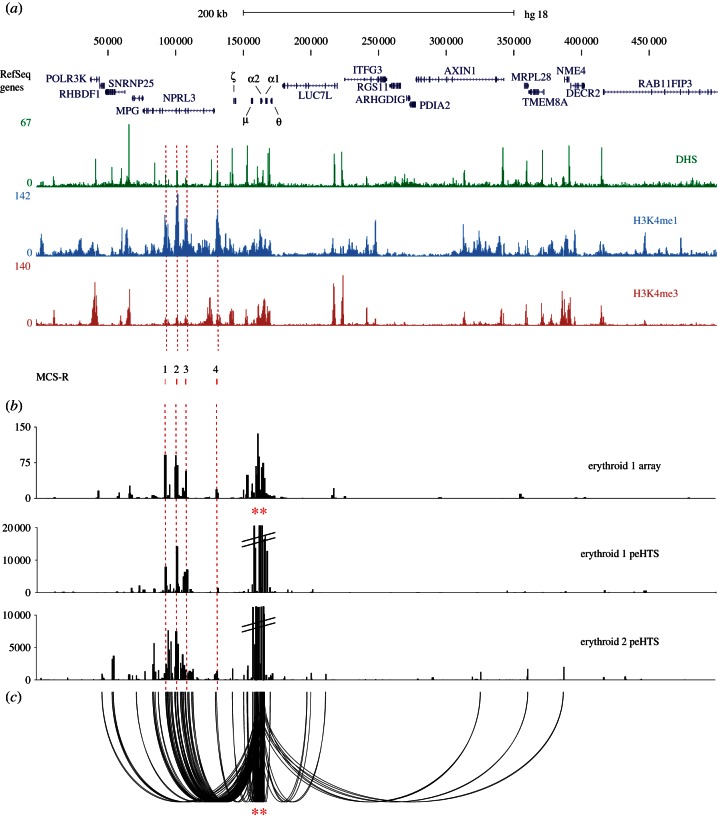
Figure 3.
Identification of cis-regulatory elements using the α-globin promoters as bait, and analysing 4C material by HTS (a) The terminal 500 kb region of chromosome 16 (HS18 16p13.3 as described in figure 2a). (b) Comparison of analysis by microarray (erythroid 1 array, signal on the y-axis is normalized for the total signal on the array) and peHTS of 4C inverse amplifications from the α-globin promoters (indicated by red asterisks). Two biological replicates of erythroid 4C libraries were analysed (erythroid 1 peHTS and erythroid 2 peHTS). The peHTS data were treated as single-end reads that were then ‘binned’ into DpnII restriction fragments (see text). (c) Shows the same data as displayed in the lower panel of (b) for erythroid 2 peHTS analysed using the sequences' paired-end relationships to identify junction fragments (one of which must map to the α-globin genes; chr16:158 550–169 250) and displayed using a custom looping glyph (promoter fragments shown as red asterisks).
Using this approach, we analysed two 4C libraries (independently derived from the primary erythroid cells of two individuals). These libraries were amplified with inverse primers (using the α-globin promoter as the bait), sonicated and prepared for paired-end sequencing. We defined an interaction as any read pair of which one end mapped to the bait and another mapped to a distal fragment in-cis or in-trans. By doing the experiment twice, we could substantially exclude random interactions or infrequent non-random interactions. The first dataset identified 338 consistently interacting regions of which 50 per cent were on chromosome 16, with other interactions distributed across the remainder of the genome. In the second dataset, there were 4172 interactions (although many of these were unique interactions recorded in a single paired-end read) of which 9 per cent mapped to chromosome 16. Although many of the interactions on chromosome 16 were consistently detected in both experiments, interactions with other chromosomes most frequently differed between experiments suggesting that they may be random.
To investigate this further, we generated two datasets (defining sequences and positions) of fragments that interacted with the α-globin promoters in both experiments. From these two datasets, we identified fragments common to both sets of data. Initially, we analysed the genomic distribution of these fragments irrespective of their number of interactions with the α-globin promoter (figure 4). These data suggest that most regions interacting with the α-globin promoters occur on chromosome 16 (presumably in cis), and that (using the α-globin promoter as bait) stable trans-interactions with this fragment are rare. We next analysed the number of sequences mapping to each of 122 interacting regions throughout the genome (identified in both experiments); in general, this should reflect the frequency of each interaction. We first analysed the number of interactions with each chromosome (regardless of the number of interacting regions on each chromosome). We excluded strong local interactions (from 157 000 to 170 000 bp on chr16; figure 4c) around the α-globin promoters to leave only those interactions representing true distal looping events. This analysis showed that 96.4 per cent of all interactions occurred on chromosome 16, although this approach does not exclude infrequent non-random interactions.
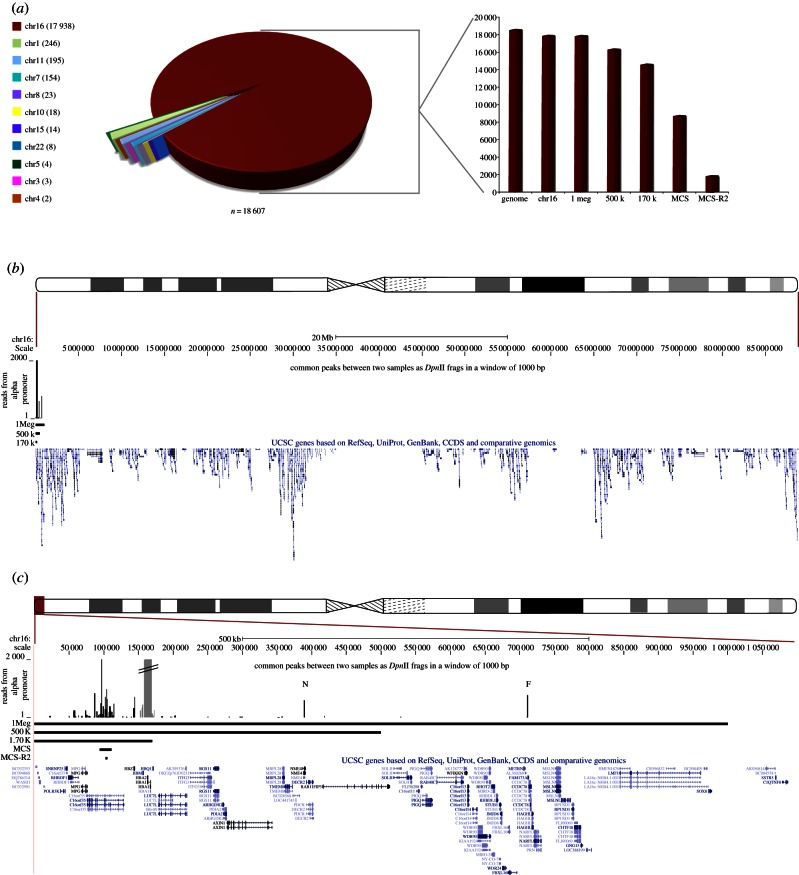
Figure 4.
Genome-wide analysis of cis- and trans-interactions with the α-globin promoter. (a) A pie chart representation of quantitative junction analysis of the two paired-end, erythroid 4C datasets, amplified from the α-globin promoter where only hits common to both datasets are represented. Each section of the pie chart represents the count (per chromosome) of all junction fragments linked to the α-globin promoter. Each interacting fragment either maps within or overlaps a 1 kb window spanning the genomic coordinates of a particular DpnII fragment in both biological replicates. Local signals around the bait (α-globin) are removed from this and all subsequent analyses (see (c) below). The numbers of junction sequences mapping to each chromosome are shown in brackets. In the adjacent bar graph, the same data are further broken down for their distribution across chromosome 16 (calculated relative to the total genome signal). The column marked ‘genome’ represents the total number of reads in this dataset; ‘chr16’ represents the sequences mapped to the whole of chromosome 16; ‘1 Mb’ represents the sequences mapped within chr16:1–1 000 000; ‘500 k’ represents the sequences mapped within chr16:1–500 000; ‘170 k’ represents the sequences mapped within chr16:1–170 000; ‘MCS' represents the sequences mapped within chr16:93 900–111 261 and ‘MCS-R2’ represents the sequences mapped within chr16: 102 493–104 848. (b) Shows the sequences mapped to chromosome 16 as a quantitative plot across its length (as displayed in UCSC). RefSeq genes and large scale regions are annotated. The region displayed is shown relative to an ideogram of chromosome 16. (c) A zoomed view to show the terminal 1 Mb region of chromosome 16p. RefSeq genes and chromosomal regions are annotated. The local signals (around the bait, α-globin) that were excluded from the genome-wide analysis are shaded as a discontinuous grey peak. The region displayed is shown relative to an ideogram of chromosome 16. The peaks of interaction corresponding to the NME4 gene and FAM173a genes are annotated by a N and F, respectively. (This is where the data are and now in §2.) To ensure that all such interactions with the α-globin promoters were observed, we relaxed the stringency of the analysis by scoring interactions with the same DpnII fragments in each dataset, but also interacting fragments in one dataset, which were within 1 kb of an interacting fragment in the other dataset.
The signal on chromosome 16 was further dissected, showing most of these interactions occur in the terminal 0.5 Mb of the short arm of the chromosome (figure 4c). This region contains the α-globin genes, their regulatory elements MCS-Rs (MCS-R1, MCS-R2, MCS-R3 and MCS-R4) and at least 12 associated prominent CTCF-bound regions. We also found two long-range interactions with genes located 300 kb (NME4) and 625 kb (FAM173A) from the α-globin cluster. We have recently shown that the closest of these two genes (NME4) is upregulated in erythroid cells and this is under the control of MCS-R2 [30]. FAM173a is not expressed in erythroid cells, and, therefore, its interaction with α-globin may represent a structural interaction.
Almost half of the consistently mapped α-globin promoter interactions are found in 0.0005 per cent of the genome containing its regulatory elements. Ten per cent of interactions occur with the major regulatory element MCS-R2. Clearly these are frequent (1859 of 18 607 mapped interactions), reproducible (present in both biological replicates) non-random interactions.
3. Discussion
The 4C approach allows analysis of chromosome conformation, in an unbiased way, without prior knowledge, to identify all sequences (in cis and trans) that interact with a genomic element of interest, and, therefore, is of considerable value in identifying the comprehensive network of regulatory elements controlling individual genes [13–15]. This addresses a common and timely problem in genome annotation, and solves the issue of assigning the functional effects of a particular sequence or structural variant (e.g. identified in genome-wide association studies analyses) to a specific gene [31].
By using a frequently cutting restriction enzyme (DpnII), tiled microarrays and HTS, cis-acting sequences were localized at a high resolution, and sequencing unequivocally identified the underlying cis-acting sequences. For the human α-globin cluster, these corresponded to previously identified regulatory elements. The use of HTS rather than microarrays considerably increased the specificity of the assay. Importantly, using paired-end reads it was possible to exclude mis-primed and non-digested sequences that do not represent true interactions with the cis-element being used as the bait. A criticism of this technique could be that the extensive rounds of amplification required and the heterogeneous sizes of the circles may blunt the dynamic range; however, the ability of the 4C technique to reproduce the known, predominant interactions in erythroid and non-erythroid cells is notable.
In addition to validating the approach used, analysis of the human α-globin cluster also revealed additional information about the interaction between the upstream regulatory elements and the α-globin promoter. It was previously known from 3C analysis that four erythroid elements (MCS-R1 to R4) interact with the α-globin promoter in erythroid cells. Furthermore, from the same 3C data, we had previously implicated CTCF/cohesin-bound regions in the establishment and/or maintenance of such loops consistent with the recent studies of others [7,24,32–36]. Preliminary experiments using direct sequence analysis of paired-end reads also identified re-circularized molecules that not only contained two or more MCS-R elements, but a number of captured sequences containing MCS-R2 (the major α-globin regulatory element) also contained the CTCF/cohesin element associated with HS-46, specifically in erythroid cells (see the electronic supplementary material, figures S3b and S4). As these sequences were found together on the same ligated molecules, this may reflect how the 4C protocol could reveal molecular interactions originating from a single locus. As this CTCF/cohesin-bound element lies between the MCS-R1 enhancer element and the α-globin promoters and appears to interact simultaneously with both of them, it appears that this CTCF-bound element does not act as either an enhancer blocker or a boundary element in this instance. Provisional analysis of these sequences showed that different interactions may be present in different individual cells suggesting that the interaction between the MCS-R elements and the α-globin promoter is dynamic. Further, deep sequencing data will be required to pursue this preliminary observation.
Other chromosome conformation studies have suggested that there may be a wide network of interactions between cis-acting elements throughout the genome and that specific trans-interactions (e.g. between α- and β-globin) occur frequently [37]. Others have suggested that trans-interactions are transient and infrequent [8,38]. Using the 4C protocol described here, we did not observe frequent, trans-interactions between the α- and β-globin loci either analysing the experiments on microarrays (see the electronic supplementary material, figure S5) or by sequencing. Within the limits of these experiments, if interactions (determined by counting the number of interacting fragments obtained by paired-end sequencing) between the α- and β-globin promoters do occur, they are at least 1000 times less frequent than the functionally relevant cis-interactions (e.g. between the globin promoters and their upstream regulatory elements).
Consistent with the 4C data presented here, Hi-C, a relatively low resolution (1 Mb) approach for full genome analysis [39] proposed that cis-elements most frequently interact with sequences located on the same chromosome in cis; trans-interactions appeared to be rare. Here, we looked for consistent very long-range (greater than 1 Mb) interactions (in cis or in-trans) by comparing all interactions with the α-globin locus throughout the entire genome. It was shown that nearly all reproducible frequent, non-random interactions with the α-globin promoter are restricted to the terminal megabase of chromosome 16 (where the α-genes are located), and more than 74 per cent of these occur within the previously defined 170 kb α-globin domain (chr16:1–170 000).
Genes located on a particular chromosome often lie within a territory occupied by other sequences on the same chromosome (chromosome territory). In the Hi-C analysis, it was suggested that the high frequency of interactions across the chromosome (in cis) could best be explained by interactions occurring within the context of a specific chromosome territory [39]. In contrast to many of the sequences analysed by Hi-C, there were relatively few consistent interactions between the α-genes and other genes located on chromosome 16 either in this study or in the published Hi-C analysis. One explanation could be that unlike many genes in the interstitial segments of chromosomes, the terminal 2 Mb region containing the α-globin locus consistently extends beyond the chromosome 16 territory in both erythroid and non-erythroid cells [40], and, therefore, may be more mobile and interact albeit inconsistently with a wide range of sequences below the detection of current analysis.
4. Material and methods
(a). Cell types
EBV-transformed lymphoblastoid (EBV B lymphocyte) cell lines were cultured in RPMI 1640 supplemented with 10% (v/v) fetal calf serum, 2 mM l-glutamine, 50 U ml−1 penicillin and 50 µg ml−1 streptomycin. Isolation and culture of primary human erythroblasts was carried out as described previously [41]. T lymphocytes were cultured as follows. Whole blood was mixed with an equal volume of RPMI and subjected to Ficoll-Paque separation (GE Heathcare Life Sciences). The interphase mononuclear layer was removed and diluted in RPMI + 10% fetal calf serum. Cells were pelleted at 700×g for 5 min and wash was repeated. Cells were then resuspended in 10 ml lysis buffer (150 mM NH4Cl, 10 mM KHCO3, 0.1 mM EDTA) and incubated on ice for 1 min. Cells were centrifuged at 700×g for 5 min and washed twice in RPMI + 5% fetal calf serum. After final wash and centrifugation at 700×g for 5 min, cells were resuspended in 30 ml RPMI + 20% fetal calf serum supplemented with 1 mg ml−1 phytohaemagglutinin and 20 U ml−1 interleukin 2. Cells were incubated at 37°C with 5 per cent CO2 for 3–4 days.
(b). Circular chromosome conformation capture
The protocol used for 4C analysis was based on Zhao et al. [9,20] with a minor modification. Following phenol/chloroform extraction and ethanol precipitation of the 4C library, DNA was resuspended in 500 μl 1× ligation buffer and 60 U high concentration T4 DNA ligase (Fermentas) and incubated for 2 h at 16°C at 1200 r.p.m. (Eppendorf Thermomixer comfort). Following phenol/chloroform extraction and ethanol precipitation, DNA was resuspended in 100 μl water, of which 10 μl was used as template in Advantage-GC PCR (Clontech) as per manufacturer's instructions using 34 cycles of amplification (cycling conditions were 94°C for 2 min; 34 cycles of 94°C for 30 s, annealing temperature for 30 s, 68°C for 5 min followed by a single extension cycle at 68°C for 8 min). Primer sequences and annealing temperatures are presented in table 1. The resulting amplified DNA was ethanol precipitated and resuspended in 20 μl water, of which 5 μl was hybridized to a custom α-globin tiled microarray using sonicated genomic DNA as input, as previously described [23]. Data from the microarray experiments are available from the GEO database under the accession no. GSE42384.
Table 1.
Primer sequences and annealing temperatures.
| target | name | sequence | position | annealing temperature (°C) |
|---|---|---|---|---|
| HBA promoter | HBA3Cf | TTCCCCACCACCAAGACCTAC | chr16:163 137–163 157 | 66 |
| chr16:166 941–166 961 | ||||
| HBA3Cr | AGAAAGTCAGCCCGCACCCC | chr16:162 497–162 516 | ||
| chr16:166 301–166 320 | ||||
| MCS-R2 (HS-40) | HS40f | CTGCTGATTACAACCTCTGGTGC | chr16:103 705–103 727 | 58 |
| HS40r | GAGCCTGGGGGAAAGGAGTAG | chr16:102 893–102 913 | ||
| HBB promoter | HBBPf1 | TGAGGTCTAAGTGATGACAGCCG | chr11:5 204 981–5 205 003 | 57 |
| HBBPr1 | TGAGGAGAAGTCTGCCGTTACTGC | chr11:5 204 787–5 204 810 | ||
| axin CTCF site | AXINf1 | GATGAGCAGAATCTGGTGATGAACAG | chr11:352 622–352 647 | 58 |
| AXINr1 | GAAGAGACAAAGGGAGCAGGGTG | chr11:351 223–351 245 |
(c). Preparation of circular chromosome conformation capture material for Solexa/Illumina sequencing
DNA obtained from the 4C amplification was reduced to an average size of 500 bp by sonication, either by shearing for 10 min using a Sonic Dismembrator 550 (cup horn, Fisher Scientific, Canada) or using a Covaris S2 sonicator (KBiosciences, Europe) using a duration of 90 s, intensity of 3 and a duty cycle of 5. A 300–600 bp fraction was gel extracted from an 8 per cent PAGE gel and prepared for paired-end sequencing using the manufacturer's recommended protocol as briefly set out here. Sonicated DNA from a 4C amplification was prepared for sequencing by end repair, A-tailing and ligation of adapters (Illumina) as outlined in the manufacturer's recommendations. Following adapter ligation, some minor modifications were made to the protocol: DNA was amplified with 18 cycles of PCR before size selection of 200–350 bp fragments from a gel. The excised library was purified using the QIAquick gel extraction kit (Qiagen) before quality checking on an Agilent bioanalyzer. DNA was quantified using the Quant-iTT dsDNA HS assay kit (Invitrogen) before dilution to 10 nM. Paired-end reads of 51 bp were generated on the Illumina GAII platform. A single lane of paired-end sequence was sufficient to produce a robust signal (table 2).
Table 2.
Raw and aligned read numbers.
| sample | total reads | number mapped | % mapped |
|---|---|---|---|
| erythroid 1 peHTS | 33 925 706 | 9 753 439 | 28.7 |
| erythroid 2 peHTS | 23 901 100 | 16 498 733 | 69.0 |
(d). Mapping of Solexa/Illumina reads
Paired-end sequences were mapped as single-end reads to overcome the built in assumptions of the relative positioning of paired-end sequences in the sequence aligning programs. The sequences were mapped to an in silico DpnII digested and repeat-masked version of HG18 (UCSC Genome browser, NCBI build 36.1). This digested version of HG18 was generated by mapping the position of all DpnII sites in the unmasked HG18 sequence. This in silico digestion had to be performed on an unmasked version of the HG18 as repeat masking would also have masked some DpnII restriction sites. These positions were then used to digest a repeat-masked version of the HG18 genome to generate a multiple Fasta file of all repeat-masked DpnII fragments in HG18. Repeat masking was performed using the latest ‘all mapped repeat’ data for HG18 (UCSC tablebrowser HG18_Human_allrmsk.txt). The sequence data were mapped to the digested genome using three different aligners, MAQ (v. 0.7.1; [42]), Exonerate (v. 2.2.1; [43]) and Novoalign (v. 2.05.13; www.novocraft.com) to exclude any aligner-specific effects, using the default stringencies for each program. The alignments were post processed, using in-house Perl scripts, for uniqueness of alignment, with reads mapping to the α-globin genes being allowed to map to three locations of chromosome 16 owing to the duplicated nature of the genes; all other reads had to align uniquely to the genome.
(e). Junction analysis
(i). Solexa/Illumina paired-end reads
The paired-end reads represent a 50 bp sequence from each end of a sonicated DNA fragment of the inverse PCR products. As sonication is mostly a random process then the paired-end reads randomly sample the sequence, at either end of approximately 500 bp intervals, across the inverse PCR products. This random sampling can be used to detect the ligation events, which underlie the 4C signal, where one end of the fragment maps to one DpnII fragment and the other end of the fragment maps to another. Although, as described above, the paired-end reads were initially mapped as single reads, the physical relationship between the paired-end reads was maintained in the naming structure of each read. Using in-house Perl scripts, this relationship could be used to call an interaction between these two fragments and ultimately count the total number of mapped interactions between all DpnII fragments in the HG18 genome. This dataset ignored all pairs that map to the same fragment (does not span a junction) and was further refined to remove interactions between fragments adjacent in the genome owing to local interactions or non-digestion. This dataset was further constrained so that each junction had to have a read in the capture fragment and hence represent a valid ligation event rather than non-specific amplification. Data from the sequencing experiments are available from the GEO database under the accession no. GSE42384.
(ii). Overlap analysis
Both biological 4C paired-end sequenced replicates (named JH1 and JH2) were analysed as described above. To identify the consistent signal between the two biological datasets, an intersection of the two results was performed such that interaction fragments that existed in both datasets were kept and the number of interactions of that fragment with the α-globin promoter were averaged. This analysis was made less stringent by the inclusion of fragments, which although they did not have exactly the same genomic coordinates, their coordinates fell within a 1 kb window of genomic distance of a peak in the other replicate. In this case, the signal for each peak was not averaged with the other, rather each was included in the final dataset with their respective coordinates and number of interactions with the α-globin promoter.
This intersection dataset was then binned into a set of genomic regions representing, each human chromosome, the chromosome 16p terminal 1 Mb (chr16:1–1 000 000), the encode region Enm008 (chr16:1–500 000), the region from the 16p telomere to the end of the globin θ gene (chr16:1–170 000), the area covering MCS-R1 to MCR-R3 (chr16:93 900–111 261) and region 1 kb either side of the MCS-R2 element (chr16:102 493–104 848).
Acknowledgements
This work was supported by the Medical Research Council (UK) and National Institute for Health Research (NIHR) Biomedical Research Center Program. J.R. is supported by the Wellcome trust. We thank Joyce Reittie for her technical assistance. We thanks David Garrick for his advice and comments on the manuscript; Nicki Gray for help preparing the manuscript; Cordelia Langford, Peter Ellis and the staff of the Wellcome Trust Sanger Institute Microarray Facility for array printing; Lorna Gregory, Yongjun Zhao and Steve Jones for sequencing support; and Zong-Pei Han of CBRG Oxford for computational and systems support. J.R.H., R.J.G. and D.R.H designed research. J.R.H., K.M.L., M.D.G., J.A.S-S and D.V. performed experiments. J.R.H., I.D. and S.T. analysed data. S.M. provided database support. J.R. provided resources. J.R.H., R.J.G. and D.R.H. wrote the manuscript. The authors declare no competing financial interest.
References
- 1.Dekker J, Rippe K, Dekker M, Kleckner N. 2002. Capturing chromosome conformation. Science 295, 1306–1311 10.1126/science.1067799 (doi:10.1126/science.1067799) [DOI] [PubMed] [Google Scholar]
- 2.Dekker J. 2006. The three ‘C’ s of chromosome conformation capture: controls, controls, controls. Nat. Methods 3, 17–21 10.1038/nmeth823 (doi:10.1038/nmeth823) [DOI] [PubMed] [Google Scholar]
- 3.Simonis M, Kooren J, de Laat W. 2007. An evaluation of 3C-based methods to capture DNA interactions. Nat. Methods 4, 895–901 10.1038/nmeth1114 (doi:10.1038/nmeth1114) [DOI] [PubMed] [Google Scholar]
- 4.Amano T, Sagai T, Tanabe H, Mizushina Y, Nakazawa H, Shiroishi T. 2009. Chromosomal dynamics at the Shh locus: limb bud-specific differential regulation of competence and active transcription. Dev. Cell 16, 47–57 10.1016/j.devcel.2008.11.011 (doi:10.1016/j.devcel.2008.11.011) [DOI] [PubMed] [Google Scholar]
- 5.Kurukuti S, Tiwari VK, Tavoosidana G, Pugacheva E, Murrell A, Zhao Z, Lobanenkov V, Reik W, Ohisson R. 2006. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc. Natl Acad. Sci. USA 103, 10 684–10 689 10.1073/pnas.0600326103 (doi:10.1073/pnas.0600326103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tiwari VK, McGarvey KM, Licchesi JD, Ohm JE, Herman JG, Schubeler D, Baylin SB. 2008. PcG proteins, DNA methylation, and gene repression by chromatin looping. PLoS Biol. 6, 2911–2927 10.1371/journal.pbio.0060306 (doi:10.1371/journal.pbio.0060306) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ling JQ, Li T, Hu JF, Vu TH, Chen HL, Qiu XW, Cherry AM, Hoffman AR. 2006. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science 312, 269–272 10.1126/science.1123191 (doi:10.1126/science.1123191) [DOI] [PubMed] [Google Scholar]
- 8.Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, van Steensel B, de Laat W. 2006. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nat. Genet. 38, 1348–1354 10.1038/ng1896 (doi:10.1038/ng1896) [DOI] [PubMed] [Google Scholar]
- 9.Zhao Z, et al. 2006. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat. Genet. 38, 1341–1347 10.1038/ng1891 (doi:10.1038/ng1891) [DOI] [PubMed] [Google Scholar]
- 10.Dostie J, et al. 2006. Chromosome conformation capture carbon copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 16, 1299–1309 10.1101/gr.5571506 (doi:10.1101/gr.5571506) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R. 2006. Interchromosomal interactions and olfactory receptor choice. Cell 126, 403–413 10.1016/j.cell.2006.06.035 (doi:10.1016/j.cell.2006.06.035) [DOI] [PubMed] [Google Scholar]
- 12.Wurtele H, Chartrand P. 2006. Genome-wide scanning of HoxB1-associated loci in mouse ES cells using an open-ended chromosome conformation capture methodology. Chromosome Res. 14, 477–495 10.1007/s10577-006-1075-0 (doi:10.1007/s10577-006-1075-0) [DOI] [PubMed] [Google Scholar]
- 13.Guo C, Gerasimova T, Hao H, Ivanova I, Chakraborty T, Selimyan R, Oltz EM, Sen R. 2011. Two forms of loops generate the chromatin conformation of the immunoglobulin heavy-chain gene locus. Cell 147, 332–343 10.1016/j.cell.2011.08.049 (doi:10.1016/j.cell.2011.08.049) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Montavon T, Soshnikova N, Mascrez B, Joye E, Thevenet L, Splinter E, de Laat W, Spitz F, Duboule D. 2011. A regulatory archipelago controls Hox genes transcription in digits. Cell 147, 1132–1145 10.1016/j.cell.2011.10.023 (doi:10.1016/j.cell.2011.10.023) [DOI] [PubMed] [Google Scholar]
- 15.Noordermeer D, Leleu M, Splinter E, Rougemont J, De Laat W, Duboule D. 2011. The dynamic architecture of Hox gene clusters. Science 334, 222–225 10.1126/science.1207194 (doi:10.1126/science.1207194) [DOI] [PubMed] [Google Scholar]
- 16.Higgs DR, Wood WG. 2008. Long-range regulation of α-globin gene expression during erythropoiesis. Curr. Opin. Hematol. 15, 176–83 10.1097/MOH.0b013e3282f734c4 (doi:10.1097/MOH.0b013e3282f734c4) [DOI] [PubMed] [Google Scholar]
- 17.Wallace HA, Marques-Kranc F, Richardson M, Luna-Crespo F, Sharpe JA, Hughes J, Wood WG, Higgs DR, Smith AJH. 2007. Manipulating the mouse genome to engineer precise functional syntenic replacements with human sequence. Cell 128, 197–209 10.1016/j.cell.2006.11.044 (doi:10.1016/j.cell.2006.11.044) [DOI] [PubMed] [Google Scholar]
- 18.Vernimmen D, De Gobbi M, Sloane-Stanley JA, Wood WG, Higgs DR. 2007. Long-range chromosomal interactions regulate the timing of the transition between poised and active gene expression. EMBO J. 26, 2041–2051 10.1038/sj.emboj.7601654 (doi:10.1038/sj.emboj.7601654) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vernimmen D, Marques-Kranc F, Sharpe JA, Sloane-Stanley JA, Wood WG, Wallace HAC, Smith AJH, Higgs DR. 2009. Chromosome looping at the human α-globin locus is mediated via the major upstream regulatory element (HS -40). Blood 114, 4253–4260 10.1182/blood-2009-03-213439 (doi:10.1182/blood-2009-03-213439) [DOI] [PubMed] [Google Scholar]
- 20.Gondor A, Rougier C, Ohlsson R. 2008. High-resolution circular chromosome conformation capture assay. Nat. Protoc. 3, 303–313 10.1038/nprot.2007.540 (doi:10.1038/nprot.2007.540) [DOI] [PubMed] [Google Scholar]
- 21.Schmiedeberg L, Skene P, Deaton A, Bird A. 2009. A temporal threshold for formaldehyde crosslinking and fixation. PLoS ONE 4, e4636. 10.1371/journal.pone.0004636 (doi:10.1371/journal.pone.0004636) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Solomon MJ, Varshavsky A. 1985. Formaldehyde-mediated DNA-protein crosslinking: a probe for in vivo chromatin structures. Proc. Natl Acad. Sci. USA 82, 6470–6474 10.1073/pnas.82.19.6470 (doi:10.1073/pnas.82.19.6470) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Gobbi M, et al. 2007. Tissue-specific histone modification and transcription factor binding in α-globin gene expression. Blood 110, 4503–4510 10.1182/blood-2007-06-097964 (doi:10.1182/blood-2007-06-097964) [DOI] [PubMed] [Google Scholar]
- 24.Splinter E, Heath H, Kooren J, Palstra RJ, Klous P, Grosveld F, Galjart N, Laat WD. 2006. CTCF mediates long-range chromatin looping and local histone modification in the β-globin locus. Genes Dev. 20, 2349–2354 10.1101/gad.399506 (doi:10.1101/gad.399506) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goh SH, Lee YT, Bhanu NV, Cam MC, Desper R, Martin BM, Moharram R, Gherman RB, Miller JL. 2005. A newly discovered human α-globin gene. Blood 106, 1466–1472 10.1182/blood-2005-03-0948 (doi:10.1182/blood-2005-03-0948) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chepelev I, Wei G, Wangsa D, Tang Q, Zhao K. 2012. Characterization of genome-wide enhancer–promoter interactions reveals co-expression of interacting genes and modes of higher order chromatin organization. Cell Res. 22, 490–503 10.1038/cr.2012.15 (doi:10.1038/cr.2012.15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li G, et al. 2012. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell 148, 84–98 10.1016/j.cell.2011.12.014 (doi:10.1016/j.cell.2011.12.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanyal A, Lajoie BR, Jain G, Dekker J. 2012. The long-range interaction landscape of gene promoters. Nature 489, 109–113 10.1038/nature11279 (doi:10.1038/nature11279) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. 2002. Looping and interaction between hypersensitive sites in the active β-globin locus. Mol. Cell 10, 1453–1465 10.1016/S1097-2765(02)00781-5 (doi:10.1016/S1097-2765(02)00781-5) [DOI] [PubMed] [Google Scholar]
- 30.Lower KM, et al. 2009. Adventitious changes in long-range gene expression caused by polymorphic structural variation and promoter competition. Proc. Natl Acad. Sci. USA 106, 21 771–21 776 10.1073/pnas.0909331106 (doi:10.1073/pnas.0909331106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cookson W, Liang L, Abecasis G, Moffatt M, Lathrop M. 2009. Mapping complex disease traits with global gene expression. Nat. Rev. Genet. 10, 184–194 10.1038/nrg2537 (doi:10.1038/nrg2537) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nativio R, et al. 2009. Cohesin is required for higher-order chromatin conformation at the imprinted IGF2-H19 locus. PLoS Genet. 5, e1000739. 10.1371/journal.pgen.1000739 (doi:10.1371/journal.pgen.1000739) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ribeiro de Almeida C, Stadhouders R, Thongjuea S, Soler E, Hendriks RW. 2012. DNA-binding factor CTCF and long-range gene interactions in V(D)J recombination and oncogene activation. Blood 119, 6209–6218 10.1182/blood-2012-03-402586 (doi:10.1182/blood-2012-03-402586) [DOI] [PubMed] [Google Scholar]
- 34.Chien R, et al. 2011. Cohesin mediates chromatin interactions that regulate mammalian β-globin expression. J. Biol. Chem. 286, 17 870–17 878 10.1074/jbc.M10.207365 (doi:10.1074/jbc.M10.207365) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Merkenschlager M. 2010. Cohesin: a global player in chromosome biology with local ties to gene regulation. Curr. Opin. Genet. Dev. 20, 555–561 10.1016/j.gde.2010.05.007 (doi:10.1016/j.gde.2010.05.007) [DOI] [PubMed] [Google Scholar]
- 36.Hou C, Dale R, Dean A. 2010. Cell type specificity of chromatin organization mediated by CTCF and cohesin. Proc. Natl Acad. Sci. USA 107, 3651–3656 10.1073/pnas.0912087107 (doi:10.1073/pnas.0912087107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schoenfelder S, et al. 2010. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat. Genet. 42, 53–61 10.1038/ng.496 (doi:10.1038/ng.496) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Laat W. 2007. Long-range DNA contacts: romance in the nucleus? Curr. Opin. Cell Biol. 19, 317–320 10.1016/j.ceb.2007.04.004 (doi:10.1016/j.ceb.2007.04.004) [DOI] [PubMed] [Google Scholar]
- 39.Lieberman-Aiden E, et al. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 10.1126/science.1181369 (doi:10.1126/science.1181369) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brown JM, Leach J, Reittie JE, Atzberger A, Lee-Prudhoe J, Wood WG, Higgs DR, Iboraa FJ, Buckle VJ. 2006. Coregulated human globin genes are frequently in spatial proximity when active. J. Cell Biol. 172, 177–187 10.1083/jcb.200507073 (doi:10.1083/jcb.200507073) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pope SH, Fibach E, Sun J, Chin K, Rodgers GP. 2000. Two-phase liquid culture system models normal human adult erythropoiesis at the molecular level. Eur. J. Haematol. 64, 292–303 10.1034/j.1600-0609.2000.90032.x (doi:10.1034/j.1600-0609.2000.90032.x) [DOI] [PubMed] [Google Scholar]
- 42.Li H, Ruan J, Durbin R. 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18, 1851–1858 10.1101/gr.078212.108 (doi:10.1101/gr.078212.108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Slater GS, Birney E. 2005. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics 6, 31. 10.1186/1471-2105-6-31 (doi:10.1186/1471-2105-6-31) [DOI] [PMC free article] [PubMed] [Google Scholar]