Abstract
CTCF has it all. The transcription factor binds to tens of thousands of genomic sites, some tissue-specific, others ultra-conserved. It can act as a transcriptional activator, repressor and insulator, and it can pause transcription. CTCF binds at chromatin domain boundaries, at enhancers and gene promoters, and inside gene bodies. It can attract many other transcription factors to chromatin, including tissue-specific transcriptional activators, repressors, cohesin and RNA polymerase II, and it forms chromatin loops. Yet, or perhaps therefore, CTCF's exact function at a given genomic site is unpredictable. It appears to be determined by the associated transcription factors, by the location of the binding site relative to the transcriptional start site of a gene, and by the site's engagement in chromatin loops with other CTCF-binding sites, enhancers or gene promoters. Here, we will discuss genome-wide features of CTCF binding events, as well as locus-specific functions of this remarkable transcription factor.
Keywords: CTCF, cohesin, transcription, chromatin loops, nuclear organization
1. Introduction
CTCF is a ubiquitously expressed and an essential protein [1], and is, in many ways, an exceptional transcription factor. It was first described as a transcriptional repressor [2], but was also found to act as a transcriptional activator [3,4]. Most strikingly, it harbours insulator activity: when positioned in between an enhancer and gene promoter, it can block their communication and prevent transcriptional activation [5–7]. Systematic chromatin immunoprecipitation experiments combined with high-throughput sequencing (ChIP-seq) have been performed to map CTCF binding events across the genome in many tissues of different species [8–10]. They show that the genome is covered with a myriad of CTCF binding sites. More than most other transcription factors CTCF appears to bind to intergenic sequences, often at a distance from the transcriptional start site (TSS) [11]. CTCF was one of the first proteins demonstrated to mediate chromatin looping between its binding sites [12,13]. Further evidence for its role in the organization of genome structure comes from observations that it frequently binds to boundaries between chromosomal regions that occupy distinct locations in the nucleus, to boundaries between regions with different epigenetic signatures and/or different transcriptional activities, and to boundaries between recently identified topological domains, which are spatially defined chromosomal units within which sequences preferentially interact with each other [14,15]. Here, we will discuss studies on CTCF and evaluate its function in genome folding and gene expression.
2. CTCF at the β-globin and the H19–Igf2 locus: a short history
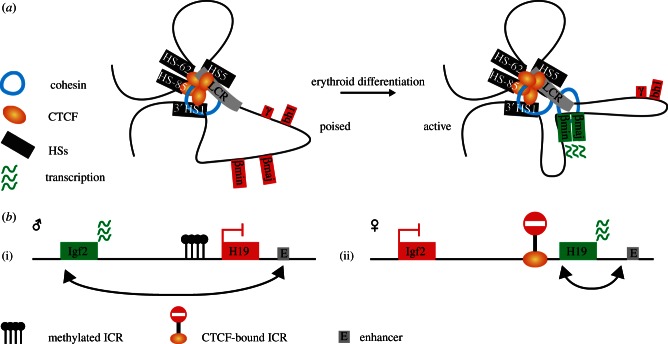
Functions of the versatile DNA-binding protein CTCF were initially explored at individual loci, in particular at the β-globin locus and the imprinted H19–Igf2 locus. The chicken β-globin locus carries a DNaseI hypersensitive site (5′HS4) at its 5′ side that separates the locus from neighbouring heterochromatin and this site was found capable of blocking enhancer activity [16]. CTCF was subsequently demonstrated to be responsible for this insulator activity of 5′HS4 [5]. The human and mouse β-globin loci are also located inside large chromosomal regions of inactive chromatin and are similarly flanked by CTCF-binding sites [17,18]. These were suspected to form a barrier for incoming heterochromatin, but their deletion did not lead to closing or inactivation of the β-globin locus [12,19]. The application of chromosome conformation capture (3C) technology enabled the demonstration that the β-globin CTCF sites physically interact with each other. They form large chromatin loops encompassing the β-globin main regulatory element, the locus control region (LCR), and its genes. These loops are erythroid-specific and are formed in erythroid progenitor cells, prior to LCR-mediated high expression of the β-globin genes (figure 1a; [12,20]). It was speculated that the CTCF loops can facilitate subsequent spatial interactions between the LCR and its target genes, but evidence for this is still lacking.
Figure 1.
CTCF, chromatin loops and transcription regulation at selected gene loci. (a) Genes at the β-globin locus are under control of the locus control region (LCR). CTCF-binding sites interact to create a chromatin hub with a loop encompassing the LCR and the β-globin genes. Upon erythroid differentiation, erythroid-specific transcription factors and cohesin enable the formation of an active chromatin hub in which the LCR contacts the genes and enhances their expression. (b) Imprinted expression of the H19 and Igf2 genes is mediated by methylation-dependent binding of CTCF at the imprinted control region (ICR). (i) On the paternal allele, methylation of the ICR prevents CTCF binding and allows expression of the Igf2 gene mediated by contacts between the distal enhancer (E) and the Igf2 promoter. (ii) CTCF binding at the ICR blocks communication between the Igf2 gene and the distal enhancer resulting in expression of the H19 gene from the maternal allele.
Another locus historically important for CTCF's reputation as an interesting transcription factor is the imprinted H19/Igf2 locus. The locus contains a differentially methylated region that is known as the imprinting control region (ICR), located in between the H19 and the Igf2 genes. The ICR determines that H19 is active on the maternal allele and that Igf2 is transcribed from the paternal allele [21,22]. CTCF entered the stage here when it was found to bind to the ICR in a methylation-dependent manner: the binding of CTCF to the unmethylated maternal ICR prevents shared enhancers near the H19 gene from reaching across and activating Igf2. On the paternal allele, CTCF cannot exert its insulator activity as DNA methylation prevents its binding to the ICR (figure 1b; [6,23]). Again, chromatin loops are formed and seem important for ICR functioning [24–26]. Allele-specific chromatin loops with both enhancers and promoters are formed by the maternal, CTCF-bound ICR, suggesting that such contacts may underlie CTCF-mediated insulator activity [26]. Collectively, the early studies on CTCF functioning at the β-globin and the H19–Igf2 locus revealed that the protein can interfere with promoter–enhancer communication. They also showed that CTCF can form chromatin loops between its binding sites, and perhaps also with other regulatory sequences.
3. CTCF binds across the genome to chromatin boundaries, enhancers and gene promoters
The systematic mapping of genome-wide binding sites by ChIP revealed that CTCF binds to tens of thousands of genomic sites [10,11,27]. Association to roughly one-third of these sites is relatively conserved across different cell types [9]. An inter-species comparison between CTCF binding profiles in the liver of five mammalian organisms uncovered approximately 5000 sites that are ultra-conserved between the species and tissues. These appear to be the high-affinity binding sites, suggesting that differences in affinity could be related to the strength of conservation [8]. The activation of retro-elements has produced species-specific expansions of CTCF-binding sites, and this form of genome evolution is still highly active in mammals [8]. Classification of CTCF binding sites based on a consensus motif score lead to similar conclusions: high occupancy sites appear to be conserved across cell types, whereas low occupancy sites are more tissue restricted [28].
The CTCF consensus binding sequence contains CpG and can, therefore, be subject to DNA methylation. CTCF is able to bind to methylated DNA sequences in vitro [29], but preferentially binds to unmethylated sequences, as seen also at the H19–Igf2 locus. In fact, DNA methylation appears to play a role in some of the tissue-specific binding events of CTCF [9]. Moreover, CTCF can influence DNA methylation by forming a complex with two enzymes related to DNA methylation: poly(ADP-ribose) polymerase 1 (PARP1) and the ubiquitously expressed DNA (cytosine-5)-methyltransferase 1 (DNMT1). CTCF activates PARP1, which then can add ADP–ribose groups to DNMT1 to inactivate this enzyme, with maintenance of methyl-free CpGs as the result [30–32].
A portion of CTCF binding sites is found enriched at transitions between active chromatin (high in H2K5Ac) and inactive chromatin domains (high in H3K27me3) [27,33]. This seems particularly true for retrotransposed CTCF binding sites [8]. CTCF sites frequently flank the so-called lamina-associated domains (LADs). LADs are chromosomal regions associated with the lamin-based protein network that coats the inner side of the nuclear envelope; these chromosomal regions tend to be transcriptionally inactive [14]. Its presence at LAD boundaries suggests that CTCF helps to organize the three-dimensional structure of chromatin. In Drosophila, the knockdown of CTCF leads to decreased levels of H3K27me3 inside inactive domains, indicating that CTCF binding at boundaries is required for the maintenance of repression [34]. Association of CTCF follows the resetting of active and inactive domains during cellular differentiation, further suggesting that it functions to separate different chromatin states [33]. Some of the LADs also dynamically change during cellular differentiation [35], but whether CTCF binds to the borders of these differential LADs is currently unclear.
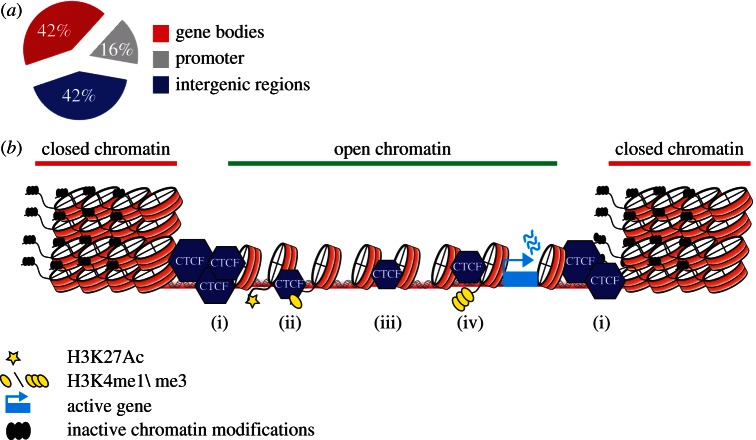
Although CTCF binding is often found distal to TSSs, it does show a strong correlation with gene density (figure 2a,b) [11]. Indeed, evidence for a direct role of CTCF in transcription regulation came from early studies on individual genes [3,37]. Genome wide, a portion of CTCF sites co-localize with the promoter-specific H3K4me3 mark and another part coincides with the enhancer mark H3K4me1 [27]. CTCF binding events at promoters tend to be conserved across tissues, whereas CTCF binding to enhancers is more tissue restricted [10].
Figure 2.
A versatile role for CTCF in chromatin biology. (a) Functional categories of CTCF binding sites across the genome, adopted from Chen et al. [36]. (b) (i) CTCF binding sites are found at boundaries that separate active and inactive domains. CTCF binding to (ii) enhancer-like sequences and (iv) gene promoters can facilitate looping between these sequences. (iii) CTCF binding in between enhancers and gene promoters can block the interaction between an enhancer and its target promoter.
4. CTCF and cohesin share DNA binding sites
An unanticipated observation was the co-localization of cohesin with many of the chromosomal binding sites of CTCF [38–42]. Cohesin has always been associated with DNA replication and sister chromatid cohesion during the S, G2 and M phase of the cell cycle [43]. It is a protein complex that contains members of a family of ‘structural maintenance of chromosomes’ proteins. The complex forms a ring-like protein structure that is thought to embrace two DNA helices. Surprisingly at the time, cohesin was also found to bind chromatin in post-mitotic cells, with half of its binding sites overlapping with CTCF sites [38–40]. Cohesin association to these sites is dependent on the presence of CTCF: without CTCF, cohesin still binds to chromatin but is no longer found at specific sequences. In contrast, CTCF does not rely on cohesin for finding its DNA binding sites. One possibility is that bound CTCF serves as a roadblock or barrier to position a sliding cohesin molecule on the chromatin template [38–42].
Its cell-cycle independent association to DNA suggests that cohesin has an additional role in gene regulation. Given its capacity to hold together two sister chromatids, cohesin is obviously also attractive as a looping factor. Indeed, at the H19–Igf2 locus, cohesin was shown to be important for CTCF-mediated chromatin loop formation and proper regulation of Igf2 transcription [44]. Similarly, at the interferon gamma (IFNG) locus, depletion of cohesin was found to disrupt chromatin loops between regulatory DNA sequences and cause a reduction in IFNG expression [45]. Also at the β-globin locus cohesin has been implicated in chromatin looping, not only between the flanking CTCF sites but also between the LCR enhancer region and the downstream β-globin target genes [46]. Conditional deletion of cohesin in thymocytes was shown to disrupt the formation of regulatory chromatin loops in the T-cell receptor-α locus, with reduced transcription and impaired V(DJ) rearrangement as a consequence [47]. Pairwise comparison between two cell types revealed that it is mostly the CTCF-independent cohesin-binding events that show cell-type specificity. At these sites, cohesin is often found co-localized with mediator and RNA polymerase II (RNAPII), indicating a CTCF-independent function at enhancer sequences. Consistent with this, these genomic sites were often found to be close to actively transcribed genes [48], and to be co-occupied by tissue-specific transcription factors [49,50]. Collectively, this shows that CTCF and cohesin have shared and independent functions at regulatory sequences in the genome. Cohesin can form chromatin loops during interphase. However, whether this occurs through its embracement of two DNA double helices still awaits formal proof.
5. CTCF and other binding partners
CTCF performs multiple roles, and in agreement the protein shares chromatin binding sites with many other factors [51–53]. Co-association events such as those with the histone deacetylase SIN3 [54], the thyroid hormone receptor [55], nucleophosmin [56], Kaiso [57] and the DEAD-box RNA helicase p68 with associated non-coding RNA [58] have been implicated in its insulator function. Interestingly, the p68 RNA–protein complex appears required for positioning cohesin at the CTCF sites of the H19–Igf2 ICR [58]. In addition, CTCF co-occupies sites with the transcription factors FOXA1 and the oestrogen receptor (ER). These sites tend to locate near ER-responsive genes, suggesting that CTCF facilitates their transcriptional activation [59]. Furthermore, CTCF recruits the basal transcription factor TAF3 to intergenic sites in embryonic stem cells (ESCs), where TAF3-dependent chromatin loop formation was shown to activate gene transcription [60]. In a study that monitored RNAPII tracking along long tumour necrosis factor-alpha responsive genes, pausing of RNAPII was observed at CTCF- and cohesin-bound sites [61]. This pausing can serve to incorporate weak exons and, therefore, facilitate alternative splicing [62]. Thus, intra- and intergenic CTCF sites can have many different roles.
6. CTCF function at individual gene loci
Given its diverse activities, it seems necessary to zoom in on individual loci to understand CTCF's local function. At the proto-oncogene Myb locus, CTCF binding occurs in the first intron of the gene, where it inhibits RNAPII elongation. Transcriptional pausing by CTCF can be overcome by upstream enhancers that bind tissue-specific transcriptional activators and loop towards the Myb promoter [63]. At the major histocompatibility complex class II (MHCII) locus, CTCF and cohesin binding to, and looping between, upstream sequences precedes transcriptional activation. Upon binding of the MHCII trans-activator CIITA to the promoter sequences, loops are induced between them and the various CTCF sites, resulting in increased expression of MHCII genes [64,65].
CTCF also binds to many sites across the immunoglobulin and T-cell receptor antigen receptor gene loci. In conditional CTCF knockout mice, V gene usage in the Igκ light chain locus was found to be altered, with increased recombination with proximal and reduced recombination with distal V segments. This was accompanied by corresponding changes in germline transcription at these locations, suggesting that CTCF, such as cohesin [47], mediates gene usage of the antigen receptor loci via the local regulation of germline transcription [66]. In this model, germline transcription increases accessibility of the region, which facilitates their selection for recombination [67]. CTCF depletion does not always result in aberrant gene expression. Using the same conditional knockout mice [1] Hoxd gene expression in the developing limb bud was unaltered after knockout of CTCF [68]. Hoxd gene expression in the limb bud is under the control of many distant regulatory sequences that physically loop towards the genes [69]. Unaltered Hoxd expression in the absence of CTCF suggests that these enhancer–promoter loops are not influenced by CTCF binding to sites in and around the locus. This raises the question whether CTCF has any impact on the three-dimensional topology of the locus. While Hoxd expression was not affected, CTCF depletion did cause massive cell death in the limb, showing that CTCF is critical for the transcriptional regulation of other genes involved in cellular homeostasis [68].
A final locus that is interesting to discuss is the protocadherin-α cluster. This cluster encodes neuronal-specific transmembrane proteins that are mono-allelically expressed and thought to be involved in the recognition and diversification of neurons. Expression of the cluster is under control of a downstream enhancer that influences the expression of the 12 isoforms, each of which is having its own alternative promoter [70]. Interestingly, the enhancer as well as each individual promoter has a binding site for CTCF. Expression of the isoforms is reduced upon conditional CTCF knockout in post-mitotic neurons. This suggests that long-range interactions are part of the regulatory process that controls transcription of these genes [70–72].
Collectively, these studies support the idea that chromatin-bound CTCF can attract many different transcription factors in a tissue- and genomic context-specific manner. Its exact function at a given genomic site is probably determined by these associated transcription factors, by the location of this site relative to the TSS of a gene, and by its engagement in chromatin loops with other CTCF-binding sites, enhancers or gene promoters.
7. Genome-wide chromatin loops mediated by CTCF
A computational intersection of the genomic-binding sites of CTCF (assessed by genome-wide ChIP) with a genome-wide DNA contact map generated by Hi-C [73] suggested that CTCF is involved in chromatin interactions between and within chromosomes across the genome [74]. Chromatin interaction analysis with paired-end tag sequencing (ChIA-PET) combines ChIP with a 3C approach and was developed to study genome-wide DNA interactions mediated by a protein of interest [75]. When targeted to CTCF, ChIA-PET uncovered roughly 1500 intra-chromosomal and around 300 inter-chromosomal interactions mediated by this protein [13]. Subsequent clustering of the regions (10–200 kb) encompassed by the intra-chromosomal loops was done based on the distribution of histone marks. This showed that CTCF loops can contain active chromatin separated from inactive chromatin outside the loops and vice versa. CTCF can also capture enhancers and promoters together in a chromatin loop [13]. Only a fraction of the roughly approximately 40 000 CTCF binding sites was found to participate in the roughly 1500 CTCF-mediated loops. This implies that either not all interactions mediated by CTCF have been identified or that most CTCF sites are not engaged in the formation of loops.
The latter may well be true, because 5C (chromosome conformation capture carbon copy) technology showed that most CTCF sites across 1 per cent of the genome do not participate in chromatin loops, no matter whether they are co-occupied by cohesin or not. CTCF-bound sequences were often skipped by gene promoters making contacts with enhancers or with other CTCF sites even further away [76].
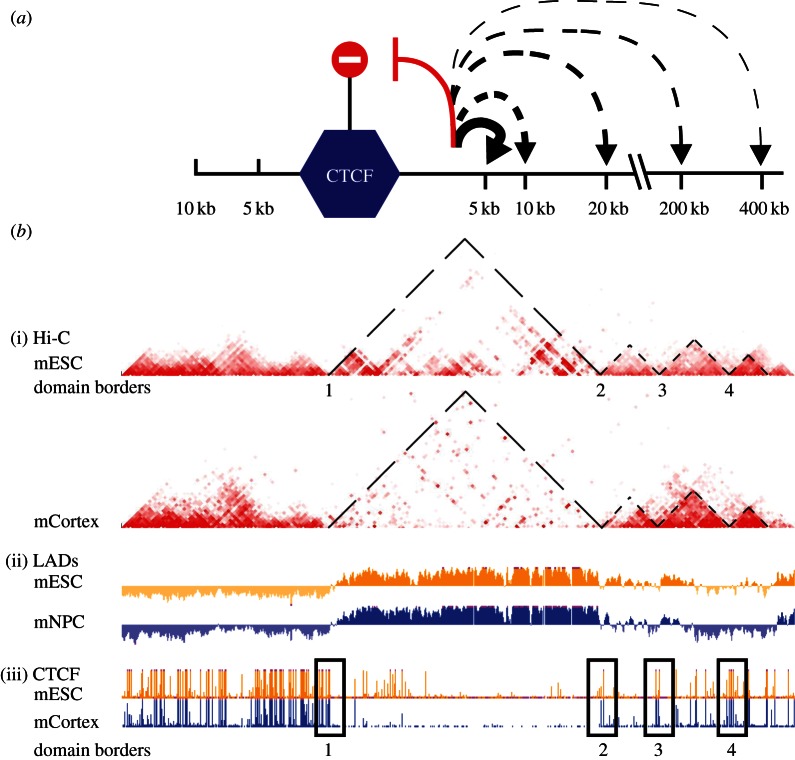
The recent availability of large genome-wide DNA interaction datasets [15,73] facilitates the assessment of CTCF's impact on chromosome topology. Sequences close on the chromosome to CTCF binding sites were shown to be biased in their DNA contacts: they interacted with other sequences on the same side of the CTCF site more than with sequences across this site (figure 3a; [77]). The same was previously shown for a different insulator protein in Drosophila: its binding to a site prevented flanking sequences to physically contact each other across this site [78]. Interestingly, this may provide an explanation for how insulators function: they can prevent spatial DNA contacts across the insulating sequence. In a particularly detailed genome-wide DNA contact study topological domains were defined; they are chromosomal regions of on average 1 Mb in size, within which sequences preferentially interact with each other [15,79]. A strong conservation of topological domains was seen between tissues and even between species, suggesting that these domains do not contribute themselves to the specific identity of cells. Interestingly, CTCF binding sites were enriched in 20 kb windows surrounding the boundaries of these domains (figure 3b), re-emphasizing its role as a chromatin organizer. In one case it was shown that disruption of a boundary led to intermingling of topological domains and caused misregulated expression of the genes involved [79]. Unlike the topological domains themselves, contacts within the domains do change during differentiation. Here to CTCF appears to play a role, probably to accomodate developmental changes in gene expression [80,81].
Figure 3.
CTCF acts as an insulator by hampering DNA contacts across its binding sites. (a) CTCF hampers sequences within 10 kb of its binding sites to reach across [77]. Decreased thickness of lines indicates a decrease in interaction probability. (b) CTCF binding sites are often found at the borders of topological domains. (i) Hi-C data in ESCs and in cortex, with colour-coded contact frequencies between sequences on mouse chromosome 12. Triangles reveal and highlight topological domains [15], being chromosomal regions within which sequences preferentially interact with each other. (ii) Lamina-associated domain (LAD) data [14,35], showing that LAD boundaries coincide with the borders of topological domains. (iii) CTCF ChIP-seq profiles, showing clusters of CTCF binding sites at the borders [10]. Note that such CTCF clusters also exist elsewhere, particularly in non-LADs. Region shown: chr12: 112.3–119.3 Mb (mm9).
8. Concluding remarks
Despite being the subject of intense research, CTCF manages to remain a mysterious transcription factor. It binds to many thousands of sites across the genome, where it can interact with a plethora of other transcription factors. It is often found engaged in chromatin loops, sometimes with and sometimes without the involvement of cohesin. It can form chromatin loops with other CTCF binding sites, but also with enhancer and promoter sequences. CTCF binds to sequences outside and away from genes, but also inside the gene body, where it appears capable of pausing the sliding polymerase molecule. Finally, CTCF binding sites still actively jump around as retrotransposable sequences, giving diversity to the CTCF binding landscape between different mammalian species.
We believe that the unifying theme that may explain the many, and sometimes opposing, functional consequences of CTCF association to chromatin is probably its ability to form chromatin loops. Depending on the sequences encompassed in the loops and those excluded from the loops, chromatin shaped by CTCF may facilitate or hamper three dimensional contacts between enhancers and target genes, with different outcomes for transcription. Many questions still remain though: why do some CTCF sites form a chromatin loop and others not? To what extent does this rely on co-associated protein factors? How does the protein manage to interact with so many other transcription factors when bound to chromatin? One possibility is that CTCF serves as a roadblock for chromatin-scanning transcription factors that somehow get trapped when encountering the bound protein. What is the relevance of CTCF-mediated interchromosomal contacts? Does CTCF block enhancer–promoter communication by preventing 3D DNA contacts? Or does insulation involve the physical interaction of the insulator sequence with both enhancers and promoters? Answers to these questions are needed to enable predicting whether a given CTCF binding event will be functionally irrelevant, will cause transcriptional activation or repression, will interfere with transcriptional activation or will create a chromatin boundary.
Acknowledgements
We would like to thank Patrick Wijchers and Peter Krijger for useful comments. This work was financially supported by grant no. 935170621 from the Dutch Scientific Organization (NWO) and a European Research Council Starting Grant (209700, ‘4C’) to W.d.L.
References
- 1.Heath H, et al. 2008. CTCF regulates cell cycle progression of αβ T cells in the thymus. EMBO J. 27, 2839–2850 10.1038/emboj.2008.214 (doi:10.1038/emboj.2008.214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lobanenkov VV, Nicolas RH, Adler VV, Paterson H, Klenova EM, Polotskaja AV, Goodwin GH. 1990. A novel sequence-specific DNA binding protein which interacts with three regularly spaced direct repeats of the CCCTC-motif in the 5′-flanking sequence of the chicken c-myc gene. Oncogene 5, 1743–1753 [PubMed] [Google Scholar]
- 3.Klenova EM, Nicolas RH, Paterson HF, Carne AF, Heath CM, Goodwin GH, Neiman PE, Lobanenkov VV. 1993. CTCF, a conserved nuclear factor required for optimal transcriptional activity of the chicken c-myc gene, is an 11-Zn-finger protein differentially expressed in multiple forms. Mol. Cell. Biol. 13, 7612–7624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Filippova GN, Fagerlie S, Klenova EM, Myers C, Dehner Y, Goodwin G, Neiman PE, Collins SJ, Lobanenkov VV. 1996. An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol. Cell. Biol. 16, 2802–2813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bell AC, West AG, Felsenfeld G. 1999. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98, 387–396 10.1016/S0092-8674(00)81967-4 (doi:10.1016/S0092-8674(00)81967-4) [DOI] [PubMed] [Google Scholar]
- 6.Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM. 2000. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405, 486–489 10.1038/35013106 (doi:10.1038/35013106) [DOI] [PubMed] [Google Scholar]
- 7.Recillas-Targa F, Pikaart MJ, Burgess-Beusse B, Bell AC, Litt MD, West AG, Felsenfeld G. 2002. Position-effect protection and enhancer blocking by the chicken β-globin insulator are separable activities. Proc. Natl Acad. Sci. USA 99, 6883–6888 10.1073/pnas.102179399 (doi:10.1073/pnas.102179399) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmidt D, et al. 2012. Waves of retrotransposon expansion remodel genome organization and CTCF binding in multiple mammalian lineages. Cell 148, 335–348 10.1016/j.cell.2011.11.058 (doi:10.1016/j.cell.2011.11.058) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang H, et al. 2012. Widespread plasticity in CTCF occupancy linked to DNA methylation. Genome Res. 22, 1680–1688 10.1101/gr.136101.111 (doi:10.1101/gr.136101.111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen Y, et al. 2012. A map of the cis-regulatory sequences in the mouse genome. Nature 488, 116–120 10.1038/nature11243 (doi:10.1038/nature11243) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, Zhang MQ, Lobanenkov VV, Ren B. 2007. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell 128, 1231–1245 10.1016/j.cell.2006.12.048 (doi:10.1016/j.cell.2006.12.048) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Splinter E, Heath H, Kooren J, Palstra RJ, Klous P, Grosveld F, Galjart N, Laat de W. 2006. CTCF mediates long-range chromatin looping and local histone modification in the β-globin locus. Genes Dev. 20, 2349–2354 10.1101/gad.399506 (doi:10.1101/gad.399506) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Handoko L, et al. 2011. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat. Genet. 43, 630–638 10.1038/ng.857 (doi:10.1038/ng.857) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guelen L, et al. 2008. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453, 948–951 10.1038/nature06947 (doi:10.1038/nature06947) [DOI] [PubMed] [Google Scholar]
- 15.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 10.1038/nature11082 (doi:10.1038/nature11082) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chung JH, Whiteley M, Felsenfeld G. 1993. A 5′ element of the chicken β-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell 74, 505–514 10.1016/0092-8674(93)80052-G (doi:10.1016/0092-8674(93)80052-G) [DOI] [PubMed] [Google Scholar]
- 17.Farrell CM, West AG, Felsenfeld G. 2002. Conserved CTCF insulator elements flank the mouse and human beta-globin loci. Mol. Cell. Biol. 22, 3820–3831 10.1128/MCB.22.11.3820-3831.2002 (doi:10.1128/MCB.22.11.3820-3831.2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bulger M, Schubeler D, Bender MA, Hamilton J, Farrell CM, Hardison RC, Groudine M. 2003. A complex chromatin landscape revealed by patterns of nuclease sensitivity and histone modification within the mouse beta-globin locus. Mol. Cell. Biol. 23, 5234–5244 10.1128/MCB.23.15.5234-5244.2003 (doi:10.1128/MCB.23.15.5234-5244.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bender MA, Byron R, Ragoczy T, Telling A, Bulger M, Groudine M. 2006. Flanking HS-62.5 and 3′ HS1, and regions upstream of the LCR, are not required for β-globin transcription. Blood 108, 1395–1401 10.1182/blood-2006-04-014431 (doi:10.1182/blood-2006-04-014431) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palstra RJ, Tolhuis B, Splinter E, Nijmeijer R, Grosveld F, de Laat W. 2003. The β-globin nuclear compartment in development and erythroid differentiation. Nat. Genet. 35, 190–194 10.1038/ng1244 (doi:10.1038/ng1244) [DOI] [PubMed] [Google Scholar]
- 21.Bartolomei MS, Webber AL, Brunkow ME, Tilghman SM. 1993. Epigenetic mechanisms underlying the imprinting of the mouse H19 gene. Genes Dev. 7, 1663–1673 10.1101/gad.7.9.1663 (doi:10.1101/gad.7.9.1663) [DOI] [PubMed] [Google Scholar]
- 22.Ferguson-Smith AC, Sasaki H, Cattanach BM, Surani MA. 1993. Parental-origin-specific epigenetic modification of the mouse H19 gene. Nature 362, 751–755 10.1038/362751a0 (doi:10.1038/362751a0) [DOI] [PubMed] [Google Scholar]
- 23.Bell AC, Felsenfeld G. 2000. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405, 482–485 10.1038/35013100 (doi:10.1038/35013100) [DOI] [PubMed] [Google Scholar]
- 24.Murrell A, Heeson S, Reik W. 2004. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat. Genet. 36, 889–893 10.1038/ng1402 (doi:10.1038/ng1402) [DOI] [PubMed] [Google Scholar]
- 25.Kurukuti S, Tiwari VK, Tavoosidana G, Pugacheva E, Murrell A, Zhao Z, Lobanenkov V, Reik W, Ohisson R. 2006. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc. Natl Acad. Sci. USA 103, 10 684–10 689 10.1073/pnas.0600326103 (doi:10.1073/pnas.0600326103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoon YS, Jeong S, Rong Q, Park KY, Chung JH, Pfeifer K. 2007. Analysis of the H19ICR insulator. Mol. Cell. Biol. 27, 3499–3510 10.1128/MCB.02170-06 (doi:10.1128/MCB.02170-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837 10.1016/j.cell.2007.05.009 (doi:10.1016/j.cell.2007.05.009) [DOI] [PubMed] [Google Scholar]
- 28.Essien K, Vigneau S, Apreleva S, Singh LN, Bartolomei MS, Hannenhalli S. 2009. CTCF binding site classes exhibit distinct evolutionary, genomic, epigenomic and transcriptomic features. Genome Biol. 10, R131. 10.1186/gb-2009-10-11-r131 (doi:10.1186/gb-2009-10-11-r131) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stadler MB, et al. 2011. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nat. 480, 490–495 [DOI] [PubMed] [Google Scholar]
- 30.Yu W, et al. 2004. Poly(ADP-ribosyl)ation regulates CTCF-dependent chromatin insulation. Nat. Genet. 36, 1105–1110 10.1038/ng1426 (doi:10.1038/ng1426) [DOI] [PubMed] [Google Scholar]
- 31.Guastafierro T, Cecchinelli B, Zampieri M, Reale A, Riggio G, Sthandier O, Zupi G, Calab P. 2008. CCCTC-binding factor activates PARP-1 affecting DNA methylation machinery. J. Biol. Chem. 283, 21 873–21 880 10.1074/jbc.M801170200 (doi:10.1074/jbc.M801170200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zampieri M, Guastafierro T, Calabrese R, Ciccarone F, Bacalini MG, Reale A, Perilli M, Passananti C, Caiafa P. 2012. ADP-ribose polymers localized on Ctcf–Parp1–Dnmt1 complex prevent methylation of Ctcf target sites. Biochem. J. 441, 645–652 10.1042/BJ20111417 (doi:10.1042/BJ20111417) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cuddapah S, Jothi R, Schones DE, Roh TY, Cui K, Zhao K. 2009. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 19, 24–32 10.1101/gr.082800.108 (doi:10.1101/gr.082800.108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Bortle K, Ramos E, Takenaka N, Yang J, Wahi JE, Corces VG. 2012. Drosophila CTCF tandemly aligns with other insulator proteins at the borders of H3K27me3 domains. Genome Res. 22, 2176–2187 10.1101/gr.136788.111 (doi:10.1101/gr.136788.111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peric-Hupkes D, et al. 2010. Molecular maps of the reorganization of genome–nuclear lamina interactions during differentiation. Mol. Cell 38, 603–613 10.1016/j.molcel.2010.03.016 (doi:10.1016/j.molcel.2010.03.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen X, et al. 2008. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133, 1106–1117 10.1016/j.cell.2008.04.043 (doi:10.1016/j.cell.2008.04.043) [DOI] [PubMed] [Google Scholar]
- 37.Vostrov AA, Quitschke WW. 1997. The zinc finger protein CTCF binds to the APBbeta domain of the amyloid beta-protein precursor promoter. Evidence for a role in transcriptional activation. J. Biol. Chem. 272, 33 353–33 359 [DOI] [PubMed] [Google Scholar]
- 38.Wendt KS, et al. 2008. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 451, 796–801 10.1038/nature06634 (doi:10.1038/nature06634) [DOI] [PubMed] [Google Scholar]
- 39.Rubio ED, Reiss DJ, Welcsh PL, Disteche CM, Filippova GN, Baliga NS, Aebersold R, Ranish JA, Krumm A. 2008. CTCF physically links cohesin to chromatin. Proc. Natl Acad. Sci. USA 105, 8309–8314 10.1073/pnas.0801273105 (doi:10.1073/pnas.0801273105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parelho V, et al. 2008. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell 132, 422–433 10.1016/j.cell.2008.01.011 (doi:10.1016/j.cell.2008.01.011) [DOI] [PubMed] [Google Scholar]
- 41.Stedman W, Kang H, Lin S, Kissil JL, Bartolomei MS, Lieberman PM. 2008. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J. 27, 654–666 10.1038/emboj.2008.1 (doi:10.1038/emboj.2008.1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiao T, Wallace J, Felsenfeld G. 2011. Specific sites in the C terminus of CTCF interact with the SA2 subunit of the cohesin complex and are required for cohesin-dependent insulation activity. Mol. Cell. Biol. 31, 2174–2183 10.1128/MCB.05093-11 (doi:10.1128/MCB.05093-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nasmyth K, Haering CH. 2009. Cohesin: its roles and mechanisms. Annu. Rev. Genet. 43, 525–558 10.1146/annurev-genet-102108-134233 (doi:10.1146/annurev-genet-102108-134233) [DOI] [PubMed] [Google Scholar]
- 44.Nativio R, et al. 2009. Cohesin is required for higher-order chromatin conformation at the imprinted IGF2-H19 locus. PLoS Genet. 5, e1000739. 10.1371/journal.pgen.1000739 (doi:10.1371/journal.pgen.1000739) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hadjur S, Williams LM, Ryan NK, Cobb BS, Sexton T, Fraser P, Fisher AG, Merkenschlager M. 2009. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature 460, 410–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chien R, et al. 2011. Cohesin mediates chromatin interactions that regulate mammalian β-globin expression. J. Biol. Chem. 286, 17 870–17 878 10.1074/jbc.M110.207365 (doi:10.1074/jbc.M110.207365) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seitan VC, et al. 2011. A role for cohesin in T-cell-receptor rearrangement and thymocyte differentiation. Nature 476, 467–471 10.1038/nature10312 (doi:10.1038/nature10312) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kagey MH, et al. 2010. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467, 430–435 10.1038/nature09380 (doi:10.1038/nature09380) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmidt D, Schwalie PC, Ross-Innes CS, Hurtado A, Brown GD, Carroll JS, Flicek P, Odam D. 2010. A CTCF-independent role for cohesin in tissue-specific transcription. Genome Res. 20, 578–588 10.1101/gr.100479.109 (doi:10.1101/gr.100479.109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Faure AJ, Schmidt D, Watt S, Schwalie PC, Wilson MD, Xu H, Ramsay RG, Odam DT, Flicek P. 2012. Cohesin regulates tissue-specific expression by stabilizing highly occupied cis-regulatory modules. Genome Res. 22, 2163–2175 10.1101/gr.136507.111 (doi:10.1101/gr.136507.111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wallace JA, Felsenfeld G. 2007. We gather together: insulators and genome organization. Curr. Opin. Genet. Dev. 17, 400–407 10.1016/j.gde.2007.08.005 (doi:10.1016/j.gde.2007.08.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zlatanova J, Caiafa P. 2009. CTCF and its protein partners: divide and rule? J. Cell Sci., 1275–1284 10.1242/jcs.039990 (doi:10.1242/jcs.039990) [DOI] [PubMed] [Google Scholar]
- 53.Lee BK, Iyer VR. 2012. Genome-wide studies of CCCTC-binding factor (CTCF) and cohesin provide insight into chromatin structure and regulation. J. Biol. Chem. 287, 30 906–30 913 10.1074/jbc.R111.324962 (doi:10.1074/jbc.R111.324962) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lutz M, et al. 2000. Transcriptional repression by the insulator protein CTCF involves histone deacetylases. Nucleic acids Res. 28, 1707–1713 10.1093/nar/28.8.1707 (doi:10.1093/nar/28.8.1707) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lutz M, et al. 2003. Thyroid hormone-regulated enhancer blocking: cooperation of CTCF and thyroid hormone receptor. EMBO J. 22, 1579–1587 10.1093/emboj/cdg147 (doi:10.1093/emboj/cdg147) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yusufzai TM, Tagami H, Nakatani Y, Felsenfeld G. 2004. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol. Cell 13, 291–298 10.1016/S1097-2765(04)00029-2 (doi:10.1016/S1097-2765(04)00029-2) [DOI] [PubMed] [Google Scholar]
- 57.Defossez PA, Kelly KF, Filion GJ, Perez-Torrado R, Magdinier F, Menoni H, Nordgaard CL, Daniel JM, Gilson E. 2005. The human enhancer blocker CTC-binding factor interacts with the transcription factor Kaiso. J. Biol. Chem. 280, 43 017–43 023 10.1074/jbc.M510802200 (doi:10.1074/jbc.M510802200) [DOI] [PubMed] [Google Scholar]
- 58.Yao H, Brick K, Evrard Y, Xiao T, Camerini-Otero RD, Felsenfeld G. 2010. Mediation of CTCF transcriptional insulation by DEAD-box RNA-binding protein p68 and steroid receptor RNA activator SRA. Genes Dev. 24, 2543–2555 10.1101/gad.1967810 (doi:10.1101/gad.1967810) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ross-Innes CS, Brown GD, Carroll JS. 2011. A co-ordinated interaction between CTCF and ER in breast cancer cells. BMC Genomics 12, 593. 10.1186/1471-2164-12-593 (doi:10.1186/1471-2164-12-593) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu Z, Scannell DR, Eisen MB, Tjian R. 2011. Control of embryonic stem cell lineage commitment by core promoter factor, TAF3. Cell 146, 720–731 10.1016/j.cell.2011.08.005 (doi:10.1016/j.cell.2011.08.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wada Y, et al. 2009. A wave of nascent transcription on activated human genes. Proc. Natl Acad. Sci. USA 106, 18 357–18 361 10.1073/pnas.0902573106 (doi:10.1073/pnas.0902573106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shukla S, Kavak E, Gregory M, Imashimizu M, Shutinoski B, Kashlev M, Oberdoeffer P, Sandberg R. 2011. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature 479, 74–79 10.1038/nature10442 (doi:10.1038/nature10442) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stadhouders R, et al. 2012. Dynamic long-range chromatin interactions control Myb proto-oncogene transcription during erythroid development. EMBO J. 31, 986–999 10.1038/emboj.2011.450 (doi:10.1038/emboj.2011.450) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Majumder P, Boss JM. 2010. CTCF controls expression and chromatin architecture of the human major histocompatibility complex class II locus. Mol. Cell. Biol. 30, 4211–4223 10.1128/MCB.00327-10 (doi:10.1128/MCB.00327-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Majumder P, Boss JM. 2011. Cohesin regulates MHC class II genes through interactions with MHC class II insulators. J. Immunol. 187, 4236–4244 10.4049/jimmunol.1100688 (doi:10.4049/jimmunol.1100688) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ribeiro de Almeida C, et al. 2011. The DNA-binding protein CTCF limits proximal Vκ recombination and restricts κ enhancer interactions to the immunoglobulin κ light chain locus. Immunity 35, 501–513 10.1016/j.immuni.2011.07.014 (doi:10.1016/j.immuni.2011.07.014) [DOI] [PubMed] [Google Scholar]
- 67.Seitan VC, Merkenschlager M. 2012. Cohesin and chromatin organisation. Curr. Opin. Genet. Dev. 22, 93–100 10.1016/j.gde.2011.11.003 (doi:10.1016/j.gde.2011.11.003) [DOI] [PubMed] [Google Scholar]
- 68.Soshnikova N, Montavon T, Leleu M, Galjart N, Duboule D. 2010. Functional analysis of CTCF during mammalian limb development. Dev. Cell 19, 819–830 10.1016/j.devcel.2010.11.009 (doi:10.1016/j.devcel.2010.11.009) [DOI] [PubMed] [Google Scholar]
- 69.Montavon T, Soshnikova N, Mascrez B, Joye E, Thevenet L, Splinter E, de Laat W, Spitz F, Duboule D. 2011. A regulatory archipelago controls Hox genes transcription in digits. Cell 147, 1132–1145 10.1016/j.cell.2011.10.023 (doi:10.1016/j.cell.2011.10.023) [DOI] [PubMed] [Google Scholar]
- 70.Kehayova P, Monahan K, Chen W, Maniatis T. 2011. Regulatory elements required for the activation and repression of the protocadherin-alpha gene cluster. Proc. Natl Acad. Sci. USA 108, 17 195–17 200 10.1073/pnas.1114357108 (doi:10.1073/pnas.1114357108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Monahan K, Rudnick ND, Kehayova PD, Pauli F, Newberry KM, Myers RM, Maniatis T. 2012. Role of CCCTC binding factor (CTCF) and cohesin in the generation of single-cell diversity of protocadherin-alpha gene expression. Proc. Natl Acad. Sci. USA 109, 9125–9130 10.1073/pnas.1205074109 (doi:10.1073/pnas.1205074109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hirayama T, Tarusawa E, Yoshimura Y, Galjart N, Yagi T. 2012. CTCF is required for neural development and stochastic expression of clustered Pcdh genes in neurons. Cell Rep. 2, 345–357 10.1016/j.celrep.2012.06.014 (doi:10.1016/j.celrep.2012.06.014) [DOI] [PubMed] [Google Scholar]
- 73.Lieberman-Aiden E, et al. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 10.1126/science.1181369 (doi:10.1126/science.1181369) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Botta M, Haider S, Leung IX, Lio P, Mozziconacci J. 2010. Intra- and inter-chromosomal interactions correlate with CTCF binding genome wide. Mol. Syst. Biol. 6, 426. 10.1038/msb.2010.79 (doi:10.1038/msb.2010.79) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fullwood MJ, et al. 2009. An oestrogen-receptor-α-bound human chromatin interactome. Nature 462, 58–64 10.1038/nature08497 (doi:10.1038/nature08497) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sanyal A, Lajoie BR, Jain G, Dekker J. 2012. The long-range interaction landscape of gene promoters. Nature 489, 109–113 10.1038/nature11279 (doi:10.1038/nature11279) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yaffe E, Tanay A. 2011. Probabilistic modeling of Hi-C contact maps eliminates systematic biases to characterize global chromosomal architecture. Nat. Genet. 43, 1059–1065 10.1038/ng.947 (doi:10.1038/ng.947) [DOI] [PubMed] [Google Scholar]
- 78.Comet I, Schuettengruber B, Sexton T, Cavalli G. 2011. A chromatin insulator driving three-dimensional Polycomb response element (PRE) contacts and Polycomb association with the chromatin fiber. Proc. Natl Acad. Sci. USA. 108, 2294–2299 10.1073/pnas.1002059108 (doi:10.1073/pnas.1002059108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nora EP, et al. 2012. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485, 381–385 10.1038/nature11049 (doi:10.1038/nature11049) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lin YC, et al. 2012. Global changes in the nuclear positioning of genes and intra- and interdomain genomic interactions that orchestrate B cell fate. Nat. Immunol. 13, 1196–1204 10.1038/ni.2432 (doi:10.1038/ni.2432) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lan X, Witt H, Katsumura K, Ye Z, Wang Q, Bresnick EH, Farnham PJ, Jin VX. 2012. Integration of Hi-C and ChIP-seq data reveals distinct types of chromatin linkages. Nucleic acids Res. 40, 7690–7704 10.1093/nar/gks501 (doi:10.1093/nar/gks501) [DOI] [PMC free article] [PubMed] [Google Scholar]