Abstract
The development of sensitive nucleic acid stains, in combination with flow cytometric techniques, has allowed the identification and enumeration of viruses in aquatic systems. However, the methods used in flow cytometric analyses of viruses have not been consistent to date. A detailed evaluation of a broad range of sample preparations to optimize counts and to promote the consistency of methods used is presented here. The types and concentrations of dyes, fixatives, dilution media, and additives, as well as temperature and length of incubation, dilution factor, and storage conditions were tested. A variety of different viruses, including representatives of phytoplankton viruses, cyanobacteriophages, coliphages, marine bacteriophages, and natural mixed marine virus communities were examined. The conditions that produced optimal counting results were fixation with glutaraldehyde (0.5% final concentration, 15 to 30 min), freezing in liquid nitrogen, and storage at −80°C. Upon thawing, samples should be diluted in Tris-EDTA buffer (pH 8), stained with SYBR Green I (a 5 × 10−5 dilution of commercial stock), incubated for 10 min in the dark at 80°C, and cooled for 5 min prior to analysis. The results from examinations of storage conditions clearly demonstrated the importance of low storage temperatures (at least −80°C) to prevent strong decreases (occasionally 50 to 80% of the total) in measured total virus abundance with time.
It has been well established that viruses are abundant and important components in aquatic ecosystems (1, 18, 32, 41, 44) since they have been shown to be a major source of mortality for bacteria and eukaryotes (4, 6, 31, 36, 38, 40). Because of their relatively short infection cycles, virus populations are highly dynamic and result in rapid changes in both total numerical abundance and diversity (5, 11, 13-15, 24, 33-35, 37, 43, 45). Assays for rapidly counting viruses with high precision are, therefore, beneficial for studies of viral ecology in the laboratory as well as the field.
Viruses have traditionally been enumerated by culture-based methods (e.g., plaque counts and most-probable-number assays) and transmission electron microscopy (1, 3, 15, 39). These techniques were either selective for viruses infectious for a specific host or very time-consuming. The introduction of high-fluorescence-yield nucleic-acid-specific stains in combination with epifluorescence microscopy (19, 20, 30) significantly improved the quantitation of viruses. With the recent introduction of flow cytometric detection and enumeration of free viruses (7, 27, 29), speed of analysis and accuracy of counting was further improved. This method no longer relies on the skills of the operator. Direct comparison showed that epifluorescence- and flow cytometry-based virus counts were highly comparable (27). Brussaard et al. (7) have shown that a variety of viruses of different morphologies and genome sizes could be detected by flow cytometry. In combination with SYBR Green I as the fluorescent stain, flow cytometry has been used successfully to count viruses from laboratory experiments (8, 9) and in natural samples (11, 12, 24, 25, 27, 43).
The methods used for flow cytometric analyses of viruses in natural samples have, however, not been consistent (7, 12, 29, 43). The few reports on methodologies suggest that optimal detection of virus particles depends on various factors. For example, the study by Marie et al. (27) indicated that the type and concentration of fluorescent stain, the solution used to dilute the sample, the incubation temperature, and the addition of a detergent may all influence the quality of the fluorescent signal. Chen et al. (12) tested the use of the fluorescent dye SYBR Gold for viruses infecting the cyanobacterium Synechococcus spp. Analyses of unfrozen samples showed that staining with SYBR Gold resulted in a higher fluorescence than SYBR Green I. Nevertheless, Marie et al. (27) reported that samples frozen in liquid nitrogen gave better results than unfrozen samples. There are no data available yet on the use of SYBR Gold in combination with freezing. Nor are there currently studies published reviewing the methods used in flow cytometric analyses of viruses without the use of antibodies.
In order to optimize the flow cytometric detection and enumeration of free viruses, a detailed evaluation of a broad range of factors (storage conditions and staining specifications) that potentially affect the quality of the fluorescent signal and subsequently the counting efficiency is presented here. Tests were performed on a variety of different viruses, including four representatives of marine phytoplankton viruses, one freshwater phytoplankton virus, one virus infecting a cyanobacterium, four characterized coliphages, three uncharacterized marine heterotrophic bacteriophages, and two natural mixed marine virus communities.
MATERIALS AND METHODS
Virus strains.
The viruses CeV-01B, MpVUF10-38, PoV-01B, and PpV-01B, which infect the photosynthetic unicellular organisms Chrysochromulina ericina, Micromonas pusilla, Pyramimonas orientalis, and Phaeocystis pouchetii were kindly provided by G. Bratbak (University of Bergen, Bergen, Norway); PBCV-1, which infects Chlorella sp., was provided by J. Van Etten (University of Nebraska); and S-PMS2, which infects Synechococcus sp., was provided by W. Wilson (Marine Biological Association, Plymouth, United Kingdom). The coliphages Lambda, T2, T4, and T7, which infect the heterotrophic bacterium Escherichia coli were kindly provided by G. Bratbak (culture collection of the Department of Microbiology, University of Bergen). The marine bacteriophages T-φHSIC A, φ16, and T-φD1B, which infect Listonella pelagia, Vibrio parahaemolyticus strain 16, and Flavobacterium sp., respectively (22, 23, 42), were kindly provided by J. Paul (University of South Florida). Natural seawater samples collected from Dutch coastal waters (NS1) and the southern North Sea (NS2) in autumn were also analyzed.
Treatments.
Samples were subjected to different treatments (Table 1), including type of fixative and percentage fixative used, storage temperature and length of storage, type and concentration of the stain and dilution solution, pH of the Tris-EDTA (TE) buffer, length and temperature of incubation, addition of detergents, and dilution factor. The treatment used as a reference was fixation with glutaraldehyde (0.5% final concentration, EM-grade; Merck) for 30 min at 4°C, followed by freezing in liquid nitrogen (N2; −196°C) and storage at −80°C (7, 27). When thawed at 35°C for a few minutes (samples should still be cool), samples were diluted in TE buffer (10 mM Tris-HCl and 1 mM EDTA, pH 8) and incubated with SYBR Green I (5 × 10−5 dilution of commercial stock; Molecular Probes) at 80°C for 10 min in the dark.
TABLE 1.
Treatments to which virus samples were subjected in order to optimize enumeration of viruses in solution by flow cytometrya
| Treatment | Levels |
|---|---|
| Fixative glutaraldehyde (%) | 0.1, 0.5, 1, or 2 |
| Fixative formaldehyde (%) | 0.4, 0.8, 2, or 4 |
| Storage temp (°C) | 4, −20, −80, or −196 (N2) |
| Storage time at 4, −20, and −80°C | 1 h or 1 mo |
| Storage time at 4°C | 1 h, 2 wk, or 1 or 6 mo |
| Dye SYBR Green I (10−5) | 0, 0.5, 1, 5, 10, 15, or 25 (of the commercial stock) |
| Dye SYBR Gold (10−5) | 5, 10, or 25 (of the commercial stock) |
| Incubation temp | 20, 40, 60, 80, or 90°C |
| Incubation time (min) | 1, 4, 7, 10, 13, 16, or 19 |
| Dilution solution | TE, Tris, PBS, dH2O, or seawater |
| pH of TE buffer | 7, 7.4, 7.8, 8, 8.4, or 8.85 |
| Dilution with seawater (%) | 0, 1, 2, 5, 10, 20, or 50 |
| Addition of detergents | Triton X-100, Tween 80, NP-40, and SDS at a 0.1% (vol/vol) final concn |
| Addition of citrate (concn [mM]) | 0, 1, 2, 5, 10, 20, or 50 |
Viruses used included four marine phytoplankton viruses (CeV, MpV, PoV, and PpV), one freshwater phytoplankton virus (PBCV), one virus infecting a cyanobacterium (S-PMS2), four characterized bacteriophages (Lambda, T2, T4, and T7), three uncharacterized marine bacteriophages (T-φHS1C A, φ16, and T-φD1B), and two natural marine virus communities. dH2O, distilled water.
Two fixatives, glutaraldehyde and formaldehyde, were tested at different concentrations. Dilution solutions tested were Tris (pH 8), TE buffer (pH 7 to 8.9), distilled water, phosphate-buffered saline buffer, and sterile seawater (0.2 μm filtered). Fluorescent stains tested were SYBR Green I and SYBR Gold (Molecular Probes). To mimic the effect of dilution of seawater virus samples in TE buffer, sterile filtered seawater was added at different final concentration. The effect of citrate and various detergents (Triton X-100, Tween 80, NP-40, and sodium dodecyl sulfate [SDS]) was also tested.
Sample analysis.
Samples were analyzed by using a FACSCalibur flow cytometer (Becton Dickinson, San Jose, Calif.) equipped with a 15-mW 488-nm air-cooled argon-ion laser and a standard filter setup. The trigger was set on green fluorescence. To avoid coincidence of viral particles (i.e., two or more particles being simultaneously within the sensing zone), the samples were diluted such that the event rate was between 100 and 1,000 viruses s−1. Yellow-green fluorescent latex microspheres (0.85 μm in diameter; Polysciences, Inc., Warrington, Pa.) were added as an internal reference to all samples. Readings were collected in logarithmic mode (at least 5,000 events per sample) and analyzed with CYTOWIN (freely available at http://www.sb-roscoff.fr/phyto/cyto.html). The data were converted to linear values and normalized to the bead internal standard. Green fluorescence (GFL), total counts, and side scatter (SSC) were recorded. Because viruses are generally smaller than the wavelength of the laser light used (7), the SSC signal cannot be used as an indicator of size and granularity but is very useful as a discriminator of the virus populations (7, 21).
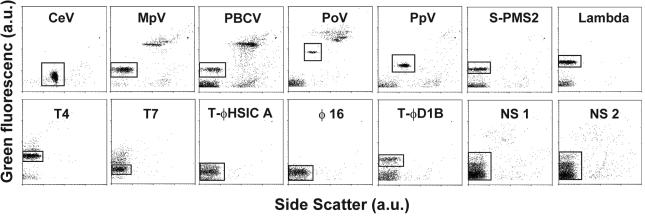
Differences in cytograms (plotting GFL versus SSC) for the different viruses tested are shown in Fig. 1. The windows indicated were used to discriminate the virus of interest and to determine the total virus count and examine specific staining characteristics. Total virus counts were obtained by correcting the measured total counts for noise (typically having the lowest GFL) with sterile and filtered (0.2-μm pore size) seawater or medium as blank. Tests with virus-free lysate (filtered to <30 kDa) showed results comparable to tests with sterile and filtered seawater or medium.
FIG. 1.
Cytograms (4 decades log scale) of green fluorescence versus side scatter (arbitrary units, a.u.) for 12 different virus cultures (see Materials and Methods) and two natural virus samples (NS 1 and 2) tested under reference conditions (glutaraldehyde fixed at 0.5% final concentration, frozen in liquid nitrogen, diluted in TE-buffer pH 8, stained for 10 min at 80°C with SYBR Green I at a final dilution of 5 × 10−5 the commercial stock). At least 2000 events are plotted for each virus type, except for NS 1 and 2, for which 5000 events were plotted. The windows mark the selected regions for analysis, and contain the viruses of interest. Total virus counts were obtained by correcting the total counts for noise (which typically has the lowest green fluorescence) with sterile and filtered (0.2 μm pore-size) seawater or medium as blank.
The marine bacteriophage yielded low fluorescence signals (Fig. 1) at the normal settings of the green fluorescence photomultiplier. Increasing the voltage of the photomultiplier improved the results slightly but simultaneously increased the noise. Consequently, there was no real improvement in the signal-to-noise ratio.
Statistical analyses of different treatments were performed by using Systat 10 software. To fulfill the assumptions of analysis of variance, all data were log transformed. Either a two-way analysis of variance (ANOVA) with viruses as a random factor (block) and treatment on 2 to 13 levels or a four- or five-way ANOVAs with interaction terms and two levels for each treatment was used. A probability of P < 0.05 was taken to conclude that the treatment differed significantly in its effect on the measured value.
RESULTS
The standard deviations for GFL, total counts, and SSC among replicates (n = 5) were <5% for the phytoplankton virus (PpV) and <12% for bacteriophages Lambda and φ16. These results show that a detailed comparison of staining characteristics and measured abundance for the different treatments and viruses is valid. Furthermore, combining four different viruses (CeV, MpV, PpV, and S-PMS2) in one sample provided identical results compared to the analysis of individual viruses. This indicates that the results obtained with individual viruses can be interpolated to mixed-virus situations such as those found in nature.
The statistical variance analyses of the different treatments are summarized in Table 2 and 3. The analysis of four treatments (freezing, fixation, heating, and addition of detergent) for all viruses in the present study indicates that there were significant differences for total virus counts, GFL, and SSC signals between the group consisting of phytoplankton viruses and the group consisting of heterotrophic bacteriophages (Table 2). Therefore, these two virus groups were also analyzed separately in order to obtain insight into the factors and extent to which the groups differed.
TABLE 2.
Statistical ANOVA of four different treatments (freezing, fixation, heating, and addition of detergent) for all viruses tested together or separated into two groups (phytoplankton viruses and heterotrophic bacteriophages)a
| Virus included in statistical analysis | Factor treatment | Levels | Log
|
||
|---|---|---|---|---|---|
| Counts | GFL | SSC | |||
| All virusesb | 2 (phytoplankton viruses and heterotrophic phages) | ** | *** | *** | |
| Fixationc | 2 (yes/no) | *** | *** | ||
| Heating at 80°Cd | 2 (yes/no) | *** | *** | *** | |
| Liquid nitrogen (N2) | 2 (yes/no) | ||||
| Triton additione | 2 (yes/no) | ||||
| Group vs 80°C | Interaction | ** | *** | ||
| Group vs fixation | Interaction | * | *** | ||
| 80°C vs fixation | Interaction | ** | |||
| Virus type in group | 13 | *** | *** | *** | |
| Virus type vs fixation | Interaction | *** | *** | ||
| Virus type vs 80°C | Interaction | ** | * | ||
| Phytoplankton viruses | Fixationc | 2 (yes/no) | *** | ||
| Heating at 80°Cd | 2 (yes/no) | * | ** | *** | |
| Liquid nitrogen (N2) | 2 (yes/no) | ** | |||
| Triton additione | 2 (yes/no) | ||||
| Virus type vs 80°C | Interaction | ||||
| Virus type vs fixation | Interaction | *** | |||
| Virus type vs N2 | Interaction | *** | |||
| 80°C vs fixation | Interaction | ||||
| 80°C vs N2 | Interaction | ||||
| Heterotrophic bacteriophages | Fixationc | 2 (yes/no) | *** | ||
| Heating at 80°Cd | 2 (yes/no) | *** | *** | ||
| Liquid nitrogen (N2) | 2 (yes/no) | ||||
| Triton additione | 2 (yes/no) | ||||
| Virus type vs 80°C | Interaction | *** | |||
| Virus type vs fixation | Interaction | *** | |||
| 80°C vs fixation | Interaction | * | |||
A four- or five-way ANOVA with interaction terms and two levels for each treatment was used. The treatments that were found to significantly affect total viral counts, GFL, or SSC of the stained virus particles are indicated by asterisks: *, P < 0.05; **, P < 0.01; and ***, P < 0.001. SYBR Green I was used as nucleic acid-specific dye (diluted ×10−5 of the commercial stock).
Virus types were a nested factor within the virus groups.
Final glutaraldehyde concentration of 0.05%.
Incubation at 80°C for 10 min, followed by cooling for 5 min or incubation at 20°C for 15 min.
Final Triton X-100 concentration of 0.1% (vol/vol).
TABLE 3.
Statistical ANOVA of different treatments tested in detail for phytoplankton viruses and heterotrophic bacteriophages together (6 to 10 viruses tested per treatment)a
| Expt | Factor treatment | Levels | Log
|
||
|---|---|---|---|---|---|
| Counts | GFL | SSC | |||
| Incubation | Temp | 5 | ** | ||
| Incubation | Time | 7 | * | ||
| Fixation | Glutaraldehyde | 4 | |||
| Fixation | Formaldehyde | 4 | * | ||
| SYBR Green 1 | Dye concn | 6 | * | ||
| SYBR dyes | Gold vs Green I | 2 (yes/no) | * | ||
| SYBR Gold | Dye concn | 3 | * | ||
| Dilution | Solution | 5 | * | ||
| TE solution | pH | 6 | |||
| Dilution factor | Addition of seawater | 7 | * | * | |
| Additives | Detergents | 5 | ** | ||
| Additives | Citrate | 7 | |||
| Storage | T = 1 h | 4 | * | ||
| Storage | T = 1 mo | 4 | *** | * | |
| Storage | 4°C | 4 | ** | ** | |
A two-way ANOVA with viruses as a random factor (block factor) and a treatment on two to seven levels was used. The treatments that were found to significantly affect total viral counts, GFL, or SSC of the stained virus particles are indicated by asterisks: *, P < 0.05; **, P < 0.01; and ***, P < 0.001. Levels are as described in Materials and Methods.
Fixatives.
Fixation had a significant positive effect on GFL and SSC compared to that of unfixed samples (Table 2). A more detailed statistical analysis showed that the improved GFL signal was due to the heterotrophic bacteriophages and that the improved SSC signal was due to the phytoplankton viruses (Table 2). The extent to which the viruses were positively affected by fixation depended on the type of virus (interaction effect). There was no difference in signal for up to 1 h of fixation (0.25, 0.5, 1, 3, and 18 h tested), and no significant difference between fixation at room temperature and 4°C was recorded. The type of fixative (glutaraldehyde or formaldehyde), as well as the final concentration of fixative (0.1 to 2% for glutaraldehyde and 0.4 to 4% for formaldehyde) were tested (Table 3). The use of formaldehyde rather than glutaraldehyde occasionally led to reduced virus counts (for example, the marine phages T-φHSIC showed a 20% reduction). The total counts were slightly reduced at concentrations of formaldehyde of >0.4% (reductions typically <10%, but a 55% reduction was recorded for T-φHSIC A). Although no significant trend was observed for the different concentrations of glutaraldehyde, reductions in virus abundance were occasionally observed at concentrations of >1% (e.g., S-PMS2 and Lambda).
Incubation temperature.
Heating of the samples to 80°C prior to analysis significantly increased the total virus count (Table 2) and improved the staining characteristics (GFL and SSC). Measured virus abundance was most enhanced for the group of heterotrophic bacteriophages (up to eightfold for φ16 and T-φD1B). Heating showed significant interactions with the type of virus and fixation (Table 2). Five different temperatures in the range of 20 to 90°C were tested in order to find the optimal temperature for staining of virus samples (Table 3). In general, heating the samples to 80 to 90°C showed the best results. Analysis of the length of the incubation period (1, 4, 7, 10, 13, 16, and 19 min) showed a significant effect for GFL (Table 3), with the signal increasing until 10 min.
Dyes.
The concentration of SYBR Green I (ranging in dilution from 5 × 10−5 to 25 × 10−5 of the commercial stock) significantly affected total virus counts (Table 2B), showing 5 to 45% reductions in measured virus abundance when 1 × 10−4 to 25 × 10−5 of the commercial stock was stained compared to the optimal dilution of 5 × 10−5 of the commercial stock. The addition of SYBR Green I after the samples were heated (instead of prior to heating) negatively affected the GFL signal and the total counts for the viruses that infect heterotrophic bacteria. For example, marine bacteriophages φ16 and T-φD1B showed a 50 to 70% reduction in GFL and a 40 to 50% reduction in counts.
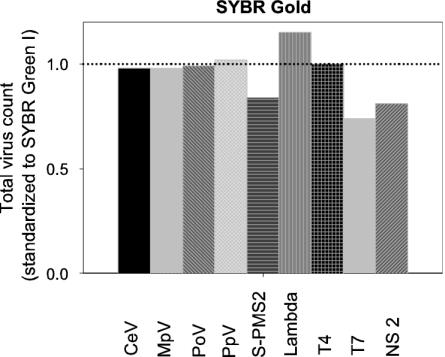
Staining with another sensitive nucleic acid-specific stain, SYBR Gold, resulted in slightly (but significantly) lower GFL signals of the stained viruses (Table 3). No significant effect was recorded for the total virus counts when all tested viruses were taken into account, but for some of the individual viruses and a natural sample, staining with SYBR Gold resulted in reduced total counts compared to staining with SYBR Green I (Fig. 2). The GFL signal decreased with increasing concentrations of dye (from 5 × 10−5 to 25 × 10−5 of the commercial stock; P < 0.05, Table 3). To allow comparison with published work on cyanophages using SYBR Gold, the virus S-PMS2 (infecting Synechococcus sp.) was specifically tested in more detail (Table 4). Staining with SYBR Gold, compared to staining with SYBR Green I, resulted in significantly lower total counts. At dilutions of >5 × 10−5 of the commercial stock, both the total virus count and the GFL signal were further reduced (Table 3). Heating the sample affected the total counts, as well as GFL and SSC signals, positively. Freezing the samples in liquid nitrogen prior to analysis also had a significant positive effect on the measured total virus abundance.
FIG. 2.
Total virus counts obtained after staining with SYBR Gold (final dilution of 5 × 10−5 the commercial stock). The data were corrected for blanks and normalized to SYBR Green I at an identical dye concentration (dotted line). NS 2 represents a natural virus sample.
TABLE 4.
Statistical ANOVA of four different treatments (type of dye, freezing, concentration of dye, and heating) tested for cyanophage S-PMS2 by using SYBR Gold as a nucleic acid-specific dyea
| Factor treatment | Levels | Log
|
||
|---|---|---|---|---|
| Counts | GFL | SSC | ||
| vs SYBR Green (dye) | 2 | *** | ||
| Dye concn | 3 | ** | *** | |
| Heating (80°C) | 2 (yes/no) | *** | *** | *** |
| Liquid nitrogen (N2) | 2 (yes/no) | *** | ||
| Dye vs concn | Interaction | |||
| Dye vs N2 | Interaction | |||
| Concn vs 80°C | Interaction | |||
| Concn vs N2 | Interaction | |||
| 80°C vs N2 | Interaction | |||
The treatments that were found to significantly affect total viral counts, GFL, or SSC of the stained virus particles are indicated by asterisks: *, P < 0.05; **, P < 0.01; and ***, P < 0.001. See Materials and Methods for a description of the levels.
Dilution solution.
To avoid coincidence during flow cytometric analysis, the virus samples were normally diluted. The choice of solution (seawater, Tris buffer, TE buffer, phosphate-buffered saline buffer, or distilled water) had a significant effect on the measured virus abundance (Table 3), showing highest counts when diluted in TE or Tris buffer. The lowest counts were obtained when the virus sample was diluted in distilled water or seawater. Dilution in TE buffer also yielded the best GFL signal, while the pH of the TE buffer (7, 7.4, 7.8, 8, 8.4, or 8.85) had no significant effect (Table 3), although individual viruses reacted very differently (e.g., PpV showed 40% reduction in total count at a pH of <8).
Additives.
The ratio of TE buffer to seawater is influenced by the dilution factor of the marine virus sample. To simulate the effect of various percentages of seawater samples at different dilutions, sterile and filtered (0.2-μm pore size) seawater at final concentrations of 1, 2, 5, 10, 20, and 50% (vol/vol) was added to highly diluted virus samples (>1,000 times diluted in TE at pH 8). With increasing concentrations of seawater, a significant decline in GFL signal and in total counts was recorded (Table 3). To test whether these effects were due to the high salt concentration, sodium chloride (NaCl) was added to the samples at final concentrations of 2, 5, and 10% (vol/vol). No significant effects were recorded as a result of the NaCl additions.
The addition of Triton X-100 (0.1% [vol/vol]) had no significant effect on total virus counts or staining characteristics (Table 2) but was occasionally found to improve the coefficient of variance of the GFL signal. It was hypothesized that detergents would facilitate the permeabilization of the viral particles and would therefore enhance the GFL signal upon staining. A significant effect on the total virus counts was recorded (Table 3) by using Triton X-100, Tween 80, NP-40, and SDS (all at 0.1% [vol/vol] final concentrations). However, there was no significant effect when the anionic surfactant SDS, negatively affecting the abundance of the heterotrophic bacteriophages (reductions up to 95%), was omitted from the statistical analysis. The problem with the addition of surfactants was the generation of high levels of noise. As long as the viruses have GFL signals well above the level of the produced noise (for example, many of the phytoplankton viruses), this will not be bothersome. However, it could present a problem when virus samples with relative low GFL levels are analyzed. Sufficient blanks should be taken to allow subtraction of the noise from the total virus counts in the samples.
Another factor that was hypothesized to affect the discrimination and enumeration of viruses by flow cytometry is citrate. No significant effect of citrate addition (final concentrations of 1, 2, 5, 10, 20, or 50 mM) was recorded in the present study (Table 3).
Storage.
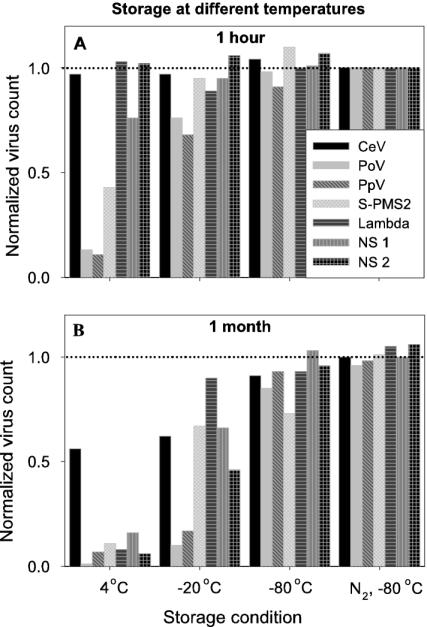
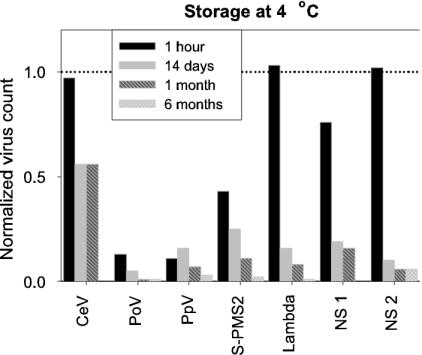
A range of temperatures—4, −20, −80, and −196°C (deep freezing in liquid nitrogen)—for storage was tested for 7 of the 12 viruses studied. Storage temperature significantly affected total virus counts (Table 3), although large differences in total count were recorded among the different viruses tested (Fig. 3). The optimal storage protocol was deep freezing in liquid nitrogen (followed by storage at −80°C for convenience). The present study clearly demonstrates that storage of virus samples should be conducted at very low temperatures (Table 3, Fig. 3 and 4). Some of the samples stored at 4°C for a few hours resulted in only ca. 10% of the measured abundance compared to samples deep frozen in liquid nitrogen and stored at −80°C (Fig. 3A). Figure 3B shows that 1 month of storage at 4 or −20°C led to high reductions in total virus counts (down to a few percent of the total count at t = 0). Also, the GFL signal was significantly reduced for the samples stored at 4°C (Table 3). Because it is not uncommon to store samples at 4°C before analysis, a separate statistical analysis was performed to test for variance in the length of storage time at this temperature. Total counts and SSC signal were negatively affected by storage duration (Table 3), showing reductions in total virus counts of 80% within 14 days (Fig. 4).
FIG. 3.
Effect of storage temperatures on virus abundance. Fixed virus samples (glutaraldehyde at 0.5% final concentration) were stored at 4°C, −20°C, and −80°C (directly or after having been deep frozen in liquid nitrogen, N2, prior to storage). Samples were analyzed after 1 h (A) and 1 month (B). Total virus counts were corrected for blanks and normalized to samples frozen in liquid nitrogen and directly analyzed (dotted line). NS 1 and 2 represent natural virus samples.
FIG. 4.
Effect of storage at 4°C on measured virus abudance (fixed with glutaraldehyde at 0.5% final concentration). Samples were stored for 1 h, 14 days, 1 month, and 6 months. Total virus counts were corrected for blanks and normalized to samples frozen in liquid nitrogen and directly analyzed (dotted line). The samples for CeV at 6 months were lost. NS 1 and 2 represent natural virus samples.
DISCUSSION
The development of sensitive nucleic acid stains, in combination with flow cytometric techniques has allowed identification and enumeration of very small particles such as viruses in aquatic systems (7, 8, 11, 12, 24, 25, 27, 29, 43). So far, however, studies using flow cytometry to count viruses in aquatic ecosystems have not used consistent methods. The present study was executed to optimize the flow cytometric enumeration of viruses and promote consistency of the methodology used. Total virus counts, green fluorescence (GFL) and side scatter (SSC) of the viral particles were recorded for a variety of different viruses (including representatives of phytoplankton viruses, cyanobacteriophages, coliphages, marine heterotrophic bacteriophages, and natural mixed marine virus communities) that where subjected to a range of different treatments potentially influencing detection and quantitation of virus particles.
Virus abundance.
In general, total virus counts were significantly and positively affected when samples were frozen in liquid nitrogen, heated at 80°C (10 min, followed by 5 min cooling down of the sample), and diluted in TE-buffer. It is recommended to add the fluorescent dye prior to heating of the sample, because staining of the heterotrophic bacteriophages was less intense when the fluorescent dye was added after heating. Marie et al. (27) did not find any effect of heating and time of addition of the dye, however this study performed the analyses with a virus infecting the phytoplankter Phaeocystis pouchetii (i.e., PpV). Consistent with these results, in the present study there was no increased counting efficiency detected upon heating for this virus. With the majority of the viruses in marine environments being bacteriophages, and two of the tested marine heterotrophic phages showing significant reduction in total virus counts at 60°C (12 to 65% reduction compared to sample heated at 80°C), even a reduction in temperature to 65°C as used by Evans et al. (17) can be expected to underestimate the total natural virus concentration.
Natural samples normally have to be diluted before flow cytometric analysis in order to avoid coincidence. Dilution in TE-buffer (pH 8) gave the best results for virus enumeration in conjunction with staining quality, as was also demonstrated by Marie et al. (27). Dilution in seawater (0.2-μm pore size filtered and autoclaved) should be avoided because of the reduction in total virus counts and a reduction in the staining signal. Since counting efficiency was reduced with increasing percentages of seawater, the dilution factor of marine virus samples in TE-buffer should be as high as possible (preferably at least > 50) while keeping statistically significant virus counts. In the present study there was a 40% reduction in estimated viral abundance for the marine heterotrophic bacteriophage and natural samples diluted 2-fold (50% final concentration of seawater). Measured virus abundance was still 15% reduced when those samples were 10-fold diluted. Studies using low dilution factors for virus samples (17) can thus be expected to underestimate the total virus concentration. Although a high dilution factor reduces the negative effect of seawater, the statistically optimal event rate should not become too low (optimal between 100 and 800 per s [27, 29]; unpublished results using different virus samples). In case the virus event rate becomes too low, one has to switch to a higher flow rate.
Staining the viruses with the nucleic-acid-specific dye SYBR Gold did not significantly enhance total virus counts compared to SYBR Green I, and in fact some individual virus samples, such as the natural sample NS 2, showed lower total counts when stained with SYBR Gold. Chen et al. (12) used SYBR Gold at a dilution of 2.5 × 10−4 the commercial stock to stain viruses (including viruses infecting Synechococcus sp.). The present study shows that at this concentration the measured virus abundance is significantly reduced compared to a dilution of 5 × 10−5 the commercial stock. This concentration is also optimal for SYBR Green I and the use of relatively high concentrations of dye is therefore not suggested for routine use. In addition to counting performance, virus staining characteristics with SYBR Green I were significantly better than with SYBR Gold, indicating the preference of using SYBR Green I for standard analysis. Other fluorescent dyes tested (7, 27) have not been shown to improve staining quality nor estimated total virus counts.
Chen et al. (12) used a high concentration of glutaraldehyde (2.5% final concentration) to fix virus samples (natural as well as viruses infecting Synechococcus sp.). This is not recommended as it was found to reduce the total cyanophage count in the present study. The phytoplankton virus abundances were lower when fixed with glutaraldehyde at final concentrations > 0.5%. The tests with glutaraldehyde versus formaldehyde in the present study were not conclusive, but the natural seawater samples showed higher GFL signals when fixation was performed with glutaraldehyde. This will allow better separation of viruses with a relative low green fluorescence upon staining. Formaldehyde is generally used in studies using epifluorescence to enumerate viruses (2, 20, 30), because it has a lower background noise on the 0.02 μm pore-sized Anodisc filters. The use of flow cytometry, however, does not restrict one to formaldehyde fixation. Because fixation with glutaraldehyde is a slower process than with formaldehyde, time should be given to fix the viral nucleic acids effectively (10). A fixation time of 15 to 60 min was found to be most efficient in the present study, as total virus counts began to decline after fixation for several hours, even at 4°C. Long fixation (12 h) of the virus samples before freezing, as is described in the study by Evans et al. (17), is therefore not advised.
The temperature at which the virus samples were stored postfixation was found to be critical. Nonfrozen samples showed a rapid loss of counts, and thus storage at 4°C is strongly discouraged. Studies using nonfrozen fixed virus samples can be expected to underestimate the total virus concentration (12), with the extent depending on the length of time at 4°C (frequently, total virus counts were down to 20% within 14 days storage). A few other studies have reported reductions of viral abundances in fixed samples upon storage at 4°C (16, 27, 46), but a comparison of storage temperatures has not been tested before. When comparing total virus counts of different studies, it is important to take the type of storage into account. Actually, only the samples that were frozen in liquid nitrogen upon fixation (whether stored at −80°C or not) did not show significant declines in total virus numbers over time (tested up to 6 months). For optimal results and to standardize methodology it is recommended for future studies to freeze virus samples in liquid nitrogen.
Staining characteristics.
Working close to the limits of staining methodology and the technical performance of a flow cytometer, the level of green fluorescence (GFL) is crucial for optimizing the detection of free viruses (7). The GFL signals of the stained viruses were positively affected by heating the samples prior to analysis, and by incubation time (optimal signal after 10 min). It is hypothesized that heat treatment affects the permeability of the viral capsid and denatures the nucleic acids (at least partly), thereby improving the staining characteristics of the dye. Fixation improved the staining of the heterotrophic bacteriophages, presumably the permeabilized membranes of the virus particles provided better access for the dye.
Additives such as citrate have been reported to improve both the staining quality and the stability of the SYBR Green I fluorescence when used for cell cycle analysis of phytoplankton (28). This could not be confirmed for viruses (present study). Addition of detergents, which potentially make the virus capsid permeable and thus allowing easier penetration of the dye into the cells (26, 27), did not significantly improve of the staining characteristics either. To the contrary, high levels of noise were generated with the addition of detergents. The use of Triton X-100 for the analysis of natural samples (12, 17), typically containing high percentages of low-staining bacteriophages, is therefore not recommended. When working with controlled mixed systems with relative high staining signals, however, the addition of detergent may improve the separation of the different virus populations.
Evaluating the effects of a broad range of variables on the staining specifications of aquatic viruses shows that there can be large variations between different viruses. Intrinsic features such as the packaging degree of the nucleic acids, and the GC content of the viral genome can be expected to affect the entry and binding of the fluorescent dye, and help to explain the different responses to the methodological variables. Whether the phage genome was circular (Lambda and T4) or linear (T7), appeared unimportant for staining characteristics.
Conclusions.
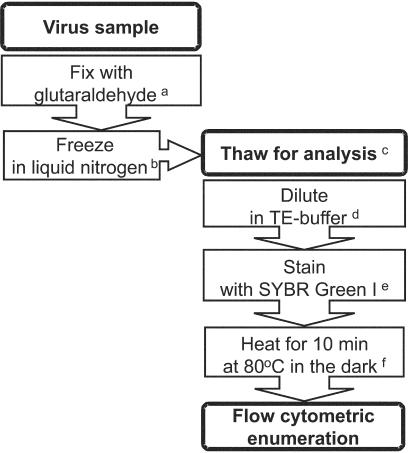
When studying a specific virus type, it is best to determine the combination of variables that results in optimal staining features and counting performance. In the field, environmental parameters may potentially affect the staining or counting efficiency and so it is recommended that investigators document the best protocol for their specific system under study (sampling location). Since this will frequently not be possible, the most practical solution is to be consistent in the methodology used to enumerate viruses. The present evaluation of sample treatments, using not only various specific cultured phytoplankton viruses and heterotrophic bacteriophages but also natural marine samples, indicates that one specific set of variables provided the best results for flow cytometric detection and enumeration (Fig. 5). In summary, virus samples are best fixed with glutaraldehyde (0.5% final concentration, 15 to 30 min at 4°C), frozen in liquid nitrogen, stored below −80°C, diluted in TE buffer (pH 8), stained with SYBR Green I at a final dilution of 5 × 10−5 the commercial stock, and incubated for 10 min in the dark at 80°C (allow cooling down of sample for 5 min prior to analysis).
FIG. 5.
Schematic overview of the optimal protocol for counting of viruses in solution. Superscript letters: a, 0.5% final concentration; b, store sample at temperatures below −80°C; c, work fast once samples are thawed since total virus count decreases with time; d, TE buffer at pH 8 and avoid low dilution factors (<10); e, final dilution SYBR Green I of 5 × 10−5 the commercial stock; f, allow sample to cool down for 5 min in the dark before analysis.
Acknowledgments
This study was supported by the European Commission through the research programs Training and Mobility of Researchers (MAST3.CT96-5033[DG12-ASAL]) and Environment and Sustainable Development (EVK3-CT-1999-00015 BIOHAB).
I thank G. Bratbak, J. Paul, C. Suttle, J. Van Etten, and W. Wilson for providing the viruses used in this study. I also thank Harry Witte for help during the final stages of the data analysis and Gerhard Herndl and two anonymous reviewers for constructive comments on the manuscript.
REFERENCES
- 1.Bergh, Ø., K. Y. Børsheim, G. Bratbak, and M. Heldal. 1989. High abundance of viruses found in aquatic environments. Nature 340:467-468. [DOI] [PubMed] [Google Scholar]
- 2.Bettarel, Y., T. Sime-Ngando, C. Amblard, and H. Laveran. 2000. A comparison of methods for counting viruses in aquatic systems. Appl. Environ. Microbiol. 66:2283-2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Børsheim, K. Y., G. Bratbak, and M. Heldal. 1990. Enumeration and biomass estimation of planktonic bacteria and viruses by transmission electron microscopy. Appl. Environ. Microbiol. 56:352-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bratbak, G., J. K. Egge, and M. Heldal. 1993. Viral mortality of the marine alga Emiliania huxleyi (Haptophyceae) and termination of algal blooms. Mar. Ecol. Prog. Ser. 93:39-48. [Google Scholar]
- 5.Bratbak, G., M. Heldal, S. Norland, and F. Thingstad. 1990. Viruses as partners in spring bloom microbial trophodynamics. Appl. Environ. Microbiol. 56:1400-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brussaard, C. P., R. S. Kempers, A. J. Kop, R. Riegman, and M. Heldal. 1996. Virus-like particles in a summer bloom of Emiliania huxleyi in the North Sea. Aquat. Microb. Ecol. 10:105-113. [Google Scholar]
- 7.Brussaard, C. P., D. Marie, and G. Bratbak. 2000. Flow cytometric detection of viruses. J. Virol. Methods 85:175-182. [DOI] [PubMed] [Google Scholar]
- 8.Brussaard, C. P. D., D. Marie, R. Thyrhaug, and G. Bratbak. 2001. Flow cytometric analysis of phytoplankton viability following viral infection. Aquat. Microb. Ecol. 26:157-166. [Google Scholar]
- 9.Brussaard, C. P. D., R. Thyrhaug, D. Marie, and G. Bratbak. 1999. Flow cytometric analyses of viral infection in two marine phytoplankton species, Micromonas pusilla (Prasinophyceae) and Phaeocystis pouchetii (Prymnesiophyceae). J. Phycol. 35:941-948. [Google Scholar]
- 10.Bullock, G. R. 1984. The current status of fixation for electron microscopy: a review. J. Microsc. 133:1-15. [Google Scholar]
- 11.Castberg, T., A. Larsen, R.-A. Sandaa, C. P. D. Brussaard, J. K. Egge, M. Heldal, R. Thyrhaug, E. J. Van Hannen, and G. Bratbak. 2001. Microbial population dynamics and diversity during a bloom of the marine coccolithophorid Emiliania huxleyi (Haptophyta). Mar. Ecol. Prog. Ser. 221:39-46. [Google Scholar]
- 12.Chen, F., J.-R. Lu, B. J. Binder, Y.-C. Liu, and R. E. Hodson. 2001. Application of digital image analysis and flow cytometry to enumerate marine viruses stained with SYBR Gold. Appl. Environ. Microbiol. 67:539-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cochlan, W. P., J. Wikner, G. F. Steward, D. C. Smith, and F. Azam. 1993. Spatial distribution of viruses, bacteria and chlorophyll a in neritic, oceanic, and estuarine environments. Mar. Ecol. Prog. Ser. 92:77-87. [Google Scholar]
- 14.Cochran, P. K., and J. H. Paul. 1998. Seasonal abundance of lysogenic bacteria in subtropical estuary. Appl. Environ. Microbiol. 64:2308-2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cottrell, M. T., and C. A. Suttle. 1995. Dynamics of a lytic virus infecting the photosynthetic marine picoflagellate Micromonas pusilla. Limnol. Oceanogr. 40:730-739. [Google Scholar]
- 16.Danovaro, R., A. Dell'Anno, A. Trucco, M. Serresi, and S. Vanucci. 2001. Determination of virus abundance in marine sediments. Appl. Environ. Microbiol. 67:1384-1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Evans, C. G. T., S. D. Archer, S. Jacquet, and A. C. Wilson. 2003. Direct measurements of the contribution of viral lysis and microzooplankton grazing to the decline of a Micromonas spp. population. Aquat. Microb. Ecol. 30:207-219. [Google Scholar]
- 18.Fuhrman, J. A., and C. A. Suttle. 1993. Viruses in marine planktonic systems. Oceanography 6:51-63. [Google Scholar]
- 19.Hara, S., K. Terauchi, and I. Koike. 1991. Abundance of viruses in marine waters: assessment by epifluorescence and transmission electron microscopy. Appl. Environ. Microbiol. 57:2731-2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hennes, K. P., and C. A. Suttle. 1995. Direct counts of viruses in natural waters and laboratory cultures by epifluorescence microscopy. Limnol. Oceanogr. 40:1050-1055. [Google Scholar]
- 21.Hercher, M., W. Mueller, and H. M. Shapiro. 1979. Detection and discrimination of individual viruses by flow cytometry. J. Histochem. Cytochem. 27:350-352. [DOI] [PubMed] [Google Scholar]
- 22.Jiang, S. C., C. A. Kellogg, and J. H. Paul. 1998. Characterization of marine temperate phage-host systems isolated from Mamala Bay, Oahu, Hawaii. Appl. Environ. Microbiol. 64:535-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kellogg, C. A., J. B. Rose, S. C. Jiang, J. M. Thurmond, and J. H. Paul. 1995. Genetic diversity of related vibriophages isolated from marine environments around Florida and Hawaii, USA. Mar. Ecol. Prog. Ser. 120:89-98. [Google Scholar]
- 24.Larsen, A., T. Castberg, R.-A. Sandaa, C. P. D. Brussaard, J. K. Egge, M. Heldal, A. Paulino, R. Thyrhaug, E. J. Van Hannen, and G. Bratbak. 2001. Population dynamics and diversity of phytoplankton, bacteria, and viruses in a seawater enclosure. Mar. Ecol. Prog. Ser. 221:47-57. [Google Scholar]
- 25.Li, W. K. W., and P. M. Dickie. 2001. Monitoring phytoplankton, bacterioplankton, and virioplankton in a coastal inlet (Bedford Basin) by flow cytometry. Cytometry 44:236-246. [DOI] [PubMed] [Google Scholar]
- 26.Li, W. K. W., J. F. Jellett, and P. M. Dickie. 1995. DNA distributions in planktonic bacteria stained with TOTO or TO-PRO. Limnol. Oceanogr. 40:1485-1495. [Google Scholar]
- 27.Marie, D., C. P. D. Brussaard, R. Thyrhaug, G. Bratbak, and D. Vaulot. 1999. Enumeration of marine viruses in culture and natural samples by flow cytometry. Appl. Environ. Microbiol. 65:45-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marie, D., F. Partensky, S. Jacquet, and D. Vaulot. 1997. Enumeration and cell cycle analysis of natural populations of marine picoplankton by flow cytometry using the nucleic acid stain SYBR Green I. Appl. Environ. Microbiol. 63:186-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marie, D., F. Partensky, D. Vaulot, and C. P. Brussaard. 1999. Enumeration of phytoplankton, bacteria, and viruses in marine samples, p. 11.11.1-11.11.15. In J. P. E. A. Robinson (ed.), Current protocols in cytometry, suppl. 10. John Wiley & Sons, Inc., New York. N.Y. [DOI] [PubMed]
- 30.Noble, R. T., and J. A. Fuhrman. 1998. Use of SYBR Green I for rapid epifluorescence counts of marine viruses and bacteria. Aquat. Microb. Ecol. 14:113-118. [Google Scholar]
- 31.Peduzzi, P., and M. G. Weinbauer. 1993. Effect of concentrating the virus-rich 2- to 200-nm size fraction of seawater on the formation of algal flocs (marine snow). Limnol. Oceanogr. 38:1562-1565. [Google Scholar]
- 32.Proctor, L. M. 1997. Advances in the study of marine viruses. Microsc. Res. Tech. 37:136-161. [DOI] [PubMed] [Google Scholar]
- 33.Sahlsten, E. 1998. Seasonal abundance in Skagerrak-Kattegat coastal waters and host specificity of viruses infecting the marine photosynthetic flagellate Micromonas pusilla. Aquat. Microb. Ecol. 16:103-108. [Google Scholar]
- 34.Steward, G. F., J. L. Montiel, and F. Azam. 2000. Genome size distributions indicate variability and similarities among marine viral assemblages from diverse environments. Limnol. Oceanogr. 45:1697-1706. [Google Scholar]
- 35.Suttle, C. A., and A. M. Chan. 1994. Dynamics and distribution of cyanophages and their effect on marine Synechococcus spp. Appl. Environ. Microbiol. 60:3167-3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suttle, C. A., A. M. Chan, and M. T. Cottrell. 1990. Infection of phytoplankton by viruses and reduction of primary productivity. Nature 347:467-469. [Google Scholar]
- 37.Tarutani, K., K. Nagasaki, and M. Yamaguchi. 2000. Viral impacts on total abundance and clonal composition of the harmful bloom-forming phytoplankton. Appl. Environ. Microbiol. 66:1916-1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Etten, J. L., L. C. Lane, and R. H. Meints. 1991. Viruses and viruslike particles of eukaryotic algae. Microbiol. Rev. 55:586-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Waterbury, J. B., and F. W. Valois. 1993. Resistance to co-occurring phages enables marine Synechococcus communities to coexist with cyanophages abundant in seawater. Appl. Environ. Microbiol. 59:3393-3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weinbauer, M. G., and M. G. Höfle. 1998. Size-specific mortality of lake bacterioplankton by natural virus communities. Aquat. Microb. Ecol. 15:103-113. [Google Scholar]
- 41.Wilhelm, S. W., and C. A. Suttle. 1999. Viruses and nutrient cycles in the sea. Bioscience 49:781-788. [Google Scholar]
- 42.Williamson, S. W., M. R. McLaughlin, and J. H. Paul. 2001. Interaction of the φHSIC virus with its host: lysogeny or pseudolysogeny? Appl. Environ. Microbiol. 67:1682-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilson, W. H., G. Tarran, and M. V. Zubkov. 2002. Virus dynamics in a coccolithophore-dominated bloom in the North Sea. Deep-Sea Res. II 49:2951-2963. [Google Scholar]
- 44.Wommack, K. E., and R. R. Colwell. 2000. Virioplankton: viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 64:69-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wommack, K. E., J. Ravel, R. T. Hill, J. Chun, and R. R. Colwell. 1999. Population dynamics of Chesapeake Bay virioplankton: total-community analysis by pulsed-field gel electrophoresis. Appl. Environ. Microbiol. 65:231-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xenopoulos, M. A., and D. F. Bird. 1997. Virus à la sauce Yo-Pro: microwave-enhanced staining for counting viruses by epifluorescence microscopy. Limnol. Oceanogr. 42:1648-1650. [Google Scholar]