Abstract
Eukaryotic cells can use the autophagy pathway to defend against microbes that gain access to the cytosol or reside in pathogen-modified vacuoles. It remains unclear if pathogens have evolved specific mechanisms to manipulate autophagy. Here we find that the intracellular pathogen Legionella pneumophila could interfere with autophagy using the bacterial effector protein RavZ to directly uncouple Atg8 proteins attached to phosphatidylethanolamine on autophagosome membranes. RavZ hydrolyzed the amide bond between the carboxyl-terminal glycine residue and an adjacent aromatic residue in Atg8 proteins, producing an Atg8 protein that could not be reconjugated by Atg7 and Atg3. Thus, intracellular pathogens can inhibit autophagy by irreversibly inactivating Atg8 proteins during infection.
Legionella pneumophila is an intracellular pathogen that manipulates evolutionarily conserved membrane transport pathways to create a specialized vacuole that supports bacterial replication in host cells (1). Legionella requires a type IV secretion system called Dot/Icm to replicate intracellularly (2, 3). Effector proteins that modulate membrane transport are translocated into host cells by the Dot/Icm system (1, 4).
The autophagy pathway is used by eukaryotic cells to sequester cytosolic proteins and organelles into a membrane-bound compartment called an autophagosome (AP), which fuses with lysosomes to promote cargo degradation (5). Autophagy can be used by plant and animal cells to target intracellular pathogens for degradation in lysosomes (6, 7). An essential step in the autophagy pathway is the coupling of an Atg8 homolog to the lipid phosphatidylethanolamine (PE) on early AP structures (8). The most widely studied Atg8 protein in mammalian cells is microtubule-associated protein light chain 3 (LC3) (9). Unconjugated LC3 (LC3-I) runs more slowly by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) than the lipidated form of LC3 (LC3-II), which means that measuring LC3-II levels by immunoblot analysis provides an indication of autophagy activity in a cell (10).
Here, we measured LC3-II levels during infection of host cells by Legionella to determine if this pathogen modulates autophagy. LC3-II levels were reduced in HEK293 cells infected with a virulent strain of Legionella pneumophila Philadelphia-1 when compared to uninfected cells (Fig. 1A). The block in LC3-II generation was more apparent when degradation of LC3-II was prevented upon treatment of cells with Bafilomycin A1 to neutralize lysosomal pH. LC3-II levels were not affected when cells were infected with an isogenic Legionella dotA mutant (11), which has a non-functional Dot/Icm system (Fig. 1A). Uninfected cells and cells infected with the dotA mutant contained punctate LC3-positive APs resulting from basal levels of autophagy, whereas, punctate LC3-positive APs were absent in most cells infected with virulent Legionella (Fig. 1, B and C). Additionally, LC3-II levels were low in primary macrophages that had been injected by the Dot/Icm system (Fig. 1D and fig. S1) and LC3 puncta were absent in these infected macrophages (Fig. 1, E and F). Thus, Legionella has a Dot/Icm-dependent mechanism to inhibit the autophagy pathway.
Fig. 1. Legionella inhibits autophagy by a Dot/Icm-dependent mechanism.
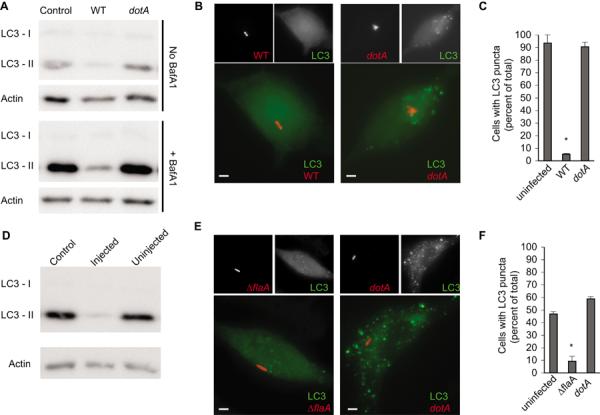
(A) Immunoblot analysis of LC3-I and LC3-II levels in uninfected cells (control) and cells infected for 2-hours with either wild type Legionella (WT) or a dotA mutant. Treatment of cells with Bafilomycin A1 is indicated. (B) Images show LC3 (green) distribution in HEK293 cells infected with the indicated Legionella strains (red) for 2-hours. Scale bar, 1 μm. (C) Graph shows percent of cells containing LC3-puncta calculated from three independent assays where a total of 100 cells were scored in each assay. Data represent the average ± s.d., *p<0.0001 compared to the uninfected control. (D) Mouse bone marrow-derived macrophages were infected for 2-hours with a Legionella ΔflaA strain producing a BlaM∷RalF fusion protein. Immunoblot analysis of was used to detect LC3-I and LC3-II levels in injected and uninjected cells (fig. S1). (E) Representative images of bone marrow-derived macrophages from GFP-LC3 transgenic mice infected with the indicated Legionella strains (red) for 2-hours. Cells were stained using an antibody specific for GFP (green). Scale bar, 1 μm. (F) Graph shows data on GFP-LC3 puncta staining for uninfected and infected mouse bone marrow-derived macrophages. The percent of cells containing GFP-LC3 puncta was calculated from three independent assays where a total of 100 cells were scored in each assay. Data represent the average ± s.d., *p<0.0001 compared to the uninfected control.
Isogenic Legionella strains having large chromosomal deletions were used to determine the genetic basis for autophagy inhibition (12). Autophagy inhibition was not observed after infection with a “pentuple” mutant having five large chromosomal deletions that eliminate 71 putative effectors (Fig. 2A). Autophagy inhibition was also lost in the Δ3 mutant, which is missing only ten of the effectors deleted in the pentuple mutant (Fig. 2A). When these ten effectors were produced individually as GFP-effector fusions in HEK293 cells, only cells producing the GFP-RavZ fusion protein displayed low LC3-II levels (Fig. 2B). GFP-RavZ also prevented the formation of LC3 puncta in transfected HEK293 cells (fig. S2) and interfered with autophagy-dependent degradation of the long-lived protein substrates (Fig. 2C). Thus, RavZ can block autophagy.
Fig. 2. The Legionella effector protein RavZ is necessary and sufficient for autophagy inhibition.
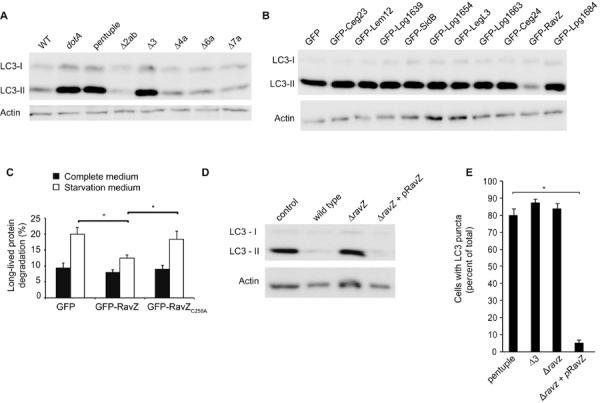
(A) Immunoblot analysis of LC3-I and LC3-II levels in HEK293 cells infected for 2-hours with either wild type Legionella (WT), a dotA mutant, the pentuple mutant, and mutants with single chromosomal deletions (pentuple, Δ2ab, Δ3, Δ4a, Δ6a, and Δ7a). (B) Immunoblot analysis of LC3-I and LC3-II levels in HEK293 cells producing the indicated Legionella effector proteins fused to GFP. (C) Long-term protein degradation was measured in cells producing GFP alone, GFP-RavZ and GFP-RavZC258A. No significant differences were observed for cells grown in complete medium (black bars). An increase in protein degradation resulting from starvation-induced autophagy was observed in cells producing GFP alone or GFP-RavZC258A but cells producing GFP-RavZ displayed a significant defect in autophagy-dependent protein degradation. Data represent average ± s.d. from 3-independent triplicates. *p<0.001. (D) Immunoblot analysis of LC3-I and LC3-II levels in HEK293 cells infected for 2-hours with wild type Legionella (WT), a dotA mutant, a ΔravZ mutant, or the complemented RavZ-deficient strain (ΔravZ + pRavZ). (E) HEK293 cells infected for 2-hours with the indicated strains of Legionella were fixed and stained for LC3. Infected cells were scored for the presence of LC3-positive puncta. The graph shows percent of cells containing LC3-puncta calculated from three independent assays where a total of 100 cells were scored in each assay. Data represent the average ± s.d., *p<0.0001 compared to the pentuple mutant control.
An in-frame deletion of the ravZ gene was constructed in a Legionella strain that contained all other effectors. The ΔravZ mutant and the dotA mutant were equally defective for autophagy inhibition as measured by LC3-II levels (Fig. 2D) and the presence of LC3 puncta in the infected cells (Fig. 2E). Complementation in both assays was achieved using a plasmid-encoded ravZ allele (Fig. 2, D and E). Likewise, introducing ravZ on a plasmid into the Δ3 strain restored autophagy inhibition during infection, whereas, the other nine presumed effectors encoded in the Δ3 region could not (fig. S3). Thus, RavZ is necessary to block autophagy during Legionella infection.
Although AP formation was restored in cells infected with a RavZ-deficient strain of Legionella, LC3-positive membranes were not recruited to vacuoles containing a ΔravZ mutant (fig. S4). The isogenic ΔravZ mutant grew equally well in control macrophages and autophagy-deficient macrophages lacking Atg5 when compared to the parental strain (fig. S4). Thus, vacuoles containing the ΔravZ mutant evade a functional autophagy system, which suggests that Legionella encodes additional effectors capable of disrupting recognition of the vacuole containing Legionella by the autophagy system.
To better understand how RavZ interferes with autophagy, purified RavZ was added to an in vitro assay that reconstitutes conjugation of Atg8 proteins to PE (13). A recombinant human Atg8 homolog called GABARAP-L1 (GR) was conjugated to PE on synthetic liposomes by a reaction requiring ATP and purified Atg7 and Atg3 (Fig. 3A). GR lipidation was strongly inhibited when RavZ was added to this reaction (Fig. 3A). RavZ inhibited GR lipidation when added at concentrations that were over 1000-fold lower than Atg7 and Atg3 (Fig. 3B), indicating that RavZ functions catalytically to interfere with GR lipidation. Thus, RavZ can inhibit autophagy by acting directly in the Atg8 lipidation pathway.
Fig. 3. RavZ functions as a cysteine protease that specifically deconjugates Atg8 proteins from phospholipid membranes.
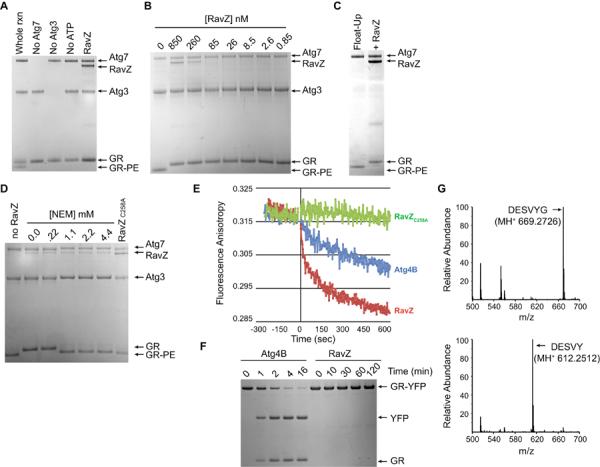
(A) Coomassie-stained SDS-PAGE gel from an in vitro GR conjugation assay containing the conjugating enzymes Atg7 and Atg3, the RavZ protein, the unconjugated GR protein and the lipid-conjugated GR protein (GR-PE). Lanes from the whole reaction (rxn) and reactions missing individual components are marked. The RavZ lane contained all the components of the whole rxn plus purified RavZ protein. (B) Indicated above each lane are the concentrations of purified RavZ added to a conjugation assay containing GR at a concentration of 12 μM. (C) Liposomes containing GR conjugated to PE were isolated on a flotation gradient (Float-Up lane) and treated with RavZ (+RavZ lane). (D) Liposomes containing GR-PE were treated with RavZ in the presence of the indicated concentrations of NEM or were treated with the RavZC258A protein. (E) Fluorescence anisotropy was used to analyze the release kinetics of Texas-Red-labeled GR from liposomes in vitro upon treatment with RavZ (red line), Atg4B (blue line), and RavZC258A (green line). (F) Stained SDS-PAGE gels show that GR-YFP was rapidly cleaved by Atg4B to generate the products GR and YFP. No cleavage of GR-YFP was observed by RavZ after a 120 min reaction. (G) LC-MS/MS chromatographs from samples containing purified GR (top graph) and GR that was isolated after RavZ-mediated deconjugation from PE-containing liposomes (bottom graph). Arrow indicates that the native C-terminal peptide in GR (DESVYG) was abundant in the untreated sample and that RavZ-mediated deconjugation resulted in a loss of the C-terminal glycine (DESVY).
The addition of RavZ to isolated liposomes (fig. S5) drove the complete conversion of lipidated GR-PE back to an apparently unmodified form of GR (Fig. 3C). Likewise, direct addition of RavZ to finished in vitro conjugation reactions also resulted in deconjugation of GR-PE (fig. S6). RavZ delipidated all Atg8 family members tested (fig. S6). Thus, RavZ is a deconjugating enzyme that targets Atg8 proteins covalently attached to PE.
Atg8 proteins are synthesized as inactive pro-forms that contain additional amino acid residues after the conserved reactive glycine located near the C-terminus (5). Atg4 is a cysteine protease that regulates autophagy by cleaving the C-terminus of pro-Atg8 after the conserved glycine, which is required for the glycine to participate in the PE conjugation reaction (5). The protease activity of Atg4 can also release Atg8 proteins from vesicles by reversing the amide bond linking the C-terminal glycine to PE (14). Based on their similar ability to deconjugate Atg8 proteins bound to PE, we asked whether RavZ was functioning as an Atg4 mimic.
The addition of N-ethylmaleimide (NEM) to the GR-PE deconjugation reaction, which inhibits cysteine proteases by covalently modifying reactive cysteine residues, interfered with RavZ-mediated deconjugation of GR in vitro (Fig 3D). Thus, RavZ does appear to use a catalytic mechanism similar to Atg4. We used anisotropy to measure the kinetics of deconjugation of fluorescently-labeled GR attached to PE on liposome membranes. RavZ deconjugated GR-PE rapidly and Atg4B deconjugation occurred more slowly (Fig. 3E). RavZ deconjugated the entire pool of GR-PE within one minute, whereas, a measureable amount of GR-PE was still detected 60-min after the addition of Atg4B (fig. S7). RavZ did not cleave a recombinant GR protein having YFP fused to the native C-terminus (Fig. 3F) or LC3 proteins having extended C-terminal regions (fig. S8), which were all cleaved efficiently by Atg4B in solution. Thus, RavZ and Atg4 have different substrate preferences, with RavZ being highly active on lipid-conjugated Atg8 proteins and having no detectable protease activity toward Atg8 proteins in solution.
Liquid chromatography tandem mass spectrometry (LC-MS/MS) analysis was used to analyze Atg8 reaction products after RavZ treatment. The C-terminal peptide (DESVYG) in GR was detected in untreated samples and also in reactions where GR in solution was incubated with RavZ (Fig. 3G). When GR was isolated after being deconjugated from PE by RavZ in vitro, the C-terminal glycine residue had been removed from the peptide (DESVY) (Fig. 3G, bottom graph). Thus, RavZ targeted the amide bond between the tyrosine and the PE-conjugated glycine residue, unlike Atg4, which targets the amide bond linking glycine to PE (Fig. S9). Thus, RavZ is a deconjugating enzyme that produces an Atg8 product that would be resistant to reconjugation by the host machinery due to the absence of the reactive C-terminal glycine.
The Otubain family of deubiquitinating enzymes are proteases having a reactive cysteine residue presented in a conserved DGNC sequence (15), which closely matched the cysteine residue at amino acid position 258 in RavZ that is presented in the context of an EGNC motif. RavZC258A derivatives were inactive in the in vitro assays that measure GRPE deconjugation (Fig. 3D), in the in vivo assays that measure LC3-II conversion and AP formation by staining LC3 puncta (fig. S10), and in the starvation-mediated long-lived protein degradation assay (Fig. 2D). GFP-RavZ localized to structures that were Atg16 positive and LC3 negative, whereas, GFP-RavZC258A localized structures that were Atg16 negative and LC3 positive (fig. S11). Thus, RavZ can bind early AP membranes, and RavZC258A represents a non-catalytic substrate-trapping mutant that remains associated with Atg8-PE on mature AP membranes.
Here we have found that the Legionella RavZ protein inhibits host autophagy by functioning as a cysteine protease that specifically targets lipid-conjugated Atg8 proteins and generates a deconjugated Atg8 product that lacks the essential C-terminal glycine required for reconjugation. RavZ can both prevent Atg8 from accumulating on the membranes of phagophores that mature into AP structures, and inactivate the Atg8 protein during the deconjugation reaction. Thus, Legionella has evolved a specific mechanism to interfere with host autophagy by directly targeting the Atg8 proteins involved in AP formation.
Supplementary Material
Acknowledgements
We thank N. Mizushima and A. Iwasaki for providing GFP-LC3 and Atg5-deficient mice, and S. Shin for providing the plasmid encoding BlaM-RalF. Support for this research came from the Science, Technology and Research Scholars Program at Yale (BM), Howard Hughes Medical Institute (T.J.O. and R.R.I.), NIH Awards AI007019 (A.C.), NS063973 (T.J.M.), AI041699 and AI048770 (C.R.R.). The data presented in this manuscript are tabulated in the main paper and in the supplementary materials.
Footnotes
An intracellular pathogen disrupts autophagy by targeting an essential host protein on the early autophagosome.
References and Notes
- 1.Hubber A, Roy CR. Modulation of host cell function by Legionella pneumophila type IV effectors. Annu Rev Cell Dev Biol. 2010;26:261–283. doi: 10.1146/annurev-cellbio-100109-104034. [DOI] [PubMed] [Google Scholar]
- 2.Segal G, Purcell M, Shuman HA. Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc Natl Acad Sci U S A. 1998;95:1669–1674. doi: 10.1073/pnas.95.4.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vogel JP, Andrews HL, Wong SK, Isberg RR. Conjugative transfer by the virulence system of Legionella pneumophila. Science. 1998;279:873–876. doi: 10.1126/science.279.5352.873. [DOI] [PubMed] [Google Scholar]
- 4.Ensminger AW, Isberg RR. Legionella pneumophila Dot/Icm translocated substrates: A sum of parts. Curr Opin Microbiol. 2009;12:67–73. doi: 10.1016/j.mib.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 6.Hayward AP, Dinesh-Kumar SP. What can plant autophagy do for an innate immune response? Annu Rev Phytopathol. 2011;49:557–576. doi: 10.1146/annurev-phyto-072910-095333. [DOI] [PubMed] [Google Scholar]
- 7.Kuballa P, Nolte WM, Castoreno AB, Xavier RJ. Autophagy and the immune system. Annu Rev Immunol. 2012;30:611–646. doi: 10.1146/annurev-immunol-020711-074948. [DOI] [PubMed] [Google Scholar]
- 8.Ichimura Y, et al. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- 9.Kabeya Y, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klionsky DJ, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nagai H, Kagan JC, Zhu X, Kahn RA, Roy CR. A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science. 2002;295:679–682. doi: 10.1126/science.1067025. [DOI] [PubMed] [Google Scholar]
- 12.O'Connor TJ, Adepoju Y, Boyd D, Isberg RR. Minimization of the Legionella pneumophila genome reveals chromosomal regions involved in host range expansion. Proc Natl Acad Sci U S A. 2011;108:14733–14740. doi: 10.1073/pnas.1111678108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jotwani A, Richerson DN, Motta I, Julca-Zevallos O, Melia TJ. Approaches to the study of Atg8-mediated membrane dynamics in vitro. Methods Cell Biol. 2012;108:93–116. doi: 10.1016/B978-0-12-386487-1.00005-5. [DOI] [PubMed] [Google Scholar]
- 14.Kirisako T, et al. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol. 2000;151:263–276. doi: 10.1083/jcb.151.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Balakirev MY, Tcherniuk SO, Jaquinod M, Chroboczek J. Otubains: a new family of cysteine proteases in the ubiquitin pathway. EMBO Rep. 2003;4:517–522. doi: 10.1038/sj.embor.embor824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pan X, Luhrmann A, Satoh A, Laskowski-Arce MA, Roy CR. Ankyrin repeat proteins comprise a diverse family of bacterial type IV effectors. Science. 2008;320:1651–1654. doi: 10.1126/science.1158160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berger KH, Isberg RR. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol Microbiol. 1993;7:7–19. doi: 10.1111/j.1365-2958.1993.tb01092.x. [DOI] [PubMed] [Google Scholar]
- 18.Ren T, Zamboni DS, Roy CR, Dietrich WF, Vance RE. Flagellin-deficient Legionella mutants evade caspase-1- and Naip5-mediated macrophage immunity. PLoS Pathog. 2006;2:e18. doi: 10.1371/journal.ppat.0020018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bardill JP, Miller JL, Vogel JP. IcmS-dependent translocation of SdeA into macrophages by the Legionella pneumophila type IV secretion system. Mol Microbiol. 2005;56:90–103. doi: 10.1111/j.1365-2958.2005.04539.x. [DOI] [PubMed] [Google Scholar]
- 20.Merriam JJ, Mathur R, Maxfield-Boumil R, Isberg RR. Analysis of the Legionella pneumophila fliI gene: intracellular growth of a defined mutant defective for flagellum biosynthesis. Infect Immun. 1997;65:2497–2501. doi: 10.1128/iai.65.6.2497-2501.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Case CL, Shin S, Roy CR. Asc and Ipaf Inflammasomes direct distinct pathways for caspase-1 activation in response to Legionella pneumophila. Infect Immun. 2009;77:1981–1991. doi: 10.1128/IAI.01382-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hara T, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 24.Shao Y, Gao Z, Feldman T, Jiang X. Stimulation of ATG12-ATG5 conjugation by ribonucleic acid. Autophagy. 2007;3:10–16. doi: 10.4161/auto.3270. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


