Summary
Alternative oxidase (AOX) is a respiratory oxidase found in certain eukaryotes and bacteria; however, its role in bacterial physiology is unclear. Exploiting the genetic tractability of the bacterium Vibrio fischeri, we explore the regulation of aox expression and AOX function. Using quantitative PCR and reporter assays, we demonstrate that aox expression is induced in the presence of nitric oxide (NO), and that the NO-responsive regulatory protein NsrR mediates the response. We have identified key amino acid residues important for NsrR function and experimentally confirmed a bioinformatically predicted NsrR binding site upstream of aox. Microrespirometry demonstrated that oxygen consumption by V. fischeri CydAB quinol oxidase is inhibited by NO treatment, whereas oxygen consumption by AOX is less sensitive to NO. NADH oxidation assays using inverted membrane vesicles confirmed that NO directly inhibits CydAB, and that AOX is resistant to NO inhibition. These results indicate a role for V. fischeri AOX in aerobic respiration during NO stress.
Keywords: Aliivibrio, Photobacterium, Euprymna scolopes, ES114
Introduction
Alternative oxidase (AOX) is a heme-independent terminal oxidase present in the respiratory chains of all plants (Vanlerberghe & McIntosh, 1997), as well as in certain fungi (Akhter et al., 2003, Umbach & Siedow, 2000), protists (Castro-Guerrero et al., 2004, Czarna & Jarmuszkiewicz, 2005), animals (McDonald & Vanlerberghe, 2004), and bacteria (McDonald & Vanlerberghe, 2005, Stenmark & Nordlund, 2003). Unlike other respiratory oxidases, AOX does not directly contribute to the generation of a proton motive force (PMF) (Vanlerberghe & McIntosh, 1997), although it may contribute to PMF indirectly when coupled with proton-pumping electron transport components. In addition, whereas other respiratory oxidases contain a heme-based reactive center, AOX utilizes a di-iron center (Berthold et al., 2002). With the exception of in thermogenic plants, where AOX is used to generate heat during flowering (Meeuse, 1975), its function is uncertain. In eukaryotes, AOX may contribute to metabolic homeostasis (Hansen, 2002, Lambers, 1982, Rasmusson et al., 2009, Vanlerberghe et al., 2009), protection from oxidative stress (Gupta et al., 2009, Millar et al., 2001, Purvis, 1997, Vanlerberghe et al., 2009, Vanlerberghe & McIntosh, 1997, Wagner & Moore, 1997), and/or virulence (Akhter et al., 2003, Clarkson et al., 1989).
AOX was discovered in bacteria through genome and environmental DNA sequencing projects (McDonald & Vanlerberghe, 2005, Stenmark & Nordlund, 2003), and the number of bacteria predicted to encode AOX continues to grow as more genomes are sequenced. Interestingly, a majority of these bacteria are marine organisms, suggesting that there may be a connection between certain environments and AOX function. Nonetheless, it remains unclear what role AOX plays in bacterial physiology, and its biological function awaits elucidation in a tractable model system.
We discovered an AOX-encoding gene in Vibrio fischeri ES114, a bioluminescent marine bacterium that forms a symbiotic relationship with the Hawaiian bobtail squid Euprymna scolopes (Visick & Ruby, 2006). V. fischeri is metabolically flexible, and it encodes both CydAB and CcoNOQP respiratory oxidases in addition to AOX. In this study, we exploited the genetic tractability of V. fischeri to investigate the regulation of aox and the physiological role of AOX. We determined that aox expression is regulated in response to nitric oxide (NO) by the NO-responsive regulatory protein NsrR, and we provide evidence that AOX constitutes a relatively NO-resistant respiratory oxidase for V. fischeri.
Results
Automated annotation of the ES114 genome sequence (Ruby et al., 2005) identified open reading frame VF_0578 as encoding AOX, and this ORF displays 53% identity over 195 of 349 amino acids to AOX in the voodoo lily (Sauromatum guttatum; accession number S30143). As with other known AOXs (Chaudhuri et al., 1998, Kumar & Soll, 1992), we found that V. fischeri AOX can functionally complement the aerobic growth defect of E. coli SASX41B (Avissar & Beale, 1989), a hemA mutant deficient in heme biosynthesis (data not shown). Presumably, as a heme-independent respiratory oxidase, AOX completes an aerobic electron transport chain in the hemA mutant with PMF generated by upstream components such as substrate-specific dehydrogenases.
Respiratory oxidases present in V. fischeri ES114
In addition to AOX, the V. fischeri genome encodes respiratory oxidases similar to the FixNOQP/CcoNOQP cbb3-type cytochrome oxidase (Cosseau & Batut, 2004, Delgado et al., 1998, Preisig et al., 1996, Thony-Meyer et al., 1994) and the CydAB quinol oxidase (Cotter et al., 1990, Kelly et al., 1990). Characterization of strains lacking one of each of these respiratory oxidases suggests that under normal laboratory aerobic conditions, CydAB is essential for growth, whereas a lack of CcoNOQP or AOX does not affect growth (data not shown). The aerobic growth defect of a cydAB mutant was complemented by introduction of cydAB or aox on a multi-copy plasmid (data not shown). In addition, when cydAB mutant cells were grown anaerobically and shifted to aerobic growth conditions, suppressor mutants frequently arose within 24 hours (32% of the colonies, +/- 4%) that restored wild-type or near wild-type aerobic growth levels. Such suppressor mutants are rarely observed in a cydAB aox double mutant, suggesting that the presence of aox is important for restoring growth in most suppressors. Because AOX is not highly expressed during growth in laboratory medium (see below), one possible explanation for the ability of the suppressor mutants to grow aerobically is that the mutations increase aox expression. Higher aox expression could increase levels of AOX and restore aerobic growth to the cydAB mutant in much the same way that introducing aox on a multi-copy plasmid restored this mutant’s growth (data not shown).
aox mRNA levels and promoter activity increase in response to NO
Microarray analysis revealed that VF_0578 transcript levels increased 82-fold in response to NO (data available under GenBank accession number GSE15522) (Y. Wang, Y.S. Dufour, H.K. Carlson, T.J. Donohue, M.A. Marletta, and E.G. Ruby, submitted), suggesting that AOX is part of a NO-responsive regulon. To confirm the influence of NO on aox transcript levels, we performed quantitative reverse transcriptase PCR (qRT-PCR). Our results demonstrated that under normal growth conditions, transcript levels are relatively low, but they are induced ∼41-fold in response to NO (Table 1). To test whether aox promoter (Paox) activity was also influenced by NO, we constructed both plasmid- and chromosome-based Paox-lacZ transcriptional reporters. Using a plasmid-based reporter that is maintained at ∼10 copies per genome (Dunn et al., 2005), we found that Paox-lacZ activity increased 4-fold in response to NO (Table 1), whereas a 30-fold increase was observed using the chromosome-based reporter strain AKD781 (Table 1). These results show that the aox promoter is up-regulated in response to NO, and the relatively high background expression from the plasmid-based reporter might suggest that a NO-responsive repressor can be titrated by multiple copies of the aox promoter.
Table 1.
Influence of nitric oxide (NO) on aoxtranscript levels and aox promoter activity in V. fischeri
| aox relative transcript levels or promoter activity | |||
|---|---|---|---|
| assay | −NO | +NO | fold-induction |
| qRT-PCR* | |||
| (ES114) | 0.03(.004)** | 1.23(0.14) | 41 |
| β-galactosidase assay*** | |||
| plasmid reporter | |||
| (ES114 pAKD500) | 15(2.2) | 65(22) | 4.3 |
| chromosomal reporter | |||
| (AKD712) | 2.9(0.2) | 86(20) | 30 |
| chromosomal reporter | |||
| (ΔnsrR, AKD785) | 876(88.1) | 735(23.2) | 0.8 |
data presented were calculated using the comparative CT method using the reference gene rpoD
number in parentheses indicates standard error of the mean
β-galactosidase activity is reported in Miller units
To determine whether increases in aox expression in response to NO correlated with increased production of AOX, a strain that expressed a translational fusion between AOX and green fluorescent protein (GFP) was exposed to NO and cellular GFP levels were examined using epifluorescence microscopy. AOX-GFP expression was only observed when cells were treated with NO (data not shown). This result corroborates the transcriptional data above and demonstrates that AOX is up-regulated in response to NO.
NsrR negatively regulates Paox activity
In a previous bioinformatics-based report (Rodionov et al., 2005), a binding site for the NO-responsive repressor NsrR (VF_2315) was predicted upstream of aox in V. fischeri, making NsrR a likely candidate for regulation of Paox activity (Figure 1). To explore this possibility, an in-frame nsrR deletion was placed in both the wild-type strain ES114, generating mutant AKD711, and in the chromosome-based Paox-lacZ reporter strain, generating mutant AKD785. In the absence of NO, Paox-lacZ activity was approximately 290-fold higher in the nsrR mutant (AKD785) relative to the nsrR+ background (AKD712) (Table 1), indicating that NsrR is a negative regulator of Paox activity. Addition of NO did not increase Paox activity in the ΔnsrR background (Table 1), indicating that NsrR is the main NO-responsive negative regulator of aox expression.
Figure 1.
A. DNA sequence upstream of aox in V. fischeri ES114 (ORF VF_0578). The sequence begins with the start codon for the oppositely transcribed gene (VF_0577) and ends with the start codon for VF_0578, which are marked with bold text. The putative NsrR binding site is underlined, with arrows indicating the inverted repeats. The mapped transcriptional start site is indicated in bold lower case. B. The putative NsrR binding site in wild type is compared to base pair changes introduced via site-directed mutagenesis (indicated by underlined lower case bold font) and insertions found in suppressor mutants (underlined bold font). These modified sequences were used in the in vivo LacZ-based binding assays (Figure 2).
Suppressor mutations compensating for poor aerobic growth of the cydAB mutant map to nsrR or the aox promoter region
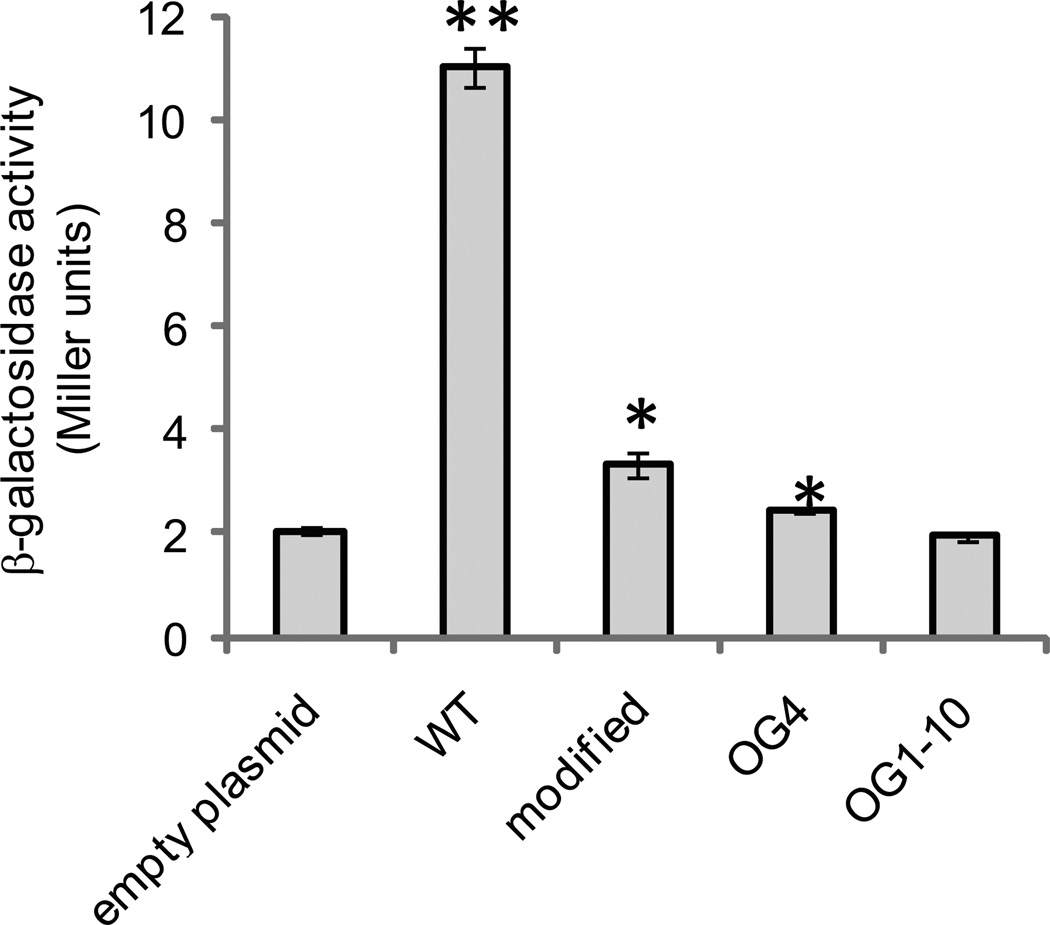
As described above, a cydAB mutant has an aerobic growth defect, but suppressor mutants arise that have substantially restored growth. To test whether the suppressor mutations influenced aox transcription, we constructed strain AKD783, which contains the chromosomal Paox-lacZ reporter in a ΔcydAB background. Suppressor mutants in this strain were tested for increased Paox activity by plating on medium containing 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-gal). Ninety six percent (195 of 204) of suppressor mutants detectably expressed β-galactosidase, whereas this reporter was undetectable in the wild-type background. To determine whether aox expression in the suppressors correlated with changes in nsrR, we sequenced nsrR in a random subset of the suppressor mutants. Twelve of the fourteen suppressors had mutations in nsrR that resulted in (i) premature translational termination, (ii) single amino acid changes, or (iii) a duplication of residues (Table 2). Premature termination resulted from GAA to TAA changes in three mutants, and the single amino acid changes varied in location and effect, including replacement of a cysteine likely required for [Fe-S] center coordination with either tyrosine or phenylalanine. For two mutants (OG4 and OG1-10), the nsrR coding and promoter sequences were identical to wild type. However, when we sequenced the DNA upstream of the coding region of aox in these two mutants, in each strain we identified a single bp insertion in the putative NsrR binding site (Table 2 and Figure 1).
Table 2.
Modifications to nsrR or the aox promoter regions in suppressor mutants that compensate for the aerobic growth phenotype of the cydAB mutant
| Suppressor mutant | Identified modification |
|---|---|
| OG1 | nsrR: 24 bp (8 amino acid) duplication (amino acids 60–67) |
| OG2 | nsrR: amino acid 96: C to Y (TGC to TAC) |
| OG3 | nsrR: amino acid 63: G to D (GGC to GAC) |
| OG4 | upstream of aox: insertion of a basepair (Fig. 1) |
| OG1-1 | nsrR: premature stop amino acid 52 (GAA to TAA) |
| OG1-2 | nsrR: amino acid 6: F to L (TTC to TTA) |
| OG1-3 | nsrR: amino acid 111: A to E (AAA to GAA) |
| OG1-5 | nsrR: amino acid 96: C to F (TGC to TTC) |
| OG1-6 | nsrR: amino acid 12: R to I (AGA to ATA) |
| OG1-7 | nsrR: premature stop amino acid 94 (GAA to TAA) |
| OG1-9 | nsrR: amino acid 111: A to E (identical to OG1-3) |
| OG1-10 | upstream of aox: insertion of a base pair (Fig. 1) |
| OG1-11 | nsrR: premature stop amino acid 94 (identical to OG1-7) |
Characterization of the putative NsrR binding site upstream of aox
To determine whether NsrR binds to the predicted site upstream of aox (Rodionov et al., 2005), we utilized a plasmid-based repressor titration assay similar to that used previously in characterizing NsrR binding in Escherichia coli (Bodenmiller & Spiro, 2006). Approximately 200 bp upstream of the translational start of aox were cloned from wild type or the suppressor mutants (OG4 and OG1-10) into plasmid pVSV209. We also cloned a fragment with four site-directed bp substitutions relative to wild type (Figure 1). These plasmids and the pVSV209 control were each introduced into AKD781, which contains the chromosomal Paox-lacZ reporter. As demonstrated in Figure 2, the plasmid containing the wild-type sequence can alleviate repression of Paox activity, consistent with titration of NsrR. However, plasmids with the modified putative NsrR binding sites do not appear to effect NsrR-mediated repression of Paox-lacZ (Figure 2). These data suggest that the inverted-repeat sequence is important for NsrR binding to the aox promoter region, as predicted. Moreover, we mapped the transcriptional start site of aox (Figure 1) to a region overlapping the putative NsrR binding site, further supporting the plausibility that NsrR acts as a negative regulator of aox expression.
Figure 2.
In vivo assay for NsrR binding to the aox promoter region. β-galactosidase assays were performed on AKD712 (chromosomal aox-lacZ fusion strain) containing either an empty control plasmid (pAKD700), pAKD700 containing approximately 200 bp of the DNA sequence upstream of the start codon encoded by aox (WT; pAKD750), pAKD700 containing a PCR-modified aox promoter region (modified; pAKD751), and pAKD700 containing the aox promoter region PCR-amplified from either suppressor mutant OG4 or OG1-10. Sequences are shown in Fig. 1. Plasmids are maintained at ∼10 copies per chromosome, and when they contain an NsrR binding site will effectively titrate out NsrR, relieving repression of aox transcription and resulting in production of LacZ. Cells were grown to mid-log phase in mineral-salts medium containing GlcNAc prior to harvesting for the assay. Data are the average of three independent cultures from one representative experiment. Error bars indicate standard error of the mean. The experiment was repeated three times. Asterisks indicate significant differences in mean activity compared to the pAKD700 control as determined using a student’s t-test. ** p < 0.01; * p<0.05
Reduced activity of NsrR encoded by suppressor mutants
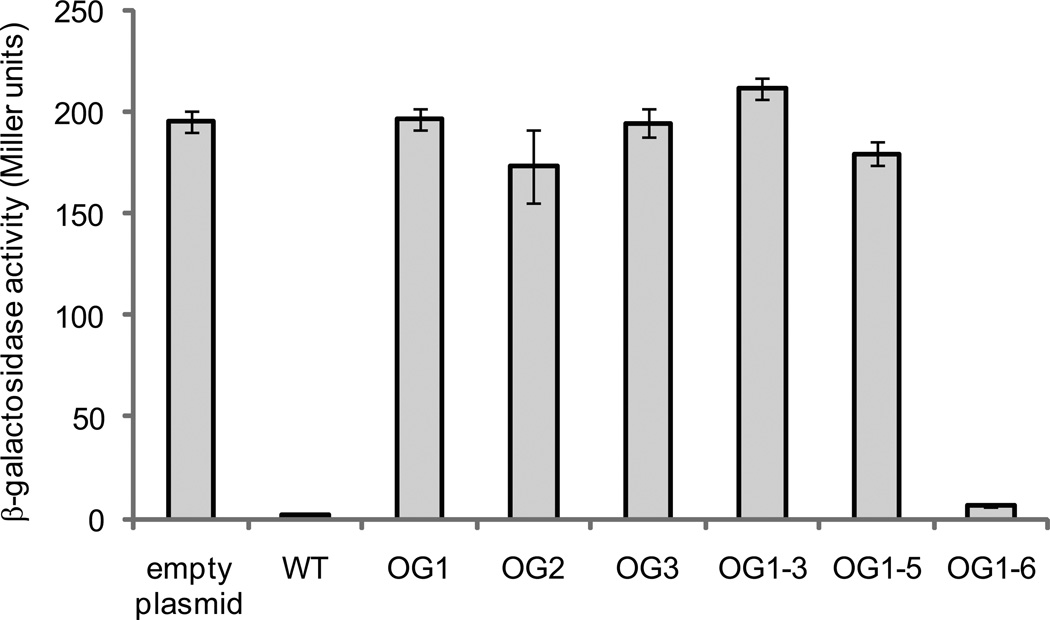
We considered the possibility that the suppressor mutants described above could contain unidentified mutations affecting Paox activity. Therefore, to test whether mutations in nsrR corresponded to the increase in Paox activity, we cloned wild-type nsrR and six unique nsrR variants into pAKD601B, placing nsrR expression under control of an isopropyl-β-D-thiogalactopyranoside (IPTG)-inducible promoter. These plasmids were then introduced into AKD712 (ΔnsrR, chromosomal Paox-lacZ), and cultures were grown with IPTG and assayed for β-galactosidase activity. Expression of wild-type NsrR in this strain repressed Paox-lacZ (Figure 3); however, for all but one of the NsrR variants, little to no repression of Paox activity was observed (Figure 3). In the one exception, strain OG1-6, the nsrR mutation either does not cause derepression of aox, or it has partial activity such that when this allele is on a multi-copy plasmid it can repress aox.
Figure 3.
In vivo assay for NsrR activity. β-galactosidase assays were performed on AKD785 (nsrR mutant, chromosomal aox-lacZ fusion strain) in which NsrR variants were overexpressed. nsrR genes from wild type and ΔcydAB suppressor mutants were cloned into pAKD601B, placing expression under the control of an IPTG-inducible promoter. Cells were grown in LBS medium overnight in the presence of kanamycin and IPTG before harvesting for the assay. High levels of β-galactosidase activity indicate a non-functional version of NsrR that can no longer bind to the aox promoter region and repress expression of lacZ. Bars are labeled with the name of the suppressor mutant from which the variant nsrR was isolated, along with the wild-type (WT) and empty plasmid controls. Data are the average of three independent cultures from one representative experiment. Error bars indicate standard error of the mean. Asterisks indicate significant differences in mean activity compared to the wild-type control as determined using a student’s t-test (p < 0.01). The experiment was repeated three times with similar results.
Influence of AOX on oxygen consumption during NO stress
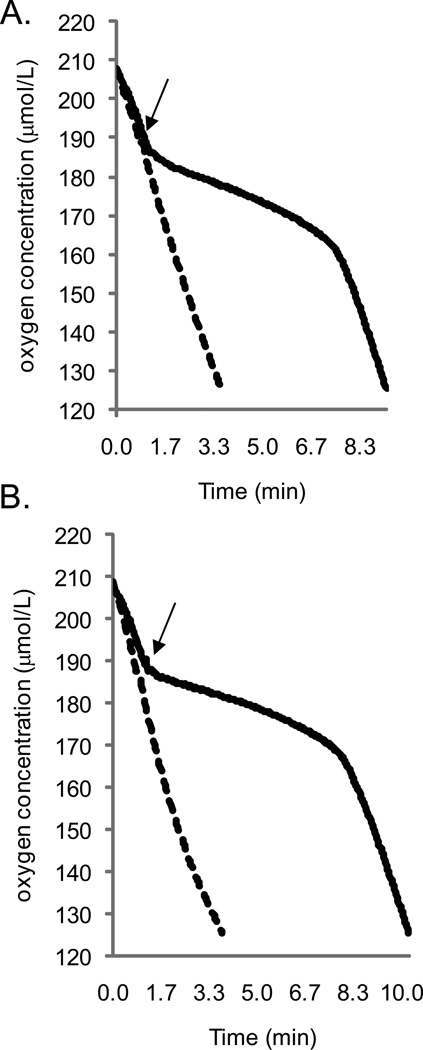
Using microrespirometry, we measured oxygen consumption patterns before, during, and after NO treatment of wild-type V. fischeri, as well as strains lacking aox and/or nsrR. NO treatment inhibited oxygen consumption of wild type, but oxygen consumption in the nsrR mutant AKD711 was not affected by NO (Figure 4A). Thus, respiration appeared to be resistant to NO when the NsrR regulon was derepressed. The aox mutant (AKD780) had an oxygen consumption profile indistinguishable from that of wild type (data not shown), and this was observed under varying conditions (NO levels, oxygen tension at NO delivery, culture density, etc.). In addition, the nsrR aox double mutant, which would have all members of the NsrR regulon excluding aox derepressed, did not display an oxygen consumption profile distinct from the nsrR mutant, AKD711 (data not shown). These data could indicate that AOX does not contribute to oxygen consumption following NO challenge; however, it is possible that the experimental conditions do not allow us to detect such a contribution of AOX. For example, other predicted members of the NsrR regulon such as Hmp (Rodionov et al., 2005) may detoxify NO and thereby protect heme-dependent oxidases (Stevanin et al., 2000), obscuring a contribution by AOX in the nsrR mutant background.
Figure 4.
Oxygen consumption profiles for V. fischeri strains demonstrating the response to treatment with 80 µM DEA NONOate. Cells were grown in mineral salts medium containing GlcNAc to mid-log phase and placed in the respirometer chamber. DEA NONOate was injected into the chamber at an oxygen concentration of 190 µmol/L (indicated by the arrow). A. Profiles for wild type (solid line; OD600 of 0.310, total protein content of 1.29 mg) and the ΔnsrR strain AKD711 (dashed line; OD600 of 0.304, total protein content of 1.28 mg). The oxygen consumption profiles for AKD780 (aox mutant) and AKD786 (aox nsrR double mutant) were not consistently different from those of the wild type and nsrR mutant, respectively (data not shown). B. Profiles for AKD788 (expressing only CydAB; solid line; OD600 of 0.299, total protein content of 1.25 mg) and AKD789 (expressing only AOX; dashed line; OD600 of 0.311, total protein content of 1.27 mg). Graphs demonstrate oxygen consumption profiles for one set of cultures and are representative of three independent experiments.
To determine whether AOX can consume oxygen in the presence of NO, we utilized suppressor mutant OG1-10. This strain contains a single bp insertion upstream of aox that disrupts the putative NsrR binding site, uncoupling AOX expression from NsrR control and resulting in NO-independent aox expression. We deleted ccoNOQP in OG1-10, which already lacks cydAB, to eliminate possible contributions of respiratory oxidases other than AOX. The ability of the resulting strain (AKD789) to consume oxygen in the presence of NO was assessed, and compared to a strain only expressing CydAB (AKD788), which has an oxygen consumption profile similar to wild type. We found that AOX can consume oxygen in the presence of NO (Figure 4B), whereas oxygen consumption by CydAB is sensitive to NO. Our results also suggest that CcoNOQP does not contribute to measurable oxygen consumption by V. fischeri under these conditions.
Interestingly, oxygen consumption by CydAB does not appear to be fully inhibited by the addition of 80 µM DEA-NONOate (Figure 4B) under these conditions. Doubling the amount of DEA-NONOate added yielded similar results, although it did increase the duration of inhibition (data not shown). For comparison, a study of E. coli CydAB indicated that at similar oxygen tensions, the half-maximal inhibitory concentration of NO was ∼400 nM (Mason et al., 2009). Although our experiments were not identical to those used by Mason et al., their results support the idea that V. fischeri CydAB is relatively resistant to inhibition by NO.
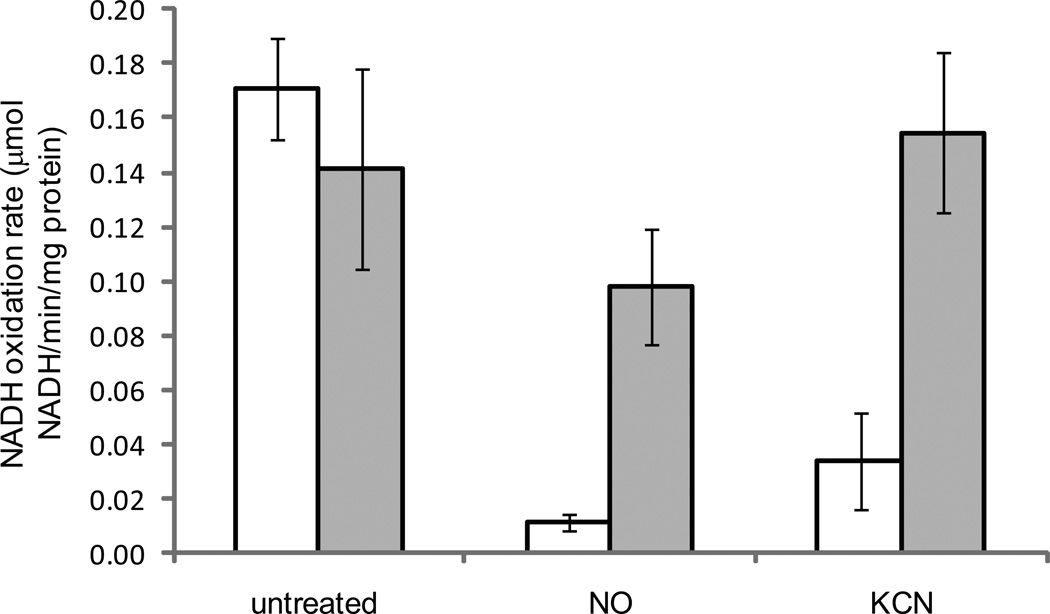
CydAB is directly inhibited by NO
The experiments described above indicate that NO inhibits CydAB-dependent oxygen consumption, but they do not distinguish whether NO inhibits production of respiratory substrates or CydAB itself. To address these possibilities, we performed NADH oxidation assays using inverted membrane vesicles from AKD788 (CydAB expression only) and AKD789 (AOX expression only). We found that NO inhibits activity of CydAB, similarly to potassium cyanide (KCN), whereas AOX is resistant to inhibition by NO or KCN (Figure 5). Although there is a somewhat lower NADH oxidation rate for AKD789 treated with NO, the difference is not significant (p>0.05).
Figure 5.
NADH oxidation assays using inverted membrane vesicles from strains AKD788 (only expressed terminal oxidase CydAB; white bars) and AKD789 (only expressed terminal oxidase AOX; gray bars). Vesicles were prepared from aerobically grown log-phase cells and assayed at 23 °C. NADH oxidation rate is reported as µmol NADH oxidized per minute, per mg of protein. Samples were either not treated with exogenous chemicals (untreated), treated with 80 µM DEA NONOate (NO), or 3 mM potassium cyanide (KCN). Data presented are the average rates from three independent sets of cultures from two separate experiments performed on different days. Error bars represent standard error of the mean. Asterisks indicate statistically significant differences (p < 0.002) from the corresponding untreated samples as determined using a student’s t-test
Discussion
This study represents the first description of a defined regulatory response of bacterial aox expression to a specific environmental condition, and connects this regulatory response to a physiological role in V. fischeri.
Our results demonstrate that aox is induced by NO and that this response is mediated by NsrR. Previous studies have revealed important characteristics of NsrR’s activity and interactions with DNA, relying on targeted amino acid or binding site modifications or in vitro mutagenesis of binding sites (Isabella et al., 2009, Partridge et al., 2009). We found that mutations affecting NsrR-mediated repression of aox conferred robust aerobic growth in a cydAB mutant, and we exploited this selection to identify key elements of NsrR and its binding site. This approach revealed both predicted and unexpected mutations affecting NsrR function. For example, substitutions that eliminated cysteines thought to be used for coordination of [Fe-S] cluster(s) (Isabella et al., 2009, Tucker et al., 2008, Yukl et al., 2008) resulted in NsrR variants that no longer repressed aox promoter activity. On the other hand, the relevance of the other amino acid substitutions in NsrR that disrupt its function are not as clear. A DNA-binding helix-turn-helix motif has been predicted for NsrR from other organisms (Tucker et al., 2010); however, the amino acid substitutions obtained in that analysis are not found in the corresponding region of V. fischeri NsrR (amino acids 26–47). Our ability to select non-functional NsrR variants, as well as binding site variants, in V. fischeri should continue to provide valuable insight into NsrR structure, function, and DNA binding, particularly once they are combined with in vitro DNA binding assays.
Our results have also provided important new insights into the regulation of aox and AOX function in V. fischeri, identifying a link between NO and AOX. Previous work by others demonstrated that NO can inhibit aerobic respiration by heme-containing oxidases in numerous organisms, including plants (Millar & Day, 1996) and bacteria (Borisov et al., 2004, Hori et al., 1996, Yu et al., 1997). In contrast, AOX proteins contain a di-iron reactive center rather than heme (Berthold et al., 2002), and AOX from plants is resistant to NO inhibition of respiration (Millar & Day, 1996). Our results using microrespirometry confirm that at relatively high oxygen tensions (190 µM), NO can inhibit oxygen consumption by CydAB in V. fischeri, whereas V. fischeri AOX appears to be resistant to NO (Figure 4). Interestingly, it was recently reported in E. coli that cytochrome bd (CydAB) has a lower sensitivity to inhibition by NO, in addition to a lower Km for oxygen, than cytochrome bo, and therefore plays a role in protecting cells from NO stress at low oxygen tensions, a condition where NO detoxification systems are predicted to be least efficient (Mason et al., 2009). It is possible that V. fischeri CydAB is not similarly NO-resistant, and that AOX is performing the role of protecting cells from NO stress at low oxygen tensions. Future work will explore the effect of NO on oxygen consumption of V. fischeri oxidases at lower oxygen tensions.
The results of this study connect the regulatory response of aox expression to a physiological role for NO-resistant oxygen consumption, but raise the question of how this relates to conditions V. fischeri encounters in the environment. V. fischeri can be found free living or associated with a eukaryotic host such as E. scolopes (Visick & Ruby, 2006). During colonization of E. scolopes, V. fischeri encounters host-produced NO (Davidson et al., 2004), suggesting a possible role for AOX during host colonization. However, strains lacking aox are fully competent for host colonization (data not shown). Moreover, we placed a gene encoding green fluorescent protein under control of the aox promoter and did not see induction of this reporter in symbiotic cells (data not shown), although we may have missed such expression if it were in a specific time and microniche in the symbiosis. These results suggest that AOX is either not required for host colonization, or the contributions of AOX to strain fitness during colonization cannot be measured using these assays (e.g. in the time frame of the experiments). Alternatively, AOX may be required for a lifestyle outside of the host, and many of the marine bacteria whose genomes encode AOX were isolated as planktonic organisms with no reported interactions with eukaryotic organisms. One possible source of NO in the environment is denitrifying bacteria, which can produce NO that affects surrounding microbes (Choi et al., 2006). Future work will attempt to identify how AOX activity benefits V. fischeri fitness, which may help answer the question of the environments in which AOX contributes to bacterial growth and survival.
The results of this study also raise the question of how aox expression and AOX function in V. fischeri relates to other aox-containing bacteria. The only other published report of AOX activity in bacteria focused on protein-expression levels in Novosphingobium aromaticivorans in response to culture conditions, but did not test the influence of NO on AOX protein levels (Stenmark & Nordlund, 2003). Future work will explore whether aox-containing bacteria regulate aox expression similarly, with corresponding physiological outcomes. Such studies will provide insight into the evolutionary history and ecological relevance of this interesting protein.
Experimental procedures
Reagents
All restriction enzymes and Quick ligase were purchased from New England Biolabs. The nitric oxide (NO) generator dietheylamine (DEA) NONOate was purchased from Cayman Chemical. DEA NONOate dissociates into NO in a pH-dependent first-order process with a half-life of approximately 16 minutes at 22–25 °C and pH 7.4, liberating 1.5 moles of NO per mole of parent compound. Stock solutions were made immediately before use by dissolving DEA NONOate in 0.01 N NaOH. PCR was performed using KOD Hifi DNA polymerase (Novagen) or Phusion DNA polymerase (New England Biolabs) and manufacturer suggested conditions. Primers were synthesized by Integrated DNA Technologies and primer sequences are available upon request. Plasmids were purified using the Zyppy™ Plasmid Miniprep Kit (Zymo Research). Chromosomal DNA was purified using the Invitrogen Easy-DNA™ kit using protocol #3 in the provided manual. All PCR-amplified DNA fragments were verified by sequencing at the University of Michigan DNA Sequencing Core Facility or the Oklahoma Medical Research Foundation DNA Sequencing Center.
Growth media
V. fischeri strains were grown at 28 °C (unless noted) in either LBS (Stabb et al., 2001), SWT (Boettcher & Ruby, 1990) wherein seawater was replaced with Instant Ocean (Aquarium Systems) or mineral salts medium (per liter: 378 µl 1M NaPO4 (pH 7.5), 50 ml 1M Tris (pH 8.0), 6 mg FeSO4-7H2O, 13.6 g MgSO4-7H2O, 0.83 g KCl, 19.5 g NaCl, 1.62 g CaCl2-2H2O) containing either 1.1 g l−1 N-acetyl glucosamine (GlcNAc) or 40 mM glycerol, one g l−1 vitamin-free casamino acids (Difco) and 0.59 g l−1 NH4Cl as carbon and nitrogen sources. When indicated, 100 µg ml−1 kanamycin or 2 mM IPTG were added to the culture medium. E. coli strains were grown at 37 °C in LB medium (Miller, 1992) with the 40 µg ml−1 kanamycin or 60 µg ml−1 ampicillin or 22.5 g l−1 HiVeg special infusion (HIMEDIA) containing 150 µg ml−1 erythromycin.
Strains and plasmids
Strains and plasmids used in this study are listed in Table 3. General cloning procedures were used in E. coli strains DH5α (Hanahan, 1983) or DH5αλpir (Dunn et al., 2005), and constructs containing the RP4 origin of transfer were introduced into V. fischeri using triparental mating (Stabb & Ruby, 2002). All Vibrio plasmids used in this study contain the pES213 origin of replication and are maintained at approximately 10 copies per chromosome (Dunn et al., 2005).
Table 3.
Strains and plasmids used in this study
| Characteristicsa | Source | |
|---|---|---|
| Strains | ||
| Vibrio fischeri | ||
| ES114 | wild-type V. fischeri | (Boettcher & Ruby, 1990) |
| AKD711 | ΔnsrR (allele exchanged from pAKD711 into ES114) |
this study |
| AKD712 | ΔnsrR, aox-lacZ chromosomal transcriptional fusion (exchanged from pAKD712 into AKD711) |
this study |
| AKD780 | Δaox (allele exchanged from pAKD780 into ES114) |
this study |
| AKD781 |
aox-lacZ chromosomal transcriptional fusion reporter (allele exchanged from pAKD712 into ES114) |
this study |
| AKD782 | ΔcydAB (allele exchanged from pAKD782 into ES114) |
this study |
| AKD783 | ΔcydAB, aox-lacZ chromosomal transcriptional fusion (exchanged from pAKD712 into AKD782) |
this study |
| AKD784 | Δaox ΔcydAB (allele exchanged from pAKD782 into AKD780) |
this study |
| AKD785 | ΔnsrR with aox-lacZ chromosomal transcriptional fusion reporter (allele exchanged from pAKD712 into AKD711) |
this study |
| AKD786 | Δaox ΔnsrR (allele exchanged from pAKD711 into AKD780) |
this study |
| AKD787 | ΔccoNOQP (allele exchanged from pAKD787 into ES114) |
this study |
| AKD788 | Δaox ΔccoNOQP (allele exchanged from pAKD787 into AKD780) |
this study |
| AKD789 | Suppressor mutant OG1-10 ΔccoNOQP (allele exchanged from pAKD787) | this study |
| Escherichia coli | ||
| DH5α | F’/endA1 hsdR17 glnV44 thi-1 recA1 gyrA relA1 Δ(lacIZYA-argF)U169 deoR (φ80dlacIΔ(lacZ)M15) |
(Hanahan, 1983) |
| DH5αλpir | λpir derivative of DH5α | (Dunn et al., 2005) |
| SASX41B | Hfr(PO2A), hemA41, relA1, spoT1, metB1 rrnB-2, mcrB1, creC510 |
E. coli genetic stock center |
| Plasmids | ||
| pVSV103 |
E. coli-V. fischeri shuttle vector, RP4 oriT pES213 and R6Kγ replication origins, lacZ, kanR |
(Dunn et al., 2006) |
| pVSV104 |
E. coli-V. fischeri shuttle vector, RP4 oriT pES213 and R6Kγ replication origins, lacZα, kanR |
(Dunn et al., 2006) |
| pVSV209 |
E. coli-V. fischeri shuttle vector, RP4 oriT pES213 and R6Kγ replication origins, promoterless gfp and chmR, constitutive rfp, kanR |
(Dunn et al., 2006) |
| pAKD601B | PCR-constructed derivative of pAKD600 this study (Dunn & Stabb, 2008) lacking gfp, contains lacIq and an IPTG-inducible promoter for overexpression of proteins in V. fischeri, kanR |
this study |
| pAKD601Baox | pAKD601B with the aox coding region | this study |
| pAKD602 | pVSV104 containing the aox coding region and the upstream intergenic region (Figure 1). |
this study |
| pAKD700 | pVSV209 digested with StuI and SanDI to remove gfp and chmR which was replaced with a StuI/SanDI lacZ fragment (lacZ PCR-amplified from pVSV103) |
this study |
| pAKD711 | ΔnsrR allele, R6Kγ and ColE1 replication origins, RP4 oriT, ermR, kanR |
this study |
| pAKD712 | aox-lacZ transcriptional fusion allele, R6Kγ and ColE1 replication origins, RP4 oriT, ermR, kanR |
this study |
| pAKD750 | pAKD700 with a 200 bp aox promoter region | this study |
| pAKD751 | pAKD750 PCR-amplified to introduce NsrR binding site modifications |
this study |
| pAKD780 | Δaox allele, R6Kγ and ColE1 replication origins, RP4 oriT, ermR, kanR |
this study |
| pAKD782 | ΔcydABybgTE allele, R6Kγ and ColE1 replication origins, RP4 oriT, ermR, kanR | this study |
| pAKD787 | ΔccoNOQP allele, R6Kγ and ColE1 replication origins, RP4 oriT, ermR, kanR |
this study |
| pEVS104 | conjugative helper plasmid, R6Kγ replication origin, kanR |
(Stabb & Ruby, 2002) |
abbreviations used: chmR, chloramphenicol resistance (cat); kanR, kanamycin resistance (aph); ermR, erythromycin resistance; rfp = red fluorescent protein
V. fischeri mutants
Strains lacking aox (VF_0578), nsrR (VF_2315), and the cydABybgTE (VF_0953-6; elsewhere shortened to cydAB) or ccoNOQP (VF_1299–1302) operons were constructed using allelic exchange (Bose et al., 2008), resulting in in-frame deletion mutants. Briefly, approximately 1.6 kb of DNA upstream of each target was PCR-amplified and fused to an approximately 1.6-kb DNA fragment downstream of each gene or operon using an engineered restriction site. The PCR products included the respective start and stop codons and in certain cases additional flanking codons, resulting in replacement of a majority of the gene or operon with a 6-bp restriction enzyme recognition site. Additional codons were included as necessary for primer design and included two codons prior to the stop in the Δaox construct, three codons after the start and three prior to the stop in the ΔnsrR construct, one codon prior to the stop codon in the ΔcydAB construct, and one codon after the start in the ΔccoNOQP construct.
Plasmid-based Paox-lacZ transcriptional reporter
The plasmid-based reporter pAKD500 was constructed by PCR amplifying approximately 200 bp upstream of the start codon encoded by aox, and directionally cloning this fragment into pVSV209. The fragment corresponds to the intergenic region between aox and the divergently transcribed upstream gene and includes the NsrR binding site.
Chromosomal aox-lacZ transcriptional fusion reporter
The chromosomal reporter was constructed using allelic exchange as described above, with a slight modification. An approximately 1.6 kb fragment terminating at the stop codon of aox was PCR amplified and fused to an approximately 1.6 kb fragment beginning with the sequence immediately downstream of the stop codon of aox. The PCR primers were engineered to introduce a restriction site between the two fragments. lacZ and the corresponding ribosome binding site were PCR-amplified from pVSV103 using primers with engineered restriction sites. The lacZ fragment was inserted into the restriction site between the previously fused PCR products, generating an allele with aox transcriptionally fused to lacZ. This plasmid was then used for allelic exchange .
Quantitative reverse transcriptase PCR
Overnight cultures of V. fischeri were grown in three ml of LBS in 18-mm tubes with agitation. The next morning, 10 µl was subcultured into three ml of ASWT and grown to an OD600 of approximately 0.3, at which point 0.75 ml was added to 15 ml of mineral salts medium containing GlcNAc in a 125-ml Erhlenmeyer flask. This culture was grown to an OD600 of approximately 0.3, and 10 ml of culture was added to a 50-ml screw cap plastic conical tube. Two conical tubes were prepared per culture. To one tube, DEA NONOate was added to a final concentration of 80 µM, while the other tube was treated with a corresponding amount of 0.01 N NaOH as a control. Samples were incubated at 28 °C for 30 minutes with shaking. Twenty ml of RNAprotect Bacteria Reagent (QIAGEN) was added to each tube and tubes were incubated for five min at room temperature. Samples were pelleted by centrifugation at 5000 x g for ten minutes at 4 °C in an Eppendorf 5810 R benchtop centrifuge. Supernatants were discarded and pellets frozen at −20 °C for no longer than 16 hours. Total RNA was prepped using the QIAGEN RNeasy Mini Kit following the manufacturer’s protocol for enzymatic lysis of bacteria using lysozyme. Residual DNA was removed by using the QIAGEN RNase-free DNase Set, with both on- and off-column digestion steps performed. cDNA was prepared for each sample from one µg of purified RNA using the manufacturer’s recommended conditions in the SYBR GreenER Two-Step qRT-PCR Kit (Invitrogen). qPCR was performed using an Applied Biosystems 7300 real time PCR machine following the qRT-PCR kit manufacturer’s recommended reaction conditions using two µl of cDNA and 58° C as the annealing/extension temperature. Primers were designed to target an internal fragment of aox or rpoD (VF_2254) as a normalizing control. PCR efficiencies were similar for both targets. Data presented are relative quantitation values calculated using the comparative CT method and rpoD as the reference gene control. qRT-PCR data presented is the average of four independent runs using RNA isolated from four independent experiments.
β- galactosidase assays of aox promoter activity
The chromosomal transcriptional aox-lacZ fusion strain AKD712 or ES114 containing pAKD750 was grown as described above for quantitative PCR, however, six ml of culture was added to six ml of mineral salts medium containing GlcNAc in a 15-ml screw-cap conical tube. Samples were treated with DEA NONOate as described above, and cells harvested by pelleting at 5000 x g for 5 minutes. Supernatant was discarded and pellets frozen at −20 °C for no longer than 24 hours. Cell pellets were then resuspended in one ml of Z buffer and β-galactosidase assays performed using a modified Miller assay as previously described (Bose et al., 2008). For each experiment three independent cultures of each strain were assayed, and each experiment was repeated at least three times. Data shown is from one representative experiment.
For in vivo titration assays for NsrR binding utilizing pVSV209 derivatives, cultures were grown overnight in LBS medium containing kanamycin, and 60 µl of this culture was added to 15 ml of mineral salts medium containing GlcNAc in a 125-ml Erhlenmeyer flask. Cultures were grown at 28° C with shaking until reaching an OD600 of approximately 0.45, at which point 10 ml of culture was harvested as described above.
For in vivo titration assays for NsrR binding utilizing pAKD601B derivatives, single colonies were used to inoculate 3 ml of LBS containing kanamycin and IPTG, and after approximately 16 hours of growth at 28° C, one ml of culture was harvested by pelleting for β-galactosidase assays.
Isolation and characterization of ΔcydAB suppressor mutants
AKD783 (ΔcydAB, aox-lacZ chromosomal fusion) was streaked from −80° C glycerol freezer stocks onto LBS medium and placed in a BD GasPak EZ anaerobic system (Becton, Dickinson, and Company) overnight at 28° C. The following morning one colony was suspended in 100 µl of LBS and 50 µl of the suspension was injected into a sealed tube containing 3 ml of anaerobic LBS. Tubes were prepared by adding medium, sealing with butyl rubber septum-like stoppers (Bellco) and aluminum seals (Wheaton), and sparging for 7 minutes with 100% N2 gas prior to autoclaving.
After incubation at 28° C with shaking for 24 hr, cultures were dilution plated onto LBS agar and incubated anaerobically using the GasPak system for 24 hr at 28° C. The number of colonies were counted and plates were placed at atmospheric oxygen tensions for 24 hr at 24° C. After anaerobic incubation colonies were small and clear in appearance. Suppressor mutants were identified by large, yellow outgrowth from the small clear colonies. The outgrowth colonies grow similarly to wild-type V. fischeri, which displays vigorous yellow colonies on LBS medium after aerobic growth. The number of outgrowth colonies was counted and colonies were patched to LBS medium containing 100 µg ml−1 X-gal (Research Products International) and incubated for approximately 14 hr at 28° C. Blue patches indicate strains in which Paox activity is no longer repressed. AKD781 was used as a control for Paox repression.
Suppressor mutant strains were characterized by PCR amplification and sequencing of nsrR or the promoter region of aox. nsrR fragments were cloned into pJet1.2, using the CloneJET™ system (Fermentas) and sequenced. For in vivo titration assays for NsrR binding, nsrR sequences were PCR amplified using primers targeting the coding sequence and cloned into pAKD601B. aox promoter regions were cloned into pVSV209 and sequenced. These plasmids were used in the in vivo NsrR titration assays along with pAKD750 (wild-type aox promoter region) and pAKD751 (modified aox promoter region). pAKD751 was constructed by using pAKD750 as a template in PCR with primers containing the modified sequence and a SalI restriction site. The PCR product was then digested and ligated, and the sequence changes verified by sequencing.
aox transcription start site mapping by primer extension
Primers (20 pmol) were end labeled using T4 polynucleotide kinase (NEB) and 100 µCi γ32P ATP (7000 Ci/mmol) (MP Biomedicals). Excess label was removed using the Qiaquick Nucleotide Removal Kit (Qiagen). The final primer concentration was 500 fmol/µl. Sequencing reactions were performed using the Thermosequenase ® Cycle Sequencing Kit (USB, Cleveland, OH) according to the radiolabeled primer cycle sequencing protocol using 500 fmol of the end labeled primer and 50 ng of a plasmid containing the promoter region and coding sequence of aox (pAKD602).
For primer annealing 10 µg of total cellular RNA from V. fischeri exposed to 80 µM DEA NONOate (as described above for quantiative PCR) was combined with 100 fmol of end labeled primer from above in a volume of 15 µl in 1× 1st strand buffer (Fermentas). The mixture was heated to 68˚C in a metal heat block for 10 minutes. The samples were allowed to slowly cool until they reached 37˚C. At this time the reaction mixture was brought to 50 µl with 1× 1st strand buffer containing 1mM each dNTP, 40 U of Ribolock™RNase inhibitor (Fermentas) and 40 U of M-MulV reverse transcriptase (Fermentas). Reactions were incubated at 37˚C for 1 hour followed by 10 minutes at 70˚C to stop the reaction. Reactions were ethanol precipitated and pellets resuspended in 4 µl of formamide loading buffer and 1 µl of 1M urea. Reactions were heated to 95˚C for 2 minutes and loaded on an 8% denaturing polyacrylamide gel alongside the appropriate sequencing reactions. Gels were exposed to a phosphorimager screen and visualized using a Storm 820 Phosphorimager (GE Healthcare).
Measurements of oxygen consumption
Microrespirometry followed previously published protocols (Stevanin et al., 2000, Yu et al., 1997). Oxygen consumption by bacterial strains was measured using a Unisense Modular Microrespiration System with 2-ml chambers modified to include injection portals. Calibration was performed using a fully aerated solution and an anoxic solution consisting of 0.1 M each of sodium ascorbate and sodium hydroxide. Both solutions were the same salinity as the culture medium. To produce cultures for analysis, 60 µl of an overnight culture grown in LBS was subcultured into 15 ml of mineral salts medium containing GlcNAc in a 125-ml Ehrlenmyer flask, and grown to an OD600 of approximately 0.3. Actual OD600 values for the presented data are reported in the respective figure legends, and the corresponding soluble protein concentration for each sample is reported. To determine protein concentration, one-ml samples of each culture were pelleted, frozen at −20 °C, and resuspended in one ml of phosphate buffer (20 mM potassium phosphate buffer, pH 7.8 with 10 mM MgSO4 and 0.2 M NaCl). Samples were sonicated using a Branson Digital Sonciator outfitted with a microtip at 20% amplitude (corresponding to 10 watts) for two 20-second bursts separated a two-minute incubation on ice. Samples were spun for 5 min at 15,000 x g in an Eppendorf 5424 microcentrifuge and the protein concentration in the supernatant was measured using the Quant-iT™ protein assay kit and Qubit fluorometer (Invitrogen).
Culture samples were pipetted into the microrespiration chambers to maximize aeration of the medium, and the chambers were placed a 28 °C water bath before introducing the oxygen probe. To prevent oxygen gradients and promote mixing , a magnetic stir bar rotating at 280 rpm was placed in each chamber. To introduce NO, 20 µl of a stock solution of DEA NONOate (1.67 mg NONOate in one ml of 0.01 N NaOH) was injected to achieve a final concentration of 80 µM DEA NONOate in the reaction chamber. Changes in oxygen concentration (µmol/L) over time were logged using the provided MicOx software, and graphs were produced using Microsoft Excel 2007. DEA NONOate was added at an oxygen tension of 190 µmol/L to represent a time when readings had stabilized and oxygen tensions represented fully aerobic growth conditions. Experiments were performed using a single culture of each strain, and experiments were repeated at least three times with independent cultures. Representative data are shown.
Preparation of inverted membrane vesicles
Inverted membrane vesicles from V. fischeri were prepared using modifications of protocols for E. coli (Imlay, 1995) and Vibrio alginolyticus (Tokuda & Unemoto, 1984). First, 200 µl of an overnight culture of either AKD788 or AKD789 was grown in LBS medium and subcultured into 100 ml of LBS medium supplemented with 0.2% glucose in a 1000-ml Ehrlenmyer flask, and grown at 28 °C to an OD600 ∼0.3. The average OD600 value for the three AKD788 samples was 0.308 +/−0.01 and for the three AKD789 samples was 0.298 +/−0.02. When cultures reached an OD600 of ∼0.3, chloramphenicol was added to a concentration of 20 µg ml−1, and cultures were allowed to incubate for 10 minutes at 28 °C with shaking prior to placement on ice. Cells were pelleted at 8,000 x g for 5 min at 4 °C in an Eppendorf 5810 centrifuge, resuspended in 50 ml of phosphate buffer (20 mM potassium phosphate buffer, pH 7.8 with 10 mM MgSO4 and 0.2 M NaCl), pelleted again, and resuspended in 5.5 ml of phosphate buffer. Samples were sonicated on ice using a Branson Digital Sonciator outfitted with a microtip at 20% amplitude (10 watts) for five 20-sec bursts separated by one min incubations on ice. The lysate was clarified by centrifugation at 18,000 x g for 20 min at 4 °C. The inverted membrane vesicles were pelleted by centrifugation at 100,000 x g for 2 hr at 4 °C, and suspended in 0.2 ml of phosphate buffer. Inverted membrane vesicles were kept on ice and used within hours of preparation.
NADH oxidation assays
Assays were performed essentially as described in (Tokuda & Unemoto, 1984); however, the assay temperature was 23 °C and 50 µg of membrane protein was used in the assay. Protein concentration was determined as described. The absorbance of NADH at 340 nm was monitored, and NADH oxidation rate was determined using a molar extinction coefficient for NADH of 6.22 × 103 liters mol−1 cm−1.
Acknowledgements
We would like to thank A.N. Septer and J.A. Imlay for helpful discussions, and three anonymous reviewers for their suggestions. This work was funded by a National Science Foundation (NSF) award (MCB-0803181) to AKD, funds provided by the University of Oklahoma to AKD, a NSF CAREER award (MCB-0347317) to EVS, by NSF award IBN0517007 to M. McFall-Ngai and EGR, and by National Institutes of Health award RR2294 to EGR. Genome information was provided by the Vibrio fischeri Genome Project, at www.ergo-light.com/ERGO, supported by the W. M. Keck Foundation.
References
- Akhter S, McDade HC, Gorlach JM, Heinrich G, Cox GM, Perfect JR. Role of alternative oxidase gene in pathogenesis of Cryptococcus neoformans. Infect. Immun. 2003;71:5794–5802. doi: 10.1128/IAI.71.10.5794-5802.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avissar YJ, Beale SI. Identification of the enzymatic basis for delta-aminolevulinic acid auxotrophy in a hemA mutant of Escherichia coli. J. Bacteriol. 1989;171:2919–2924. doi: 10.1128/jb.171.6.2919-2924.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthold DA, Voevodskaya N, Stenmark P, Graslund A, Nordlund P. EPR studies of the mitochondrial alternative oxidase. Evidence for a diiron carboxylate center. J. Biol. Chem. 2002;277:43608–43614. doi: 10.1074/jbc.M206724200. [DOI] [PubMed] [Google Scholar]
- Bodenmiller DM, Spiro S. The yjeB (nsrR) gene of Escherichia coli encodes a nitric oxide-sensitive transcriptional regulator. J. Bacteriol. 2006;188:874–881. doi: 10.1128/JB.188.3.874-881.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettcher KJ, Ruby EG. Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J. Bacteriol. 1990;172:3701–3706. doi: 10.1128/jb.172.7.3701-3706.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borisov VB, Forte E, Konstantinov AA, Poole RK, Sarti P, Giuffre A. Interaction of the bacterial terminal oxidase cytochrome bd with nitric oxide. FEBS Lett. 2004;576:201–204. doi: 10.1016/j.febslet.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Bose JL, Rosenberg CS, Stabb EV. Effects of luxCDABEG induction in Vibrio fischeri: enhancement of symbiotic colonization and conditional attenuation of growth in culture. Arch. Microbiol. 2008;190:169–183. doi: 10.1007/s00203-008-0387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro-Guerrero NA, Krab K, Moreno-Sanchez R. The alternative respiratory pathway of euglena mitochondria. J. Bioenerg. Biomembr. 2004;36:459–469. doi: 10.1023/B:JOBB.0000047328.82733.ef. [DOI] [PubMed] [Google Scholar]
- Chaudhuri M, Ajayi W, Hill GC. Biochemical and molecular properties of the Trypanosoma brucei alternative oxidase. Mol. Biochem. Parasitol. 1998;95:53–68. doi: 10.1016/s0166-6851(98)00091-7. [DOI] [PubMed] [Google Scholar]
- Choi PS, Naal Z, Moore C, Casado-Rivera E, Abruna HD, Helmann JD, Shapleigh JP. Assessing the impact of denitrifier-produced nitric oxide on other bacteria. Appl. Environ. Microbiol. 2006;72:2200–2205. doi: 10.1128/AEM.72.3.2200-2205.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson AB, Jr, Bienen EJ, Pollakis G, Grady RW. Respiration of bloodstream forms of the parasite Trypanosoma brucei brucei is dependent on a plant-like alternative oxidase. J. Biol. Chem. 1989;264:17770–17776. [PubMed] [Google Scholar]
- Cosseau C, Batut J. Genomics of the ccoNOQP-encoded cbb3 oxidase complex in bacteria. Arch. Microbiol. 2004;181:89–96. doi: 10.1007/s00203-003-0641-5. [DOI] [PubMed] [Google Scholar]
- Cotter PA, Chepuri V, Gennis RB, Gunsalus RP. Cytochrome o (cyoABCDE) and d (cydAB) oxidase gene expression in Escherichia coli is regulated by oxygen, pH, and the fnr gene product. J. Bacteriol. 1990;172:6333–6338. doi: 10.1128/jb.172.11.6333-6338.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czarna M, Jarmuszkiewicz W. Activation of alternative oxidase and uncoupling protein lowers hydrogen peroxide formation in amoeba Acanthamoeba castellanii mitochondria. FEBS Lett. 2005;579:3136–3140. doi: 10.1016/j.febslet.2005.04.081. [DOI] [PubMed] [Google Scholar]
- Davidson SK, Koropatnick TA, Kossmehl R, Sycuro L, McFall-Ngai MJ. NO means 'yes' in the squid-vibrio symbiosis: nitric oxide (NO) during the initial stages of a beneficial association. Cell Microbiol. 2004;6:1139–1151. doi: 10.1111/j.1462-5822.2004.00429.x. [DOI] [PubMed] [Google Scholar]
- Delgado MJ, Bedmar EJ, Downie JA. Genes involved in the formation and assembly of rhizobial cytochromes and their role in symbiotic nitrogen fixation. Adv. Microb. Physiol. 1998;40:191–231. doi: 10.1016/s0065-2911(08)60132-0. [DOI] [PubMed] [Google Scholar]
- Dunn AK, Martin MO, Stabb EV. Characterization of pES213, a small mobilizable plasmid from Vibrio fischeri. Plasmid. 2005;54:114–134. doi: 10.1016/j.plasmid.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Dunn AK, Millikan DS, Adin DM, Bose JL, Stabb EV. New rfp- and pES213-derived tools for analyzing symbiotic Vibrio fischeri reveal patterns of infection and lux expression in situ. Appl. Environ. Microbiol. 2006;72:802–810. doi: 10.1128/AEM.72.1.802-810.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn AK, Stabb EV. The twin arginine translocation system contributes to symbiotic colonization of Euprymna scolopes by Vibrio fischeri. FEMS Microbiol. Lett. 2008;279:251–258. doi: 10.1111/j.1574-6968.2007.01043.x. [DOI] [PubMed] [Google Scholar]
- Gupta KJ, Zabalza A, van Dongen JT. Regulation of respiration when the oxygen availability changes. Physiol. Plant. 2009;137:383–391. doi: 10.1111/j.1399-3054.2009.01253.x. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Hansen LD, Church JN, Matheson S, McCarlie VW, Thygerson T, Criddle RS, Smith BN. Kinetics of plant growth and metabolism. Thermochim. Acta. 2002;388:415–425. [Google Scholar]
- Hori H, Tsubaki M, Mogi T, Anraku Y. EPR study of NO complex of bd-type ubiquinol oxidase from Escherichia coli. J. Biol. Chem. 1996;271:9254–9258. doi: 10.1074/jbc.271.16.9254. [DOI] [PubMed] [Google Scholar]
- Imlay JA. A metabolic enzyme that rapidly produces superoxide, fumarate reductase of Escherichia coli. J. Biol. Chem. 1995;270:19767–19777. [PubMed] [Google Scholar]
- Isabella VM, Lapek JD, Jr, Kennedy EM, Clark VL. Functional analysis of NsrR, a nitric oxide-sensing Rrf2 repressor in Neisseria gonorrhoeae. Mol. Microbiol. 2009;71:227–239. doi: 10.1111/j.1365-2958.2008.06522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly MJ, Poole RK, Yates MG, Kennedy C. Cloning and mutagenesis of genes encoding the cytochrome bd terminal oxidase complex in Azotobacter vinelandii: mutants deficient in the cytochrome d complex are unable to fix nitrogen in air. J. Bacteriol. 1990;172:6010–6019. doi: 10.1128/jb.172.10.6010-6019.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar AM, Soll D. Arabidopsis alternative oxidase sustains Escherichia coli respiration. Proc. Natl. Acad. Sci. U. S. A. 1992;89:10842–10846. doi: 10.1073/pnas.89.22.10842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambers H. Cyanide-resistant respiration: a non-phosphorylating electron transport pathway acting as an energy over-flow. Physiol. Plant. 1982;55:478–485. [Google Scholar]
- Mason MG, Shepherd M, Nicholls P, Dobbin PS, Dodsworth KS, Poole RK, Cooper CE. Cytochrome bd confers nitric oxide resistance to Escherichia coli. Nat. Chem. Biol. 2009;5:94–96. doi: 10.1038/nchembio.135. [DOI] [PubMed] [Google Scholar]
- McDonald A, Vanlerberghe G. Branched mitochondrial electron transport in the Animalia: presence of alternative oxidase in several animal phyla. IUBMB Life. 2004;56:333–341. doi: 10.1080/1521-6540400000876. [DOI] [PubMed] [Google Scholar]
- McDonald AE, Vanlerberghe GC. Alternative oxidase and plastoquinol terminal oxidase in marine prokaryotes of the Sargasso Sea. Gene. 2005;349:15–24. doi: 10.1016/j.gene.2004.12.049. [DOI] [PubMed] [Google Scholar]
- Meeuse JBD. Thermogenic respiration in aroids. Annu. Rev. Plant. Physiol. 1975;26:117–126. [Google Scholar]
- Millar AH, Day DA. Nitric oxide inhibits the cytochrome oxidase but not the alternative oxidase of plant mitochondria. FEBS Lett. 1996;398:155–158. doi: 10.1016/s0014-5793(96)01230-6. [DOI] [PubMed] [Google Scholar]
- Millar H, Considine MJ, Day DA, Whelan J. Unraveling the role of mitochondria during oxidative stress in plants. IUBMB Life. 2001;51:201–205. doi: 10.1080/152165401753311735. [DOI] [PubMed] [Google Scholar]
- Miller JH. In: A short Course in Bacterial Genetics. New York: Cold Spring Harbor Laboratory Press; 1992. p. 456. [Google Scholar]
- Partridge JD, Bodenmiller DM, Humphrys MS, Spiro S. NsrR targets in the Escherichia coli genome: new insights into DNA sequence requirements for binding and a role for NsrR in the regulation of motility. Mol. Microbiol. 2009;73:680–694. doi: 10.1111/j.1365-2958.2009.06799.x. [DOI] [PubMed] [Google Scholar]
- Preisig O, Zufferey R, Thony-Meyer L, Appleby CA, Hennecke H. A high-affinity cbb3-type cytochrome oxidase terminates the symbiosis-specific respiratory chain of Bradyrhizobium japonicum. J. Bacteriol. 1996;178:1532–1538. doi: 10.1128/jb.178.6.1532-1538.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purvis AC. Role of the alternative oxidase in limiting superoxide production by plant mitochondria. Physiol. Plant. 1997;100:165–170. [Google Scholar]
- Rasmusson AG, Fernie AR, van Dongen JT. Alternative oxidase: a defence against metabolic fluctuations? Physiol. Plant. 2009;137:371–382. doi: 10.1111/j.1399-3054.2009.01252.x. [DOI] [PubMed] [Google Scholar]
- Rodionov DA, Dubchak IL, Arkin AP, Alm EJ, Gelfand MS. Dissimilatory metabolism of nitrogen oxides in bacteria: comparative reconstruction of transcriptional networks. PLoS Comput. Biol. 2005;1:e55. doi: 10.1371/journal.pcbi.0010055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby EG, Urbanowski M, Campbell J, Dunn A, Faini M, Gunsalus R, Lostroh P, et al. Complete genome sequence of Vibrio fischeri: a symbiotic bacterium with pathogenic congeners. Proc. Natl. Acad. Sci. U. S. A. 2005;102:3004–3009. doi: 10.1073/pnas.0409900102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stabb EV, Reich KA, Ruby EG. Vibrio fischeri genes hvnA and hvnB encode secreted NAD(+)-glycohydrolases. J. Bacteriol. 2001;183:309–317. doi: 10.1128/JB.183.1.309-317.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stabb EV, Ruby EG. RP4-based plasmids for conjugation between Escherichia coli and members of the Vibrionaceae. Methods Enzymol. 2002;358:413–426. doi: 10.1016/s0076-6879(02)58106-4. [DOI] [PubMed] [Google Scholar]
- Stenmark P, Nordlund P. A prokaryotic alternative oxidase present in the bacterium Novosphingobium aromaticivorans. FEBS Lett. 2003;552:189–192. doi: 10.1016/s0014-5793(03)00920-7. [DOI] [PubMed] [Google Scholar]
- Stevanin TM, Ioannidis N, Mills CE, Kim SO, Hughes MN, Poole RK. Flavohemoglobin Hmp affords inducible protection for Escherichia coli respiration, catalyzed by cytochromes bo' or bd, from nitric oxide. J. Biol. Chem. 2000;275:35868–35875. doi: 10.1074/jbc.M002471200. [DOI] [PubMed] [Google Scholar]
- Thony-Meyer L, Beck C, Preisig O, Hennecke H. The ccoNOQP gene cluster codes for a cb-type cytochrome oxidase that functions in aerobic respiration of Rhodobacter capsulatus. Mol. Microbiol. 1994;14:705–716. doi: 10.1111/j.1365-2958.1994.tb01308.x. [DOI] [PubMed] [Google Scholar]
- Tokuda H, Unemoto T. Na+ is translocated at NADH:quinone oxidoreductase segment in the respiratory chain of Vibrio alginolyticus. J. Biol. Chem. 1984;259:7785–7790. [PubMed] [Google Scholar]
- Tucker NP, Hicks MG, Clarke TA, Crack JC, Chandra G, Le Brun NE, Dixon R, et al. The transcriptional repressor protein NsrR senses nitric oxide directly via a [2Fe-2S] cluster. PloS one. 2008;3:e3623. doi: 10.1371/journal.pone.0003623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker NP, Le Brun NE, Dixon R, Hutchings MI. There's NO stopping NsrR, a global regulator of the bacterial NO stress response. Trends Microbiol. 2010 doi: 10.1016/j.tim.2009.12.009. In press. [DOI] [PubMed] [Google Scholar]
- Umbach AL, Siedow JN. The cyanide-resistant alternative oxidases from the fungi Pichia stipitis and Neurospora crassa are monomeric and lack regulatory features of the plant enzyme. Arch. Biochem. Biophys. 2000;378:234–245. doi: 10.1006/abbi.2000.1834. [DOI] [PubMed] [Google Scholar]
- Vanlerberghe GC, Cvetkovska M, Wang J. Is the maintenance of homeostatic mitochondrial signaling during stress a physiological role for alternative oxidase? Physiol. Plant. 2009;137:392–406. doi: 10.1111/j.1399-3054.2009.01254.x. [DOI] [PubMed] [Google Scholar]
- Vanlerberghe GC, McIntosh L. ALTERNATIVE OXIDASE: From Gene to Function. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1997;48:703–734. doi: 10.1146/annurev.arplant.48.1.703. [DOI] [PubMed] [Google Scholar]
- Visick KL, Ruby EG. Vibrio fischeri and its host: it takes two to tango. Curr. Opin. Microbiol. 2006;9:632–638. doi: 10.1016/j.mib.2006.10.001. [DOI] [PubMed] [Google Scholar]
- Wagner AM, Moore AL. Structure and function of the plant alternative oxidase: its putative role in the oxygen defence mechanism. Biosci. Rep. 1997;17:319–333. doi: 10.1023/a:1027388729586. [DOI] [PubMed] [Google Scholar]
- Yu H, Sato EF, Nagata K, Nishikawa M, Kashiba M, Arakawa T, Kobayashi K, et al. Oxygen-dependent regulation of the respiration and growth of Escherichia coli by nitric oxide. FEBS Lett. 1997;409:161–165. doi: 10.1016/s0014-5793(97)00494-8. [DOI] [PubMed] [Google Scholar]
- Yukl ET, Elbaz MA, Nakano MM, Moenne-Loccoz P. Transcription Factor NsrR from Bacillus subtilis Senses Nitric Oxide with a 4Fe-4S Cluster. Biochemistry. 2008;47:13084–13092. doi: 10.1021/bi801342x. [DOI] [PMC free article] [PubMed] [Google Scholar]