Abstract
Neuroblastoma is a malignancy of the developing sympathetic nervous system that often presents with widespread metastatic disease, resulting in survival rates of less than 50%1. To determine the spectrum of somatic mutation in high-risk neuroblastoma, we studied 240 cases using a combination of whole exome, genome and transcriptome sequencing as part of the Therapeutically Applicable Research to Generate Effective Treatments (TARGET) initiative. Here we report a low median exonic mutation frequency of 0.60 per megabase (0.48 non-silent), and remarkably few recurrently mutated genes in these tumors. Genes with significant somatic mutation frequencies included ALK (9.2% of cases), PTPN11 (2.9%), ATRX (2.5%, an additional 7.1% had focal deletions), MYCN (1.7%, a recurrent p.Pro44Leu alteration), and NRAS (0.83%). Rare, potentially pathogenic germline variants were significantly enriched in ALK, CHEK2, PINK1, and BARD1. The relative paucity of recurrent somatic mutations in neuroblastoma challenges current therapeutic strategies reliant upon frequently altered oncogenic drivers.
Neuroblastoma is an embryonal malignancy of early childhood with a poor prognosis for patients diagnosed at over 18 months of age with disseminated disease, overall accounting for 12% of childhood cancer-related deaths1,2. Despite multimodal chemo- and immuno-therapeutic strategies that improved the survival of patients with high-risk disease3,4, a disproportionate number of these patients will die or suffer profound treatment-related morbidity5. Novel therapeutic approaches are needed to improve cure rates while minimizing toxicity.
Highly penetrant, heritable mutations in the ALK or PHOX2B genes account for the majority of familial neuroblastomas6–9. For patients with sporadic disease, genome-wide association studies have identified multiple DNA polymorphisms in genes that influence neuroblastoma susceptibility and clinical phenotype10–15. Somatically acquired amplification of MYCN, and hemizygous deletions of 1p and 11q are highly recurrent and are associated with poor prognosis16. While these latter aberrations are useful as prognostic biomarkers of patient outcome, there remain few known oncogenic drivers of the malignant process.
Three recent studies have together reported genome or exome analysis of 162 neuroblastoma cases17–19. Molenaar and colleagues reported an overall low somatic mutation count (12 per tumor), few recurrent mutations beyond ALK (7% of cases) and TIAM1 (3%), a high frequency of chromothripsis in stage 3 and 4 tumors (18%), and frequent mutation of Rac/Rho pathway genes regulating neuritogenesis17. Cheung and colleagues found ATRX loss-of-function mutations and deletions associated with neuroblastoma in adolescents and young adults18. Sausen and colleagues uncovered recurrent mutation or focal deletion of ARID1A and ARID1B in 11% of cases using a low coverage WGS and targeted sequencing strategy19. Given the genetic heterogeneity described in neuroblastoma, we sought to build upon these studies through a focused analysis of a large cohort of high-risk stage 4 neuroblastomas, where the need for translational advances are most pressing, using several genomic approaches.
Here we examined 240 matched tumor/normal (blood leukocyte) pairs from patients older than 18 months of age at diagnosis with metastatic (Stage 4) disease by whole exome sequencing (WES; 221 cases), whole genome sequencing (WGS; 18 cases; one using two different sequencing platforms), or both (1 case; Supplementary Table 1; Supplementary Note). WES of ~33 megabases of coding sequence yielded an average 124X coverage with 87% of bases suitable for mutation detection (Supplementary Figure 1, Supplementary Table 2). We used two different WGS approaches, Illumina20 (10 cases, 29.7X average coverage) and Complete Genomics21 (10 cases, 59.9X average coverage), to interrogate structural variation and supplement mutation detection (powered to detect mutations at 86% and 94% of mappable exonic bases, respectively). To assess expression of mutations and fusion transcripts, over 10 Gbp of RNA-seq data was generated for the ten Illumina WGS cases.
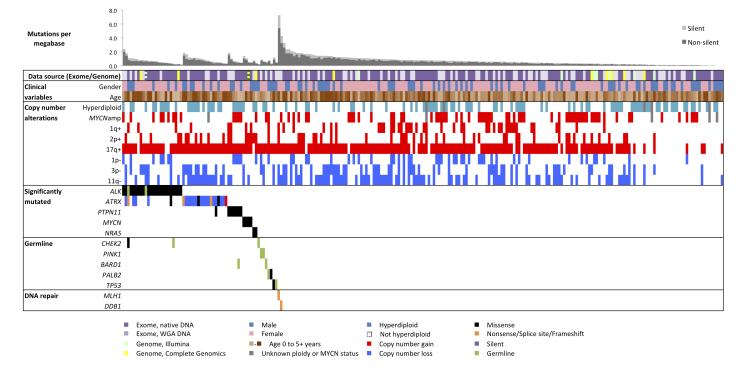
Across the coding regions of 240 cases, we detected 5,291 candidate somatic mutations in 3,960 genes (Supplementary Table 3). A median of 18 candidate exomic mutations (17 substitutions, 1 indel) was found per tumor (range 0-218), of which 14 were non-silent mutations predicted to alter protein sequences (range 0-158, median 12 missense, 1 nonsense, 1 indel, 0 splice site, Supplementary Table 1). This corresponds to a median frequency of 0.60 mutations per megabase (0.48 non-silent per megabase), considering only exonic bases with sufficient data for mutation detection (Figure 1). This frequency is consistent with unselected neuroblastomas17–19, medulloblastoma22, and hematopoietic malignancies23,24, twice that of pediatric rhabdoid cancer25, and significantly less than adult solid tumors24,26,27, particularly those with strong environmental contribution24,28–31. We verified 241 of 282 coding candidate somatic substitutions (85%, 525 of 605 including non-coding) and 26/41 coding indels (63%, 27/79 including non-coding) using mass-spectrometric genotyping or PCR-based re-sequencing (Supplementary Text).
Figure 1. Landscape of genetic variation in neuroblastoma.
Data tracks (rows) facilitate comparison of clinical and genomic data across neuroblastoma cases (columns). Data source: sequencing technology used, purple = WES from WGA (light) and native DNA (dark), green = Illumina WGS, yellow = Complete Genomics Inc. WGS. Striped blocks indicate cases analyzed using two approaches. Clinical variables: gender (male/female = blue/pink) and age (brown spectrum). Copy number alterations: ploidy measured by flow cytometry (hyperdiploid = DNA index > 1) and clinically relevant copy number alterations derived from sequence data. Significantly mutated: genes with statistically significant mutation counts given the background mutation rate, gene size, and expression in neuroblastoma. Germline: genes with significant numbers of germline ClinVar or loss-of-function cancer gene variants in our cohort. DNA repair: Genes that may be associated with increased mutation frequency in two apparently hyper-mutated tumors. Predicted effects of somatic mutations are color-coded by the provided legend.
We did not observe a correlation between mutation frequency and age at diagnosis (p=0.28, Spearman) or other clinical variables (Supplementary Table 4). Consistent with a postulated limited environmental contribution to neuroblastoma development1, context-specific transition and transversion rates were not elevated compared to other cancers (Supplementary Figure 2) and we did not detect significant sequencing reads corresponding to pathogenic viruses (Supplementary Table 5). Two tumors with markedly increased mutation frequencies (7.27 and 4.29 mutations per megabase) harbored alterations of DNA repair genes (nonsense mutation and deletion of MLH1 and nonsense mutation of DB1).
Using the MutSig algorithm32, we identified six genes mutated at a significant frequency in the 240 tumors (q<0.1; Table 1; Supplementary Table 6). A seventh gene, NRAS, was implicated by restricting this analysis to genes listed in the Catalogue of Somatic Mutations in Cancer (COSMIC, v48)33. Using neuroblastoma data from our RNA-seq cohort (10 cases), the TARGET RNA microarray project (250 cases), and a publically-available RNA microarray project (416 cases)34, we determined that OR5T1 and PDE6G have very low or absent mRNA expression in neuroblastoma (Supplementary Figure 3). Therefore, we focused our analysis on five genes with statistical and biological rationale for neuroblastoma involvement: ALK, PTPN11, ATRX, MYCN, and NRAS.
Table 1.
Genes with significant frequency of somatic mutation across 240 neuroblastomas
| Gene | Description | Mutations | Patients | Unique sites |
Missense | Loss of function* |
q-value | Expressed in neuroblastoma# |
|---|---|---|---|---|---|---|---|---|
| ALK | anaplastic lymphoma receptor tyrosine kinase |
22 | 22 | 7 | 22 | 0 | <1.8×10−7 | Yes |
| PTPN11 | protein tyrosine phosphatase, non-receptor type 11 |
7 | 7 | 6 | 7 | 0 | 1.8×10−5 | Yes |
| ATRX | alpha thalassemia/mental retardation syndrome X-linked |
6 | 6 | 6 | 3 | 3 | 0.031 | Yes |
| OR5T1 | olfactory receptor, family 5, subfamily T, member 1 |
3 | 2 | 3 | 3 | 0 | 0.040 | No |
| PDE6G | phosphodiesterase 6G, cGMP- specific, rod, gamma |
2 | 2 | 2 | 2 | 0 | 0.052 | No |
| MYCN | v-myc myelocytomatosis viral related oncogene, neuroblastoma |
4 | 4 | 1 | 0 | 0 | 0.093 | Yes |
| NRAS | neuroblastoma RAS viral (v-ras) oncogene homolog |
2 | 2 | 2 | 2 | 0 | 0.017 (COSMIC only) |
Yes |
Nonsense, splice site, or frameshift
very low or absent mRNA expression in RNA-seq or microarray data sets
ALK and PTPN11 were previously reported to be mutated in up to 10% and 3.4% of neuroblastoma cases respectively8,9,35–37 consistent with our screen here. All 22 somatic ALK mutations (9.2%) were restricted to the kinase domain and all 7 PTPN11 mutations (2.9%) have been previously reported33,37–40. While no pathogenic germline PTPN11 variants were found, two patients had germline ALK variants: pathogenic, activating p.Arg1275Gln and the likely benign, kinase-dead p.IIle1250Thr. Activating ALK variants were not associated with MYCN amplification (p=0.28). Contrary to a prior report41, we did not observe p.Phe1174 mutations in a higher proportion of MYCN-amplified cases than wild-type ALK cases (p=0.53). Notably, ALK was the only significantly mutated gene with an association with clinical outcome, as mutation positive cases had a decreased overall survival probability (p=0.0103, Supplementary Figure 4).
Loss-of-function mutations or deletions of RNA-helicase ATRX have recently been described in neuroblastoma17,18. We observed putative loss of function ATRX alterations in 9.6% of cases (6 mutations, 17 multi-exon deletions; Supplementary Figure 5). We confirmed prior observations18 that alterations of ATRX and MYCN were mutually exclusive and that ATRX alterations were enriched in older children (p=0.0021, Supplementary Figure 6). One case had an apparent gain of exons 18-26 of unclear functional effect.
High-level MYCN amplification has long been known as a negative prognostic indicator in neuroblastoma42, but activating mutations have not been described. In our cohort, four cases without MYCN amplification had an identical p.Phe44Leu alteration. All four tumors had regional single-copy gain of chromosome 2p, three with gain of the mutant allele. In a tumor with matched RNA-seq data, the mutant allele was expressed at a level twice that of wild-type. This mutation has been documented in single cases of glioblastoma, medulloblastoma, and pancreatic adenocarcinoma33, and is scored as functional by PolyPhen243 (score 0.971), SIFT44 (score 0), MutationTaster45 (score 1.0), and AlignGVGD46 (score C65). The residue is highly conserved across the MYC superfamily (pfam01056; Supplementary Figure 7)47, and an additional tumor had a mutation in the homologous domain of MYC, p.Thr58Ile, a common mutation in Burkitt lymphoma48. Despite the relative infrequency of MYCN mutations in neuroblastoma, these mutations may be clinically relevant if they confer myc dependency similar to high-level amplification.
We next searched, as previously described22,27, for enrichment of somatic mutations in components of canonical pathways49, chromatin modifiers, or splice factors50–52. Of 857 gene sets, 12 were enriched for somatic mutation (q<0.1, Supplementary Table 7), 8 implicating RAS/MAPK signaling components. Contrary to Molenaar and colleagues17, we did not see any mutations in TIAM1 nor any enrichment of mutations in genes regulating neuritogenesis and GTPase activity through the Rac/Rho pathway (q>0.275, Supplementary Text). However, an analysis of their mutation list using our methods recapitulated their finding of significant mutation frequencies in guanine nucleotide exchange factors (q=6.26×10−3) and GTPase activating proteins (q=3.15×10−5), but none of the 12 pathways identified by analysis of our cohort (q≥0.848). This comparison suggests limitations to current gene set and pathway analysis methods, especially when mutation frequencies are low.
Molenaar and colleagues reported somatic mutations in FANCM and FAN1 in two cases with chromothripsis17. While we observed 3 cases with FANCM mutations, Fanconi-Anemia genes were not enriched for somatic mutation (q=0.764, Supplementary Table 7), nor did we detect any exonic breakpoints in cases with FANCM mutations. This is perhaps not surprising given the relatively small portion of the genome queried by exome sequencing, so we cannot rule out an association of FANCM mutations and chromothripsis at this time.
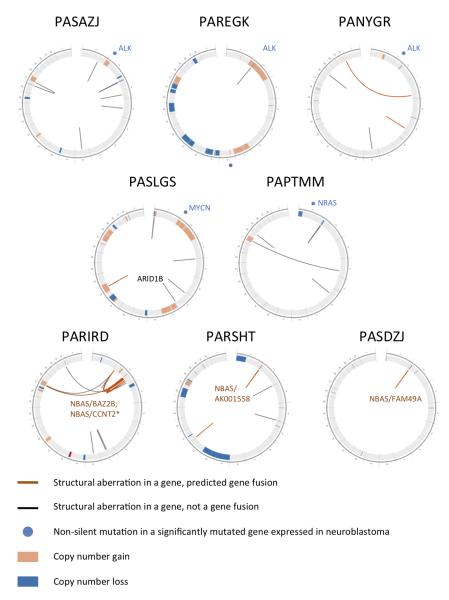
Of the five recurrently mutated genes reported by Sausen and colleagues, we found mutations in ALK (see above), ARID1A (p.G1139V, p.G1942D) and VANGL1 (p.Gly308Trp). Two cases had focal deletions of ARID1B: PASLGS had an exon 2 deletion (Figure 2, Supplementary Table 10) and PARGKK had loss of exons 1-3. Of the 113 genes with apparent hemizygous mutations on 1p, 3p, and 11q (arms frequently lost in neuroblastoma), only PBRM1 showed loss-of-function mutations in two cases, all others were singleton variations (Supplementary Note).
Figure 2. Structural variation in neuroblastoma genomes.
CIRCOS68 plots of cases with recurrent somatic alterations, labeled using TARGET identifiers. Chromosomes are arranged end-to-end in the outer-most ring. Mutations in significantly mutated genes are depicted in light blue outside of each diagram. The inside ring shows somatic copy number gains and losses (high-level gains are red, low-level gains are orange, losses are blue) detected by WGS. The innermost arcs depict genic structural aberrations (gene fusions are orange, all others are black) detected by RNA-seq and confirmed by local reassembly of WGS reads. Non-genic rearrangements are not shown. The top five cases have mutations in significantly mutated genes ALK, MYCN, and NRAS. The bottom three cases each have several rearrangements of NBAS, with expressed fusion transcripts as annotated.
To identify rare germline variants predisposing to neuroblastoma, we searched for enrichment of clinically-annotated variants from the ClinVar database and loss-of-function variants in cancer genes53–55 in the blood-derived DNA samples from our WES cohort, compared to normal DNAs from 1,974 European American individuals sequenced by the Exome Sequencing Project (ESP)56 (see Methods, Supplementary Tables 8 and 9, and Supplementary Figure 8). This approach nominated five genes with candidate germline pathogenic variants: ALK, CHEK2, PINK1, TP53 and BARD1 (Table 2). The ALK p.Arg1275Gln variant has been reported as the most common pathogenic variant in familial neuroblastoma8,9. Three CHEK2 germline variants destabilize the protein57,58 and are reported cancer predisposition alleles58,59 not previously described in neuroblastoma. The TP53 p.Pro219Ser variant has been associated with Li-Fraumeni syndrome60, consistent with prior reports of neuroblastomas occurring in these families61. Two PINK1 variants are associated with Parkinson disease62–64, and this gene is known to be transcriptionally regulated by myc proteins65. Finally, two loss of function variants in BARD1, a recently discovered neuroblastoma susceptibility locus14, support the concept that rare variants may exist at loci where common polymorphisms impact disease occurrence. Another member of the BRCA complex, PALB2, had a germline variant predicted to ablate a splice site in one case (Table 2) and a somatic missense mutation in another (Figure 1, Supplementary Table 3). Taken together, our conservative approach to identifying putatively pathogenic germline variants suggest that these events may play a larger role in neuroblastoma initiation than previously suspected.
Table 2.
Candidate pathogenic germline variants in 222 neuroblastoma WES cases
| Gene | Subject Identifier |
Genome position (build hg19) |
cDNA change |
Protein change |
|---|---|---|---|---|
| ALK | PARVLK | chr2:29432664 | c.3824G>A | p.Arg1275Gln |
| CHEK2 | PAKXDZ | chr22:29121242 | c.433C>T | p.Arg145Tro |
| CHEK2 | PAPTFZ | chr22:29121015 | c.542G>A | p.Arg181His |
| CHEK2 | PARJMX | chr22:29121018 | c.539G>A | p.Arg180His |
| PINK1 | PANYBL | chr1:20972133 | c.1040T>C | p.Leu437Pro |
| PINK1 | PATINJ | chr1:20971042 | c.836G>A | p.Arg279His |
| BARD1 | PAHYWC | chr2:215657051 | c.334C>T | p.Arg112* |
| BARD1 | PATGWT | chr2:215595215 | c.1921C>T | p.Arg641* |
| TP53 | PAICGF | chr17:7578194 | c.655C>T | p.Pro219Ser |
| PALB2 | PAPZYZ | chr16:23646182 | c.1684+1C>A | Splice at p.Gly562 |
Structural analysis of the 19 WGS cases identified a median of 41.5 breakpoints per case (range 29 to 143, Figure 2, Supplementary Figure 9). Overall, 83 rearrangements affected 97 genes, 22 of which had evidence from RNA-seq data (Supplementary Table 10). While 11q;17q translocations were found in 3 of 19 cases (1 case with two events), we did not observe any recurrent fusion transcripts in our cohort. NBAS, located near MYCN on the short arm of chromosome 2, was the most commonly rearranged gene and harbored 11 distinct events in three MYCN-amplified cases (Figure 2). Substantial local rearrangement was seen in three cases, all affecting the vicinity of MYCN and NBAS loci, but the numerous complex copy number states and retention of heterozygosity in lower copy number regions are more consistent with an episomal model66 than chromothripsis67 in the 19 cases evaluated here (Supplementary Figure 10). No other areas of clustered chromosomal breakpoints suggestive of chromothripsis were identified in the WGS cases, nor were any clusters evident within coding regions of 142 WES cases sequenced from native DNA.
High-risk neuroblastomas harbor a very low frequency of recurrent somatic mutations. We do not expect that significant numbers of mutations went undetected as tumor purities were high and identical methods have identified significant mutations in other tumor types22,23,26,27,30. The relative paucity of recurrent mutations challenges the concept that druggable targets can be defined in each patient by DNA sequencing alone. Our data suggest that the majority of high-risk neuroblastomas may be driven by rare germline variants and/or by copy number alterations and epigenetic modifications during tumor evolution. The striking lack of precisely defined genomic causes of this highly aggressive pediatric neoplasm reinforces the need to understand the interplay of host genetic factors, somatic mutations, chromosomal abnormalities, and epigenetic alterations in the context of nervous system development.
Online Methods
Summary
Paired tumor/normal DNA from 240 high-risk neuroblastoma cases were identified from the Children Oncology Group biobank on the basis of subjects having metastatic disease and preferably being between 18 months and 5.5 years of age at diagnosis. Whole genome sequences were generated for 19 pairs using two technologies: 9 Illumina sequencing-by-synthesis20, 9 Complete Genomics probe-anchor-ligation21, and 1 using both. RNA-seq data were generated for the 10 Illumina WGS cases. Whole exome sequences of 222 pairs were generated using in-solution hybrid capture69 followed by Illumina sequencing. Phi29-based whole genome amplification was used to generate sufficient tumor and matched normal DNA template from 80 cases. Reads were aligned to build hg19/GRCh37 of the human genome reference sequence using Burrows-Wheeler Aligner70 and somatic mutations were detected using SNVmix71 (Illumina genomes and RNA-seq), muTect27 (exomes) and version 2 of the Complete Genomics’ custom caller21,72 (Complete Genomics genomes). Mutations were annotated using Oncotator and MutSig32 was used to identify genes mutated at significant frequencies. PathSeq73 was used to query exome data sets for reads supporting viral infection. These and other tools used for exome sequence analysis are described on the Broad Institute Cancer Genome Analysis website. Rearrangements were detected from whole genome data using trans-ABySS de novo assembly74 and Complete Genomics’ custom software75. Somatic mutations and structural alterations were confirmed by mass-spectrometric genotyping (Sequenom) or PCR followed by Sanger or Illumina sequencing.
Sample selection and preparation
This study focused on high-risk neuroblastoma, and we attempted to reduce heterogeneity by restricting eligibility to subjects with stage 4 (metastatic) disease and preferably between 1.5 and 5.5 years of age at diagnosis (median 3.4 years, range 1.5 – 16.5 years) (Supplementary Table 1). All specimens were obtained at original diagnosis after informed consent at Children’s Oncology Group member institutions. Males outnumbered females 149 to 91 (62%). Amplification of the MYCN oncogene was seen in 77 tumors (32%) by fluorescence in situ hybridization and 131 (55%) had a diploid DNA index by flow cytometry. Flash frozen tumor samples were analyzed for percent tumor content by histopathology and samples with <75% tumor content were excluded.
Genome sequencing and analysis
Illumina sequencing technology (BC Cancer Agency)
Whole genome and transcriptome libraries of 10 cases sequenced using Illumina technology at the BC Cancer Agency were constructed from input amounts of 2-4 μg DNA and 3-10 μg DNaseI-treated total RNA, respectively, following previously described protocols76,77. The sequencing was carried out using Illumina GAIIx (Illumina, Hayward, CA, USA) instruments as per the manufacturer’s instructions. Paired end reads generated from genome and transcriptome sequencing were aligned to the hg19/GRCh37 reference human genome assembly78 using BWA70 version 0.5.7. RNA-seq reads were processed as previously described79,80.
Single nucleotide variant (SNV) detection in Illumina tumor genome and transcriptome data was performed using SNVMix2 with filtering to include SNVs such that combined probability of either heterozygous or homozygous SNV was greater than 0.9971. Reads flagged as poor quality according to Illumina chastity filter, duplicate reads, and reads aligned with a mapping quality < 40 were excluded from SNV calling. The somatic status of SNV calls was determined using read evidence from the SAMtools version 0.1.13 pileup81 constructed at the variant positions in the matched normal genome. Positions with normal genome coverage by least 5 unique reads supporting the reference allele were considered somatic. The candidate somatic SNV calls were inspected using IGV82, and only those calls confirmed by visual inspection were used in the analysis.
Short insertions and deletions were detected in the tumor and normal Illumina WGS bam files using two software programs, Pindel83 and SAMtools81. The mean and standard deviation of read pair insert sizes were calculated for all samples to be ~400 bp, and this value was used in each Pindel run. The Pindel short insertion output was filtered to select events that mapped to annotated genes (Ensembl59). Candidate somatic short insertion events that recurred in at least two cases were manually reviewed using the Integrative Genomics Viewer82. The output from SAMtools pileup and varFilter functionality81 run separately on normal and tumor libraries were filtered to identify somatic events. In the normals, any event with a total coverage of less than 8 was discarded. In the tumor libraries, only indels supported by at least 16% of reads at a locus were considered. After the filtering, any indel present in one or more normal libraries was flagged as germline. None of the candidate somatic coding indels from the Pindel or SAMtools analysis were confirmed by manual inspection using IGV82, consistent with the low frequency of somatic indels in the rest of the cohort (median 1 across all other WGS and WES cases, 86 with no indels).
Copy number analysis of the Illumina WGS data was conducted using a previously described hidden Markov model (HMM) method84. Briefly, 50 million reads with mapping qualities >10 were randomly selected from matched tumor and normal data. Reads were divided into bins of 200 adjacent alignments and the ratio of tumor/normal reads was calculated for each bin. These ratios were then normalized by subtracting the median of these ratios across the whole genome. This resulted in a metric of relative read density from the tumors and matched normals in bins of variable length along the genome, where bin width was inversely proportional to the number of mapped reads in the normal genome. GC bias correction was applied, and an HMM was used to classify and segment the tumor genome into continuous regions of somatic copy number loss (HMM state 1), neutrality (HMM state 2), slight gain (HMM state 3), gain (HMM state 4) or high gain (HMM state 5).
To identify candidate transcript rearrangements, we used ABySS85 to perform de novo transcriptome assembly of ten RNA-seq datasets. To identify known and novel transcript structures, the assembled contigs were aligned to the hg19 (GRCh37) human reference genome assembly and compared to annotated transcript models using the trans-ABySS pipeline74. This approach identified all contigs with two discrete genomic BLAT alignments. The top five scoring alignments were manually inspected to remove likely false positive events primarily due to few supporting reads. Local rearrangements were identified from contigs with single, gapped BLAT alignments and supporting read evidence from manual review. Targeted assembly of the candidate rearrangement regions was performed to validate the events in the genomic data.
Complete Genomics sequencing technology
Whole genome sequencing libraries of 10 cases were constructed from 3.5 ug of DNA and sequenced using Complete Genomics Inc. (CGI) technology21. Sequencing and alignment of reads to hg19/GRCh37 reference human genome assembly was performed by the CGI Cancer Sequencing service, analytic pipeline version 1 (See Complete Genomics Analysis Tools website). Mutation call files provided by this service were used to extract somatic mutations using the criteria in Supplementary Table 11. CGI also provided flat files containing candidate rearrangements and segmental relative copy number ratios derived from normalized read counts from matched tumor and normal samples. Copy number calls were converted to the five HMM states described above using the criteria listed in Supplementary Table 12.
Exome sequencing and analysis
The generation, sequencing, and analysis of 222 pairs of exome libraries at the Broad Institute was performed using a previously described protocol27. Due to the small quantities of DNA available, 81 DNA samples were amplified using Phi29-based multiple-strand displacement whole genome amplification (Repli-g service, QIAgen). Exonic regions were captured by in-solution hybridization using RNA baits similar to those described27 but supplemented with additional probes capturing additional genes listed in ReqSeq78 in addition to the original Consensus Coding Sequence (CCDS)78 set. In total, ~33 Mb of genomic sequence was targeted, consisting of 193,094 exons from 18,863 genes annotated by the CCDS86 and RefSeq86 databases as coding for protein or micro-RNA (accessed November 2010). Sequencing of 76 bp paired-end reads was performed using Illumina Genome Analyzer IIx and HiSeq 2000 instruments. Reads were aligned to the hg19/GRCh37 build of the reference human genome sequence78using BWA70. PCR duplicates were flagged in the bam files for exclusion from further analysis using the Picard MarkDuplicates tool. To confirm sample identity, copy number profiles derived from sequence data were compared with those derived from microarray data when available. Candidate somatic base substitutions were detected using muTect (previously referred to as muTector27) and insertions and deletions were detected using IndelGenotyper27. Segmental copy number ratios were calculated as the ratio of tumor fraction read-depth to the average fractional read-depth observed in normal samples for that region.
Removal of oxoG library preparation artifact
Cases sequenced using WGA and native DNA were sequenced more than eight months apart by the Sequencing Platform at the Broad Institute. Initial comparison of candidate mutation calls from these two data sets identified a preponderance of apparent G>T or C>A substitutions of low allele fraction (<0.15) and within specific sequence contexts (Supplementary Figure 2A). We subsequently characterized this artifact and developed a method to detect and remove these events. In brief, these artifacts are introduced at the DNA shearing step of the library construction process and arise from the oxidation of guanine bases (oxoG) by high-energy sonication. During downstream PCR, oxoG bases preferentially pair with thymine rather than cytosine, resulting in apparent G>T or C>A substitutions of low allele fraction and enriched within specific sequence contexts (Supplementary Figure 2B). Consistent with this mechanism, the intensity of the sonication process was increased with the introduction of a new 150 bp shearing protocol between preparation of the WGA and native DNA samples.
The number of artifacts in a library was apparently sample-dependent (Supplementary Figure 2C) and these events were found in unmatched tumor and normal libraries. In some cases, thousands of candidate mutations were called in cases with a heavily affected tumor sample and an unaffected normal. However, nearly every sample had at least one such artifact and we have observed similar events in publically available data sets from other centers, suggesting a common artifact mode that was exacerbated in some of our samples. To address this problem, we devised a method to differentiate oxoG artifacts from bona fide mutations.
Due to the modification of only one strand of a G:C base-pair (i.e. only the G base), reads supporting the artifact have characteristic read-orientation conferred upon adapter ligation. Therefore, all reads supporting an artifact were almost exclusively derived from the first or second read of the Illumina HiSeq instrument. Bona fide variants are supported by near-equal numbers of first and second reads. We made use of the skewed read-orientation combinations and low allele fractions characteristic of this artifact to identify and remove oxoG artifacts from mutation calls in our cohort (i.e. removal of all variants with allele fraction <0.1 or exclusively supported by a single read orientation). This method restored the mutation pattern and frequency seen in earlier sequencing of WGA cases (Supplementary Figure 2D).
Verification of somatic mutations and rearrangements
We used a combination of genotyping and sequencing technologies to verify random candidate mutations (PCR/Sanger and PCR/HiSeq sequencing of candidates from Complete Genomics and BC Cancer Agency Illumina WGS and RNA-seq data), as well as mutations supportive of our significance analyses (Sequenom and PCR/MiSeq of WES and WGS data). Combining all of the validation experiments resulted in overall validation rates of 87% for substitutions (525/605 candidates, 241/282 coding) and 34% for indels (27/79 candidates, 26/41 coding). Some mutations were verified using multiple technologies and therefore the total number of candidate mutations verified is lower than the sum total of mutations described in the Supplementary Note. See Supplementary Note for details and cross-platform comparisons.
Integrated analysis of somatic variation from exome and genome data sets
Somatic mutations detected in WGS, WES, and RNA-seq data sets were annotated using Oncotator (See Broad Institute Cancer Genome Analysis webpage). Genes mutated at a statistically significant frequency were identified using MutSig32, a method that identifies genes with mutation frequencies greater than expected by chance, given detected background mutation rates, gene length and callable sequence in each tumor/normal pair. The relationship between mutation frequency and age of diagnosis was tested using the Spearman rank test. The implementation of the Kolmogorov-Smirnov test in R version 2.11.1 (ks.test) was used to test differences in mutation frequency distributions of several clinical variables (Supplementary Table 4).
To identify frequently mutated groups of genes, we applied the MutSig algorithm to sets of genes rather than individual genes. These gene sets consisted of 853 “canonical pathways” curated by Gene Set Enrichment Analysis49 as well as a lists of chromatin modifiers and splice factors curated from the literature50–52 (Supplementary Table 6 “CHROMATIN_MODIFIERS”, “EPIGENETIC_COMPLEXES”, “SPLICE_FACTORS”, and “DNA_METHYLATION”). Significance analysis of mutations and pathways reported by Molenaar et al17 are provided in the Supplementary Note.
Expression analysis of significantly mutated genes
Alignments of RNA-seq data were used to estimate gene expression levels. Gene coverage analysis was based on Ensembl gene annotations (homo_sapiens_core_59_37d). These annotations were collapsed into a single gene model containing the union of exonic bases from all annotated transcripts of the gene. The analysis used SAMtools pileup to get the per-base coverage depths, and excluded reads with mapping quality < 10 and reads flagged as poor quality according to the Illumina chastity filter. Duplicate reads were kept in this analysis. The reads per kilobase of exon model per million mapped reads (RPKM) metric was used to estimate gene expression level87. RPKM was calculated using the following formula: where
(NORM_TOTAL x sum of the lengths of all exons in the gene) NORM_TOTAL = the total number of reads that are mapped to non-mitochondrial exons The expression threshold for each RNA-seq library was determined as the 95th percentile of the distribution of the expression levels of silent intergenic regions computed and defined as described on the ALEXA platform website88. Using this threshold, we determined that ALK, PTPN11, ATRX, MYCN, and NRAS were expressed above background in each of the 10 cases with available RNA-seq data. In contrast, OR5T1 and PDE6G were not expressed above background in at least 9 out of 10 cases in our cohort.
The TARGET neuroblastoma Affymetrix Human Exon Array data (manuscript in preparation) of 250 primary diagnostic tumor specimens was normalized by quantile normalization and summarized using robust multichip average (Affymetrix Power Tools software package version 1.12). This dataset includes samples from 220 patients with high risk and 30 with low risk disease. The transcript level data of core probe sets for each sample were averaged based on gene symbol annotations provided by the manufacturer (17,422 unique genes). To identify relative expression of genes in neuroblastomas, the percentile values of ALK, PTPN11, ATRX, MYCN, NRAS, OR5T1, and PDE6G were computed from the cumulative distribution function calculated for each sample’s gene profile. Same analysis was conducted on Agilent 44K microarray data (19,528 unique genes) of 416 neuroblastoma tumors from the MicroArray Quality Control (MAQC)-II study (GEO GSE16716)88. This data set includes tumors from patients diagnosed with high risk (n=135), intermediate risk (n=34), or low risk (n=247) neuroblastoma. Su et al89 demonstrated that individual tissues express 30-40% of all genes by comparing microarray expression levels across panels of human and mouse tissues. The median percentile levels for ALK, PTPN11, ATRX, MYCN, and NRAS in both data sets are well within the percentile range of genes that are likely expressed in a tissue. The low median percentile levels for OR5T1 and PDE6G (less than 40%ile and 25%ile in TARGET and MAQC-II data) suggest low expression levels in neuroblastoma tumors (Supplementary Figure 3).
Germline variant analysis
Detection of pathogenic germline variation at base-pair resolution in a cohort of cancer patients is complicated by selection of an appropriately matched and sized control population, relatively high carrier frequencies for unrelated disorders, and complex genetics underlying cancer predisposition. To nominate germline variants predisposing to neuroblastoma, we searched for enrichment of putative functional variants in the blood-derived DNA samples from our WES cohort compared to normal DNAs from 1,974 European American individuals sequenced by the National Heart, Lung, and Blood Institute Grand Opportunity Exome Sequencing Project (ESP)56. As indel calls from the ESP cohort were not publically available at the time of our study, we did not include them in our analysis.
To ensure consistency and accuracy of germline variant detection, all neuroblastoma WES cases were called simultaneously with 800 WES cases from the 1000Genomes project using the UnifiedGenotyper from the Genome Analysis Toolkit. A principal component analysis of the genotype calls was performed to determine the ethnic background of our cases (Supplementary Figure 7) with respect to three 1000Genomes populations. As over 80% of our cohort was Caucasian or ad-mixed Caucasian, we downloaded genotyping calls and coverage information from 1,974 European American individuals available on the ESP website to serve as a control population. To focus our analysis on rare variation consistent with the low prevalence of neuroblastoma, we removed from both data sets all variants present in individuals sequenced as part of the 1000 Genomes project. Next, we generated two lists of rare variants: overlaps with clinically-reported variants recorded in ClinVar (downloaded 4/27/2012, 284 variants in neuroblastoma, 2,947 in ESP) and loss-of-function variants in any of 924 genes listed in the Cancer Gene Census53, Familial Cancer database54, or a list of DNA repair genes55 (86 neuroblastoma, 1,068 ESP). We then tested each gene for significant enrichment of variants in the neuroblastoma compared to the ESP cohort (1-tailed Fisher’s exact test, Supplementary Tables 7 and 8).
The germline ClinVar analysis uncovered four genes of significance driven by single variants seen at greater frequency in neuroblastoma compared to ESP: CYP2D6, NOD2, SLC34A3, and HPD. All of these variants are present at low frequency in an expanded European American ESP cohort (rs5030865 in 1/8,524 chromosomes, rs104895438 in 5/8600, rs121918239 in 14/8514, and rs137852868 in 11/8600), suggesting they are benign polymorphisms. Note that, while candidates detected by this approach are not significant after correction for multiple testing, we believe there is sufficient biological rationale and supporting evidence for validation in larger cohorts. We also looked for overlap with sites recorded in COSMIC33. This analysis identified a TP53 variant associated with Li-Fraumeni syndrome60.
Supplementary Material Supplementary tables 1-10 are provided as separate Microsoft Excel files.
Supplementary Note: Additional verification data, significance analyses, and discussion of complex rearrangements
Supplementary Table 1, Master data table: Clinical and molecular data for all neuroblastoma cases including identifiers from other databases, sequencing technologies used, clinical and biological covariates, and matrix of mutation calls
Supplementary Table 2, Coverage: Fraction of bases in each exon with sufficient coverage for mutation detection
Supplementary Table 3, Full mutation list: All coding somatic mutations called in all cases
Supplementary Table 4, Mutation frequency correlates: Statistical comparison of mutation frequency distributions (Kolmogorov-Smirnov) when comparing cases by clinical and biological variables
Supplementary Table 5, Pathogens: Counts of sequencing reads in exome capture libraries corresponding to known viruses
Supplementary Table 6, MutSig: Significance analysis of somatic mutation frequency in all genes and a focused set of genes listed in the Catalogue of Somatic Mutations in Cancer
Supplementary Table 7, Gene set significance analysis: Full list of pathways, member genes, mutated genes, and significance values as calculated by MutSig with and without significantly mutated genes
Supplementary Table 8, Significance analysis of germline ClinVar variation: List of all genes tested for enrichment in neuroblastoma of ClinVar variants
Supplementary Table 9, Significance analysis of germline loss-of-function variants in Cancer Census, cancer syndrome, or DNA repair genes
Supplementary Table 10, Structural rearrangements: All structural variants detected in neuroblastoma genomes or transcriptomes
Supplementary Table 11, Criteria used to identify somatic mutations in call files provided by the Complete Genomics Cancer Sequencing service
Supplementary Table 12, Criteria used to identify copy number alterations in call files provided by the Complete Genomics Cancer Sequencing service
Supplementary Table 13. Primer sequences used for DNA verification of structural variants and gene fusions in WGS cases
Supplementary Table 14. Primer sequences used for RNA verification of structural variants and gene fusions detected in WGS cases
Supplementary Material
| Therapeutically Applicable Research to Generate Effective Treatments (TARGET) |
http://target.cancer.gov |
| Broad Institute Cancer Genome Analysis tools |
http://www.broadinstitute.org/cancer/cga |
| ClinVar | http://www.ncbi.nlm.nih.gov/clinvar |
| Familial Cancer Database | http://www.facd.info |
| Human DNA repair genes | http://sciencepark.mdanderson.org/labs/wood/DNA_Repair_Genes.html |
| NHLBI Grand Opportunity Exome Sequencing Project |
https://esp.gs.washington.edu/drupal/ |
| International Agency for Research on Cancer TP53 Database |
http://p53.iarc.fr |
| Complete Genomics Analysis Tools | http://www.completegenomics.com/analysis-tools/cgatools/ |
| ALEXA RNA-seq analysis tools | http://www.alexaplatform.org/ |
| Picard analysis tools | http://picard.sourceforge.net |
| The R Project for Statistical Computing | http://www.r-project.org/ |
Acknowledgements
The investigators thank the Children’s Oncology Group for the collection and annotation of samples for this study and all TARGET co-investigators for scientific support of this project. Funding was provided by National Institutes of Health grants CA98543 and CA98413 to the Children’s Oncology Group, RC1MD004418 to the TARGET consortium, CA124709 (J.M.M.) and CA060104 (R.C.C.), and NHGRI U54HG003067 (E.S.L., D.A., S.B.G., G.G., M.M.) as well as a contract from the National Cancer Institute, National Institute of Health (HHSN261200800001E). Additional support included a Canadian Institutes of Health Research Fellowship (T.J.P.), a Roman M. Babicki Fellowship in Medical Research at the University of British Columbia (O.M.), the Canada Research Chair in Genome Science (M.A.M.), the Giulio D’Angio Endowed Chair (J.M.M.), the Alex’s Lemonade Stand Foundation (J.M.M.), the Arms Wide Open Foundation (J.M.M.) and the Cookies for Kids Foundation (J.M.M.). The authors would like to thank Elizabeth Nickerson, Sheridon Channer, Karen Novik, Cecelia Suragh, and Robyn Roscoe for project management support. We also thank the staff of the Genome Sciences Centre Biospecimen Core, Library Construction, Sequencing, and Bioinformatics teams, and the staff of the Broad Institute Biological Samples, Genome Sequencing and Genetic Analysis Platforms for their expertise in genomic processing of samples and generating the sequencing data used in this analysis.
Footnotes
Author Contributions J.M.M., J.K., R.C.S., D.S.G. and M.A.S. conceived and led the project. M.A.M. and M.M. conceived of and supervised all aspects of the sequencing work at the BC Cancer Agency Genome Sciences Centre and Broad Institute, respectively. T.J.P. and O.M performed the analyses and interpreted the results. E.F.A., S.A., J.S.W., K.A.C., M.D., S.J.D., A.C.W., Y.P.M, L.J, T.B., Y.M., J.G-F and M.D.H. selected and characterized samples, provided disease-specific expertise in data analysis, and edited the manuscript. R.S. and W.L. provided statistical support and analyses of clinical covariates. D.A, E.S., C.S., M.D., and J.M.G.A. provided overall project management and quality control support. S.L.C., K.C., M.H., A.K, J.K., M.S.L., L.L, A.M., A.H.R., A.S., and C.S. supported analysis of somatic and germline alterations in the exome sequencing data. C-S.P. performed the pathogen discovery analysis. I.B., K.L.M., R.C., S.D.J., and J.Q. performed de novo assembly of Illumina sequencing data. Y.Z. led the library construction effort for the Illumina libraries. A.T. and Y.Z. planned the sequencing verification, and A.A. and B.K. performed the experiments. R.D.C. performed copy number analysis of genome sequencing data. M.K. performed verification of candidate rearrangements. N.T. performed gene- and exon-level quantification analysis of RNA-seq data. A.L. and A.H.K. helped interpret data provided by Complete Genomics. R.A.M. and M.H. led the sequencing effort for the Illumina genome and transcriptome libraries. S.B.G. and E.S.L. led the sequencing effort for the exome sequencing libraries. G.G. and S.J.M.J. supervised the bioinformatics group at the Broad Institute and BC Cancer Agency Genome Sciences Centre, respectively. T.J.P, O.M., D.J.G., M.A.M., M.M. and J.M.M. co-wrote the manuscript with input from all co-authors.
Author Information M.M. is a paid consultant for and equity holder in Foundation Medicine, a genomics-based oncology diagnostics company, and is a paid consultant for Novartis.
URLs
References
- 1.Maris JM. Recent advances in neuroblastoma. N. Engl. J. Med. 2010;362:2202–2211. doi: 10.1056/NEJMra0804577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith MA, et al. Outcomes for children and adolescents with cancer: challenges for the twenty-first century. J. Clin. Oncol. 2010;28:2625–2634. doi: 10.1200/JCO.2009.27.0421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matthay KK, et al. Treatment of high-risk neuroblastoma with intensive chemotherapy, radiotherapy, autologous bone marrow transplantation, and 13-cis-retinoic acid. Children’s Cancer Group. N. Engl. J. Med. 1999;341:1165–1173. doi: 10.1056/NEJM199910143411601. [DOI] [PubMed] [Google Scholar]
- 4.Yu AL, et al. Anti-GD2 antibody with GM-CSF, interleukin-2, and isotretinoin for neuroblastoma. N. Engl. J. Med. 2010;363:1324–1334. doi: 10.1056/NEJMoa0911123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oeffinger KC, et al. Chronic health conditions in adult survivors of childhood cancer. N. Engl. J. Med. 2006;355:1572–1582. doi: 10.1056/NEJMsa060185. [DOI] [PubMed] [Google Scholar]
- 6.Trochet D, et al. Germline mutations of the paired-like homeobox 2B (PHOX2B) gene in neuroblastoma. Am. J. Hum. Genet. 2004;74:761–764. doi: 10.1086/383253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mosse YP, et al. Germline PHOX2B mutation in hereditary neuroblastoma. Am. J. Hum. Genet. 2004;75:727–730. doi: 10.1086/424530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mosse YP, et al. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature. 2008;455:930–935. doi: 10.1038/nature07261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Janoueix-Lerosey I, et al. Somatic and germline activating mutations of the ALK kinase receptor in neuroblastoma. Nature. 2008;455:967–970. doi: 10.1038/nature07398. [DOI] [PubMed] [Google Scholar]
- 10.Diskin SJ, et al. Common variation at 6q16 within HACE1 and LIN28B influences susceptibility to neuroblastoma. Nat. Genet. 2012;44:1126–1130. doi: 10.1038/ng.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang K, et al. Integrative genomics identifies LMO1 as a neuroblastoma oncogene. Nature. 2011;469:216–220. doi: 10.1038/nature09609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nguyễn LB, et al. Phenotype Restricted Genome-Wide Association Study Using a Gene-Centric Approach Identifies Three Low-Risk Neuroblastoma Susceptibility Loci. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1002026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diskin SJ, et al. Copy number variation at 1q21.1 associated with neuroblastoma. Nature. 2009;459:987–991. doi: 10.1038/nature08035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Capasso M, et al. Common variations in BARD1 influence susceptibility to high-risk neuroblastoma. Nat Genet. 2009;41:718–723. doi: 10.1038/ng.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maris JM, et al. Chromosome 6p22 Locus Associated with Clinically Aggressive Neuroblastoma. N Engl J Med. 2008;358:2585–2593. doi: 10.1056/NEJMoa0708698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deyell RJ, Attiyeh EF. Advances in the understanding of constitutional and somatic genomic alterations in neuroblastoma. Cancer Genetics. 2011;204:113–121. doi: 10.1016/j.cancergen.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 17.Molenaar JJ, et al. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature. 2012;483:589–593. doi: 10.1038/nature10910. [DOI] [PubMed] [Google Scholar]
- 18.Cheung N-KV. Association of Age at Diagnosis and Genetic Mutations in Patients With Neuroblastoma. JAMA: The Journal of the American Medical Association. 2012;307:1062. doi: 10.1001/jama.2012.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sausen M, et al. Integrated genomic analyses identify ARID1A and ARID1B alterations in the childhood cancer neuroblastoma. Nature Genetics. 2012 doi: 10.1038/ng.2493. doi:10.1038/ng.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bentley DR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drmanac R, et al. Human Genome Sequencing Using Unchained Base Reads on Self-Assembling DNA Nanoarrays. Science. 2010;327:78–81. doi: 10.1126/science.1181498. [DOI] [PubMed] [Google Scholar]
- 22.Pugh TJ, et al. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature. 2012;488:106–110. doi: 10.1038/nature11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang L, et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N. Engl. J. Med. 2011;365:2497–2506. doi: 10.1056/NEJMoa1109016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Network TCGAR. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee RS, et al. A remarkably simple genome underlies highly malignant pediatric rhabdoid cancers. Journal of Clinical Investigation. 2012 doi: 10.1172/JCI64400. doi:10.1172/JCI64400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Banerji S, et al. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature. 2012;486:405–409. doi: 10.1038/nature11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chapman MA, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471:467–472. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hodis E, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Imielinski M, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–1120. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stransky N, et al. The Mutational Landscape of Head and Neck Squamous Cell Carcinoma. Science. 2011;333:1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nikolaev SI, et al. Exome sequencing identifies recurrent somatic MAP2K1 and MAP2K2 mutations in melanoma. Nat. Genet. 2012;44:133–139. doi: 10.1038/ng.1026. [DOI] [PubMed] [Google Scholar]
- 32.Getz G, et al. Comment on ‘The Consensus Coding Sequences of Human Breast and Colorectal Cancers’. Science. 2007;317:1500. doi: 10.1126/science.1138764. [DOI] [PubMed] [Google Scholar]
- 33.Forbes SA, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Research. 2010;39:D945–D950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi L, et al. The MicroArray Quality Control (MAQC)-II study of common practices for the development and validation of microarray-based predictive models. Nat. Biotechnol. 2010;28:827–838. doi: 10.1038/nbt.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.George RE, et al. Activating mutations in ALK provide a therapeutic target in neuroblastoma. Nature. 2008;455:975–978. doi: 10.1038/nature07397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen Y, et al. Oncogenic mutations of ALK kinase in neuroblastoma. Nature. 2008;455:971–974. doi: 10.1038/nature07399. [DOI] [PubMed] [Google Scholar]
- 37.Bentires-Alj M, et al. Activating mutations of the noonan syndrome-associated SHP2/PTPN11 gene in human solid tumors and adult acute myelogenous leukemia. Cancer Res. 2004;64:8816–8820. doi: 10.1158/0008-5472.CAN-04-1923. [DOI] [PubMed] [Google Scholar]
- 38.Tartaglia M, et al. Mutations in PTPN11, encoding the protein tyrosine phosphatase SHP-2, cause Noonan syndrome. Nat Genet. 2001;29:465–468. doi: 10.1038/ng772. [DOI] [PubMed] [Google Scholar]
- 39.Tartaglia M, et al. Somatic mutations in PTPN11 in juvenile myelomonocytic leukemia, myelodysplastic syndromes and acute myeloid leukemia. Nat Genet. 2003;34:148–150. doi: 10.1038/ng1156. [DOI] [PubMed] [Google Scholar]
- 40.Sarkozy A, et al. A novel PTPN11 gene mutation bridges Noonan syndrome, multiple lentigines/LEOPARD syndrome and Noonan-like/multiple giant cell lesion syndrome. Eur. J. Hum. Genet. 2004;12:1069–1072. doi: 10.1038/sj.ejhg.5201290. [DOI] [PubMed] [Google Scholar]
- 41.De Brouwer S, et al. Meta-analysis of neuroblastomas reveals a skewed ALK mutation spectrum in tumors with MYCN amplification. Clin. Cancer Res. 2010;16:4353–4362. doi: 10.1158/1078-0432.CCR-09-2660. [DOI] [PubMed] [Google Scholar]
- 42.Brodeur GM, Seeger RC, Schwab M, Varmus HE, Bishop JM. Amplification of N-myc in untreated human neuroblastomas correlates with advanced disease stage. Science. 1984;224:1121–1124. doi: 10.1126/science.6719137. [DOI] [PubMed] [Google Scholar]
- 43.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schwarz JM, Rödelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nature Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 46.Tavtigian SV, et al. Comprehensive statistical study of 452 BRCA1 missense substitutions with classification of eight recurrent substitutions as neutral. J. Med. Genet. 2006;43:295–305. doi: 10.1136/jmg.2005.033878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marchler-Bauer A, et al. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:D225–229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Love C, et al. The genetic landscape of mutations in Burkitt lymphoma. Nature Genetics. 2012 doi: 10.1038/ng.2468. doi:10.1038/ng.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. PNAS. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barbosa-Morais NL, Carmo-Fonseca M, Aparício S. Systematic genome-wide annotation of spliceosomal proteins reveals differential gene family expansion. Genome Res. 2006;16:66–77. doi: 10.1101/gr.3936206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat. Rev. Mol. Cell Biol. 2009;10:741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gabut M, Chaudhry S, Blencowe BJ. SnapShot: The splicing regulatory machinery. Cell. 2008;133:192.e1. doi: 10.1016/j.cell.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 53.Futreal PA, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sijmons RH. Encyclopaedia of tumour-associated familial disorders. Part I: from AIMAH to CHIME syndrome. Hered Cancer Clin Pract. 2008;6:22–57. doi: 10.1186/1897-4287-6-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wood RD, Mitchell M, Lindahl T. Human DNA repair genes, 2005. Mutat. Res. 2005;577:275–283. doi: 10.1016/j.mrfmmm.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 56.Tennessen JA, et al. Evolution and Functional Impact of Rare Coding Variation from Deep Sequencing of Human Exomes. Science. 2012;337:64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sodha N, Mantoni TS, Tavtigian SV, Eeles R, Garrett MD. Rare Germ Line CHEK2 Variants Identified in Breast Cancer Families Encode Proteins That Show Impaired Activation. Cancer Res. 2006;66:8966–8970. doi: 10.1158/0008-5472.CAN-06-1990. [DOI] [PubMed] [Google Scholar]
- 58.Lee SB, et al. Destabilization of CHK2 by a missense mutation associated with Li-Fraumeni Syndrome. Cancer Res. 2001;61:8062–8067. [PubMed] [Google Scholar]
- 59.Dong X, et al. Mutations in CHEK2 Associated with Prostate Cancer Risk. Am J Hum Genet. 2003;72:270–280. doi: 10.1086/346094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Petitjean A, et al. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum. Mutat. 2007;28:622–629. doi: 10.1002/humu.20495. [DOI] [PubMed] [Google Scholar]
- 61.Birch JM, et al. Relative frequency and morphology of cancers in carriers of germline TP53 mutations. Oncogene. 2001;20:4621–4628. doi: 10.1038/sj.onc.1204621. [DOI] [PubMed] [Google Scholar]
- 62.Choi JM, et al. Analysis of PARK genes in a Korean cohort of early-onset Parkinson disease. Neurogenetics. 2008;9:263–269. doi: 10.1007/s10048-008-0138-0. [DOI] [PubMed] [Google Scholar]
- 63.Marongiu R, et al. PINK1 heterozygous rare variants: prevalence, significance and phenotypic spectrum. Hum. Mutat. 2008;29:565. doi: 10.1002/humu.20719. [DOI] [PubMed] [Google Scholar]
- 64.Klein C, et al. PINK1, Parkin, and DJ-1 mutations in Italian patients with early-onset parkinsonism. Eur. J. Hum. Genet. 2005;13:1086–1093. doi: 10.1038/sj.ejhg.5201455. [DOI] [PubMed] [Google Scholar]
- 65.Fredlund E, Ringnér M, Maris JM, Påhlman S. High Myc pathway activity and low stage of neuronal differentiation associate with poor outcome in neuroblastoma. PNAS. 2008;105:14094–14099. doi: 10.1073/pnas.0804455105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Storlazzi CT, et al. Gene amplification as double minutes or homogeneously staining regions in solid tumors: origin and structure. Genome Res. 2010;20:1198–1206. doi: 10.1101/gr.106252.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stephens PJ, et al. Massive Genomic Rearrangement Acquired in a Single Catastrophic Event during Cancer Development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Krzywinski M, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gnirke A, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotech. 2009;27:182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Goya R, et al. SNVMix: predicting single nucleotide variants from next-generation sequencing of tumors. Bioinformatics. 2010;26:730–736. doi: 10.1093/bioinformatics/btq040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Carnevali P, et al. Computational techniques for human genome resequencing using mated gapped reads. J. Comput. Biol. 2012;19:279–292. doi: 10.1089/cmb.2011.0201. [DOI] [PubMed] [Google Scholar]
- 73.Kostic AD, et al. PathSeq: software to identify or discover microbes by deep sequencing of human tissue. Nature Biotechnology. 2011;29:393–396. doi: 10.1038/nbt.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Robertson G, et al. De novo assembly and analysis of RNA-seq data. Nat Meth. 2010;7:909–912. doi: 10.1038/nmeth.1517. [DOI] [PubMed] [Google Scholar]
- 75.Lee W, et al. The mutation spectrum revealed by paired genome sequences from a lung cancer patient. Nature. 2010;465:473–477. doi: 10.1038/nature09004. [DOI] [PubMed] [Google Scholar]
- 76.Morin RD, et al. Somatic mutation of EZH2 (Y641) in Follicular and Diffuse Large B-cell Lymphomas of Germinal Center Origin. Nat Genet. 2010;42:181–185. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Morin RD, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476:298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pruitt KD, Tatusova T, Klimke W, Maglott DR. NCBI Reference Sequences: current status, policy and new initiatives. Nucleic Acids Res. 2009;37:D32–36. doi: 10.1093/nar/gkn721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Morozova O, et al. System-Level Analysis of Neuroblastoma Tumor–Initiating Cells Implicates AURKB as a Novel Drug Target for Neuroblastoma. Clinical Cancer Research. 2010;16:4572–4582. doi: 10.1158/1078-0432.CCR-10-0627. [DOI] [PubMed] [Google Scholar]
- 80.Morin R, et al. Profiling the HeLa S3 transcriptome using randomly primed cDNA and massively parallel short-read sequencing. BioTechniques. 2008;45:81–94. doi: 10.2144/000112900. [DOI] [PubMed] [Google Scholar]
- 81.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Robinson JT, et al. Integrative genomics viewer. Nat Biotech. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25:2865–2871. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shah SP, et al. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature. 2009;461:809–813. doi: 10.1038/nature08489. [DOI] [PubMed] [Google Scholar]
- 85.Simpson JT, et al. ABySS: a parallel assembler for short read sequence data. Genome Res. 2009;19:1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pruitt KD, et al. The consensus coding sequence (CCDS) project: Identifying a common protein-coding gene set for the human and mouse genomes. Genome Res. 2009;19:1316–1323. doi: 10.1101/gr.080531.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 88.Griffith M, et al. Alternative expression analysis by RNA sequencing. Nat Meth. 2010;7:843–847. doi: 10.1038/nmeth.1503. [DOI] [PubMed] [Google Scholar]
- 89.Su AI, et al. Large-scale analysis of the human and mouse transcriptomes. Proc. Natl. Acad. Sci. U.S.A. 2002;99:4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.