Abstract
Microbial community diversity and heterogeneity in saturated and unsaturated subsurface soils from Abbott's Pit in Virginia (1.57, 3.25, and 4.05 m below surface) and Dover Air Force Base in Delaware (6.00 and 7.50 m below surface) were analyzed using a culture-independent small-subunit (SSU) rRNA gene (rDNA)-based cloning approach. Four to six dominant operational taxonomic units (OTUs) were identified in 33 to 100 unique SSU rDNA clones (constituting about 40 to 50% of the total number of SSU rDNA clones in the clone library) from the saturated subsurface samples, whereas no dominant OTUs were observed in the unsaturated subsurface sample. Less than 10% of the clones among samples from different depths at the same location were identical, and the proportion of overlapping OTUs was lower for the samples that were vertically far apart than for adjacent samples. In addition, no OTUs were shared between the Abbott's Pit and Dover samples. The majority of the clones (80%) had sequences that were less than 5% different from those in the current databases. Phylogenetic analysis indicated that most of the bacterial clones were affiliated with members of the Proteobacteria family (90%), gram-positive bacteria (3%), and members of the Acidobacteria family (3%). Principal component analysis revealed that samples from different geographic locations were well separated and that samples from the same location were closely grouped together. In addition, the nonsaturated subsurface samples from Abbott's Pit clustered together and were well separated from the saturated subsurface soil sample. Finally, the overall diversity of the subsurface samples was much lower than that of the corresponding surface soil samples.
Soil microbial communities are among the most complex, diverse, and important assemblages in the biosphere. Because of such high-level diversity, soil microbial communities are among the most difficult to phenotypically and genetically characterize. DNA-based molecular techniques have the potential to provide a comprehensive picture of soil microbial community diversity and composition, because both culture-grown and non-culture-grown components of a community can be surveyed. Investigations using small-subunit (SSU) rRNA gene (rDNA)-based cloning and sequencing approaches have revealed astonishing diversity in soils and other environments (cf. 1, 2, 4, 5, 7, 10-14, 17, 20, 21, 23, 26, 29, 33-35, 38-40, 42, 45, 46, 51). Many studies using single soil samples showed that very little repetition has been observed among the SSU rRNA sequences obtained (3-5, 13, 29, 33, 40, 45, 51, 53). However, due to the complexity of soil communities and the efforts required for the cloning and sequencing-based approach, only a limited number of soil environments have been surveyed. Thus, our understanding of the extent of microbial diversity in soil environments is still very limited. More studies of a variety of soil types and habitats are needed to obtain a more comprehensive view of microbial community diversity and structure in soil environments.
Studies of subsurface environments have received attention because they are important to human health, ecosystem functions, agriculture, and environmental management (22, 32). Since microorganisms play essential roles in subsurface geology, hydrology, and ecology, knowledge about microbial community structure and composition is important to improve our conceptual and predictive understanding of subsurface ecosystem processes, functions, and management. However, subsurface microbial communities are often difficult to investigate because subsurface environments are characterized by very high levels of spatial and temporal variability in subsurface hydrological and biogeochemical processes. Considerable spatial variability of microbial distribution in subsurface environments was also observed using traditional microbiological methods (19, 43, 48).
In contrast to studies of surface microbial communities, studies of subsurface environments have not adequately characterized microbial community composition and diversity because the cost and difficulty of obtaining a large number of samples have been prohibitive. Relatively few studies (e.g., 6, 8, 9) have used culture-independent molecular approaches to obtain specific information on the composition of the subsurface microbial community. Thus, it is not clear whether the subsurface microbial community is closely tied to the surface soil microbial community or is an independent ecosystem with a distinct assemblage of microorganisms. Also, subsurface communities are isolated from each other to a greater degree than are surface soil communities. In surface soil, aeolian transport, flooding, and other mechanisms can move and mix soils over significant distances. There is much less opportunity for this type of transport in the subsurface. Thus, community differences between sites separated even by relatively small distances are potentially greater for subsurface communities.
In a previous study, Zhou et al. used SSU rRNA-based restriction fragment length polymorphism (RFLP) analysis to examine 29 soil samples taken from the surface, vadose, and saturated zones of four geographically distinct locations (52). Zhou et al. also examined the phylogenetic diversity of two humid-region, sandy surface soils from Virginia and Delaware (53). The results indicated that the surface soil microbial community is extremely diverse, which is consistent with the observations reported from many other studies (cf. 4, 5, 12, 13, 16, 20, 23, 24, 26, 30, 34, 39, 51). Zhou et al. hypothesized that spatial isolation and resource heterogeneity are key mechanisms controlling microbial community diversity and structure (53). Based on these hypotheses, we predict that levels of phylogenetic diversity of saturated subsurface environments with low carbon content will be low. In this paper, we therefore extend our previous work by further examining the phylogenetic diversity of the microbial communities of five sandy subsurface soils from two graphically separated locations (Delaware and Virginia). Our results indicate that microbial distribution is heterogeneous in the subsurface environments but that the level of microbial diversity is much lower than that observed in the surface soils.
MATERIALS AND METHODS
Site description and soil sample collection.
Five subsurface soil samples were collected from previously described sites (52) near the Atlantic Coast of northern Virginia (Abbott's Pit [sites Ab-4, Ab-5, and Ab-10]) and central Delaware (Dover Air Force Base [sites D1-4 and D1-8]) (Table 1). Soil coring was done with a sonic drilling technique and using steam-cleaned Lexan core liners. Soil from the center of the core was removed by a sterile spatula, collected into sterile Whirl-Pak bags, and immediately frozen.
TABLE 1.
SSU rDNA clone diversity in subsurface soil samples from Abbott's Pit (Virginia) and Dover Air Force Base (Delaware)
| Sample | Sampling depth in m (sampling site) | Total no. of SSU, clones | No. of unique SSU, rDNA clones | No. (%) of unique clones, resolved by:
|
|
|---|---|---|---|---|---|
| 1st pair of enzymesa | 2nd pair of enzymesb | ||||
| AVZ | 1.57 (Ab-3) | 54 | 33 | 29 (87.9) | 4 (22.1) |
| AAW | 3.25 (Ab-5) | 74 | 36 | 30 (83.3) | 6 (16.7) |
| ABW | 4.05 (Ab-10) | 252 | 85 | 65 (76.5) | 20 (23.5) |
| DBW1 | 6.00 (D1-4) | 216 | 100 | 80 (80.0) | 20 (20.0) |
| DBW2 | 7.50 (D1-8) | 218 | 79 | 57 (72.2) | 22 (27.8) |
MspI plus RsaI.
HhaI plus HaeII.
The soils from these sites were very similar except for the presence of a low level of organic contaminants at the Dover sites. The soils from Abbott's Pit were medium- and fine-grain sand derived from beach and upper shore surface sediments. The water table was 3.75 m below land surface at the time of sampling but may fluctuate annually up to a meter. The total organic carbon (TOC) levels ranged from 0.27 to 1.83 mg/g of sample (52). The water table at the Dover site at the time of sampling was at 5.5 m. The TOC levels were 0.84 and 0.51 mg/g for D1-4 and D1-8, respectively (52). The Dover site was contaminated with low levels of chloroethenes and petroleum hydrocarbons. Groundwater contaminants in well IR-7 (located near the Dover sites) were measured at 15.6 μg of perchloroethylene/liter, 504 μg of trichloroethylene/liter, 1,190 μg of cis-dichloroethylene/liter, and 596 μg of vinyl chloride/liter (31). Benzene, ethyl benzene, toluene, and zylene concentrations in this region in 1996 were reported to be 17 μg/liter (unpublished data); in general, these concentrations (where present) were below 200 μg/liter (47). The sediment texture was more variable at the Dover site than at the Abbott's Pit site, ranging from coarse sands to clay (see, e.g., reference 47). More details on the sites can be found in previously published papers (e.g., references 36 and 36a).
DNA extraction.
Combined methods (grinding, freezing and thawing, and the use of sodium dodecyl sulfate) were employed to directly extract bulk community DNA from 10 g of soil for cell lysis (50). The grinding process was performed under sterile conditions in the presence of liquid nitrogen. The crude DNA was purified by a Mini Column purification method as described previously (50) except that the DNA was eluted twice from the resin column with 50 μl of hot water (80°C) each time.
SSU rDNA amplification, cloning, RFLP analysis, and sequencing.
PCR amplification, cloning, screening, and sequencing of SSU rDNA were performed exactly as described previously (53) except that the DNA template concentration for PCR amplification ranged from 300 to 700 pg. All of the white colonies were picked and screened for SSU rDNA inserts, which were amplified (20 μl) with the primers specific to the polylinker of the vector pCR II (51). One-third of the remaining 18 μl of PCR-amplified products was digested overnight at 37°C with 0.1 U of each endonuclease in the pair MspI plus RsaI and the pair HhaI plus HaeIII (Gibco BRL Life Technologies, Gaithersburg, Md.). The resulting RFLP products were separated by gel electrophoresis in 3.5% Metaphor agarose (FMC Bioproducts, Rockland, Maine). RFLP patterns were compared using Molecular Analyst software (Bio-Rad, Hercules, Calif.). Clones with unique patterns are reported as operational taxonomic units (OTUs); their frequencies were used as an indicator of abundance, but they may not reflect the true number of these organisms in the environment.
The unique rDNA clones were sequenced with primer 529R (5′CGCGGCTGCTGGCAC3′) and a BigDye Terminator kit (Applied Biosystems, Foster City, Calif.) and using a 3700 DNA analyzer (Perkin-Elmer, Wellesley, Mass.) at a 5:1 dilution according to the instructions of the manufacturer. For the clones whose rDNAs were 85% or less similar to the SSU rDNAs in current databases, nearly full sequences were obtained using an additional nine forward primers (Escherichia coli SSU rRNA positions of 342 to 357, 519 to 533, 787 to 802, and 1099 to 114) and reverse primers (E. coli SSU rRNA positions of 357 to 342, 529 to 515, 802 to 787, and 115 to 1100) as described previously (49). Sequences were obtained for all clones that appeared more than once in the samples and for a selection of the unique clones.
Sequence analysis.
The partial DNA sequences obtained with the primer 529R were edited with SeqEd software (PE Biosystems). Only the regions (120 to 450 bp) having good quality were used for phylogenetic analysis. For the clones whose sequences were determined with multiple primers, all of the sequences from different primers were assembled with phredPhrap and Consed software (University of Washington, Seattle) without editing with SeqEd. Chimeric sequences were detected by using CHECK_CHIMERA software (28), by using the branching order discrepancies in phylogenetic trees inferred independently with the 5′ and 3′ end sequences, and by comparing all of the sequences obtained in this study. The clones whose partial sequences were available were subjected to heteroduplex analysis with polyacrylamide gel electrophoresis as described previously (37).
Preliminary sequence analysis and the affiliation of the clones were carried out as described previously (53). Appropriate subsets of SSU rDNA sequences were selected on the basis of the initial phylogenetic results and subjected to final phylogenetic analysis using the maximum-likelihood method (fastDNAml software; Ribosomal Database Project) and the neighbor-joining method (ARB software) with bootstrap analyses. The phylogenetic trees established by maximum likelihood were constructed with a transition/transversion ratio of 2.0 by using jumbled orders of 10 for the addition of taxa.
Statistical methods.
Data analysis was carried out using SigmaPlot software and Statistical Analysis System (SAS) for Windows software (version 8.02) (SAS Institute, Cary, N.C.). The rarefaction analysis was performed with SigmaPlot software. An exponential model, y = a × [1 − exp(−b × x)], was used with SigmaPlot 8.0 nonlinear regression software to fit the clone distribution data. SAS was used primarily for principal component (PC) analysis (PCA) of the clone data. For this analysis the clones were categorized at the genus level, yielding 33 units of bacteria for the analysis. Most of the clones were recognized genera (e.g., Pseudomonas); for a few clones, however, there were no close matches in the databases or the closest matches were with sequences that were not assigned to a recognized genus. In these cases, the unit was given an arbitrary designation. These units were used with standard SAS methods for PCA. Because no sequences of the unique clones were obtained for the Abbott's Pit samples taken just above the water table (AAW samples) and the Abbott's Pit vadose zone (AVZ) samples, the PCA was also performed by removing the unique clones from all samples to assess the potential bias towards the samples that had sequences from the unique clones. As a result, 11 bacterial units were used in the PCA.
Nucleotide sequence accession number.
The rDNA sequences of the sequenced clones are available under the following GenBank accession numbers: AY456755 to AY456883 and AY456885 to AY456903.
RESULTS
SSU rDNA RFLP analysis.
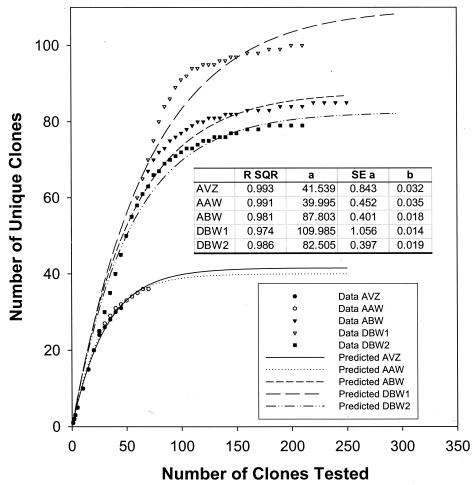
A total of 54 to 252 clones containing entire SSU rRNA inserts were obtained from the DNA isolated from each of the different subsurface soils (Table 1). A range of 33 to 100 unique SSU rDNA fragments from each soil sample were detected with four tetrameric enzymes. While a majority of the unique SSU rDNA clones among these samples were detected with the restriction enzymes MspI plus RsaI, an additional 17 to 28% of the unique patterns were obtained by a secondary enzyme digestion (Table 1) with the restriction enzymes HhaI plus HaeII. The frequency distributions of the intact SSU rDNA clones among these OTUs for each sample were determined. OTUs were ranked in the order of abundance. While almost no dominant OTUs were found in the AVZ sample, one clone was represented more than 10 times in the AAW sample. Four to six OTUs were represented more than 10 times in the other samples (data not shown). These dominant clones accounted for 40 to 50% of the SSU rDNA clones. The remaining OTUs were present at low levels. Rarefaction curves (plots of the cumulative number of OTUs as a function of a clone number) indicated that the majority of the OTUs in the sample were detected (Fig. 1). A significant (P = 0.05) decrease in the rate of OTU detection with increasing numbers of clones examined was observed in rarefaction curves in all of these samples (Fig. 1). While more than 80% of the OTUs were detected within the first 30 SSU rDNA clones for the AVZ and AAW samples and within the first 80 clones for the ABW and DBW2 samples, less than 20% of the total OTUs were detected among the remaining 50 to 70% of the clones. All of the dominant clones were detected in the first 15 clones analyzed in all samples. Nonlinear regression analysis suggests saturation at 42, 40, 88, 110, and 83 for the AVZ, AAW, ABW, DBW1, and DBW2 samples, respectively. This suggests that the level of analysis was sufficient to detect the level of community diversity and infer the level of distribution within these communities.
FIG. 1.
Evaluation of the representation of the clones obtained from the subsurface soil samples by rarefaction analysis. (A) Abbott's Pit samples. (B) Dover samples. The SSU rDNA clones were numbered on the basis of their order of initial detection, which is assumed to be stochastic relative to the distribution of clones in the clone library.
There were differences between the Abbott's Pit samples and the Dover samples at both the clone and genus level. About 10% of the clones were identical between the AVZ (1.57-m depth) and AAW (3.25-m) samples and between the AAW and ABW (4.05-m) samples, whereas about 5% of the clones were identical between the AVZ and ABW samples (Table 2). About 8% of the clones were identical between these two Dover soil samples. Several clones (e.g., those from ABW1, ABW12, ABW126, DBW120, and DBW2) were dominant in the samples from different depths. However, no identical OTUs were found between the Abbott's Pit and Dover samples. At the genus level, 100% overlap was observed between the AVZ and AAW samples (which each had only four genus level groups). The next greatest similarity at the genus level was between the two Dover samples, between which there was a 42% overlap. Not surprisingly, there was more overlap between the Dover and Abbott's Pit samples at the genus level (approximately 17 to 23%) than was observed at the clone level, where there was no overlap.
TABLE 2.
The number and percentages of OTUs and genera overlapping among different subsurface soil samplesa
| Sample | No. (%) overlapping with:
|
||||
|---|---|---|---|---|---|
| AVZb | AAWc | ABWd | DBW1e | DBW2f | |
| AVZ | 7 (10.1) | 6 (5.1) | 0 (0) | 0 (0) | |
| AAW | 4 (100) | 13 (10.7) | 0 (0) | 0 (0) | |
| ABW | 3 (15) | 3 (15) | 0 (0) | 0 (0) | |
| DBW1 | 3 (18.75) | 3 (18.75) | 5 (17) | 15 (8.4) | |
| DBW2 | 3 (23.5) | 3 (23.08) | 5 (19.23) | 8 (42) | |
Upper right portion of the table is the overlap based on OTU. Lower left portion of the table is the overlap based on genus. For genus comparison, percent overlap is the number of overlapping genera between the two samples divided by the total number of genera at the two sites.
n = 33 (OTU); n = 4 (genus).
n = 36 (OTU); n = 4 (genus).
n = 85 (OTU); n = 19 (genus).
n = 100 (OTU); n = 15 (genus).
n = 79 (OTU); n = 12 (genus).
Phylogenetic analysis.
To determine the phylogenetic diversity, representative SSU rDNA clones of OTUs that occurred more than once in a given library, as well as representatives of some of the unique OTUs, were partially sequenced. Most of the clones whose sequences showed less than 85% similarity to the sequences in the current databases were fully sequenced. The majority (80.7%) of the clones sequenced had sequences that were less than 5% different from those in the current databases. The sequences of about 12% of the clones differed by 5 to 10% from the sequences in the databases, those of 3.3% of the clones differed by 10 to 15%, and those of only 4% of the clones differed by more than 15%.
The phylogenetic distribution of the Abbott's Pit and Dover soil samples was established with a bootstrap neighbor-joining method with the sequences from all known and candidate divisions (5, 53). These sequences fell into 6 of the 35 to 42 putative main phylogenetic divisions. While a few clones were affiliated with the Acidobacterium family (2.7%) and gram-positive bacteria (2.7%), the majority (90%) of the clones were affiliated with the Proteobacteria family. Within the Proteobacteria family, the majority of the clones were affiliated with the γ-Proteobacteria (50.3%) classification, with a substantial portion of the clones falling into the α-Proteobacteria (20.1%) and β-Proteobacteria (19.5%) classifications. About 0.7% of the clones were affiliated with green sulfur bacteria of the genus Thermonema and the novel candidate division AD1 (53).
The phylogenetic distributions of the sequences between Abbott's Pit and Dover subsurface soil samples were similar for some dominant divisions. For example, members of the γ-Proteobacteria family were the most prevalent in both Abbott's Pit (53.2%) and Dover (47.2%) samples. However, the relative levels of abundance of members of the α- and β-Proteobacteria families were reversed between Abbott's Pit and Dover subsurface samples. In the Abbott's Pit samples, a total of 30% of the clones (primarily of the genus Agrobacterium) belonged to the α-Proteobacteria family and 7.8% belonged to the β-Proteobacteria family. In the Dover samples, 31.9% of the clones (primarily of the genus Ralstonia) belonged to the β-Proteobacteria family and 13.9% belonged to the α-Proteobacteria family. The proportion of members of the Acidobacteria family was low in both the Abbott's Pit and Dover samples (6.5% and 4.2%). Members of the novel candidate division AD1 were observed in the Dover samples (1.3%) but not in the Abbott's Pit samples. Similarly, clones from green sulfur bacteria occurred in the Dover samples but not in the Abbott's Pit samples. The reverse was true for the Thermonema clones, which only appeared in the Abbott's Pit samples.
At the genus level, the similarities and differences among these sites were pronounced (Table 3). Clones similar to Pseudomonas species were clearly dominant (representing nearly 48% of the total at the five sites) and ubiquitous (occurring at all five sites). The distribution of clones similar to Agrobacterium species was comparable in that they occurred in all five samples but at lower levels. Clones similar to Ralstonia species were also dominant at Dover but were not present at Abbott's Pit. Clones similar to Halomonas and Staphylococcus species were primarily found in the Abbott's samples. Clones similar to Stentrophomonas and Burkholderia species were generally found in the Dover samples.
TABLE 3.
Summary of the units of bacteria found that were present at the 5 sites at a mean abundance of >0.05%a
| Genus | Group | % of units of bacteria in sample
|
Mean % | No. of diffb | PCc | Loadingd | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| DBW2 | DBW1 | AAW | AVZ | ABW | ||||||
| Pseudomonas | Gamma | 19.8 | 50.5 | 76.8 | 48.2 | 44.5 | 47.96 | 20 | 2 | −0.234 |
| Agrobacterium | Alpha | 16.6 | 10.8 | 1.9 | 3.7 | 31.4 | 12.88 | 1 | 1 | 0.206 |
| Ralstonia | Beta | 33.4 | 15.3 | 0 | 0 | 0 | 9.74 | 1 | 2 | 0.268 |
| Halomonas | Gamma | 1 | 0.5 | 13.6 | 16.8 | 4.3 | 7.24 | 2 | 2 | −0.326 |
| Staphylococcus | Low G+C | 0 | 0 | 4.1 | 3.7 | 0 | 1.56 | 1 | 2 | −0.317 |
| Stenotrophomonas | Gamma | 3.4 | 0.5 | 0 | 0 | 0 | 0.78 | 1 | 3 | −0.260 |
| Burkholderia | Beta | 0.5 | 1 | 0 | 0 | 1.2 | 0.54 | 4 | 2 | 0.243 |
Data are presented as percentage of each sample.
diff, the number of different strains represented.
The PC on which the unit loads most heavily.
Loading values represent the results of PCA of the 33 units.
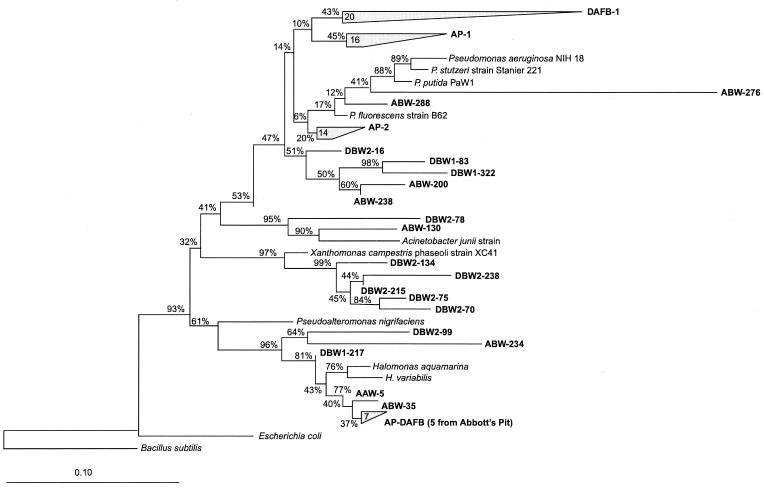
All of the clones affiliated with the γ-Proteobacteria family fell into four major groups. The most abundant group (76% of the total clones in the γ subdivision) was affiliated with Pseudomonas species (Fig. 2). Within this group, four subgroups existed (Fig. 2): AP-1 and AP-2 occurred in only the Abbott's Pit samples whereas subgroup DAFB-1 existed only in the Dover samples. Five clones (ABW-200, ABW-322, DBW1-83, DBW1-322, and DBW2-16) were well separated from the three subgroups (AP-1, AP-2, and DAFB-1) described above and were observed in both locations. Nine clones exhibited 94 to 99% similarity to Halomonas species. Three clones (ABW-234, DBW1-217, and DBW2-99) were also clustered with Halomonas species, but they were less similar (84 to 89% similarity). Five clones were associated with Xanthomonas species (90 to 93% similarity), and these clones were only found in the Dover samples. Two clones were associated with Acinetobacter junii, with a similarity of 87 to 91%.
FIG. 2.
Phylogenetic relationships of the SSU rDNA sequences present in these samples with respect to the γ-Proteobacteria family. The tree was constructed using a neighbor-joining method. Bootstrap analyses with 10,000 replicates were performed to provide confidence estimates for tree topologies.
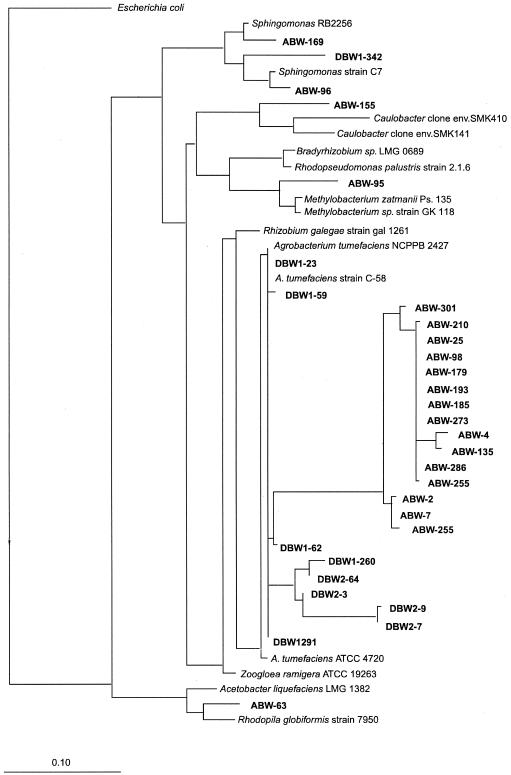
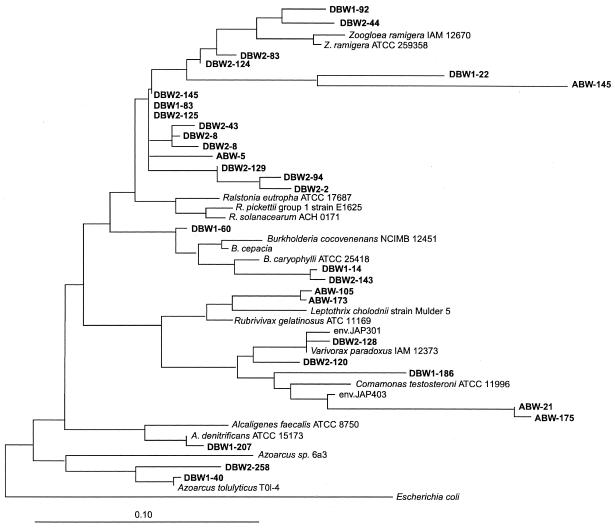
Clones associated with the α-Proteobacteria family were the second most dominant, falling into five different groups. The most abundant group (80%) was closely related to Agrobacterium species, with levels of similarity ranging from 87 to 99%. This group can be divided into three subgroups (Fig. 3): two subgroups were observed in the Dover subsurface soil samples, whereas the other subgroup was only found in the Abbott's Pit samples. Three clones were related to Sphingomonas; one clone each was clustered with Rhodopila globiformis, clone SMK141 (related to Caulobacter species), and Methylobacterium species. The majority of the clones (50%) in the β subdivision of the Proteobacteria family were closely related to Ralstonia eutropha, with similarity of 95 to 98% (Fig. 4). All of these Ralstonia species-related clones (except for ABW-145) were from the Dover surface samples. Seven clones (23.3%) from both the Abbott's Pit and Dover subsurface samples were related to Rubrivivax, Variovorax, and Comamonas species. Small numbers of clones were related to Burkholderia species (three clones), the anaerobic toluene-degrading bacteria Azoarcus tolulyticus (two clones), and Alcaligenes denitrificans (one clone).
FIG. 3.
Phylogenetic relationships of the SSU rDNA sequences present in these samples with respect to the α-Proteobacteria family. The tree was constructed using a maximum-likelihood method.
FIG. 4.
Phylogenetic relationships of the SSU rDNA sequences present in these samples with respect to the β-Proteobacteria family. The tree was constructed using a maximum-likelihood method.
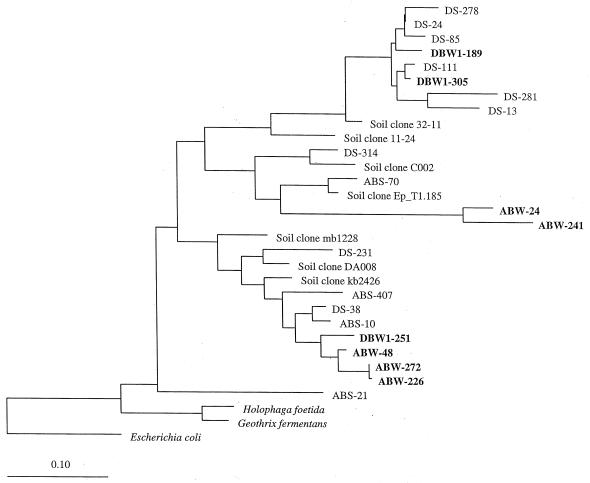
Phylogenetic analysis indicates that members of the Acidobacterium family are also found in the subsurface environment and that all of the eight clones fell into three of the eight subdivisions, which may represent a novel subdivision (Fig. 5). Four clones were affiliated with subdivision 6 of the Acidobacterium family (Fig. 5). The clones in this subdivision were closely related to the soil clones ABS-10 and DS-38 (91 to 95% similarity), which were recovered from the surface soil samples (53). While two clones from Abbott's Pit were affiliated with subdivision 1, the other two clones were grouped with subdivision 4 (Fig. 5). The clones in subdivision 1 were related to the forest soil clone Ep_T1.185 and to clone ABS-70, and the clones in subdivision 4 were similar to soil clone 32-11.
FIG. 5.
Phylogenetic relationships of the SSU rDNA sequences present in these samples with respect to the Acidobacterium family. The tree was constructed using a maximum-likelihood method.
Both PCAs (performed with 33 and 11 genus-level units each) captured virtually all of the variability in the clone data (data not shown). Approximately 99% of the variability in the 33 bacterial units used in the analysis was represented in three PCs. The first PC (PC 1) captured 50.5% of the variability, and PC 2 and PC 3 captured 26.5 and 22.2%, respectively. Very similar results were obtained for the PCA with the 11 units. In the following section, therefore, we focus on describing the results for the PCA that used 33 bacterial units.
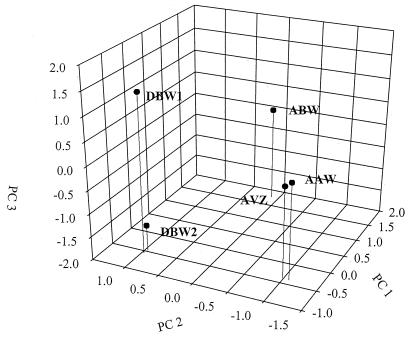
A broad range of bacterial units, including clones similar to Bacillus and Sphingomonas species and an unidentified clone (ABS130) (53), load heavily on the PC 1, and these are among the many units that represent less than 0.5% of the total clones sequenced. Also, the eight rarest units all loaded heavily on the PC 1. The only unit that loaded heavily on PC 1 and whose frequency of appearance was higher than 0.5% was that associated with Agrobacterium (Table 3), but it loaded almost as heavily on PC 2 (0.175) as on PC 1 (0.206). PC 1 primarily separated the ABW sample from the others (Fig. 6). Thus, the bacteria loading on PC 1 likely represented key differences between ABW samples and the samples from the other sites.
FIG. 6.
PCA of the 33-unit case, with the stations plotted in component space.
Many of the more commonly occurring units loaded heavily on the PC 2, either negatively (clones similar to Pseudomonas, Halomonas, and Staphylococcus species) or positively (clones similar to Ralstonia and Burkholderia species) (Table 3). PC 2 separated two of the Abbott's Pit samples (AVZ and AAW) from the Dover samples (Fig. 6). These results suggest that these sites differ significantly in many of these commonly occurring groups.
Among the most common units present, only clones similar to Stenotrophomonas species loaded heavily on PC 3 (Table 3). The other units loading heavily on PC 3 included clones similar to Zoogloea, Rhizomonas, and Variovorax species and several different unidentified clones. These units presented less than 0.1 to 0.2% of the total. The PC 3 served to separate the two Dover samples from each other and from those from the Abbott's Pit sites (Fig. 6). Thus, it appears that of the moderately common species, only Stenotrophomonas species are important in differentiating between the two Dover sites and that several relatively rare species separated the Dover sites from each other and from the Abbott's Pit sites.
In the three-dimensional PCA graph, the samples from Dover were well separated from the samples from Abbott's Pit (Fig. 6). These results suggested that the overall community structures were significantly different between these two sites. Also, AAW and AVZ, the two nonsaturated Abbott's Pit samples, were more closely clustered together than they were with ABW, the saturated Abbott's Pit sample, suggesting that the overall microbial community structures were more similar between the unsaturated samples than they were to the structures seen with the saturated sample.
DISCUSSION
Microbial diversity is much greater in soil than elsewhere, apparently because the soil matrix can promote and sustain diversification. Many recent studies of surface soils have indicated that the soil microbial community is extremely diverse (cf. 4, 5, 12, 13, 20, 23, 24, 26, 30, 34, 39). Results determined by Zhou et al. (on the basis of molecular approaches that used SSU rRNA) with respect to the surface soils support the general notion of extremely high-level diversity of soil microbial communities (52, 53). In contrast, we found in this study that the microbial community in the subsurface environment is much less diverse than in the surface environment. For example, the sequences in surface soils fell into 13 different divisions (53) whereas the sequences in the subsurface samples were clustered into 6 different divisions. Also, the majority (82%) of the clones from the surface soil samples had sequences that were less than 15% different from those in the current databases whereas only 4% of the clones from the subsurface soil samples were more than 15% different.
The microbial community structures are distinctly different in the surface and subsurface soils. In the surface soils, no dominant OTUs were observed and most of the clones remained at low levels of abundance. Zhou et al. referred to this phenomenon as a noncompetitive diversity pattern (52). In contrast, the saturated subsurface samples exhibited dominance of one or a few community members and much less diversity of OTUs. We refer to this as a competitive diversity pattern. Both spatial isolation and resource heterogeneity could explain the differences in microbial community diversity patterns and structures observed with the surface and subsurface samples (44, 52).
Two hypotheses could explain how the noncompetitive and competitive diversity patterns are formed (52). First, spatial isolation (because of low moisture) in the surface samples could allow for maintenance of diverse types of microorganisms and lead to a high level of diversity. At the surface, levels of water content differ and are usually low. Water film is transient, existing only after a rainfall. As gravity removes the water film, the connectivity of soil particles decreases and a high degree of spatial isolation results. As a result, microbial species that would normally be lost by competitive exclusion are able to persist. In saturated soils, excess water allows for a high level of connectivity. Consequently, there is ample opportunity for competition due to the movement of nutrients and microbes. Under these conditions, the microorganisms best able to scavenge nutrients or migrate to a nutrient source outgrow less-fit types. Thus, some bacteria become dominant, which leads to a decrease in diversity. Both mathematical and experimental studies have indicated that spatial isolation is a key mechanism in controlling microbial community diversity patterns of these soil samples (44, 52). A key prediction of the spatial isolation hypothesis is that microbial community will be less diverse in competitive environments than in noncompetitive environments. The observation that microbial communities in subsurface environments are much less diverse than those at the surface supports the spatial isolation hypothesis.
Reports of previous studies by Zhou et al. with respect to subsurface samples with various levels of carbon content suggest that carbon resources also play a role in controlling microbial diversity and structure (52). Greater resource heterogeneity of the surface soil allows for the maintenance of high-level microbial diversity. At many sites, it is likely that both TOC levels and the variety of carbon types decrease with increasing soil depth due to the input of carbon at the surface from primary production. For example, the carbon content at these sites is 10- to 20-fold lower in the subsurface soils than in the surface soils. Thus, multiple resources at the surface soil could create a variety of microhabitats that support a diverse collection of species. In the saturated zone, the community is farther from the primary nutrient resource and many labile substrates may be absent. The lack of diverse carbon sources would result in fewer species. The low level of diversity in the saturated subsurface samples (as indicated by the results of sequence-based phylogenetic analysis) supports the hypothesis that resource heterogeneity might also play important roles in shaping microbial community diversity and structure.
Subsurface environments are chemically and physically heterogeneous. Two important issues are how such heterogeneous environments affect microbial distribution and behavior and how microbial community diversity and structure are spatially different. In samples that were only a few meters apart vertically in this study, more than 90% of OTUs were different. The proportion of the overlapping OTUs was lower between the samples vertically far apart than that seen between adjacent samples. In addition, no identical clones were observed among the subsurface and surface soil samples. While no dominant populations were observed in surface soil samples, the subsurface soils were dominated by members of the Proteobacteria family (90%). These results suggest that microbial community composition is heterogeneous in the subsurface environments.
The microbial heterogeneity in the subsurface is evident despite our having taken samples in an environment that would be considered geochemically and physically very uniform, porous, and conducive to the vertical flux of water. The sediments were uniformly and predominately sands, with small changes in grain size (18, 47). There were no clay layers sampled, and there were no large changes in redox or oxygen between samplings (unpublished data). The sites receive large amounts of rainfall: e.g., an annual mean of 1.12 m for the years 1870 to 1995 for Dover, Delaware, and an annual mean of 1.02 m for 1961 to 1990 for Oyster, Virginia, as found at the WorldClimate website. Despite the uniformity of the environment, the microbial communities at the clone level changed substantially with depth. As indicated by the results seen with all three PCs in the analyses with both 11 and 33 units (Fig. 6), the samples from the shallowest nonsaturated Abbott's Pit location, AVZ and AAW, were quite different from the sample deepest saturated location (ABW). This result contrasts with that of an analysis of lipid samples from the vadose zone of Dover Air Force Base (36a), where there were apparently relatively small differences in the communities detectable over short vertical distances. However, the resolution of the lipid technique with respect to determining differences in communities is poorer than that offered by the techniques used here. In addition, it is expected that the overall microbial community structures are more similar among the samples within a site than among those taken between sites because the geochemical and physical environments appear to be more similar in the former case than in the latter case. As expected, our results showed that the overall community structures were more similar for the Abbott's Pit samples than for the Dover samples. The general consistency between the community structures and geochemical environments also implied that the differences observed among different samples appear to be due to site differences rather than sample differences. However, further studies are needed with more samples from a variety of subsurface environments.
The results presented here support the idea that the distribution of soil microorganisms is geographically unique. Microorganisms are small and can be readily transported globally by wind, birds, and human activity. In addition, the soil matrix that maintains diversity can promote an ongoing diversification when the rate of local genetic change exceeds the rate of microbial dispersal. Thus, one basic issue is whether soil microbial communities are geographically unique. Previous studies with 3-chlorobenzoic acid-degrading bacteria suggested that soil microorganisms are basically geographically unique rather than cosmopolitan (15, 41). The results of this study showed that no identical OTUs were observed in comparisons of the geographically separated Abbott's Pit samples to the Dover subsurface soil samples. Sequence analysis indicated the clones from each site tend to be more closely clustered together than the clones from different sites. This finding is also supported by the previous studies by Zhou et al. of surface soil samples from Virginia and Dover (52, 53); there were no common OTUs found to be shared between the two surface soil samples.
While the phylogenetic distribution characteristics at the division level were very similar between the Abbott's Pit and Dover samples, there was a striking difference in the members of the β-Proteobacteria family from these two environments. About 8% of the clones in Abbott's Pit samples belonged to the β-Proteobacteria family, whereas about 33% of clones in the Dover samples belonged to the β-Proteobacteria family. Many OTUs in Dover samples were clustered with aromatic compound-degrading bacteria such as Azoarcus, Burkholderia, and Ralstonia species, whereas no OTUs from the Abbott's Pit samples were grouped with these bacteria. This could be because the Dover samples were slightly contaminated with chloroethenes and hydrocarbon; these bacteria could have been selected by these contaminants.
Phylogenetic analyses have resulted in the description of over 40 major lineages (divisions), with new lineages being described as environmental sampling with culture independent techniques is expanded. We found one sequence (ABW1-256) from a subsurface sample that was affiliated with AD1 (53), a candidate division whose putative species had been originally found in Virginia and Delaware surface samples. The association of the subsurface soil sequence types with the AD1 candidate division expands the known diversity for this division and strengthens its division-level status.
The Acidobacterium family is a recently recognized bacterial division with only three cultivated representatives. The majority of sequences that make up this division are from environmental clones. Eight monophyletic subdivisions were identified on the basis of the available sequences (24). This group of bacteria appears to be widely distributed and was observed in our samples. SSU rDNA-based molecular analysis indicates that they exist in a variety of soil environments from the tropics to the tundra (cf. 2, 8, 13, 23-25, 27, 30, 53). About 3% of the clones in these subsurface samples were associated with three of the eight subdivisions of Acidobacterium. These results further indicate that the members of the Acidobacterium family are widely distributed in natural environments. The broad diversity of the rDNA sequences in this division may reflect diverse physiology and ecological adaptation in the group. The widespread occurrence of this group in natural environments also suggests that members of this group are functionally important in many different ecosystems. However, little is known about their ecological functions due to the lack of members of the division in cultivation.
The division-level phylogenetic diversity of Abbott's Pit and Dover samples was much lower than seen in a study using similar techniques at Wurtsmith Air Force Base in Michigan (8), where the geochemistry was quite different. Among the 94 sequence types detected in that study, about 11% of the clones had no close phylogenetic association with known taxonomic divisions; they were phylogenetically classified as six novel divisions, WS1 to WS6. Additionally, a large proportion (22%) of the sequences belonged to the recently described candidate divisions OP5, OP8, OP10, and OP11 (23), and 67% of the sequences are associated with 10 well-recognized divisions, e.g., Acidobacterium, Cytophagales, low- and high-G+C-content gram-positive bacteria, Proteobacteria, Spirochaetes, non-green sulfur bacteria, and Verrucomicrobia. The abundant bacterial sequence type belongs to the delta subdivision of Proteobacteria and Syntrophus species, which produce energy from anaerobic oxidation of organic acids with the production of acetate and hydrogen. A total of 150 sequence types were observed with both Abbott's Pit and Dover samples. These sequences fell into only six known main phylogenetic divisions, however, and the majority of the clones (90%) were associated with the γ-, α-, and β-Proteobacteria families. Such differences can most likely be explained by differences in geographic locations, geological history, and geochemistry. These differences in phylogenic diversity may be related to the striking differences between the Abbott's Pit and Dover sites and the Wurtsmith site with respect to carbon and oxygen levels. The carbon content of the Abbott's Pit and Dover samples was much lower than that seen with the Wurtsmith samples, which contained an average of 13.65 mg/g of sample (8). While the Abbott's Pit and Dover samples remained largely oxygenic, the Wurtsmith samples were collected from anaerobic methanogenic and iron- and sulfate-reducing zones (8). Thus, more diverse phylogenetical and physiological groups could be supported in Wurtsmith subsurface environments. Overall, the results of the molecular studies appear to be consistent with the environmental conditions extant at the sites.
Acknowledgments
We thank Susan Pfiffner for providing the samples and Lynn Kszos for editorial assistance.
This research was supported by the U.S. Department of Energy under the Natural and Accelerated Bioremediation Research Program of the Office of Biological and Environmental Research, Office of Science. Oak Ridge National Laboratory is managed by UT-Battelle, LLC, for the DOE under contract DE-AC05-00OR22725.
REFERENCES
- 1.Amann, R. I., W. Ludwig, and K. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barns, S. M., S. L. Takala, and C. R. Kuske. 1999. Wide distribution and diversity of members of the bacterial kingdom Acidobacterium in the environment. Appl. Environ. Microbiol. 65:1731-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bintrim, S. B., T. J. Donohue, J. Handelsman, G. P. Roberts, and R. M. Goodman. 1997. Molecular phylogeny of archaea from soil. Proc. Natl. Acad. Sci. USA 94:277-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borneman, J., and E. W. Triplett. 1997. Molecular microbial diversity in soils from eastern Amazonia: evidence for unusual microorganisms and microbial population shifts associated with deforestation. Appl. Environ. Microbiol. 63:2647-2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borneman, J., P. W. Skroch, K. M. O'Sullivan, J. A. Palus, N. G. Rumjanek, J. L. Jansen, J. Nienhuis, and E. W. Triplett. 1996. Molecular microbial diversity of an agricultural soil in Wisconsin. Appl. Environ. Microbiol. 62:1935-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chandler, D. P., F. J. Brockman, T. J. Bailey, and J. K. Fredrickson. 1998. Phylogenetic diversity of archaea and bacteria in a deep subsurface paleosol. Microb. Ecol. 36:37-50. [DOI] [PubMed] [Google Scholar]
- 7.Delong, E. F. 1992. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. USA 89:5685-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dojka, M. A., P. Hugenholtz, S. K. Haack, and N. R. Pace. 1998. Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl. Environ. Microbiol. 64:3869-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dojka, M. A., J. K. Harris, and N. R. Pace. 2000. Expanding the known diversity and environmental distribution of an uncultured phylogenetic division of bacteria. Appl. Environ. Microbiol. 66:1617-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunbar, J., S. Takala, S. M. Barns, J. A. Davis, and C. R. Kuske. 1999. Levels of bacterial community diversity in four arid soils compared by cultivation and 16S rRNA gene cloning. Appl. Environ. Microbiol. 65:1662-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunbar, J., L. O. Ticknor, and C. R. Kuske. 2000. Assessment of microbial diversity in four southwestern United States soils by 16S rRNA gene terminal restriction fragment analysis. Appl. Environ. Microbiol. 66:2943-2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dunbar, J., S. M. Barns, L. O. Ticknor, and C. R. Kuske. 2002. Empirical and theoretical bacterial diversity in four Arizona soils. Appl. Environ. Microbiol. 68:3035-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Felske, A., A. Wolterink, R. V. Lis, and A. D. L. Akkermans. 1998. Phylogeny of the main bacterial 16S rRNA sequences in Drentse A grassland soils (the Netherlands). Appl. Environ. Microbiol. 64:871-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuhrman, J. A., K. McCallum, and A. A. Davis. 1993. Phylogenetic diversity of subsurface marine microbial communities from Atlantic and Pacific oceans. Appl. Environ. Microbiol. 59:1294-1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fulthorpe, R. R., A. N. Rhodes, and J. M. Tiedje. 1998. High levels of endemicity of 3-chlorobenzoate-degrading soil bacteria. Appl. Environ. Microbiol. 64:1620-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Girvan, M. S., J. Bullimore, J. N. Pretty, A. M. Osborn, A. S. Ball. 2003. Soil type is the primary determinant of the composition of the total and active bacterial communities in arable soils. Appl. Environ. Microbiol. 69:1800-1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Groβkopf, R., P. H. Janssen, and W. Liesack. 1998. Diversity and structure of the methanogenic community in anoxic rice paddy soil microcosms as examined by cultivation and direct 16S rRNA gene sequence retrieval. Appl. Environ. Microbiol. 64:960-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grosso, N. R., L. Plante Leitzinger, and C. Bartlett. 1999. Site characterization of area 6, Dover Air Force Base, in support of natural attenuation and enhanced anaerobic bioremediation projects. EPA/600/R-99/044. NTIS no. PB99-166456/XAB. Environmental Protection Agency, Washington, D.C.
- 19.Halderman, D. L., and P. S. Amy. 1993. Bacterial heterogeneity in deep subsurface tunnels at Rainier Mesa, Nevada, test site. Microb. Ecol. 25:183-194. [DOI] [PubMed] [Google Scholar]
- 20.Hengstmann, U., K. J. Chin, P. H. Janssen, and W. Liesack. 1999. Comparative phylogenetic assignment of environmental sequences of genes encoding 16S rRNA and numerically abundant culturable bacteria from an anoxic rice paddy soil. Appl. Environ. Microbiol. 65:5050-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirons, W. D., B. A. Methe, S. A. Nierzwicki-Bauer, and J. P. Zehr. 1997. Bacterial diversity in Adirondack Mountain lakes as revealed by 16S rRNA gene sequences. Appl. Environ. Microbiol. 63:2957-2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoyle, B. L., and E. L. Arthur. 2000. Biotransformation of pesticides in saturated-zone materials. Hydrogeology J. 8:89-103. [Google Scholar]
- 23.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hugenholtz, P., C. Pitulle, K. L. Hershberger, and N. R. Pace. 1998. Novel division level bacterial diversity in a Yellowstone hot spring. J. Bacteriol. 180:366-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janssen, P. H., P. S. Yates, B. E. Grinton, P. M. Taylor, and M. Sait. 2002. Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl. Environ. Microbiol. 68:2391-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuske, C. R., L. O. Ticknor, M. E. Miller, J. M. Dunbar, J. A. Davis, S M. Barns, and J. Belnap. 2002. Comparison of soil bacterial communities in rhizospheres of three plant species and the interspaces in an arid grassland. Appl. Environ. Microbiol. 68:1854-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuske, C. R., S. M. Barns, and J. D. Busch. 1997. Diverse uncultivated bacterial groups from soils of the arid Southwestern United States that are present in many geographic regions. 63:3614-3621. [DOI] [PMC free article] [PubMed]
- 28.Larsen, N., G. J. Olsen, B. L. Maidak, M. J. McCaughey, R. Overbeek, T. J. Macke, T. L. Marsh, and C. R. Woese. 1993. The ribosomal database project. Nucleic Acids Res. 21(Suppl.):3021-3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liesack, W., and E. Stackebrandt. 1992. Occurrence of novel groups of the domain Bacteria as revealed by analysis of genetic material isolated from an Australian terrestrial environment. J. Bacteriol. 174:5072-5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liles, M. R., B. F. Manske, S. B. Bintrim, J. Handelsman, and R. M. Goodman. 2003. A census of rRNA genes and linked genomic sequences within a soil metagenomic library. Appl. Environ. Microbiol. 69:2684-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lollar, B. S., G. F. Slater, B. Sleep, M. Witt, G. M. Klecka, M. Harkness, and J. Spivack. 2001. Stable carbon isotope evidence for intrinsic bioremediation of tetrachloroethene and trichloroethene at area 6, Dover Air Force Base. Environ. Sci. Technol. 35:261-269. [DOI] [PubMed] [Google Scholar]
- 32.Looney, B. B., and R. W. Falta. 2000. Vadose zone: science and technological solutions, vol. I and II. Battelle Press, Columbus, Ohio.
- 33.Ludwig, W., S. H. Bauer, M. Bauer, I. Held, G. Kirchhof, R. Schulze, I. Huber, S. Spring, A. Hartmann, and K. H. Schleifer. 1997. Detection and in situ identification of representatives of a widely distributed new bacterial phylum. FEMS Microbiol. Lett. 153:181-190. [DOI] [PubMed] [Google Scholar]
- 34.Norris, T. B., J. M. Wraith, R. W. Castenholz, and T. R. McDermott. 2002. Soil microbial community structure across a thermal gradient following a geothermal heating event. Appl. Environ. Microbiol. 68:6300-6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nusslein, K., and J. M. Tiedje. 1998. Characterization of the dominant and rare members of a young Hawaiian soil bacterial community with small-subunit ribosomal DNA amplified from DNA fractionated on the basis of its guanine and cytosine composition. Appl. Environ. Microbiol. 64:1283-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pfiffner, S. M., P. A Sobecky, T. J. Phelps, and A. V. Palumbo. 2002. Microbiology of Atlantic coastal plain aquifers and other unconsolidated subsurface sediments, p. 2028-2042. In Gabriel Bitton (ed.), Encyclopedia of microbiology, vol 4. John Wiley & Sons, New York, N.Y.
- 36a.Pfiffner, S. M., A. V. Palumbo, B. L. Kinsall, A. D. Peacock, D. C. White, and T. J. Phelps. 2001. Microbial heterogeneity implications for bioremediation, p. 73-80. In V. Magar, T. Vogel, C. Aelion, and A. Leeson (ed.), Innovative methods in support of bioremediation. Battelle Press, Columbus, Ohio.
- 37.Qiu, X., L. Wu, H. Huang, P. E. McDonal, A. V. Palumbo, J. M. Tiejde and J.-Z. Zhou. 2001. Evaluation of PCR-generated chimeras, mutations, and heteroduplexes with 16S rRNA gene-based cloning. Appl. Environ. Microbiol. 67:880-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reysenbach, A., G. S. Wickham, and N. R. Pace. 1994. Phylogenetic analysis of the hyperthermophilic pink filament community in Octopus Spring, Yellowstone National Park. Appl. Environ. Microbiol. 60:2113-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosch, C., A. Mergel, and H. Bothe. 2002. Biodiversity of denitrifying and dinitrogen-fixing bacteria in an acid forest soil. Appl. Environ. Microbiol. 68:3818-3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stackebrandt, E., W. Liesack, and B. M. Goebel. 1993. Bacterial diversity in a soil sample from a subtropical Australian environment as determined by SSU rDNA analysis. FASEB J. 7:232-236. [DOI] [PubMed] [Google Scholar]
- 41.Tiedje, J. M., J.-C. Cho, A. Murray, D. Treves, B.-C. Xia, and J.-Z. Zhou. 2001. Soil teaming with life: new frontiers for soil science, p. 393-412. In R. M. Rees, B. C. Ball, C. D. Campbell, and C. A. Watson (ed.), Sustainable management of soil organic matter. CABI Publishing, Cambridge, Mass.
- 42.Tiedje, J. M., J.-Z. Zhou, K. Nusslein, C. L. Moyer, and R. R. Fulthorpe. 1997. Extent and patterns of soil microbial diversity. p. 35-41. In M. T. Martins et al. (ed.), Progress in microbial ecology. Brazilian Society for Microbiology, Sao Paulo, Brazil.
- 43.Tobin, K. J., T. C. Onstott, M. F. DeFlaun, F. S. Colwell, and J. Fredrickson. 1999. In situ imaging of microorganisms in geologic material. J. Microbiol. Methods 37:201-213. [DOI] [PubMed] [Google Scholar]
- 44.Treves, D. S., B. Xia, J. Zhou, and J. M. Tiedje. 2003. A two-species test of the hypothesis that spatial isolation influences microbial diversity in soil. Microb. Ecol. 45:20-28. [DOI] [PubMed] [Google Scholar]
- 45.Ueda, T., Y. Suga, and T. Matsuguchi. 1995. Molecular phylogenetic analysis of a soil microbial community in a soil field. Eur. J. Soil. Sci. 46:415-421. [Google Scholar]
- 46.Wise, M. G., J. V. McArthur, and L. J. Shimkets. 1997. Bacterial diversity of a Carolina bay as determined by 16S rRNA gene analysis: confirmation of novel taxa. Appl. Environ. Microbiol. 63:1505-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Witt, M. E., G. M. Klecka, E. J. Lutz, T. A. Ei, N. R. Grosso, and F. H. Chapelle. 2002. Natural attenuation of chlorinated solvents at Area 6, Dover Air Force Base: groundwater biogeochemistry. J. Contam. Hydrol. 57:61-80. [DOI] [PubMed] [Google Scholar]
- 48.Zhang, C., A. V. Palumbo, T. J. Phelps, J. J. Beauchamp, F. J. Brockman, C. J. Murray, B. S. Parsons, and D. J. P. Swift. 1998. Grain-size and depth constraints on microbial variability in coastal plain subsurface sediments. Geomicrobiology 15:171-185. [Google Scholar]
- 49.Zhou, J.-Z., M. R. Fries, J. Chee-Sanford, and J. M. Tiedje. 1995. Phylogenetic analyses of a new group of denitrifiers capable of anaerobic growth on toluene: Description of Azoarcus tolulyticus sp. nov. Int. J. Syst. Bacteriol. 45:500-506. [DOI] [PubMed] [Google Scholar]
- 50.Zhou, J.-Z., M. A. Bruns, and J. M. Tiedje. 1996. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62:461-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou, J.-Z., M. E. Davey, J. B. Figueras, E. Rivkina, D. Gilichinsky, and J. M. Tiedje. 1997. Phylogenetic diversity of a bacterial community determined from Siberian tundra soil DNA. Microbiology 143:3913-3919. [DOI] [PubMed] [Google Scholar]
- 52.Zhou, J.-Z., B. C. Xia, D. S. Treves, L.-Y. Wu, T. L. Marsh, R. V. O'Neill, A. V. Palumbo, and J. M. Tiedje. 2002. Spatial and resource factors influencing high soil microbial diversity. Appl. Environ. Microbiol. 68:326-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou, J.-Z., X. Beicheng, H. Huang, D. S. Treves, L. J. Hauser, R. J. Mural, A. V. Palumbo, and J. M. Tiedje. 2003. Bacterial phylogenetic diversity and a novel candidate division of two humid region, sandy surface soils. Soil Biol. Biochem. 35:915-924. [Google Scholar]