Abstract
Background
A skewed assemblage of two epi-, meso- and bathypelagic fish families makes up the order Myctophiformes – the blackchins Neoscopelidae and the lanternfishes Myctophidae. The six rare neoscopelids show few morphological specializations whereas the divergent myctophids have evolved into about 250 species, of which many show massive abundances and wide distributions. In fact, Myctophidae is by far the most abundant fish family in the world, with plausible estimates of more than half of the oceans combined fish biomass. Myctophids possess a unique communication system of species-specific photophore patterns and traditional intrafamilial classification has been established to reflect arrangements of photophores. Myctophids present the most diverse array of larval body forms found in fishes although this attribute has both corroborated and confounded phylogenetic hypotheses based on adult morphology. No molecular phylogeny is available for Myctophiformes, despite their importance within all ocean trophic cycles, open-ocean speciation and as an important part of neoteleost divergence. This study attempts to resolve major myctophiform phylogenies from both mitogenomic sequences and corroborating evidence in the form of unique mitochondrial gene order rearrangements.
Results
Mitogenomic evidence from DNA sequences and unique gene orders are highly congruent concerning phylogenetic resolution on several myctophiform classification levels, corroborating evidence from osteology, larval ontogeny and photophore patterns, although the lack of larval morphological characters within the subfamily Lampanyctinae stands out. Neoscopelidae is resolved as the sister family to myctophids with Solivomer arenidens positioned as a sister taxon to the remaining neoscopelids. The enigmatic Notolychnus valdiviae is placed as a sister taxon to all other myctophids and exhibits an unusual second copy of the tRNA-Met gene – a gene order rearrangement reminiscent of that found in the tribe Diaphini although our analyses show it to be independently derived. Most tribes are resolved in accordance with adult morphology although Gonichthyini is found within a subclade of the tribe Myctophini consisting of ctenoid scaled species. Mitogenomic sequence data from this study recognize 10 reciprocally monophyletic lineages within Myctophidae, with five of these clades delimited from additional rearranged gene orders or intergenic non-coding sequences.
Conclusions
Mitogenomic results from DNA sequences and unique gene orders corroborate morphology in phylogeny reconstruction and provide a likely scenario for the phylogenetic history of Myctophiformes. The extent of gene order rearrangements found within the mitochondrial genomes of myctophids is unique for phylogenetic purposes.
Keywords: Myctophiformes, Myctophidae, Neoscopelidae, Phylogeny, Mitogenomics, Gene rearrangements, Non-coding sequence
Background
Lanternfishes (Myctophidae) and blackchins (Neoscopelidae) comprise the only two families in the order Myctophiformes, superorder Scopelomorpha [1], a group currently including some 250 species [2]. The 32 genera of Myctophidae contain about 98% of the species diversity, and morphological characters have clearly shown Myctophidae more derived compared to the more generalized Neoscopelidae [3-6]. Myctophids are exclusively marine and pelagic, occupying depths from surface waters down to the upper 1000 meters of the bathypelagic layer (1000–4000 meters). The group as a whole constitutes fairly small fishes ranging in size from 20 to 300 millimeters, with larger members being confined to the bathypelagic realm. They are ubiquitous in the World’s oceans and exhibit extremely high abundances with one estimate as high as 65% of all pelagic deep-sea fish biomass [7].
The most striking feature of myctophids is their often species-specific patterns of photophores (light-organs), which have earned them their popular name lanternfishes. Photophores are named according to specific patterns present in various degrees throughout different myctophid lineages (Figure 1), making photophore patterns important when reconstructing the evolution of myctophids. It is generally believed that species-specific flash communication underpins the evolution of photophore patterns although myctophids are also macrosmatic, i.e. possessing large olfactory organs, indicating a complicated communication system not fully understood [8]. Myctophid fishes are believed to communicate in a firefly-like manner by using duration- and intensity-variable flashes of light produced by species-specific and sexually dimorphic patterned photophores [3,9]. One argument against flash communication is the almost identical photophore patterns observed between some species, making it virtually impossible to discriminate between flashes except within very close range [10]. Although bioluminescence has evolved multiple times across open-ocean organisms [11], the possibility of using this feature in tracing the natural history of an entire clade is, however, unique within open-ocean and deep-sea animals. The mechanism behind myctophid bioluminescence has caused some debate since bioluminescent bacteria of the strain Vibrio were found by Foran [12] using hybridization probes and later challenged by Haygood et al. [13] who confidently excluded both symbiotic bacteria and bacterial luciferase as the light source. It is now generally accepted that a coelenterazine system is responsible for myctophid bioluminescence [11,14], contrary to e.g. the famous deep-sea anglerfishes that possess family-specific symbiotic photobacterial strains of Vibrio in the esca [15,16]. Most myctophids that have been examined concerning their visual spectrum show a tendency towards longer wavelengths for absorption and emission, resulting in a dim blue bioluminescence as typically found in deep-sea fishes [17]. The latter study found a relatively confined spectral range of absorption in myctophids compared to other mesopelagic fishes. However, Hasegawa et al. [18] found that Myctophum nitidulum also possessed longer wavelength retinal pigments enabling them to detect a broader spectrum of wavelengths produced by other animals, and/or their own light emission and down-welling sunlight. Extended spectral range is familiar from loose-jaw dragonfishes well-known for emitting long-wavelength red light from orbital photophores, enabling them to communicate or detect prey at wavelengths invisible to other deep-sea organisms [19]. Turner et al. [17] note that possession of a relatively limited spectral range could be the result of a shared evolutionary history more than multiple instances of visual adaptation to the mesopelagic environment.
Figure 1.
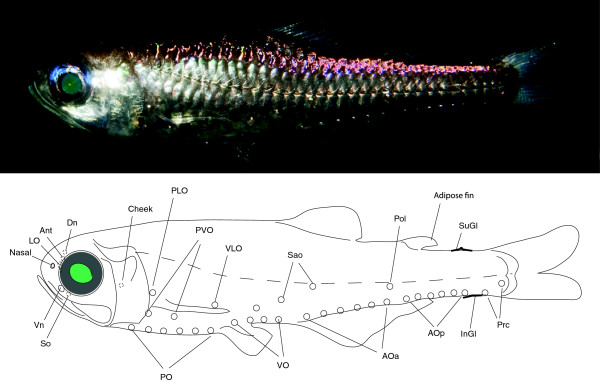
Benthosema glaciale, © Rudolf Svendsen, Høgsfjorden, Norway. Standard myctophid photophore terminology noted with photophores/luminescent organs truncated if found in other myctophids than B. glaciale. Abbreviations: Ant – Anterior orbital (antorbital), AOa – Anal organs anterior, AOp – Anal organs posterior, Dn – Dorsal nasal (dorsonasal), InGl – Infracaudal gland, LO – Luminous organ, PO – Pectoral organs, Pol – Posterior organ lateral, PLO – Pectoral lateral organ, Prc – Precaudal organs (precaudals), PVO – Pectoral ventral organs, Sao – Supraanal organs, So – Suborbital, Gl – Supracaudal gland, VLO – Ventral lateral organ, Vn – Ventral nasal (ventronasal), VO – Ventral organs.
Diel vertical migration (DVM) is a prominent feature of myctophids although not all myctophid species engage in this activity [20]. DVM prompted the famous “false bottom” (or the deep scattering layer) discovery, turning out to be massive occurrences of the swimbladder-possessing myctophid Ceratoscopelus maderensis among other deep-sea animals [21]. DVM is common across many animal groups and is generally believed to involve feeding and/or reduced predation risk associated with reduced light intensity [22]. Herring [23] notes that if populations are biased towards horizontal layers, as is often the case with myctophids, DVM might be an effective way to channel intra-specific encounters and could be important in mate recognition. A phylogenetic component in DVM has not been explored although DVM is clearly more pronounced in some groups of myctophids than others, e.g. the slendertails of the tribe Gonichthyini including the genera Gonichthys, Centrobranchus, Tarletonbeania and Loweina.
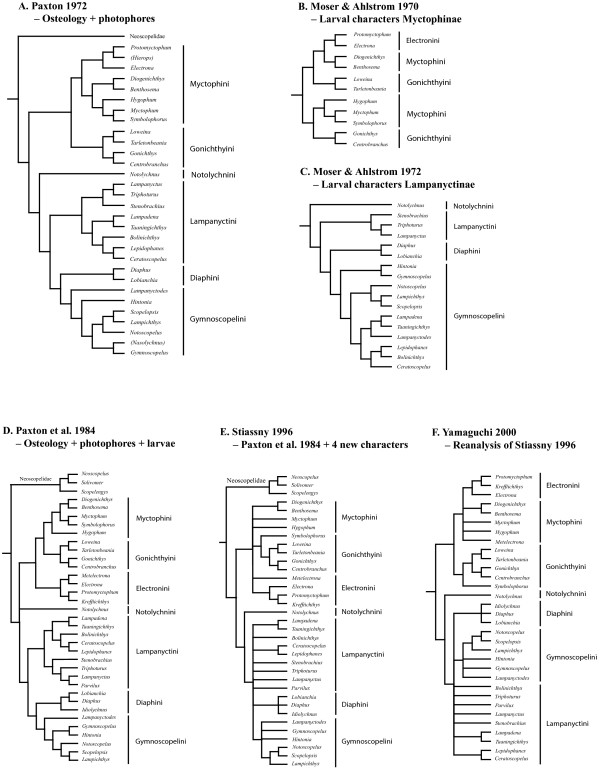
Stiassny [5] and Yamaguchi [24] have most recently reviewed myctophiform synapomorphies and there is little doubt that the order constitutes a monophyletic assemblage. Intrarelationships of Neoscopelidae (3 genera and 6 species) are not well resolved [5] with generic relationships within Myctophidae better understood bearing on the classification initially by Bolin [6], subsequently Fraser-Brunner [4], and finally Paxton [3]. They classified the order into two families, divided the myctophids into two subfamilies that together include six tribes, based on osteology and photophore patterns (Figure 2A). This classification is still in use today, except for the lineage Electronini also being recognized as a distinct tribe as originally designated by Wisner [25]. Additional corroborating evidence for this classification has been presented partly from urodermal bones [26] and notably a broad range of larval body forms [27-30] (Figure 2B-D). Myctophid larvae show some of the most diverse forms within any teleost order and characters associated with early ontogeny have proven valuable in phylogenetics [30]. Stiassny [5] added new data and reviewed existing characters, resulting in a topology little resolved below subfamily level (Figure 2E). Yamaguchi [24] carried out the most recent analysis using the characters provided by the previous studies, which again resulted in an unresolved topology (Figure 2F). Morphological work on photophore patterns, adult osteology and larval ontogeny have clearly established their utility in myctophid phylogenetics, although missing larval characters in the subfamily Lampanyctinae have been a significant source of discrepancy between morphological hypotheses [29]. Larval characters corroborate parts of adult phylogenetic reconstruction (Figure 2) although the larvae of the large genus Diaphus have proven exceptionally difficult to identify [30]. The most persistent issue concerning myctophid phylogenetics has been the placement of Notolychnus valdiviae, a monotypic, diminuitive, putatively plesiomorphic myctophid showing dorsally located photophores, weak ossification and several characters difficult to assess even on subfamily level [3]. Recent cladistic analyses of morphological characters have converged on Notolychnus as sister taxon to the remaining lampanyctines (Figure 2) as initially suggested by Paxton [3].
Figure 2.
Phylogenetic hypotheses from previous morphological studies. A) Adult osteology [3]. Parenthesized genera no longer recognized. B) Larval development (Myctophinae only) with topology constructed from written considerations by Moser & Ahlstrom [27] and tribes inferred according to [3]. C) Larval development (Lampanyctinae only) [28]. D) Adult osteology + larval development [30]. E) Unordered multistate parsimony analysis of adult + larval characters including 4 new characters [5]. F) Ordered multistate parsimony reanalysis of all adult and larval characters [24].
Surprisingly little molecular work has been carried out on Myctophiformes considering their abundance and at times problematic species identification. Yamaguchi et al. [31] tested photophore patterns, biogeographical distributions and diverse larval eye morphology within Hygophum in a phylogenetic context. The results indicated all three attributes as somewhat informative concerning speciation pattern, although clear conclusions were hard to extract due to few characters (partial 16S rRNA gene sequences) and incomplete taxon sampling. However, with an increasing understanding of myctophiform phylogeny, this type of study will become highly valuable in terms of assessing open-ocean speciation patterns in myctophids as well as other open-ocean fish components [32].
Miya et al. [33] produced the first mitochondrial (mt) genome mapping of myctophiform fishes, showing that the two myctophids Diaphus splendidus and Myctophum affine possess non-typical vertebrate mt gene arrangements, whereas the neoscopelid fish Neoscopelus microchir exhibits the typical gene arrangement for vertebrates [34]. Their findings indicated that mt gene arrangements of myctophid fishes have the potential to provide evidence concerning clade delimitation at various levels. Clade-specific gene orders are considered strong evidence for monophyly since the process of gene duplication and subsequent deletions are considered random and rare events [35-39]. However, accumulating evidence shows positive selection on some rearranged mt gene orders [40] to be much more common than initially expected [41]. Intergenic non-coding (INC) regions are likewise believed to provide evidence for phylogenetic history, although this group of markers has been poorly explored mainly because of insufficient taxon sampling.
This study is the first to use mitogenomic DNA sequences combined with additional information from extensively rearranged gene orders and INC-sequences to explore myctophiform phylogeny.
Methods
Taxon sampling
Twenty-three genera and a total of 41 species of myctophiform fishes are included in this study – 38 species representing new data and three from a previous mitogenomic study on higher teleostean phylogeny [33]. Forty-six outgroup taxa ranging from early euteleost groups (root) to ophidiiform and lophiiform fishes (Percomorpha) were chosen with all putative myctophiform sister groups included. Taxonomic sampling focused on tribal classification sensu Paxton [3] representing the distinct myctophiform lineages. We sampled all non-monotypic tribes with multiple genera although only a single representative of Gonichthyini is included due to technical difficulties in amplification of DNA. A large number of taxa within the genus Diaphus were included since about 1/3 of all myctophid species are placed within this genus. Voucher specimens were obtained from sampling onboard Research Vessel Pâmiut (Greenland Institute of Natural Resources) around Greenland (JYP), from the MAR-ECO cruise (IB), catalogued and uncatalogued material at NSMT (TPS and GS) and institutional loans from ASIZP, BSKU, KU, MCZ, SIO, WU and ZMUC (Table 1). Voucher specimens were examined and photographed with e-vouchers available online [42].
Table 1.
87 taxa included in this study
| Clade | Species | Acc. # | Tissue # | Voucher Museum # | Study |
|---|---|---|---|---|---|
|
Esociformes |
Esox lucius |
AP004103 |
|
|
Ishiguro et al. 2003 [43] |
| |
Dallia pectoralis |
AP004102 |
|
|
Ishiguro et al. 2003 [43] |
|
Salmoniformes |
Coregonus lavaretus |
AB034824 |
|
|
Miya & Nishida 2000 [44] |
| |
Thymallus thymallus |
FJ853655 |
|
|
Yasuike et al. 2010 |
| |
Salvelinus alpinus |
AF154851 |
|
|
Doiron et al. 2002 |
|
Argentiniformes |
Bathylagus ochotensis |
AP004101 |
|
|
Ishiguro et al. 2003 [43] |
| |
Nansenia ardesiaca |
AP004106 |
|
|
Ishiguro et al. 2003 [43] |
| |
Glossanodon semifasciatus |
AP004105 |
|
|
Ishiguro et al. 2003 [43] |
| |
Opistophroctus soleatus |
AP004110 |
|
|
Ishiguro et al. 2003 [43] |
|
Osmeriformes |
Retropinna retropinna |
AP004108 |
|
|
Ishiguro et al. 2003 [43] |
| |
Salangichthys microdon |
AP004109 |
|
|
Ishiguro et al. 2003 [43] |
| |
Mallotus villosus |
HM106491 |
|
|
Li et al. 2010 |
| |
Plecoglossus altivelis |
AB047553 |
|
|
Ishiguro et al. 2001 |
|
Stomiiformes |
Diplophos taenia |
AB034825 |
|
|
Miya & Nishida 2000 [44] |
| |
Chauliodus sloani |
AP002915 |
|
|
Miya et al. 2001 [33] |
| |
Gonostoma gracile |
AB016274 |
|
|
Miya & Nishida 1999 [45] |
|
Galaxiiformes |
Galaxiella nigrostriata |
AP006853 |
|
|
Miya et al. (unpubl.) |
| |
Galaxias maculates |
AP004104 |
|
|
Ishiguro et al. 2003 [43] |
| |
Galaxias gollumoides |
HM106487 |
|
|
Li et al. 2010 |
|
Aulopiformes |
Synodus variegatus |
AY524977 |
|
|
Chen & Wu (unpubl.) |
| |
Chlorophthalmus agassizi |
AP002918 |
|
|
Miya et al. 2001 [33] |
| |
Aulopus japonicus |
AB047821 |
|
|
Kawaguchi et al. 2001 [33] |
| |
Harpadon microchir |
AP002919 |
|
|
Miya et al. 2001 [33] |
| |
Saurida undosquamis |
AP002920 |
|
|
Miya et al. 2001 [33] |
|
Ateleopodiformes |
Ateleopus japonicus |
AP002916 |
|
|
Miya et al. 2001 [33] |
| |
Ijimaia dofleini |
AP002917 |
|
|
Miya et al. 2001 [33] |
|
Lampriformes |
Lampris guttatus |
AP002924 |
|
|
Miya et al. 2001 [33] |
| |
Zu cristatus |
AP002926 |
|
|
Miya et al. 2001 [33] |
| |
Trachipterus trachypterus |
AP002925 |
|
|
Miya et al. 2001 [33] |
|
Myctophiformes |
Neoscopelus macrolepidotus |
AP012238 |
KU 3297 |
Uncatalogued |
This study |
| |
Neoscopelus microchir |
AP002921 |
|
|
Miya et al. 2001 [33] |
| |
Scopelengys tristis |
AP012228 |
NSMT-P 99997 |
NSMT-P 99997 |
This study |
| |
Solivomer arenidens |
AP012249 |
NSMT |
Uncatalogued |
This study |
| |
Benthosema fibulatum |
AP012253 |
NSMT-P 75816 |
NSMT-P 75816 |
This study |
| |
Benthosema glaciale |
AP012264 |
ZMUC #8160 |
ZMUC P2393965 |
This study |
| |
Benthosema pterotum |
AP012260 |
NSMT-P 75855 |
NSMT-P 75855 |
This study |
| |
Bolinichthys distofax |
AP012232 |
NSMT-P 97624 |
NSMT-P 97624 |
This study |
| |
Bolinichthys pyrsobolus |
AP012261 |
NSMT-P 92273 |
NSMT-P 92273 |
This study |
| |
Centrobranchus choerocephalus |
AP012237 |
NSMT-P 102902 |
NSMT-P 102902 |
This study |
| |
Ceratoscopelus maderensis |
AP012259 |
ZMUC #6600 |
ZMUC P2393929 |
This study |
| |
Diaphus chrysorhynchus |
AP012230 |
NSMT-P 79902 |
NSMT-P 79902 |
This study |
| |
Diaphus gigas |
AP012235 |
NSMT-P 91530 |
NSMT-P 91530 |
This study |
| |
Diaphus luetkeni |
AP012231 |
ASIZP 911514 |
ASIZP66276 |
This study |
| |
Diaphus splendidus |
AP002923 |
|
|
Miya et al. 2001 [33] |
| |
Diaphus theta |
AP012240 |
KU 2135 |
KU 27971 |
This study |
| |
Diogenichthys atlanticus |
AP012233 |
SIO 09320 |
SIO 09320 |
This study |
| |
Electrona antarctica |
AP012248 |
ZMUC #7552 |
ZMUC P2393962 |
This study |
| |
Gymnoscopelus nicholsi |
AP012250 |
ZMUC #7806 |
ZMUC P2393963 |
This study |
| |
Krefftichthys anderssoni |
AP012236 |
ZMUC #7550 |
ZMUC P2393964 |
This study |
| |
Lampadena anomala |
AP012227 |
ME 4201#3 |
ZMUB 18019 |
This study |
| |
Lampadena urophaos atlantica |
AP012251 |
ME 7371#4 |
ZMUB 18050 |
This study |
| |
Lampadena yaquinae |
AP012257 |
NSMT-P 72341 |
NSMT-P 72341 |
This study |
| |
Lampanyctus macdonaldi |
AP012241 |
ZMUC #8077 |
ZMUC P2393967 |
This study |
| |
Lampanyctus crocodilus |
AP012258 |
ZMUC #8076 |
ZMUC P2393968 |
This study |
| |
Lobianchia gemellarii |
AP012242 |
ME 4239#6 |
ZMUB O.1725 |
This study |
| |
Myctophum affine |
AP002922 |
|
|
Miya et al. 2001 [33] |
| |
Myctophum asperum |
AP012234 |
NSMT-P 91490 |
NSMT-P 91490 |
This study |
| |
Myctophum nitidulum |
AP012255 |
NSMT-P 92345 |
NSMT-P 92345 |
This study |
| |
Myctophum orientale |
AP012254 |
NSMT-P 77350 |
NSMT-P 77350 |
This study |
| |
Myctophum punctatum |
AP012239 |
ZMUC #6594 |
ZMUC P2393969 |
This study |
| |
Nannobrachium ritteri |
AP012247 |
KU 2237 |
KU 28276 |
This study |
| |
Notoscopelus caudispinosus |
AP012256 |
KU 5301 |
MCZ 161883 |
This study |
| |
Notoscopelus japonicus |
AP012252 |
BSKU 103772 |
BSKU 103772 |
This study |
| |
Notoscopelus kroyeri |
AP012262 |
ZMUC #8078 |
ZMUC P2393970 |
This study |
| |
Notolychnus valdiviae |
AP012229 |
NSMT-P 102930 |
NSMT-P 102930 |
This study |
| |
Protomyctophum arcticum |
AB648909-10 |
ZMUC #8476 |
ZMUC P2393971 |
This study |
| |
Stenobrachius leucopsarus |
AP012245 |
NSMT-P 78748 |
NSMT-P 78748 |
This study |
| |
Symbolophorus californiensis |
AP012246 |
NSMT-P 92257 |
NSMT-P 92257 |
This study |
| |
Taaningichthys minimus |
AP012244 |
ME 5633 |
ZMUB 18049 |
This study |
| |
Triphoturus nigrescens |
AP012243 |
NSMT-P 102934 |
NSMT-P 102934 |
This study |
|
Ophidiiformes |
Cataetyx rubrirostris |
AP004407 |
|
|
Miya et al. 2003 [46] |
| |
Carapus bermudensis |
AP004404 |
|
|
Miya et al. 2003 [46] |
| |
Bassozetus zenkevitchi |
AP004405 |
|
|
Miya et al. 2003 [46] |
|
Lophiiformes |
Sladenia gardineri |
AB282827 |
|
|
Miya et al. 2010 [47] |
| |
Coelophrys brevicaudata |
AB282834 |
|
|
Miya et al. 2010 [47] |
| |
Thaumatichthys pagidostomus |
AB282847 |
|
|
Miya et al. 2010 [47] |
|
Percopsiformes |
Percopsis transmontana |
AP002928 |
|
|
Miya et al. 2001 [33] |
| |
Aphredoderus sayanus |
AP004403 |
|
|
Miya et al. 2003 [46] |
|
Polymixiiformes |
Polymixia japonica |
AB034826 |
|
|
Miya & Nishida 2000 [44] |
| |
Polymixia lowei |
AP002927 |
|
|
Miya et al. 2001 [33] |
|
Gadiformes |
Stylephorus chordatus |
AB280688 |
|
|
Miya et al. 2007 |
| |
Bregmaceros transmontana |
AP004411 |
|
|
Miya et al. 2003 [46] |
| |
Trachyrincus murrayi |
AP008990 |
|
|
Satoh et al. 2006 [48] |
| |
Lota lota |
AP004412 |
|
|
Miya et al. 2003 [46] |
|
Zeiformes |
Zeus faber |
AP002941 |
|
|
Miya et al. 2001 [33] |
| |
Parazen pacificus |
AP004433 |
|
|
Miya et al. 2003 [46] |
| Zenion japonicum | AP004434 | Miya et al. 2003 [46] |
Taxa included in this study listed according to ordinal classification. Mitogenome accession number, tissue number, specimen + institute number and study noted if information available.
Production of mitogenomic data
Genomic DNA was extracted using the Qiagen Puregene extraction kit following the manufacturer´s protocol and used directly for long and accurate amplification PCR (LA PCR) of the entire mitochondrial genome [49]. Depending on tissue quality and LA primer fidelity, a variable number of LA PCR reactions were needed for complete coverage, with a minimum of two and a maximum of six for some species. The LA PCR product was diluted with ddH2O according to success of amplification and nested short PCR was subsequently performed on the LA product with a broad range of fish universal and species specific primers (>200). Double stranded PCR products were cleaned with Exo-Sap at 60°C for 60 minutes and used as template for direct cycle-sequencing with dye labeled terminators (Applied Biosystems). All fragments were sequenced on automated DNA sequencers according to length of labelled fragments. Quality of tissue was shown to be a prominent obstacle regarding successful amplifications. Issues regarding amplification necessitated multiple PCR protocols and tailor-made primer sets for different taxa. Variable number tandem repeats found in the control region and various INC-regions added to the technical issues. Primer information and protocols regarding PCR and sequencing for particular species are available by request (JYP). Newly determined mitogenome sequences are available as [DDBJ, EMBL or GenBank AP012227-62, AP0122264 and AB648909-10] (Table 1).
Sequence editing, alignment and analyses
Raw sequence data were processed and formatted with the software 4peaks Ver. 1.7.2 [50], Textwrangler Ver. 3.5.3 [51] and MacClade Ver. 4.06 [52] at various stages before alignment. Mitogenome assembly was performed using Sequencher Ver. 4.10.1 [53] by concatenation of overlapping sequences. Numerous single nucleotide repeat regions were concatenated from the L- and H-strand sequences assuming the repeat region to be identical in the two strands. This was verified by electrophoresis of the sequenced PCR products showing bands with equal size to the sequenced fragments. All genes were initially annotated by alignment to those from closely related fishes already available in GenBank [54]. Subsequently, protein coding genes were examined by reference to ORFs and the 22 tRNA genes were examined for secondary structure by tRNAscan-SE Ver. 1.21 [55,56]. The two ribosomal RNA genes (12S and 16S) were annotated as the sequences between flanking tRNA Phe, Val and LeuUUR in addition to alignment using previously determined myctophid RNA genes [33].
Alignment of the 12 protein coding genes was done by eye whereas the 22 tRNA genes and 12S+16S rRNA genes were aligned with a probabilistic multiple alignment approach using ProAlign Ver. 0.5 [57]. The ND6 gene was excluded due to heterogeneous base composition [44] leaving 10908 protein coding nucleotide characters (3636 amino acids). Ambiguous alignment of the tRNA and rRNA genes were discarded from the analyses based on the 90% minimum posterior probability of correct alignment implemented in ProAlign, leaving 938 and 1474 nucleotide characters from 22 tRNA and 12S+16S rRNA genes, respectively. All genes were aligned separately and subsequently concatened into one dataset consisting of six partitions as suggested by PartitionFinder [58] (see Additional file 1); amino acids, 12S+16S rRNA genes and 22 concatenated tRNA genes (designated as 123ARTn). Missing nucleotides, e.g. tRNA-Thr and Pro genes for some species, were coded as missing characters. Levels of mutational saturation were calculated from p-distances for 1st, 2nd and 3rd codon positions using PAUP* Ver. 4.0b10 [59] (see Additional file 2). Since no mutational saturation was detected when including only myctophiforms, we constructed a second dataset consisting of only nucleotides from the 41 myctophiform species. This dataset included all mitogenomic nucleotides except the CR and ProAlign ambiguous alignment as already noted above (123nRTn).
Heuristic maximum likelihood phylogenetic analyses with 1000 non-parametric bootstrap replications for all nodes were performed in a single run with the sequential version of RAxML Ver. 7.2.8 [60,61] under the GTR+Γ+I substitution model [62] as chosen by Akaike and Bayesian information criteria [63,64] implemented in Modelgenerator [65]. A final ML optimization of every 5th bootstrapped tree for the highest scoring ML-tree is performed by RAxML under the specified GTR+Γ+I substitution model [61]. Partitions constructed from amino acids were analyzed with the mtREV model [66] chosen by Modelgenerator.
Bayesian analysis was performed on the datasets with MrBayes Ver. 3.2.1 [67] using the same partitions and models as described above. Maximum likelihood analyses were conducted on a Quad-core Mac Pro desktop computer whereas the Bayesian analyses were computed on the Bioportal computer cluster University of Oslo [68]. Convergence of chains and burn-in were determined using TRACER Ver. 1.5 [69].
Additional analyses and diaphini tRNA-met pseudogenes
All 41 myctophiform mitogenomes were scrutinized for mt features such as rearranged gene orders and INC-regions. All INC-sequences were analyzed for sequence similarity in relation to possible duplicated regions in the mitochondrial genome by using NCBI Blast [54], tRNAscan [55] and a series of decreasing assembly parameters in Sequencher [53]. Subsequently, alignments of INC-regions were examined for secondary folding patterns with LocARNA Ver. 1.5.2 [70] and RNAshapes Ver. 2.1.6 [71] in order to detect conserved regions. Secondary tRNA-structures were drawn with VARNA Ver. 3.8 [72]. In order to examine a putative associated gene order found in Notolychnus and species within the tribe Diaphini, neighbour-joining analyses were carried out including multiple myctophid tRNA-Met genes, putative duplicated diaphinid tRNA-Met genes (pseudogenes) and the two tRNA-Met genes observed in Notolychnus. Pairwise sequence variation of the pseudogenes and detected tRNA-Met genes in Diaphini were compared in order to validate the INC-regions as actual tRNA-Met pseudogenes, the latter expected to show higher substitution rates.
Testing alternative phylogenetic hypotheses
Topology comparison from morphological phylogenies and this study were tested by using the approximately unbiased log-likelihood test [73] implemented in CONSEL [74]. Constrained trees were produced with Mesquite [75] and per-site log-likelihood scores were produced using RAxML using the f –g option with best scoring ML-trees passed via –z option. Per-site log-likelihood scores and tree topologies were used as direct input into CONSEL outputting tree-specific ML-scores and associated p-values.
Results
Mitogenome organization
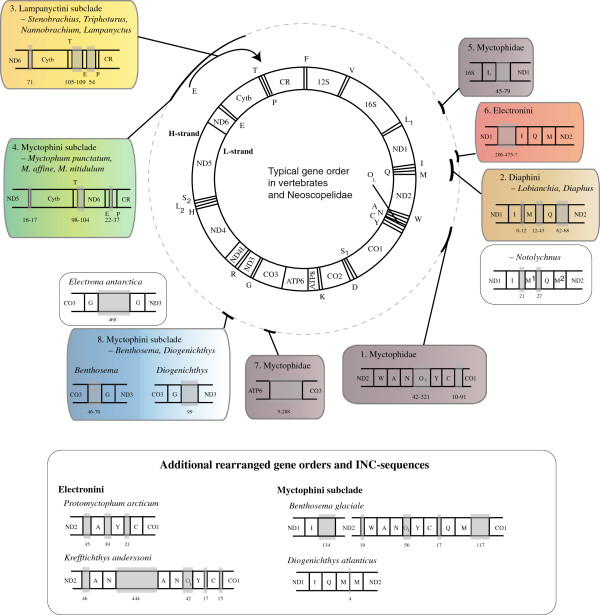
The organization of the mitogenome is unique in all fishes of the family Myctophidae, showing rearranged gene orders for either the whole family, tribal or subtribal clades, whereas Neoscopelidae show the canonical vertebrate gene order. All these features are summarized in Figure 3 with numbers 1–8 corresponding to rearranged gene orders (1–4) and INC-sequences (5–8).
Figure 3.
Typical gene orders in vertebrates and Neoscopelidae with eight unique gene orders and INC-regions found in Myctophidae. 1–4 show rearranged gene orders and 5–8 show INC-sequences with all eight features treated as clade-defining synapomorphies. Taxon-specific gene rearrangements are shown at the bottom. All INC-regions are shown in grey with number of base pairs noted if known. Mt information depicted is provided from all included taxa within the clades except for the rearrangement in 3 (information from N. ritteri and L. crocodilus), in 4 (information from M. affine and M. nitidulum) and in 6 (information from K. anderssoni and E. antarctica). The missing information for some included taxa is due to unsuccessful amplification of particular regions. All eight features are discussed in the text. Abbreviations follow common mt usage and colorcoding is signifying the same clades and defining gene orders throughout all figures in this article.
1. WANYC-gene order found in all myctophids. The tRNA-Cys and Tyr have shifted positions and the putative OL-region was much longer than the usual vertebrate sequence in this region of about 30–40 base pairs (56–321 in myctophids included in this study). We were not able to find any conserved sequence blocks or secondary cloverleaf structure typical of tRNA genes [76] for this INC-region (see Additional file 3).
2. IMQMψ gene order found in the tribe Diaphini. The tRNA-Gln and Met have shifted positions with the typical L- and H-strand coding intact (tRNA-Gln is coded on the L-strand and tRNA-Met coded on the H-strand). An INC-region (tRNA-Met pseudogene denoted as Mψ) is present between tRNA-Gln and ND1 in all diaphinid taxa included. A similar gene order was also observed for Notolychnus valdiviae although the INC-region is a tRNA-Met gene for this taxon (IMQM gene order). This particular gene rearrangement was analyzed and is discussed in detail below.
3. Relocation of tRNA-Glu resulting in gene order Cytb/T/E/P in a Lampanyctini subclade. Species included within Lampanyctus, Nannobrachium, Stenobrachius and Triphoturus all show tRNA-Glu pseudogenes in the canonical position between ND6 and Cytb, verified from sequence comparisons and partial secondary cloverleaf structures (see Additional file 3). We were only able to determine the position of the tRNA-Glu gene in two of the five taxa, Nannobrachium ritteri and Lampanyctus crocodilus, all showing the novel position of tRNA-Glu between tRNA-Thr and Pro. A highly variable region downstream the CR and multiple nucleotide repeats prevented us from determining this sequence in Lampanyctus macdonaldi, Stenobrachius leucopsarus and Triphoturus nigrescens.
4. Cytb/T/ND6/E/P gene order in a Myctophini subclade. Myctophum punctatum, M. affine and M. nitidulum showed this rearranged gene order whereas M. asperum and M. orientale showed the canonical gene order. Unknown technical issues prevented us from determining the sequence of ND6 in M. punctatum although the gene rearrangement is evident also in this species from ND5/Cytb contiguous sequences as opposed to the canonical ND5/ND6/E/Cytb gene order.
5. INC-sequence between LeuUUR and ND1 found in all myctophids. This spacer ranged from 45–79 base pairs and showed no secondary structure or sequence similarity to any other mt genes (see Additional file 3).
6. INC-sequence between ND1 and tRNA-Ile found in Electronini. This spacer showed significant length variation and we were unable to determine the whole sequence in Protomyctophum arcticum.
7. INC-sequence between ATP6 and CO3 found in all myctophids. This spacer was 288 base pairs long in Notolychnus valdiviae and ranged from 9–43 base pairs in all other myctophids. We found no mt gene similarity or secondary structure for this INC-region (see Additional file 3).
8. INC-sequence associated with tRNA-Gly found in a Myctophini subclade. This INC-region is treated as a character state for the clade comprising Benthosema and Diogenichthys although the exact position varies between the two genera. The three species of Benthosema included showed the INC-region located upstream the tRNA-Gly whereas in Diogenichthys atlanticus, the spacer was located downstream tRNA-Gly. Electrona antarctica showed a different organization with two detected tRNA-Gly genes separated by a large INC-region. We found no sequence similarity to other mt genes for this region or any secondary folding structures (see Additional file 3).
Additional gene orders and INC-sequences were observed for various myctophid species although none of the rearrangements were distributed across multiple taxa at this point. These additional gene order rearrangements and INC-sequences are also presented in Figure 3. Detailed information for rearranged gene orders such as length of INC-sequences and missing regions is presented in the supplementary material (see Additional file 4). The highly polymorphic control region (CR) was consistently problematic to amplify or sequence, resulting in parts of the CR missing for most species as well as flanking tRNA-Thr and -Pro for some species.
Mitogenome sequences
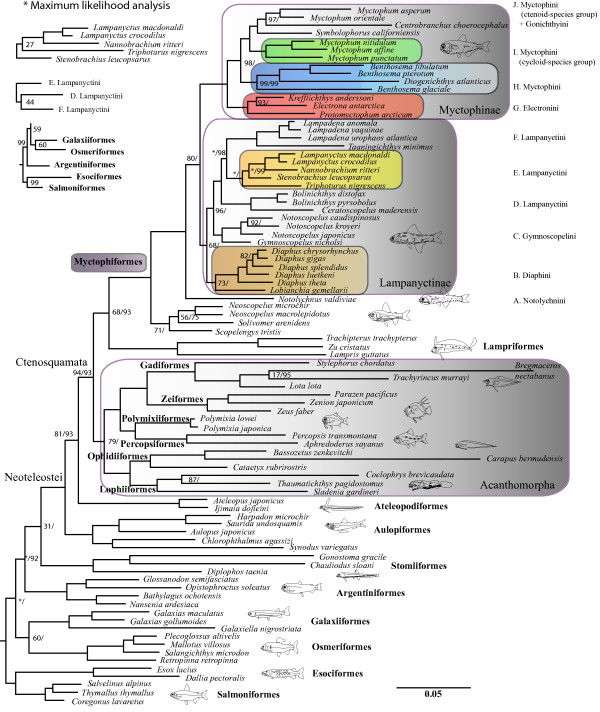
ML- and Bayesian analyses of the mitogenomic dataset consisting of 87 taxa resolved a phylogenetic tree of eu- and neoteleostean orders as presented in Figure 4. Myctophiformes was resolved as the sister clade to Lampriformes supported by a BS-value of only 68% whereas the BPP-value was 93%. Both analyses conducted for this study showed this relationship; however, considering the long basal branches of Lampriformes and Myctophiformes, we conclude that the phylogenetic position of Myctophiformes, from mitogenomic evidence, in relation to basal neoteleostean orders and Acanthomorpha remains ambiguous. This is supported by comparison to previous mitogenomic studies [33,43,46,77] and nuclear genes [78]. Although we have included all neoteleostean orders in this study (Figure 4), this issue requires the inclusion of additional mitogenomes from all neoteleostean orders, and analyses carried out using newly developed models accounting for site-specific modulations of the amino acid replacement process, such as for example the CAT mixture model [79] implemented in phylobayes [80]. However, the latter program does not implement mixture of characters preventing the use of all information included in the mt genome, making this analysis premature and is therefore not employed in this study.
Figure 4.
Mitogenomic Neoteleostei phylogeny (123ARTn) rooted with Euteleostei. Branch support from 1000 Bootstrap replicates and Bayesian posterior probabilities noted only if values are less than 100 – separated by a slash (/), respectively. Three topological differences between the Bayesian- and ML-analyses are noted with bootstrap values included. Ten distinct myctophid lineages are noted (A-J) with respect to current tribal classification.
The myctophiform ingroup topology from the two datasets analyzed (87 taxa: 123ARTn and 41 taxa: 123nRTn) showed differences related to lampanyctine tribal relationships (Figures 4 and 5). ML- and Bayesian analyses of the 87 taxa also showed incongruence concerning the phylogenetic position of Triphoturus nigrescens (Figure 4).
Figure 5.
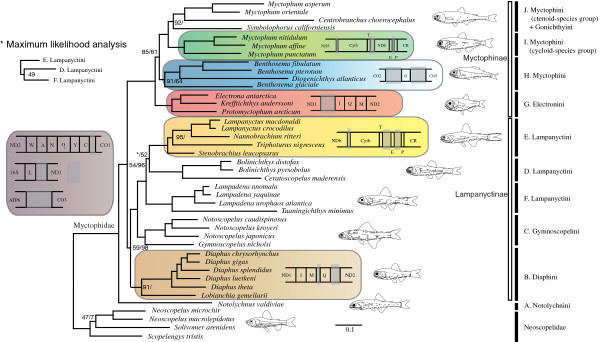
Mitogenomic Myctophiformes phylogeny (123nRTn) rooted with Neoscopelidae. Branch support from 1000 Bootstrap replicates and Bayesian posterior probabilities noted only if values are less than 100 – separated by a slash (/), respectively. One topological difference between the Bayesian- and ML-analyses is noted with bootstrap value included. Ten distinct myctophid lineages are noted (A-J) with respect to current tribal classification.
Neoscopelidae and Myctophidae were both recovered as monophyletic with Scopenlengys tristis recovered as the sister taxon to the remaining neoscopelids (low support) and Notolychnus valdiviae was recovered as the sister taxon to the remaining myctophids (high support). Myctophine and lampanyctine subfamilies were also recovered as monophyletic in all analyses. The tribe Diaphini was recovered as the sister lineage to the other lampanyctines, found in all analyses although with low support values, despite the unresolved tribal relationships within the remaining lampanyctines. The tribe Electronini was recovered as the sister lineage to the other myctophines with maximum levels of support and a clade consisting of Benthosema spp. and Diogenichthys atlanticus was resolved as the sister lineage to the tribes Gonichthyini and Myctophini. Ten myctophid lineages are recognized from this study and are noted according to current tribal classification as A-J in Figures 4 and 5. We note that deep myctophiform- and tribe-defining nodes were consistently found in all analyses, although support values indicate uncertainty concerning basal neoscopelid-, basal lampanyctine- and to a lesser degree basal myctophine phylogenetic relationships. Tribal relationships within Lampanyctinae (clades D, E and F) proved to be the most problematic part of myctophid phylogenetics from mitogenomics, showing incongruent results between datasets employed and mode of analysis (Figures 4 and 5).
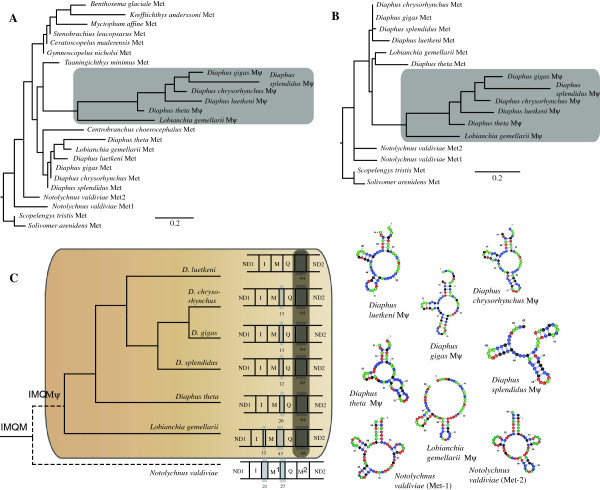
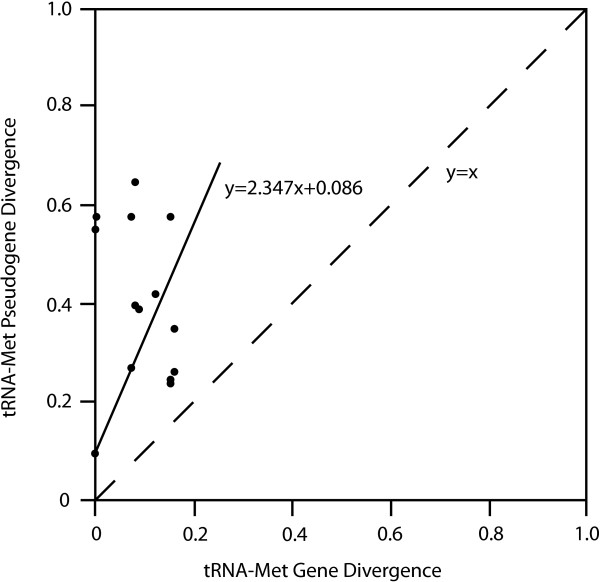
Diaphini tRNA-Met pseudogenes
All taxa within the tribe Diaphini show an IMQ-gene order in addition to an INC-region upstream from ND2 ranging from 62 to 68 base pairs. The INC-region shows varying degrees of sequence similarity to detected tRNA-Met genes with most tRNA secondary structures missing only the D- and variable loops of the typical tRNA gene cloverleaf secondary structure (Figure 6). Pairwise distances for tRNA-Met pseudogenes (INC-regions) were all much higher than for the detected tRNA-Met genes (Figure 7). In view of the IMQMψ gene order and putative tRNA-Met duplication in Diaphini, an interesting finding is observed in the monotypic basal branching taxon Notolychnus valdiviae, exhibiting two identified tRNA-Met genes resulting in an IMQM gene order. The two tRNA-Met genes both show the CAU-anticodon although they are highly divergent with respect to sequence similarity (~20% p-distance). Neigbour-joining distance method analyses of the tRNA-Met genes and their pseudogene counterparts showed clear phylogenetic structure in the diaphinid pseudogenes (Figure 6). Including the two tRNA-Met genes observed in Notolychnus (IMQM gene order) in the analyses, in order to examine a possible connection to Diaphini (IMQMψ gene order) concerning this gene-order rearrangement, showed no phylogenetic relationship between these taxa (Figure 6).
Figure 6.
K2P-distance analyses of tRNA-Met genes and the tRNA-Met pseudogenes found in Diaphini and Notolychnus. A. Analysis done with randomly selected myctophid tRNA-Met genes using a neoscopelid outgroup. B. Analysis done with only diaphinid taxa and Notolychnus using a neoscopelid outgroup. Note the better phylogenetic structure in the pseudogenes compared to their counterpart tRNA-Met genes. C. Mitogenomic cladogram showing the IMQMψ – region in Diaphini. The truncated branch of Notolychnus signifies a phylogenetic position not corroborated from DNA sequences or tRNA-Met/tRNA-Met pseudogene distance analyses. Shaded areas represent INC-regions with number of base pairs noted for each taxon. Note the “gradual” removal of the INC-regions in diaphinid taxa except the tRNA-Met pseudogene (dark shaded). Secondary folding structures are presented for Notolychnus tRNA-Met1 and Met2 and all diaphinid tRNA-Met pseudogenes.
Figure 7.
Fifteen pairwise sequence divergences of tRNA-Met and tRNA-Met pseudogenes of the six diaphinid taxa included in this study. Note the statistical significant higher sequence divergences in the tRNA-Met pseudogenes compared to the tRNA-met genes (t-test, df=13, P < 0.05).
Testing alternative phylogenetic hypotheses
Comparing constrained topologies according to morphology with the best scoring ML-tree from this study rejected a Notolychnini-Lampanyctinae relationship (P = 0.024; Table 2) as found from most morphological studies (Figure 2). A Notolychnini-Myctophinae relationship could not be rejected (P = 0.151) and the topology with Notolychnini constrained within Diaphini as indicated from the tRNA-Met duplication event was clearly not rejected (P = 0.407).
Table 2.
Statistical comparisons between phylogenetic hypotheses using the AU-test (41 taxa Myctophiformes dataset)
| Topology | -ln L | Δ ln L | AU* |
|---|---|---|---|
| (Notolychnus (Lampanyctinae, Myctophinae)) |
221112.283340 |
Best |
0.733 |
| ((Notolychnus, Lampanyctinae) Myctophinae)) |
221130.952336 |
18.668996 |
0.024* |
| (Notolychnus, Myctophinae) Lampanyctinae) |
221125.980254 |
13.696914 |
0.151 |
| (((Notolychnus, Diaphini) Lampanyctinae) Myctophinae) | 221118.943061 | 06.659721 | 0.407 |
*P < 0.05 = constrained topology rejected at the 5% confidence level.
Discussion
Monophyly of myctophiformes and the two families
This study recovered a monophyletic Myctophiformes and monophyletic families with Neoscopelidae as the sister family of the highly diversified Myctophidae (Figures 4 and 5; [3,5,81,82] although see Rosen [83]). We found Scopelengys tristis was a sister taxon to the remaining neoscopelids, including the monotypic endemic Sulu Sea taxon Solivomer arenidens[84], in congruence with morphology (Figure 2). Stiassny [5] presented characters in support for Scopelengys as sister taxa to the remaining neoscopelids, although she did note a possibility of apparently derived characters in Neoscopelus and Solivomer being secondarily reduced in Scopelengys. Analyses from the present study are congruent on this matter although support values are alarmingly low concerning the phylogenetic position of Solivomer arenidens. We found Notolychnus valdiviae to be the sister taxon to the remaining myctophids; however, morphological character states of Notolychnus such as dorsally located photophores, small larvae without pigmentation and a reduced otolith, have been extremely difficult to interpret concerning early myctophid divergence. Photophores are present in all myctophids except Taaningichthys paurolychnus and present only in one Neoscopelidae genus Neoscopelus. Our results show both of these genera nested within their respective families and the evolution of photophores within Myctophiformes is difficult to assess, i.e. whether photophores have evolved once or twice within ancestral myctophiforms. This question is pertinent from morphological considerations on photophore structure in Myctophidae and Neoscopelidae, as it is quite different in the two groups [85,86]. The phylogenetic positions of Scopelengys/Solivomer and Notolychnus in this study revives an interesting observation by Paxton [3] concerning parallel evolutionary trends for Solivomer/Lampanyctinae and for Neoscopelus/Myctophinae, regarding changes in jaw length and characters possibly associated with these changes. These characters have been difficult to interpret in terms of polarity and this study supports long jaw length as the plesiomorphic state in Neoscopelidae and Myctophidae from the basal branchings of Scopelengys/Solivomer and Notolychnus. Also, Stiassny [5] noted that important myctophiform character states like elongated jaw length and raised photophores show ambiguity concerning early myctophiform divergence.
A monophyletic Myctophidae was strongly supported by mitogenome sequences, a WANYC gene order and an INC-spacer between tRNA-LeuUUR and ND1 (Figures 3 and 4). Rearrangement of the canonical vertebrate WANCY gene order was shared by all myctophids with the tRNA-Cys relocated downstream resulting in a synapomorphic WANYC gene order (Figure 5). A prolonged sequence (putative OL + INC-sequence) in the canonical position of the putative origin of L-strand replication was found within all myctophids (see Additional file 4), although fishes within the tribe Electronini were difficult to determine due to additionally rearranged gene orders in this region (Figure 3). An INC-region between the tRNA-LeuUUR and ND1 also represents synapomorphic evidence for Myctophidae (Figure 3). The size of this spacer ranged from 45–79 base pairs and we found no obvious tendency regarding size change for this INC-region (see Additional file 4). Neither the OL + INC-region nor the LeuUUR/ND1 spacers showed sequence similarity in any species to any other mitochondrial genes. Neither could they be folded into clover shape secondary structures, so their origins are elusive. On the other hand, assembly of the INC-spacers showed clear sequence similarity within genera indicating the INC-regions to be inherited synapomorphic characters within the family. Smith et al. [87] reported a length expansion for a similar small INC-sequence located between tRNA-Val and 12S rRNA in a clade of serranid fairy basslet fishes (Serranidae). They showed the INC-sequence to be homologous for all genera from simultaneous analysis of nucleotides and gene order – a very difficult task for myctophids considering multiple gene order rearrangements and putative associated INC-sequences. The third myctophid synapomorphy from this study is an INC-region between ATP6 and CO3 ranging from 288 base pairs in Notolychnus to 9–43 base pairs in the remaining myctophids included (see Additional file 4). No conserved sequence motifs were found in this region. This is a peculiar spacer in the sense that the canonical vertebrate gene order for this region is ATP6/CO3 with no tRNA gene acting as punctuation mark for RNA processing.
Monophyly of the two subfamilies and five tribes
The monophyly of the two subfamilies Lampanyctinae and Myctophinae was recovered from all analyses, although the low support values associated with this split are incomprehensible from morphological characters unambigously delimiting these two groups. As with the Myctophidae, the two subfamilies are fully supported by adult and larval morphology and no discussion is needed – character states are presented by Moser & Ahlstrom [28], Paxton [3] and Stiassny [5]. A well supported result from this study, differing from morphology, is the phylogenetic position of the tribe Diaphini within Lampanyctinae, resolved as sister tribe to the other lampanyctines. Adult and larval morphology within Lampanyctinae show incongruences and both hypotheses are different compared to our molecular results (Figures 2, 4 and 5).This shows problematic morphological characters, lampanyctine larval characters are noted as few and tentative in previous larval studies [30,88] and of little use above genus level, as well as molecular characters, no unique gene orders connect tribes and the mitogenomic phylogenetic sequence information is insufficient, as witnessed from the three different resolved Lampanyctinae topologies from this study (Figures 4 and 5).
Monophyly of the tribes Notolychnini (clade A in Figures 4 and 5), Diaphini (B), Gymnoscopelini (C), Lampanyctini (D+E+F), Electronini (G) and two subclades of Myctophini (H and I+J) are strongly supported from all mitogenomic DNA sequence analyses and various mt gene orders discussed below. Only the node delimiting the tribe Diaphini (B) show relatively low support values. The tribe Gonichthyini is tentatively placed from a single taxon as nested within a Myctophini subclade consisting of ctenoid-scaled species. Mitogenomic DNA sequence data from this study supports recognition of 10 mutually exclusive clades within Myctophidae (Figures 4 and 5).
Phylogeny of tribes and genera
Students of lanternfish phylogenetics will note the position of Notolychnus (clade A) resolved as sister taxon to the remaining family Myctophidae from mitogenome sequences. Paxton [3] tentatively placed Notolychnus as sister taxon to the subfamily Lampanyctinae (Figure 2A). Subsequent parsimony analysis of the same data by Paxton et al. [30], including additional larval characters [27-29], placed Notolychnus in an unresolved Myctophinae-Notolychnini-Lampanyctinae trifurcation (Figure 2D). Subsequently, Stiassny [5] added four additional characters and reanalyzed the aforementioned data with a similar result, showing Notolychnus as the sister taxon to the remaining lampanyctines (Figure 2E). Most recently, Yamaguchi [24] scrutinized previously published characters (Figure 2A-E) once again and reanalyzed the data, resulting in a largely unresolved myctophid tree (Figure 2F). However, the position of Notolychnus was congruent with Paxton [3] and Stiassny [5]. The novel phylogenetic position of Notolychnus from this study, as the basal branching in Myctophidae, highlight the past issues concerning subfamilial affiliation and partly explains the lack of phylogenetically informative morphological characters in relation to the two subfamilies.
A monophyletic Diaphini (clade B) was well supported from both sequences and a unique IMQ-gene order (Figure 3), all in complete agreement with morphology, showing Lobianchia (2 species) as sister group of the very speciose Diaphus (78 species) [89-91]. Four species-groups of Diaphus have been suggested from the presence of Dn-Vn, Ant, Suo and So photophores (for photophore terminology, see Figure 1) [91]; however, our five species of Diaphus included were insufficient to validate anything on this matter. One very rare monotypic taxon Idiolychnus urolampus, missing from our study, was transferred from Diaphus to Lobianchia by Bolin [92] and most lately erected into its own genus Idiolychnus by the position of VO3 and possession of two SAOs [93].
Gymnoscopelini (clade C) sensu Paxton [3] was found to be monophyletic although not fully congruent with Ahlstrom et al. [88]. Unfortunately, the monotypic taxon Lampanyctodes hectoris is not included in this study, a myctophid showing ambiguous morphological characters concerning tribal placement. Paxton [3] noted Lampanyctodes as a possible early divergence based on several characters shared with the most plesiomorphic diaphinid genus Lobianchia. Conversely, Ahlstrom et al. [88] noted Lampanyctodes as being specialized within Gymnoscopelini from a similar larval form and development to that found within Lampadena, arguing that characters reflect habitat instead of true phylogeny.
Ceratoscopelus and Bolinichthys formed a clade (D) as either sister group to the remaining lampanyctines or included within them (Figures 4 and 5, respectively). Paxton [3] included these genera within Lampanyctini as closely related, whereas Ahlstrom et al. [88] placed them within Gymnoscopelini. Our study supports inclusion in Lampanyctini despite unresolved intrarelationships of this tribe. The clade itself is very distinct in terms of mt sequences and supported by both osteology [3] and larval development [30].
A tRNA-Glu gene could be detected in the canonical vertebrate position between ND6 and Cytb in all myctophids included in this study, except in a Lampanyctini subclade (E) comprising Stenobrachius, Triphoturus, Nannobrachium and Lampanyctus (Figures 3). In Nannobrachium ritteri and Lampanyctus crocodilus, we were able to determine a novel location of tRNA-Glu in the highly polymorphic region downstream Cytb including the tRNA-Thr and Pro genes, the CR and various INC-regions. Sequence similarities between tRNA-Glu from N. ritteri and L. crocodilus and the INC-regions between ND6 and Cytb were evident from sequence comparisons and putative secondary structures (see Additional file 3). Interestingly, this INC-region showed no sign of redundancy since all five taxa have retained a fragment equal in size to the typical length of tRNA genes (68–73 base pairs). We tentatively assign this gene order to the lampanyctine subclade E and note that this clade is congruent with Paxton [3] and Paxton et al. [30], although the latter notes that no synapomorphic character is present to actually define the clade. We expect this feature to be found within the unsampled genus Parvilux[94] as well. A new position of tRNA-Glu and retention of an INC-sequence between ND6 and Cytb has also been found in other vertebrates, e.g. the amphisbaenian reptile family Bipedidae [95], and seems to be one of the most common gene order rearrangements in vertebrates.
A clade consisting of Lampadena and Taaningichthys (F) is found monophyletic and completely congruent with morphology. Taaningichthys was previously a part of the genus Lampadena[4] although it was separated by Bolin [92] based on characters such as reduced number of photophores and flaccid body structure. Nafpaktitis and Paxton [96] noted that Lampadena chavesi, L. dea and L. speculigera were more closely related to Taaningichthys than the remaining species of Lampadena based on expanded neural arches on the anterior vertebrae as found in Taaningichthys. In fact, Paxton [3] noted that discriminating characters between the two genera were hard to find. Lampadena and especially Taaningichthys are among the largest and deepest living myctophids, with the latter showing reduction of the lateral line and in the number of body photophores. This apparent reductional trend has resulted in one taxon, Taaningichthys paurolychnus, having lost all body photophores although retained the supra- and infracaudal glands. Lampadena yaquinae is found nested within the Lampadena species included and corroborates Paxton [97] in synonymizing Dorsadena[98] with Lampadena. Likewise, the subgenera Lampadena and Lychnophora proposed by Fraser-Brunner [4], supported by only an elevated PO4[96], are not supported from this study. Increased mitogenomic taxon sampling of these two genera is interesting concerning evolution of photophores within these deep dwelling genera showing atypical character states within the Myctophidae [99].
The tribe Electronini (G), erected from low-leveled PVO and PLO photophores [25], composes a subset of the tribe Myctophini as recognized by Paxton [3]. Our results clearly support Electronini as a separate tribe [100] from both DNA sequences and long (>200 base pairs) synapomorphic INC-regions upstream the tRNA-Ile gene (Figures 3 and 5). We recovered a sister relationship of Electronini to the rest of Myctophinae, a result also presented by Paxton et al. [30], although cladistic analyses of morphological characters failed to recover this result (Figures 2E-F).
The IQM-regions of species included in the genera Benthosema and Diogenichthys (clade H) were peculiar and the IQM-region and WANYC-region of both Electronini and Benthosema-Diogenichthys were highly differentiated compared to all other myctophids. Benthosema glaciale showed new positions of tRNA-Gln and -Met to the downstream region of the WANYC-region (Figure 3) representing a novel gene order in vertebrate mitogenomes. Diogenichthys atlanticus showed an IQMM gene order and the canonical vertebrate gene order was observed within Benthosema pterotum and B. fibulatum. Taxon sampling prevents us from discussing rearrangements for these taxa at this point; however, a close relationship between the two genera is consistent with morphology. Diogenichthys was separated from Benthosema by ventrally leveled Prc 1 and 2 in addition to various jaw differences and hooked dentary teeth, a character state only found within lampanyctines. Similar to Notolychnus, D. atlanticus is a diminutive species with adult maximum size < 30 millimeters and could represent a problematic branch within this clade considering character states related to miniaturization. Diminutive species are also found within the genus Diaphus. The Benthosema-Diogenichthys clade was supported from DNA sequences and an INC-region up- or downstream tRNA-Gly; all three species of Benthosema showed an INC-region flanked by an upstream CO3 gene and downstream tRNA-Gly. In addition, B. glaciale showed an INC-region after tRNA-Gly. In D. atlanticus, however, the INC-spacer was located downstream from the tRNA-Gly gene (Figure 3). Interestingly, two tRNA-Gly genes separated by a large INC-region were observed in this same region in Electrona antarctica, although none of the INC-regions could be shown to have any sequence similarity to each other or to tRNA-Gly. We have assumed an INC-sequence associated with tRNA-Gly as a character state for the Benthosema-Diogenichthys clade, although with reservations. In view of E. antarctica possesing two tRNA-Gly genes we note that an association of this region in E. antarctica, D. atlanticus and Benthosema spp. is plausible and should be elucidated with increased mitogenomic taxon sampling.
The tribe Myctophini was not supported from this study as the sister clade to Electronini, because the single representative of the tribe Gonichthyini (Centrobranchus choerocephalus) included in this study renders Myctophini paraphyletic. We note that broader taxonomic representation of the genera Gonichthys, Centrobranchus, Tarletonbeania and Loweina is necessary concerning phylogenetic position of Gonichthyini within Myctophinae. However, evidence from mt sequences and a unique gene order in part of Myctophini (Figure 5) is consistent with the results from Paxton [3] stating that”..A number of characters suggest that the Myctophum-Symbolophorus line gave rise to the slendertails [ed. tribe Gonichthyini]..”. Additional evidence for this result comes from the non-monophyly of Gonichthyini based on larval characters [27] (Figure 2C) and the analyses by Stiassny [5] and Yamaguchi [24] (Figure 2E-F) showing Symbolophorus a basal branching within Gonichthyini. This study confidently resolves the genus Myctophum into two different clades from mitogenome sequences and also from a unique gene order involving ND6, tRNA-Glu and Cytb (Figures 3, 5). Myctophum affine, M. punctatum and M. nitidulum (clade I) showed this particular gene arrangement whereas M. asperum and M. orientale showed the typical myctophid gene order also found in Centrobranchus choerocephalus and Symbolophorus californiensis, the latter four forming a well supported clade J (Figures 4 and 5). Two species-groups are currently recognized in the genus Myctophum based on cycloid versus ctenoid scales [101] corresponding to the two separated groups of Myctophum found in this study. Moser and Ahlstrom [27] discussed eye shape within Myctophum and noted this genus as difficult to distinguish phylogenetically, with stalked eyes usually absent, although present in e.g. Myctophum nitidulum and M. punctatum corresponding to the cycloid-scaled Myctophum lineage from this study. This result is, however, complicated by stalked eyes also being found in all species of Symbolophorus[27]. Clearly, mitogenome sequences and a unique gene order in the cycloid-scaled species of Myctophum strongly suggest that the two groups are in fact different lineages within Myctophini and that the tribe Gonichthyini is nested within the ctenoid-scaled group. This indicates ambiguous relationships from adult and larval morphology or convergent evolution within Myctophini lineages. Paxton [3] noted that no additional characters are present within Myctophum to support a split between cycloid- and ctenoid-scaled Myctophum species. The term”near-surface” myctophids has been used for a group of myctophids represented within the genera Loweina, Tarletonbeania, Gonichthys, Centrobranchus, Symbolophorus and Myctophum with most species showing DVM behaviour and most being easily netted in surface layers at night time [102]. Denser taxon sampling within Gonichthyini, and the ctenoid-scaled Myctophini group as recognized from this study, should clarify if DVM patterns and phylogeny are correlated.
Diaphini tRNA-Met pseudogenes
Our included taxa in the tribe Diaphini opens up for a unique empirical case of a putative duplication event that can be observed through time for retention and deletion of various genes/pseudogenes in separate diaphinid taxa. All tRNA-Met pseudogenes were retained with 64–68 base pairs whereas other INC-regions between the tRNA genes in this segment (grey areas in Figure 6) seem to be in the process of being removed. Phylogenetic congruence between the mitogenomic sequences and the diaphinid pseudogenes (Figures 5 and 6, respectively) is clear despite the short pseudogene sequence lengths. Only one other vertebrate group, scarrid parrotfishes, shows a similar pattern of gene rearrangement of the IQM-region and retention of the pseudogene [103], although the pattern of retention and deletion of INC-regions in parrotfishes is much less pronounced (see Additional file 5). We note that the tribe Electronini and other taxa within the subfamily Myctophinae exhibit gene rearrangements, albeit different in organization, associated with this particular region (Figure 3, see Additional file 4). We acknowledge the missing discussion of this region in the subfamily Myctophinae; however, our results concerning this particular region in Myctophinae awaits increased taxon sampling. Topology testing using the AU-test could not reject a Notolychnini-Diaphini relationship (P=0.407; Table 2) in contrast to morphology, mitogenomic sequences and our NJ-analyses of the INC-regions (Figures 2, 5 and 6, respectively).
We note a third case of various anuran putative duplication events of the tRNA-Met gene [104] and although this case is reminiscent of the low sequence similarity between the two detected tRNA-Met genes as seen in Notolychnus, the tRNA-Met anticodon is repeatedly detected in anurans making this situation different to lantern- and parrotfishes.
Mt gene order rearrangements in vertebrates
Observations of unique gene rearrangements in Myctophidae, combined with their utility as clade defining synapomorphies, are quite numerous compared to similar findings within other vertebrate groups [105]. It should be noted that whereas mt sequences and gene order rearrangements resolve myctophiform relationships in a highly corroborative manner in this study (Figure 5), taxon sampling and unknown selection on gene order and INC-sequences are problematic when inferring phylogeny. Clearly, gene order rearrangement hotspots are present in vertebrate mt genomes and selective constraints could plausibly show convergent evolution to be the reason in some cases [106]. Almost all gene order rearrangements and INC-regions presented for Myctophidae have also been found within other vertebrate groups. For example, the INC-sequence present between ATP6 and CO3 is found within Gonostoma gracile (order Stomiiformes) [45] and in various deep-sea anglerfishes [47]. Birds (class Aves) all have a clade-defining rearranged gene order encompassing a fragment ranging from ND6 to the CR [107], although multiple variations have subsequently occurred on top of this synapomorphy [40]. Plethodontid salamanders show multiple gene rearrangements with likely concerted evolution on duplicated regions [108]. The latter also suggested an INC-sequence between tRNA-Thr and -Pro possibly functioning as an alternative initiation site for replication. All myctophiforms successfully determined for this mt site show an INC-sequence between tRNA-Thr and -Pro, although the length of this spacer in myctophids is highly variable (see Additional file 4). Bakke et al. [109] postulated a selective advantage as a protein-binding site for this spacer, supported by a conservative motif sequence in codfishes. Concerted evolution on CR-duplicates is also reported from snakes [110] and mantellid frogs [111], with the latter strongly indicating recombination associated with the CR in various taxa exhibiting duplication events. Varanids and monitors show events in the variable region upstream of the CR [112] and acrodont lizards show a QIM gene order [113]. As mentioned, scarid parrotfishes all have an IMQ-gene order showing similar patterns as found in Diaphini, with an INC-sequence (pseudogene) retained upstream the ND2 gene from a putative duplication of tRNA-Met [103]. The latter suggested that the tRNA-Met pseudogene neighbouring ND2 retained a function as punctuation mark for ND2 mRNA processing. Mt tRNA transcriptional efficiency in relation to distance of initiation of replication was explored by Satoh et al. [114], who found support for highly expressed genes positioned closer to the control region. Selection on INC-regions in relation to bordering genes is an important theoretical caveat when using INC-regions as phylogenetic markers. Satoh et al. [48] initiated mt gene order comparisons within codfishes (Gadiformes), showing rearranged gene orders and INC-regions in a possible similar fashion to Myctophidae. Most notably one species, Bathygadus antrodes, possessed all of the gadiform rearrangements although somewhat differently arranged from other subclades. This indicates early rearrangement events subsequently modified into presently observed synapomorphic gene orders for specific gadiform clades. The most basal branching in Myctophidae, Notolychnus, shows mt rearrangements such as two tRNA-Met genes and a comparatively large INC-sequence between ATP6 and CO3. These features are either modified or lost in other myctophids. On the other hand, several rearranged gene orders found in various myctophid tribes are not traceable in Notolychnus. Gene order rearrangements within Myctophidae provide a unique example of the phylogenetic utility of mt gene orders and the problems associated with this group of markers.
Conclusions
Mitogenomes from myctophiform fishes provide a unique empirical case of mt DNA sequences and gene order rearrangements that corroborate evolutionary history. Future mitogenomic determination of the 200+ remaining species within Myctophidae will most certaninly increase our knowledge concerning issues associated with gene order rearrangement events and their utility as phylogenetic markers. Multiple different gene orders found throughout the mt genome, utility as phylogenetic markers and the unknown causes and mechanisms associated with gene duplication and subsequent retention and/or removal of sequences, calls for the determination of complete mt genomes over single genes. Our results support the use of osteology and photophores in phylogeny of Myctophiformes, the latter being a very unique feature of any deep-sea group of organisms. Larval ontogeny is also informative albeit difficult to use on higher phylogenetic levels and particularly within the subfamily Lampanyctinae.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
All authors contributed in preparation of the study. JYP performed molecular lab work, initial analyses and drafted the original manuscript. DR, HT and TPS continually reviewed and discussed lab-procedures. JYP and EW finalized the analyses. JYP, IB, EW and MM finalized the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Table T2. Partitioning schemes used in the present study suggested by PartitionFinder.
Saturation plots of pairwise distances of 1st, 2nd and 3rd codon positions in the 12 protein coding genes employed in the analyses. Vertical axis shows p-distance for each codon position against total p-distance (horizontal axis). Saturation from RY-coding of the 3rd codon positions included (only transversions valued) although not used in the present study from early onset of saturation.
Secondary structure of INC-regions. Sequence alignments and secondary tRNA structures of INC-regions are shown for synapomorphic spacers pertaining to gene order rearrangements 1, 3, 5 and 8 presented in Figure 3. The remaining INC-regions presented in Figure 3 are relatively short sequences and are not included.
Base pair details of INC-regions found in Myctophidae. Eight mt regions correspond to gene order and INC-region synapomorphies presented in Figure 3 with slash (/) denoting bordering genes. The last column includes mt information from the polymorphic region from tRNA-Thr and downstream towards the control region (CR). Numbers of INC-sequence base pairs are parenthesized next to flanking genes. Greater-than sign (>) and hyphen (−) denote partial and no sequence, respectively, from failure to complete the sequences.
IMQMψ gene order in a clade of scarid parrotfishes. Taxa constituting a monophyletic clade in Scaridae all show similar patterns in the IMQ-region as observed within Diaphini. Grey areas show INC-regions with number of base pairs noted for each taxon and the arrow indicates possible gradual removal of INC-regions.
Contributor Information
Jan Y Poulsen, Email: Jan.Poulsen@bm.uib.no.
Ingvar Byrkjedal, Email: Ingvar.Byrkjedal@bm.uib.no.
Endre Willassen, Email: Endre.Willassen@bm.uib.no.
David Rees, Email: David.Rees@bm.uib.no.
Hirohiko Takeshima, Email: takeshim@aori.u-tokyo.ac.jp.
Takashi P Satoh, Email: satotaka@kahaku.go.jp.
Gento Shinohara, Email: s-gento@kahaku.go.jp.
Mutsumi Nishida, Email: mnishida@aori.u-tokyo.ac.jp.
Masaki Miya, Email: miya@chiba-muse.or.jp.
Acknowledgements
No ethical issues
The authors declare that no ethical issues were present for this study.
JYP thanks K. Mabuchi, T. Kon, Y. Yamanoue, M. Nakatani, M. Seimiya, M. Yorifuji, S. Hayeun, M. Takada, M. Kawaguchi, T. Shuto, M. Kato, A. Kinjo, T. Nagasaki, (AORI), K. Sünksen, N. Hammeken, S. Hvass, H. Lund (GN-Pinngortitaleriffik) and all crew onboard R/V Pâmiut.
For providing tissue and access to vouchers we thank Center for Molecular Biodiversity Research (NSMT), M. Moku (AORI), H.J. Walker (SIO), S.W. Knudsen, M. Kragh, T. Menne, P. Møller, J. Nielsen (ZMUC), K. Hartel (MCZ), A.C. Bentley (KU), C. Kenealey (WU), A. Graham (CSIRO), G. Herrera, L. Rodriguez-Grana, (Chile & Uruguay, respectively), J. Paxton, A. Hay (AUS), K-T. Shao, L.Y-C. Liao, P-L. Lin (ASIZP), N.V. Gordeeva (IOGEN). A big thanks goes to H. Endo and N. Nakayama (BSKU) for onboard sampling for this study.
For general logistics we thank G. Langhelle, L. Emdal (ZMUB), L. Lindblom, K. Meland (UIB) and Rudolf Svendsen for contributing UW-photograph.
Two anonymous reviewers and editor provided highly constructive reviews of this manuscript before publication. Funding was provided by JSPS Research grants (17207007, 19207007, and 22370035) and Bergen University Museum Ph.D.- and travel grants for JYP.
References
- Rosen DE. In: Interrelationships of Fishes. Greenwood PH, Miles RS, Patterson C, editor. London: Academic Press; 1973. Interrelationships of higher euteleostean fishes; pp. 397–513. [Google Scholar]
- Nelson JS. Fishes of the world. Hoboken, New Jersey: John Wiley & Sons; 2006. [Google Scholar]
- Paxton JR. Osteology and relationships of the lanternfishes (family Myctophidae) Bull Nat Hist Mus Los Angeles Cty. 1972;13:1–81. [Google Scholar]
- Fraser-Brunner A. A classification of the fishes of the family myctophidae. Proc Zool Soc London. 1949;118:1019–1106. [Google Scholar]
- Stiassny MLJ. In: Interrelationships of Fishes. Stiassny MLJ, Parenti LR, Johnson GD, editor. San Diego: Academic; 1996. Basal Ctenosquamate relationships and the interrelationships of the myctophiform (Scopelomorph) fishes; pp. 405–426. [Google Scholar]
- Bolin RJ. A review of the myctophid fishes of the pacific coast of the unites states and of lower California. Stanford Ichthyological Bull. 1939;1:89–156. [Google Scholar]
- Hulley AP. In: Encyclopedia of Fishes 2. Paxton J, Eschmeyer WN, editor. San Diego: Academic; 1998. Myctophidae; pp. 127–128. [Google Scholar]
- Lawry JV. The olfactory epithelium of the lantern fish, Tarletonbeania crenularis (Myctophidae) Z Zellforsch. 1973;138:31–39. doi: 10.1007/BF00307076. [DOI] [PubMed] [Google Scholar]
- Stanger-Hall KF, Lloyd JE, Hillis DM. Phylogeny of north American fireflies (coleoptera: lampyridae): implications for the evolution of light signals. Mol Phylogenet Evol. 2007;45:33–49. doi: 10.1016/j.ympev.2007.05.013. [DOI] [PubMed] [Google Scholar]
- Mensinger AF, Case FJ. Luminescent Properties of fishes from nearshore California Basins. J Exp Biol Ecol. 1997;210:75–90. doi: 10.1016/S0022-0981(96)02719-0. [DOI] [Google Scholar]
- Widder EA. Bioluminescence in the ocean: origins of biological, chemical, and ecological diversity. Science. 2010;328:704–708. doi: 10.1126/science.1174269. [DOI] [PubMed] [Google Scholar]
- Foran. Evidence of luminous bacterial symbionts in the light organs of myctophid and stomiiform fishes. J Evo Zool. 1991;259:1–8. doi: 10.1002/jez.1402590102. [DOI] [PubMed] [Google Scholar]
- Haygood MG, Edwards DB, Mowlds G, Rosenblatt RH. Bioluminescence of myctophid and stomiiform fishes is not due to bacterial luciferase. J Exp Zool. 1994;270:225–231. doi: 10.1002/jez.1402700212. [DOI] [Google Scholar]
- Shimomura O. Bioluminescence. Chemical Principles and Methods. World Scientific Publishing Singapore; 2006. [Google Scholar]
- Leisman G, Cohn DH, Nealson KH. Bacterial origin of bioluminescence in marine animals. Science. 1980;208:1271–1273. doi: 10.1126/science.208.4449.1271. [DOI] [PubMed] [Google Scholar]
- Haygood MG, Distel DL. Bioluminescent symbionts of flashlight fishes and deep-sea anglerfishes from unique lineages related to the genus Vibrio. Nature. 1993;363:154–156. doi: 10.1038/363154a0. [DOI] [PubMed] [Google Scholar]
- Turner JR, White EM, Collins MA, Partridge JC, Douglas RH. Vision in lanternfish (myctophidae): adaptations for viewing bioluminescence in the deep-sea. Deep-sea Res 1. 2009;56:1003–1017. doi: 10.1016/j.dsr.2009.01.007. [DOI] [Google Scholar]
- Hasegawa EI, Sawada K, Abe K, Watanabe K, Uchikawa K, Okazaki Y, Toyama M, Douglas RH. The Visual pigments of a deep-sea myctophid fish Myctophum nitidulum Garman; an HPLC and spectroscopic description of a non-paired rhodopsin-porphyropsin system. J Fish Biol. 2008;72:937–945. doi: 10.1111/j.1095-8649.2007.01772.x. [DOI] [Google Scholar]
- Douglas RH, Partridge JC, Marshall NJ. The eyes of deeep-sea fish 1: Lens pigmentation, tapeta and visual pigments. Prog Retinal Eye Res. 1998;17:597–636. doi: 10.1016/S1350-9462(98)00002-0. [DOI] [PubMed] [Google Scholar]
- Watanabe H, Moku M, Kawaguchi K, Ishimaru K, Ohno A. Diel vertical migration of myctophid fishes (family Myctophidae) in the transitional waters of the western North Pacific. Fisheries Oceanography. 1999;8:115–127. doi: 10.1046/j.1365-2419.1999.00103.x. [DOI] [Google Scholar]
- Backus RH, Craddock JE, Haedrich RL, Shores DL, Teal JM, Wing AS, Mead GW, Clarke WD. Ceratoscopelus maderensis: peculiar sound-scattering layer identified with this myctophid fish. Science. 1968;160:991–993. doi: 10.1126/science.160.3831.991. [DOI] [PubMed] [Google Scholar]
- Kerfoot WC. Adaptive value of vertical migration: comments on the predation hypothesis and some alternatives. Contrib Mar Sci. 1985;27:91–113. [Google Scholar]
- Herring PJ. Species abundance, sexual encounter and bioluminescent signaling in the deep sea. Phil Trans Royal Soc London B. 2000;355:1273–1276. doi: 10.1098/rstb.2000.0682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M. Phylogenetic analyses of myctophid fishes using morphological characters: Progress, problems, and future perspectives. Japanese J Ichthyol. 2000;47:87–107. [Google Scholar]
- Wisner RL. A new genus and species of myctophid fish from the South-Central Pacific Ocean, with notes on related genera and the designation of a new tribe, Electronini. Copeia. 1963;1:24–28. [Google Scholar]
- Miyake T, Uyeno T. The urodermals in lanternfish family myctophidae (Pisces: myctophiformes) Copeia. 1987;1:176–181. [Google Scholar]
- Moser HG, Ahlstrom EH. Development of lanternfishes (family myctophidae) in the California current. Part 1. Species with narrow-eyed larvae. Bull Nat Hist Mus Los Angeles Cty. 1970;7:1–145. [Google Scholar]
- Moser HG, Ahlstrom EH. Development of the lanternfish, Scopelopsis multipunctatus Brauer 1906, with a discussion of its phylogenetic position in the family Myctophidae and its Role in a proposed mechanism for the evolution of photophore patterns in lanternfishes. Fish Bull. 1972;70:541–564. [Google Scholar]
- Moser HG, Ahlstrom EH. Role of larval stages in systematic investigations of marine teleosts: a case study. Fish Bull. 1974;72:391–413. [Google Scholar]
- Paxton J, Ahlstrom EH, Moser HG. Ontogeny and Systematics of Fishes. Special Publication 1, ASIH; 1984. Myctophidae: relationships; pp. 239–244. [Google Scholar]
- Yamaguchi M, Miya M, Okiyama M, Nishida M. Molecular phylogeny and larval morphological diversity of the lanternfish genus Hygophum (Teleostei: Myctophidae) Mol Phylogenet Evol. 2000;15:103–114. doi: 10.1006/mpev.1999.0726. [DOI] [PubMed] [Google Scholar]
- Miya M, Nishida M. Speciation in the open ocean. Nature. 1997;389:803–804. doi: 10.1038/39774. [DOI] [Google Scholar]
- Miya M, Kawaguchi A, Nishida M. Mitogenomic exploration of higher teleostean phylogenies: a case study for moderate-scale evolutionary genomics with 38 newly determined complete mitochondrial DNA sequences. Mol Biol Evol. 2001;18:1993–2009. doi: 10.1093/oxfordjournals.molbev.a003741. [DOI] [PubMed] [Google Scholar]
- Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Boore JL. Animal mitochondrial genomes. Nuc Acids Res. 1999;27:1767–1780. doi: 10.1093/nar/27.8.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boore JL, Brown WM. Big trees from little genomes: mitochondrial gene order as a phylogenetic tool. Curr Opinion in Gen Dev. 1998;8:668–674. doi: 10.1016/S0959-437X(98)80035-X. [DOI] [PubMed] [Google Scholar]
- Macey JR, Larson A, Ananjeva NB, Fang Z, Papenfuss TJ. Two novel gene orders and the role of light-strand replication in rearrangement of the vertebrate mitochondrial genome. Mol Biol Evol. 1997;14:91–104. doi: 10.1093/oxfordjournals.molbev.a025706. [DOI] [PubMed] [Google Scholar]
- Moritz C, Brown WM. Tandem duplications in animal mitochondrial DNAs: variation in incidence and gene content among lizards. Proc Natl Acad Sci USA. 1987;84:7183–7187. doi: 10.1073/pnas.84.20.7183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boore JL. In: Computational Biology Series Volume 1. Sankoff D, Nadeau J, editor. Dordrecht (The Netherlands): Kluwer Academic Publishing; 2000. The duplication/random loss model for gene rearrangement exemplified by mitochondrial genomes of deuterostome animals; pp. 133–147. [Google Scholar]
- Mindell DP, Sorenson MD, Dimcheff DE. Multiple independent origins of mitochondrial gene order in birds. Proc Nat Acad Sci. 1998;95:10693–10697. doi: 10.1073/pnas.95.18.10693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavrov DV, Boore JL, Brown WM. Complete mtDNA Sequences of two millipedes suggest a new model for mitochondrial gene rearrangements: duplication and nonrandom loss. Mol Biol Evol. 2002;19:163–169. doi: 10.1093/oxfordjournals.molbev.a004068. [DOI] [PubMed] [Google Scholar]
- Personal webpage Jan Yde Poulsen. http://jypichthyology.info.
- Ishiguro NB, Miya M, Nishida M. Basal euteleostean relationships: a mitogenomic perspective on the phylogenetic reality of the “Protacanthopterygii”. Mol Phylogenet Evol. 2003;27:476–488. doi: 10.1016/S1055-7903(02)00418-9. [DOI] [PubMed] [Google Scholar]
- Miya M, Nishida M. Use of mitogenomic information in teleostean molecular phylogenetics: a tree-based exploration under the maximum-parsimony optimality criterion. Mol Phylogenet Evol. 2000;17:437–455. doi: 10.1006/mpev.2000.0839. [DOI] [PubMed] [Google Scholar]
- Miya M, Nishida M. Organization of the mitochondrial genome of a deep-sea fish, Gonostoma gracile (Teleostei: Stomiiformes): first example of transfer RNA gene rearrangements in bony fishes. Mar Biotech. 1999;1:416–426. doi: 10.1007/PL00011798. [DOI] [PubMed] [Google Scholar]
- Miya M, Takeshima H, Endo H, Ishiguro NB, Inoue JG, Mukai T, Satoh TP, Yamaguchi M, Kawaguchi A, Mabuchi K, Shirai SM, Nishida M. Major patterns of higher teleostean phylogenies: a new perspective based on 100 complete mitochondrial DNA sequences. Mol Phylogenet Evol. 2003;26:121–138. doi: 10.1016/S1055-7903(02)00332-9. [DOI] [PubMed] [Google Scholar]
- Miya M, Pietsch TW, Orr JW, Arnold RJ, Satoh TP, Shedlock AM, Ho H-C, Shimazaki M, Yabe M, Nishida M. Evolutionary history of anglerfishes (Teleostei:Lophiiformes): A mitogenomic perspective. BMC Evol Biol. 2010;10:58. doi: 10.1186/1471-2148-10-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh TP, Miya M, Endo H, Nishida M. Round and pointed-head grenadier fishes (Actinopterygii: Gadiformes) represent a single sister group: evidence from the complete mitochondrial genome sequences. Mol Phylogenet Evol. 2006;40:129–138. doi: 10.1016/j.ympev.2006.02.014. [DOI] [PubMed] [Google Scholar]
- Cheng S, Higuchi R, Stoneking M. Complete mitochondrial genome amplification. Nat Gen. 1994;7:350–351. doi: 10.1038/ng0794-350. [DOI] [PubMed] [Google Scholar]
- 4peaks version 1.7.2. http://www.mekentosj.com/science/4peaks.
- TextWrangler version 3.5.3. http://barebones.com/products/textwrangler.
- Maddison DR, Maddison WP. MacClade Version 4. Sunderland, Massachusetts: Sinauer Associates; 2003. [Google Scholar]
- Sequencher version 4.10.1. http://genecodes.com.
- NCBI. http://www.ncbi.nlm.nih.gov.
- Lowe TM, Eddy SR. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schattner P, Brooks AN, Lowe TM. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2005;33:686–689. doi: 10.1093/nar/gki366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löytynoja A, Milinkovitch MC. A hidden Markov model for progressive multiple alignment. Bioinformatics. 2003;19:1505–1513. doi: 10.1093/bioinformatics/btg193. [DOI] [PubMed] [Google Scholar]
- Lanfear R, Calcott B, Ho SYW, Guindon S. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 2012;29:1695–1701. doi: 10.1093/molbev/mss020. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. Sunderland, Massachusetts: Sinauer Associates; 2003. [Google Scholar]
- Stamatakis A. Phylogenetic models of rate heterogeneity: a high performance computing perspective. Proc IPDPS. 2006.
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Yang Z. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: approximate methods. J Mol Evol. 1994;39:306–314. doi: 10.1007/BF00160154. [DOI] [PubMed] [Google Scholar]
- Akaike H. A new look at the statistical model identification. IEEE Trans Automatic Control. 1974;19:716–723. doi: 10.1109/TAC.1974.1100705. [DOI] [Google Scholar]
- Schwarz GE. Estimating the dimension of a model. Ann Stat. 1978;6:461–464. doi: 10.1214/aos/1176344136. [DOI] [Google Scholar]
- Keane TM, Creevey CJ, Pentony MM, Naughton TJ, McInerney JO. Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evol Biol. 2006;6:29. doi: 10.1186/1471-2148-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adachi J, Hasegawa M. Model of amino acid substitution in proteins encoded by mitochondrial DNA. J Mol Evol. 1996;42:459–468. doi: 10.1007/BF02498640. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Kumar S, Skjæveland Å, Russell RJS, Enger P, Ruden T, Mevik BH, Burki F, Botnen A, Shalchian-Tabrizi K. AIR: A batch-oriented web program package for construction of supermatrices ready for phylogenomic analyses. BMC Bioinformatics. 2009;10:357. doi: 10.1186/1471-2105-10-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A, Drummond AJ. Tracer version 1.5. http://tree.bio.ed.ac.uk/software/tracer.
- Smith C, Heyne S, Richter AS, Will S, Backofen R. Freiburg RNA Tools: a web server integrating IntaRNA, ExpaRNA and LocARNA. Nucleic Acids Res. 2010;38:373–377. doi: 10.1093/nar/gkq316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen P, Voss B, Rehmsmeier M, Reeder J, Giegerich R. RNAshapes: an integrated RNA analysis package based on abstract shapes. Bioinformatics. 2006;22:500–503. doi: 10.1093/bioinformatics/btk010. [DOI] [PubMed] [Google Scholar]
- Darty K, Denise A, Ponty Y. VARNA: Interactive drawing and editing of the RNA secondary structure. Bioinformatics. 2009;25:1974–1975. doi: 10.1093/bioinformatics/btp250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimodaira H. An approximately unbiased test of phylogenetic tree selection. Syst Biol. 2002;51:492–508. doi: 10.1080/10635150290069913. [DOI] [PubMed] [Google Scholar]
- Shimodaira H, Hasegawa M. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics. 2001;17:1246–1247. doi: 10.1093/bioinformatics/17.12.1246. [DOI] [PubMed] [Google Scholar]
- Maddison WP, Maddison DR. Mesquite: a modular system for evolutionary analysis. Version 2.75. http://mesquiteproject.org.
- Holley RW, Apgar J, Everett GA, Madison JT, Marquisee M, Merrill SH, Penswick JR, Zamir A. Structure of a ribonucleic acid. Science. 1965;147:1462–1465. doi: 10.1126/science.147.3664.1462. [DOI] [PubMed] [Google Scholar]
- Miya M, Satoh TP, Nishida M. The Phylogenetic position of toadfishes (order Batrachoidiformes) in the higher ray-finned fish as inferred from partitioned Bayesian analysis of 102 mitochondrial genome sequences. Bio J Linn Soc. 2005;85:289–306. doi: 10.1111/j.1095-8312.2005.00483.x. [DOI] [Google Scholar]
- Near TJ, Eytan RI, Dornburg A, Kuhn KL, Moore JA, Davis MP, Wainwright PC, Friedman M, Smith WL. Resolution of ray-finned fish phylogeny and timing of diversification. PNAS. 2012;109:13698–13703. doi: 10.1073/pnas.1206625109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartillot N, Philippe H. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol Biol Evol. 2004;21:1095–1109. doi: 10.1093/molbev/msh112. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Lepage T, Blanquart S. Phylobayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics. 2009;25:2286–2288. doi: 10.1093/bioinformatics/btp368. [DOI] [PubMed] [Google Scholar]
- Miyashita T, Fujita K. Two series of parapophyses in neoscopelid fishes (Teleostei: Myctophiformes) Ichthyol Res. 2000;47:143–148. doi: 10.1007/BF02684234. [DOI] [Google Scholar]
- Schwarzhans W. Otolith-morphology and its usage for higher systematical units, with special reference to the Myctophiformes s.l. Meded Tert Kwart Geol. 1978;15:167–185. [Google Scholar]
- Rosen DE. An essay on euteleostean classification. Am Mus Novit. 1985;2827:1–45. [Google Scholar]
- Miller RR. A new genus and species of deep-sea fish of the family Myctophidae from the Philippine Islands. Proc US Nat Mus. 1947;97:81–90. doi: 10.5479/si.00963801.97-3211.81. [DOI] [Google Scholar]
- Bolin RL. Interim account of family myctophidae. In fishes of the Western North Atlantic. Mem Sears Foundation Mar Res 1. 1966;5:190–191. [Google Scholar]
- Herring PJ, Morin JG. In: Bioluminescence in action. Herring PJ, editor. London: Academic; 1978. Bioluminescence in fishes; pp. 273–329. [Google Scholar]
- Smith L, Smith KR, Wheeler WC. Mitochondrial intergenic spacer in fairy basslets (Serranidae: Anthiinae) and the simultaneous analysis of nucleotide and rearrangement data. Am Mus Novit. 2009;3652:1–10. [Google Scholar]
- Ahlstrom EH, Moser HG, O’Toole M. Development and distribution of larvae and early juveniles of the commercial lanternfish, Lampanyctodes hectoris (günther), off the west coast of southern Africa with a discussion of phylogenetic relationships of the genus. Bull Southern California Acad Sci. 1976;75:138–152. [Google Scholar]
- Nafpaktitis BG. Systematics and distribution of lanternfishes of the genera Lobianchia and Diaphus (Myctophidae) in the Indian Ocean. Bull Nat His Mus Los Angeles County. 1978;30:1–92. [Google Scholar]
- Nafpaktitis BG. Taxonomy and distribution of the lanternfishes, genera Lobianchia and Diaphus, in the North Atlantic. Dana-Report. 1968;73:1–134. [Google Scholar]
- Kawaguchi K, Shimizu H. Taxonomy and distribution of the lanternfishes, genus Diaphus (Pisces, Myctophidae) in the Western Pacific, Eastern Indian Oceans and the Southeast Asian Seas. Bull Ocean Res Inst Univ Tokyo. 1978;10:1–142. [Google Scholar]
- Bolin RL Iniomi. Myctophidae from the “Michael sars” north Atlantic deep-Sea expedition Rept Sci Res “Michael Sars” Deep-Sea Expd 1910 195941–45.10724406 [Google Scholar]
- Nafpaktitis BG, Paxton JR. Idiolychnus, a new genus of Myctophidae based on Diaphus urolampus. Copeia. 1978;3:492–497. [Google Scholar]
- Hubbs CL, Wisner RL. Parvilux, a new genus of myctophid fishes from the northeastern Pacific, with two new species. Zool Mededelingen. 1964;39:445–463. [Google Scholar]
- Macey JR, Papenfuss TJ, Kuehl JV, Fourcade HM, Boore JL. Phylogenetic relationships among amphisbaenian reptiles based on complete mitochondrial genomic sequences. Mol Phylogenet Evol. 2004;33:22–31. doi: 10.1016/j.ympev.2004.05.003. [DOI] [PubMed] [Google Scholar]
- Nafpaktitis BG, Paxton JR. Review of the lanternfish genus Lampadena with a description of a new species. Contributions Sci. 1968;138:1–29. [Google Scholar]
- Paxton JR. Nominal genera and species of lanternfishes (Family Myctophidae) Contributions Sci Nat Hist Mus Los Angeles County. 1979;322:1–28. [Google Scholar]
- Coleman LR, Nafpaktitis BG. Dorsadena yaquinae, a new species and genus of myctophid fish from the Eastern North Pacific Ocean. Contributions Sci. 1972;225:1–11. [Google Scholar]
- Davy B. A Review of the lanternfish genus Taaningichthys (family Myctophidae) with the description of a new species. Fish Bull. 1972;70:67–78. [Google Scholar]
- Hulley PA. In: Fishes of Antarctica: A Biological Overview. Prisco GD, Pisano E, Clarke A, editor. Milano: Springer; 1998. Preliminary investigations on the evolution of the tribe Electronini (Myctophiformes, Myctophidae) pp. 75–86. [Google Scholar]
- Kawaguchi K, Aioi K. Myctophid fishes of the genus Myctophum in the Pacific and Indian Oceans. J Oceanographical Soc Japan. 1972;28:161–175. doi: 10.1007/BF02108760. [DOI] [Google Scholar]
- Bekker VE, Borodulina OD. New species of lanternfish of the genus Myctophum (Myctophidae, Pisces) Voprosy Ikhtiologii. 1971;11:418–426. [Google Scholar]
- Mabuchi K, Miya M, Satoh TP, Westneat MW, Nishida M. Gene rearrangements and evolution of tRNA pseudogenes in the mitochondrial genome of the parrotfish (Teleostei: Perciformes: Scaridae) J Mol Evol. 2004;59:287–297. doi: 10.1007/s00239-004-2621-z. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Zhang JY, Zheng RQ, Yu BG, Yang G. Complete nucleotide sequence and gene organization of the mitochondrial genome of Paa spinosa (Anura: Ranoidae) Gene. 2009;447:86–96. doi: 10.1016/j.gene.2009.07.009. [DOI] [PubMed] [Google Scholar]
- Inoue JG, Miya M, Tsukamoto K, Nishida M. Evolution of the deep-sea gulper eel mitochondrial genomes: Large-scale gene rearrangements originated within the eels. Mol Biol Evol. 2003;20:1917–1924. doi: 10.1093/molbev/msg206. [DOI] [PubMed] [Google Scholar]
- Mauro DS, Gower DJ, Zardoya R, Wilkinson M. A Hotspot of gene order rearrangement by tandem duplication and random loss in the vertebrate mitochondrial genome. Mol Bio Evol. 2006;23:227–234. doi: 10.1093/molbev/msj025. [DOI] [PubMed] [Google Scholar]
- Desjardins P, Morais R. Sequence and gene organization of the chicken mitochondrial genome: a novel gene order in higher vertebrates. J Mol Biol. 1990;212:599–634. doi: 10.1016/0022-2836(90)90225-B. [DOI] [PubMed] [Google Scholar]
- Mueller RL, Boore JL. Molecular mechanisms of extensive mitochondrial gene rearrangement in plethodontid salamanders. Mol Biol Evol. 2005;22:2104–2112. doi: 10.1093/molbev/msi204. [DOI] [PubMed] [Google Scholar]
- Bakke IG, Shields F, Johansen S. Sequence characterization of a unique intergenic spacer in Gadiformes mitochondrial DNA. Mar Biotech. 1999;1:411–415. doi: 10.1007/PL00011797. [DOI] [PubMed] [Google Scholar]
- Kumazawa Y, Ota H, Nishida M, Ozawa T. Gene Rearrangements in snake mitochondrial genomes: Highly concerted evolution of control-region-like sequences duplicated and inserted into a tRNA gene cluster. Mol Biol Evol. 1996;13:1242–1254. doi: 10.1093/oxfordjournals.molbev.a025690. [DOI] [PubMed] [Google Scholar]
- Kurabayashi A, Sumida M, Yonekawa H, Glaw F, Vences M, Hasegawa M. Phylogeny, recombination, and mechanisms of stepwise mitochondrial genome reorganization in mantellid frogs from Madagascar. Mol Biol Evol. 2008;25:874–891. doi: 10.1093/molbev/msn031. [DOI] [PubMed] [Google Scholar]
- Kumazawa Y, Endo H. Mitochondrial genome of the komodo dragon: efficient sequencing method with reptile-oriented primers and novel gene rearrangements. DNA Res. 2004;11:115–125. doi: 10.1093/dnares/11.2.115. [DOI] [PubMed] [Google Scholar]
- Okajima Y, Kumazawa Y. Mitochondrial genomes of acrodont lizards: Timing of gene rearrangements and phylogenetic and biogeographic implications. BMC Evol Biol. 2010;10:141. doi: 10.1186/1471-2148-10-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh TP, Sato Y, Masuyama N, Miya M, Nishida M. Transfer RNA gene arrangement and codon usage in vertebrate mitochondrial genomes: A new insight into gene order conservation. BMC Genomics. 2010;11:479. doi: 10.1186/1471-2164-11-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table T2. Partitioning schemes used in the present study suggested by PartitionFinder.
Saturation plots of pairwise distances of 1st, 2nd and 3rd codon positions in the 12 protein coding genes employed in the analyses. Vertical axis shows p-distance for each codon position against total p-distance (horizontal axis). Saturation from RY-coding of the 3rd codon positions included (only transversions valued) although not used in the present study from early onset of saturation.
Secondary structure of INC-regions. Sequence alignments and secondary tRNA structures of INC-regions are shown for synapomorphic spacers pertaining to gene order rearrangements 1, 3, 5 and 8 presented in Figure 3. The remaining INC-regions presented in Figure 3 are relatively short sequences and are not included.
Base pair details of INC-regions found in Myctophidae. Eight mt regions correspond to gene order and INC-region synapomorphies presented in Figure 3 with slash (/) denoting bordering genes. The last column includes mt information from the polymorphic region from tRNA-Thr and downstream towards the control region (CR). Numbers of INC-sequence base pairs are parenthesized next to flanking genes. Greater-than sign (>) and hyphen (−) denote partial and no sequence, respectively, from failure to complete the sequences.
IMQMψ gene order in a clade of scarid parrotfishes. Taxa constituting a monophyletic clade in Scaridae all show similar patterns in the IMQ-region as observed within Diaphini. Grey areas show INC-regions with number of base pairs noted for each taxon and the arrow indicates possible gradual removal of INC-regions.