Abstract
Kidney nephrons are comprised of proximal and distal tubule segments that perform unique roles in excretion. The developmental pathways that establish nephron segment identities from renal progenitors are poorly understood. Here, we used the zebrafish pronephros to study nephron segmentation. We found that zebrafish nephron progenitors undergo elaborate spatiotemporal expression changes of many genes before adopting a segment fate. Initially, two domains of nephron progenitors are established, then are subdivided and demarcate individual nephron segments. Using genetic and chemical genetic models of retinoic acid (RA) deficiency, we discovered that RA modulates rostral progenitor formation. To delineate downstream pathways, we knocked down the irx3b transcription factor and found it regulates proximal tubule segment size and distal segment differentiation. Our results suggest a model whereby RA patterns the early field of nephron progenitors, with subsequent factors like irx3b acting to refine later progenitor subdomains and ensure activation of segment-specific gene programs.
Keywords: kidney organogenesis, nephron, segmentation, zebrafish
INTRODUCTION
Vertebrate kidney development is characterized by the progressive formation of several kidney types. For example, mammals form three kidney structures of increasing complexity: the pronephros, the mesonephros, and finally the metanephros, which serves as the adult organ (Dressler, 2006). Other vertebrates, like fish and amphibians, initially utilize a pronephros and later form a mesonephros that functions during larval and adult stages (Dressler, 2006). Despite the serial changes in organ complexity, the nephron is the universal structural and functional unit of all vertebrate kidneys. In most cases, nephrons consist of three parts that have discrete activities: (1) a sieve (the glomerulus) comprised of an intricate arrangement of fenestrated capillaries surrounded by a capsule of podocytes that filters the blood, (2) an epithelial tubule that receives the filtrate and modifies it by reabsorbing and secreting metabolites, and (3) a duct that performs limited solute exchange while transporting the waste for excretion (Dressler, 2006). The tubule epithelium is patterned along its length into functionally specialized segments that express unique gene expression profiles of solute transporters (El-Dahr, et al., 2008; Kopan et al., 2007; Schedl, 2007). Proximal tubule segments perform the bulk of metabolite recovery for amino acids, salts, and sugars, while distal tubule segments are specialized for fine-tuning salt levels (Hebert et al., 2001). Interestingly, vertebrates that reside in diverse habitats ranging from the terrestrial to the aquatic possess a largely similar proximodistal complement of nephron tubule segments. Recent molecular analyses have demonstrated that zebrafish, frog, mouse, and human nephrons contain functionally comparable proximal and distal tubule segments and express many of the same solute transporter and transcription factor genes (Wingert and Davidson, 2008). Differences in segment gene expression likely reflect physiological contrasts between species.
Nephron segmentation is the process through which segment identities are produced during nephrogenesis. Several key signals have been identified that are essential for the formation of certain nephron segments, but a comprehensive understanding of segment patterning in any one model organism has not yet been achieved (Schedl, 2007; Wingert and Davidson, 2008). For example, metanephric nephron segmentation in mammals requires Notch signaling for proximal tubule formation and Pou3f3/Brn1, a POU-domain containing transcription factor, for distal tubule development (Nakai, et al., 2003; Cheng, et al., 2007). Notch ligands and receptors are regionally expressed in the nephron anlagen (Cheng and Al-Awqati, 2005), and conditional inactivation of Notch2 results in a loss of proximal tubule segments (Cheng, et al., 2007). Conversely, genetic inactivation of Pou3f3/Brn1 leads to metanephric nephrons that display a dramatic reduction in distal segments (Nakai, et al., 2003). Recent studies of pronephric nephron formation in lower vertebrates have identified roles for retinoid signaling and the Iroquois homeobox transcription factor Irx3. During zebrafish pronephros formation, retinoic acid (RA) is necessary for specifying proximal tubule fates, and the abrogation of RA synthesis leads to an expansion of the distal tubule and duct segments (Wingert, et al., 2007). In Xenopus, knockdown of Irx3 eliminates the formation of at least one distal segment, and Irx3 has been proposed to act as a master regulator in the specification of this segment (Alarcon, et al., 2008; Reggiani, et al., 2007). While it remains unclear if the signals that pattern vertebrate nephrons can be integrated into one universal model of nephrogenesis, trends in the regional expression of Notch genes, Pou3f3/Brn1, and Irx3 support the enticing suggestion that nephron patterning is conserved (Wingert and Davidson, 2008).
The zebrafish pronephros is an excellent model for nephron segment patterning studies. Zebrafish embryos form an anatomically simple pronephros: it is comprised of two nephrons that derive from the intermediate mesoderm that can be visualized throughout their ontogeny (Drummond, et al., 1998; Drummond, 2003). Nephron segment lengths and boundaries can be quantified at a high resolution relative to the adjacent somites, which provide ‘landmarks’ along the body axis (Wingert, et al., 2007). Zebrafish pronephric nephrons possess at least eight discrete cell populations in common with mammals: the podocytes (P) which contribute to the glomerulus, a neck segment (N) that connects the glomerulus and tubule, two proximal segments (the proximal convoluted tubule (PCT) and proximal straight tubule (PST)), two distal segments (the distal early (DE) and distal late (DL)), and a pronephric duct (PD) that empties into the cloaca (C) (Wingert, et al., 2007). The parallels between fish and mammalian nephron components suggest that the zebrafish studies can generate insights into the conserved mechanisms of nephrogenesis.
In this study, we have examined the domains of gene expression in zebrafish embryo renal progenitors prior to the appearance of proximal and distal tubule segments. We discovered that an intricate, nested pattern of transcription factor domains precedes the appearance of mature tubule segments in the nephron, and that this pattern undergoes a series of complex spatiotemporal alterations. Using lib, a newly isolated genetic model of RA deficiency, we found that retinoid signals influence the earliest spatial patterns of transcription factor domains. We discovered that a loss of the paraxial mesoderm disrupts the expression of the RA-biosynthesis gene aldh1a2 and induces nephron segment defects similar to lib, thus demonstrating that the paraxial mesoderm is a key source of RA for nephron patterning. In addition, we found that irx3b is required at relatively late stages of nephron patterning for the differentiation of the first distal segment. These findings suggest a stepwise model whereby the sequential actions of RA and irx3b orchestrate segmentation of the zebrafish pronephros.
RESULTS
Expression domains are dynamic within the nephron progenitor territory
To study the origins of nephron segments, we analyzed the expression of transcription factors and signaling molecules between the time when the intermediate mesoderm is first detected around the 3 somite stage to the emergence of mature nephron segments around 24 hours post fertilization (hpf; equivalent to the 28 somite stage) (Wingert, et al., 2007). For each kidney gene, we determined the precise gene expression domain relative to the somites by performing double whole mount in situ hybridization with an age-appropriate somite marker (myogenic differentiation 1 (myoD) for embryos <15 somites and myosin heavy chain (mhc) for embryos >15 somites). Consistent with our previously published observations, we found that nephron progenitors displayed uniform expression of several transcription factors including pax2a and pax8 until approximately the 5 somite stage (data not shown; Wingert, et al., 2007). Between the 6 and 8 somite stages, the nephron territory was subdivided into two molecularly distinct adjacent regions that showed subtle dynamic alterations. At the 6 somite stage, we observed a rostral domain, located adjacent to somites 2–5 and marked by expression of the Notch ligand genes delta-c (dlc) and jagged-2 (jag2), as well as a caudal domain marked by expression of the zinc finger transcription factor mecom (also known as evi1) that began adjacent to somite 6 (data not shown; Wingert, et al., 2007). At the 8 somite stage, the dlc and mecom domains remained mutually exclusive, however the jag2 domain expanded such that it overlapped with mecom at somites 6–7 (Fig. 1A, 1C, 3). Thus, a set of overlapping rostral and caudal identities is established among nephron progenitors during early somitogenesis, and likely represents the influences of early proximo-distal patterning signals occurring at this time.
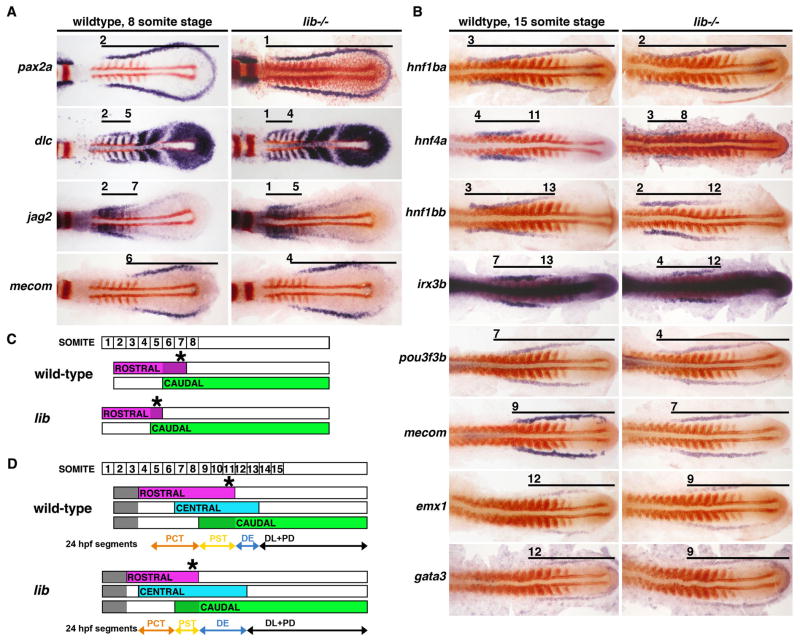
Figure 1. Pronephros progenitors are delineated into a series of molecularly distinct regions during early somitogenesis that are RA-dependent.
Gene expression patterns in the nephron territory in wildtype embryos and lib mutants at the (A) 8 somite stage (B) 15 somite stage and schematized respectively (C–D). Embryos were flat-mounted to remove the yolk and are shown in dorsal views with anterior to the left. Whole mount in situ hybridization was used to mark kidney gene expression (purple) and the somites with myoD (red). Black lines indicate areas of kidney gene expression and numbers correspond to the somite position. (A) In 8 somite wildtypes, pax2a transcripts marked all nephron progenitors while dlc and jag2 expression was restricted proximally and mecom expression was restricted distally; a short stretch of overlap between the jag2 and mecom domains was evident. lib embryos had reduced dlc and jag2 domains, and an expanded mecom domain. (B) In 15 somite wildtype embryos, hnf1ba marked all nephron progenitors, and the other transcription factors were restricted to subsets of cells in the rostral (hnf4a, hnf1bb), central (irx3b), or caudal (pou3f3b, mecom, emx1, gata3) areas of the nephron territory. lib had a shortened rostral hnf4a area while the central and caudal were expanded. (C–D) Schematic depictions of nephron progenitor domains: (top) labeled boxes indicate somite number, and (below) summary of rostral (purple), central (blue) and caudal (green) areas of the nephron progenitors. At 8 somites, the overlap between rostral and caudal areas is depicted as dark purple-colored subset of the rostral area. At 15 somites, subsets of the caudal area are depicted as dark green and light green; gray indicates divergence of non-tubule populations (future podocytes and neck segment cells) at the most anterior regions of the renal progentors. Asterisk marks the posterior limit of the rostral area, which shifted anteriorly in lib at the 8 and 15 somite stages.
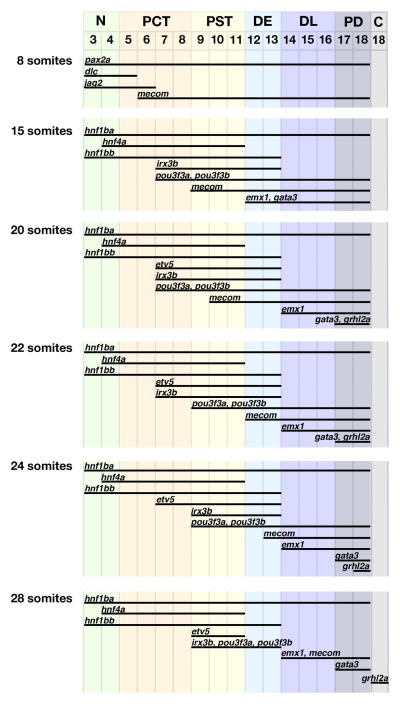
Figure 3. Analysis of gene expression domain dynamics in the pronephros progenitors reveals that progressive refinements ultimately correlate with specific segment fates.
(Foreground) Gene expression domains (represented by black bars) are schematized in the pronephros territory between 8 and 28 somite stages. (Background) Nephron segment identities form adjacent to particular somites (demarcated by numbered columns, top), and each segment region is uniquely color-coded to envisage a comparison between gene expression domains at each timepoint with the eventual nephron segment which arises at any given physical location in the pronephros territory. Segments and color codes are N (neck)-green, PCT (proximal convoluted tubule)-orange, PST (proximal straight tubule)-yellow, DE (distal early)-light blue, DL (distal late)-dark blue, PD (pronephric duct)-dark gray, and C (cloaca)-light gray.
By the 15 somite stage, we found a complex, nested arrangement of transcription factor domains within the intermediate mesoderm. A number of gene transcripts were expressed by all nephron progenitors, such as hnf1ba, pax2a, and pax8 (Fig. 1B; data not shown). Genes with restricted expression domains broadly marked either the rostral, central, and/or caudal regions of the nephron field, though the precise extent of each gene varied within these partially overlapping areas. Expression of hnf4a was found rostrally, whereas hnf1bb transcripts were found throughout both the rostral and central areas (Fig. 1B, 1D). We found that irx3b transcripts marked the central area, and that several genes marked portions of the central as well as the entire caudal area, namely pou3f3a, pou3f3b, mecom, emx1, and gata3 (Fig. 1B, 1D; data not shown). Interestingly, we noted that some genes at this time-point have an expression boundary that is coincident with the mature segment boundaries that appear at the 28 somite stage. For example, the anterior limit of the mecom expression domain (somite 9) and the posterior limit of hnf4a (somite 11) coincide with the eventual PST segment boundaries (somites 9–11) (Fig. 1D, 3). In addition, the posterior limits of hnf1bb and irx3b coincide with the posterior boundary of the DE segment (at somite 13) (Fig. 3). Other gene expression domains though, like the anterior limits of irx3b, pou3f3a, and pou3f3b, were not coincident with subsequent segment boundaries and instead spanned the eventual boundary of the adjacent PCT and PST segments (Fig. 3). These data suggest that the boundaries of some segments can be predicted as early as the 15 somite stage, while others are defined at later stages.
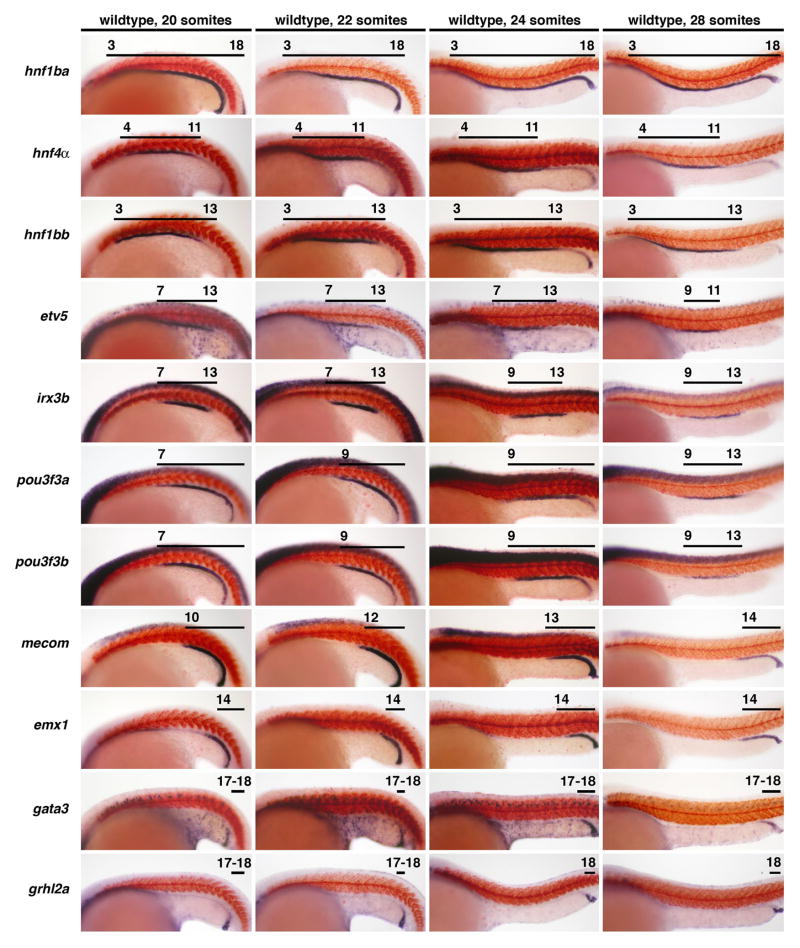
The nested arrangement of transcription factor domains in nephron progenitors persisted at subsequent stages, and was still evident at the 28 somite stage, the time-point when segment-specific solute transporters mark the differentiated PCT, PST, DE, and DL tubule segments (Fig. 3). However, we found that spatiotemporal changes in gene expression occurred between the 20 and 28 somite stages. Interestingly, there was a common trend whereby genes that mark the rostral region showed no change after 15 somites, while genes that mark the central and caudal regions underwent dynamic alterations at subsequent time-points. For example, the domain of the rostral region gene hnf4a remained constant from the 15 somite stage onward (Fig. 1B, 2, 3). In contrast, the domains of the central and caudal region genes etv5, irx3b, pou3f3a, pou3f3b, mecom, emx1, gata3, and grhl2a showed one or more alterations at different times between 15 and 28 somites (Fig. 1B, 2, 3). In general, these changes can be summarized as posterior shifts in the anterior expression boundary of each gene, although some genes also exhibited contractions at their posterior boundary. For example, the genes irx3b, pou3f3a, and pou3f3b initially shared a common anterior boundary adjacent to somite 7 until the 20 somite stage. This boundary retracted to somite 9 for pou3f3a and pou3f3b at the 22 somite stage, followed by a similar retraction for irx3b slightly later, at the 24 somite stage. The expression domain of pou3f3a/3b underwent further refinement by the 28 somite stage, with a down-regulation of transcripts posteriorly. The net result of these changes is that irx3b, pou3f3a and pou3f3b become restricted to the PST and DE (somites 9–13) at 28 somites (Fig. 2, 3).
Figure 2. Dynamic spatiotemporal expression changes during late somitogenesis precede the appearance of segment identities in the pronephros.
Gene expression patterns of transcription factors in nephron progenitors in wildtype embryos at the 20, 22, 24, and 28 somite stage. Embryos are shown in lateral views with anterior to the left, and have been stained by whole mount in situ hybridization to mark kidney gene expression (purple) and the somites with mhc (red). Black lines indicate areas of high gene expression, and numbers correspond to the somite position. The spatial distributions of hnf1ba, hnf4a, hnf1bb, emx1, and gata3 transcripts were unaltered between 20–28 somites of development, though note that the proximal boundary of the emx1 and gata3 domains at 20 somites changed since the 15 somite stage (compare to Figure 1B). The expression domains of pou3f3a and pou3f3b were altered at 22 and 28 somites, the irx3b and grhl2a expression domains changed at 24 somites, and the etv5 domain was restricted at the 28 somite stage. The expression domain of mecom exhibited multiple alterations between 20–28 somites.
We found a striking correlation between the pattern of transcription factor expression domains and the location of nephron segment boundaries at the 28 somite stage (Fig. 2, 3). Surprisingly, we found only one transcription factor, etv5, restricted to a single tubule segment (the PST), while the others marked two or more nephron segments. The transcription factors formed an overlapping, nested fashion such that each tubule segment possessed a unique combination of transcripts. For example, the PST contained hnf1ba, hnf4a, hnf1bb, etv5, irx3b, pou3f3a and pou3f3b transcripts, while the DE expressed a subset of this medley that excluded hnf4a and etv5 (Fig. 2, 3).
Cell proliferation is not essential for nephron segmentation during somitogenesis
One possible explanation for the dynamic spatiotemporal alterations in gene expression domains is that regional changes in cell proliferation generate shifts in the physical arrangement of nephron tubule progenitors. To explore this possibility, we treated embryos from the 15, 20, 22, or 24 somites stages with camptothecin or nocodazole to prevent cell division, and then examined the final location of each tubule segment at 24 hpf (Fig. S1). We found no evidence that an inhibition of cell proliferation altered the normal positioning of the proximal and distal segments. Treatment with proliferation-blocking compounds at younger stages was prohibitive to embryo development and precluded the ability for segmentation markers to be assessed (data not shown). Based on these experiments, we conclude that the emergence of nephron tubule segment identities occurs independently of growth after 15 somites.
Mutation of the lightbulb gene disrupts proximo-distal patterning of nephron segments
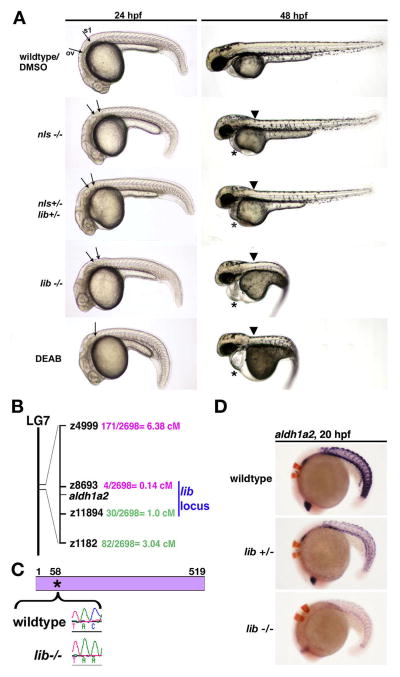
We next sought to gain insight into the genetic pathways that establish nephron segment fates. We isolated the lightbulb (lib) mutant in a forward genetic screen of ENU-mutagenized zebrafish for recessive embryonic kidney defects. lib embryos have a truncated cervical region at 24 hpf based on the abnormally close proximity of the otic vesicle to the first somite compared to wildtypes (Fig. 4A). At 48 hpf, lib embryos exhibited a kink in the cervical region, pericardial edema, lacked pectoral fins, and had a pronounced trunk curvature along with a distorted lightbulb-shaped yolk (Fig. 4A). These morphological characteristics of lib are very similar to the hallmarks of RA deficiency in zebrafish embryos, leading us to hypothesize that lib had a deficit in RA bioavailability (Alexa, et al., 2009; Begemann, et al., 2001; Begemann, et al., 2004; Grandel, et al., 2002).
Figure 4. lib is the most severe genetic model of aldh1a2-deficiency in the zebrafish.
(A) Lateral views of living 24 and 48 hpf wildtype embryos, nls homozygotes, nls/lib compound heterozygotes, lib homozygotes, and DEAB-treated wildtypes. aldh1a2 mutations are associated with a truncation of the cervical region evident at 24 hpf (arrows indicate the caudal boundary of the otic vesicle (OV) and the rostral boundary of the first somite (S1)). DEAB-treated embryos lack this region, with the OV and S1 located adjacent to each other (indicated by single arrow). At 48hpf, aldh1a2 mutations are associated with a kink at the head-trunk boundary (black arrow) and pericardial edema (*). Also at 48 hpf, lib and DEAB-treated embryos show pronounced body curvature and develop a lightbulb-shaped yolk compared to wildtypes and other aldh1a2 mutants. (B) Meiotic mapping placed lib in a genetic interval on linkage group (LG) 7 between z8693 and z11894 that includes aldh1a2. (C) RT-PCR and sequence analysis of aldh1a2 transcripts from lib mutants detected a C -> A transversion at nucleotide 174 that is predicted to introduce a Stop codon in lieu of a Tyrosine residue at amino acid position 58 in the 519 amino acid raldh2 enzyme. (D) Whole mount in situ hybridization analysis of aldh1a2 expression (purple) and krox20 expression (red) shows decreased aldh1a2 transcripts both in lib mutants and lib heterozygote embryos, consistent with nonsense mediated decay.
In the developing embryo, RA bioavailability is controlled by enzymatic production and degradation (Duester, 2008). RA is produced by the sequential action of retinol dehydrogenases (RDHs) and aldehyde dehydrogenase (Aldh or Raldh) synthesizing enzymes, and degraded by the action of cytochrome P450 enzymes belonging to the Cyp26 family (Duester, 2008). We compared the phenotype of lib mutant embryos to neckless (nls), which have a point mutation in the catalytic domain of aldh1a2, and wildtype embryos treated with diaminobenzaldehyde (DEAB), a reversible chemical inhibitor of Aldh enzymes. nls embryos are thought to be Aldh1a2-deficient but have less severe tissue defects compared to DEAB treatment, perhaps due to maternal Aldh1a2 activity present in the yolk (Alexa, et al., 2009; Begemann, et al., 2004). At 48 hpf, nls embryos had a less severe cervical kink, pericardial edema, and body curvature compared to lib, while DEAB-treated embryos and lib were more similar (Fig. 4A). Next, nephron segmentation in lib, nls, and DEAB-treated embryos was evaluated using double whole mount in situ hybridization with segment-specific solute transporters and mhc to mark the somites. Compared to wildtypes, 24 hpf lib embryos had reduced expression of the podocyte markers, wt1b, wt1a, and mafb, and the podocytes were located more rostrally, suggesting that podocyte development was partially affected (Fig. 5A; data not shown). At 48 hpf, lib embryos had reduced expression of both wt1b and nephrin, suggesting that podocyte numbers were not restored as development progressed (data not shown). lib embryos also formed shorter proximal segments, as shown by the PCT and PST markers slc20a1a and trpm7 (Fig. 5A). In addition, lib formed expanded distal segments, as shown by the DE, DL and PD markers slc12a1, slc12a3, and gata3, respectively (Fig. 5B). nls embryos had a milder nephron segment phenotype than lib mutants, with reduced podocytes located at the correct location along the trunk, a slightly reduced PCT segment, and a slightly expanded DE segment (Fig. 5). DEAB treated embryos had nephrons comprised entirely of the distal segments (DE, DL, and PD) that were each expanded in length (Fig. 5). These data indicate that lib mutants have a nephron patterning defect more severe than nls, but less severe than DEAB treatment.
Figure 5. Genetic and chemical models of RA biosynthesis deficiency exhibit a similar nephron segment phenotype of reduced proximal fates and expanded distal fates, and lib is the most severely affected aldh1a2 mutant.
Whole mount in situ hybridization analysis for nephron segment markers (purple) and mhc (red) at the 28 somite stage in aldh1a2 mutants and DEAB-treated wildtype embryos. Embryos are shown in lateral views with anterior to the left, with the exception of dorsal views to show podocytes. Black brackets and arrows indicated expression domains, and numbers correspond to somite position. (A) Proximal fates of podocytes, the PCT and PST, were all reduced in the absence of RA biosynthesis, while (B) distal fates of the DE, DL, CS, and PD were expanded. (C) Summary of nephron segmentation with respect to embryo somite number. Each color represents a different epithelial population: podocytes (green), PCT (orange), PST (yellow), DE (light blue), CS (red lettering indicates location among nephron tubule cells), DL (dark blue) and PD (black), and the overlap between DL and PD-expressed genes is indicated in white. The nephron segment phenotypes among RA-deficient zebrafish embyros showed a graded severity such that: nls homozygote < nls/lib compound heterozygotes < lib homozygotes < DEAB-treated wildtype embryos.
lib is a new model of Aldh1a2 deficiency
We next undertook a positional cloning strategy to determine the genetic defect in lib. Using bulk segregant analysis, the lib gene was mapped to zebrafish chromosome 7 (Fig. 4B). Linkage analysis of 2698 meioses placed the lib locus between the SSLP markers z8693 and z11894, a region that includes the aldh1a2 gene. This was surprising as the kidney phenotype of lib mutants is more severe than nls mutants, and because nls and its phenotypically similar alleles no-fin and aldh1a2um22 are all considered to represent null alleles of aldh1a2 in the zebrafish (Alexa, et al., 2009; Begemann, et al., 2001; Begemann, et al., 2004; Grandel, et al., 2002). Nevertheless, based on the mapping and phenotype, we hypothesized that lib was a novel, more severe mutant allele of aldh1a2.
To investigate if lib was an aldh1a2 mutation, we mated lib and nls adult heterozygotes and found that the mutations failed to complement one another (Fig. 4A). Compound nls/lib heterozygote embryos had morphological characteristics most similar to nls mutant embryos, with a kink in the cervical region, pericardial edema, and absent pectoral fins (Fig. 4A). In addition, compound nls/lib heterozygote embryos had a nephron segment pattern of intermediate severity between the nls and lib phenotypes. Compared to nls, compound nls/lib heterozygotes had a slightly shorter PST and slightly larger DE, but had no change in DL size or podocyte location as seen in lib (Fig. 5). Next, RT-PCR was used to isolate aldh1a2 cDNA transcripts from lib mutants. Sequence analysis identified a C to A transversion at nucleotide 174, predicting a Tyr -> Stop nonsense mutation at amino acid 58 (Fig. 4C). The wildtype zebrafish aldh1a2 peptide is 518 amino acids in length, and includes a series of nucleotide binding domains, tetramerization domains, and catalytic domain (Alexa, et al., 2009). The lib mutation is predicted to truncate the first nucleotide binding domain and eliminate all subsequent domains. Interestingly, examination of aldh1a2 expression in lib revealed that transcripts were markedly reduced both in lib homozygous and heterozygote embryos, consistent with nonsense-mediated decay (Fig. 4D).
To provide further evidence that the lib phenotype is caused by a RA synthesis defect, we investigated whether elevating RA levels could rescue lib. Embryos from lib heterozygous incrosses were injected at the 1-cell stage with wildtype aldh1a2 cRNA or treated with exogenous RA from 60% epiboly to the 28 somite stage, then scored for morphology and pectoral fin development at 48 hpf (Fig. S2A). Overexpression of aldh1a2 was sufficient to rescue lib morphology (Figs. S2A, B). Interestingly, exposure to a high dose of RA (10−8M) completely rescued lib mutant embryos, but a lower RA dose (10−9M) was insufficient to rescue lib (Fig. S2B). In contrast, the lower RA dose was able to fully rescue nls and nls/lib compound heterozygote embryos (Fig. S2B). These results support the notion that lib quantitatively possesses the most severe RA deficiency among these genotypes and that RA dosage is responsible for the graded phenotypes in lib, nls/lib, and nls mutants.
Next, we determined if exogenous RA application from 60% epiboly to the 28 somite stage was sufficient to rescue proximal nephron segments in lib. Nephron segmentation was evaluated at 24 hpf using double whole mount in situ hybridization with segment-specific solute transporters and the somite marker mhc. RA-treated lib mutants developed expanded proximal segments (PCT, PST) and reduced distal segments (DE, DL, PD) (Fig. S2C, D). In addition, RA treatment rescued podocyte number and positioning in lib embryos (Fig. S2C, D). These results demonstrate that nephron progenitors in lib mutants are competent to respond to an RA signal(s), and suggest that a paucity of retinoid ligands underlies the lib phenotype. Taken together, our analysis of the lib and nls mutants highlights that RA production by Aldh1a2 is essential for proximo-distal patterning of the pronephros.
lib mutants have altered renal progenitor domains during early somitogenesis
To investigate how RA deficiency in lib mutants influenced nephron segment patterning, we examined the spatiotemporal expression of renal genes between the 8–28 somite changes. At the 8 somite stage, lib mutant embryos had slightly shortened domains of dlc and jag2 expression that were shifted anteriorly compared to wild-types (Fig. 1A, 1C). In addition, lib embryos had a mecom expression domain that was slightly expanded anteriorly compared to wildtypes (Fig. 1A). Overall, the combination of the dlc, jag2, and mecom domains in lib mutants still formed an overlapping rostral-caudal pattern similar to wildtypes, but shifted anteriorly (Fig. 1A, 1C). Since nephron progenitor patterning is altered in lib mutants at the earliest stages that regional differences can be detected, it suggests that RA regulates the initial proximodistal patterning of nephron progenitors, consistent with our previous observations (Wingert, et al., 2007).
More significant alterations in nephron progenitor patterning were detected in lib mutants at the 15 somite stage. Notable changes included anterior shifts in the rostral (hnf4a), central (irx3b), and caudal (mecom) expression domains (Fig. 1B, D). These shifts were not equivalent for each domain, with hnf4a shifting anteriorly by 1 somite, irx3b by 3 somites, and mecom by 2 somites. The posterior boundaries of the hnf4a and irx3b domains were similarly shifted anteriorly, and also by unequal extents (3 somites for hnf4a and 1 somite for irx3b; Fig. 1B, D). The net effect of these changes was a reduction in the rostral domain and expansion in both the central and caudal domains (Fig. 1D). Despite these patterning changes, the correlations between the rostral-central-caudal domains at 15 somites and the positions of the final nephron segments that we noted in wildtype embryos were likewise the case in lib mutants. For instance, in both wildtypes and lib mutants, the boundaries of the PST segment correlate to the region of overlap between the rostral (hnf4a) and caudal (mecom) domains, despite this region being smaller and anteriorly shifted in lib mutants. These results provide further support for the notion that renal transcription factor domains during early pronephros development are predictive of nephron segment fates at later stages.
Between the 20–28 somite stages, lib embryos exhibited similar dynamic spatiotemporal shifts in central and caudal domain markers that we observed in wildtypes (Fig. S4, S5). For example, the anterior expression boundary of the central domain gene irx3b and caudal domain marker mecom retracted in lib and wildtypes (Fig. S4, S5). However, there was one exception: the caudal markers pou3f3a and pou3f3b failed to retract their anterior expression boundaries and remained ectopically expressed in the PCT and PST segments. Taken together, these results provide genetic evidence that RA has an early role in establishing the rostral and caudal domains within the intermediate mesoderm, and that RA is also required at later time-points to refine the expression patterns of genes such as pou3f3a and pou3f3b.
Paraxial mesoderm is necessary for proximo-distal nephron patterning
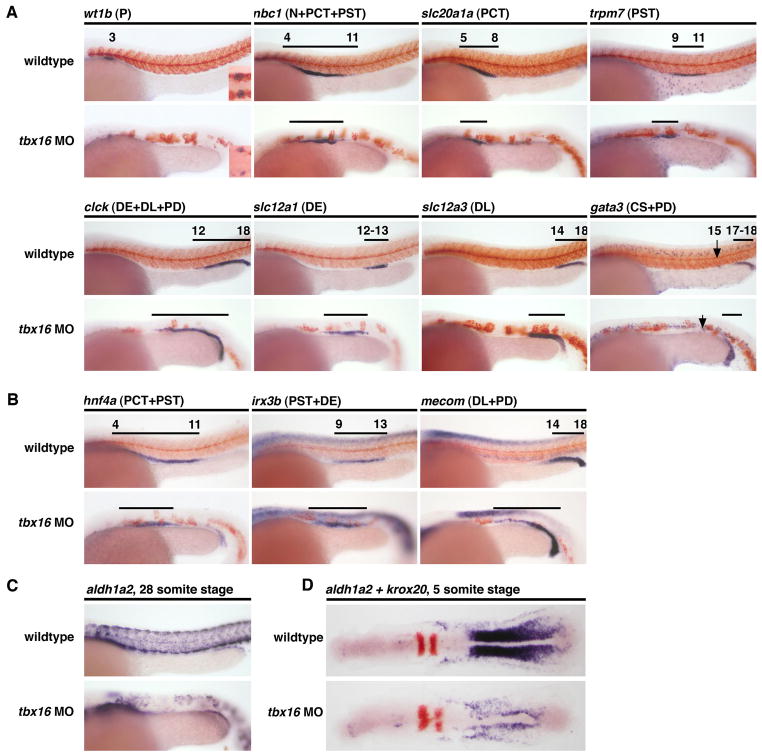
Based on our previous work showing that aldh1a2 was highly expressed in developing somites, we hypothesized that the paraxial mesoderm was a major source of RA responsible for establishing the proximodistal pattern of the pronephros (Wingert, et al., 2007). To test this, we knocked down tbx16/spadetail, which encodes a T-box transcription factor required for paraxial mesoderm formation (Amacher, et al., 2002; Morley, et al., 2009), and then examined the segmentation pattern of the pronephros at 24 hpf. A deficiency in tbx16 was associated with reduced podocytes and proximal tubule segments (Fig. 6A). Conversely, tbx16 morphants formed an expanded DE segment, as shown by the marker slc12a1, while the DL (slc12a3+) and PD (gata3+) segments were not significantly altered (Fig. 6A). These patterning changes were associated with corresponding alterations in the sizes and/or positioning of the rostral, central, and caudal domains. For instance, the rostral domain marker hnf4a was reduced, while the central domain marker irx3b and the caudal domain marker mecom were expanded anteriorly (Fig. 6B). Precise mapping of the segment size differences relative to somite number was not possible in tbx16 morphants due to defective somitogenesis (Fig. 6C). Analysis of aldh1a2 expression in tbx16 morphants at the 5 and 28 somite stages showed a significant reduction in transcript levels, consistent with the loss of paraxial mesoderm that characterizes the tbx16 mutant phenotype (Amacher, et al., 2002). The persistent presence of aldh1a2 transcripts in tbx16 morphants likely represents expression in the lateral plate mesoderm along with a small number of muscle precursors that form in tbx16 mutants (Amacher, et al., 2002). These data reveal that tbx16 deficiency induces similar proximodistal nephron patterning defects to lib/aldh1a2 mutants, thus supporting our hypothesis that the paraxial mesoderm provides a major source of the RA that patterns the pronephros.
Figure 6. tbx16-dependent paraxial mesoderm formation is essential for proximo-distal nephron patterning.
Whole mount in situ hybridization analysis for nephron segment markers (purple) and mhc (red) at the 28 somite stage in wildtype embryos and tbx16 MO-injected embryos. Embryos are shown in lateral views with anterior to the left. Black brackets and arrows indicate expression domains, and numbers correspond to somite position. (A) tbx16 knockdown in wildtypes was associated with formation of reduced podocyte numbers, shown by reduced wt1b expression, and shorter proximal segments, shown by smaller nbc1, slc10a1a and trpm7 gene expression domains; conversely, distal segments were expanded, shown by longer clck, slc12a1, and slc12a3 expression domains. The DL (gata3-expresing segment) was unaffected. (B) tbx16 morpholino injected animals have a reduced rostral domain (hnf4a), and shifts in the central (irx3b), and caudal (mecom) domains at 28 somites that are consistent with shorter proximal and expanded distal segments. (C–D) tbx16 morpholino injected animals have dramatically reduced expression of aldh1a2 at 28 somites (C) and 5 somites (D).
RA receptor activity is also essential for nephron segment patterning
Retinoid ligands transduce their effects via heterodimeric complexes of nuclear receptors that consist of a retinoic acid receptor (RAR) and retinoid X receptor (RXR) (Duester, 2008). To explore whether modulations in retinoid receptor activity can alter nephron segmentation, we tested the effects of retinoid receptor antagonists and agonists in wildtype zebrafish embryos. Wildtype embryos treated with an RAR-α antagonist from 60% epiboly to the 28 somite stage formed shortened proximal segments (PCT, PST) and expanded distal segments (DE, DL, PD) (Fig. S3). Conversely, treatment with an RAR-α agonist from 60% epiboly to the 28 somite stage had the opposite effect, and led to expanded proximal segments along with absent or dramatically reduced distal segments (Fig. S3). These findings provide independent evidence that RA signaling is essential for nephron segmentation, and suggest that RAR-α may be a key functional component in nephron patterning.
irx3b acts downstream of RA to specify the DE segment in the zebrafish pronephros
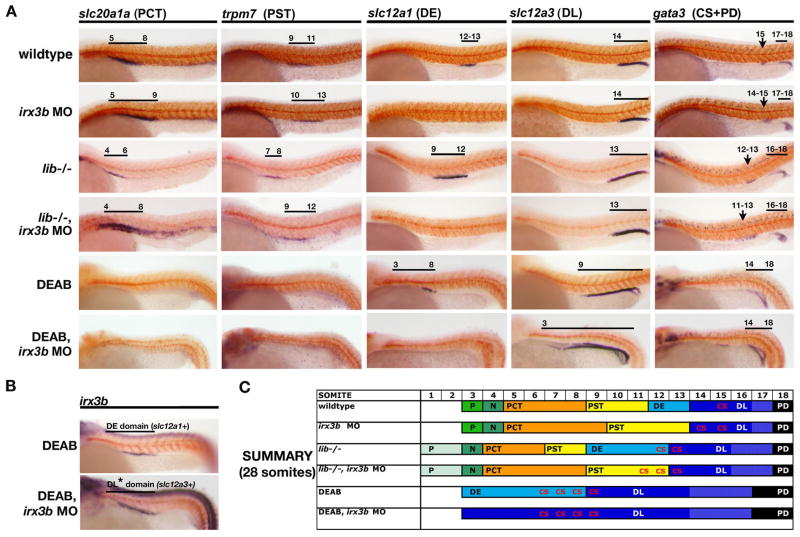
A striking feature of lib mutants is the significantly enlarged DE segment. Previous studies in Xenopus have demonstrated that Irx3 is needed to specify the distal segments during frog pronephros nephrogenesis (Alarcon, et al., 2008; Reggiani, et al., 2007). While the function of the zebrafish orthologue irx3b has not been assessed, our analysis of early transcription factor expression domains traced irx3b expression in medial nephron progenitors and showed a shift of this expression in lib mutants. To explore the functional importance of irx3b for specifying DE segment fate downstream of RA, we used morpholinos to knockdown this factor and examined nephron segmentation compared to mismatch control-injected animals. We observed pericardial edema in irx3b morphants at 72 hpf, suggestive of renal failure or salt imbalance (data not shown), and analyzed nephron segmentation in irx3b morphants by whole mount in situ hybridization. irx3b-deficient embryos lacked a mature DE segment, as revealed by the absence of slc12a1 and romk expression at 24 hpf (Fig. 7; data not shown). In place of the DE segment, the proximal segments were slightly expanded, shown by lengthened domains of the PCT marker slc20a1a and PST marker trpm7, such that the boundary between these segments was shifted caudally along the trunk (Fig. 7A, 7C). In addition, the corpuscle of Stannius (CS), a gland derived from the tubular epithelium between the DE and DL segments and marked by gata3, was dramatically expanded (Fig. 7A, 7C). These results indicate that irx3b is essential for the expression of DE-specific markers and suggest that irx3b regulates the PCT/PST boundary, as well as restrict CS fate.
Figure 7. irx3b activity is requisite for DE segment identity, and the absence of RA and irx3b activity generates nephrons with a DL-only tubule identity.
(A) Whole mount in situ hybridization analysis for nephron segment markers (purple) and mhc (red) at the 28 somite stage in wildtype embryos, irx3b MO-injected embryos, lib homozygous mutants, lib homozygous mutants injected with irx3b MO, DEAB-treated embryos, and DEAB-treated embryos injected with irx3b MO. Embryos are shown in lateral views with anterior to the left. Black brackets and arrows indicated expression domains, and numbers correspond to somite position. irx3b knockdown in wildtypes and lib mutants was associated with DE segment abrogation, expansion of the PCT and PST segments as well as the CS population. In DEAB-treated embryos, which normally only form DE and DL tubule segments, irx3b knockdown was associated with expansion of the DL while the PD is unchanged. (B) In DEAB-treated embryos, irx3b expression marked the DE domain that becomes defined by expression of slc12a1. DEAB-treated embryos injected with irx3b MO showed expression of irx3b in the same region of the pronephros, suggesting that DE regionalization had been established, but that in the absence of irx3b DE-specific solute transporter expression does not ensue. (C) Summary of nephron segmentation with respect to embryo somite number. Asterisks in the proximal DL of DEAB-treated wildtypes injected with irx3b MO indicate partial regionalization that is blocked/prevented in these animals.
Consistent with the onset of irx3b expression at 15 somites, irx3b-deficient embryos showed no changes in renal gene expression until late somitogenesis. At 28 somites, we found changes in transcription factor domains that correlated with the alterations in the mature nephron segmentation pattern at 28 somites (Fig. S6). For example, irx3b morphants possessed an expanded domain of hnf4a that coincided with the expansion of PCT and PST segments (Fig. S6B). In addition, the domains of pou3f3a and pou3f3b were shifted posteriorly, such that they were located next to somite 10 compared to somite 9 in wildtype embryos, also corresponding with the shifted position of the PST (Fig. S6). Interestingly, the posterior boundary of pou3f3a was located at somite 14, coincident with the location of the expanded CS population observed in irx3b morphants, and possibly representing ectopic expression in these cells. Lastly, the domains of mecom, emx1, and gata3 were unchanged in irx3b morphants, consistent with the normal location and size of the DL and PD segments (Fig. S6). The finding that transcription factor domains were not altered until approximately the 28 somite stage suggests that irx3b function is required late, likely between the 24 and 28 somite stages, to establish a DE-specific gene program and restrict the expression of proximal tubule markers.
The observation that the PCT/PST boundary was posteriorly shifted in irx3b morphants led us to test whether irx3b knockdown could rescue the shortened shortened proximal segment(s) at the expense of the expanded DE in lib mutants. lib heterozygous incrosses were injected with either an irx3b MO or mismatch control, and then nephron segmentation was examined at the 28 somite stage. Knockdown of irx3b in lib abrogated the DE segment, and significantly expanded the length of both the PCT and PST segments (Fig. 7A, 7C). In particular, the PST segment was doubled in length in lib/irx3b doubly-deficient animals, occupying the portion of the nephron that would otherwise have formed the DE in lib mutants. These findings indicated that while RA signaling is needed to establish the initial PCT and PST progenitor domains, the ultimate size of these segments is not irrevocably set during early nephron patterning and can be altered by modulating the activity of the later-acting gene irx3b.
Recent work in Xenopus has shown that overexpression of Irx3 induces ectopic expression of mature distal tubule markers (Alarcon, et al., 2008; Reggiani, et al., 2007). However, it is not clear whether Irx3 plays an early patterning role during nephron development to specify the DE domain or whether it acts later within this segment to upregulate or restrict gene expression. This issue is challenging to address in the zebrafish as early markers specific to DE progenitors have not been identified and irx3b transcripts are found in both PST and DE segments. To resolve this, we made use of DEAB to create embryos with nephrons comprised of DE and DL tubule segments only (Wingert, et al., 2007) (Fig. 7A). Because the PST segment is absent in DEAB-treated embryos (Wingert, et al., 2007), expression of irx3b in these animals will specifically mark the DE progenitors. Consistent with this, we found irx3b transcripts in the same region of the nephron as the DE marker slc12a1 at the 28 somite stage (Fig. 7B). Next we investigated whether irx3b+ DE progenitors were still present in DEAB-treated embryos that were also injected with the irx3b morpholino. Expression of irx3b was found in DEAB-treated/irx3b-deficient embryos in a region equivalent to where the DE segment would normally form, despite this region now expressing the DL marker slc12a3 (Fig. 7A, B). These results suggest that two distal domains are patterned in DEAB-treated/irx3b-deficient embryos, but that in the absence of irx3b activity the DE region fails to express DE-specific solute transporters and instead expresses DL markers. Thus, we conclude that irx3b in zebrafish is unlikely to be involved in the early patterning of the DE segment but rather plays a role in its terminal differentiation by regulating the expression DE-specific genes and/or repressing the gene programs of neighboring segments.
DISCUSSION
There are many gaps in our knowledge about the mechanisms that accomplish nephron patterning during kidney development. Here, we show that the nephron segments in the zebrafish pronephros arise following an elaborate series of spatiotemporal gene expression changes in the intermediate mesoderm. We have constructed a detailed transcription factor map that describes this process and is predictive of the mature segmentation pattern of the pronephros. Spatiotemporal expression domains in nephron progenitors during mid- to late-somitogenesis were unaffected by blocking cell proliferation, suggesting that cellular turnover does not significantly account for the dynamic alterations that we detected. Furthermore, our genetic evidence demonstrates that RA is essential for delineating the earliest rostrocaudal patterning events in the intermediate mesoderm, and suggest that RA dosage influences the size and positioning of subsequent transcription factor domains and thereby the mature nephron segmentation pattern. We also show that irx3b, acting at relatively late stages of nephron development, is involved in determining the position of the PST/DE boundary and modulating gene expression in the DE segment. Together, our study provides a new molecular map that can be utilized to guide future interrogations into the workings of nephrogenesis, and adds additional insights to previous reports about the ways that RA and irx3b influences nephron formation.
RA signals demarcate an early subdivision of pronephros progenitors
Several lines of evidence now indicate that retinoid signaling is crucial for pronephros patterning, and that the location of RA, time of production, and dosage are all crucial. We recently demonstrated that the caudal homeobox genes regulate the locale of RA by establishing the expression domains of the aldh1a2 and cyp26a1 genes (Wingert, et al., 2007). Axial shifts in the expression of these enzymes were associated with shifts in pronephros position along the embryo trunk, suggesting that the position of RA synthesis had important inductive consequences for nephron patterning (Wingert, et al., 2007). aldh1a2 expression in the paraxial mesoderm has been suggested to serve as the source of retinoids that pattern the pronephros along with other nearby tissues, like the hindbrain, and our analysis of tbx16 morphants provides further support for this hypothesis. It should be noted, however, that the disruption of the formation of the paraxial mesoderm via tbx16 knockdown alters both aldh1a2 expression and leads to other broad defects, thus we cannot eliminate the possibility that the change in nephron patterning in tbx16 morphants is affected by dirsuptions in global patterning. Further studies are needed to delineate between RA-specific and other changes. Given the outcomes of genetic or chemical alterations in RA signaling, we favor the hypothesis that RA produced by paraxial mesoderm acts to pattern nephron progenitors. Proximal tubule segment identities rely on RA for their specification, and dramatic alterations in segment fates ensue when RA levels are greatly elevated or repressed: zebrafish embryos treated with exogenous RA develop nephrons made entirely of proximal segments, while the abrogation of RA production with DEAB leads to nephrons composed entirely of distal segments (Wingert, et al., 2007). Interestingly, segmentation changes only occur if RA bioavailability was altered from the onset of gastrulation through to early somitogenesis stages, establishing an early role for retinoid signaling in pronephros development.
In this study, our analysis of the lib (presumptive null allele) and nls (hypomorph allele) mutants provides genetic evidence that modulating RA levels produces graded effects on the nephron segmentation pattern. By morphological criteria, lib mutants exhibit a more severe RA-deficiency phenotype than nls, and form a slightly shorter PST segment concomitant with an expanded DE segment. The rescue of the lib mutant phenotype accordingly requires treatment with higher levels of exogenous RA than the nls mutant. Taken together, these lines of evidence suggest that RA dosage determines the size of the proximal tubule domain. Intriguingly, partial effects of RA were also observed when embryos were treated with an inhibitor that targets RAR-α activity. Interference with RAR-α led to reduced proximal segments and expanded distal tubule segments, but these changes were not nearly as dramatic as DEAB exposure. This finding could reflect the involvement of other RA receptors during nephron progenitor patterning, though such an interpretation may be complicated by partial drug efficacy in zebrafish cells as compared to other organisms for this particular compound. Future studies, such as RA receptor knockdown experiments, are needed to tease out the specific roles of RA receptors during nephrogenesis, and to address fully whether RA receptor activity is necessary in nephron progenitors themselves.
Our studies raise several intriguing questions about how RA actually affects the nephron progenitors. Cell-autonomy has yet to be assessed regarding RA activity in the nephron progenitors. Another pressing question is whether RA and its receptors cooperate to directly or indirectly modulate the expression of renal genes. Interestingly, a recent study provided evidence that RA participates actively to trigger gene expression of the kidney podocyte lineage. Podocyte precursors express the Wilm’s tumor 1a (wt1a) gene, and wt1a promoter analysis discovered the presence of functional RARE elements that were shown to mediate responsiveness of the promoter to RA in vivo (Bollig, et al., 2009). Abrogation of RA synthesis in the lib mutant (this study) and with chemical inhibitors to aldh1 enzymes (Wingert, et al., 2007) blocks or reduces podocyte formation, respectively. These findings illuminate the possibility that RA may act directly, by activating genes required for proximal tubule fate and also by repressing distal (caudal domain) genes. However, these data do not rule out the notion that indirect mechanisms are also operative.
Interestingly, changes in RA synthesis are associated with alterations in the earliest regionalization of the intermediate mesoderm. Nephron progenitors display at least two regional identities between the 6–8 somite stages, which we have termed rostral and caudal. We found a fascinating, partial overlap of these domains at the 8 somite stage, perhaps suggestive of a more complex arrangement of molecular identities that will be better understood as more early renal markers are discovered. Nevertheless, in the lib setting of decreased RA production, the rostral nephron progenitor population is reduced while the caudal population is expanded, and we noted previously that DEAB treatment abrogates the rostral domain entirely (Wingert, et al., 2007). These data support the notion that RA ultimately works to induce a rostral progenitor identity among nephron progenitors. While the genes involved in nephron patterning downstream of RA are still unclear, the notch ligand genes dlc and jag2 are excellent candidates, given the role of the Notch pathway in promoting proximal tubule fates during mammalian kidney development (Cheng, et al., 2005; Cheng, et al., 2007).
Conserved and unique elements of Irx3 function during zebrafish and frog nephrogenesis
Irx genes encode homeodomain transcription factors, and members of this gene family have functions in the development of many tissues. In diverse settings ranging from imaginal discs in flies to the vertebrate neural plate, Irx genes act to pattern regional differences amongst cell territories and can both to activate and repress target gene expression (Cavodeassi, et al., 2001). Irx3 has recently been identified as a requisite player in two aspects of Xenopus pronephros development: first, it maintains the pronephric territory at early stages, and second, it is required to form slc12a1-expressing distal tubule cells (Alarcon, et al., 2008; Reggiani, et al., 2007).
In this work, we show a conserved requirement for irx3b in distal tubule differentiation in the zebrafish pronephros. Knockdown of irx3b results in an absence of DE marker expression and concomitant expansions in the lengths of the PCT and PST segments. This phenotype does not appear to be caused by a failure to correctly pattern the nephron into a discrete DE domain as irx3b transcripts, which mark presumptive DE progenitors, are still localized to a DE-sized subdomain in DEAB-treated/irx3b-deficient embryos. Instead, we propose that in the absence of Irx3b, the DE segment adopts either a PST expression profile, in the case of irx3b knockdown in wildtype embryos, or a DL expression profile, in the case of DEAB-treated/irx3b-deficient embryos. Consistent with a late role for irx3b during nephron development, alterations in gene expression domains were only detectable in irx3b morphants at time-points that immediately preceded the onset of the segment-specific solute transporter genes. Thus, the transcriptional targets of Irx3b may include these transporters (acting as an activator of DE-specific transporters and/or repressor of PST and DL transporters) as well as equivalent transcription factors acting in neighboring segments. In support of this, there is precedence in other tissues for such diverse targets of Irx factors; regional patterning in the developing vertebrate brain utilizes a system of transcription factor antagonism between Irx3 and Six3, while Irx4 and Irx5 in myocardial cells alters potassium ion channel gene expression (Constantini, et al., 2005; He, et al., 2009; Kobayashi, et al., 2002). We have identified putative Iroquois binding sites upstream of zebrafish slc12a1, consistent with the possibility that irx3b regulates the expression of this DE-specific transporter (Wingert and Davidson, unpublished).
In contrast, the role of Irx3 in the maintenance of the pronephros territory does not appear to be conserved in zebrafish, highlighting one functional difference between the frog and zebrafish Irx3 genes. In zebrafish, at least four irx orthologues are expressed in the zebrafish pronephros, irx2a, irx3b, irx4a, and irx5a, which may explain the difference between these species (Lecaudey, et al., 2005). Further, Irx3 expression in Xenopus embryos appears to be positively regulated by retinoids, as fewer transcripts are detected in the absence of retinoids while exogenous RA is associated with elevated transcript numbers (Alarcon, et al., 2008). These alterations appear linked to early changes in the mesoderm destined to form the pronephros rather than later proximodistal patterning events (Alarcon, et al., 2008). While RA similarly appears to function upstream of irx3b in the zebrafish pronephros, changes in the irx3b domain correlate with altered nephron patterning.
Insights into conserved nephron segmentation mechanisms among vertebrates
This study has identified critical roles for RA and irx3b during zebrafish pronephros segmentation, but other vital signals have yet to be determined. Our characterization of transcription factor gene expression in the intermediate mesoderm has revealed a complex molecular profile that is spatially and temporally dynamic, and future functional analysis of these genes will reveal key determinants of segment identity and cross-regulatory interactions. A similar level of transcriptional complexity is beginning to be realized during mammalian nephrogenesis, largely in part due to the efforts of the GenitoUrinary Development Molecular Anatomy Project (GUDMAP), which has mapped the expression of mouse genes to different compartments of the developing metanephric kidney (Brunskill, et al, 2008; Georgas, et al., 2009; McMahon, et al., 2008). Findings from the GUDMAP, as well as previous studies, have documented a set of genes with regionalized domains within murine nephrons. For example, Pou3f3 is restricted to prospective distal regions of the nephron, similar to our observations of the zebrafish orthologues pou3f3a and pou3f3b (Nakai, et al., 2003). It is likely that many of the same transcriptional pathways underlie proximodistal nephron patterning in all vertebrates. Therefore our work, establishing a detailed molecular description of gene expression during nephrogenesis, should provide a rich source of data for generating hypothesis-driven experiments in other model organisms such as the mouse. A greater knowledge of the molecular basis of vertebrate nephron development will help elucidate the causes of congenital kidney defects, which are often poorly understood, as well as further our understanding of the regulatory networks that maintain nephron function in the adult.
EXPERIMENTAL PROCEDURES
Zebrafish husbandry, genetic strains and lib mutant mapping
Zebrafish were maintained and staged as described (Kimmel, et al., 1995). Tübingen strain embryos were used for all experiments, and collected from natural matings. lib was recovered in a diploid ENU (N-ethyl-N-nitrosurea) screen for mutations affecting kidney organogenesis, performed as described (Solnica-Krezel, et al., 1994). Families were screened to isolate mutants with kidney development defects, as manifested by edema between 24 and 72 hpf. lib was cloned using meiotic mapping. lib mapping strains were generated by mating lib mutant carriers, generated and maintained on the Tübingen strain, were to the polymorphic WIK strain. Embryos were collected from pairwise matings of lib Tü/WIK heterozygotes, and scored at 72 hpf for edema. A panel of 239 CA simple sequence length polymorphisms (SSLP) markers were used to scan chromosomes and establish linkage as described (Wingert, et al., 2005). aldh1a2 transcripts were isolated from wild-types and lib by reverse transcription PCR, using the following primers: forward ATGACCTCCAGTG-AAGTTGAACTG and reverse, TTAAGACGTCTTGCCTGACATCTT, and subcloned into pGEMT-easy (Promega) for sequence analysis. For lib rescues (and aldh1a2 expression studies, see below), full-length wild-type aldh1a2 was subcloned into the pCS2 expression vector using the EcoRI and XhoI sites. WISH aldh1a2 probe was generated by linearizing the plasmid with EcoRI and performing antisense RNA synthesis with T7 RNA polymerase (Roche).
Embryo chemical treatments, morpholinos and in situ hybridization analysis
Reported expression patterns show representative results as gathered from analysis of between 10 and 15 animals per each timepoint and genotype (mutant studies and knockdown). Gene expression domains as reported by somite boundaries were based on counts of at least 5 separate samples to ensure accurate documentation.
For chemical treatments, wild-type embryos were incubated with camptothecin and nocodazole, as described (Murphey, et al., 2006). Briefly, cell cycle inhibitors (Tocris) were dissolved to make stock concentrations of 10 mg/mL dissolved in 100% dimethyl sulfoxide (DMSO), and embryos treated at the established effective dose (Murphey, et al., 2006). For lib rescue studies, embryos from lib or nls heterozygous incrosses were incubated in RA treatment, as described (Wingert, et al., 2007). Briefly, all-trans retinoic acid (Sigma) was dissolved in 100% DMSO to make a 1 M stock, aliquots stored at −80°C, and embryos treated with 1 × 10−8M or 1 × 10−9M RA/DMSO, which were made by performing serial dilution of the 1M stock in E3 embryo water. For RA agonist and antagonist studies, RO 41-5232 and AM-580 (Tocris) were similarly dissolved in 100% DMSO to make a 1 M stock, aliquots stored at −80°C, and wildtype embryos were incubated in 1 × 10−8M RO 41-5232/DMSO or 1 × 10−7M AM-580/DMSO diluted in E3 embryo water. All chemical treatments were fully penetrant and produced consistent results at the doses and treatment windows that were examined.
For Aldh1a2 rescue studies in lib, the full-length open reading frame of wildtype and lib aldh1a2 alleles were subcloned into the EcoR1/XhoI sites of the pCS2 expression vector. Plasmids were linearized with Not1, and synthetic capped RNA was synthesized using SP6 mMessage machine (Ambion). Embryos were injected with 100 pg of cRNA at the 1-cell stage.
For all morpholino experiments, morpholinos were solubulized as recommended by Gene-Tools at and stored at a 4mM concentration. Dilution of morpholino stocks were injected into 1-cell stage wild-type embryos. The tbx16 morpholino (AAGACAAGTACTCACCTCTGATAGC) targets the exon 1 donor splicing site and produces consistent results at the injected dose (Burns, et al., 2009). irx3b morpholinos were designed to target the 5′ untranslated region and start site morpholinos, and gave similar results upon injection of ~500 pl of a 0.15 mM working dose and 0.2 mM dose, respectively: (ACCGGGAGGACTGCGGGGAAACTCG) and (ATAGCCTAGCTGCGGGAGAGACATG), respectively. A 5 base-pair mismatch start site morpholino (ATTGCCTACCTGGGGGACAGAGATG) was injected for a control comparison. Whole mount in situ hybridization for cdh17, clck, dlc, gata3, jag2, mecom, mhc, myoD, nbc1, pax2a, pax8, raldh2, wt1a and wt1b were performed as described (Wingert, et al., 2007). Expression of emx1, etv5, grhl2a, hnf1ba, hnf1bb, hnf4a, irx3b, pou3f3a, pou3f3b were previously reported (Hauptmann and Gerster, 2000; Kawahara and Dawid, 2002; Lecaudey, et al., 2005; Thisse, et al., 2001).
Supplementary Material
Whole mount in situ hybridization analysis for nephron segment markers (purple) and mhc (red) at the 28 somite stage in wildtype embryos treated with DMSO (vehicle alone) or camptothecin starting at the 24, 22, 20, 18 and 15 somite stage or (B) nocodazole starting at the 22 somite stage. Embryos are shown in lateral views with anterior to the left. Black brackets and arrows indicated expression domains, and numbers correspond to somite position. Embryos in all chemical treatments formed PCT, PST, DE, and DL segments at the same somite position as wildtype embryos, consistent with a negligible role for cell proliferation in the establishment of segment domains after the 15 somite stage. The segments detected using the following transcripts for all embryos except the 15 somite stage treatment: PCT by slc20a1a, PST by trpm7, DE by slc12a1, DE by slc12a3. The 15 somite stage camptothecin-treated embryos were significantly delayed by the drug treatment and did not show trpm7 or slc12a1 transcripts (data not shown). To probe the existence of these areas, the overlap between hnf4a and irx3b was ascertained, as indicated by a single asterisk for hnf4a and double asterisk for irx3b transcripts. The overlap between hnf4a and irx3b showed the presence of the PCT identity (cells that expressed hnf4a only) and PST (region of cells that expressed both hnf4a and irx3b), a correlation based on that seen in wildtype embryos at this stage.
(A) Lateral views of living 28 somite wildtype and lib embryos compared to embryos injected with aldh1a2 cRNA transcripts. lib embryos formed a normal body trunk without curvature and a kink between the head and trunk, though mild pericardial edema still developed. (B) Table of rescue percentages in offspring from nls, nls/lib, and lib heterozygous crosses after treatment with DMSO (vehicle alone), all-trans RA at 1× 10−9M, all-trans RA at 1×10−8M, and wildtype aldh1a2 cRNA injection. Rescue evaluation was determined by presence of pectoral fins in mutant embryos. lib homozygote embryos were rescued only by higher RA or aldh1a2 overexpression, while nls homozygotes and nls/lib compound heterozygotes were rescued at the lower RA dosage. (C) Whole mount in situ hybridization analysis for nephron segment markers (purple) and mhc (red) at the 28 somite stage in wildtype and lib mutants after exogenous treatment with all-trans RA at 1× 10−9M. Embryos are shown in lateral views with anterior to the left, with the exception of dorsal views to show podocytes. Black brackets and arrows indicated expression domains, and numbers correspond to somite position. RA treatment restored the podocyte population in lib, and expanded proximal tubule fates (PCT and PST), while the distal fates (DE, DL, PD) were all reduced, showing pronephros progenitors were competent to respond to elevated exogenous RA. Similar phenotype trends were observed in wildtypes exposed to this dosage of RA. (D) Summary of nephron segmentation with respect to embryo somite number in wildtype and lib with DMSO or all-trans RA 1× 10−9M exposure.
(A, B) Whole mount in situ hybridization analysis for nephron segment markers (purple) and mhc (red) at the 28 somite stage in wildtype embryos treated with DMSO (vehicle only), an RAR-α antagonist, or RAR-α agonist. Embryos are shown in lateral views with anterior to the left, with the exception of dorsal views to show podocytes. Black brackets and arrows indicated expression domains, and numbers correspond to somite position. RAR-α antagonist treatment reduced proximal segments and expanded distal segments, while RAR-α agonist treatment expanded proximal segments at the expense of distal fates. (C) Summary of nephron segmentation with respect to embryo somite number for each treatment.
Gene expression patterns of transcription factors in nephron progenitors in wild-type embryos compared to lib mutant embryos at the 20, 22, 24, and 28 somite stage. Embryos are shown in lateral views with anterior to the left, and have been stained by whole mount in situ hybridization to mark kidney expression (purple) and the somites with mhc (red). Black lines indicate areas of high gene expression, and numbers correspond to the adjacent somite position. The spatial distributions of hnf1a, hnf4a, and hnf1bb transcripts were unaltered between 20–28 somites (28 somite stage not shown). The distributions of etv5 and irx3b changed in wildtypes and lib at the 28 and 24 somite stages, respectively.
Gene expression patterns of transcription factors in nephron progenitors in wild-type embryos compared to lib mutant embryos at the 20, 22, 24, and 28 somite stage. Embryos are shown in lateral views with anterior to the left, and have been stained by whole mount in situ hybridization to mark kidney expression (purple) and the somites with mhc (red). Black lines indicate areas of high gene expression, and numbers correspond to the adjacent somite position. The spatial distributions of pou3f3a and pou3f3b transcripts in lib mutants were continually expressed in rostral regions between 20–28 somites (28 somite stage not shown). The distribution of mecom changed progressively in both wildtypes and lib throughout the 20–28 somite stages. The distribution of emx1 and gata3 transcripts was unaltered in wildtypes between the 20–28 somite stages, whereas lib had progressive restrictions of these transcripts to caudal populations.
(A) Gene expression patterns of transcription factors in nephron progenitors in wildtype embryos and irx3b MO injected animals at the 28 somite stage. Embryos are shown in lateral views with anterior to the left, and have been stained by whole mount in situ hybridization to mark kidney gene expression (purple) and the somites with mhc (red). Black lines indicate areas of high gene expression, and numbers correspond to the somite position. In irx3b morphants, the domains of hnf1ba, hnf1g, irx3b, mecom, emx1, and gata3 were unchanged, while the expression of hnf4a was expanded slightly and the rostral boundary of pou3f3a and pou3f3b were shifted distally by one somite. In addition, the pou3f3a domain was expanded to somite 14. (B) The domain of transcription factor gene expression in wildtype and irx3b morphant embryos correlates closely to the segment pattern. (Foreground) Summary of transcription factor expression domains (represented by black bars) are schematized in the nephrons at the 28 somite stages (Note: wildtype schema is repeated from Figure 3 for easy comparison). (Background) Nephron segment identities form adjacent to particular somites (demarcated by numbered columns, top), and each segment region is color-coded to envisage a comparison between the transcription factor expression domains and the nephron segments. Segment color codes are N-green, PCT-orange, PST-yellow, DE-light blue, DL-dark blue, and PD-gray.
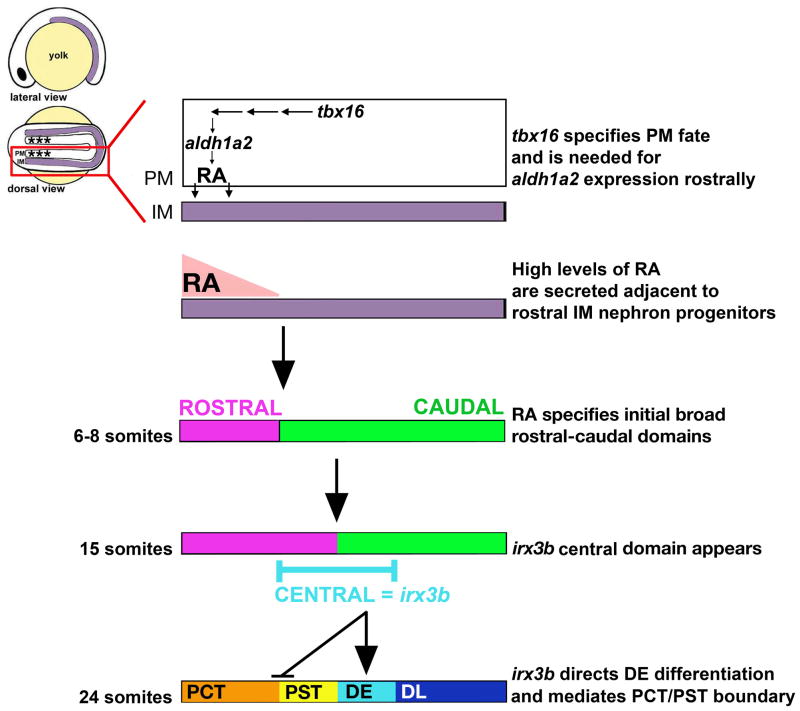
Figure 8. Model of how RA and irx3b signals sequentially pattern the pronephros territory to generate discrete nephron segments.
Nephron progenitors, derived from the intermediate mesoderm (IM) are situated adjacent to the paraxial mesoderm (PM). The rostral PM (indicated by asterisks, *) provides an early local source of RA, which is thought to diffuse to nearby tissues. Subsequent to RA signaling, nephron progenitors are broadly regionalized into rostral and caudal domains by the 6–8 somite stages. At the 15 somite stage, a nested pattern of transcription factor domains suggests the continued presence of broad rostral and caudal domains as well as an overlapping medial domain, defined by irx3b expression. Activity of irx3b sets the DE identity given its dynamic spatiotemporal shifts in this vicinity during later somitogenesis stages. Segment epithelia have unique solute transporter expression patterns at the 28 somite stage, concomitant with an end to many spatiotemporal transcription factor domain shifts in the nephron progenitors.
Acknowledgments
RAW was supported by Polycystic Kidney Foundation Fellowship Grant 160a2f, a Harvard Stem Cell Institute Seed Grant, and an NIH-NIDDK award K01DK083512. RAW wishes to thank the staff of the Center for Zebrafish Research at the University of Notre Dame for providing ongoing husbandry of the lib zebrafish strain. AJD was supported by NIH-NIDDK award R01DK077186 and grants from the Harvard Stem Cell Institute.
Abbreviations
- Zebrafish nephron segments were abbreviated as follows
C, cloaca
- CS
corpuscle of Stannius
- DE
distal early
- DL
distal late
- G
glomerulus
- N
neck
- P
podocytes
- PCT
proximal convoluted tubule, PST, proximal straight tubule
References
- Alarcon P, Rodriquez-Seguel E, Fernandez-Gonzalez A, Rubio R, Gomez-Skarmeta JL. A dual requirement for Iroquois genes during Xenopus kidney development. Development. 2008;135:3197–3207. doi: 10.1242/dev.023697. [DOI] [PubMed] [Google Scholar]
- Alexa K, Choe SK, Hirsch N, Etheridge L, Laver E, Sagerström CG. Maternal and zygotic aldh1a2 activity is required for pancreas development in zebrafish. PLoS ONE. 2009;4(12):e8261. doi: 10.1371/journal.pone.0008261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amacher SL, Draper BW, Summers BR, Kimmel CB. The zebrafish T-box genes no tail and spadetail are required for development of trunk and tail mesoderm and medial floor plate. Development. 2002;129:3311–3323. doi: 10.1242/dev.129.14.3311. [DOI] [PubMed] [Google Scholar]
- Begemann G, Schilling TF, Rauch GJ, Geisler R, Ingham PW. The zebrafish neckless mutation reveals a requirement for raldh2 in mesodermal signals that pattern the hindbrain. Development. 2001;128:3081–3094. doi: 10.1242/dev.128.16.3081. [DOI] [PubMed] [Google Scholar]
- Begemann G, Marx M, Mebus K, Meyer A, Bastmeyer M. Beyond the neckless phenotype: influence of reduced retinoic acid signaling on motor neuron development in the zebrafish hindbrain. Dev Biol. 2004;271:119–129. doi: 10.1016/j.ydbio.2004.03.033. [DOI] [PubMed] [Google Scholar]
- Bollig F, Perner B, Besenbeck B, Köthe S, Ebert C, Taudien S, Englert C. A highly conserved retinoic acid responsive element controls wt1a expression in the zebrafish pronephros. Development. 2009;136:2883–2892. doi: 10.1242/dev.031773. [DOI] [PubMed] [Google Scholar]
- Brunskill EW, Aronow BJ, Georgas K, Rumballe B, Valerius MT, Aronow J, Kaimal V, Jegga AG, Yu J, Grimmond S, et al. Atlas of gene expression in the developing kidney at microanatomic resolution. Dev Cell. 2008;15:781–791. doi: 10.1016/j.devcel.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns CE, Galloway JL, Smith AC, Keefe MD, Cashman TJ, Paik EJ, Mayhall EA, Amsterdam AH, Zon LI. A genetic screen in zebrafish defines a hierarchical network of pathways required for hematopoietic stem cell emergence. Blood. 2009;113 (23):5776–5782. doi: 10.1182/blood-2008-12-193607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavodeassi F, Modolell J, Gomez-Skarmeta JL. The Iroquois family of genes: from body building to neural patterning. Development. 2001;128:2847–2855. doi: 10.1242/dev.128.15.2847. [DOI] [PubMed] [Google Scholar]
- Cheng L, Al-Awqati Q. Segmental expression of Notch and Hairy genes in nephrogenesis. Am J Physiol Renal Physiol. 2005;288:F939–F952. doi: 10.1152/ajprenal.00369.2004. [DOI] [PubMed] [Google Scholar]
- Cheng HT, Kim M, Valerius MT, Surendran K, Schuster-Gossler K, Gossler A, McMahan AP, Kopan R. Notch2, but not Notch1, is required for proximal fate acquisition in the mammalian nephron. Development. 2007;134:801–811. doi: 10.1242/dev.02773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantini DL, Arruda EP, Agarwal P, Kim KH, Zhu Y, Zhu W, Lebel M, Cheng CW, Park CY, et al. The homeodomain transcription factor Irx5 establishes the mouse cardiac ventricular repolarization gradient. Cell. 2005;123:347–358. doi: 10.1016/j.cell.2005.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dressler GR. The cellular basis of kidney development. Annu Rev Cell Dev Biol. 2006;22:509–529. doi: 10.1146/annurev.cellbio.22.010305.104340. [DOI] [PubMed] [Google Scholar]
- Drummond IA, Majumdar A, Hentschel H, Elger M, Solnica-Krezel L, Schier AF, Neuhauss SC, Stemple DL, Zwartkruis F, Rangini Z, et al. Early development of the zebrafish pronephros and analysis of mutations affecting pronephric function. Development. 1998;125:4655–4667. doi: 10.1242/dev.125.23.4655. [DOI] [PubMed] [Google Scholar]
- Drummond IA. Making a zebrafish kidney: a tale of two tubes. Trends Cell Biol. 2003;13:357–365. doi: 10.1016/s0962-8924(03)00124-7. [DOI] [PubMed] [Google Scholar]
- Duester G. Retinoic acid synthesis and signaling during early organogenesis. Cell. 2008;134:921–931. doi: 10.1016/j.cell.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Dahr SS, Aboudehen K, Saifudeen Z. Transcriptional control of terminal nephron differentiation. Am J Physiol Renal Physiol. 2008;294:F1273–F1278. doi: 10.1152/ajprenal.00562.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgas K, Rumballe B, Valerius MT, Chiu HS, Thiagarajan RD, Lesieur E, Aronow BJ, Brunskill EW, Combes AN, Tang D, et al. Analysis of early nephron patterning reveals a role for the distal RV proliferation in fusion to the ureteric tip via a cap mesenchyme-derived connecting segment. Dev Biol. 2009;332:273–286. doi: 10.1016/j.ydbio.2009.05.578. [DOI] [PubMed] [Google Scholar]
- Grandel H, Lun K, Rauch GJ, Rhinn M, Piotrowski T, Houart C, Sordino P, Küchler AM, Schulte-Merker S, Geisler R, et al. Retinoic acid signaling in the zebrafish embryo is necessary during pre-segmentation stages to pattern the anterior-posterior axis of the CNS and to induce a pectoral limb bud. Development. 2002;129:2851–2865. doi: 10.1242/dev.129.12.2851. [DOI] [PubMed] [Google Scholar]
- Hauptmann G, Gerster T. Combinatorial expression of zebrafish Brn-1 and Brn-2-related POU genes in the embryonic brain, pronephric primordium, and pharyngeal arches. Dev Dyn. 2000;218(2):345–58. doi: 10.1002/(SICI)1097-0177(200006)218:2<345::AID-DVDY8>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- He W, Jia Y, Takimoto K. Interaction between transcription factors Iroquois proteins 4 and 5 controls cardiac potassium channel Kv4.2 gene transcription. Cardiovascular Research. 2009;81:64–71. doi: 10.1093/cvr/cvn259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert SC, Reilly RF, Kriz W. Structural-functional relationships in the kidney. In: Schrier RW, editor. Diseases of the Kidney and Urinary Tract. 7. Lippincott Williams and Wilkins; Philadelphia: 2001. pp. 3–57. [Google Scholar]
- Houweling AC, Dildrop R, Peters T, Mummenhoff J, Moorman AFM, Ruther U, Christoffels VM. Gene and cluster-specific expression of the Iroquois family members during mouse development. Mech Dev. 2001;107:169–174. doi: 10.1016/s0925-4773(01)00451-8. [DOI] [PubMed] [Google Scholar]
- Kawahara A, Dawid IB. Developmental expression of zebrafish emx1 during early embryogenesis. Gene Expr Patterns. 2002;2(3–4):201–206. doi: 10.1016/s1567-133x(02)00062-5. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kobayashi D, Kobayashi M, Matsumoto K, Ogura T, Nakafuku M, Shimamura K. Early subdivisions in the neural plate define distinct competence for inductive signals. Development. 2002;129:83–93. doi: 10.1242/dev.129.1.83. [DOI] [PubMed] [Google Scholar]
- Kopan R, Cheng HT, Surendran K. Molecular insights into segmentation along the proximal-distal axis of the nephron. J Am Soc Nephrol. 2007;18:2014–2020. doi: 10.1681/ASN.2007040453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecaudey V, Anselme I, Dildrop R, Rüther U, Schneider-Maunoury S. Expression of the zebrafish iroquois genes during early nervous system formation and patterning. J Comp Neurol. 2005;492:289–302. doi: 10.1002/cne.20765. [DOI] [PubMed] [Google Scholar]
- McMahon A, Aronow BJ, Davidson DR, Davies JA, Gaido KW, Grimmond S, Lessard JL, Little MH, Potter SS, Wilder EL, et al. GUDMAP: the genitourinary developmental molecular anatomy project. J Am Soc Nephrol. 2008;19:667–671. doi: 10.1681/ASN.2007101078. [DOI] [PubMed] [Google Scholar]
- Morley RH, Lachani K, Keefe D, Gilchrist MJ, Flicek P, Smith JC, Wardle FC. A gene regulatory network directed by zebrafish No tail accounts for its roles in mesoderm formation. Proc Natl Acad Sci USA. 2009;106(10):3829–3834. doi: 10.1073/pnas.0808382106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphey RD, Stern HM, Straub CT, Zon LI. A chemical genetic screen for cell cycle inhibitors in zebrafish embryos. Chem Biol Drug Des. 2006;68:213–219. doi: 10.1111/j.1747-0285.2006.00439.x. [DOI] [PubMed] [Google Scholar]
- Nakai S, Sugitani Y, Sato H, Ito S, Miura Y, Ogawa M, Nishi M, Jishage K, Minowa O, Noda T. Crucial roles of Brn1 in distal tubule formation and function in mouse kidney. Development. 2003;130:4751–4759. doi: 10.1242/dev.00666. [DOI] [PubMed] [Google Scholar]
- Reggiani L, Raciti D, Airik R, Kispert A, Brändli AW. The prepattern transcription factor Irx3 directs nephron segment identity. Genes Dev 2007. 2007;21:2358–2370. doi: 10.1101/gad.450707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schedl A. Renal abnormalities and their developmental origin. Nat Rev Genetics. 2007;8:791–802. doi: 10.1038/nrg2205. [DOI] [PubMed] [Google Scholar]
- Solnica-Krezel L, Schier AF, Driever W. Efficient recovery of ENU-induced mutations from the zebrafish germline. Genetics. 1994;136(4):1401–1420. doi: 10.1093/genetics/136.4.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse B, Pflumio S, Fürthauer M, Loppin B, Heyer V, Degrave A, Woehl R, Lux A, Steffan T, Charbonnier XQ, Thisse C. ZFIN Direct Data Submission. 2001. Expression of the zebrafish genome during embryo-genesis. [Google Scholar]
- Wingert RA, Galloway JL, Barut B, Foott H, Fraenkel P, Axe JL, Weber GJ, Dooley K, Davidson AJ, et al. Deficiency of glutaredoxin 5 reveals Fe-S clusters are required for vertebrate haem synthesis. Nature. 2005;18;436(7053):1035–1039. doi: 10.1038/nature03887. [DOI] [PubMed] [Google Scholar]
- Wingert RA, Selleck R, Yu J, Song HD, Chen Z, Song A, Zhou Y, Thisse B, Thisse C, McMahon AP, Davidson AJ. The cdx genes and retinoic acid control the positioning and segmentation of the zebrafish pronephros. PLoS Genet. 2007;3:e189. doi: 10.1371/journal.pgen.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingert RA, Davidson AJ. The zebrafish pronephros: a model to study nephron segmentation. Kid Int. 2008;73 (10):1120–1127. doi: 10.1038/ki.2008.37. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Whole mount in situ hybridization analysis for nephron segment markers (purple) and mhc (red) at the 28 somite stage in wildtype embryos treated with DMSO (vehicle alone) or camptothecin starting at the 24, 22, 20, 18 and 15 somite stage or (B) nocodazole starting at the 22 somite stage. Embryos are shown in lateral views with anterior to the left. Black brackets and arrows indicated expression domains, and numbers correspond to somite position. Embryos in all chemical treatments formed PCT, PST, DE, and DL segments at the same somite position as wildtype embryos, consistent with a negligible role for cell proliferation in the establishment of segment domains after the 15 somite stage. The segments detected using the following transcripts for all embryos except the 15 somite stage treatment: PCT by slc20a1a, PST by trpm7, DE by slc12a1, DE by slc12a3. The 15 somite stage camptothecin-treated embryos were significantly delayed by the drug treatment and did not show trpm7 or slc12a1 transcripts (data not shown). To probe the existence of these areas, the overlap between hnf4a and irx3b was ascertained, as indicated by a single asterisk for hnf4a and double asterisk for irx3b transcripts. The overlap between hnf4a and irx3b showed the presence of the PCT identity (cells that expressed hnf4a only) and PST (region of cells that expressed both hnf4a and irx3b), a correlation based on that seen in wildtype embryos at this stage.
(A) Lateral views of living 28 somite wildtype and lib embryos compared to embryos injected with aldh1a2 cRNA transcripts. lib embryos formed a normal body trunk without curvature and a kink between the head and trunk, though mild pericardial edema still developed. (B) Table of rescue percentages in offspring from nls, nls/lib, and lib heterozygous crosses after treatment with DMSO (vehicle alone), all-trans RA at 1× 10−9M, all-trans RA at 1×10−8M, and wildtype aldh1a2 cRNA injection. Rescue evaluation was determined by presence of pectoral fins in mutant embryos. lib homozygote embryos were rescued only by higher RA or aldh1a2 overexpression, while nls homozygotes and nls/lib compound heterozygotes were rescued at the lower RA dosage. (C) Whole mount in situ hybridization analysis for nephron segment markers (purple) and mhc (red) at the 28 somite stage in wildtype and lib mutants after exogenous treatment with all-trans RA at 1× 10−9M. Embryos are shown in lateral views with anterior to the left, with the exception of dorsal views to show podocytes. Black brackets and arrows indicated expression domains, and numbers correspond to somite position. RA treatment restored the podocyte population in lib, and expanded proximal tubule fates (PCT and PST), while the distal fates (DE, DL, PD) were all reduced, showing pronephros progenitors were competent to respond to elevated exogenous RA. Similar phenotype trends were observed in wildtypes exposed to this dosage of RA. (D) Summary of nephron segmentation with respect to embryo somite number in wildtype and lib with DMSO or all-trans RA 1× 10−9M exposure.
(A, B) Whole mount in situ hybridization analysis for nephron segment markers (purple) and mhc (red) at the 28 somite stage in wildtype embryos treated with DMSO (vehicle only), an RAR-α antagonist, or RAR-α agonist. Embryos are shown in lateral views with anterior to the left, with the exception of dorsal views to show podocytes. Black brackets and arrows indicated expression domains, and numbers correspond to somite position. RAR-α antagonist treatment reduced proximal segments and expanded distal segments, while RAR-α agonist treatment expanded proximal segments at the expense of distal fates. (C) Summary of nephron segmentation with respect to embryo somite number for each treatment.
Gene expression patterns of transcription factors in nephron progenitors in wild-type embryos compared to lib mutant embryos at the 20, 22, 24, and 28 somite stage. Embryos are shown in lateral views with anterior to the left, and have been stained by whole mount in situ hybridization to mark kidney expression (purple) and the somites with mhc (red). Black lines indicate areas of high gene expression, and numbers correspond to the adjacent somite position. The spatial distributions of hnf1a, hnf4a, and hnf1bb transcripts were unaltered between 20–28 somites (28 somite stage not shown). The distributions of etv5 and irx3b changed in wildtypes and lib at the 28 and 24 somite stages, respectively.
Gene expression patterns of transcription factors in nephron progenitors in wild-type embryos compared to lib mutant embryos at the 20, 22, 24, and 28 somite stage. Embryos are shown in lateral views with anterior to the left, and have been stained by whole mount in situ hybridization to mark kidney expression (purple) and the somites with mhc (red). Black lines indicate areas of high gene expression, and numbers correspond to the adjacent somite position. The spatial distributions of pou3f3a and pou3f3b transcripts in lib mutants were continually expressed in rostral regions between 20–28 somites (28 somite stage not shown). The distribution of mecom changed progressively in both wildtypes and lib throughout the 20–28 somite stages. The distribution of emx1 and gata3 transcripts was unaltered in wildtypes between the 20–28 somite stages, whereas lib had progressive restrictions of these transcripts to caudal populations.
(A) Gene expression patterns of transcription factors in nephron progenitors in wildtype embryos and irx3b MO injected animals at the 28 somite stage. Embryos are shown in lateral views with anterior to the left, and have been stained by whole mount in situ hybridization to mark kidney gene expression (purple) and the somites with mhc (red). Black lines indicate areas of high gene expression, and numbers correspond to the somite position. In irx3b morphants, the domains of hnf1ba, hnf1g, irx3b, mecom, emx1, and gata3 were unchanged, while the expression of hnf4a was expanded slightly and the rostral boundary of pou3f3a and pou3f3b were shifted distally by one somite. In addition, the pou3f3a domain was expanded to somite 14. (B) The domain of transcription factor gene expression in wildtype and irx3b morphant embryos correlates closely to the segment pattern. (Foreground) Summary of transcription factor expression domains (represented by black bars) are schematized in the nephrons at the 28 somite stages (Note: wildtype schema is repeated from Figure 3 for easy comparison). (Background) Nephron segment identities form adjacent to particular somites (demarcated by numbered columns, top), and each segment region is color-coded to envisage a comparison between the transcription factor expression domains and the nephron segments. Segment color codes are N-green, PCT-orange, PST-yellow, DE-light blue, DL-dark blue, and PD-gray.