Abstract
Glutaraldehyde (GTA) is a potent virucidal disinfectant whose exact mode of action against enteroviruses is not understood. Earlier reports showed that GTA reacts preferentially with the VP1 capsid protein of echovirus 25 and poliovirus 1 and that GTA has affinity for exposed lysine residues on proteins. To investigate further the inactivation of enteroviruses by GTA, seven strains were selected on the basis of differences in their overall number and the positions of lysine residues in the amino acid sequences of the VP1 polypeptide. Inactivation kinetics experiments were performed with 0.10% GTA. The viruses grouped into three clusters and exhibited significantly different levels of sensitivity to GTA. The results were analyzed in the light of current knowledge of the three-dimensional structure of enteroviruses and the viral life cycle. The differences observed in sensitivity to GTA were related to the number of lysine residues and their locations in the VP1 protein. The overall findings suggest that the BC and DE loops, which cluster at the fivefold axis of symmetry and are the most exposed on the outer surface of the virions, are primary reactive sites for GTA.
Glutaraldehyde (GTA) is a saturated 5-carbon dialdehyde with powerful biocidal activity against bacteria and their spores, fungi, and viruses. Two-percent-GTA-based products have been used to disinfect flexible endoscopes for about 30 years worldwide (24). Although GTA-containing disinfectants are currently discouraged in endoscope disinfection in some countries because of prion resistance and toxicological problems, GTA is still widely used in heat-sensitive medical devices and on environmental surfaces (1, 9). Considerable progress has been made in understanding the mechanisms of its bactericidal and sporicidal action. In contrast, data on the precise mechanism by which GTA inactivates viruses are still lacking. Current knowledge of the interaction between viral components and GTA suggests that the cross-linking properties of GTA may play a role in its virucidal activity (for a review, see reference 21).
In picornaviruses, the virion particle consists of a single-stranded positive-sense RNA genome enclosed in a nonenveloped protein shell. Despite significant differences observed in amino acid sequences, the capsid architecture is conserved among these viruses. The atomic structure of the capsid has been determined for representatives of the Enterovirus genus, including the three serotypes of polioviruses (10, 14, 19), coxsackievirus B3 (22), coxsackievirus A9 (13), and echovirus 1 (11). The capsid is composed of 60 protomers, each consisting of one copy of the four viral proteins VP1 to VP4, assembled with icosahedral symmetry. The major proteins VP1, VP2, and VP3 are partly located on the outer surface of the capsid and have a common structural motif composed of an eight-stranded antiparallel β-barrel and two α-helices. In enterovirus strains, structural differences are primarily found in surface-exposed loops and in the amino and carboxyl termini, located on the inner surface of the capsid.
RNA genome and capsid proteins are potential targets for GTA in enterovirus particles. In previous works, we showed that the genome is an unfavorable reactive site and that GTA reacts strongly with the VP1 structural polypeptide (2, 5). When comparing GTA virucidal activity against isolates and reference strains of poliovirus 1 and echovirus 25, we also observed significant intertypic and intratypic differences (6, 7). Numerous reports investigating the process of binding between GTA and various proteins and enzymes have shown that only the ɛ-amino group of lysine reacts to any significant extent with GTA (8, 16, 17, 20). In light of these findings, we made the assumption that the differences previously observed between enterovirus strains were related to variations in the locations of lysine residues in the capsid proteins, in particular in externalized regions of the VP1 protein. The aim of the present study was to test this hypothesis. We investigated the GTA sensitivity of seven enteroviruses selected on the basis of differences in the number and position of lysine groups located in the VP1 capsid protein.
MATERIALS AND METHODS
Virus strains and cells.
All the enterovirus prototype strains used in the study were obtained from the World Health Organization Collaborating Center, National Center for Enterovirus, Lyon, France. The TH222 echovirus 25 clinical isolate was recovered in France in 1986. The structural features of this echovirus 25 variant have been characterized (3, 23). The echovirus strains and coxsackievirus B3 were propagated in MRC-5 cells (human lung embryonic fibroblasts; Bio-Mérieux, Marcy l'Etoile, France) used for passages 23 to 24, and in Vero cells (African green monkey kidney cells; Flow Laboratories, Inc., McLean, Va.), used for passages 154 to 158, respectively. The cells were grown in Eagle's minimal essential medium (EMEM). Cell culture procedures have been detailed elsewhere (2, 7).
Nucleotide sequence determination.
Isolation of viral genomic RNAs from virus particles was performed with the QIAamp viral RNA mini kit (Qiagen S.A., Courtaboeuf, France). Synthesis of cDNAs was performed with random hexamers as primers and a cDNA synthesis kit (Stratagene). The complete VP1 coding sequence of the 15 echovirus prototype strains was amplified with oligonucleotide primers ecox1 (5′-GC GGA TCC GCG GCC GCG AGC TCI GCR TGC AAY GAY TTY TCW G-3′) and ecox2 (5′-GCT GCA GGG CGC GCC TCT AGA RTC YCT RTT RTA RTC YTC CCA-3′) (I, inosine; R, A or G; Y, C or T; W, A or T) by a previously described method (4). The amplification reactions were performed with 2 μl of the cDNA in a mixture containing 200 nM concentrations (each) of primers ecox1 and ecox2, a 200 μM concentration of each of the four deoxynucleotides, and 1.75 U of the enzyme mix. Thermal cycling comprised 40 cycles as follows: 1 cycle of denaturation for 2 min at 94°C, followed by 38 cycles of denaturation for 15 s at 94°C, annealing for 20 s at 50°C, and elongation for 50 s at 68°C, and a last cycle of 10 min at 68°C. An Omnigene thermocycler (Hybaid) was used for amplifications. Forward and reverse sequencing reactions were performed with the oligonucleotide primers 5′-GC GGA TCC GCG GCC GCG AGC-3′ and 5′-GCT GCA GGG CGC GCC TCT AGA-3′. Two computer programs were used to analyze and compare the VP1 sequences determined: BioEdit (T. A. Hall, available at http://www.mbio.ncsu.edu/BioEdit/) and Genedoc (K. B. Nicholas and H. B. Nicholas Jr., available at http://www.psc.edu.biomed/genedoc/).
Virus stocks for inactivation assays.
High-titer stocks of viruses were produced, concentrated, and purified as follows. The virus stock suspensions were frozen (−80°C) and thawed (37°C) twice. Cellular material in viral suspensions was removed by differential centrifugation at 4°C. The first centrifugation was carried out at 1,000 × g for 10 min in a Jouan GT 422 centrifuge, and the second and third centrifugations were performed at 60,000 × g for 45 min and at 120,000 × g for 4 h, respectively, with an SW28 rotor in an L8-50 M/E ultracentrifuge (Beckman). The virus pellets were suspended in phosphate-buffered saline (PBS), and the virus suspensions were centrifuged at 10,000 × g for 4 min at 4°C. Final purification was performed by isopycnic centrifugation in CsCl in an SW55TI rotor (Beckman) at 200,000 × g for 40 h at 4°C. Desalting and concentration of virus-containing fractions recovered from CsCl gradients and determination of the protein concentration were performed as previously described (7). The virus titer was determined as described below, and the purified virus suspensions were divided into aliquots (20 μl) and stored at −80°C until used in GTA inactivation assays.
GTA virucidal efficacy testing.
A single stock of purified GTA (reagent grade 1, 25% aqueous solution; Sigma, Saint Quentin Fallavier, France) was used throughout the study. A 10% GTA solution was prepared in PBS, aliquoted, and stored at −80°C. The chemical stability of GTA was checked by UV absorption during storage and before each inactivation assay (2). Virucidal efficacy testing was carried out with a suspension test as previously described (6). Briefly, the purified viral suspensions were mixed with the appropriate GTA dilution in PBS to obtain a final concentration of 0.10% (wt/vol) and incubated at 25°C for 15, 30, 60, and 120 min. All inactivation assays were carried out under the same controlled conditions at 25°C and pH 7.2. GTA activity was stopped by dilution in ice-cold EMEM immediately at the end of the contact periods. Viral infectivity was determined in microtiter plates by the endpoint dilution method with fourfold dilution rate and 11 replicates per dilution for all titration assays. The wells were scored microscopically as positive or negative for virus growth on day 5. The most probable number of cytopathic units (MPNCU) was calculated with an in-house computer program. Details of this method have been reported elsewhere (2).
Statistical techniques.
A cluster analysis was performed with the Centroid method to compare the GTA inactivation processes of the seven enterovirus strains. The test was computed with STATGRAPHICS version 4 for Windows.
Amino acid sequence accession numbers.
The EMBL/GenBank accession numbers of the sequences reported in this paper are AJ241422 to AJ241436.
RESULTS
Characterization of the VP1 sequence in 15 echovirus prototype strains.
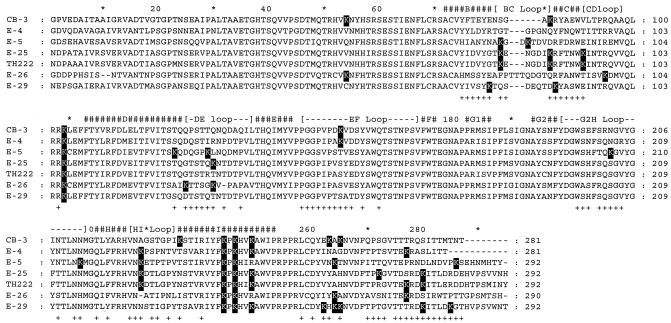
To explore further the role of the lysine residues in the interaction of GTA with the VP1 capsid protein of enteroviruses, the sequence of the protein was determined in 15 echovirus prototype strains (serotypes 1, 3, 4, 5, 7, 13, 14, 15, 19, 20, 24, 26, 29, 31, and 33). The amino acid sequences were aligned with each other and with that of coxsackievirus B3, used as a reference (data not shown). Of the 15 prototype strains analyzed, 4 echovirus strains with major and complementary differences in the location of the lysine residues were selected for GTA efficacy testing (Fig. 1). The echovirus 4 Du Toit strain was chosen because only three of the seven lysine residues present in the VP1 protein were observed in amino acid positions that are putatively exposed on the surface of the virion particle. The echovirus 29 JV-10 strain was chosen because of the presence of two lysine residues, one each in β-strands B and C, and five residues in the C terminus of VP1. The other two echovirus strains (echovirus 5 Noyce and echovirus 26 Coronel prototype strains) were selected because of their complementarity to each other. In the echovirus 5 and 26 prototype strains, two lysine residues were observed within or in the vicinity of the DE loop, and both strains had two residues in the VP1 C terminus. In addition, in the echovirus 5 strain, lysine residues were observed in five other positions: two in the BC loop, two in the G2H loop, and one in the HI loop.
FIG. 1.
Amino acid sequences of the VP1 capsid proteins of five echovirus prototype strains (echoviruses 4, 5, 25, 26, and 29) and one clinical isolate (echovirus 25 TH222) selected on the basis of differences in the number of lysine residues located in exposed regions on the outer surface of the VP1 protein. The sequences were aligned with that of coxsackievirus B3, whose virion structure was solved by X-ray crystallography (22). β-Strands (#) and connecting loops in coxsackievirus B3 were assigned manually. Amino acid residues exposed on the outer surface of the virus particle in coxsackievirus B3 are indicated under the sequences (+). Differences observed in positions containing lysine (K) residues are shown against a black background.
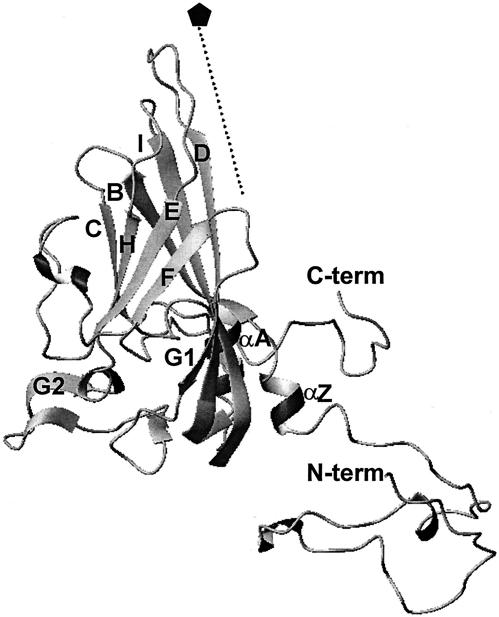
The activity of GTA was tested on the four selected echovirus prototype strains. Three virus controls were also tested by the virucidal test, the coxsackievirus B3 Nancy and echovirus 25 JV-4 prototype strains and the echovirus 25 TH222 clinical isolate. Coxsackievirus B3 was chosen because it is the most closely related enterovirus for which the three-dimensional structure is known (Fig. 2) (22), and the two echovirus 25 strains were chosen because they had been studied previously in our laboratory.
FIG. 2.
Ribbon diagram illustrating the secondary structure elements of the coxsackievirus B3 VP1 protein. The fivefold symmetry axis is shown by a dotted line and labeled with a pentagon. β-Strands (B, C, D, E, F, G1, G2, H, and I) and the N and C termini are labeled. Flanking helices are prefixed with α. The figure was generated with the program MOLMOL (18).
Inactivation kinetics and statistical analysis.
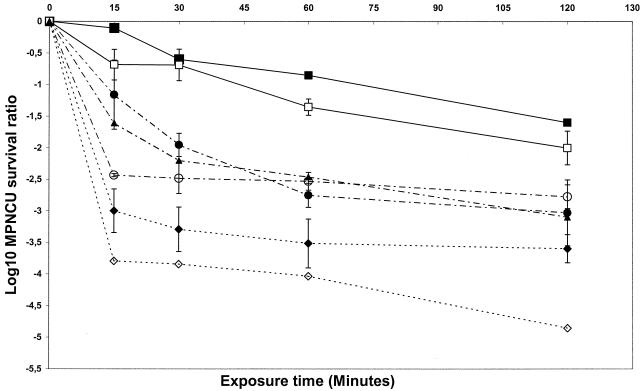
A single virus stock was prepared for each of the seven selected enteroviruses and used throughout the study. The comparison of the seven virus stocks was based on the infectious titer and the total protein concentration. Homogeneity was estimated by the relative infectious titer. The relative infectious titers were 6.5 (echovirus 29 and coxsackievirus B3), 7.0 (echovirus 5, echovirus 25 TH222 clinical isolate, and echovirus 26), 7.4 (echovirus 4), and 7.6 (echovirus 25 JV-4 prototype strain) log10 MPNCU per ml per μg of protein. Kinetics experiments were performed with 20-μl aliquots of the virus stocks to compare the activity of GTA at 0.10%. The virus survival ratio, defined as the reduction in virus concentration and expressed as log10, was plotted versus time of exposure to GTA (Fig. 3). Each plotted point is the mean of two survival ratios (for coxsackievirus B3, echovirus 4, and echovirus 5) or three survival ratios (for echovirus 25 TH222, echovirus 25 JV-4, echovirus 26, and echovirus 29).
FIG. 3.
Kinetics of inactivation of seven enteroviruses by GTA. The strains included one prototype coxsackievirus B3 (▴), five prototype echoviruses (echovirus 4 [▪], echovirus 5 [◊], echovirus 25 [○], echovirus 26 [⧫], and echovirus 29 [□]) and one clinical isolate of echovirus 25, TH222 (•). The virucidal test was performed by a suspension method. The purified virus-containing samples were incubated at 25°C in PBS with 0.10% GTA for various times. Inactivation was stopped by 100-fold dilution in cold EMEM. The survival ratio is the reduction of the viral concentration and is expressed as log10 (T/T0), where T is the titer at elapsed time t and T0 is the titer at the outset of exposure to GTA (time zero). The results are expressed as means ± the standard deviations.
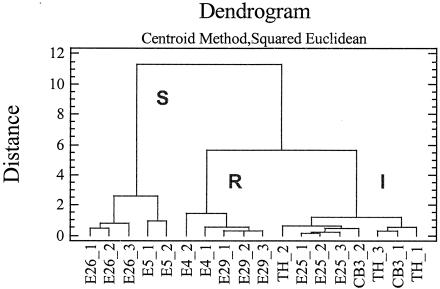
The comparative analysis of the inactivation kinetics suggested that the enteroviruses tested clustered in three groups with different rates of inactivation. The first group comprised echoviruses 4 and 29, which were the most resistant viruses at the concentration of GTA tested. The second group comprised the two echovirus 25 isolates and coxsackievirus B3, which exhibited intermediate sensitivity, and the third group comprised echoviruses 5 and 26, which had the highest sensitivity. These results were supported by statistical analysis of the experimental data. A cluster analysis was performed to determine groups of observations with similar characteristics (Fig. 4). For each virus tested, statistical observation included the virus survival ratios determined at the four exposure times. Of the seven enteroviruses tested, 18 observations were used to analyze the data with the centroid method. Three clusters (sensitive [S], resistant [R], and intermediate [I]) were observed at distance metric 5. The clusters identified by the statistical analysis corresponded to those observed in Fig. 3. The sensitive viruses grouped in the S cluster, whereas the resistant viruses were in the R cluster; viruses with intermediate GTA sensitivity were in the I cluster. The overall results are summarized in Table 1, which shows the numbers of lysine residues and their distribution on putative VP1 secondary structures for the seven enteroviruses investigated.
FIG. 4.
Cluster analysis performed with GTA virucidal test data by the centroid method computed by STATGRAPHICS version 4. The distance metric used was the squared Euclidean distance. Eighteen observations (virus survival ratios at the four exposure times 15, 30, 60, and 120 min) were analyzed. Two or three observations (labeled 1, 2, and 3) were available for each virus. To form the clusters, the procedure began with each observation in a separate group and then combined the two observations which were closest by the centroid method to form a new group. After the distance was computed, based on the squared Euclidean metric between the groups, the two groups that were then closest together were combined. This process was repeated until only one group remained. Viruses with the highest sensitivity to GTA inactivation, viruses with intermediate sensitivity, and those with the greatest resistance were designated clusters S, I, and R, respectively. E, echovirus; CB3, coxsackievirus B3; TH, echovirus 25 TH222.
TABLE 1.
Susceptibility of enteroviruses to inactivation by GTA and locations of lysine residues in VP1 proteina
| GTA sensitivity | Virus strainb | No. of lysine residues in the indicated secondary structural element of VP1
|
Total no. of K residues | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N terminus | B* | BC loop | C* | CD loop | D* | DE loop | EF loop | G2H loop | HI loop | I* | C terminus | |||
| Cluster R | E-4 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 3 | 1 | 7 |
| E-29 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 3 | 5 | 11 | |
| Cluster I | CB-3 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 4 | 2 | 10 |
| TH222 | 0 | 0 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 1 | 3 | 2 | 10 | |
| E-25 | 0 | 0 | 2 | 0 | 2 | 0 | 1 | 0 | 0 | 1 | 3 | 2 | 11 | |
| Cluster S | E-26 | 1 | 0 | 0 | 0 | 2 | 0 | 2 | 0 | 0 | 0 | 2 | 2 | 9 |
| E-5 | 0 | 0 | 2 | 0 | 1 | 1 | 1 | 0 | 2 | 1 | 2 | 2 | 12 | |
β-Strands are marked with an asterisk. The virus strains are grouped according to the data obtained with 0.10% GTA. R, resistance; I, intermediate sensitivity; S, high sensitivity.
E, echovirus; CB3, coxsackievirus B3; TH222, echovirus 25 isolate.
DISCUSSION
This is the first study to report the interaction between specific structures in the enterovirus VP1 capsid protein and GTA, an interaction which results in inactivation of the infectious properties of virus particles. The development of antiviral compounds with activity against picornaviruses has contributed to a greater understanding of the structural and functional properties of the capsid and of the viral cell entry pathways (12, 13, 15). Studies of the mode of virucidal action of chemical disinfectants such as GTA could be of interest in the design of new compounds.
It is generally assumed that the reaction between GTA and proteins does not bear any simple relationship to the number of lysine residues and depends closely on the accessibility of the reactive amino acids over the surface of the protein molecule (17). This is borne out by the results of the GTA virucidal test performed in this study. It is clear that the differences in sensitivity to GTA observed in kinetics experiments are not related to the total number of lysine residues in the VP1 protein. For example, the VP1 protein of echovirus 29, which was among the most resistant strains, contained 11 lysine residues, whereas echovirus 26, with nine lysine residues, was among the most sensitive strains (Table 1).
The comparative analysis of the amino acid sequences and inactivation kinetics provided information about the nature of the VP1 structures, which are the preferential binding sites for GTA. The seven enteroviruses tested clustered into three groups and had significantly different levels of sensitivity to GTA, as supported by the statistical analysis (Fig. 4). In the R cluster, lysine groups were found in β-strands B, C, and I, in the CD, EF, and HI loops, and in the C terminus. Since these two viruses were resistant to GTA inactivation, these VP1 structures are unlikely to be involved in the GTA inactivation process. However, we cannot rule out the possibility that the GTA molecule binds to these structures without altering infectivity. The resistant viruses had another common feature in having no lysine residues in the N terminus, β-strand D, and the BC, DE, and G2H loops. Unlike the viruses in the R cluster, the viruses in the I and S clusters exhibited lysine residues in the BC loop and in the DE loop, respectively. However, echoviruses 25 and 5, which do not belong to the same cluster, both contained two lysine residues in the BC loop and one in the DE loop. The great sensitivity of echovirus 5 could be explained by the presence of two reactive lysines in the G2H loop. In addition, this virus also had one lysine residue in a segment of β-strand D, which is exposed in coxsackievirus B3 (Fig. 2). Overall, the highest level of sensitivity to GTA inactivation correlated with the presence of lysine residues on exposed VP1 regions, the BC, DE, and G2H loops and β-strand D, which are directly accessible for GTA binding. Among these four structures, the DE loop seems to play a major role in the GTA sensitivity of a given virus. This can be seen by comparing TH222 (no lysine in the DE loop), echovirus 25 (one lysine residue), and echovirus 26 (two lysine residues).
The overall results of this study support our initial hypothesis and corroborate that the lysine residues located on the outermost loops of VP1, which cluster at the fivefold axes of symmetry, are major reactive sites for GTA binding. In coxsackievirus B3, of the three BC, DE, and HI loops around the fivefold axes, the HI loop is the innermost, and the DE loop is more prominent than the BC loop (Fig. 2). The high-resolution structures of several enteroviruses showed that the DE loop runs closest and parallel to the fivefold axis and as a result makes extensive interactions with its icosahedral fivefold-symmetry-related DE loops (10, 11, 13, 14, 19, 22). Hence, the size and the orientation of the DE loop could explain why it has more opportunity to interact with GTA than the BC loop. The lysine residues located at the tops of the DE loops are very close to one another and as a consequence are easily accessible to GTA over the surface of the virion. This combined vicinality and accessibility may allow GTA to produce cross-links between the five VP1 molecules at each prominent fivefold peak of the capsid. Such cross-links could account for the higher sensitivity to GTA inactivation observed in echoviruses 25, 26, and 5. This interpretation is supported by previously reported observations. Electrophoretic and immunoblot analyses of the structural proteins of the echovirus 25 prototype strain after treatment of the virions with GTA revealed the presence of complexes involving the VP1 polypeptide, which indicates the formation of intermolecular cross-links (6). In this earlier report, we also demonstrated intratypic differences in echovirus 25 strains, suggesting that, unlike the JV-4 prototype strain, only minor intermolecular cross-links were produced by GTA binding in viral particles of the TH222 isolate.
The general conclusion to emerge from the present study is that outside lysine residues located in the BC and the DE loops are important sites for the virucidal action of GTA on enteroviruses. Although the seven enteroviruses tested are close genetic relatives and belong to the human enterovirus B species, they display significant differences in the loop regions, particularly in the BC loop. Hence, it cannot be excluded that the positions and steric arrangements of individual lysine residues in the loops are involved in GTA reactivity.
Crystallographic studies of the VP1 protein and our current knowledge of the early stages of picornavirus infection suggest that the uncoating and release of the RNA genome require a degree of capsid flexibility. By producing cross-links via lysine residues located on the VP1 loops, GTA probably makes the capsid structure more rigid, and this may prevent viral attachment to a cellular receptor and halt the exit of the RNA genome.
Acknowledgments
We are grateful to Jeffrey Watts for revision of the English manuscript.
The laboratory (EA2148) is supported by grants from the Ministère de l'Education Nationale, de la Recherche et de la Technologie, France.
REFERENCES
- 1.Ayliffe, G. 2000. Decontamination of minimally invasive surgical endoscopes and accessories. J. Hosp. Infect. 45:263-277. [DOI] [PubMed] [Google Scholar]
- 2.Bailly, J. L., M. Chambon, H. Peigue-Lafeuille, H. Laveran, C. De Champs, and D. Beytout. 1991. Activity of glutaraldehyde at low concentrations (less than 2%) against poliovirus and its relevance to gastrointestinal endoscope disinfection procedures. Appl. Environ. Microbiol. 57:1156-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bailly, J. L., M. Chambon, H. Peigue-Lafeuille, and F. Charbonne. 1994. Replication of echo virus type 25 JV-4 reference strain and wild type strains in MRC5 cells compared with that of poliovirus type 1. Arch. Virol. 137:327-340. [DOI] [PubMed] [Google Scholar]
- 4.Bailly, J. L., A. Beguet, M. Chambon, C. Henquell, and H. Peigue-Lafeuille. 2000. Nosocomial transmission of echovirus 30: molecular evidence by phylogenetic analysis of the VP1 encoding sequence. J. Clin. Microbiol. 38:2889-2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chambon, M., J. L. Bailly, and H. Peigue-Lafeuille. 1992. Activity of glutaraldehyde at low concentrations against capsid proteins of poliovirus type 1 and echovirus type 25. Appl. Environ. Microbiol. 58:3517-3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chambon, M., J. L. Bailly, and H. Peigue-Lafeuille. 1994. Comparative sensitivity of the echovirus type 25 JV-4 prototype strain and two recent isolates to glutaraldehyde at low concentrations. Appl. Environ. Microbiol. 60:387-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chambon, M., C. Jallat-Archimbaud, J. L. Bailly, J. M. Gourgand, F. Charbonne, C. Henquell, F. Fuchs, and H. Peigue-Lafeuille. 1997. Comparative sensitivities of Sabin and Mahoney poliovirus type 1 prototype strains and two recent isolates to low concentrations of glutaraldehyde. Appl. Environ. Microbiol. 63:3199-3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang, L. S., S. R. Lin, and C. C. Yang. 2001. Glutaraldehyde cross-linking alters the environment around Trp29 of cobrotoxin and the pathway for regaining its fine structure during refolding. J. Peptide Res. 58:173-179. [DOI] [PubMed] [Google Scholar]
- 9.Cowan, R. E. 1998. Cleaning and disinfection of equipment for gastrointestinal endoscopy. Report of a working party of the British Society of Gastroenterology Endoscopy Committee. Gut 42:585-593. [PMC free article] [PubMed] [Google Scholar]
- 10.Filman, D. J., R. Syed, M. Chow, A. J. Macadam, P. D. Minor, and J. M. Hogle. 1989. Structural factors that control conformational transitions and serotype specificity in type 3 poliovirus. EMBO J. 8:1567-1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Filman, D. J., M. W. Wien, J. A. Cunningham, J. M. Bergelson, and J. M. Hogle. 1998. Structure determination of echovirus 1. Acta Crystallogr. D Biol. Crystallogr. 54:1261-1272. [DOI] [PubMed] [Google Scholar]
- 12.Hadfield, A. T., W. Lee, R. Zhao, M. A. Oliveira, I. Minor, R. R. Rueckert, and M. G. Rossmann. 1997. The refined structure of human rhinovirus 16 at 2.15 Å resolution: implications for the viral life cycle. Structure 5:427-441. [DOI] [PubMed] [Google Scholar]
- 13.Hendry, E., H. Hatanaka, E. Fry, M. Smyth, J. Tate, G. Stanway, J. Santti, M. Maaronen, T. Hyypia, and D. Stuart. 1999. The crystal structure of coxsackievirus A9: new insights into the uncoating mechanisms of enteroviruses. Structure Fold Des. 7:1527-1538. [DOI] [PubMed] [Google Scholar]
- 14.Hogle, J. M., M. Chow, and D. J. Filman. 1985. Three-dimensional structure of poliovirus at 2.9 Å resolution. Science 229:1358-1365. [DOI] [PubMed] [Google Scholar]
- 15.Hogle, J. M. 2002. Poliovirus cell entry: common structural themes in viral cell entry pathways. Annu. Rev. Microbiol. 56:677-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hopwood, D., C. R. Callen, and M. McCabe. 1970. The reactions between glutaraldehyde and various proteins. An investigation of their kinetics. Histochem. J. 2:137-150. [DOI] [PubMed] [Google Scholar]
- 17.Hopwood, D. 1972. Theoretical and practical aspects of glutaraldehyde fixation. Histochem. J. 4:267-303. [DOI] [PubMed] [Google Scholar]
- 18.Koradi, R., M. Billeter, and K. Wuthrich. 1996. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 14:29-32, 51-55. [DOI] [PubMed] [Google Scholar]
- 19.Lentz, K. N., A. D. Smith, S. C. Geisler, S. Cox, P. Buontempo, A. Skelton, J. DeMartino, E. Rozhon, J. Schwartz, V. Girijavallabhan, J. O'Connell, and E. Arnold. 1997. Structure of poliovirus type 2 Lansing complexed with antiviral agent SCH48973: comparison of the structural and biological properties of three poliovirus serotypes. Structure 5:961-978. [DOI] [PubMed] [Google Scholar]
- 20.Martino, M., and A. M. Tamburro. 2001. Chemical synthesis of cross-linked poly(KGGVG), an elastin-like biopolymer. Biopolymers 59:29-37. [DOI] [PubMed] [Google Scholar]
- 21.McDonnell, G., and D. Russell. 1999. Antiseptics and disinfectants: activity, action, and resistance. Clin. Microbiol. Rev. 12:147-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muckelbauer, J. K., M. Kremer, I. Minor, G. Diana, F. J. Dutko, J. Groarke, D. C. Pevear, and M. G. Rossmann. 1995. The structure of coxsackievirus B3 at 3.5 Å resolution. Structure 3:653-667. [DOI] [PubMed] [Google Scholar]
- 23.Peigue-Lafeuille, H., J. L. Bailly, F. Fuchs, M. Chambon, and M. Aymard. 1991. Heterogeneity of capsid proteins of echovirus type 25 wild-type strain and prototype strain, studied by using sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting. J. Clin. Microbiol. 29:1780-1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Russell, A. D. 1994. Glutaraldehyde: current status and uses. Infect. Control Hosp. Epidemiol. 15:724-733. [DOI] [PubMed] [Google Scholar]