Abstract
In extreme thermal environments such as hot springs, phages are the only known microbial predators. Here we present the first study of prokaryotic and phage community dynamics in these environments. Phages were abundant in hot springs, reaching concentrations of a million viruses per milliliter. Hot spring phage particles were resistant to shifts to lower temperatures, possibly facilitating DNA transfer out of these extreme environments. The phages were actively produced, with a population turnover time of 1 to 2 days. Phage-mediated microbial mortality was significant, making phage lysis an important component of hot spring microbial food webs. Together, these results show that phages exert an important influence on microbial community structure and energy flow in extreme thermal environments.
Phages, viruses that infect and kill bacteria, are important components of all known microbial food webs. The influences of phages on ecosystem dynamics are best understood in the context of the marine microbial food web, the consortium of heterotrophic and autotrophic prokaryotes, as well as their predators that inhabit the Earth's oceans and seas. The marine microbial food web regulates the transfer of energy and nutrients to higher trophic levels and greatly influences global carbon and nutrient cycles (6, 32, 43). Heterotrophic production by prokaryotes within the marine microbial food web accounts for ≈50% of the oceanic carbon fixed by photosynthesis every day (5). These heterotrophs, in turn, are controlled in a top-down fashion by protozoa and phages (23, 44). Phages are also important mediators of genetic exchange in the environment via generalized (29, 41, 42) and specialized (1, 21, 50) transduction.
In extreme thermal environments above the upper temperature limit for eukaryotic life, phages are the only known predators of prokaryotes. Despite their potential importance, very little is known about the influences of phages on the microbial communities in these ecosystems. Phage particles in hot springs have been observed by electron microscopy (40), and phages have been cultured on Bacteria and Archaea isolated from these ecosystems (4, 8, 17, 35, 39, 45, 46, 56-58). However, no effort has been made to determine the abundance or dynamics of naturally occurring phage communities or to quantify the effects of these phages on the microbial populations in extreme thermal environments.
Here we show that phages are abundant and active components of hot springs capable of killing a significant proportion of the resident microbial populations. In addition, the resistance of the phage particles to temperature shifts implies that phages can laterally transfer DNA from these extreme environments.
MATERIALS AND METHODS
Direct counts of prokaryotes and VLP.
Prokaryotes (Bacteria and Archaea) and virus-like particles (VLP) were counted by filtering samples fixed in 2% paraformaldehyde onto a 0.02-μm Anodisc (Whatman), staining with SYBR Gold (Molecular Probes, Inc.), and direct counting by epifluorescent microscopy (37). Direct counts of viruses were performed at all the hot springs sampled.
Transmission electron microscopy of hot spring samples.
Samples from Little Hot Creek site 4 were fixed in 2% paraformaldehyde and absorbed onto Formvar-coated copper grids for 2 h. The grids were then stained with 2% uranyl acetate-1% acetic acid for 10 s. Images were collected on Kodak electron film (SO-163) with a Phillips EM410A transmission electron microscope at an accelerating voltage of 60 kV.
Prokaryotic production rates.
All prokaryotic production measurements were performed on water samples from Little Hot Springs site 4 and the outflow of Imperial Spa. Prokaryotic production rates were measured with both [4,5-3H]leucine and [methyl-3H]thymidine incorporation methods (22, 30, 47). For each experiment, triplicate 11-ml water samples were placed into warmed borosilicate scintillation vials. The vials were incubated with 20 nM [4,5-3H]leucine or [methyl-3H]thymidine (Amersham Biosciences) for 2 h in the spring from which they were collected (i.e., ambient temperature). Experiments were also performed in which the water samples were incubated at a lower temperature or with a 0.2% glucose addition in order to determine the effects of temperature or energy supplements on growth rate. Incorporation was stopped by adding trichloroacetic acid (TCA) to a final concentration of 5%. Carryover controls to which 5% TCA was added prior to incubation with the radiolabeled compounds were also performed. The carryover controls were used to determine how much unincorporated radiolabeled [4,5-3H]leucine or [methyl-3H]thymidine was still associated with the samples after processing in the absence of cellular assimilation. At the Imperial Spa site, salmon sperm DNA was added to the samples at the same time as the TCA as a carrier to facilitate precipitation. The TCA precipitates were stored on ice until they were returned to the laboratory, where they were pelleted and washed once with cold 5% TCA and once with 80% ethanol. The washed TCA precipitates were resuspended in scintillation cocktail, vortexed vigorously, and radioassayed. Production rates were calculated with average conversion factors of 2 × 1018 cells per mol of [methyl-3H]thymidine incorporated and 1.5 × 1017 cells per mol of [4,5-3H]leucine incorporated (15, 22, 31). The turnover time of the prokaryotic population was calculated by dividing the average number of prokaryotic cells by the production rate.
Phage production rates.
Phage production was measured at Little Hot Creek sites 3 and 4 and Imperial Spa with fluorescently labeled viruses as tracers (36). Phage particles were concentrated directly from each hot spring site by using 100-kDa tangential flow filters (Amersham Biosciences). Prokaryotes were then removed by filtering the concentrate through a 0.2-μm Sterivex filter (Millipore). The final concentrates contained between 400,000 and 1,600,000% of the natural concentration of viruses (200 to 1,000 liters were concentrated to ≈50 ml). An aliquot of the concentrated viruses was labeled with SYBR Gold (Molecular Probes, Inc.) at a final concentration of 2.5% (vol/vol) for 3.5 h in the dark. After labeling, the fluorescently labeled viruses were diluted into 1 liter of 100-kDa filtrate from the hot spring and reconcentrated with tangential flow filters. This process was repeated three times to ensure that all unbound SYBR Gold was removed. The fluorescently labeled viruses were then added at tracer levels (≈10% of the original total virus concentration) to triplicate dark Nalgene bottles containing 1 liter of unfiltered hot spring water. In addition, fluorescently labeled virus tracers were added to a bottle containing 1 liter of 100-kDa-filtered hot spring water that was incubated in the same manner in order to determine the decay rate of fluorescently labeled viruses in the absence of a microbial community. The bottles with fluorescently labeled virus tracers were then incubated in the hot spring to maintain natural temperatures. Samples were collected at 0, 4.5, and 20.5 h, fixed in paraformaldehyde, and stored in the dark at 4°C until viruses were counted by epifluorescent microscopy. In a pilot experiment, we determined that fluorescently labeled viruses stained with SYBR Gold can be stored in this manner for at least 2 weeks without a noticeable decline in particle number (data not shown). Phage production and decay rates were calculated as described by Noble and Fuhrman, with an average burst size of 20 (36).
Phage stability at different temperatures.
The stability of the phage communities at different temperatures was determined by incubating water samples collected from various hot springs at lower temperatures, on ice (≈0°C), or in a pot of boiling water (≈105°C). Before the various incubations, the water samples were filtered (0.2 μm) to remove the prokaryotes, thus preventing the production of new phage particles. The number of phages in controls that were fixed at time zero were counted, and this value was set at 100%. The experimental samples were incubated for ≈20 h, fixed in 2% paraformaldehyde, and stored in the dark at 4°C until phages were counted by epifluorescent microscopy.
Mitomycin C experiments.
To determine if extremophilic microbes in the hot springs carried prophages that could be induced by DNA-damaging agents, water samples from Little Hot Creek sites 3 and 4 were incubated at their natural temperatures for 1 h with 100 μg of mitomycin C ml−1. Negative control samples without mitomycin C addition were prepared and incubated in parallel. After the incubations, samples were fixed in 2% paraformaldehyde and kept at 4°C until the viruses were counted by epifluorescence microscopy.
RESULTS AND DISCUSSION
Sampling sites.
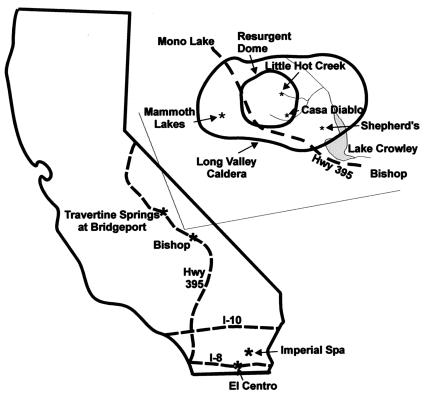
All of the hot spring study sites were located in California (Fig. 1). The Long Valley caldera is a 15- by 30-km oval, volcanically active depression located along the east side of the Sierra Nevada (Fig. 1, inset). The caldera was originally formed 760,000 years ago when the roof above the underlying magma chamber collapsed. Approximately 100,000 years after that eruption, increasing pressure within the magma chamber formed a resurgent dome. The area around the resurgent dome is geothermally active, with a number of hot springs, including Little Hot Creek and Casa Diablo (Fig. 1 and 2). Casa Diablo (37o38.776′ N, 118o51.489′ W) is a large, deep pool with a temperature of 82°C and pH of 8.16. The pool is disturbed every 10 to 15 min by a geyser-like release of water in the deepest part of the spring. The Little Hot Creek site (37o41.465′ N, 118o50.633′ W) consists of numerous springs that flow together to form a stream (Fig. 2). The hottest water (79 to 84°C) is found in four different pools with black bottoms (sites 1 to 3 and 10). One of these pools, Little Hot Creek site 10, was not present when Little Hot Creek was first sampled in July 2001. The pH, temperature, and bottom color (Fig. 2) change as the water flows away from these pools.
FIG. 1.
California hot springs sampled in the study: Travertine Springs (Bridgeport), several hot springs located in the Long Valley caldera (Little Hot Creek, Casa Diablo, and Shepherd's Pool), and Imperial Spa (part of the Desert Hot Springs geothermal field). The line drawing of the caldera was adapted from an image at http://lvo.wr.usgs.gov/images/maps/index.html.
FIG. 2.
Photographs of the hot springs sampled in this study. Direct counts of viruses and prokaryotes were performed at all sites (Imperial Spa, Travertine Springs, Casa Diablo, Shepherd's Pool, and Little Hot Creek). Different sampling sites along the Little Hot Creek system are indicated by numbers on the central overview map, with lines leading to expanded pictures detailing each site. Viral and microbial production measurements were performed at Little Hot Creek sites 3 and 4 and the spigot at the Imperial Spa where the water was emerging from the ground (not the pool).
Most of the work described below was performed at Little Hot Creek sites 3 (82 to 84°C, pH 7.50) and 4 (73 to 74°C, pH 7.70), which are separated by ≈3 m. The surface flow rate between these two sites was ≈0.2 m s−1. A small stream of input water from an independent source also joined the springs right before Little Hot Creek site 4. Shepherd's Pool, located southeast of Little Hot Creek, is a 45°C, ≈1.5-m2 pool that is about 0.5 m deep and contains very large algal mats (Fig. 2). Travertine Springs (38°14′N, 119°12′W) is located near Bridgeport, Calif., and consists of a long stream that runs down the center of the formation to enter a large bathing pool (Fig. 2).
The second major sampling area was Desert Hot Springs geothermal field, located in the Imperial Valley just east of the Salton Sea (Fig. 1 and 2). The shallow aquifer supplying the hot springs is fed by a deep convection system (1.2 to 1.7 km and temperatures of >135°C) (28). The Imperial Spa site was 63°C, with a pH of 7.2. Flow rates at the Imperial Spa site were >100 liters min−1.
VLP abundance in hot springs.
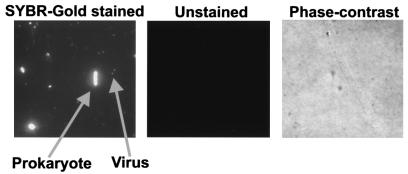
Upon staining with SYBR Gold, prokaryotes and viruses were seen in each of the hot spring samples (Fig. 3, SYBR Gold stained). Controls were performed by filtering down an aliquot of each sample and looking for autofluorescence. No signal was observed in the absence of SYBR Gold stain when slides were examined by epifluorescence microscopy (Fig. 3, unstained panel). The phase-contrast panel in Fig. 3 shows that there were cells in the unstained sample and that the filter was in focus. The presence of free viruses in the hot springs was also confirmed by electron microscopy (Fig. 4). VLP were present in every hot spring pool examined, with concentrations ranging from 0.07 × 106 to 7.0 × 106 VLP ml−1 (Table 1). There were statistically fewer VLP in hot springs with temperatures above the upper limit for eukaryotic life (i.e., >62°C) (12, 13) than in the cooler springs (P < 0.05, Mann-Whitney U test). However, there were a number of exceptions to this general trend, and several high-temperature springs displayed high VLP counts (e.g., ≈3 × 106 VLP ml−1 at Casa Diablo at 82°C and Little Hot Creek site 4 at 73°C). Since the temperature of these springs was greater than the known upper temperature limit for eukaryotic life, the VLP present are probably phages and not viruses that infect eukaryotes.
FIG. 3.
Example of SYBR Gold staining of prokaryotic cells and VLP in the hot springs samples. SYBR Gold stained, typical sample from Little Hot Creek site 4, which was fixed with 2% paraformaldehyde, filtered onto a 0.02-μm Anodisc, stained with SYBR Gold, and viewed by epifluorescent microscopy. Unstained, aliquot of the same sample viewed under epifluorescent microscopy in the absence of SYBR Gold staining. No autofluorescence of the samples was observed. The phase-contrast panel was the same field of view as the unstained sample, viewed under phase contrast to show that the filter was in focus and contained cells.
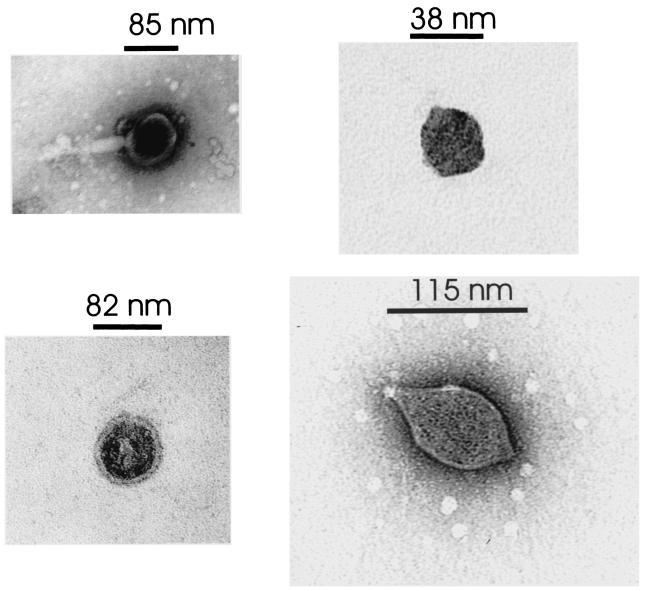
FIG. 4.
Representative electron micrographs of VLP observed in the hot spring water from Little Hot Creek site 4.
TABLE 1.
Number of VLP and prokaryotes in hot springs as determined by epifluorescence microscopya
| Sample site | Date (mo/day/yr) | Temp (°C) | No. of phages ml−1 (105) | No. of prokaryotes ml−1 (105) |
|---|---|---|---|---|
| Casa Diablo | 7/15/01 | 82 | 30 | 5.7 |
| 7/21/02 | 82 | 4.6 | ND | |
| Imperial Spa* | 1/8/03 | 63 | 10.8 | 1.0 |
| LHC site 1 | 7/15/01 | 79 | 2.0 | 0.63 |
| 7/21/02 | 79 | 1.3 | ND | |
| LHC site 10 | 7/21/02 | 78 | 2.8 | ND |
| LHC site 2 | 7/21/02 | 80 | 2.2 | ND |
| LHC site 3* | 7/15/01 | 84 | 3.6 | 1.8 |
| 7/21/02 | 82 | 6.6 | ND | |
| LHC site 4* | 7/15/01 | 73 | 34 | 3.9 |
| 7/21/02 | 74 | 7.0 | 1.4 | |
| LHC site 5 | 7/15/01 | 68 | 19 | 5.3 |
| 7/21/02 | 68 | 5.8 | ND | |
| LHC site 7 | 7/15/01 | 55 | 13 | 3.4 |
| LHC site 8 | 7/15/01 | 40 | 17 | 1.9 |
| 7/21/02 | 57 | 7.1 | ND | |
| LHC site 9 | 7/15/01 | 29 | 68 | ND |
| 7/21/02 | 39 | 30 | ND | |
| Travertine site 1 | 7/15/01 | 59 | 9.4 | ND |
| Travertine site 2 | 7/15/01 | 59 | 6.8 | 1.5 |
| Travertine site 3 | 7/15/01 | 67 | 0.70 | 0.59 |
| Shepherd's Hot Spring | 7/15/01 | 45 | 70 | 12 |
There was great temporal variation in the numbers of VLP observed in some of the hot springs. The number of VLP in Casa Diablo varied by 6.5-fold at the two time points that were sampled (Table 1). On average for all the hot springs, there were 5.2 VLP per prokaryotic cell, which is similar to the range observed in many other environments (2, 3, 7, 9, 10, 14, 18-20, 25-27, 34, 38, 54, 55).
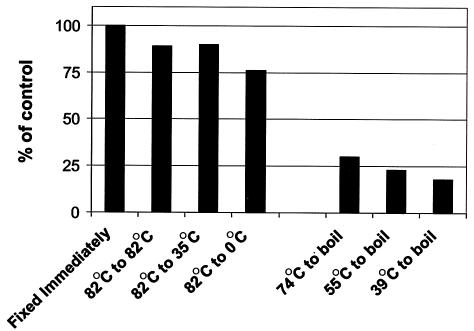
Phage particle stability at different temperatures.
Phage communities from the hottest pools were placed in 50-ml tubes and moved to cooler pools or onto ice. Greater than 75% of the hot spring VLP remained intact even when incubated on ice (Fig. 5). The particles were more sensitive to boiling, with only 18 to 30% of the VLP remaining intact after treatment. The overall resistance to temperature shifts shows that phages can be transported from hot springs to cooler ecosystems. As phages move between these biomes, they may be efficient shuttles of genetic material via transduction. In support of this hypothesis, Chiura has demonstrated that VLP from a hot spring were capable of generalized gene transfer to Escherichia coli and Bacillus subtilis (16).
FIG. 5.
Temperature shift experiments demonstrated that hot spring phage particles were relatively resistant to lower temperatures but sensitive to boiling. Water samples collected from Little Hot Creek site 3 (82°C) were incubated for ≈20 h at various temperatures to determine the stability of the phage particles at different temperatures. Similarly, samples from Little Hot Creek site 4 (74°C), Little Hot Creek site 8 (55°C), and Little Hot Creek site 9 (39°C) were incubated in a pot of boiling water (≈105°C) for ≈20 h. The number of intact phage particles observed by epifluorescent microscopy in the samples that were fixed immediately was set at 100%, and the number of intact VLP after the incubations is expressed as a percentage of that control value.
Production of phages in hot springs.
Phages were produced at a rate of 1.0 × 109 to 1.5 × 109 VLP liter−1 day−1 at both the Imperial Spa and Little Hot Creek sites (Table 2). At these rates, the turnover time for the phage communities was 1.2 to 2.2 days (Table 2). These turnover times are comparable to those of nearshore marine and freshwater lake phage communities (36, 51). Combined with the direct counts, these results show that phage communities are abundant, active, and important components of hot springs.
TABLE 2.
Phage production and decay rates in the Little Hot Creek (LHC) and Imperial Spa hot springsa
| Site (temp, °C) | Viral production rate (109 VLP liter−1 day−1) | Viral decay/ removal rate (109 VLP liter−1 day−1) | Viral community turnover time (days) |
|---|---|---|---|
| LHC site 3 (82) | 1.3 | 1.9 | 1.2 |
| LHC site 4 (74) | 1.0 | 1.5 | 1.7 |
| Imperial Spa (66) | |||
| Expt 1 | 1.1 | 1.9 | 2.2 |
| Expt 2 | 1.5 | 2.1 | 1.7 |
Two independent experiments were performed at the Imperial Spa site.
Induction of prophages by mitomycin C.
To determine if the extremophilic prokaryotes contained prophages that could be induced with DNA-damaging agents, samples were incubated with mitomycin C. This treatment increased the number of VLP by 1.2- and 1.4-fold at Little Hot Creek sites 3 (82°C) and 4 (74°C), respectively, suggesting that lysogeny may be a viable lifestyle in hot springs. By assuming a burst size of 20, an estimated 1 to 9% of the microbes were carrying a prophage. Future studies with longer incubation times need to be performed in order to determine the significance of lysogeny in these environments (53).
Prokaryotic production in hot springs.
While the overall numbers of prokaryotes at both sampling sites were similar (1.0 × 108 to 1.8 × 108 cells liter−1), prokaryotic production at Little Hot Creek site 4 (average, 1.75 × 108 cells liter−1day−1) was approximately 20-fold higher than at the Imperial Spa site (average, 8.0 × 106 cells liter−1 day−1), as determined by [4,5-3H]leucine and [methyl-3H]thymidine incorporation (Table 3). The turnover time for the prokaryotic community at Little Hot Creek site 4 was less than 1 day (0.73 to 0.86 day), while the turnover time for the prokaryotic community at Imperial Spa was 9.28 to 19.98 days (Table 3). The low microbial production rates measured in the Imperial Spa samples may reflect the rapid decrease in temperature as the water moves to the surface, yielding an underestimate of microbial growth (e.g., see the effect of temperature shifts in Table 3). Alternatively, the different values may simply reflect variations between the sampling sites, or it is possible that a portion of the prokaryotic community was unable to incorporate the radiolabeled compounds, leading to an underestimation of prokaryotic production. Energy supplements in the form of d-glucose (0.2%, wt/vol) had no measurable effects on prokaryotic production at the Imperial Spa site. Shifting the community to a lower temperature (66 to 39°C) reduced prokaryotic production by 86%, demonstrating that the microbial community was adapted to growth at higher temperatures. This suggests that the necessary incubations may have underestimated microbial production because the communities were better adapted to grow at higher temperatures and/or pressures.
TABLE 3.
Prokaryotic production in the Little Hot Creek and Imperial Spa hot springsa
| Site (temp, °C) | Prokaryotic production rate (107 cells liter−1 day−1) with:
|
Prokaryotic community turnover time (days) with:
|
||
|---|---|---|---|---|
| Leucine | Thymidine | Leucine | Thymidine | |
| LHC site 4 (74) | 16 | 19 | 0.73 | 0.86 |
| Imperial Spa (66) | ||||
| Expt 1 | 1.1 | 0.5 | 9.28 | 19.98 |
| 0.2% glucose | 1.1 | ND | 9.28 | ND |
| Shifted to 39°C | 0.15 | ND | 8.81 | ND |
[methyl-3H]thymidine and [4,5-3H]leucine incorporation was measured in separate samples. All samples were measured in triplicate. ND, not determined.
Viral effects on hot spring microbial communities.
Phage-mediated killing accounted for 26 to 32% of the microbial community production at Little Hot Creek site 4 and 490 to 1,500% of the total prokaryotic production at Imperial Spa (Table 4). It is typical for these estimates to range widely, reflecting the Lotka-Volterra (predator-prey) behavior of phage-host relationships (49). As mentioned above, prokaryotic production may have been underestimated at the Imperial Spa site. Additionally, we measured prokaryotic production only at the beginning of the fluorescently labeled virus incubations. At the end of the Imperial Spa fluorescently labeled virus experiment, a noticeable increase in the number of prokaryotic cells was observed in the sample. Therefore, it is possible that prokaryotic production increased during the fluorescently labeled virus incubation due to bottle effects. This would result in an overestimation of phage versus bacterial production, explaining the higher phage-mediated mortality observed at this site. For future experiments, we recommend modifying the fluorescently labeled virus technique to measure prokaryotic production throughout the phage production incubations.
TABLE 4.
Effects of phages on prokaryotic populations and carbon cycling in hot springsa
| Site (temp, °C) | % of prokaryotic community killed
|
Carbon released by phage predation (μg of C liter−1 day−1) | |
|---|---|---|---|
| Leucine | Thymidine | ||
| LHC site 4 (74) | 26 | 31 | 1.0 |
| Imperial Spa (66) | |||
| Expt 1 | 490 | 1,055 | 1.0 |
| Expt 2 | 711 | 1,532 | 1.5 |
This study constitutes the first examination of thermophilic phage ecology and provides data that support the conclusion that these phages replicate and are released in geothermal environments. Based on this evidence, we propose that phages are important components of thermophilic communities. A possible source of these phages are the prokaryotes within the microbial mats. However, our data suggest that both bacteria and phages are also replicating in the water. Given the flowing-stream nature of the environments studied, some of the phages may have originated from the subsurface. Future efforts should be aimed at differentiating between these possibilities. Together, these results indicate that phages can play an important role as predators of hot spring microbial communities and influence carbon and nutrient cycling in these extreme environments.
Possible implications for microbial dynamics in the deep hot biosphere.
The results presented here show that phages are important components of surface hot springs. We propose that phages may also be major players in the microbial ecology of all extreme thermal ecosystems, including the deep hot biosphere. Gold estimated that this biome contains ≈1019 liters of pore space (24). If the production rates measured in this study are representative of this biome, then 3.7 × 1029 prokaryotic cells and 3.7 × 1030 phages would be produced in this biosphere every year. These rates are similar to those estimated by a completely independent approach based on data from groundwater (52). Our calculated rates of phage and prokaryotic production are probably an underestimation of the in situ rates (e.g., the source aquifers and/or associated biofilms). Although there are undoubtedly great variances in the pH, temperature, nutrient, and microbial composition of the terrestrial subsurface, at the rates observed in this study, carbon released as a result of new phage production in the deep hot biosphere would be 3.7 gigatones of C year−1. This value is similar to the estimated impact of phages in the world's oceans (≈5 to 10 gigatones of C year−1) (11, 48). Thus, the phage communities found in extreme thermal environments may be important components of global carbon cycles.
Acknowledgments
We thank David Mead and Anca Segall for helpful suggestions, Jan and Gary Ahrens at Imperial Spa RV for their hospitality, and Mark Swift and Lance Washington for help with the electron microscopy.
This work was supported by NSF03-16518 (F.R.), NSF01-09756 (T.S.), the San Diego State University (SDSU) Foundation, and the SDSU College of Sciences. Mya Breitbart was funded by the National Center for Environmental Research (NCER) STAR Program, Environmental Protection Agency.
REFERENCES
- 1.Acheson, D. W. K., J. Reidl, X. Zhang, G. T. Keusch, J. J. Mekalanos, and M. K. Waldor. 1998. In vivo transduction with Shiga toxin 1-encoding phage. Infect. Immun. 66:4496-4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, N. G. (ed.). 1967. Isolation of viral particles from large volumes. Interscience Publishers, New York, N.Y.
- 3.Anderson, N. G., and G. B. Cline. 1967. New centrifugal methods for virus isolation, p. 137-178. In M. K. Maramorosch and H. Koprowski (ed.), Methods in virology, vol. II. Academic Press, New York, N.Y.
- 4.Arnold, H. P., W. Zillig, U. Ziese, I. Holz, M. Crosby, T. Utterback, J. F. Weidmann, J. K. Kristjanson, H. P. Klenk, K. E. Nelson, and C. M. Fraser. 2000. A novel lipothrixvirus, SIFV, of the extremely thermophilic crenarchaeon Sulfolobus. Virology 267:252-266. [DOI] [PubMed] [Google Scholar]
- 5.Azam, F. 1998. Microbial control of oceanic carbon flux: the plot thickens. Science 280:694-696. [Google Scholar]
- 6.Azam, F., T. Fenchel, J. G. Field, J. S. Gray, L. A. Meyer-Reil, and F. Thingstad. 1983. The ecological role of water-column microbes in the sea. Mar. Ecol. Prog. Ser. 10:257-263. [Google Scholar]
- 7.Bergh, Ø., K. Y. Børsheim, G. Bratbak, and M. Heldal. 1989. High abundance of viruses found in aquatic environments. Nature 340:467-468. [DOI] [PubMed] [Google Scholar]
- 8.Blum, H., W. Zillig, J. Mallok, H. Domaley, and D. Prangishvili. 2001. The genome of the archaeal virus SIRV1 has features in common with genomes of eukaryal viruses. Virology 281:6-9. [DOI] [PubMed] [Google Scholar]
- 9.Børsheim, K. 1993. Native marine bacteriophages. FEMS Microbiol. Ecol. 102:141-159. [Google Scholar]
- 10.Børsheim, K. Y., G. Bratbak, and M. Heldal. 1990. Enumeration and biomass estimation of planktonic bacteria and viruses by transmission electron microscopy. Appl. Environ. Microbiol. 56:352-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bratbak, G., M. Heldal, T. F. Thingstad, B. Riemann, and O. H. Haslund. 1992. Incorporation of viruses into the budget of microbial C-transfer. A first approach. Mar. Ecol. Prog. Ser. 83:273-280. [Google Scholar]
- 12.Brock, T. D. 1994. Life at high temperatures. Yellowstone Association for Natural Science, History and Education, Inc., Yellowstone National Park, Wyo.
- 13.Brock, T. D. 2001. The origins of research on thermophiles, p. 1-8. In R. Mancinelli (ed.), Thermophiles: biodiversity, ecology and evolution. Kluwer Academic/Plenum Publishers, New York, N.Y.
- 14.Bruttin, A., F. Desiere, N. D'Amico, J.-P. Guerin, J. Sidoti, B. Huni, S. Lucchini, and H. Brussow. 1997. Molecular ecology of Streptococcus thermophilus bacteriophage infections in a cheese factory. Appl. Environ. Microbiol. 63:3144-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chin-Leo, G., and D. L. Kirchman. 1988. Estimating bacterial production in marine waters from the simultaneous incorporation of thymidine and leucine. Appl. Environ. Microbiol. 54:1934-1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chiura, H. X. 2002. Broad host range xenotrophic gene transfer by virus-like particles from a hot spring. Microb. Environ. 17:53-58. [Google Scholar]
- 17.Chiura, H. X., H. Yamamoto, D. Koketsu, H. Naito, and K. Kato. 2002. Virus-like particle derived from a bacterium belonging to the oldest lineage of the domain Bacteria. Microb. Environ. 17:48-52. [Google Scholar]
- 18.Cochlan, W. P., J. Wikner, G. F. Steward, D. C. Smith, and F. Azam. 1993. Spatial distribution of viruses, bacteria, and chlorophyll a in neritic, oceanic, and estuarine environments. Mar. Ecol. Prog. Ser. 92:77-87. [Google Scholar]
- 19.Danovaro, R., and M. Serresi. 2000. Viral density and virus-to-bacterium ratio in deep-sea sediments of the Eastern Mediterranean. Appl. Environ. Microbiol. 66:1857-1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Demuth, J., H. Neve, and K.-P. Witzel. 1993. Direct electron microscopy study on the morphological diversity of bacteriophage populations in Lake Plussee. Appl. Environ. Microbiol. 59:3378-3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Faruque, S. M., Asadulghani, M. M. Rahman, M. K. Waldor, and D. A. Sack. 2000. Sunlight-induced propagation of the lysogenic phage encoding cholera toxin. Infect. Immun. 68:4795-4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fuhrman, J. A., and F. Azam. 1982. Thymidine incorporation as a measure of heterotrophic bacterioplankton production in marine surface waters: evaluation and field results. Mar. Biol. 66:109-120. [Google Scholar]
- 23.Fuhrman, J. A., and R. T. Noble. 1995. Viruses and protists cause similar bacterial mortality in coastal seawater. Limnol. Oceanogr. 40:1236-1242. [Google Scholar]
- 24.Gold, T. 1992. The deep hot biosphere. Proc. Natl. Acad. Sci. USA 89:6045-6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guixa-Boixareu, N., J. I. Calderón-Paz, M. Heldal, G. Bratbak, and C. Pedrós-Alió. 1996. Viral lysis and bacterivory as prokaryotic loss factors along a salinity gradient. Aquat. Microb. Ecol. 11:21-227. [Google Scholar]
- 26.Hara, S., K. Terauchi, and I. Koike. 1991. Abundance of viruses in marine waters: assessment by epifluorescence and transmission electron microscopy. Appl. Environ. Microbiol. 57:2731-2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hewson, I., J. M. O'Neil, J. A. Fuhrman, and W. C. Dennison. 2001. Virus-like particle distribution and abundance in sediments and overlying waters along eutrophication gradients in two subtropical estuaries. Limnol. Oceanogr. 46:1734-1746. [Google Scholar]
- 28.Hunter, C. 1998. An investigation of the hot mineral spa geothermal resources, Riverside and Imperial counties, California, p. 93-108. In L. Lindsay and W. Hample (ed.), Geology and geothermal resources of the Imperial and Mexicali Valleys. San Diego Association of Geologists, San Diego, Calif.
- 29.Jiang, S. C., and J. H. Paul. 1996. Occurrence of lysogenic bacteria in marine microbial communities as determined by prophage induction. Mar. Ecol. Prog. Ser. 142:27-38. [Google Scholar]
- 30.Kirchman, D., E. K'Nees, and R. Hodson. 1985. Leucine incorporation and its potential as a measure of protein synthesis by bacteria in natural aquatic systems. Appl. Environ. Microbiol. 49:599-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kirchman, D. L. 1992. Incorporation of thymidine and leucine in the subarctic Pacific: application to estimating bacterial production. Mar. Ecol. Prog. Ser. 82:301-309. [Google Scholar]
- 32.Kirchman, D. L. 1994. The uptake of inorganic nutrients by heterotrophic bacteria. Microb. Ecol. 28:255-271. [DOI] [PubMed] [Google Scholar]
- 33.Lee, S. H., and J. A. Fuhrman. 1990. DNA hybridization to compare species compositions of natural bacterioplankton assemblages. Appl. Environ. Microbiol. 56:739-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maranger, R., and D. F. Bird. 1996. High concentrations of viruses in the sediments of Lake Gilbert, Quebec. Microb. Ecol. 31:141-151. [DOI] [PubMed] [Google Scholar]
- 35.Martin, A., S. Yeats, D. Janekovic, W. D. Reiter, W. Aicher, and W. Zillig. 1984. SAV-1 a temperate UV inducible DNA virus-like particle from the archaebacterium Sulfolobus acidocaldarius isolate B-12. EMBO J. 3:2165-2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Noble, R., and J. Fuhrman. 2000. Rapid virus production and removal as measured with fluorescently labeled viruses as tracers. Appl. Environ. Microbiol. 66:3790-3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Noble, R. T., and J. A. Fuhrman. 1998. Use of SYBR Green I for rapid epifluorescence counts of marine viruses and bacteria. Aquat. Microb. Ecol. 14:113-118. [Google Scholar]
- 38.Oren, A., G. Bratbak, and M. Heldal. 1997. Occurrence of virus-like particles in the Dead Sea. Extremophiles 1:143-149. [DOI] [PubMed] [Google Scholar]
- 39.Palm, P., C. Schleper, B. Grampp, S. Yeats, P. McWilliam, W. D. Reiter, and W. Zillig. 1991. Complete nucleotide sequence of the virus SSV1 of the archaebacterium Sulfolobus shibatae. Virology 185:242-250. [DOI] [PubMed] [Google Scholar]
- 40.Patel, B. K. C., J. P. Chalcroft, H. W. Morgan, and R. M. Daniel. 1989. In situ morphologies of some bacteria from New Zealand hot springs. Syst. Appl. Microbiol. 11:187-193. [Google Scholar]
- 41.Paul, J. H. 1999. Microbial gene transfer: an ecological perspective. J. Mol. Microbiol. Biotechnol. 1:45-50. [PubMed] [Google Scholar]
- 42.Paul, J. H., M. E. Frischer, and J. M. Thurmond. 1991. Gene transfer in marine water column and sediment microcosms by natural plasmid transformation. Appl. Environ. Microbiol. 57:1509-1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pomeroy, L. R. 1974. The ocean's food web, a changing paradigm. BioScience 24:499-504. [Google Scholar]
- 44.Proctor, L. M., and J. A. Fuhrman. 1988. Marine bacteriophages and bacterial mortality. EOS J. Immunol. Immunopharmacol. 69:1111-1112. [Google Scholar]
- 45.Rice, G., K. Stedman, J. Snyder, B. Wiedenheft, D. Willits, S. Brumfield, T. McDermott, and M. J. Young. 2001. Viruses from extreme thermal environments. Proc. Natl. Acad. Sci. USA 98:13341-13345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sakaki, Y., and T. Oshima. 1975. Isolation and characterization of a bacteriophage infectious to an extreme thermophile, Thermus thermophilus strain HB-8. J. Virol. 15:1449-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simon, M., and F. Azam. 1989. Protein content and protein synthesis rates of planktonic marine bacteria. Mar. Ecol. Prog. Ser. 51:201-213. [Google Scholar]
- 48.Steward, G. F., J. Wikner, W. P. Cochlan, D. C. Smith, and F. Azam. 1992. Estimation of virus production in the sea: II. Field results. Mar. Microb. Food Webs 6:79-90. [Google Scholar]
- 49.Thingstad, T. F. 2000. Elements of a theory for the mechanisms controlling abundance, diversity, and biogeochemical role of lytic bacterial viruses in aquatic systems. Limnol. Oceanogr. 45:1320-1328. [Google Scholar]
- 50.Waldor, M. K., and J. J. Mekalanos. 1996. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272:1910-1914. [DOI] [PubMed] [Google Scholar]
- 51.Weinbauer, M. G., and M. G. Hofle. 1998. Significance of viral lysis and flagellate grazing as factors controlling bacterioplankton production in a eutrophic lake. Appl. Environ. Microbiol. 64:431-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Whitman, W., D. Coleman, and W. Wiebe. 1998. Prokaryotes: the unseen majority. Proc. Natl. Acad. Sci. USA 95:6578-6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wilcox, R. M., and J. A. Fuhrman. 1994. Bacterial viruses in coastal seawater: lytic rather than lysogenic production. Mar. Ecol. Prog. Ser. 114:35-45. [Google Scholar]
- 54.Wommack, K., and R. Colwell. 2000. Virioplankton: viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 64:69-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wommack, K. E., R. T. Hill, M. Kessel, E. Russek-Cohen, and R. Colwell. 1992. Distribution of viruses in the Chesapeake Bay. Appl. Environ. Microbiol. 58:2965-2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zillig, W., A. Kletzin, C. Schleper, I. Holz, D. Janekovic, J. Hain, M. Lanzendoerfer, and J. K. Kristjansson. 1994. Screening for Sulfolobales, their plasmids and their viruses in Icelandic solfataras. Syst. Appl. Microbiol. 16:619-628. [Google Scholar]
- 57.Zillig, W., D. Prangishvilli, C. Schleper, M. Elferink, I. Holz, S. Albers, D. Janekovic, and D. Gotz. 1996. Viruses, plasmids and other genetic elements of thermophilic and hyperthermophilic Archaea. FEMS Microbiol. Rev. 18:225-236. [DOI] [PubMed] [Google Scholar]
- 58.Zillig, W., K. O. Stetter, S. Wunderl, W. Schulz, H. Priess, and I. Holz. 1980. The Sulfolobus-“Caldariella” group: taxonomy on the basis of the structure of DNA-dependent RNA polymerases. Arch. Microbiol. 135:259-269. [Google Scholar]