Abstract
Background
Retinal dystrophies (RD) are a group of hereditary diseases that lead to debilitating visual impairment and are usually transmitted as a Mendelian trait. Pathogenic mutations can occur in any of the 100 or more disease genes identified so far, making molecular diagnosis a rather laborious process. In this work we explored the use of whole exome sequencing (WES) as a tool for identification of RD mutations, with the aim of assessing its applicability in a diagnostic context.
Methodology/Principal Findings
We ascertained 12 Spanish families with seemingly recessive RD. All of the index patients underwent mutational pre-screening by chip-based sequence hybridization and resulted to be negative for known RD mutations. With the exception of one pedigree, to simulate a standard diagnostic scenario we processed by WES only the DNA from the index patient of each family, followed by in silico data analysis. We successfully identified causative mutations in patients from 10 different families, which were later verified by Sanger sequencing and co-segregation analyses. Specifically, we detected pathogenic DNA variants (∼50% novel mutations) in the genes RP1, USH2A, CNGB3, NMNAT1, CHM, and ABCA4, responsible for retinitis pigmentosa, Usher syndrome, achromatopsia, Leber congenital amaurosis, choroideremia, or recessive Stargardt/cone-rod dystrophy cases.
Conclusions/Significance
Despite the absence of genetic information from other family members that could help excluding nonpathogenic DNA variants, we could detect causative mutations in a variety of genes known to represent a wide spectrum of clinical phenotypes in 83% of the patients analyzed. Considering the constant drop in costs for human exome sequencing and the relative simplicity of the analyses made, this technique could represent a valuable tool for molecular diagnostics or genetic research, even in cases for which no genotypes from family members are available.
Introduction
Retinal dystrophies (RD), comprising the wide spectrum of retinal degeneration, are rare genetic conditions leading to visual deficiency and in some instances to blindness [1]. These diseases affect roughly one person out of 4,000 and are the result of the progressive loss or dysfunction of photoreceptors, the light-sensing cells of the eye [2]. RD are caused by mutations in more than 100 different genes (RetNet; https://sph.uth.edu/Retnet/home.htm). Mutations causing RD can occur in any of these genes, as well as in other disease genes that still await identification, and can be transmitted as dominant, recessive, or X-linked alleles. This very elevated genetic heterogeneity is possibly the highest detected so far among all Mendelian diseases and leads to a very high carrier frequency of heterozygous mutant alleles, i.e. possibly more than 1 in 4–5 individuals [3]. Molecular diagnosis is therefore an extremely daunting task since unrelated patients with the similar clinical presentations are likely to have defects in different genes, each of which with a small chance to be found positive for mutations. In other cases, accurate molecular diagnosis is hampered by limited clinical information, which is critical for efficiently deciding which genes have to be analyzed. To bypass the drawbacks of classical exon-by-exon PCR screenings, associated with very long processing times, the use of mutation- or gene-specific microarrays has been recently adopted [4]–[12], with different results reported by us and others to be largely dependent on the characteristics of the population screened [13]–[19].
Whole exome sequencing (WES) is a procedure that allows the purification by sequence capture of all exonic regions of a genome and their further processing by next-generation sequencing (NGS) [20]. Since the selection of the target regions to be sequenced by WES spans all of the genome, this method is not conventionally used to interrogate a specific DNA fragment or a limited number of genes, but conversely it is adopted to investigate genetic features at a genome scale. Moreover, in virtue of their general applicability, WES protocols and kits have diffused rapidly and have become more and more affordable, to the point of being in some instances less expensive than targeted DNA capture/NGS projects interrogating a lower number of DNA features.
In this work, we evaluate the possibility of using WES as a tool for routine molecular diagnoses in patients with apparently recessive RD. Although we specifically selected families with multiple affected members to allow validation of the findings, we willingly ignored the information related to the pedigree, to simulate the use of this technique for the large majority of people with RD, i.e. isolate patients.
Materials and Methods
Ethics Statement
This study was carried out in accordance with the tenets of the Declaration of Helsinki and was approved by the Institutional Review Boards of the University of Lausanne and the Clinical Research Ethics Committee of the Fundacion Jimenez Diaz. Written informed consent for WES analyses was obtained from the subjects who participated in this study and donated their blood for research. Each individual was anonymized by assigning to him/her a numeric ID; confidentiality and protection of data were ensured by applying international recommendations and current Spanish legislation (Ley de Investigacion Biomedica 14/2007 and LOPD).
Patients
All patients were previously tested and all resulted to be negative for known autosomal recessive retinitis pigmentosa (ARRP) or Leber Congenital Amaurosis (LCA) mutations by microarray screening, based on the Arrayed Primer EXtension (APEX) technology [Asper Ophthalmics, Tartu, Estonia] [21]. Seven families (RP-0137, RP-0298, RP-0461, RP-1102, RP-1116, RP-1164, RP-1263) were also analyzed by whole genome homozygosity mapping using SNP arrays from Affymetrix (Genome Wide Human SNP array 6.0 and GeneChip Human Mapping 500K Array Set) or Illumina (HumanLinkage V Panel Set or Omni Whole Genome arrays HumanCytoSNP-12), as previously described [22]. Affymetrix genotyping services were provided by the Spanish “Centro Nacional de Genotipado” (CEGEN-ISCIII). No significant homozygous regions larger than 1 Mb were found in these families. Only index patients from each family were analyzed by WES, except for family RP-0235, for which all 5 members underwent WES analyses.
Sample Preparation
Genomic DNA was extracted from 7 ml of whole blood using an automated DNA extractor (BioRobot EZ1 Qiagen, Hilden, Germany) following the manufacturer’s instructions.
Library Preparation and Sequencing
Purification of target sequences, library preparation, and NGS processing was performed by BGI-Shenzen (Shenzhen, China), as previously described [23].
Alignment and Analysis of Reads
Reads were mapped to the human genome reference sequence (NCBI build 36.1) and call of variants was performed according to previously published algorithms and procedures [24]. These procedures were performed by BGI, under a fee-for-service agreement.
Filtering Procedures of Detected Variants
DNA variants identified following mapping and evaluation of the reads were assessed to represent actual RD mutations by a 5-step filtering procedure, mainly by the use of simple Perl scripts developed in house and available upon request and/or by the use of standard electronic spreadsheets. First, all variants that were not part of coding sequences or represented silent changes were eliminated, to produce a set of DNA changes that would include only missense, nonsense, and indel events, as well as substitutions affecting exon splicing signals. A second step consisted of selecting only variations found to affect 160 known RD genes. The number of remaining variants was then further reduced by removing known SNPs that were present in dbSNP version 130 (a version that is devoid of data from large-scale endeavors) and having an allelic frequency higher than 0.02. Subsequently, a fourth filtering procedures consisted in ascertaining RD genes carrying two of these variants (or the same variant, but homozygously) to account for the recessive mode of inheritance. At this point, all candidate variants were carefully checked for previous description in the literature and databases, including dbSNP version 137.
PCR and Sanger Sequencing
Variants detected by NGS and suspected to be pathogenic were re-amplified by PCR by using as a template an aliquot of the same DNA samples that were used for WES. Cycling conditions and primers used are listed in Table S1. PCR products were enzymatically purified using ExoSAP-it (USB, Affymetrix, Santa Clara, CA) and sequenced on both strands using the Big Dye Terminator Cycle Sequencing Kit v3.1 Kit (Applied Biosystems). The sequence products were purified on a 96-well multiscreen filter plate (Montage SEQ96 Sequencing Reaction Cleanup Kit, Millipore, Bedford, MA) and resolved on a 3130xl ABI instrument (Applied Biosystems). Chromatograms were interpreted and aligned to the human reference sequence using the STADEN package [25].
Results
Ascertainment of Patients
Twelve Spanish families with recessive RD were ascertained. Diagnoses were based on ophthalmologic examination and pedigree data, according to previously described/established clinical and classification criteria [26]–[30]. Clinical findings of these patients are summarized in Table S2.
Detection and Filtering of DNA Variants
The number of variants remaining after the application of each of the five filtering processes is summarized in Table S3. In short, we detected on average 67,000 DNA variants per genome. Of these, only ∼12,000 represented changes that could potentially alter the sequence or the structure of coding transcripts. Among them, 108 to 143 variants were present in the 160 genes that were previously found to cause RD and 18 to 34 were rare changes (less than 2% in frequency) or were not registered in dbSNP 130. The few variants that were identified as 2 alleles (either as a compound heterozygote with another variant or as a homozygote) in a single RD gene and thus could account for the recessive inheritance pattern were further analyzed.
Detection of Mutations in Known RD Genes
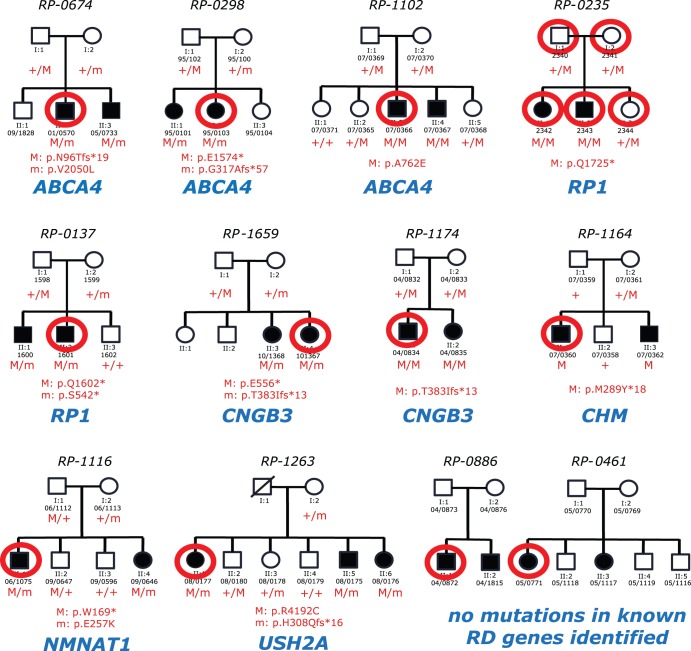
Ten of the 12 index patients analyzed by WES were found to be either homozygous or compound heterozygous for variants in known RD genes that would satisfy our filtering criteria (Table 1, Figure 1).
Table 1. RD mutations identified by WES analyses.
| FAMILY ID | INDEX PATIENT ID | GENE (OMIM entry) | NUCLEOTIDE CHANGE | PROTEIN CHANGE | NOVEL/KNOWN | REFERENCE |
| RP-0674 | 01-0570 | ABCA4 | c.287delA | p.N96Tfs*19 | novel | |
| (601691) | c.6148G>C | p.V2050L | known | [43] | ||
| RP-0298 | 95-0103 | ABCA4 | c.4720G>T | p.E1574* | known | [44] |
| c.950delG | p.G317Afs*57 | novel | ||||
| RP-1102 | 07-0366 | ABCA4 | c.2285C>A (homoz) | p.A762E | known | [45] |
| RP-1164 | 07-0360 | CHM(300390) | c.863dupA | p.M289Y*18 | novel | |
| RP-1263 | 08-0177 | USH2A | c.920_923dupGCCA | p.H308Qfs*16 | known | [46] |
| (608400) | c.12574C>T | p.R4192C | novel | |||
| RP-1659 | 10-1367 | CNGB3 | c.1148delC | p.T383Ifs*13 | known | [31] |
| (605080) | c.1666G>T | p.E556* | novel | |||
| RP-1174 | 04-0834 | CNGB3 | c.1148delC (homoz) | p.T383Ifs*13 | known | [31] |
| RP-0137 | 1601 | RP1 | c.1625C>G | p.S542* | novel | |
| (603937) | c.4804C>T | p.Q1602* | novel | |||
| RP-0235 | 2343 | RP1 | c.5173C>T (homoz) | p.Q1725* | novel | |
| RP-1116 | 06-1075 | NMNAT1 | c.507G>A | p.W169* | known | [47] |
| (608700) | c.769G>A | p.E257K | known | [47] |
Figure 1. Pedigrees of patients analyzed and mutations identified in this work.
The family ID is given above the pedigree, while the individuals’ IDs are indicated below the symbols depicting them. Red circles indicate individuals whose DNA underwent WES analysis. The name of the RD gene identified as causative of the disease is given in blue. M/M, homozygous mutation; M/m compound heterozygous mutations.
More specifically, 3 patients/families were positive for mutations in ABCA4, 2 had mutations in the RP1 gene, and 2 others in the CNGB3 gene. The remainder of these carried variants in CHM, USH2A, and NMNAT1. All of these mutations cosegregated perfectly with the disease in all families, according to a recessive or, in the case of family RP-1164, X-linked pattern of inheritance, as ascertained by exon-PCR and Sanger sequencing (Figure 1). Of the 15 different mutations identified, 8 were never described previously and included 7 changes that were clearly deleterious alleles (frameshifts or nonsenses, Table 1). The remaining mutation was a missense change (c.12574C>T, p.R4192C) identified in the USH2A gene. This was considered pathogenic in conjunction with another clear-cut mutation (c.920_923dupGCCA, p.H308Qfs*16) based on the following reasons. First, cosegregation analysis in the mother and 5 other siblings (2 affected and 3 unaffected) was statistically significant, with a p-value of 0.006 (Figure 1). Second, the mutation was not found in 100 ethnically-matched healthy controls or any other public database, including the one from the 1000 Genomes Project. Lastly, in silico assessment of the consequence of the missense mutation predicted the change to affect the function of the translated protein (SIFT and Polyphen).
All patients identified with disease-associated mutations had no other genes/variants that fulfilled our filtering criteria, except for individual 04/0834. This patient carried the previously-reported homozygous CNGB3 mutation p.T383Ifs*13 [31] but was also found to have two novel missense variations in the USH2A gene. Sanger sequencing failed to detect one of such USH2A variants (thus representing a false negative result) while the presence of the homozygous mutation in CNGB3 was confirmed.
The remaining 2 index patients from families RP-0886 and RP-0461 were not identified with pathogenic RD mutations. They were therefore all considered as patients for whom molecular diagnosis could not be achieved in absence of additional information.
Discussion
Because of the elevated genetic and allelic heterogeneity displayed by hereditary retinal degenerations, molecular diagnoses are in general a rather complicated task. Although precise clinical information or family history can facilitate this procedure, the number of known mutations that medical geneticists have to consider in the screening process is likely in the range of a few thousands, over more than 100 different genes. For example, approximately 600 variants/mutations in the USH2A gene alone have been reported so far (the USH2A mutations database: http://www.lovd.nl/USH2A). Furthermore, novel RD mutations are constantly being discovered, reducing the value of semi-automated analyses interrogating a specific set of DNA variants.
Starting from the evidence that costs of whole-exome sequencing are constantly lowering and that this technique, although not too sensitive, allows an unsupervised analysis of all of the coding sequences of the human genome, we reasoned that WES could in fact be used in routine DNA-based diagnosis of RD. Our results show an 83% success rate, over 12 families with seemingly recessive RD. This high success rate can partially be explained by the nature itself of recessive conditions, for which two mutations have to be present in the same gene to cause disease. This event alone enables a drastic reduction of the noise associated with WES, since the likelihood that two false negative results (e.g. real DNA changes with no pathogenic effects or simple sequencing errors) affect by chance the same gene is very low. In dominantly inherited cases, for which such a filter cannot be applied, there would be a priori no such a way of distinguishing pathogenic variants form rare DNA changes or sequencing false calls. However, although with lower efficiency, WES could in principle still be applied. Important success factors in dominant investigations would be represented by the detection of clear-cut mutations, such as indels and nonsense variants, in known disease genes and the use of control population data analyzed with WES, in order to systematically subtract the noise and enhance the signal throughout the genome. Additionally, the identification of less striking but previously-reported pathogenic changes would also suffice to provide molecular diagnoses to dominant RD cases. For instance, we could efficiently detect a novel frameshift mutation in the CHM gene in a male patient with choroideremia. Since CHM is located on the chromosome X, this patient and his affected brother do in fact carry a single causative mutation, as in individuals affected by autosomal dominant conditions. Isolated RD cases would be, in the vast majority of the circumstances, either unrecognized recessive cases or de novo dominant cases, and therefore could be analyzed by following the same procedures.
NGS has been applied previously to screen for specific forms of RD, with variable results [32]–[34]. For example, NGS provided genetic diagnosis in 36% (36 out of 100) of patients with retinitis pigmentosa [33]. After correcting for the mutation detection rate of NGS and by taking into consideration previously solved cases, the diagnostic yield increased to ∼50%. The comparatively favorable results of the current study that identified 83% of the causes of RD may be attributable to a few specific elements, including the size of the cohort analyzed, the methods and extensiveness of the genetic screenings carried out in the past, the clinical phenotype of the patients, and/or the sequencing/mapping methods of NGS itself.
Another element of note is that out of the 15 mutations identified, 8 (∼50%) were novel, despite the genes in which they were identified were extensively screened for mutations in other cohorts of patients. This finding confirms, unfortunately, the limited value of mutation-based microarrays or sequencing procedures (such as pyrosequencing) in the context of genetics of RD. We also found that wide phenotypic variability of RD and limited clinical information could preclude accurate clinical diagnosis that is essential for current targeted screening methods. In this context, the “screen all RD genes” approach has certainly benefitted three of our cases, initially diagnosed as undefined RD (patient 10/1367), LCA (patient 04/0834), or RP (patient 07/0360), but having mutations in CNGB3 and CHM causing achromatopsia and choroideremia, respectively, that would be missed by ARRP or LCA genotyping microarray. Following such findings, these patients were re-evaluated clinically, and confirmed to have achromatopsia and choroideremia.
Despite the power of WES, validation by Sanger sequencing and, if possible, of additional genetic investigation in family members of the patient remain key elements for the success of this screening technique. The value of familial analyses is particularly evident when we consider the two families for which a DNA diagnosis could not be reached. In these pedigrees, the number of candidate mutations in non-reported RD genes was simply too elevated and, in absence of an additional filtering based on co-segregation of disease and DNA changes in affected relatives, could not be reduced. Moreover, due to limitations that are intrinsic to the exome sequencing procedure, our analyses were underpowered to score DNA copy number variations (CNVs). It is therefore possible that these unsolved cases may in fact carry pathogenic CNVs in RD genes that we were unable to detect.
Methodologically, the procedures used were in general rather simple. We outsourced the sequencing to a large sequencing company, which provided us not only with the raw sequencing reads, but also with an annotated list of variants. These services represent nowadays a standard offer of many private sequencing centers as well as of internal genetic facilities, since most WES protocols and downstream analyses can be streamlined to what closely resembles routine procedures. Computer-based data analysis was also not particularly complicated; we mostly used simple text-parsing scripts and/or commercial spreadsheet programs with filtering capabilities, on common desktop computers.
In our screening we could also gain a few elements of information concerning the molecular pathology of RD mutations. Specifically, we could confirm that missense mutations in USH2A, a gene found to be mutated in patients suffering from the blindness-deafness disease known as Usher syndrome, cause in general retinitis pigmentosa without hearing loss (patient 08–177) [35]. We also confirmed that RP1, a gene that has been for a long time associated with dominant retinitis pigmentosa, can in fact carry recessive mutations, as previously reported [22], [36]–[42].
In summary, in our work we show that whole-exome sequencing could represent a useful tool for molecular diagnosis of hereditary retinal degeneration, a disease for which standard screening procedures still struggle to produce information in a cost- or time-efficient manner. Further reduction in prices and improvement in sensitivity and specificity, already in progress, would probably make of WES the technique of choice for all future routine DNA diagnoses.
Supporting Information
PCR primers used in this study.
(PDF)
Clinical findings of patients.
(XLSX)
Number of variants identified in patients after each filtering process.
(PDF)
Acknowledgments
We would like to acknowledge the Spanish families of patients for taking part in this research and Dr. Carlos Vazquez, Miguel Ángel López, Paola Benaglio and Ma Belen Benavides for their contributions to this work.
Funding Statement
The authors would like to acknowledge the Swiss National Science Foundation (grants 320030-121929 and 310030_138346), the Gebert Rüf Foundation, Switzerland (Rare Diseases - New Technologies grant), the Centre for Biomedical Network Research on Rare Diseases (CIBERER), the FIS (Fondo de Investigaciones Sanitarias, grant PS09/00459) from the Spanish Ministry of Health (ISCIII), the Spanish National Organization of the Blind (ONCE) and the Fighting Blindness Spanish Foundation (FUNDALUCE) grants. MC is supported by CIBERER, SDT by Fundación Conchita Rábago de Jiménez Díaz, AAV by CIBERER and FIS PI09/90047. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Berson EL (1993) Retinitis pigmentosa. The Friedenwald Lecture. Invest Ophthalmol Vis Sci 34: 1659–1676. [PubMed] [Google Scholar]
- 2. Ayuso C, Millan JM (2010) Retinitis pigmentosa and allied conditions today: a paradigm of translational research. Genome Med 2: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nishiguchi KM, Rivolta C (2012) Genes associated with retinitis pigmentosa and allied diseases are frequently mutated in the general population. PLoS One 7: e41902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clark GR, Crowe P, Muszynska D, O’Prey D, O’Neill J, et al.. (2010) Development of a diagnostic genetic test for simplex and autosomal recessive retinitis pigmentosa. Ophthalmology 117: 2169–2177 e2163. [DOI] [PubMed]
- 5. Zernant J, Kulm M, Dharmaraj S, den Hollander AI, Perrault I, et al. (2005) Genotyping microarray (disease chip) for Leber congenital amaurosis: detection of modifier alleles. Invest Ophthalmol Vis Sci 46: 3052–3059. [DOI] [PubMed] [Google Scholar]
- 6. Cremers FP, Kimberling WJ, Kulm M, de Brouwer AP, van Wijk E, et al. (2007) Development of a genotyping microarray for Usher syndrome. J Med Genet 44: 153–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pomares E, Marfany G, Brion MJ, Carracedo A, Gonzalez-Duarte R (2007) Novel high-throughput SNP genotyping cosegregation analysis for genetic diagnosis of autosomal recessive retinitis pigmentosa and Leber congenital amaurosis. Hum Mutat 28: 511–516. [DOI] [PubMed] [Google Scholar]
- 8. Henderson RH, Waseem N, Searle R, van der Spuy J, Russell-Eggitt I, et al. (2007) An assessment of the apex microarray technology in genotyping patients with Leber congenital amaurosis and early-onset severe retinal dystrophy. Invest Ophthalmol Vis Sci 48: 5684–5689. [DOI] [PubMed] [Google Scholar]
- 9. den Hollander AI, Lopez I, Yzer S, Zonneveld MN, Janssen IM, et al. (2007) Identification of novel mutations in patients with Leber congenital amaurosis and juvenile RP by genome-wide homozygosity mapping with SNP microarrays. Invest Ophthalmol Vis Sci 48: 5690–5698. [DOI] [PubMed] [Google Scholar]
- 10. Bowne SJ, Sullivan LS, Koboldt DC, Ding L, Fulton R, et al. (2011) Identification of disease-causing mutations in autosomal dominant retinitis pigmentosa (adRP) using next-generation DNA sequencing. Invest Ophthalmol Vis Sci 52: 494–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Simpson DA, Clark GR, Alexander S, Silvestri G, Willoughby CE (2011) Molecular diagnosis for heterogeneous genetic diseases with targeted high-throughput DNA sequencing applied to retinitis pigmentosa. J Med Genet 48: 145–151. [DOI] [PubMed] [Google Scholar]
- 12. Coppieters F, De Wilde B, Lefever S, De Meester E, De Rocker N, et al. (2012) Massively parallel sequencing for early molecular diagnosis in Leber congenital amaurosis. Genet Med 14: 576–585. [DOI] [PubMed] [Google Scholar]
- 13. Yzer S, Leroy BP, De Baere E, de Ravel TJ, Zonneveld MN, et al. (2006) Microarray-based mutation detection and phenotypic characterization of patients with Leber congenital amaurosis. Invest Ophthalmol Vis Sci 47: 1167–1176. [DOI] [PubMed] [Google Scholar]
- 14. Koenekoop RK, Lopez I, den Hollander AI, Allikmets R, Cremers FP (2007) Genetic testing for retinal dystrophies and dysfunctions: benefits, dilemmas and solutions. Clin Experiment Ophthalmol 35: 473–485. [DOI] [PubMed] [Google Scholar]
- 15. Simonelli F, Ziviello C, Testa F, Rossi S, Fazzi E, et al. (2007) Clinical and molecular genetics of Leber’s congenital amaurosis: a multicenter study of Italian patients. Invest Ophthalmol Vis Sci 48: 4284–4290. [DOI] [PubMed] [Google Scholar]
- 16. Vallespin E, Cantalapiedra D, Riveiro-Alvarez R, Wilke R, Aguirre-Lamban J, et al. (2007) Mutation screening of 299 Spanish families with retinal dystrophies by Leber congenital amaurosis genotyping microarray. Invest Ophthalmol Vis Sci 48: 5653–5661. [DOI] [PubMed] [Google Scholar]
- 17. Coppieters F, Casteels I, Meire F, De Jaegere S, Hooghe S, et al. (2010) Genetic screening of LCA in Belgium: predominance of CEP290 and identification of potential modifier alleles in AHI1 of CEP290-related phenotypes. Hum Mutat 31: E1709–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Avila-Fernandez A, Cantalapiedra D, Aller E, Vallespin E, Aguirre-Lamban J, et al. (2010) Mutation analysis of 272 Spanish families affected by autosomal recessive retinitis pigmentosa using a genotyping microarray. Mol Vis 16: 2550–2558. [PMC free article] [PubMed] [Google Scholar]
- 19. Blanco-Kelly F, Garcia-Hoyos M, Corton M, Avila-Fernandez A, Riveiro-Alvarez R, et al. (2012) Genotyping microarray: mutation screening in Spanish families with autosomal dominant retinitis pigmentosa. Mol Vis 18: 1478–1483. [PMC free article] [PubMed] [Google Scholar]
- 20. Ng SB, Turner EH, Robertson PD, Flygare SD, Bigham AW, et al. (2009) Targeted capture and massively parallel sequencing of 12 human exomes. Nature 461: 272–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tõnisson N, Lõhmussaar E, Metspalu A (2000) Arrayed Primer Extension on the DNA chip method and applications. In: Schena M, editor. Microarray Biochip Technology. Natick, MA: Eaton Publishing Company. 247–263.
- 22. Avila-Fernandez A, Corton M, Nishiguchi KM, Munoz-Sanz N, Benavides-Mori B, et al. (2012) Identification of an RP1 prevalent founder mutation and related phenotype in Spanish patients with early-onset autosomal recessive retinitis. Ophthalmology 119: 2616–2621. [DOI] [PubMed] [Google Scholar]
- 23. Shi Y, Li Y, Zhang D, Zhang H, Lu F, et al. (2011) Exome sequencing identifies ZNF644 mutations in high myopia. PLoS Genet 7: e1002084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zuchner S, Dallman J, Wen R, Beecham G, Naj A, et al. (2011) Whole-exome sequencing links a variant in DHDDS to retinitis pigmentosa. Am J Hum Genet 88: 201–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Staden R, Beal KF, Bonfield JK (2000) The Staden package, 1998. Methods Mol Biol 132: 115–130. [DOI] [PubMed] [Google Scholar]
- 26. Ayuso C, Garcia-Sandoval B, Najera C, Valverde D, Carballo M, et al. (1995) Retinitis pigmentosa in Spain. The Spanish Multicentric and Multidisciplinary Group for Research into Retinitis Pigmentosa. Clin Genet 48: 120–122. [PubMed] [Google Scholar]
- 27. De Laey JJ (1991) Leber’s congenital amaurosis. Bull Soc Belge Ophtalmol 241: 41–50. [PubMed] [Google Scholar]
- 28. Moore AT (1992) Cone and cone-rod dystrophies. J Med Genet 29: 289–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Marmor MF, Aguirre GD, Arden GB (1983) Retinitis pigmentosa. A symposium on terminology and methods of examination. Ophthalmology 90: 126–131. [PubMed] [Google Scholar]
- 30.Traboulsi EI (1998) Genetic diseases of the eye; Oxford, editor.
- 31. Sundin OH, Yang JM, Li Y, Zhu D, Hurd JN, et al. (2000) Genetic basis of total colourblindness among the Pingelapese islanders. Nat Genet 25: 289–293. [DOI] [PubMed] [Google Scholar]
- 32. Wang X, Wang H, Cao M, Li Z, Chen X, et al. (2011) Whole-exome sequencing identifies ALMS1, IQCB1, CNGA3, and MYO7A mutations in patients with Leber congenital amaurosis. Hum Mutat 32: 1450–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Neveling K, Collin RW, Gilissen C, van Huet RA, Visser L, et al. (2012) Next-generation genetic testing for retinitis pigmentosa. Hum Mutat 33: 963–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Licastro D, Mutarelli M, Peluso I, Neveling K, Wieskamp N, et al. (2012) Molecular diagnosis of Usher syndrome: application of two different next generation sequencing-based procedures. PLoS One 7: e43799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rivolta C, Sweklo EA, Berson EL, Dryja TP (2000) Missense mutation in the USH2A gene: association with recessive retinitis pigmentosa without hearing loss. Am J Hum Genet 66: 1975–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu Q, Collin RW, Cremers FP, den Hollander AI, van den Born LI, et al. (2012) Expression of wild-type Rp1 protein in Rp1 knock-in mice rescues the retinal degeneration phenotype. PLoS One 7: e43251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Singh HP, Jalali S, Narayanan R, Kannabiran C (2009) Genetic analysis of Indian families with autosomal recessive retinitis pigmentosa by homozygosity screening. Invest Ophthalmol Vis Sci 50: 4065–4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Khaliq S, Abid A, Ismail M, Hameed A, Mohyuddin A, et al. (2005) Novel association of RP1 gene mutations with autosomal recessive retinitis pigmentosa. J Med Genet 42: 436–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Riazuddin SA, Zulfiqar F, Zhang Q, Sergeev YV, Qazi ZA, et al. (2005) Autosomal recessive retinitis pigmentosa is associated with mutations in RP1 in three consanguineous Pakistani families. Invest Ophthalmol Vis Sci 46: 2264–2270. [DOI] [PubMed] [Google Scholar]
- 40. Chen LJ, Lai TY, Tam PO, Chiang SW, Zhang X, et al. (2010) Compound heterozygosity of two novel truncation mutations in RP1 causing autosomal recessive retinitis pigmentosa. Invest Ophthalmol Vis Sci 51: 2236–2242. [DOI] [PubMed] [Google Scholar]
- 41. Al-Rashed M, Abu Safieh L, Alkuraya H, Aldahmesh MA, Alzahrani J, et al. (2012) RP1 and retinitis pigmentosa: report of novel mutations and insight into mutational mechanism. Br J Ophthalmol 96: 1018–1022. [DOI] [PubMed] [Google Scholar]
- 42. Siemiatkowska AM, Astuti GD, Arimadyo K, den Hollander AI, Faradz SM, et al. (2012) Identification of a novel nonsense mutation in RP1 that causes autosomal recessive retinitis pigmentosa in an Indonesian family. Mol Vis 18: 2411–2419. [PMC free article] [PubMed] [Google Scholar]
- 43. Allikmets R, Singh N, Sun H, Shroyer NF, Hutchinson A, et al. (1997) A photoreceptor cell-specific ATP-binding transporter gene (ABCR) is mutated in recessive Stargardt macular dystrophy. Nat Genet 15: 236–246. [DOI] [PubMed] [Google Scholar]
- 44. Maia-Lopes S, Aguirre-Lamban J, Castelo-Branco M, Riveiro-Alvarez R, Ayuso C, et al. (2009) ABCA4 mutations in Portuguese Stargardt patients: identification of new mutations and their phenotypic analysis. Mol Vis 15: 584–591. [PMC free article] [PubMed] [Google Scholar]
- 45. Aguirre-Lamban J, Riveiro-Alvarez R, Cantalapiedra D, Vallespin E, Avila-Fernandez A, et al. (2008) Gene symbol: ABCA4. Disease: Macular dystrophy. Hum Genet 123: 546. [PubMed] [Google Scholar]
- 46. Weston MD, Eudy JD, Fujita S, Yao S, Usami S, et al. (2000) Genomic structure and identification of novel mutations in usherin, the gene responsible for Usher syndrome type IIa. Am J Hum Genet 66: 1199–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chiang PW, Wang J, Chen Y, Fu Q, Zhong J, et al. (2012) Exome sequencing identifies NMNAT1 mutations as a cause of Leber congenital amaurosis. Nat Genet 44: 972–974. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PCR primers used in this study.
(PDF)
Clinical findings of patients.
(XLSX)
Number of variants identified in patients after each filtering process.
(PDF)