Abstract
A novel salicylate-degrading Streptomyces sp., strain WA46, was identified by UV fluorescence on solid minimal medium containing salicylate; trace amounts of gentisate were detected by high-pressure liquid chromatography when strain WA46 was grown with salicylate. PCR amplification of WA46 DNA with degenerate primers for gentisate 1,2-dioxygenase (GDO) genes produced an amplicon of the expected size. Sequential PCR with nested GDO primers was then used to identify a salicylate degradation gene cluster in a plasmid library of WA46 chromosomal DNA. The nucleotide sequence of a 13.5-kb insert in recombinant plasmid pWD1 (which was sufficient for the complete degradation of salicylate) showed that nine putative open reading frames (ORFs) (sdgABCDEFGHR) were involved. Plasmid pWD1 derivatives disrupted in each putative gene were transformed into Streptomyces lividans TK64. Disruption of either sdgA or sdgC blocked salicylate degradation; constructs lacking sdgD accumulated gentisate. Cell extracts from Escherichia coli DH5α transformants harboring pUC19 that expressed each of the sdg ORFs showed that conversions of salicylate to salicylyl-coenzyme A (CoA) and salicylyl-CoA to gentisyl-CoA required SdgA and SdgC, respectively. SdgA required CoA and ATP as cofactors, while NADH was required for SdgC activity; SdgC was identified as salicylyl-CoA 5-hydroxylase. Gentisyl-CoA underwent spontaneous cleavage to gentisate and CoA. SdgA behaved as a salicylyl-CoA ligase despite showing amino acid sequence similarity to an AMP-ligase. SdgD was identified as a GDO. These results suggest that Streptomyces sp. strain WA46 degrades salicylate by a novel pathway via a CoA derivative. Two-dimensional polyacrylamide gel electrophoresis and reverse transcriptase-PCR studies indicated that salicylate induced expression of the sdg cluster.
Streptomycetes are one of the most abundant microbial genera in soil and are well known for their ability to produce biologically active small molecules, such as antibiotics. The biosynthesis of secondary metabolites has been investigated thoroughly by biochemical and genetic analyses, and many different pathways have been characterized. More than 5% of the genes of streptomycetes appear to be involved in the production of small molecules (3, 46). In contrast, although streptomycetes are saprophytic and known to play important roles in biotransformation and biodegradation in nature, the metabolic pathways have been little studied (7, 26, 32).
Bacterial biodegradation has been extensively characterized in gram-negative bacteria; Pseudomonas species are capable of degrading many organic compounds, including chlorinated polyaromatics (35, 65). Among the gram-positive bacteria, Rhodococcus spp. are effective in the biodegradation of a wide range of xenobiotics (11, 13, 58, 60, 63). Comparative studies have indicated that there are often differences between biodegradation processes in gram-positive and -negative bacteria (11, 17, 54, 58).
Salicylate is widely produced by plants and some bacterial genera, e.g., Pseudomonas and Vibrio spp. (23, 59), and has many biochemical functions (19, 49, 56). The compound and its derivatives (for example, acetylsalicylate) have been employed medicinally for thousands of years. In some bacteria, salicylate is active as a siderophore (23) and also plays a role in gene regulation, such as the expression of antibiotic resistance (10). It has been identified as an intermediate in the bacterial degradation of naphthalene and naphthoquinone (5, 44, 65).
Salicylate degradation has been studied in Pseudomonas spp., and catechol and gentisate are key intermediates in the two known pathways (9, 24, 27, 65). Salicylate is converted to catechol and gentisate by salicylate hydroxylase (4, 6) and salicylate 5-hydroxylase (22, 27), respectively. The gentisate pathway appears to be less common than the catechol pathway. The benzene ring of catechol is cleaved by a 1,2-dioxygenase or 2,3-dioxygenase (1, 40), whereas gentisate is cleaved by gentisate 1,2-dioxygenase (GDO) (8, 18, 21, 62, 67). Investigations of salicylate degradation in gram-positive bacteria, including Streptomyces spp., have revealed the presence of both pathways (26). However, no molecular studies of these processes have been reported. We describe the isolation of salicylate-degrading streptomycetes and the molecular and functional analysis of the associated degradation pathways.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
Streptomyces sp. strain WA46 was isolated from a cornfield in Ohio. Streptomyces sp. strains DSM40278 and DSM40302 were provided by R. Eichenlaub. Streptomyces lividans TK64 came from the laboratory collection. Competent cells of Escherichia coli DH5α and DH10B and plasmids pUC18, pUC19, and pZErO-2 were purchased from Invitrogen. The plasmids used are listed in Table 1. pWHM601, a Streptomyces/E. coli shuttle vector, was provided by C. R. Hutchinson. Minimal salts-yeast extract medium (MSYE) contained (per liter) 0.1 g of (NH4)2SO4, 0.1 g of NaCl, 0.2 g of MgSO4 · 7H2O, 0.01 g of CaCl2, 1 g of K2HPO4, 0.5 g of KH2PO4, and 0.5 g of yeast extract. Freeze broth contained (per liter) 5 g of yeast extract, 10 g of tryptic soy broth (TSB), 5 g of NaCl, 6.3 g of K2HPO4 · 3H2O, 1.8 g of KH2PO4, 0.46 g of sodium citrate, 0.9 g of (NH4)2SO4, and 4.5% glycerol.
TABLE 1.
Plasmids used in this study
| Plasmid | Description | Source or Reference |
|---|---|---|
| pUC18 | Cloning and expression vector | 64 |
| pUC19 | Cloning and expression vector | 64 |
| pZErO-2 | Cloning vector | Invitrogen |
| pWHM601 | 16.5-kb bifunctional cloning and expression vector, Apr, multiple cloning site in lacZα | C. R. Hutchinson |
| pWD1 | 13.5-kb fragment from strain WA46 inserted into pWHM601 | This study |
| pWD2 | 21.4-kb fragment from strain WA46 inserted into pWHM601 | This study |
| pWD4 | 21.3-kb fragment from strain WA46 inserted into pWHM601 | This study |
| pWD1H1 | 17.2-kb HindIII fragment from pWD1 | This study |
| pWD1H2 | 12.9-kb HindIII fragment from pWD1 inserted into pUC18 | This study |
| pZErOH2 | 12.9-kb HindIII fragment from pWD1 inserted into pZErO-2 | This study |
| pWD4E4 | 3.5-kb EcoRI fragment from pWD4 inserted into pUC18 | This study |
| pWD1H2D1 | sdgABCGH-disrupted pWD1H2 | This study |
| pWD1H2D2 | sdgDEF-disrupted pWD1H2 | This study |
| pWD4E4SX | 4.7-kb SstI/XbaI fragment from pWD4E4 | This study |
| pWD1S1 | 4.7-kb SstI fragment from pWD1 inserted into pUC18 | This study |
| pWD1D1 | pWD1 with sdgABCGH disrupted by ClaI | This study |
| pWD1D2 | pWD1 with sdgDEF disrupted by FseI | This study |
| pWD1D4 | pWD1 with sdgD disrupted by deletion at ScaI site | This study |
| pWD1D5 | pWD1 with sdgE disrupted by deletion at SgfI site | This study |
| pWD1D6 | pWD1 with sdgF disrupted by deletion at BamHI and XhoI sites | This study |
| pWD1D7 | pWD1 with sdgC disrupted by deletion at Bsp119I site | This study |
| pWD1D8 | pWD1 with sdgB disrupted by Mva1269I site | This study |
| pWD1D9 | pWD1 with sdgA disrupted by deletion at BglII site | This study |
| pWD1D10 | pWD1 with sdgG disrupted by deletion at Bpu1102I site | This study |
| pWD1D11 | pWD1 with sdgH disrupted by deletion at StuI site | This study |
| p18sdgA-p18sdgF | PCR fragment containing each sdgA-sdgF inserted into pUC18 to express inverted gene | This study |
| p19sdgA-p19sdgF | PCR fragment containing each sdgA-sdgF inserted into pUC19 to express correct gene | This study |
Soil sampling and strain isolation.
Soil samples were collected from various sites and air dried for several days at room temperature; 1 g was suspended in 5 ml of sterile water, mixed by vortexing (30 s), and diluted 100-fold, and 50 μl was spread on ISP4 agar (Difco) containing 20 μg of cycloheximide and 60 μg of benomyl (to inhibit fungal growth) per ml. Following incubation at 30°C for up to 9 days, sporulating colonies were picked and purified on ISP4. Genus identification was performed by PCR with primers for 16S ribosomal DNA (rDNA) and sequencing of the amplified DNA. The primers are shown in Table 2. Amplification conditions were denaturation at 96°C for 3 min; 30 cycles of denaturation at 96°C for 3 min, annealing at 60°C for 30 s, and polymerization at 72°C for 90 s; and final polymerization at 72°C for 10 min. The 1.1-kb amplicon was purified by agarose gel electrophoresis and cloned with a TOPO TA cloning kit (Invitrogen).
TABLE 2.
Primers used in this study
| Primer | Sequence (5′-3′) | Purpose |
|---|---|---|
| 16S-440F | AGCAGGGAAGAAGCG(A/T/C)(A/G)AGT | Forward primer for 16S rDNA |
| 16S-1491R | CGGCTACCTTGTTACGACTTC | Reverse primer for 16S rDNA |
| GDOF1 | CC(C/G)GG(C/G)GAGIT(C/G)GC(C/G)CC | Forward degenerate primer for GDO |
| GDOR3 | GTC(C/G)AI(C/G)CCGTC(C/G)AGCCA | Reverse degenerate primer for GDO |
| GDOF6 | CTCACACCGGCACAGCCA | Forward nested primer for GDO |
| GDOR4 | GCCATCGGCTCGTCGGTGA | Reverse nested primer for GDO |
| sdgAEXF | AAAAGCTTTCATATGACGCGTGAGGGATTCGTG | Forward primer for sdgA expression |
| sdgAEXR | AATCTAGATTGATCACACCGCCTCGACGGAGT | Reverse primer for sdgA expression |
| sdgBEXF | AAAAGCTTTCATATGACCGCCCTGGACCTGCTG | Forward primer for sdgB expression |
| sdgBEXR | AATCTAGATTGATCACGCGTCATCGGATCCTCC | Reverse primer for sdgB expression |
| sdgCEXF | AAAAGCTTTCATATGAAAGTTGCCTGCATCGGCG | Forward primer for sdgC expression |
| sdgCEXR | AATCTAGATTGATCACTTGGTGCCTCCGACGA | Reverse primer for sdgC expression |
| sdgDEXF | AAAAGCTTTCATATGACCGACACCACCAGCGAAC | Forward primer for sdgD expression |
| sdgDEXR | AATCTAGATTGATCACTTGTGTTCTCCTCGGGA | Reverse primer for sdgD expression |
| sdgEEXF | AAAAGCTTTCATATGAAGCTCGCCACCATCCGG | Forward primer for sdgE expression |
| sdgEEXR | AATCTAGATTGATCACTGTGTCTCCCGGCGGCA | Reverse primer for sdgE expression |
| sdgFEXF | AAAAGCTTTCATATGAGCGCACGCCCGGCGGC | Forward primer for sdgF expression |
| sdgFEXR | AATCTAGATTGATCACAGCCACGCGGGCAGTGT | Reverse primer for sdgF expression |
| sdgGEXF | AAAAGCTTTCATATGCTCAGCTTCGGTCACCTC | Forward primer for sdgG expression |
| sdgGEXR | AATCTAGATTGATCATTCCCCTTCGCAGATGCC | Reverse primer for sdgG expression |
| sdgHEXF | AAAAGCTTTCATATGTCACCGCGCAACCAGACC | Forward primer for sdgH expression |
| sdgHEXR | AATCTAGATTGATCAGGCGTGGTGCTTGTCGA | Reverse primer for sdgH expression |
| sdgRF | GATGCTCGTCTTCGAGGGATTC | Forward primer for sdgR detection |
| sdgRR | CTCCTGGAGATTGATGGCGTAG | Reverse primer for sdgR detection |
| sdgAF | CCATGACGACTACGAGTACAAC | Forward primer for sdgA detection |
| sdgAR | CCTGCGTCTCGATCTTGATGTC | Reverse primer for sdgA detection |
| sdgBF | ATCTCAACAACCCGGTAGACAC | Forward primer for sdgB detection |
| sdgBR | CACTTCCGTCTTGTGCAGGATC | Reverse primer for sdgB detection |
| sdgCF | ATCTGGTTCGGCACCACTTACA | Forward primer for sdgC detection |
| sdgCR | GTTCTGGATCCTCTCGACCTTG | Reverse primer for sdgC detection |
| sdgDF | AGGTTCGTCGTCGAAGGCGAA | Forward primer for sdgD detection |
| sdgDR | TGGCGATCTCGAAGACCTTGTC | Reverse primer for sdgD detection |
| sdgEF | TCGTGTGTGTCGGTCTCAACTA | Forward primer for sdgE detection |
| sdgER | GGAGTGATCCGTCCTGCAGATA | Reverse primer for sdgE detection |
| sdgFF | ACGCACTCGTCAATTTGCTGAC | Forward primer for sdgF detection |
| sdgFR | TCGAAGCCTGTGTCGATGTCCA | Reverse primer for sdgF detection |
| sdgGF | TCACCATCATCAACACCTTTGTC | Forward primer for sdgG detection |
| sdgGR | TGCCCTCTACTGCGAACTTCTG | Reverse primer for sdgG detection |
| sdgHF | CTCCTCGATCCAGAGCAAGATC | Forward primer for sdgH detection |
| sdgHR | CTTGTCGAGGAAGTCGGTCATG | Reverse primer for sdgH detection |
Colony screening for salicylate degradation.
Salicylate fluoresces strongly under UV transillumination (312 nm), and this fluorescence is readily detected on MSYE agar. Gentisate could also be detected, but not catechol. Streptomyces strains were incubated on MSYE agar containing 0.1% salicylate at 30°C for several days, and salicylate degradation was detected by UV transillumination. Positive strains were confirmed by loss of salicylate, as detected by high-pressure liquid chromatography (HPLC) analyses following incubation in MSYE containing 1 mM salicylate.
DNA preparation.
Genomic DNA from Streptomyces sp. strain WA46 was prepared as described in Practical Streptomyces Genetics (36) with minor modifications. WA46 was incubated in TSB (20 ml) for 4 days at 30°C, and the cells were collected by centrifugation and frozen at −20°C. After thawing, 5 ml of Tris-EDTA (TE) buffer and 5 ml of methanol-chloroform (1:2) were added and the mixture was shaken gently for 30 min. After centrifugation, the organic and water layers were removed and the organic solvent evaporated at 50°C for 30 min. Five milliliters of TE and 0.5 ml of 1 M Tris-HCl (pH 9.0) containing 10 mg of lysozyme were added, and the mixture was incubated at 37°C overnight. One-tenth volume of 10% sodium dodecyl sulfate-polyacrylamide and 0.01 volume of 10-mg/ml proteinase K were added to the mixture, and incubation continued at 55°C for 4 h. Phenol-chloroform (1:1) was added and the mixture was centrifuged. The aqueous layer was removed and reextracted, and the DNA was precipitated with isopropanol, dissolved in TE, digested with RNase, and precipitated as described above.
Construction of a genomic DNA library of Streptomyces sp. strain WA46.
Genomic DNA was partially digested with Sau3AI and electrophoresed on an agarose gel (0.7%). DNA fragments of 10 to 23 kb were isolated with the Qiaex II gel extraction kit (Qiagen), ligated with BamHI-digested pWHM601 DNA (dephosphorylated with shrimp alkaline phosphatase), and electroporated into E. coli DH10B. Transformants were plated on Luria-Bertani (LB) medium containing 50 μg of apramycin and 100 μg of isopropyl-β-d-thiogalactopyranoside (IPTG) per ml and incubated at 37°C overnight; 3,840 colonies were picked into 384-well microtiter plates with Freeze broth containing 50 μg of apramycin per ml, incubated at 37°C overnight, and stored at −80°C.
PCR amplification of the GDO gene.
The degenerate primers GDOF1 and GDOR3 (Table 2) were designed from the known sequence of GDO. A 198-bp fragment was PCR amplified from strain WA46 chromosomal DNA and cloned by the TOPO TA cloning method. Nucleotide sequence analysis and BLAST search confirmed that the amplicon was derived from a GDO. Based on this sequence, the nested primers GDOF6 and GDOR4 were used to amplify a 163-bp fragment of DNA from strain WA46.
PCR screening for the salicylate degradation cluster.
A sequential PCR procedure with nested GDO primers was employed to screen the strain WA46 clone library. The first PCR step identified which microtiter plate possessed a positive clone. The second PCR step detected a lane in the plate with a positive clone. The third PCR step identified positive clones. Half of the contents of each 384-well plate of the gene library was pooled, and total plasmid DNA was isolated with a Qiagen plasmid minikit (Qiagen). Each DNA isolate was therefore a mixture of 192 different plasmids. The plasmid DNA mixture was then used as the template in the next PCR with the nested primers. The colonies of each pair of lanes were pooled, and total plasmid DNA was extracted by boiling. These DNA preparations were used in the second PCR step. Finally, plasmid DNA was extracted by boiling from each clone in the positive lane, and the positive clones were identified by a final PCR amplification.
Gene disruption procedure.
To identify individual gene functions within the sdgABCDEFGH cluster, various manipulations of pWD1 were carried out (summarized in Table 1). As an example, pWD1D1 (sdgC to sdgH disruption) was prepared as follows. pWD1 DNA was digested with HindIII to generate two fragments, pWD1H1 (a 17.2-kb fragment containing pWHM601 and a 0.7-kb insert) and a 12.9-kb fragment that includes most of the sdg cluster. The 12.9-kb fragment was subcloned into pUC18 to give pWD1H2, which was digested with ClaI to remove the fragment from sdgC to sdgH. A 10.8-kb ClaI fragment containing pUC18 and sdgRDEF was self-ligated to give pWD1H2D1. An 8.1-kb HindIII fragment from pWD1H2D1 was ligated with pWD1H1 to provide pWD1D1. The orientation of the insert in pWD1D1 was confirmed by PCR to be identical to that in pWD1. All other sdg genes were manipulated in the same manner (see Table 1).
Transformation of S. lividans TK64 with plasmid DNA.
Protoplasts of S. lividans TK64 were transformed with plasmid pWD1 and its derivatives and pWD4 by previously described protocols (37). The transformants were regenerated on R2YE agar medium at 30°C for 14 h, overlaid with soft nutrient agar containing apramycin (final concentration, 50 μg/ml), and incubated at 30°C for 2 to 4 days. The size of the insert was confirmed in all cases by colony PCR with appropriate primers.
HPLC analysis of salicylate degradation by transformants.
S. lividans TK64 transformants were incubated in TSB at 30°C for 4 days. Mycelia were centrifuged at 6,000 × g for 10 min and washed with N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid (TES) buffer. The mycelial preparation was incubated in MSYE containing 1 mM salicylate and 50 μg of apramycin per ml at 30°C with shaking. Samples were taken at different incubation times for HPLC analysis.
Preparation of cell extracts.
Strains were incubated in TSB at 30°C for 4 days. Cells were pelleted by centrifugation, washed with TES buffer, and incubated in MSYE containing 1 mM salicylate at 30°C for 4 h with shaking. As a negative control, MSYE without salicylate was used. Cells were collected by centrifugation, washed with TES buffer, suspended in a buffer containing 10 mM Tris-HCl (pH 7.2), 1 mM EDTA, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, and 10% glycerol, and disrupted by sonication (three times for 30 s each). Cell extracts were centrifuged at 100,000 × g for 30 min, and the supernatants were stored at −80°C.
Expression of sdg genes in E. coli.
Individual sdg genes were amplified by PCR with Pfx polymerase, and the start codons were engineered (as necessary) to ATG (Table 2). HindIII and NdeI sites were added at the 5′ side of the start codon in the forward primers, and XbaI and BclII sites were added at the 5′ side of the stop codon in the reverse primers. The amplified sdg genes were digested with HindIII and XbaI, ligated into pUC18, and transformed into E. coli DH5α. The absence of mutations in the amplified sdg genes was confirmed by nucleotide sequencing. Plasmid DNA preparations of different sdg genes were double digested with HindIII and XbaI, and the fragments were ligated into pUC19 and transformed into E. coli DH5α. Constructs were designed so that the sdg genes were expressed only when ligated into pUC19 but not when inverted in pUC18. Transformants were incubated in LB medium containing 100 μg of ampicillin per ml at 37°C overnight, centrifuged, and incubated with LB medium containing 100 μg of IPTG per ml at room temperature for 4 h. After incubation, cells were collected by centrifugation, washed with TES buffer, and resuspended in buffer containing 10 mM Tris-HCl (pH 7.2), 1 mM EDTA, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, and 10% glycerol. Cells were disrupted by sonication (three times for 30 s each), and the extracts were centrifuged at 100,000 × g for 30 min; the supernatants were stored at −80°C.
Enzyme assays.
The conversion of salicylate to salicylyl-CoA was performed in a buffer containing 75 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 0.1 mM salicylate, 0.125 mM coenzyme A (CoA), and 1 mM ATP, to which 50 μl of cell extract was added, and the mixture (1 ml) was incubated at room temperature. Samples were analyzed by HPLC at different incubation times to detect salicylate and salicylyl-CoA.
Determination of the conversion to gentisate was performed similarly. NADH was added to a final concentration of 1 mM together with 50 μl of cell extract, and the mixture was incubated at room temperature. Samples were analyzed by HPLC for salicylyl-CoA, gentisyl-CoA, and gentisate. The conversion from gentisate to maleylpyruvate (GDO catalysis) was assayed by incubating 50 μl of cell extract in a buffer containing 0.1 mM phosphate (pH 7.4) and 0.1 mM gentisate at room temperature. GDO catalysis was monitored by the increase in absorbance at 330 nm, as described by Zhou et al. (67).
HPLC analysis.
Culture media were filtered and diluted five times with water, and 10 ml was applied to an HPLC column (Beckman Ultrasphere, 46 by 75 mm). Cell extract reaction mixtures were injected without dilution. HPLC conditions for salicylate, gentisate, salicylyl-CoA, and gentisyl-CoA separation were as follows: temperature, 40°C; UV, 215 nm; flow rate, 1 ml/min. Three different elution conditions were used. (i) In the methanol-0.1% phosphoric acid method of gradient elution, we used 25% methanol at 0 min to 60% methanol at 6 min. Under these conditions, the retention times of salicylate and gentisate are 5.2 min and 2.1 min, respectively. (ii) In the acetonitrile (MeCN)-0.1% phosphoric acid method of gradient elution, we used 0% MeCN at 0 min to 55% MeCN at 6 min. The retention times for gentisate, salicylate, gentisyl-CoA, salicylyl-CoA, and CoA were 4.0, 5.5, 4.0, 4.6, and 3.0 min, respectively. (iii) In the MeCN-0.1% phosphoric acid method of gradient elution, we used 0% MeCN at 0 min to 30% MeCN at 6 min. The retention times of gentisate, gentisyl-CoA, and CoA were 4.8, 5.3, and 3.5 min, respectively.
Proteome analysis.
Two-dimensional polyacrylamide gel electrophoresis and liquid chromatographic (LC)/mass spectroscopic (MS) analyses of cell extracts were performed by the Proteome Centre, University of Victoria, Victoria, British Columbia, Canada.
Preparation of total RNA and RT-PCR.
Liquid cultures of strain WA46 were incubated and induced by salicylate as described under “Preparation of cell extracts”. After induction, total RNA was isolated by published procedures (38). DNase digestion was carried out until no band could be amplified by PCR. The cDNA was prepared with random hexamers and the ThermoScript reverse transcription (RT)-PCR system (Invitrogen) with total RNA. RT-PCR was performed with gene-specific primers for each sdg gene, as shown in Table 2. DNA amplicons were obtained from sdgR (442 bp), sdgA (451 bp), sdgB (533 bp), sdgC (501 bp), sdgD (587 bp), sdgE (570 bp), sdgF (304 bp), sdgG (285 bp), and sdgH (394 bp).
Comparative studies of salicylate-degrading Streptomyces spp.
Detection of salicylate-degrading genes among different Streptomyces spp. was performed by PCR with the primer sets shown in Table 2.
DNA sequencing.
BigDye terminator cycle sequencing with an ABI300 automatic sequencer was done at the Nucleic Acid and Protein Sequencing Unit of the University of British Columbia; 27,521 bp of contiguous sequence was obtained from three plasmids, pWD1, pWD2, and pWD4. Open reading frames (ORFs) were identified by FramePlot 2.3.2 (31) (www.nih.gov.jp/≈jun/cgi-bin/frameplot.pl).
Nucleotide sequence accession number.
The complete sequence obtained from plasmids pWD1, pWD2, and pWD4 has been deposited in the DNA Data Bank of Japan with accession no. AB112586.
RESULTS
Isolation of salicylate-degrading streptomycetes.
Soil extracts and strains were plated on MSYE containing 0.1% salicylate and examined under a UV light source. Streptomyces sp. strain WA46 degraded salicylate and 4-hydroxybenzoate but not benzoate, 3-hydroxybenzoate, gentisate, protocatechuate, or catechol; only trace amounts of gentisate were detected after incubation with salicylate. A 16S rDNA sequence analysis showed 99% sequence similarity to Streptomyces resistomycificus; the two strains differed in morphology, growth characteristics, and ability to utilize salicylate.
Cloning the salicylate degradation genes.
A library of Sau3A-digested fragments of strain WA46 genomic DNA was made in the Streptomyces/E. coli shuttle vector pWHM601 in E. coli DH10B. A sequential PCR procedure with a set of degenerate primers designed from the known GDO sequence was applied successfully to identify three E. coli transformants, pWD1, pWD2, and pWD4, from a 3,840-clone library; this required less than 100 PCRs (see Materials and Methods).
Salicylate degradation by S. lividans TK64 transformants.
E. coli DH10B transformants carrying pWD1 (13.5 kb), pWD2 (21.4 kb), or pWD4 (21.3 kb) showed no evidence of salicylate degradation in liquid or solid medium. When pWD1 carrying the 13.5-kb insert was introduced into S. lividans TK64, which is deficient in salicylate degradation, it conferred the ability to degrade salicylate and produce trace amounts of gentisate during growth on salicylate, as shown for Streptomyces sp. strain WA46.
DNA sequence analysis of salicylate degradation genes.
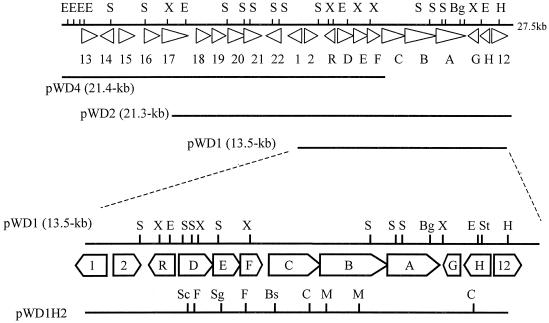
The strain WA46 DNA fragments in pWD1, pWD2, and pWD4 were sequenced, and 22 putative ORFs in a 27,521-bp stretch of DNA were identified, as shown in Fig. 1. pWD1 contained 12 putative ORFs, of which a cluster of nine genes (sdg genes) appeared to be responsible for the ability of this plasmid to confer salicylate degradation on S. lividans transformants, as demonstrated by growth studies and BLAST analysis. The coordinates, G+C content, product size, putative function, and related genes for the sdg-encoded proteins are shown in Table 3. The ORFs identified have a high G+C content typical of streptomycetes, and 7 of the 22 appear to have GTG start codons. In the cluster, two groups, sdgDEF and sdgCBA, have short overlapping reading frames that may indicate translational coupling between neighboring genes. This arrangement appears to be a common feature in streptomycetes (57).
FIG. 1.
Physical map of sdg gene cluster from Streptomyces sp. strain WA46. Bg, BglII; Bs, Bsp119I; C, ClaI; E, EcoRI; F, FseI; H, HindIII; M, Mva1269I; S, SstI; Sc, ScaI; Sg, SgfI; St, StuI; X, XhoI.
TABLE 3.
Identification of sdg gene functions
| Gene | Proposed function | Start-stop (positions and codons) | G+C % at 3rd position | Product size (no. of residues; kDa) | Related gene (accession number [% identity]) (reference) |
|---|---|---|---|---|---|
| sdgR | Transcriptional regulator | 16472ATG-15698TGA | 71.3 (88.0) | 257; 27.5 | Operon regulator (Arthrobacter keyseri 12B, AAK16539 [36%]) (14) |
| Regulatory protein (Terrabacter sp. strains DBF63, BAB55883 [36%]) (33) | |||||
| PcaR (Rhodococcus opacus ICP, AAC38247 [26%]) (17) | |||||
| sdgD | Gentisate 1,2-dioxygenase | 16591GTG-17667TGA | 71.2 (92.2) | 358; 39.7 | Gentisate 1,2-dioxygenase (Corynebacterium glutamicumATCC 13032, BAC00417 [60%]) |
| 1,2-Dioxygenase (Corynebacterium efficiensYS-314, BAC19670 [58%]) | |||||
| Gentisate 1,2-dioxygenase (Ralstonia solanacearum, CAD14789 [38%]) (51) | |||||
| sdgE | Hydrolase? | 17664GTG-18494TGA | 69.8 (91.0) | 276; 29.9 | 2-Hydroxyhepta-2,4-diene-1,7-dioate isomerase (Bacillus halodurans C-125, BAB05719 [49%]) (55) |
| 2-Keto-4-pentenoate hydratase/2-oxohepta-3-ene-1,7-dioic acid hydratase (catechol pathway) (Corynebacterium glutamicumATCC 13032, BAC00416 [44%]) | |||||
| Conserved hypothetical protein (Corynebacterium efficiensYS-314, BAC19669 [46%]) | |||||
| sdgF | Unknown | 18491GTG-19216TGA | 72.7 (85.1) | 241; 25.9 | Hypothetical protein (Corynebacterium glutamicumATCC 13032, BAC00415 [30%]) |
| sdgC | Salicylyl-CoA 5-hydroxylase | 19233ATG-20786TGA | 69.6 (90.0) | 517; 56.6 | Bifunctional hydroxylase/oxidoreductase (S. coelicolor A3(2), CAC44594 [42%]) (3) |
| Oxydoreductase protein (Ralstonia solanacearum, CAD15528 [41%]) (51) | |||||
| 2-Aminobenzoyl-CoA monooxygenase/reductase (Azoarcus evansii, AAL02063 [40%]) (52) | |||||
| sdgB | Salicylyl-CoA synthetase | 20783GTG-22840TGA | 73.4 (86.4) | 685; 70.2 | Acetyl-CoA synthetase (Halobacterium sp. strain NRC-1, NP 444202 [31%]) (45) |
| Acyl-CoA synthetase (Salmonella enterica serovar Typhi CT18, CAD05835 [24%]) (47) | |||||
| Acetyl-CoA synthetase (Salmonella enterica serovar Typhimurium LT2, AAL21545 [24%]) (41) | |||||
| sdgA | Salicylyl-AMP ligase | 22830ATG-24491TGA | 70.8 (88.6) | 553; 59.3 | Aromatic AMP ligase MxcE (Stigmatella aurantiaca Sg a15, AAG31128 [48%]) (53) |
| Pristinamycin I synthetase I (S. pristinaespiralis, CAA67140 [49%]) (12) | |||||
| Salicyl-AMP ligase PchD (Pseudomonas aeruginosa, AAD55799 [44%]) (50) | |||||
| sdgG | Unknown | 24925ATG-24554TGA | 65.3 (89.5) | 123; 13.5 | Hypothetical transmembrane protein (Ralstonia solanacearum, CAD15552 [39%]) (51) |
| sdgH | Hydrolase? | 25804ATG-25010TGA | 72.1 (92.1) | 264; 27.7 | EtbD1 (Rhodococcus sp. strain RHA1, BAA31163 [33%]) (63) |
| EtbD2 (Rhodococcus sp. strain RHA1, BAA31164 [33%]) (63) | |||||
| Putative 3-oxoadipate enol-lactone hydrolase/4-carboxymuconolactone decarboxylase [S. coelicolor A3(2), CAB45571] [30%] (3) |
SdgR was similar to several known regulatory proteins, including PhtR and PcaR, which are involved in the bacterial biodegradation of hydroxyaromatics (13, 14). We suggest that sdgR may play a role in the control of salicylate degradation in Streptomyces sp. strain WA46. SdgA showed similarity to benzoate-AMP ligases from Stigmatella aurantiaca Sga15 (53) and Streptomyces pristinaespiralis (12) and to salicylyl-AMP ligase from Pseudomonas aeruginosa (50), which catalyze the addition of AMP to an aromatic substrate. SdgB had amino acid similarity to acetyl-CoA synthetases and thus appears to be involved in CoA addition in the degradation of salicylate by strain WA46.
Although SdgC showed sequence similarity to several large bifunctional enzymes, the relationship was limited to the amino-terminal region; it was similar to hydroxylases from Streptomyces avermitilis (46) and Streptomyces coelicolor (3) and VioD from Chromobacterium violaceum (2). Since gentisate is an intermediate in salicylate degradation in strain WA46, we propose that SdgC participates in aromatic hydroxylation and is likely the salicylate 5-hydroxylase required for the conversion of salicylate to gentisate.
SdgD is similar in sequence to the GDOs from two Corynebacterium spp. and Ralstonia solanacearum (67). SdgE shows similarity to enzymes from Bacillus halodurans C-125 (55) and Corynebacterium glutamicum ATCC 13032, which metabolize hepta-1,7-dienoates such as maleylpyruvate. SdgH is similar to the 2-hydroxy-6-oxohepta-2,4-dienoate hydrolase from Rhodococcus sp. strain RHA1 (54). Therefore, it is suggested that SdgE and SdgH are required to metabolize short-chain fatty acids such as maleylpyruvate. No function could be assigned to SdgF and SdgG based on sequence comparisons. Complete nucleotide sequencing of pWD2 and pWD4 revealed at least 10 additional ORFs that encode putative regulatory components, transporters, and dehydrogenases (Fig. 1).
Gene disruption studies.
To confirm the assignments listed above, each of the sdg ORFs in pWD1 was disrupted by partial deletion and transformed into S. lividans TK64, and the transformants were analyzed for product formation after incubation in the presence of salicylate (see Materials and Methods). Transformants carrying pWD1D1, pWD1D7, and pWD1D9 did not degrade salicylate. These results suggest that sdgA and sdgC participate in the conversion from salicylate to gentisate as the first step of salicylate degradation. On the other hand, transformants carrying pWD1D2 and pWD1D4 accumulated gentisate. Thus, sdgD would appear to be involved in the degradation of gentisate. Since no intermediates of salicylate degradation were detected by HPLC analysis of sdgB, sdgE, sdgF, sdgG, and sdgH disruptants, we are not able to assign functions to their translation products at this time.
Formation of salicylyl-CoA from salicylate is catalyzed by SdgA.
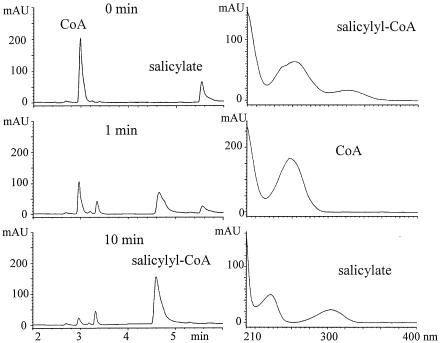
After each of the putative ORFs was expressed in E. coli, cell extracts were made and incubated in reaction mixtures containing potential substrates, and the products were examined by HPLC. Extracts of E. coli DH5α/p19sdgA produced a new HPLC peak in the presence of ATP concomitant with a decrease in salicylate and CoA (Fig. 2). On the basis of the UV spectrum and MS analysis indicating the expected m/z 908 [M+Na-H]−, this product was shown to be salicylyl-CoA (61). Cell extracts of E. coli DH5α/p18sdgA incubated similarly did not produce salicylyl-CoA. Thus, although the BLAST analysis implies that SdgA is an AMP ligase, experimental data indicate that SdgA is essential for the production of salicylyl-CoA. The substrate specificity of SdgA was tested with benzoate, 3-hydroxybenzoate, 4-hydroxybenzoate, gentisate, 2-aminobenzoate, salicylamide, salicylaldoxime, and 2-hydroxyphenyl acetate in the place of salicylate, but no CoA adducts were detected.
FIG. 2.
Conversion of salicylate to salicylyl-CoA by SdgA. At 1 and 10 min after the reaction was started, the reaction mixture was analyzed by HPLC. With the elution conditions given in the text (ii; MeCN-0.1% phosphoric acid; gradient elution, 0% MeCN at 0 min to 55% MeCN at 6 min), salicylate was detected at 5.5 min, salicylyl-CoA was detected at 4.6 min, and CoA was detected at 3.0 min. Salicylyl-CoA was produced by SdgA, as shown by the decrease in CoA and salicylate. ATP was required for this conversion. mAU, milli-arbitrary units.
SdgC catalyzes the conversion of salicylyl-CoA to gentisyl-CoA.
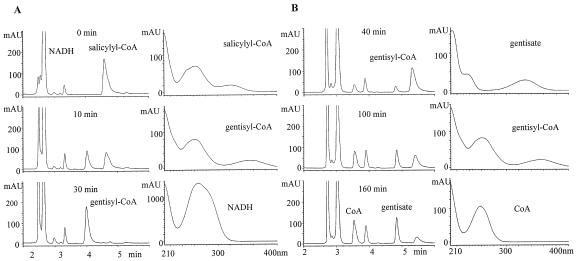
Cell extracts of E. coli DH5α/p19sdgC were incubated with salicylyl-CoA obtained in the reactions mentioned above. Analysis by HPLC revealed a novel peak at 4.0 min in the presence of NADH with a reduction in the amount of salicylyl-CoA (Fig. 3A). The peak at 4.0 min decreased on continued incubation, with the generation of gentisate (4.8 min) and CoA (3.5 min) (Fig. 3B). Cell extracts of E. coli DH5α/p18sdgC did not catalyze this conversion; neither NADPH nor flavin adenine dinucleotide could replace NADH in these reactions.
FIG. 3.
(A) Conversion of salicylyl-CoA to gentisyl-CoA by SdgC. At 10 and 30 min after addition of NADH and SdgC, the reaction mixture was analyzed by HPLC. With the elution conditions given in the text (ii; MeCN-0.1% phosphoric acid; gradient elution, 0% MeCN at 0 min to 55% MeCN at 6 min), salicylyl-CoA was detected at 4.6 min and gentisyl-CoA was detected at 4.0 min. Gentisyl-CoA was generated by SdgC, as shown by decreasing salicylyl-CoA. NADH was required for this conversion. (B) Gentisate and CoA generation from gentisyl-CoA. With the elution conditions given in the text (iii; MeCN-0.1% phosphoric acid; gradient elution, 0% MeCN at 0 min to 30% MeCN at 6 min), gentisate was detected at 4.8 min, gentisyl-CoA was detected at 5.3 min, and CoA was detected at 3.5 min. Gentisyl-CoA was hydrolyzed spontaneously; gentisate and CoA were generated from the conversion of salicylyl-CoA to gentisyl-CoA by the action of SdgC. mAU, milli-arbitrary units.
SdgA, SdgC, ATP, CoA, and NADH are necessary to convert salicylate to gentisate, and the direct conversion of salicylate to gentisate by SdgC was not observed. This suggests strongly that SdgC is a salicylyl-CoA 5-hydroxylase.
SdgD is a GDO.
UV spectral analysis of the products of incubation of cell extracts of E. coli DH5α/p19sdgD with gentisate indicated a shift in the UV spectrum from 320 nm (gentisate) to 330 nm (maleylpyruvate) (62, 67); E. coli DH5α/p18sdgD did not exhibit this conversion. We conclude that SdgD is a GDO. The substrate specificity was strong, since salicylate, catechol, protocatechuic acid, homogenetisate, 2,3-dihydroxybenzoate, or 5-aminosalicylate could not substitute for gentisate in the ring cleavage reaction.
Regulation of expression of the sdg gene cluster by salicylate.
Streptomyces sp. strain WA46 was incubated in liquid medium with salicylate; cell extracts were prepared and analyzed for the activity of the pathway enzymes. Extracts of induced cells possessed GDO activity, and maleylpyruvate was generated. Noninduced cell extracts did not show this activity. However, neither CoA ligation nor hydroxylation of salicylate was detected. Thus, it appears that SdgD (GDO) production is induced by salicylate in strain WA46.
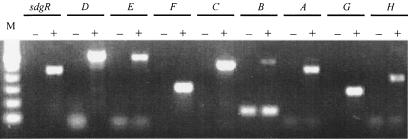
The proteome of salicylate-induced cell extracts was examined by two-dimensional polyacrylamide gel electrophoresis analysis; although several spots appeared, only one protein spot could be identified at ≈30 kDa and pI 4.7. The protein was excised and digested with trypsin, and the peptides were subjected to LC-MS analysis, revealing sequences corresponding to the proposed amino acid sequence of SdgE (results not shown). RT-PCR studies of mRNA production in the presence and absence of salicylate revealed that transcription of the sdg genes occurred only in the presence of salicylate (Fig. 4).
FIG. 4.
RT-PCR showed that salicylate induced all sdg genes. The expected fragment sizes of sdgR, sdgD, sdgE sdgF, sdgC, sdgB, sdgA, sdgG, and sdgH are 442, 587, 570, 304, 501, 533, 451, 285, and 394 bp, respectively. −, no induction; +, salicylate induction. Lane M, 100-bp ladder. The 150-bp band in the absence of induction with sdgB is nonspecific.
Comparison of sdg clusters from different salicylate-degrading streptomycetes.
Streptomyces sp. strain WA18, which also degraded salicylate via gentisate, was isolated from the same soil sample as strain WA46 and is closely related; the 16S rDNA sequences of the two strains differ by one nucleotide base. The growth properties of strain WA46 were different from those of WA18 (results not shown). PCRs carried out with primers for the strain WA46 sdg genes identified a related gene cluster in strain WA18 (results not shown). Grund et al. described the isolation and characterization of two Streptomyces strains, DSM40278 and DSM40302, which degrade salicylate via gentisate and catechol, respectively (26). The presence of all sdg genes except sdgF and sdgH was detected in strain DSM40278, while only sdgG could be detected in strain DSM40302.
DISCUSSION
Salicylate is produced by many plants and microbes and is widely distributed in nature. It has a variety of biological activities, most notably its role in human medicine as a member of an important anti-inflammatory drug class. Salicylate acts as a regulatory molecule for bacteria (49). The biosynthesis and natural degradation pathways of this simple organic molecule have been well studied. In this work, we have identified a salicylate degradation cluster (sdgABCDEFGHR) in Streptomyces sp. strain WA46 and shown that salicylate is initially converted to gentisate via salicylyl-CoA and gentisyl-CoA (Fig. 2 and 3A). Salicylate is presumably converted to salicylyl-CoA via salicylyl-AMP by the action of SdgA and SdgB. AMP ligases are key components in the biosynthesis of antibiotics and siderophores, in which specific substrates are activated by AMP addition (12, 20, 34, 50, 53); the activated adduct binds to the thiol group of biosynthetic enzymes or to an acyl carrier protein. We were unable to detect salicylyl-AMP in Streptomyces sp. strain WA46; salicylyl-CoA was made in the presence or absence of SdgB. We speculate that in the heterologous hosts S. lividans TK64 and E. coli DH5α, spontaneous conversion of salicylyl-AMP to salicylyl-CoA occurs. It has been suggested that acyl-AMP is an intermediate in acyl-CoA synthesis by propionyl-CoA synthetase (30). Taking into account the possibility of translational coupling of SdgA and SdgB, we propose that salicylate is converted to salicylyl-AMP by SdgA and subsequently to salicylyl-CoA by SdgB in strain WA46; if correct, this is the first report of the participation of an AMP ligase in aerobic biodegradation of salicylate.
Gentisyl-CoA cleavage in strain WA46 may or may not be enzyme catalyzed, because gentisyl-CoA appears to undergo spontaneous hydrolysis under the reaction conditions used in our studies. Salicylyl-CoA was also slowly hydrolyzed to salicylate and CoA in the reaction mixture, although salicylyl-CoA was more stable than gentisyl-CoA (data not shown). The instability of salicylyl-CoA could be due to the ortho-hydroxy group (phenolate-assisted water attack) (61). The additional hydroxy group on gentisate may explain the greater lability of gentisyl-CoA compared to salicylyl-CoA.
Although gentisate was converted to maleylpyruvate by SdgD, further degradation of maleylpyruvate could not be detected. Two pathways for maleylpyruvate degradation have been reported. One is direct cleavage to maleate and pyruvate (29, 48), and the other is isomerization to fumarylpyruvate, followed by hydrolysis to fumarate and pyruvate (67). SdgE and SdgH may be involved, but we are unable to make definitive functional assignment based on sequence comparisons. The pathway of degradation of maleylpyruvate by strain WA46 remains to be revealed.
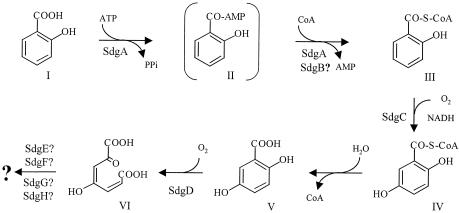
With the above facts in mind, we propose a novel salicylate degradation pathway in Streptomyces sp. strain WA46, as shown in Fig. 5. Catechol and gentisate are the common intermediates in salicylate degradation, but direct fission (28) and CoA addition have also been identified (4). In the direct fission pathway, salicylate is converted to 2-oxohepta-3.5-dienedioate by the action of a GDO with a broad substrate range. On the other hand, CoA adduct formation occurs anaerobically in the pseudomonad-like strain S100, in which salicylate is converted to salicylyl-CoA in the presence of CoA and ATP, followed by reduction of the aromatic ring (4). Another mechanism of CoA addition occurs in the aerobic degradation of salicylate by Rhodococcus sp. strain B4, in which the conversion of salicylate to gentisate requires CoA, ATP, and NADPH (25). In Streptomyces sp. strain WA46, the conversion from salicylate to gentisate via CoA addition takes place under aerobic conditions, and NADH is required instead of NADPH.
FIG. 5.
Proposed pathway of salicylate degradation by Streptomyces sp. strain WA46. I, salicylate; II, salicylyl-AMP; III, salicylyl-CoA; IV, gentisyl-CoA; V, gentisate; VI, maleylpyruvate. Salicylyl-AMP is suggested to be an intermediate in the conversion of salicylate to salicylyl-CoA. SdgB rather than SdgA may be involved in the conversion from salicylyl-AMP to salicylyl-CoA.
CoA addition is performed by CoA ligase in the presence of ATP and CoA, releasing AMP (4, 15, 16, 39, 42, 43, 66). For example, in benzoate degradation, CoA addition has been found under both anaerobic and aerobic conditions. In most cases of degradation involving CoA adducts, the CoA is not removed as acetyl-CoA until the final step. This does not appear to be the case for salicylate degradation in Streptomyces sp. strain WA46.
The BLAST results showed that the most significant protein sequence similarities for SdgD, SdgE, and SdgF were found in both Corynebacterium glutamicum ATCC 13032 and Corynebacterium efficiens YS-314. The G+C content of strain WA46 sdgDEF is 71.2%, while that of the two Corynebacterium strains is approximately 60%, with the average G+C contents of the third codon position being 89.4% and 60%, respectively. The order of the genes was identical in the three organisms. A putative transcription regulation gene similar to sdgR is found upstream of the gene encoding GDO in the Corynebacterium strains.
Although salicylate induced the production of GDO in strain WA46, gentisate degradation was not observed; only salicylate was degraded when strain WA46 was incubated with both salicylate and gentisate. These results might imply that gentisate does not enter the cells.
Comparisons of the salicylate degradation genes of several Streptomyces strains showed that the gentisate pathway is associated with related gene clusters. Thus, Streptomyces sp. strain DSM40278 possesses a homolog of the sdgABCDEGR gene cluster which uses the CoA addition pathway for salicylate degradation. This may be the preferred pathway for salicylate degradation in streptomycetes. The streptomycetes are high-G+C (>70%) microbes of cosmopolitan distribution, well known for their ability to produce biologically active small molecules such as antibiotics. They have been largely ignored for their biotransformation capabilities; however, we suggest that the streptomycetes and related high-G+C bacteria have great potential for the discovery and application of novel and efficient biodegradation and biocatalytic processes.
Acknowledgments
We thank C. R. Hutchinson for pWHM601and R. Eichenlaub for Streptomyces sp. strains DSM40278 and DSM40302. We are grateful to D. Davies for preparation of the manuscript.
This work was funded by Kaken Pharmaceutical Co. Ltd., Tokyo, Japan, and the National Science and Engineering Research Council of Canada.
REFERENCES
- 1.An, H. R., H. J. Park, and E. S. Kim. 2001. Cloning and expression of thermophilic catechol 1,2-dioxygenase gene (catA) from Streptomyces setonii. FEMS Microbiol. Lett. 195:17-22. [DOI] [PubMed] [Google Scholar]
- 2.August, P. R., T. H. Grossman, C. Minor, M. P. Draper, I. A. MacNeil, J. M. Pemberton, K. M. Call, D. Holt, and M. S. Osburne. 2000. Sequence analysis and functional characterization of the violacein biosynthetic pathway from Chromobacterium violaceum. J. Mol. Microbiol. Biotechnol. 2:513-519. [PubMed] [Google Scholar]
- 3.Bentley, S. D., K. F. Chater, A. M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 4.Bonting, C. F., and G. Fuchs. 1996. Anaerobic metabolism of 2-hydroxybenzoic acid (salicylic acid) by a denitrifying bacterium. Arch. Microbiol. 165:402-408. [DOI] [PubMed] [Google Scholar]
- 5.Bosch, R., E. García-Valdés, and E. R. Moore. 2000. Complete nucleotide sequence and evolutionary significance of a chromosomally encoded naphthalene-degradation lower pathway from Pseudomonas stutzeri AN10. Gene 245:65-74. [DOI] [PubMed] [Google Scholar]
- 6.Bosch, R., E. R. B. Moore, E. García-Valdés, and D. H. Pieper. 1999. NahW, a novel, inducible salicylate hydroxylase involved in mineralization of naphthalene by Pseudomonas stutzeri AN10. J. Bacteriol. 181:2315-2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chow, K. T., M. Pope, and J. Davies. 1999. Characterization of a vanillic acid non-oxidative decarboxylation gene cluster from Streptomyces sp. D7. Microbiology 145:2393-2403. [DOI] [PubMed] [Google Scholar]
- 8.Chua, C. H., Y. Feng, C. C. Yeo, H. E. Khoo, and C. L. Poh. 2001. Identification of amino acid residues essential for catalytic activity of gentisate 1, 2-dioxygenase from Pseudomonas alcaligenes NCIB 9867. FEMS Microbiol. Lett. 204:141-146. [DOI] [PubMed] [Google Scholar]
- 9.Civilini, M., M. de Bertoldi, and G. Tell. 1999. Molecular characterization of Pseudomonas aeruginosa 2NR degrading naphthalene. Lett. Appl. Microbiol. 29:181-186. [DOI] [PubMed] [Google Scholar]
- 10.Cohen, S. P., S. B. Levy, J. Foulds, and J. L. Rosner. 1993. Salicylate induction of antibiotic resistance in Escherichia coli: activation of the mar operon and a mar-independent pathway. J. Bacteriol. 175:7856-7862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dean-Ross, D., J. D. Moody, J. P. Freeman, D. R. Doerge, and C. E. Cerniglia. 2001. Metabolism of anthracene by a Rhodococcus species. FEMS Microbiol. Lett. 204:205-211. [DOI] [PubMed] [Google Scholar]
- 12.de Crécy-Lagard, V., V. Blanc, P. Gil, L. Naudin, S. Lorenzon, A. Famechon, N. Bamas-Jacques, J. Crouzet, and D. Thibaut. 1997. Pristinamycin I biosynthesis in Streptomyces pristinaespiralis. J. Bacteriol. 179:705-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Di Gennaro, P., E. Rescalli, E. Galli, G. Sello, and G. Bestetti. 2001. Characterization of Rhodococcus opacus R7, a strain able to degrade naphthalene and o-xylene isolated from a polycyclic aromatic hydrocarbon-contaminated soil. Res. Microbiol. 152:641-651. [DOI] [PubMed] [Google Scholar]
- 14.Eaton, R. W. 2001. Plasmid-encoded phthalate catabolic pathway in Arthrobacter keyseri 12B. J. Bacteriol. 183:3689-3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Egland, P. G., J. Gibson, and C. S. Harwood. 1995. Benzoate-coenzyme A ligase, encoded by badA, is one of three ligases able to catalyze benzoyl-coenzyme A formation during anaerobic growth of Rhodopseudomonas palustris on benzoate. J. Bacteriol. 177:6545-6551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elshahed, M. S., V. K. Bhupathiraju, N. Q. Wofford, M. A. Nanny, and M. J. McInerney. 2001. Metabolism of benzoate, cyclohex-1-ene carboxylate, and cyclohexane carboxylate by Syntrophus aciditrophicus strain SB in syntrophic association with H2-using microorganisms. Appl. Environ. Microbiol. 67:1728-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eulberg, D., S. Lakner, L. A. Golovleva, and M. Schlömann. 1998. Characterization of a protocatechuate catabolic gene cluster from Rhodococcus opacus 1CP: evidence for a merged enzyme with 4-carboxymuconolactone-decarboxylating and 3-oxoadipate enol-lactone-hydrolyzing activity. J. Bacteriol. 180:1072-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng, Y., H. E. Khoo, and C. L. Poh. 1999. Purification and characterization of gentisate 1,2-dioxygenases from Pseudomonas alcaligenes NCIB 9867 and Pseudomonas putida NCIB 9869. Appl. Environ. Microbiol. 65:946-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feys, B. J., and J. E. Parker. 2000. Interplay of signaling pathways in plant disease resistance. Trends Genet. 16:449-455. [DOI] [PubMed] [Google Scholar]
- 20.Fiedler, H. P., P. Krastel, J. Muller, K. Gebhardt, and A. Zeeck. 2001. Enterobactin: the characteristic catecholate siderophore of Enterobacteriaceae is produced by Streptomyces species. FEMS Microbiol. Lett. 196:147-151. [DOI] [PubMed] [Google Scholar]
- 21.Fu, W., and P. Oriel. 1998. Gentisate 1,2-dioxygenase from Haloferax sp. D1227. Extremophiles 2:439-446. [DOI] [PubMed] [Google Scholar]
- 22.Fuenmayor, S. L., M. Wild, A. L. Boyes, and P. A. Williams. 1998. A gene cluster encoding steps in conversion of naphthalene to gentisate in Pseudomonas sp. strain U2. J. Bacteriol. 180:2522-2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gaille, C., P. Kast, and D. Haas. 2002. Salicylate biosynthesis in Pseudomonas aeruginosa. J. Biol. Chem. 277:21768-21775. [DOI] [PubMed] [Google Scholar]
- 24.Goetz, F. E., and L. J. Harmuth. 1992. Gentisate pathway in Salmonella typhimurium: metabolism of m-hydroxybenzoate and gentisate. FEMS Microbiol. Lett. 76:45-49. [DOI] [PubMed] [Google Scholar]
- 25.Grund, E., B. Denecke, and R. Eichenlaub. 1992. Naphthalene degradation via salicylate and gentisate by Rhodococcus sp. strain B4. Appl. Environ. Microbiol. 58:1874-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grund, E., C. Knorr, and R. Eichenlaub. 1990. Catabolism of benzoate and monohydroxylated benzoates by Amycolatopsis and Streptomyces spp. Appl. Environ. Microbiol. 56:1459-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hickey, W. J., G. Sabat, A. S. Yuroff, A. R. Arment, and J. Pérez-Lesher. 2001. Cloning, nucleotide sequencing, and functional analysis of a novel, mobile cluster of biodegradation genes from Pseudomonas aeruginosa strain JB2. Appl. Environ. Microbiol. 67:4603-4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hintner, J.-P., C. Lechner, U. Riegert, A. E. Kuhm, T. Storm, T. Reemtsma, and A. Stolz. 2001. Direct ring fission of salicylate by a salicylate 1,2-dioxygenase activity from Pseudaminobacter salicylatoxidans. J. Bacteriol. 183:6936-6942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hopper, D. J., P. J. Chapman, and S. Dagley. 1968. Enzyme formation of d-malate. Biochem. J. 110:798-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horswill, A. R., and J. C. Escalante-Semerena. 2002. Characterization of the propionyl-CoA synthetase (PrpE) enzyme of Salmonella enterica: residue Lys592 is required for propionyl-AMP synthesis. Biochemistry 41:2379-2387. [DOI] [PubMed] [Google Scholar]
- 31.Ishikawa, J., and K. Hotta. 1999. FramePlot: a new implementation of the frame analysis for predicting protein-coding regions in bacterial DNA with a high G+C content. FEMS Microbiol. Lett. 174:251-253. [DOI] [PubMed] [Google Scholar]
- 32.Iwagami, G. S., K. Yang, and J. Davies. 2000. Characterization of the protocatechuic acid catabolic gene cluster from Streptomyces sp. strain 2065. Appl. Environ. Microbiol. 66:1499-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kasuga, K., H. Habe, J. S. Chung, T. Yoshida, H. Nojiri, H. Yamane, and T. Omori. 2001. Isolation and characterization of the genes encoding a novel oxygenase component of angular dioxygenase from the gram-positive dibenzofuran-degrader Terrabacter sp. strain DBF63. Biochem. Biophys. Res. Commun. 283:195-204. [DOI] [PubMed] [Google Scholar]
- 34.Keating, T. A., C. G. Marshall, and C. T. Walsh. 2000., Vibriobactin biosynthesis in Vibrio cholerae: VibH is an amide synthase homologous to nonribosomal peptide synthetase condensation domains. Biochemistry 39:15513-15521. [DOI] [PubMed] [Google Scholar]
- 35.Khan, A. A., and S. K. Walia. 1991. Expression, localization, and functional analysis of polychlorinated biphenyl degradation genes cbpABCD of Pseudomonas putida. Appl. Environ. Microbiol. 57:1325-1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Preparation and analysis of genomic and plasmid DNA, p. 162-170. In Practical Streptomyces genetics. The John Innes Foundation, Norwich, England.
- 37.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Introduction of DNA into Streptomyces, p. 229-252. In Practical Streptomyces genetics. The John Innes Foundation, Norwich, England.
- 38.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. RNA methods, p. 353-360. In Practical Streptomyces genetics. The John Innes Foundation, Norwich, England.
- 39.Koch, J., W. Eisenreich, A. Bacher, and G. Fuchs. 1993. Products of enzymatic reduction of benzoyl-CoA, a key reaction in anaerobic aromatic metabolism. Eur. J. Biochem. 211:649-661. [DOI] [PubMed] [Google Scholar]
- 40.Lee, J., K. R. Min, Y. C. Kim, C. K. Kim, J. Y. Lim, H. Yoon, K. H. Min, K. S. Lee, and Y. Kim. 1995. Cloning of salicylate hydroxylase gene and catechol 2,3-dioxygenase gene and sequencing of an intergenic sequence between the two genes of Pseudomonas putida KF715. Biochem. Biophys. Res. Commun. 211:382-388. [DOI] [PubMed] [Google Scholar]
- 41.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 42.Mohamed, M. E.-S. 2000. Biochemical and molecular characterization of phenylacetate-coenzyme A ligase, an enzyme catalyzing the first step in aerobic metabolism of phenylacetic acid in Azoarcus evansii. J. Bacteriol. 182:286-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mohamed, M. E.-S., A. Zaar, C. Ebenau-Jehle, and G. Fuchs. 2001. Reinvestigation of a new type of aerobic benzoate metabolism in the proteobacterium Azoarcus evansii. J. Bacteriol. 183:1899-1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muller, U., and F. Lingens. 1988. Degradation of 1,4-naphthoquinones by Pseudomonas putida. Biol. Chem. Hoppe Seyler 369:1031-1043. [DOI] [PubMed] [Google Scholar]
- 45.Ng, W. V., S. P. Kennedy, G. G. Mahairas, B. Berquist, M. Pan, H. D. Shukla, S. R. Lasky, N. S. Baliga, V. Thorsson, J. Sbrogna, S. Swartzell, D. Weir, J. Hall, T. A. Dahl, R. Welti, Y. A. Goo, B. Leithauser, K. Keller, R. Cruz, M. J. Danson, D. W. Hough, D. G. Maddocks, P. E. Jablonski, M. P. Krebs, C. M. Angevine, H. Dale, T. A. Isenbarger, R. F. Peck, M. Pohlschroder, J. L. Spudich, K.-W. Jung, M. Alam, T. Freitas, S. Hou, C. J. Daniels, P. P. Dennis, A. D. Omer, H. Ebhardt, T. M. Lowe, P. Liang, M. Riley, L. Hood, and S. DasSarma. 2000. Genome sequence of Halobacterium species NRC-1. Proc. Natl. Acad. Sci. USA 97:12176-12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Omura, S., H. Ikeda, J. Ishikawa, A. Hanamoto, C. Takahashi, M. Shinose, Y. Takahashi, H. Horikawa, H. Nakazawa, T. Osonoe, H. Kikuchi, T. Shiba, Y. Sakaki, and M. Hattori. 2001. Genome sequence of an industrial microorganism Streptomyces avermitilis: deducing the ability of producing secondary metabolites. Proc. Natl. Acad. Sci. USA 98:12215-12220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parkhill, J., G. Dougan, K. D. James, N. R. Thomson, D. Pickard, J. Wain, C. Churcher, K. L. Mungall, S. D. Bentley, M. T. Holden, M. Sebaihia, S. Baker, D. Basham, K. Brooks, T. Chillingworth, P. Connerton, A. Cronin, P. Davis, R. M. Davies, L. Dowd, N. White, J. Farrar, T. Feltwell, N. Hamlin, A. Haque, T. T. Hien, S. Holroyd, K. Jagels, A. Krogh, T. S. Larsen, S. Leather, S. Moule, P. O'Gaora, C. Parry, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848-852. [DOI] [PubMed] [Google Scholar]
- 48.Poh, C. L., and R. C. Bayly. 1980. Evidence for isofunctional enzymes used in m-cresol and 2,5-xylenol degradation via the gentisate pathway in Pseudomonas alcaligenes. J. Bacteriol. 143:59-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Price, T. D. C., I. R. Lee, and J. E. Gustafson. 2000. The effects of salicylate on bacteria. Int. J. Biochem. Cell Biol. 32:1029-1043. [DOI] [PubMed] [Google Scholar]
- 50.Quadri, L. E., T. A. Keating, H. M. Patel, and C. T. Walsh. 1999. Assembly of the Pseudomonas aeruginosa nonribosomal peptide siderophore pyochelin: in vitro reconstitution of aryl-4,2-bisthiazoline synthetase activity from PchD, PchE, and PchF. Biochemistry 38:14941-14954. [DOI] [PubMed] [Google Scholar]
- 51.Salanoubat, M., S. Genin, F. Artiguenave, J. Gouzy, S. Mangenot, M. Arlat, A. Billault, P. Brottier, J. C. Camus, L. Cattolico, M. Chandler, N. Choisne, C. Claudel-Renard, S. Cunnac, N. Demange, C. Gaspin, M. Lavie, A. Moisan, C. Robert, W. Saurin, T. Schiex, P. Siguier, P. Thebault, M. Whalen, P. Wincker, M. Levy, J. Weissenbach, and C. A. Boucher. 2002. Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 415:497-502. [DOI] [PubMed] [Google Scholar]
- 52.Schühle, K., M. Jahn, S. Ghisla, and G. Fuchs. 2001. Two similar gene clusters coding for enzymes of a new type of aerobic 2-aminobenzoate (anthranilate) metabolism in the bacterium Azoarcus evansii. J. Bacteriol. 183:5268-5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Silakowski, B., B. Kunze, G. Nordsiek, H. Blöcker, G. Höfle, and R. Müller. 2000. The myxochelin iron transport regulon of the myxobacterium Stigmatella aurantiaca Sg a15. Eur. J. Biochem. 267:6476-6485. [DOI] [PubMed] [Google Scholar]
- 54.Suemori, A., K. Nakajima, R. Kurane, and Y. Nakamura. 1995. o-, m- and p-hydroxybenzoate degradative pathways in Rhodococcus erythropolis. FEMS Microbiol. Lett. 125:31-36. [DOI] [PubMed] [Google Scholar]
- 55.Takami, H., K. Nakasone, C. Hirama, Y. Takaki, N. Masui, F. Fuji, Y. Nakamura, and A. Inoue. 1999. An improved physical and genetic map of the genome of alkaliphilic Bacillus sp. C-125. Extremophiles 3:21-28. [DOI] [PubMed] [Google Scholar]
- 56.Tegeder, I., J. Pfeilschifter, and G. Geisslinger. 2001. Cyclooxygenase-independent actions of cyclooxygenase inhibitors. FASEB J. 15:2057-2072. [DOI] [PubMed] [Google Scholar]
- 57.Thompson, J., S. Rae, and E. Cundliffe. 1984. Coupled transcription-translation in extracts of Streptomyces lividans. Mol. Gen. Genet. 195:39-43. [DOI] [PubMed] [Google Scholar]
- 58.Uz, I., Y. P. Duan, and A. Ogram. 2000. Characterization of the naphthalene-degrading bacterium, Rhodococcus opacus M213. FEMS Microbiol. Lett. 185:231-238. [DOI] [PubMed] [Google Scholar]
- 59.Visca, P., A. Ciervo, V. Sanfilippo, and N. Orsi. 1993. Iron-regulated salicylate synthesis by Pseudomonas spp. J. Gen. Microbiol. 139:1995-2001. [DOI] [PubMed] [Google Scholar]
- 60.Warhurst, A. M., and C. A. Fewson. 1994. Biotransformations catalyzed by the genus Rhodococcus. Crit. Rev. Biotechnol. 14:29-73. [DOI] [PubMed] [Google Scholar]
- 61.Webster, L. T., J. J. Mieyal, and U. A. Siddiqui. 1974. Benzoyl and hydroxybenzoyl esters of coenzyme A. J. Biol. Chem. 249:2641-2645. [PubMed] [Google Scholar]
- 62.Werwath, J., H.-A. Arfmann, D. H. Pieper, K. N. Timmis, and R.-M. Wittich. 1998. Biochemical and genetic characterization of a gentisate 1,2-dioxygenase from Sphingomonas sp. strain RW5. J. Bacteriol. 180:4171-4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yamada, A., H. Kishi, K. Sugiyama, T. Hatta, K. Nakamura, E. Masai, and M. Fukuda. 1998. Two nearly identical aromatic compound hydrolase genes in a strong polychlorinated biphenyl degrader, Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 64:2006-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 65.Yen, K. M., and C. M. Serdar. 1988. Genetics of naphthalene catabolism in pseudomonads. Crit. Rev. Microbiol. 15:247-268. [DOI] [PubMed] [Google Scholar]
- 66.Zaar, A., W. Eisenreich, A. Bacher, and G. Fuchs. 2001. A novel pathway of aerobic benzoate catabolism in the bacteria Azoarcus evansii and Bacillus stearothermophilus. J. Biol. Chem. 276:24997-25004. [DOI] [PubMed] [Google Scholar]
- 67.Zhou, N.-Y., S. L. Fuenmayor, and P. A. Williams. 2001. nag genes of Ralstonia (formerly Pseudomonas) sp. strain U2 encoding enzymes for gentisate catabolism. J. Bacteriol. 183:700-708. [DOI] [PMC free article] [PubMed] [Google Scholar]