Abstract
PCR-based assays for detecting enterohemorrhagic Escherichia coli serogroups O26 and O113 were developed by targeting the wzx (O-antigen flippase) and the wzy (O-antigen polymerase) genes found in the O-antigen gene cluster of each organism. The PCR assays were specific for the respective serogroups, as there was no amplification of DNA from non-O26 and non-O113 E. coli serogroups or from other bacterial genera tested. Using the PCR assays, we were able to detect the organisms in seeded apple juice inoculated at concentration levels as low as ≤10 CFU/ml. The O26- and O113-specific PCR assays can potentially be used for typing E. coli O26 and O113 serogroups; these assays will offer an advantage to food and environmental microbiology laboratories in terms of identifying these non-O157 serogroups by replacing antigen-based serotyping.
Escherichia coli O26 strains, first isolated from cases of infantile diarrhea, have been implicated in causing hemolytic uremic syndrome (HUS) (13) and serious enteric disorders in humans in the United Kingdom (12), Germany (17), Poland (9), Spain (2), and Finland (7). Among the non-O157 Shiga toxin-producing E. coli (STEC) isolates, O26 has been the most common serogroup, composing 18% (1,066 of 5,913) of the total number of STEC isolates reported from 1997 to 1999 (6). E. coli O26 strains have been found to be genetically diverse with unique virulence profiles (19). An eae-negative O113:H21 STEC strain was responsible for an HUS outbreak in South Australia (10). Since traditional E. coli growth and isolation methods show all non-O157 STEC to be phenotypically similar to nonpathogenic E. coli, detection of specific STEC serogroups is problematic. There is no rapid method presently available for detecting specific STEC strains.
The O antigen is part of the lipopolysaccharide present in the outer membrane of gram-negative bacteria and consists of many repeats of an oligosaccharide unit (O unit). The O antigen is the major contributor of antigenic variability on the cell surface, and on this basis different O types have been designated. The genes involved in the biosynthesis of O antigens in E. coli are generally clustered and flanked by the galF and gnd genes at the 5′ and 3′ ends, respectively. O-antigen gene clusters including O26 and O113 have been cloned and sequenced (4, 11). Analyses of each of the genes in the cluster by National Center for Biotechnology Information genome BLAST and gene alignment software programs showed that the O-unit flippase gene (wzx) and the polymerase gene (wzy) were unique for E. coli O26 as well as for O113 antigens. Therefore, these genes were targeted for developing PCR assays for detecting these serogroups.
All E. coli strains used in the study were from the bacterial collection of the Gastroenteric Disease Center at The Pennsylvania State University. Reference standard strains, E. coli O26:H− (H31b) and E. coli O113:H21 (6182-50) from the World Health Organization (8), were used for developing the assays. The 179 World Health Organization O reference standard strains with different O serogroups (O1 to O30, O32 to O46, O48 to O71, O73 to O92, O95 to O121, O123 to O175, X6, X9, X10, X13, X18, X19, X21, X23, X25, X28, X38, and X43) (8) and other E. coli strains (n = 50) belonging to different serogroups isolated from animals, chickens, and environmental sources were used for examining specificities of the PCR assays. Fifty cultures each of O26 and O113 serogroups from humans, animals, and environmental sources were also used for examining specificities. All cultures were grown in Luria-Bertani (LB) agar medium.
Template DNA from the bacteria was prepared by mixing a colony from the LB agar in sterile distilled water and heating at 100°C for 20 min in a heating block. The suspension was centrifuged at 13,000 × g for 5 min, and the supernatant containing the DNA was used for the PCR. Since PCR assays based on using primers for the E. coli wzx and wzy genes have been found to be specific for several serogroups (5, 14, 15, 16), PCR assays were developed using the primers listed in Table 1, which were designed by using the Primer3 software program. These assays were intended to perform amplifications of regions in the wzx and wzy genes in the O-antigen gene clusters of E. coli O26 (4) and O113 (11). Reaction contents for each PCR (total reaction volume, 11 μl) consisted of 3 μl of template DNA, a 0.5 μM concentration of primers (Integrated DNA Technologies, Inc., Coralville, Iowa), a 0.18 mM concentration of each of the four deoxynucleoside triphosphates, 2 mM MgCl2 (for the O113 PCR assays) and 3 mM MgCl2 (for the O26 PCR assays), 0.4 U of TaqDNA polymerase (PGC Scientific, Gaithersburg, Md.), 50 mM Tris (pH 8.3), 250 μg of bovine serum albumin per ml, 2% sucrose, and 0.1 mM Cresol Red. The PCR was performed in a RapidCycler (Idaho Technologies, Inc., Salt Lake City, Utah) by using a rapid-cycle DNA amplification method (18). This method consisted of initial denaturing at 94°C for 30 s followed by 30 cycles of template denaturation at 94°C (0 s); primer annealing at 54°C (0 s) for O26 wzx and 60°C (0 s) for O26 wzy and O113 wzx and wzy; and extension at 72oC for 10 s for O26 wzx and 12 s for O26 wzy and O113 wzx and wzy. The amplified products were electrophoresed in 1% agarose gels at 200 V for 1 h, stained with ethidium bromide, and visualized under UV light. Positive samples were identified based on the presence of bands of appropriate sizes compared to those of positive O26 and O113 control strains.
TABLE 1.
Oligonucleotide primers used for amplification of the E. coli O26 and O113 wzx and wzy genes
| Target gene | Sequencea | Amplicon size (bp) | GenBank accession no. |
|---|---|---|---|
| O26 wzx | F, GCGCTGCAATTGCTTATGTA | 152 | AF529080 |
| R, TTTCCCCGCAATTTATTCAG | |||
| O26 wzy | F, TAAATTGCGGGGAAAGAATG | 276 | AF529080 |
| R, GACTTCATGGGTACCGCCTA | |||
| O113 wzx | F, GGGTTAGATGGAGCGCTATTGAGA | 771 | AF172324 |
| R, AGGTCACCCTCTGAATTATGGCAG | |||
| O113 wzy | F, GCATGTATGATGCATAGCTTCGCC | 419 | AF172324 |
| R, TGATATCGTTCGCTAACCACCCA |
F, forward; R, reverse.
PCR assays were tested by using 50 strains belonging to serogroup O26 and another 50 belonging to serogroup O113 obtained from humans, animals, and environmental sources. All O26 and O113 isolates were positive for the presence of the respective wzx and wzy genes. The PCR assays did not produce bands with DNA lysates from 178 non-O26 or non-O113 serogroups or from cultures from different environmental sources that belonged to different O serogroups, thus showing 100% specificity for E. coli cultures belonging to O26 and O113 serogroups (Table 2). Several other non-E. coli bacterial strains from several sources including the American Type Culture Collection were also tested for the presence of these genes. Shigella boydii, Shigella flexneri, Shigella dysenteriae, Vibrio cholerae, Staphylococcus aureus, Salmonella enterica serovar Typhimurium, Klebsiella pneumoniae, Listeria monocytogenes, Citrobacter freundii, Yersinia enterocolitica, and Pseudomonas aeruginosa were tested and exhibited negative reactions for the presence of O26 and O113 wzx and wzy genes.
TABLE 2.
PCR assays tested against E. coli of different O groups and other bacterial generaa
| Strain (no.) | Source | O type | Assay results for:
|
|||
|---|---|---|---|---|---|---|
| O26 wzx | O26 wzy | O113 wzx | O113 wzy | |||
| E. coli (50) | Humans, cows, chickens, ferrets | 26 | + | + | ND | ND |
| E. coli (50) | Humans, cows, pigs, avian, horses, tigers, water | 113 | ND | ND | + | + |
| E. coli (50) | Pigs, avian, antelopes, horses, cows, unknown | 50 different serogroupsb | − | − | − | − |
| Shigella flexneri (1) | Unknown | Unknown | − | − | − | − |
| Vibrio cholerae (1) | Human | Unknown | − | − | − | − |
| Staphylococcus aureus (1) | ATCC 51740 | Unknown | − | − | − | − |
| Shigella boydii (1) | Unknown | Unknown | − | − | − | − |
| Shigella dysenteriae (1) | Field isolate | Unknown | − | − | − | − |
| Shigella dysenteriae (1) | ATCC 29029 | Unknown | − | − | − | − |
| Salmonella enterica serovar Typhimurium (1) | ATCC 13311 | Unknown | − | − | − | − |
| Klebsiella pneumoniae (1) | ATCC 27736 | Unknown | − | − | − | − |
| Listeria monocytogenes (1) | Unknown | Unknown | − | − | − | − |
| Citrobacter freundii (1) | ATCC 8090 | Unknown | − | − | − | − |
| Yersinia enterocolitica (1) | Unknown | Unknown | − | − | − | − |
| Pseudomonas aeruginosa (1) | Unknown | Unknown | − | − | − | − |
+, positive; −, negative; ND, not done.
Other than O26 and O113.
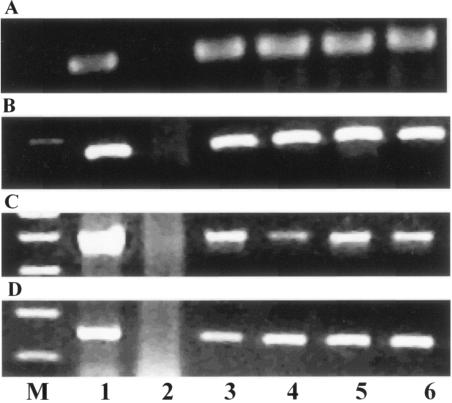
The optimized PCR assays were evaluated for detecting E. coli O26 or O113 strains in apple juice, since E. coli infections in humans by STEC that caused HUS have been linked to the consumption of contaminated apple juice (1, 3). Pasteurized apple juice (25 ml) purchased from a local supermarket was diluted in 225 ml of Trypticase soy broth, and the samples were inoculated with 1 ml of serial 10-fold dilutions of E. coli O26:H− (H31B) or E. coli O113:H21 (6182-50). The enrichments were incubated at 37°C for 18 h in a rotary shaker at 150 rpm. The concentration of each inoculum was determined by plating 100 μl of serially diluted samples onto LB agar. A 0.4-ml volume of enrichment was centrifuged to deposit cellular material and was washed once with deionized water. DNA (3 μl) templates were prepared, and wzx and wzy PCR assays were conducted by following the methods described above. Positive results were obtained with apple juice samples inoculated with 10 CFU/ml. Bands of the expected sizes for the E. coli O26 and O113 wzx and wzy genes were visualized by agarose gel electrophoresis (Fig. 1).
FIG. 1.
PCR products in agarose gel showing the presence of E. coli O26 (panels A and B) and O113 (panels C and D) DNA in apple juice. The procedure was performed by using O26 wzx primers, fragment size 152 bp (A), using O26 wzy primers, fragment size 276 bp (B), using O113 wzx primers, fragment size 771 bp (C), using O113 wzy primers, fragment size 419 bp (D). Lane M, standard molecular weight size markers; lane 1, positive control; lane 2, negative control; lane 3, 10 CFU/ml concentration; lane 4, 102 CFU/ml concentration; lane 5, 103 CFU/ml concentration; lane 6, 104 CFU/ml concentration.
The PCR assays specific for E. coli O26 and O113 targeting the wzx and wzy genes are rapid, sensitive, and specific and can be employed for confirmation of E. coli O26 and O113 serogroups and potentially for detecting these strains, as the PCR assays are less labor intensive and more rapid than is conventional O serotyping, and they do not require the use of antisera. We are now developing PCR assays for detecting other serogroups with an ultimate objective of developing microarray-based methods for distinguishing among the different O serogroups of E. coli.
REFERENCES
- 1.Anonymous. 1996. Outbreak of Escherichia coli O157:H7 infections associated with drinking unpasteurized commercial apple juice—British Columbia, California, Colorado and Washington, October 1996. Morb. Mortal. Wkly. Rep. 45:975.. [PubMed] [Google Scholar]
- 2.Blanco, M., J. E. Blanco, J. Blanco, E. A. Gonzalez, M. P. Alonso, H. Mass, and W. H. Jansen. 1996. Prevalence and characteristics of human and bovine verotoxigenic Escherichia coli strains in Galicia (northwestern Spain). Eur. J. Epidemiol. 12:13-19. [DOI] [PubMed] [Google Scholar]
- 3.Cody, S. H., M. K. Glynn, J. A. Farrar, K. L. Cairns, P. M. Griffin, J. Kobayashi, M. Fyfe, R. Hoffman, A. S. King, J. H. Lewis, B. Swaminathan, R. G. Bryant, D. J. Vugia. 1999. An outbreak of Escherichia coli O157:H7 infection from unpasteurized commercial apple juice. Ann. Intern. Med. 130:202-209. [DOI] [PubMed] [Google Scholar]
- 4.D'Souza, J. M., L. Wang, and P. Reeves. 2002. Sequence of the Escherichia coli O26 O antigen gene cluster and identification of O26 specific genes. Gene 297:123-127. [DOI] [PubMed] [Google Scholar]
- 5.Fratamico, P. M., C. E. Briggs, D. Needle, C. Chen, and C. DebRoy. 2003. Sequence of the Escherichia coli O121 O antigen gene cluster and detection of E. coli O121 by PCR amplification of the wzx and wzy genes. J. Clin. Microbiol. 41:3379-3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Infectious Agents Surveillance Center, National Institute of Infectious Diseases. 2000. The status of enterohemorrhagic Escherichia coli infection, 1998-March 2000. Infect. Agents Surveillance Rep. 21:92-93. [Google Scholar]
- 7.Keskimaki, M., M. Saari, T. Heiskanen, and A. Shitonen. 1998. Shiga toxin-producing Escherichia coli in Finland from 1990 through 1997: prevalence and characteristics of isolates. J. Clin. Microbiol. 36:3641-3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Orskov, I., F. Orskov, and K. Jann. 1977. Serology, chemistry and genetics of O and K antigens of Escherichia coli. Bacteriol. Rev. 41:667-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paciorek, J. 2002. Virulence properties of Escherichia coli faecal strains isolated in Poland from healthy children and strains belonging to serogroups O18, O26, O44, O86, O126 and O127 isolated from children with diarrhea. J. Med. Microbiol. 51:548-556. [DOI] [PubMed] [Google Scholar]
- 10.Paton, A. W., M. C. Woodrow, M. Doyle, J. A. Lanser, and J. C. Paton. 1999. Molecular characterization of a Shiga-toxigenic Escherichia coli O113:H21 strain lacking eae responsible for a cluster of cases of hemolytic-uremic syndrome. J. Clin. Microbiol. 37:3357-3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paton, A. W., and J. C. Paton. 1999. Molecular characterization of the locus encoding biosynthesis of the lipopolysaccharide O antigen of Escherichia coli serotype O113. Infect. Immun. 67:5930-5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scotland, S., G. Willshaw, H. Smith, B. Said, N. Stokes, and B. Rowe. 1993. Virulence properties of Escherichia coli strains belonging to serogroups O26, O111 and O128 isolated in the United Kingdom in 1991, from patients with diarrhea. Epidemiol. Infect. 111:429-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sramkova, L., M. Bielaszewska, J. Janda, K. Blahova, and O. Hausner. 1990. Vero cytotoxin producing strains of Escherichia coli in children with hemolytic uremic syndrome and diarrhea in Czechoslovakia. Infection 18:204-209. [DOI] [PubMed] [Google Scholar]
- 14.Wang, L., C. E. Briggs, D. Rothemund, P. Fratamico, J. B. Luchansky, and P. R. Reeves. 2001. Sequence of the E. coli O104 antigen gene cluster and identification of O104 specific genes. Gene 270:231-236. [DOI] [PubMed] [Google Scholar]
- 15.Wang, L., H. Curd, W. Qu, and P. R. Reeves. 1998. Sequencing of Escherichia coli O111 O-antigen gene cluster and identification of O111-specific genes. J. Clin. Microbiol. 36:3182-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang, L., and P. R. Reeves. 1998. Organization of Escherichia coli O157 O antigen gene cluster and identification of its specific genes. Infect. Immun. 66:3545-3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Werber, D., A. Fruth, A. Liesegang, M. Littmann, U. Buchholz, R. Prager, H. Karch, T. Breuer, H. Tschape, and A. Ammon. 2002. A multistate outbreak of shiga-toxin producing Escherichia coli O26:H11 infections in Germany, detected by molecular subtyping surveillance. J. Infect. Dis. 186:419-422. [DOI] [PubMed] [Google Scholar]
- 18.Wittwer, C. T., G. B. Reed, and K. M. Ririe. 1994. Rapid cycle DNA amplification, p. 174-181. In K. B. Mullis, F. Ferré, and R. A. Gibbs (ed.), The polymerase chain reaction. Birkauser, Boston, Mass.
- 19.Zhang, W. L., M. Bielaszewska, A. Liesegang, H. Tschäpe, H. Schmidt, M. Bitzan, and H. Karch. 2000. Molecular characteristics and epidemiological significance of shiga toxin-producing Escherichia coli O26 strains. J. Clin. Microbiol. 38:2134-2140. [DOI] [PMC free article] [PubMed] [Google Scholar]