Abstract
Heat stress resistance and response were studied in strains of Lactobacillus plantarum. Stationary-phase cells of L. plantarum DPC2739 had decimal reduction times (D values) (D value was the time that it took to reduce the number of cells by 1 log cycle) in sterile milk of 32.9, 14.7, and 7.14 s at 60, 72, and 75°C, respectively. When mid-exponential-phase cells were used, the D values decreased. The temperature increases which caused a 10-fold reduction in the D value ranged from 9 to 20°C, depending on the strain. Part of the cell population treated at 72°C for 90 s recovered viability during incubation at 7°C in sterile milk for 20 days. When mid-exponential- or stationary-phase cells of L. plantarum DPC2739 were adapted to 42°C for 1 h, the heat resistance at 72°C for 90 s increased ca. 3 and 2 log cycles, respectively. Heat-adapted cells also showed increased growth at pH 5 and in the presence of 6% NaCl. Two-dimensional gel electrophoresis of proteins expressed by control and heat-adapted cells revealed changes in the levels of expression of 31 and 18 proteins in mid-exponential- and stationary-phase cells, respectively. Twelve proteins were commonly induced. Nine proteins induced in the heat-adapted mid-exponential- and/or stationary-phase cells of L. plantarum DPC2739 were subjected to N-terminal sequencing. These proteins were identified as DnaK, GroEL, trigger factor, ribosomal proteins L1, L11, L31, and S6, DNA-binding protein II HlbA, and CspC. All of these proteins have been found to play a role in the mechanisms of stress adaptation in other bacteria. Antibodies against GroES detected a protein which was induced moderately, while antibodies against DnaJ and GrpE reacted with proteins whose level of expression did not vary after heat adaptation. This study showed that the heat resistance of L. plantarum is a complex process involving proteins with various roles in cell physiology, including chaperone activity, ribosome stability, stringent response mediation, temperature sensing, and control of ribosomal function. The physiological mechanisms of response to pasteurization in L. plantarum are fundamental for survival in cheese during manufacture.
Lactobacillus plantarum, proposed as Streptobacterium plantarum by Orla-Jensen in 1919, is a widely distributed species in most fermented products of animal or plant origin, where it either is used in controlled fermentation or is derived from the environment and emerges after manufacture (12).
L. plantarum is one of a group of mesophilic lactobacilli, referred to as the nonstarter lactic acid bacteria (NSLAB), which become the dominant microorganisms in several types of cheese during ripening. Starter lactic acid bacteria (e.g., lactococci) grow rapidly in cheese milk and curd during manufacture, reaching concentrations of 8.0 to 9.0 log CFU g−1, but the levels subsequently decline to approximately 1% of the maximum levels within a few weeks (43). In contrast, adventitious NSLAB usually grow from a low concentration (<2.0 log CFU g−1) in fresh curd to dominate the microflora of mature cheese (48). High numbers of L. plantarum have been found in Italian (e.g., Pecorino), Spanish (e.g., Manchego, Cabrales, and Roncal), Portuguese (e.g., Picante), Greek (e.g., Feta), British, Irish, and United States (e.g., Cheddar) cheeses (12).
The role of NSLAB in cheese ripening has yet to be resolved satisfactorily. To have beneficial activities, NSLAB must (i) be competitive against other adventitious microorganisms; (ii) adapt under the physiological and nutritional conditions in the cheese-curd ecosystem; and (iii) have a complementary effect with starter lactic acid bacteria. Cheddar cheese manufactured in the absence of NSLAB by using aseptic cheese vats is thought to lack full mature flavor (14). The inclusion of some strains of NSLAB with the starter lactococci (37), the use of raw milk (44), and the use of blends of raw and pasteurized milk (55) in Cheddar cheese manufacture have indicated that NSLAB are involved positively in the release of free amino acids and fatty acids. In addition, the flavor and texture of other semihard cheeses were improved or ripening was accelerated by using L. plantarum as an adjunct starter (13, 25). In particular, peptidase activities of L. plantarum are well adapted to the hostile environmental conditions of cheese during ripening and make a major contribution to mature cheese flavor after primary proteolysis by chymosin, plasmin, and proteinases of starter bacteria (24). Overall, NSLAB possess amino acid-catabolizing enzymes, which also affect the turnover of amino acids and, consequently, the intensity of cheese flavor (62).
What remains to be established is the primary source of NSLAB in cheese. Theoretically, these organisms may survive pasteurization or enter the milk or curd as postpasteurization contaminants. Practically, the use of closed cheese vats and more hygienic cheese-making practices slightly reduces the level of NSLAB contamination. Therefore, it may be assumed that the initial NSLAB in cheese milk are due mainly to a few strains that withstand pasteurization.
There have been only a few reports describing the physiological stress responses in lactic acid bacteria, particularly Lactobacillus species which have a broad biodiversity (for reviews, see references 15 and 60). In spite of the extensive use of lactic acid bacteria, there is a paucity of information concerning the stress-induced mechanisms for improving the survival of these organisms during food processing. A better understanding of the adaptive responses of lactic acid bacteria is important because dairy processing often subjects these microorganisms to adverse environmental conditions, including temperature extremes. The effect of heat shock and the induction of a stress response in Lactobacillus spp. have been studied for Lactobacillus delbrueckii subsp. bulgaricus and Lactobacillus paracasei (18, 23, 27, 28), Lactobacillus acidophilus, Lactobacillus casei and Lactobacillus helveticus (9), Lactobacillus collinoides (38), Lactobacillus sakei (54), Lactobacillus johnsonii (64), Lactobacillus rhamnosus (51), and Lactobacillus salivarius (23). To our knowledge, in the only studies in which L. plantarum was examined the researchers considered heat inactivation by differential scanning calorimetry (39) and flow cytometry (56) and characterized the heat resistance parameters (33).
In this study, the heat resistance and adaptation of L. plantarum strains were studied, and two-dimensional electrophoresis (2DE) and identification of stress proteins by N-terminal sequencing were used to highlight the response of this cheese-related species to pasteurization.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
L. plantarum DPC2739 and DPC2741, which were isolated from Cheddar cheese (Culture Collection of the University College, Cork, Ireland), and strains DC400 and 18E, which were isolated from sourdoughs (Culture Collection of the Dipartimento di Protezione delle Piante e Microbiologia Applicata, University of Bari, Bari, Italy) were used. Strains were routinely propagated and cultivated in MRS broth (Oxoid Ltd., Basingstoke, United Kingdom) at 30°C for 24 h.
Kinetics of cell growth at different temperatures.
Twenty-four-hour-old cells of L. plantarum strains were harvested by centrifugation at 9,000 × g for 10 min at 4°C, washed twice with 50 mM sterile potassium phosphate buffer (pH 7.0), resuspended in sterile distilled water to a final optical density at 620 nm (OD620) of 2.5 (corresponding to ca. 9.0 log CFU ml−1), inoculated (4%, vol/vol) into MRS broth, and incubated at 30, 42, 45, or 48°C for 24 h. Growth was monitored directly by plating on MRS agar after 48 h of incubation at 30°C or indirectly by measuring the OD620. Growth data were modeled by using the Gompertz equation, as modified by Zwietering et al. (65): y = K + A exp{−exp[(μmax e/A) (λ − t) + 1]}, where y is the extent of growth expressed in log CFU per milliliter at time t (in hours), k is the size of the initial cell population expressed in log CFU per milliliter, A is the difference in the number of cells between the inoculum and the stationary phase, μmax is the maximum growth rate expressed as the change in log CFU per milliliter per hour, and λ is the length of the lag phase in hours. The experimental data were modeled by using the nonlinear regression procedure of the statistics package Statistica for Windows (Statsoft, Tulsa, Okla.).
Heat resistance.
Cells of L. plantarum DPC2739 and DC400 were cultivated in MRS broth at the optimum growth temperature (30°C) until they reached the mid-exponential phase of growth (5 h; OD620, ca. 1.0) or the stationary phase of growth (12 h; OD620, ca. 2.5). Cells were harvested by centrifugation at 9,000 × g for 10 min at 4°C, washed twice with 50 mM sterile potassium phosphate buffer (pH 7.0), and resuspended in sterile distilled water to an OD620 of 2.5 (corresponding to ca. 9.0 log CFU ml−1). Each suspension was used to inoculate (4%, vol/vol) 5 ml of sterile milk. Samples (0.5 ml) of inoculated milk were transferred to capillary glass tubes (catalog no. 0893.01830; Carlo Erba Reagenti, Milan, Italy) and heated in a water bath at 60 to 72°C for 0 to 150 s. After heat treatment, samples were chilled on ice for 5 min, diluted, plated on MRS agar, and incubated at 30°C for 48 to 96 h. By using the same protocol, heat challenge experiments were carried out in MRS broth. Each experiment was repeated three times, and the average and standard deviation were calculated. At each temperature, a best-fit straight line was obtained by regression analysis, and the decimal reduction time (D value) (the time that it took to reduce the number of cells by 1 log cycle) and Z value (the temperature increase which caused a 10-fold reduction in the D value) were calculated (57).
Recovery of heat-injured cells.
As described above, a suspension of mid-exponential-phase L. plantarum DPC2739 or DC400 cells was heated at 72°C for 90 s and then incubated at 7°C for 20 days. Concomitantly, a suspension of mid-exponential-phase cells, used as a control, was inoculated into sterile (ultrahigh temperature) milk purchased from a market and diluted sufficiently to have no detectable cells in 1 ml as determined by plating on MRS agar; samples of this suspension were also incubated at 7°C for 20 days. During incubation, both cell suspensions were plated on MRS agar at 3-day intervals.
Heat adaptation.
Cells of L. plantarum DPC2739 which had been cultivated at the optimum temperature, 30°C, were harvested in the mid-exponential or stationary phase of growth, washed, and resuspended in sterile milk at a final concentration of ca. 7.5 log CFU ml−1. To induce heat adaptation (adapted cells), the cell suspensions were incubated for 1 h at 42, 45, or 48°C. Controls for mid-exponential- and stationary-phase cells were incubated for 1 h at 30°C. Chloramphenicol (1 mg ml−1) was used during the adaptation phase according to the protocol described by Hartke et al. (30). After incubation, control and adapted cells, with or without chloramphenicol, were treated at 72°C for 90 s. After heat treatment, cells were chilled on ice for 5 min and enumerated on MRS agar by incubation at 30°C for 48 h. The heat resistance of L. plantarum strains after adaptation was expressed as the thermotolerance factor (6). This corresponded to the ratio of the average values for adapted and control cells recovered after lethal challenge. The values reported below are the means obtained from at least three separate experiments. By using the same protocol, heat adaptation experiments were also carried out in MRS broth.
Growth at low pH and in the presence of NaCl.
Growth at pH 5 was assayed by sudden transfer of control or adapted cells of L. plantarum DPC2739 (ca. 7.5 log CFU ml−1) to sterile milk and further incubation for 24 h at the same temperature. The ability of L. plantarum DPC2739 to grow at a high salt concentration was examined by inoculating, under the same conditions, control or adapted cells into sterile milk containing 0, 1.5, 3.0, or 6% of NaCl and incubating the preparations at 30°C for 24 h.
Protein extraction and 2DE.
For 2DE analyses, cells were adapted and heat treated in MRS broth. Before and after heat treatment, control and adapted L. plantarum DPC2739 cells at the mid-exponential or stationary phase of growth were harvested by centrifugation at 9,000 × g for 10 min at 4°C, resuspended in 0.05 M Tris-HCl (pH 7.5) at a density of ca. 9.4 log CFU ml−1, centrifuged at 15,000 × g for 15 min at 4°C, and frozen or directly resuspended in denaturing buffer containing 8 M urea, 4% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 40 mM Tris base, and 65 mM dithiothreitol. To extract total proteins, cells (ca. 9.4 log CFU ml−1) were disrupted by three cycles of sonication (1 min each) with a Branson model B15 Sonifier. After unbroken cells were pelleted by centrifugation at 15,000 × g for 15 min at 4°C, the protein content of the supernatant was measured by the method of Bradford (7). Two-dimensional gel electrophoresis was performed by using the immobiline-polyacrylamide system, essentially as described by Görg et al. (26) and Hochstrasser et al. (32), and a Pharmacia 2D-EF system. The same amount of total protein (30 μg) was used for each electrophoretic run. Isoelectric focusing was carried out on immobiline strips that provided a nonlinear gradient from pH 3 to 10 (IPG strips; Amersham Pharmacia Biotech) by IPG-phore at 15°C. The voltage was increased from 300 to 5,000 V during the first 5 h and then increased to 8,000 V and kept at that value for 8 h. After electrophoresis, the IPG strips were equilibrated for 12 min against a solution containing 6 M urea, 30% (vol/vol) glycerol, 2% (wt/vol) sodium dodecyl sulfate (SDS), 0.05 M Tris-HCl (pH 6.8), and 2% (wt/vol) dithiothreitol and for 5 min against a solution containing 6 M urea, 30% (vol/vol) glycerol, 2% (wt/vol) SDS, 0.05 M Tris-HCl (pH 6.8), 2.5% (vol/vol) iodoacetamide, and 0.5% (wt/vol) bromophenol blue. For the second dimension, homogeneous SDS—12.5% polyacrylamide gel electrophoresis gels were used. The gels were calibrated with two molecular mass markers, as follows: (i) comigration of the cell extracts with markers for two-dimensional electrophoresis (pI range, 7.6 to 3.8; molecular mass range, 17 to 89 kDa) from Sigma Chemical Co., and (ii) migration of human serum proteins for the molecular mass range from 200 to 10 kDa and markers from Pharmacia Biotech for low molecular masses (16.9, 14.4, 10.7, 8.2, 6.2, and 2.5 kDa). The electrophoretic coordinates used for serum proteins were determined by the method Bjellqvist et al. (4). The gels were silver stained, as described by Hochstrasser et al. (32) and Oakley et al. (46). The protein maps were scanned with a laser densitometer (Molecular Dynamics 300s) and were analyzed by using the Image Master 2D elite computer software (Pharmacia). Three gels were analyzed, and spot intensities were normalized as described by Bini et al. (3). In particular, the spot quantification for each gel was calculated as a relative volume; the relative volume was the volume of each spot divided by the total volume for the whole image. In this way, differences in the color intensities among the gels were eliminated (1). The induction factor for individual proteins was expressed as the ratio of the spot intensity of a protein in the adapted cells to the spot intensity of the same protein in the nonadapted cells. All the induction factors were calculated based on the average of the spot intensities on each of the three gels, and standard deviations were calculated.
N-terminal amino acid sequencing.
Spots from 2DE gels were transferred to polyvinylidene difluoride membranes by passive absorption, as described by Messer et al. (45), with some modifications. Protein bands were excised and dried with a Speed-Vac for 30 min; then the gel pieces were reswollen in 0.03 ml of 2% (wt/vol) SDS in 0.2 M Tris-HCl (pH 8.4) for 30 min. After swelling, 0.15 ml of high-performance liquid chromatography-grade water was added, and then a piece (4 by 4 mm) of prewetted (methanol) polyvinylidene difluoride membrane (Problott Applied Biosystems) was added to the solution. The preparation was incubated for 2 days at room temperature (23°C) with gentle vortexing. At the end of this transfer time, the gel piece and the solution were clear and the membrane was blue. The membrane was washed five times with 1 ml of 10% methanol, with vortexing. N-terminal Edman sequencing was performed with an Applied Biosystems Procise 494HT by using reagents and methods recommended by the manufacturer. Sequence comparison was performed by using SWALL and BLAST at the National Center for Biotechnology Information nonredundant databases.
Immunoblot analysis.
Gels were electroblotted onto nitrocellulose membranes as described by Towbin et al. (58) and were further processed by standard procedures, modified as described by Bini et al. (2) and Magi et al. (40). Briefly, before immunodetection, the membranes were stained with 0.2% (wt/vol) Ponceau S in 3% (wt/vol) trichloroacetic acid for 3 min, and the positions of selected landmark spots were marked on the membranes to assist subsequent matching of the immunoblots with the silver-stained map. Immunoreactive spots were detected by overnight incubation at room temperature with antibodies for DnaK, GroEL, DnaJ, and GrpE diluted 1:1,000 and antibody for GroES diluted 1:800, followed by incubation with conjugated peroxidase (Sigma Chemical Co., St. Louis, Mo.) diluted 1:3,500, and the spots were revealed by staining with 4 chloro-1-naphthol (Sigma).
RESULTS
Kinetics of cell growth at different temperatures.
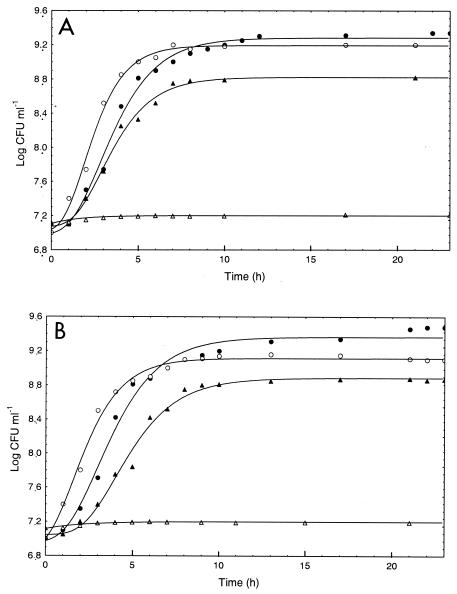
Preliminarily, the four strains of L. plantarum were cultivated at 30, 42, 45, or 48°C. Growth parameters were calculated by using the Gompertz equation. Two strains isolated from Cheddar cheese (DPC2739 and DPC2741) and two strains isolated from bread sourdoughs (DC400 and 18E) were used to compare the behaviors of microorganisms isolated from different sources. The growth kinetics of strains DPC2739 and DC400 are shown in Fig. 1. In both cases, the highest cell yield was obtained at 30°C (9.34 and 9.48 log CFU ml−1 for DPC2739 and DC400, respectively), while the μmax values were slightly higher at 42°C than at 30°C (0.75 and 0.48 Δlog CFU ml−1 h−1, respectively, for DPC2739 and 0.54 and 0.46 Δlog CFU ml−1 h−1, respectively, for DC400). Consistent with these findings, the length of the lag phase was slightly longer at 30°C than at 42°C (e.g., 1.06 versus 0.64 h for strain DPC2739). Both strains showed reduced growth at 45°C and did not grow at 48°C. Strains DPC2741 and 18E behaved like the strains isolated from the same sources (data not shown).
FIG. 1.
Growth kinetics of L. plantarum DPC2739 (A) and DC400 (B) at 30°C (•), 42°C (○), 45°C (▴), and 48°C (▵).
Heat resistance.
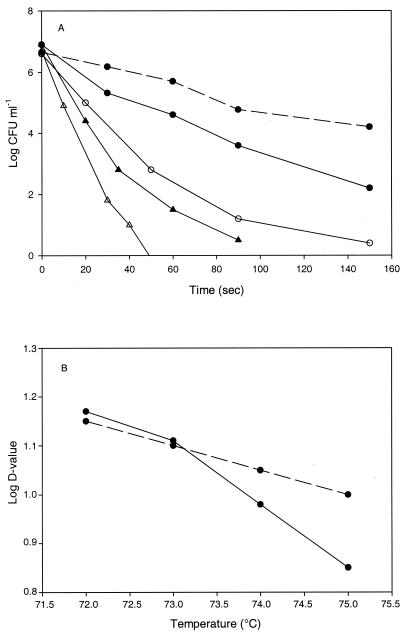
Mid-exponential- and stationary-phase cells of L. plantarum DPC2739 and DC400 were heat treated at 60 to 75°C for 0 to 150 s in sterile milk or MRS broth. For stationary-phase cells of L. plantarum DPC2739, the D values in sterile milk were 32.9, 14.7, and 7.14 s at 60, 72, and 75°C, respectively (Fig. 2A). When mid-exponential-phase cells were used, the D value at 60°C decreased to 24.4 s. L. plantarum DC400 was characterized by a higher D value at 60°C (58.5 s), and about the same increase was observed for the other heat challenges. To determine the Z values, the log of the D values was plotted against temperature and the negative inverse of the slope was determined (Fig. 2B). The Z values were 9 and 20°C for L. plantarum DPC2739 and DC400, respectively. When cell suspensions in MRS broth were subjected to the same heat challenges, the D and Z values were slightly lower than those obtained with sterile milk (data not shown).
FIG. 2.
(A) Thermal death curves for stationary-phase cells of L. plantarum DPC2739 at 60°C (• with solid line), 72°C (▴), and 75°C (▵), for stationary-phase cells of L. plantarum DC400 at 60°C (• with dashed line), and for mid-exponential-phase cells of L. plantarum DPC2739 at 60°C (○). (B) Plot of log of D value versus temperature for stationary-phase cells of L. plantarum DPC2739 (solid line) and DC400 (dashed line). The heat challenges were carried out in sterile milk.
Recovery of heat-injured cells.
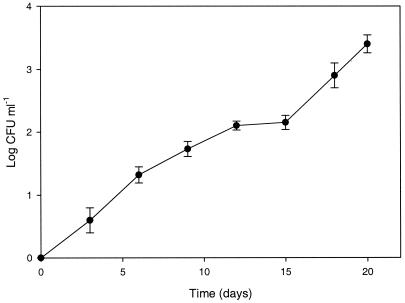
After treatment of mid-exponential-phase cells of L. plantarum DPC2739 at 72°C for 90 s, cells were not detected in 1 ml of sterile milk when it was plated on MRS agar. The same result was obtained when the suspension of unheated mid-exponential-phase cells was diluted in sterile milk (control). The two samples were incubated at 7°C for 20 days. After incubation, no cells were detected in the control. This showed that when a cell suspension was diluted so that the concentration was less than 1 CFU ml−1, cells were not detectable by plating when the preparation was incubated at 7°C for 20 days. In contrast, the concentration of the heat-treated suspension increased to ca. 3.5 log CFU ml−1 (Fig. 3), showing that heat-stressed cells recovered from the injury. The same results were obtained with L. plantarum DC400 (data not shown).
FIG. 3.
Recovery of mid-exponential-phase cells of L. plantarum DPC2739 (ca. 7.0 log CFU ml−1) heated at 72°C for 90 s and then incubated in sterile milk at 7°C for 20 days.
Heat adaptation.
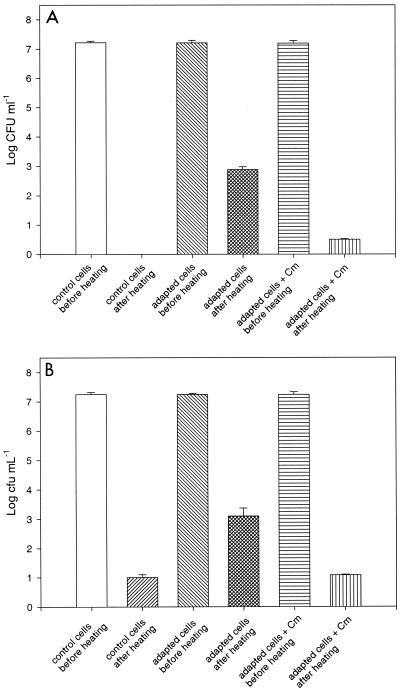
To induce heat adaptation, mid-exponential- or stationary-phase cells of L. plantarum DPC2739 and DC400, grown at 30°C, were treated at 42, 45, or 48°C for 1 h (adapted cells); control cells were kept at 30°C for 1 h. Adaptation was carried out in sterile milk or in MRS broth, with or without chloramphenicol (1 mg ml−1). Control and adapted cells were then heated at 72°C for 90 s. Compared to control cells, adaptation of mid-exponential-phase cells of strain DPC2739 to 42°C for 1 h in sterile milk increased the thermotolerance factor ca. 3 log cycles (Fig. 4A). Control cells at the stationary phase of growth showed survival of ca. 1 log cycle, probably due to inherent heat resistance (Fig. 4B). The increase in the thermotolerance factor for the adapted cells of strain DPC2739 was only ca. 2 log cycles. Adaptation for 1 h at 42°C in the presence of chloramphenicol prevented cell survival almost totally in both cases (Fig. 4). Similar results were obtained with L. plantarum DC400 cells (data not shown). Adaptation at 45 or 48°C did not result in the same increase in cell survival. Similar results were obtained when the adaptation experiments were carried out in MRS broth (data not shown). Further experiments were carried out in MRS broth to facilitate cell recovery and 2DE analysis of proteins.
FIG. 4.
Densities of mid-exponential-phase (A) and stationary-phase (B) cells of L. plantarum DPC2739 subjected to 72°C for 90 s. Cm, chloramphenicol.
2DE analysis.
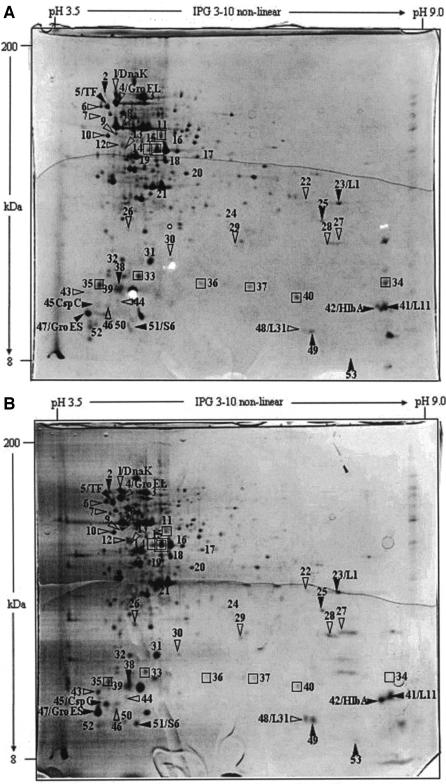
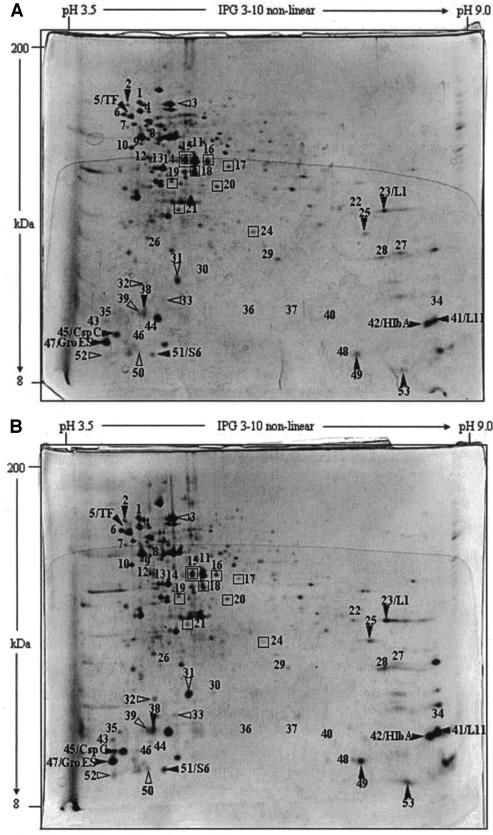
After characterization of heat resistance and adaptation, we decided to use only L. plantarum DPC2739, since this strain is a cheese-related strain and since only small interstrain differences were found. As shown by 2DE analysis (Fig. 5 and Table 1), the whole-cell protein extracts of mid-exponential-phase cells adapted to 42°C for 1 h showed that there was an increase in the level of expression of 31 proteins compared to the level of expression in control cells. These proteins were distributed over a large range of pI values and molecular masses. Compared to the level of expression in control cells, the level of expression of 18 proteins increased in stationary-phase cells adapted to 42°C for 1 h (Fig. 6 and Table 1). In this case, with the exception of three proteins, the proteins had molecular masses less than 30 kDa. Twelve proteins were induced in both mid-exponential- and stationary-phase cells. On the other hand, the level of expression of nine and eight proteins decreased in mid-exponential- and stationary-phase adapted cells, respectively, compared to the level of expression in the corresponding controls (Fig. 5 and 6). The level of expression of only one protein decreased under both conditions.
FIG. 5.
2DE analysis of protein expression in L. plantarum DPC2739. (A) Mid-exponential-phase cells grown in MRS broth at 30°C and kept at 30°C for 1 h (control cells); (B) mid-exponential-phase cells grown in MRS broth at 30°C and heat adapted for 1 h at 42°C (adapted cells). Symbols: ▿ and □, proteins whose amounts were increased and decreased, respectively, compared to the amounts in nonadapted cells; ▾ and ▪, proteins whose amounts were increased or decreased, respectively, compared to the amounts in nonadapted cells in the mid-exponential and stationary phases. The positions of the following proteins are indicated: DnaK, GroEL, GroES, CspC, trigger factor (TF), ribosomal protein L1 (L1), ribosomal protein L11 (L11), DNA-binding protein II (HlbA), ribosomal protein L31 (L31), and ribosomal protein S6 (S6).
TABLE 1.
Properties of heat-induced proteins in L. plantarum DPC3970
| Spota | Estimated pI | Estimated Mr (103) | Induction factor (mean ± SD)b
|
|
|---|---|---|---|---|
| Mid-exponential-phase cells | Stationary-phase cells | |||
| 1 | 4.64 | 66.50 | 2.00 ± 0.05 | 0 |
| 2 | 4.57 | 66.20 | 2.00 ± 0.05 | 2.35 ± 0.10 |
| 3 | 4.94 | 66.3 | 0 | 2.00 ± 0.06 |
| 4 | 4.63 | 59.50 | 2.45 ± 0.07 | 0 |
| 5 | 4.60 | 58.60 | 2.00 ± 0.04 | 2.00 ± 0.11 |
| 6 | 4.53 | 58.20 | 2.10 ± 0.08 | 0 |
| 7 | 4.57 | 54.00 | 3.90 ± 0.10 | 0 |
| 8 | 4.72 | 45.20 | 2.00 ± 0.08 | 0 |
| 9 | 4.65 | 45.00 | 2.00 ± 0.05 | 0 |
| 10 | 4.58 | 43.00 | 2.00 ± 0.10 | 0 |
| 12 | 4.65 | 39.50 | 6.35 ± 0.16 | 0 |
| 13 | 4.77 | 38.70 | 3.6 ± 0.07 | 0 |
| 22 | 7.89 | 29.00 | 4.45 ± 0.21 | 0 |
| 23 | 8.15 | 28.90 | 2.00 ± 0.05 | 3.55 ± 0.23 |
| 25 | 8.05 | 24.80 | 2.00 ± 0.05 | 2.90 ± 0.04 |
| 26 | 4.79 | 22.40 | 7.50 ± 0.44 | 0 |
| 27 | 8.22 | 22.30 | 2.00 ± 0.06 | 0 |
| 28 | 8.20 | 21.70 | 2.00 ± 0.05 | 0 |
| 29 | 6.60 | 21.60 | 2.23 ± 0.11 | 0 |
| 30 | 5.70 | 19.90 | 3.25 ± 0.29 | 0 |
| 31 | 5.25 | 19.90 | 0 | 2.00 ± 0.06 |
| 32 | 4.76 | 18.90 | 0 | 2.30 ± 0.42 |
| 38 | 4.76 | 12.38 | 3.57 ± 0.14 | 2.00 ± 0.10 |
| 39 | 4.61 | 12.38 | 0 | 4.77 ± 0.35 |
| 41 | 8.90 | 12.25 | 2.00 ± 0.07 | 2.00 ± 0.08 |
| 42 | 8.81 | 12.00 | 3.65 ± 0.05 | 2.00 ± 0.07 |
| 43 | 4.48 | 12.10 | 9.40 ± 0.38 | 0 |
| 44 | 4.69 | 12.00 | 5.87 ± 0.29 | 0 |
| 45 | 4.55 | 11.75 | 9.05 ± 0.43 | 2.00 ± 0.07 |
| 46 | 4.63 | 11.75 | 6.60 ± 0.31 | 0 |
| 47 | 4.48 | 10.75 | 2.35 ± 0.18 | 2.00 ± 0.05 |
| 48 | 7.87 | 8.53 | 14.0 ± 0.51 | 0 |
| 49 | 7.95 | 8.50 | 4.30 ± 0.20 | 2.70 ± 0.27 |
| 50 | 4.61 | 8.75 | 0 | 7.20 ± 0.26 |
| 51 | 4.82 | 8.40 | 3.30 ± 0.15 | 3.13 ± 0.10 |
| 52 | 4.50 | 8.10 | 0 | 4.90 ± 0.28 |
| 53 | 8.50 | 8.00 | 10.0 ± 0.50 | 6.00 ± 0.21 |
The induction factor is defined as the ratio of the spot intensity of a protein in the adapted cells to the spot intensity of the same protein in the control cells. All of the induction factors were calculated based on the means of the spot intensities on three gels, and ± SD are shown.
FIG. 6.
2DE analysis of protein expression in L. plantarum DPC2739. (A) Stationary-phase cells grown in MRS broth at 30°C and kept at 30°C for 1 h (control cells); (B) stationary-phase cells grown in MRS broth and heat adapted for 1 h at 42°C (adapted cells). Symbols: ▿ and □, proteins whose amounts were increased and decreased, respectively, compared to the amounts in nonadapted cells; ▾ and ▪, proteins whose amounts were increased and decreased, respectively, compared to the amounts in mid-exponential- and stationary-phase adapted cells. The positions of the following proteins are indicated: DnaK, GroEL, GroES, CspC, trigger factor (TF), ribosomal protein L1 (L1), ribosomal protein L11 (L11), DNA-binding protein II (HlbA), ribosomal protein L31 (L31), and ribosomal protein S6 (S6).
Identification of proteins by N-terminal sequencing and Western blotting.
Proteins 5, 23 41, 42, 45, and 51, which were induced in the adapted mid-exponential- and stationary-phase cells of L. plantarum DPC2739, and proteins 1, 4, and 48, which were induced specifically in the adapted mid-exponential-phase cells, were subjected to N-terminal sequencing. The sequences are shown in Table 2. The N-terminal sequences of the nine proteins exhibited 100% of identity with the deduced amino acid sequences determined from the complete genome sequence of L. plantarum WCFS1, as reported by Kleerebezem et al. (36). The proteins were identified as follows: spot 1, DnaK (93% identity with DnaK of Lactobacillus sanfranciscensis; accession number Q8KML6); spot 4, GroEL (93% identity with GroEL of Enterococcus durans; accession number Q8GBC4); spot 5, trigger factor (75% identity with trigger factor of Staphylococcus epidermidis ATCC 12228; accession number Q8CNY4); spot 23, ribosomal protein L1 (66% identity with L1 of Bacillus cereus ATCC 14579; accession number NC-004722); spot 41, L11 (86% identity with 50S ribosomal L11 of Streptococcus mutans UA159; accession number Q8DSX9); spot 48, L31 (80% identity with L31 of Corynebacterium glutamicum ATCC 13032; accession number Q8NS12); spot 51, S6 (81% identity with 30S ribosomal protein S6 of Oceanobacillus iheyensis HTE831; accession number Q8EKV4); spot 42, DNA-binding protein II HlbA (83% identity with DNA-binding protein II HlbA of L. delbrueckii subsp. bulgaricus; accession number Q8KQE1); and spot 45, CspC.
TABLE 2.
Proteins identified by N-terminal sequencing
| Spot or homologous proteina | N-terminal sequence | Identity
|
Reference or accession no.c | |
|---|---|---|---|---|
| %b | Organism | |||
| 1 | ASNKIIGIDLGTTNS | This study | ||
| DnaK | ASNKIIGIDLGTTNS | 100 | L. plantarum WCFS1 | Q88VM0 |
| ASNKVIGIDLGTTNS | 93 | L. sanfranciscensis | Q8KML6 | |
| 4 | AKELKFSEDARSAML | This study | ||
| GroEL | AKELKFSEDARSAML | 100 | L. plantarum WCFS1 | Q88YM5 |
| GroEL | AKELKFSEDARAAML | 93 | E. durans | Q8GBC4 |
| 5 | AAKWEKKEGNQGELT | This study | ||
| Trigger factor | AAKWEKKEGNQGELT | 100 | L. plantarum WCFS1 | Q88VE1 |
| Trigger factor | ATWEKKEGNEGVLT | 75 | S. epidermidis ATCC 12228 | Q8CNY4 |
| 23 | AKKSKQYQDAAKLVD | This study | ||
| Ribosomal protein L1 | AKKSKQYQDAAKLVD | 100 | L. plantarum WCFS1 | Q88YW9 |
| Ribosomal protein L1 | AKRGKKYVEAAKLVD | 66 | B. cereus ATCC 14579 | NC_004722.1 |
| 41 | AKKVANVVKLQIPAG | This study | ||
| Ribosomal protein L11 | AKKVANVVKLQIPAG | 100 | L. plantarum WCFS1 | NP_784387.1 |
| 50S ribosomal L11 protein | AKKVENIVKLQIPAG | 86 | S. mutans UA159 | Q8DSX9 |
| 42 | ANKAELVNNVAAATN | This study | ||
| DNA-binding protein II | ANKAELVNNVAAATN | 100 | L. plantarum WCFS1 | Q88VZ8 |
| HlbA | MANKAELVSEVAAKTK | 83 | L. delbrueckii subsp. bulgaricus | Q8KQE1 |
| 45 | MEHGTVKWFNADKGF | This study | ||
| CspC | MEHGTVKWFNADKGF | 100 | L. plantarum WCFS1 | Q88Y09 |
| CspC | MEHGTVKWFNADKGF | 100 | L. plantarum | Q9FCV6 |
| 48 | MQKGIHPDYHLVVFQ | This study | ||
| Ribosomal protein L31 | MQKGIHPDYHLVVFQ | 100 | L. plantarum WCFS1 | Q88Z52 |
| Ribosomal protein L31 | MKKDIHPDYHAVVFQ | 80 | C. glutamicum ATCC 13032 | Q8NS12 |
| 51 | SESKKYEITYIVRPD | This study | ||
| Ribosomal protein S6 | SESKKYEITYIVRPD | 100 | L. plantarum WCFS1 | Q890K2 |
| 30S ribosomal protein S6 | KYEIMYIIRPD | 81 | O. iheyensis HTE831 | Q8EKV4 |
Protein labeled in Fig. 5 and 6 or similar amino acid sequence found in the databases. The similarity searches were done by using BLAST at the National Center for Biotechnology Information nonredundant databases and the SWALL database.
Based on the number of matching amino acids in the test sequence.
Accession number in the SwissProt and Trembl databases.
Antibody against GroES detected a single protein (spot 47) with an experimental molecular mass of 10.75 kDa and a pI of 4.48. This protein was induced moderately in the adapted mid-exponential- and stationary-phase cells. Antibodies against DnaK and GroEL reacted with spots 1 and 4, respectively, while antibodies against DnaJ and GrpE reacted with individual proteins which did not show variation in the level of expression after adaptation.
Growth at low pH and in the presence of NaCl.
Mid-exponential-phase cells of L. plantarum DPC2739 were adapted at 42°C for 1 h and assayed for growth in sterile milk at pH 5 and in the presence of 0 to 6% NaCl, which represent restrictive conditions found in cheese during ripening. Adapted cells exhibited moderately higher growth either at pH 5 or in the presence of 6% NaCl (ca. 0.5 to 1 log cycle) compared to the growth of control cells (data not shown). The same results were obtained with stationary-phase cells.
DISCUSSION
After animals are milked, lactobacilli from the environment rapidly contaminate raw milk, but compared to the growth of other bacteria, the growth of these organisms is usually limited. The concentration of NSLAB, including L. plantarum, is estimated to range from >1.0 to 50 CFU ml−1 in single-herd milk of good hygienic quality and to be ca. 3.0 log CFU ml−1 in bulked herd market milk (12). Starting from this low value, NSLAB dominate the microbial population of cheese after several weeks of ripening and contribute strongly to secondary proteolysis and catabolism of amino acids (14, 62). Since more hygienic cheese-making practices have negligible effects on NSLAB contamination, survival during pasteurization is the most probable entry route for the few NSLAB cells found in cheese during early ripening. Based on these considerations, in this study we aimed to identify the main physiological mechanisms of heat shock response in L. plantarum, one of the most important NSLAB species in cheese during ripening.
L. plantarum DPC2739 showed D values that were in substantial agreement with the values reported by other workers (33) for other strains isolated from Cheddar cheese. The same was true for Z values. Nevertheless, the heat resistance of L. plantarum was low compared to that of other NSLAB species, such as L. paracasei (15, 33). Under our experimental conditions, a heat treatment consisting of 72°C for 90 s reduced the survival of strain DPC2739 from ca. 7.0 log CFU ml−1 to less than 1 cell ml−1. It was shown that some of the heat-treated cells may recover during incubation in sterile milk at 7°C; nevertheless, the number of cultivable cells after 20 days was still ca. 3.0 log CFU ml−1.
Overall, heat shock resistance of bacteria is affected by genetic differences among species, the physiological status of the cells, and environmental factors, such as pH, water activity, salt content, and preservatives (10). When mid-exponential-phase cells of L. plantarum DPC2739 were subjected to adaptation at 42°C for 1 h, the thermotolerance increased by ca. 3 log cycles compared to the thermotolerance of nonadapted cells (control). The same was true for the viability of L. delbrueckii subsp. bulgaricus RD546 cells, which increased by ca. 3 log cycles when the cells were heated at 50°C for 30 min before treatment at 65°C for 10 min (27). A moderate heat shock increased the survival of exponential-phase cells of L. acidophilus NCFM, L. casei LC301, and L. helveticus LH212 after challenge at a high temperature for 20 min by ca. 27-, 5-, and 11-fold, respectively (9). The heat shock tolerance of L. plantarum DPC2739 depended mainly on induction of protein synthesis. Tolerance to 72°C for 90 s decreased markedly when a bacteriostatic concentration of chloramphenicol (1 mg ml−1) was added during adaptation. Similar results were obtained with Lactococcus lactis and L. sanfranciscensis CB1 (16, 53). Adaptation at 42°C for 1 h increased the thermotolerance of stationary-phase L. plantarum DPC2739 cells by ca. 2.0 log cycles. As shown for L. lactis (60, 63), stationary-phase cells showed an inherent resistance to heat shock which was not related to adaptation.
2DE analyses showed that there were increases in the levels of expression of 31 and 18 proteins of adapted mid-exponential- and stationary-phase cells of L. plantarum DPC2739, respectively, compared to the levels of expression in the corresponding controls. Twelve proteins were induced under both conditions. 2DE analyses were carried out with cells grown in MRS broth, and, as shown for heat resistance, cultivation in sterile milk may have slightly affected the response. Nevertheless, all the proteins identified were shown to have a role in the stress response mechanisms of other bacteria.
The adaptive response during the exponential phase of growth usually involves induction of specific groups of genes or regulons to cope with specific stress conditions, while the onset of the stationary phase may lead to multiple-stress resistance. Several studies have shown that stress-induced adaptation occurs during the stationary phase (21, 35, 42). Indeed, several proteins (e.g., spots 3, 31, 32, etc.) were specifically induced during adaptation of stationary-phase cells of L. plantarum DPC2739. Examination of the heat responses of lactobacilli by 2DE has been used widely. Different numbers of proteins were induced depending on the species; 24 proteins were induced in L. acidophilus NCFM, 15 proteins were induced in L. casei LC301, 18 proteins were induced in L. helveticus LH212 (9), and 36 proteins were induced in L. collinoides (38). Analysis of the effect of heat shock on L. plantarum by differential scanning calorimetry showed that the thermal stability of Escherichia coli ribosomes is greater than that of L. plantarum ribosomes (39), and a flow cytometry analysis showed the effect of heat stress on the subsequent duration of the lag time of individual cells (56).
The N-terminal sequences of nine proteins induced during adaptation were determined. All these proteins have been found to play a role in the stress response mechanisms in other bacteria, and for the first time in this study some of them were shown to be involved in the heat response of lactobacilli or, more generally, the heat response of lactic acid bacteria. The main functions of these proteins are described below. DnaK and GroEL were induced only in the adapted mid-exponential-phase cells. By using a specific antibody, spot 47 was identified as GroES, and it was induced under both conditions. These proteins are part of the two major groups of molecular chaperones that are well known as heat shock proteins, which are in the 70-kDa family (DnaK) and the 60-kDa family (GroEL) and function with accessory proteins as chaperone machines. The components of the DnaK chaperone are typically DnaK, DnaJ, and GrpE, while the GroEL chaperone is composed of GroEL and GroES. In situ reconstitution experiments have shown that the DnaK-DnaJ and GroES-GroEL chaperones may interact successively to bind, refold, and release a chemically denatured protein, demonstrating that the two machines are responsible for sequential stages of the same chaperone process (8). As shown for three other species of dairy lactobacilli, the expression of heat shock proteins may vary depending on the species, and, as we found for DnaJ and GrpE, some components of the chaperone machines may not be specifically induced in response to heat shock (9). A cluster of four ribosomal proteins were identified as L1, L11, L31, and S6. As shown for some gram-negative bacteria, ribosomes or ribosome-associated factors are involved in sensing stress (59). Protein L1 plays an important role in both the structure and biological activity of E. coli ribosomes. L1 is among a small group of ribosomal proteins that interact directly with rRNA during translation processes, including the process for resetting the pool of cytoplasmic proteins during the heat stress response (20). The rplK gene, which codes for ribosomal L11 in E. coli and C. glutamicum, was shown to be essential for activation of the RelA protein and, consequently, for synthesis of the stringent mediator (p)ppGpp upon amino acid starvation (22, 61). A putative role for ppGpp during acid stress resistance of L. lactis has been described (52), showing that it is associated with the major signal involved in saving cell energy (11). Ribosomal protein L31 is encoded by the rpmE gene and may be present in two different copies, consisting of 66 to 97 amino acid residues, which are components of the large 50S ribosomal subunit. This protein contains four conserved cysteine residues and rapidly forms intracellular disulfide bonds in vitro. Two of these cysteine residues are arranged as a CXXC motif, which is the sequence commonly found in thiol-disulfide redox proteins, such as thioredoxin. Ribosomal protein S6 was found to be induced in heat-shocked Myxococcus xanthus (47) and was identified as a cold shock protein in E. coli (49) and Bacillus subtilis (29), suggesting that 30S ribosomal protein S6 may play a unique role in sensing temperature differences to control ribosome function. The trigger factor is a peptidylprolyl isomerase which catalyzes protein folding, associates with nascent polypeptides on ribosomes, and cooperates with the GroEL chaperone in promoting the degradation of some unstable proteins (19, 31). The trigger factor also has a role in adaptation of B. subtilis and E. coli to low temperature (29, 34). An HlbA protein with DNA-binding activity and a CspC protein were also identified. DNA-binding protein II, also called HU-type protein, is a small basic protein which binds DNA without sequence specificity but recognizes special DNA structures, such as cruciforms and bulges (50). It plays a role in cell division and modulates the interaction of regulatory proteins with their specific sites on DNA (5). Low-molecular-mass (ca. 7-kDa) cold-induced proteins are often distinguished from the other proteins since they putatively belong to the cold shock protein family. The cspL, cspP, and cspC genes encode small 66-amino-acid proteins in L. plantarum (17, 41), and the amount of these proteins fluctuates strongly during the life cycle, showing that cold shock proteins are not only involved in cold adaptation.
The results of this study showed that the heat resistance of L. plantarum is a complex process, which is also related to cold shock and general stress responses. Heat resistance induced the synthesis of proteins which have various roles in cell physiology, including chaperone activity, ribosome stability, stringent response mediation, temperature sensing, and control of ribosomal function. To our knowledge, this is the first study in which 2DE analyses and identification of heat stress proteins were combined to study the physiological mechanisms of a response to pasteurization in L. plantarum which are fundamental for survival in cheese during manufacture.
Acknowledgments
This work was supported by the Italian Ministry of Agriculture and Forest through the project Strategie innovative per il miglioramento della sicurezza e per la differenziazione di prodotti lattiero-caseari (D.M. no. 41775/2001) and by the project M.I.U.R. “Programmi di Ricerca di Interesse Nazionale Anno 2002. Proteomics to study the environmental adaptation in lactic acid bacteria used in food production.”
We thank Antonio Limitone for technical support.
REFERENCES
- 1.Appel, D., and D. F. Hochstrasser. 1999. Computer analysis of 2-D images. Methods Mol. Biol. 112D:431-443. [DOI] [PubMed] [Google Scholar]
- 2.Bini, L., S. Liberatori, B. Magi, B. Marzocchi, R. Raggiaschi, and V. Pallini. 1999. Protein blotting and immunoblotting, p. 127-141. In T. Rabilloud (ed.), Proteome research: two-dimensional gel electrophoresis and identification methods. Springer. New York, N.Y.
- 3.Bini, L., B. Magi, B. Marzocchi, F. Arcuri, S. Tripodi, M. Cintorino, J. C. Sanchez, S. Frutiger, G. Hughes, V. Pallini, D. F. Hochstrasser, and P. Tosi. 1997. Protein expression profiles in human breast ductal carcinoma and histologically normal tissue. Electrophoresis 18:2832-2841. [DOI] [PubMed] [Google Scholar]
- 4.Bjellqvist, B., G. J. Hughes, C. Pasquali, N. Paquet, F. Ravier, J. C. Sanchez, S. Fructiger, and D. Hochstrasser. 1993. The focusing positions of polypeptides in immobilized pH gradients can be predicted from their amino acid sequences. Electrophoresis 14:1023-1031. [DOI] [PubMed] [Google Scholar]
- 5.Bonnefoy, E., and J. Rouvière-Yaniv. 1992. HU, the major histone-like protein of E. coli, modulates the binding of IHF to oriC. EMBO J. 11:4489-4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boutibonnes, P., C. Tranchard, A. Hartke, B. Thammavongs, and Y. Auffray. 1992. Is thermotolerance correlated to heat shock protein synthesis in Lactococcus lactis subsp. lactis? Int. J. Food Microbiol. 16:227-236. [DOI] [PubMed] [Google Scholar]
- 7.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 8.Broadbent, J. R., C. J. Oberg, and L. Wei. 1998. Characterization of the Lactobacillus helveticus groESL operon. Res. Microbiol. 149:247-253. [DOI] [PubMed] [Google Scholar]
- 9.Broadbent, J. R., J. C. Oberg, H. Wang, and L. Wei. 1997. Attributes of the heat shock response in three species of dairy Lactobacillus. Syst. Appl. Microbiol. 20:12-19. [Google Scholar]
- 10.Casadei, M. A., R. Ingram, E. Hitchings, J. Archer, and J. E. Gaze. 2001. Heat resistance of Bacillus cereus, Salmonella typhimurium and Lactobacillus delbrueckii in relation to pH and ethanol. Int. J. Food Microbiol. 63:125-134. [DOI] [PubMed] [Google Scholar]
- 11.Chatterji, D., and A. K. Ojha. 2001. Revisiting the stringent response, ppGpp and starvation signaling. Curr. Opin. Microbiol. 4:160-165. [DOI] [PubMed] [Google Scholar]
- 12.Corsetti, A., and M. Gobbetti. 2002. Lactobacillus plantarum, p. 1501-1507. In H. Proginsli, J. W. Fuquay, and P. F. Fox (ed.), Encyclopedia of dairy sciences. Academic Press Ltd., New York, N.Y.
- 13.Corsetti, A., M. Gobbetti, E. Smacchi, M. De Angelis, and J. Rossi. 1998. Accelerated ripening of Pecorino Umbro cheese. J. Dairy Res. 65:631-642. [Google Scholar]
- 14.Crow, V., B. Curry, and M. Hayes. 2001. The ecology of non-starter lactic acid bacteria (NSLAB) and their use as adjuncts in New Zealand Cheddar. Int. Dairy J. 11:275-283. [Google Scholar]
- 15.De Angelis, M., and M. Gobbetti. 2004. Environmental stress responses in Lactobacillus: a review. Proteomics 4:106-122. [DOI] [PubMed]
- 16.De Angelis, M., L. Bini, V. Pallini, P. S. Cocconcelli, and M. Gobbetti. 2001. The acid stress response in Lactobacillus sanfranciscensis CB1. Microbiology 147:1863-1873. [DOI] [PubMed] [Google Scholar]
- 17.Derzelle, S., B. Hallet, K. P. Francis, T. Ferain, J. Delcour, and P. Hols. 2000. Changes in cspL, cspP, and cspC mRNA abundance as a function of cold shock and growth phase in Lactobacillus plantarum. J. Bacteriol. 182:5105-5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Desmond, C., C. Stanton, G. F. Fitzgerald, K. Collins, and R. P. Ross. 2001. Environmental adaptation of probiotic lactobacilli towards improvement of performance during spray drying. Int. Dairy J. 11:801-808. [Google Scholar]
- 19.Deuerling, E., A. Schulze-Specking, T. Tomoyasu, A. Mogk, and B. Bukau. 1999. Trigger factor and DnaK cooperate in folding of newly synthesized proteins. Nature 400:693-696. [DOI] [PubMed] [Google Scholar]
- 20.Drygin, D., and A. R. Zimmermann. 2000. Magnesium ions mediate contacts between phosphoryl oxygens at positions 2122 and 2176 of the 23S rRNA and ribosomal protein L1. RNA 6:1714-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duwat, P., B. Cesselin, S. Sourice, and A. Gruss. 2000. Lactococcus lactis, a bacterial model for stress responses and survival. Int. J. Food Microbiol. 55:83-86. [DOI] [PubMed] [Google Scholar]
- 22.Friesen, J. D., N. P. Fiil, J. M. Parker, and W. A. Haseltine. 1974. A new relaxed mutant of Escherichia coli with an altered 50S ribosomal subunit. Proc. Natl. Acad. Sci. 71:3465-3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gardiner, G. E., E. O'Sullivan, J. Kelly, M. A. E. Auty, G. F. Fitzgerald, J. K. Collins, R. P. Ross, and C. Stanton. 2000. Comparative survival rates of human-derived probiotic Lactobacillus paracasei and L. salivarius strains during heat treatment and spray drying. Appl. Environ. Microbiol. 66:2605-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gobbetti, M., R. Lanciotti, M. De Angelis, M. R. Corbo, R. Massini, and P. Fox. 1999. Study of the effects of temperature, pH, NaCl, and aw on the proteolytic and lipolytic activities of cheese-related lactic acid bacteria by quadratic response surface methodology. Enzyme Microb. Technol. 25:795-809. [Google Scholar]
- 25.Gomez, M. J., P. Gaya, M. Nunez, and M. Medina. 1996. Effect of Lactobacillus plantarum as adjunct starter on the flavour and texture of a semi-hard cheese made from pasteurised cow's milk. Lait 76:461-472. [Google Scholar]
- 26.Görg, A., W. Postel, and S. Gunther. 1988. The current state of two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis 9:531-546. [DOI] [PubMed] [Google Scholar]
- 27.Gouesbert, G., G. Jan, and P. Boyaval. 2001. Lactobacillus delbrueckii subsp. bulgaricus thermotolerance. Lait 81:301-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gouesbert, G., G. Jan, and P. Boyaval. 2002. Two-dimensional electrophoresis study of Lactobacillus delbrueckii subsp. bulgaricus thermotolerance. Appl. Environ. Microbiol. 68:1055-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Graumann, P., K. Schroder, R. Schmid, and M. A. Marahiel. 1996. Cold shock stress-induced proteins in Bacillus subtilis. J. Bacteriol. 178:4611-4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hartke, A., S. Bouchè, J. C. Giard, A. Benachour, P. Boutibonnes, and Y. Auffray. 1996. The lactic acid stress response of Lactococcus lactis subsp. lactis. Curr. Microbiol. 33:194-199. [DOI] [PubMed] [Google Scholar]
- 31.Hesterkamp, T., and B. Bukau. 1996. The Escherichia coli trigger factor. FEBS Lett. 389:32-34. [DOI] [PubMed] [Google Scholar]
- 32.Hochstrasser, D. F., M. G. Harrington, A. C. Hochstrasser, M. J. Miller, and C. R. Merril. 1988. Methods for increasing the resolution of two dimensional protein electrophoresis. Anal. Biochem. 173:424-435. [DOI] [PubMed] [Google Scholar]
- 33.Jordan, K. N., and T. M. Cogan. 1999. Heat resistance of Lactobacillus spp. isolated from Cheddar cheese. Lett. Appl. Microbiol. 29:136-140. [DOI] [PubMed] [Google Scholar]
- 34.Kandror, O., and A. L. Goldberg. 1997. Trigger factor is induced upon cold shock and enhances viability of Escherichia coli at low temperatures. Proc. Natl. Acad. Sci. 94:4978-4981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim, W., J. Ren, and N. Dunn. 1999. Differentiation of Lactococcus lactis subspecies lactis and subspecies cremoris strains by their adaptive response to stresses. FEMS Microbiol. Lett. 171:57-63. [DOI] [PubMed] [Google Scholar]
- 36.Kleerebezem M., J. Boekhorst, R. van Kranenburg, D. Molemaar, O. P. Kiupers, R. Leer, R. Tarchini, S. A. Peters, H. M. Sandbrink, M. W. E. J. Fiers, W. Stiekema, R. M. Klein Lankhorst, P. A. Bron, S. M. Hoffer, M. N. Nierop Groot, R. Kerkhoven, M. de Vries, B. Ursing, W. M. De Vos, and R. J. Siezen. 2003. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. 100:1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lane, C. N., and P. F. Fox. 1996. Contribution of starter and adjunct lactobacilli to proteolysis in Cheddar cheese during ripening. Int. Dairy J. 6:728-751. [Google Scholar]
- 38.Laplace, J. M., N. Sauvageot, A. Harke, and Y. Auffray. 1999. Characterization of Lactobacillus collinoides response to heat, acid and ethanol treatments. Appl. Microbiol. Biotechnol. 51:659-663. [Google Scholar]
- 39.Lee, J., and G. Kaletunc. 2002. Evaluation of the heat inactivation of Escherichia coli and Lactobacillus plantarum by differential scanning calorimetry. Appl. Environ. Microbiol. 68:5379-5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Magi, B., L. Bini, B. Marzocchi, S. Liberatori, R. Raggiaschi, and V. Pallini. 1999. Immunoaffinity identification of 2-DE separated proteins. Methods Mol. Biol. 112D:431-443. [DOI] [PubMed] [Google Scholar]
- 41.Mayo, B., S. Derzelle, L. Fernandez, C. Leonard, T. Ferain, P. Hols, J. E. Suarez, and J. Delcour. 1997. Cloning and characterization of cspL and cspP, two cold-inducible genes from Lactobacillus plantarum. J. Bacteriol. 179:3039-3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McMohan, C., C. Byrne, J. J. Sheridan, D. A. McDowell, J. S. Blair, and T. Hegarty. 2000. The effect of culture growth phase on induction of the heat shock response in Yersinia enterocolitica and Listeria monocytogenes J. Appl. Microbiol. 89:198-206. [DOI] [PubMed] [Google Scholar]
- 43.McSweeney, P. L. H., E. M. Walsh, P. F. Fox, T. M. Cogan, F. D. Drinan, and M. Castelo-Gonzalez. 1994. A procedure for the manufacture of Cheddar cheese under controlled bacteriological conditions and the effect of adjunct lactobacilli on cheese quality. Irish J. Agric. Food Res. 33:183-192. [Google Scholar]
- 44.McSweeney, P. L. H., P. F. Fox, J. A. Lucey, K. N. Jordan, and T. M. Cogan. 1993. Contribution of the indigenous microflora to the maturation of Cheddar cheese. Int. Dairy J. 3:613-634. [Google Scholar]
- 45.Messer, M., M. Griffiths, P. D. Rismiller, and D. C. Shaw. 1997. Lactose synthesis in a monotreme, the echidna (Tachyglossus aculeatus): isolation and amino acid sequence of echidna α-lactalbumin. Comp. Biochem. Physiol. 118B:403-410. [DOI] [PubMed] [Google Scholar]
- 46.Oakley, B. R., D. R. Kirsch, and R. Morris. 1980. A simplified ultrasensitive silver stain for detecting proteins in polyacrylamide gels. Anal. Biochem. 105:361-363. [DOI] [PubMed] [Google Scholar]
- 47.Otani, M., J. Tabata, T. Ueki, K. Sano, and S. Inouve. 2001. Heat-shock-induced proteins from Myxococcus xanthus. J. Bacteriol. 183:6282-6287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peterson, S. D., and R. T. Marshall. 1990. Non-starter lactobacilli in Cheddar cheese: a review. J. Dairy Sci. 73:1395-1410. [Google Scholar]
- 49.Phadtare, S., K. Yamanaka, and M. Inouye. 2000. The cold stress response, p. 35-45. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.
- 50.Pontiggia, A., A. Negri, M. Beltrame, and M. E. Bianchi. 1993. Protein HU binds specifically to kinked DNA. Mol. Microbiol. 7:343-350. [DOI] [PubMed] [Google Scholar]
- 51.Prasad, J., P. McJarrow, and P. Gopal. 2003. Heat and osmotic stress responses of probiotic Lactobacillus rhamnosus HN001 (DR20) in relation to viability after drying. Appl. Environ. Microbiol. 69:917-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rallu, F., A. Gruss, S. D. Ehrlich, and E. Maguin. 2000. Acid- and multistress-resistant mutants of Lactococcus lactis: identification of intracellular stress signals. Mol. Microbiol. 35:517-528. [DOI] [PubMed] [Google Scholar]
- 53.Rallu, F., A. Gruss, and E. Maguin. 1996. Lactococcus lactis and stress. Antonie Leeuwenhoek 70:243-251. [DOI] [PubMed] [Google Scholar]
- 54.Schmidt, G., C. Hertel, and W. P. Hammes. 1999. Molecular characterisation of the danK operon of Lactobacillus sakei LTH681. Syst. Appl. Microbiol. 22:321-328. [DOI] [PubMed] [Google Scholar]
- 55.Shakeel-Ur-Rehman, P. L. H. McSweeney, J. M. Banks, E. Y. Brechany, D. D. Muir, and P. F. Fox. 2000. Ripening of Cheddar cheese made from blends or raw and pasteurised milk. Int. Dairy J. 10:33-44. [Google Scholar]
- 56.Smelt, J. P. G. D. Otten, and A. P. Bos. 2002. Modelling the effect of sublethal injury on the distribution of the lag times of individual cells of Lactobacillus plantarum. Int. J. Food Microbiol. 73:207-212. [DOI] [PubMed] [Google Scholar]
- 57.Stumbo, C. R. 1965. Thermobacteriology in food processing. Academic Press, New York, N.Y.
- 58.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Van Bogelen, R., and F. C. Neidhardt. 1990. Ribosomes as sensor of heat and cold shock in Escherichia coli. Proc. Natl. Acad. Sci. 87:5589-5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Van de Guchte, M., P. Serror, C. Chervaux, T. Smokvins, S. D. Ehrlich, and E. Maguin. 2002. Stress response in lactic acid bacteria. Antonie Leeuwenhoek 82:187-216. [PubMed] [Google Scholar]
- 61.Wehmeier, L., O. Brockmann-Gretza, A. Pisabarro, A. Tauch, A. Pühler, J. F. Martin, and J. Kalinowski. 2001. A Corynebacterium glutamicus mutant with a defined deletion within the rplk gene is impaired in (p)ppGpp accumulation upon amino acid starvation. Microbiology 147:691-700. [DOI] [PubMed] [Google Scholar]
- 62.Williams, A. G., J. Noble, J. Tammam, D. Lloyd, and J. M. Banks. 2002. Factors affecting the activity of enzymes involved in peptide and amino acid catabolism in non starter lactic acid bacteria isolated from Cheddar cheese. Int. Dairy J. 12:841-852. [Google Scholar]
- 63.Wouters, J. A., B. Jeynov, F. M. Rombouts, W. M. de Vos, O. P. Kiupers, and T. Abe. 1999. Analysis of the role of 7 KDa proteins of Lactococcus lactis MG 1363 in cryoprotection. Microbiology 145:3185-3194. [DOI] [PubMed] [Google Scholar]
- 64.Zink, R., C. Walker, G. Schmidt, M. Elli, D. Pridmore, and R. Reniero. 2000. Impact of multiple stress factors on the survival of dairy lactobacilli. Sci. Aliments 20:119-126. [Google Scholar]
- 65.Zwietering, M. H., I. Jongeberger, F. M. Roumbouts, and K. van't Riet. 1990. Modelling of bacterial growth curve. Appl. Environ. Microbiol. 56:1875-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]