Abstract
Background
Prevention of rectal HIV transmission is a high priority goal for vaccines and topical microbicides because a large fraction of HIV transmissions occurs rectally. Yet, little is known about the specific target cell milieu in the human rectum other than inferences made from the colon.
Methods
We conducted a comprehensive comparative in situ fluorescence study of HIV target cells (CCR5-expressing T cells, macrophages and putative dendritic cells) at 4 and 30 cm proximal of the anal canal in 29 healthy individuals, using computerized analysis of digitized combination stains.
Results
Most strikingly, we find that more than three times as many CD68+ macrophages express the HIV co-receptor CCR5 in the rectum than the colon (p=0.0001), and as such rectal macrophages appear biologically closer to the HIV-susceptible CCR5high phenotype in the vagina than the mostly HIV-resistant CCR5low phenotype in the colon. Putative CD209+ dendritic cells are generally enriched in the colon compared to the rectum (p=0.0004), though their CCR5 expression levels are similar in both compartments. CD3+ T cell densities and CCR5 expression levels are comparable in the colon and rectum.
Conclusions
Our study establishes the target cell environment for HIV infection in the human distal gut and demonstrates in general terms that the colon and rectum are immunologically distinct anatomical compartments. Greater expression of CCR5 on rectal macrophages suggests that the most distal sections of the gut may be especially vulnerable to HIV infection. Our findings also emphasize that caution should be exercised when extrapolating data obtained from colon tissues to the rectum.
Introduction
HIV infection frequently occurs through anal intercourse, in both men having sex with men and in women,1–7 so the distal gut is an important target organ for achieving HIV control through topical microbicides or vaccination. The design of effective prevention strategies depends on knowing where HIV penetrates the gut mucosa and establishes infection most successfully and what the target cell composition is at that site. In the macaque model, SIV penetration through the rectal mucosa, followed by rapid dissemination to local lymphatic tissues, has been shown.8 In humans, however, it remains unclear which anatomical sections of the distal gut (anal canal, rectum or sigmoid colon) are most vulnerable to infection upon luminal contact with HIV.
A recent study showed that surrogates of cell-free and cell-associated HIV (99mTc labeled sulfur colloid particles and 111In-oxine labeled autologous leukocytes, respectively) introduced into the rectum through simulated intercourse reached their highest concentrations 10–20 cm from the anus, where the rectum transitions into the sigmoid colon.9 The occurrence of HIV infection in humans cannot be directly observed in vivo but in general depends highly on the likelihood of virion penetration into the mucosa and the local availability of susceptible target cells.10 For the colon, target cell availability has been relatively well established: T cells tend to express CCR5 and be highly susceptible to HIV,11–20 while macrophages express little to no CCR5 and are mostly resistant.21–25. Myeloid dendritic cells (DCs) and epithelial cells were shown to enhance infection by disseminating HIV to T cells in the colon.26–28 In contrast to the colon, less is known about target cell density and cell-virus interactions within the rectum. The rectum contains many CCR5+ T cells,29 but macrophages and DCs have not been studied at this site. Thus, whether immunological variation - and potentially differences in HIV susceptibility - exists between the colon and the rectum remains unexplored. A better understanding of the rectal mucosa could be important for the design of rectal microbicide gels to prevent anal HIV transmission. Moreover, as intestinal immune responses to HIV vaccination are currently measured in the colon, the extent to which colon biopsies are fully representative of the rectum needs to be addressed.
In this study, we took advantage of the unique opportunity of having biopsies available from both the rectum (taken 4 cm proximal to the pectinate line in the anal canal) and colon (~30 cm) of 29 healthy men who were enrolled in an exploratory follow-up study of a phase 2B HIV vaccine trial (Step Study).30, 31 This allowed us to compare with high statistical reliability the absolute and relative densities of the three main potential target cell populations for HIV infection - T cells, macrophages and DCs - as well as their CCR5 expression levels in the rectum and colon. Our results show that many more macrophages express CCR5 in the rectum than the colon, indicating a potential increase in susceptibility to HIV infection toward the most distal part of the large bowel.
Materials and Methods
Study Subjects
The 29 study subjects from HVTN 905 Project 01, a study examining long-term cellular immune responses at mucosal sites following immunization with HIV antigens previously delivered using an adenovirus serotype 5 vector, MRK HIV-1 gag/pol/nef (Step Study).30, 31 No investigational agents were administered in HVTN 905 Project 01. The median time period between administration of the last investigational vaccine dose during the Step Study and the intestinal biopsies taken from the 29 subjects in this study was 866 days (mean 918, σ 168). The 29 study subjects were healthy US men who have sex with men aged 25 to 53 years (median and mean 38). At the time of entry into the original Step Study the subjects were at high risk of HIV-1 acquisition on the basis of reported unprotected anal intercourse with a male partner or anal intercourse with two or more male partners in the 6 months before enrollment.30 Risk was not an eligibility criteria for enrollment into the HVTN 905 protocol and thus was not assessed again before 905 study entry. Additional information on study subjects, human subjects protection and clinical specimens is given in Supplemental Methods.
Biopsy Staining and Analysis
Biopsies were processed and stained for the combinations CD3/CCR5, CD68/CCR5 or CD209/CCR5 as described in detail in Supplemental Methods. A sequence of slightly overlapping 20x images covering each stained tissue section in its entirety was acquired on a Zeiss Imager Z2 microscope, and overlapping images stitched together, using TissueFAXS (TissueGnostics, Vienna, Austria) (Figure 1A). Each stained marker was acquired separately as a monochrome image. Monochrome images were superimposed and rendered in pseudo-colors using TissueQuest Analysis Software (TissueGnostics) as depicted in Figure 1B. Individual cells were identified based on the nuclear counter-stain, with comparable accuracy of nuclei counts by the software and a human operator (Figure 1C and D). Cell outlines were superimposed on the acquired images of the violet and far red stains and assessed for expression of the phenotypic markers (CD3, CD68 or CD209) and CCR5. Measurements of individual cell fluorescence were displayed in two-dimensional dot plots, with each cell defined by a combination of its specific mean (x axis) and maximum (y axis) fluorescence intensity (Figure 2B). Positive-negative cut-offs for phenotypic marker or CCR5 expression were based on isotype control stains. Only cells above the positive-negative cut-off for both mean and maximum fluorescence intensity were considered positive, ensuring a conservative assessment of marker positivity. Cells positive for either CD3, CD68 or CD209 were gated and displayed on a downstream scatterplot depicting their mean and maximum fluorescence intensity in the CCR5 staining channel (Figures 3A and 4A). Cell counting was performed automatically on the entire slide for each tissue section, permitting a more robust and objective analysis than traditional manual screening and cell counting methods that rely on the analysis of a limited number of user-selected fields of view.
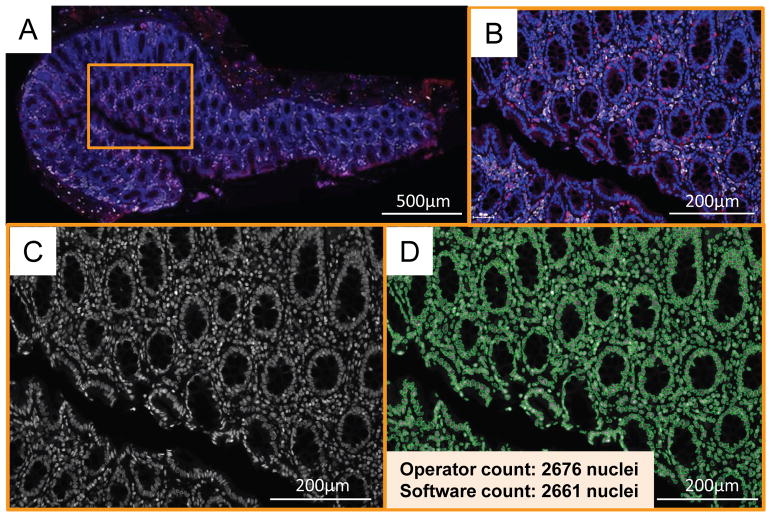
Figure 1.
Tissue stitching and nuclear recognition. A–D: A representative colon section stained as described in the Methods with anti-CCR5 (detected by AlexaFluor350 and pseudo-colored in red), anti-CD68 (detected by AlexaFluor647 and pseudo-colored in white) and the nuclear stain Sytox Orange (pseudo-colored in blue) is depicted in its entirety. TissueFAXS software (TissueGnostics) stitched together this image of the complete colon section on the slide by aligning all overlapping 20x fields-of-view that had been sequentially acquired on the microscope (A). One 20x field-of-view is marked in A by the orange rectangle, which is then shown magnified in B–D. B: Three-color stain of cells in colon tissue, depicting CCR5 in red, CD68 in white and nuclei in blue. C: Nuclear stain Sytox Orange in monochrome grayscale. D: Recognition of the individual nuclei by the algorithm of the TissueQuest (TissueGnostics) software. Operator and software cell counts are indicated.
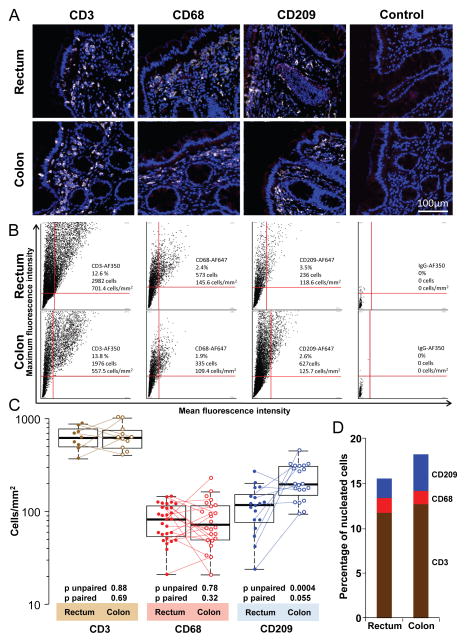
Figure 2.
Density of T cells, macrophages and putative dendritic cells (DCs) in the human rectum and colon. A: Representative 20x pseudo-color images in one individual of CD3 (T cells), CD68 (macrophages) and CD209 (DCs) in the rectum and colon. Tissue sections were stained as described in Methods for three-color immunofluorescence: anti-CD3, anti-CD68 or anti-CD209 in one fluorescence channel (pseudo-colored in white), mouse IgG isotype control for anti-CCR5 staining in a second fluorescence channel (red), and the nuclear counter-stain Sytox Orange in the third channel (blue). Note that these sections therefore also depict representative single isotype controls for the CCR5 staining data presented in Figures 3 and 4. The two control images in A are double isotype controls with no marker-specific primary antibodies. B: Dot plots corresponding to biopsies in panel A, but showing data from all cells in the entire tissue sections. Each dot corresponds to one cell, defined by its specific mean and maximum fluorescence intensity. Quadrant cutoffs were determined by isotype control stains. Only cells in the upper right quadrant were judged as positive. The numbers and percentages of cells in the upper right quadrants, and their corresponding cell densities, are specified in each plot. C: Summary of CD3+, CD68+ and CD209+ cell densities in all biopsies analyzed. The horizontal line in each box denotes the median, the ends of the box denote the 25th and 75th percentiles, and the whiskers extend to the extreme data points that are no more than 1.5 x the interquartile range. Paired samples, i.e., where the data were derived from a colon and rectum biopsy obtained from the same individual at the same time point, are connected by dotted lines. Unpaired (Mann-Whitney U test) and paired (Wilcoxon signed-rank test) p values for differences between rectum and colon are listed below the corresponding boxes. D: Median percentages of CD3+, CD68+ and CD209+ cells among all nucleated cells.
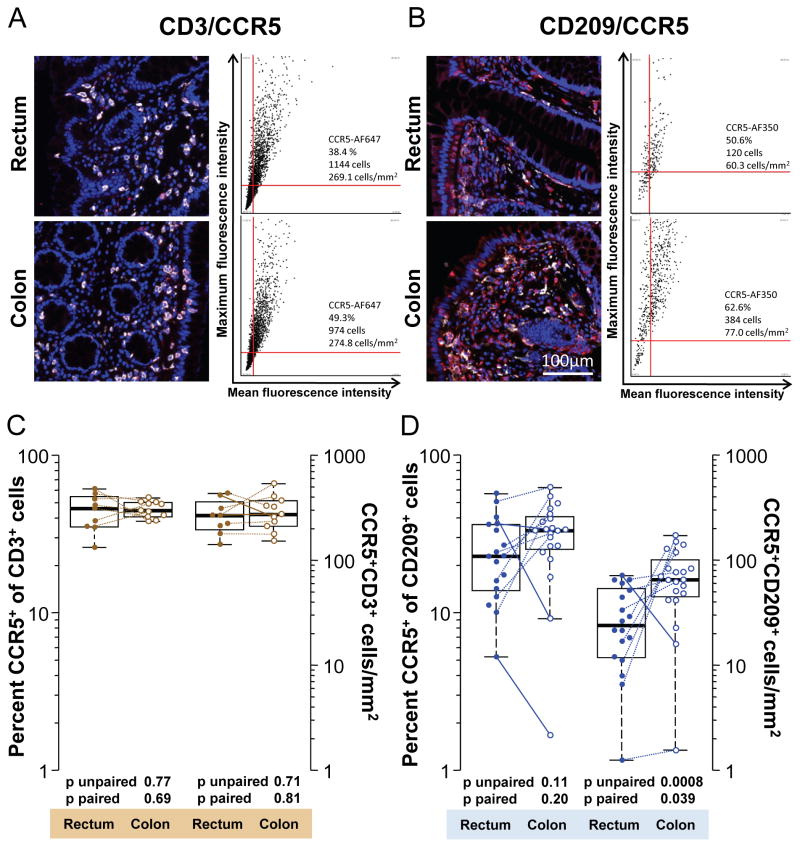
Figure 3.
CCR5 expression by T cells and putative DCs in the human rectum and colon. A: Representative 20x pseudo-color images in one individual of CD3 (white) and CCR5 (red) co-staining in the rectum and colon. Tissue sections were stained as described in Methods. Mouse IgG isotype controls for CCR5 staining are shown in Figure 2A. Corresponding dot plots are shown next to each image and depict CCR5 mean and maximum fluorescence intensities of cells that were first gated to stain positive for CD3. Thus, cells within the upper right quadrants of the dot plots co-express CD3 and CCR5. The numbers of CD3+ CCR5+ cells in the upper right quadrants and their corresponding cell densities are specified in each plot. Also given is the percentage of CD3+ T cells co-expressing CCR5. B: Representative 20x pseudo-color images and dot plots in one individual of CD209 (white) and CCR5 (red) co-staining in the rectum and colon. C: Summary of percentages (left y axis) of CD3+ T cells co-expressing CCR5 and their cell densities (right y axis) in all biopsies analyzed. D: Summary of percentages (left y axis) of CD209+ cells co-expressing CCR5 and their cell densities (right y axis) in all biopsies analyzed. In C and D, paired samples, i.e., where the data were derived from a colon and rectum biopsy obtained from the same individual at the same time point, are connected by lines. Differences going against the prevailing trend are indicated by solid lines, others by dotted lines.
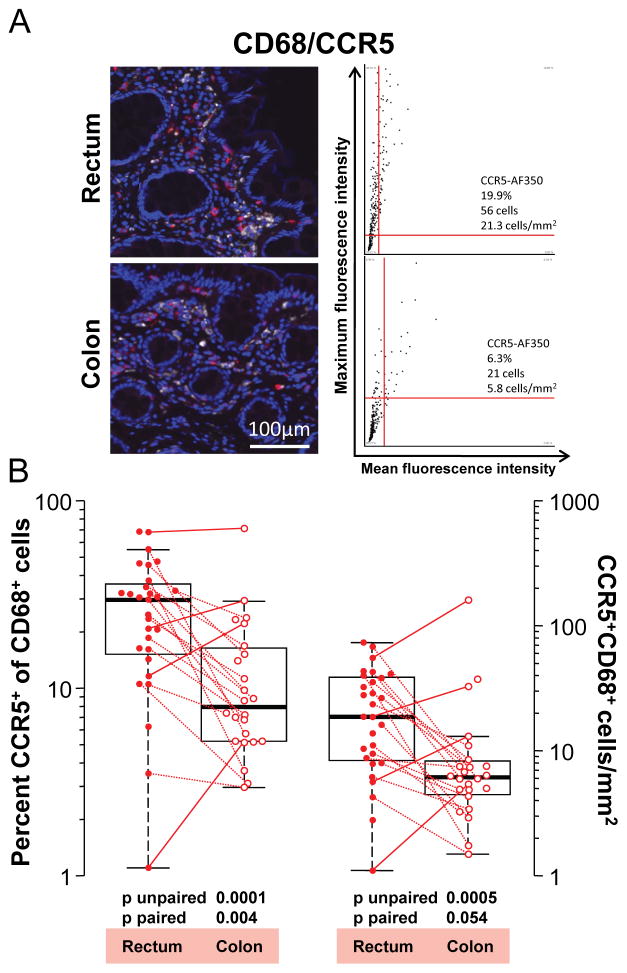
Figure 4.
CCR5 expression by macrophages in the human rectum and colon. A: Representative 20x pseudo-color images in one individual of CD68 (white) and CCR5 (red) co-staining in the rectum and colon. Tissue sections were stained as described in Methods. Mouse IgG isotype controls for CCR5 staining are shown in Figure 2A. Corresponding dot plots are shown next to each image and depict CCR5 mean and maximum fluorescence intensities of cells that were first gated to stain positive for CD68. Thus, cells within the upper right quadrants of the dot plots co-express CD68 and CCR5. The numbers of CD68+ CCR5+ cells in the upper right quadrants and their corresponding cell densities are specified in each plot. Also given is the percentage of CD68+ macrophages co-expressing CCR5. B: Summary of percentages (left y axis) of CD68+ macrophages co-expressing CCR5 and their cell densities (right y axis) in all biopsies analyzed. Paired samples, i.e., where the data were derived from a colon and rectum biopsy obtained from the same individual at the same time point (n=18), are connected by lines. Differences going against the prevailing trend are indicated by solid lines, others by dotted lines.
Isotype control slides were used to determine the overall background in our data set. Double isotype controls were negative (Figure 2A), and single isotype controls for the phenotypic markers also showed only negligible background (not shown). Single isotype controls for CCR5, a representative set of which is shown in Figure 2A, displayed the following background levels across all control slides tested: (1) CCR5 isotype-positive cells among CD3+ T cells (n=11 slides): median 0.155% (mean 0.146, σ 0.134) in colon, and median 0.103% (mean 0.171, σ 0.16) in rectum. (2) CCR5 isotype-positive cells among CD68+ macrophages (n=30 slides): median 0% (mean 0.117, σ 0.343) in colon, and median 0% (mean 0.534, σ 1.075) in rectum. (3) CCR5 isotype-positive cells among CD209+ cells (n=24 slides): median 0.176% (mean 0.403, σ 0.688) in colon, and median 0% (mean 0.642, σ 1.095) in rectum.
Results
T cell, macrophage and CD209+ cell (putative DC) densities in rectum and colon
To assess how potential target cells for HIV-1 infection distribute in the colon and the rectum, we stained formalin fixed tissue sections from both sites (~30 and 4 cm proximal of the pectinate line in the anal canal) for markers characteristic of T cells (CD3), putative DCs (CD209) and macrophages (CD68). Representative images of cell type staining and corresponding dot plots depicting the total cells counted in all 20× fields-of-view of each of these samples, are shown in Figure 2A and 2B. The median density of T cells in rectum biopsy sections (n=9) was 610 cells/mm2 (mean 617, σ 162). The median density of CD3+ T cells in the colon (n=11) was 613/mm2 (mean 648, σ 223) (Figure 2C). Seven of these samples were paired, i.e., rectum and colon biopsies taken from the same individuals on the same day. CD3+ T cell densities did not differ between the rectum and the colon (p unpaired=0.88; p paired=0.69).
The median density of CD68+ macrophages in the rectum (n=29) was 81/mm2 (mean 83, σ 36) and in the colon (n=24) it was 71/mm2 (mean 85, σ 46) (Figure 2C). Eighteen of the CD68-stained samples were paired. Like the T cells, CD68+ macrophages resided at comparable densities in the rectum and colon (p unpaired=0.78; p paired=0.32).
CD209-expressing cells were present at higher densities in the colon than in the rectum. The median density in the rectum (n=17) was 116/mm2 (mean 118, σ 65), whereas in the colon (n=21) it was 194/mm2 (mean 220, σ 85) (Figure 2C). This difference was markedly significant by unpaired testing of all biopsy donors (p=0.0004) but less so in the nine donors with paired staining (p=0.055).
The same pattern can be seen in the frequencies of T cells, macrophages and DCs among all nucleated cells evaluated in each biopsy section (Figure 2D). A median of 11.7% (mean 11.7, σ 3.2) and 10.7% (mean 12.7, σ 3.9) of all cells in the rectum and colon mucosa, respectively, were CD3+ T cells (p unpaired=0.7; p paired=0.69). A median of 1.5% (mean 1.6, σ 0.67) and 1.3% (mean 1.5, σ 0.87) of all cells were CD68+ macrophages (p unpaired=0.49; p paired=0.56), and a median of 2% (mean 2.2, σ 1.2) and 3.6% (mean 4.1, σ 1.8) of all cells were CD209+ cells (p unpaired=0.001; p paired=0.13) in the rectum and colon, respectively. In summary, the colon mucosa was enriched in CD209+ putative DCs, but not in T cells and macrophages, compared to the rectum mucosa. T cells remained by far the most abundant of the three cell populations we measured in the rectum as well as in the colon.
Percentages of CCR5-expressing T lymphocytes and putative DCs are similar between rectum and colon, but the colon is generally enriched in putative DCs
In most cases HIV transmission occurs with viruses using the chemokine receptor CCR5 to enter and infect CD4+ host cells. The distribution pattern of CCR5 expression at potential mucosal sites of HIV transmission is therefore of particular interest. While CCR5 expression in the human colon has been investigated before, we found no information detailing its distribution on different leukocyte populations in the rectum immediately proximal to the anal canal. We therefore compared the expression of CCR5 on potential target cells for HIV between the colon (~30 cm) and the distal rectum (4 cm) by co-staining for phenotypic markers and CCR5 (Figures 3 and 4). We analyzed the same number of total and paired biopsies as specified above for the single phenotypic stains. We found that a median of 46.1% (mean 44.5, σ 11.6) and 44.6% (mean 45.5, σ 5.38) of CD3+ T cells expressed CCR5 in the rectum and colon, respectively (Figure 3A and C) (p unpaired=0.77; p paired=0.69). This corresponded to a median density of CCR5+ CD3+ T cells of 267 cells/mm2 (273, σ 303) in the rectum and of 275 cells/mm2 (mean 298, σ 112) in the colon (p unpaired=0.71; p paired=0.81).
The median percentage of CCR5-expressing CD209+ cells was 22.9% (mean 25.8, σ 14.4) in the rectum and 33.3% (mean 32.9, σ 14.2) in the colon (p unpaired=0.11; p paired=0.20) (Figure 3D). The median density of CCR5+ CD209+ cells was 24 cells/mm2 (mean 31.4, σ 22.7) in the rectum and 65.2 cells/mm2 (mean 74.9, σ 44.9) in the colon (p unpaired=0.0008; p paired=0.039). Thus, by percentage, neither T cells nor putative DCs differed significantly between the rectum and the colon. Likewise, CCR5+ CD3+ T cells populated both compartments at comparable densities. In contrast, the general enrichment of putative DCs in the colon compared to the rectum, as seen in Figure 2C, extended to significantly higher densities of CCR5+ putative DCs in the colon as well (Figure 3D).
Percentages and densities of CCR5-expressing macrophages are significantly higher in the rectum than the colon
A median of 29.8% (mean 24.7, σ 17.5) of macrophages stained positive for CCR5 in the rectum but only 8% (mean 13.3, σ 14.4) stained positive for CCR5 in the colon (p unpaired = 0.0001; p paired = 0.004) (Figure 4). Similarly, the median density of CCR5+ CD68+ macrophages was 18.7 cells/mm2 (24.8, σ 19.5) in the rectum but only 6.1 cells/mm2 (14.7, σ 32.2) in the colon (p unpaired = 0.0005; p paired = 0.054) (Figure 4). Thus, CCR5 was expressed on many more macrophages in the rectum than in the colon.
T cells contribute the largest number of CCR5+ cells in the rectum and in the colon
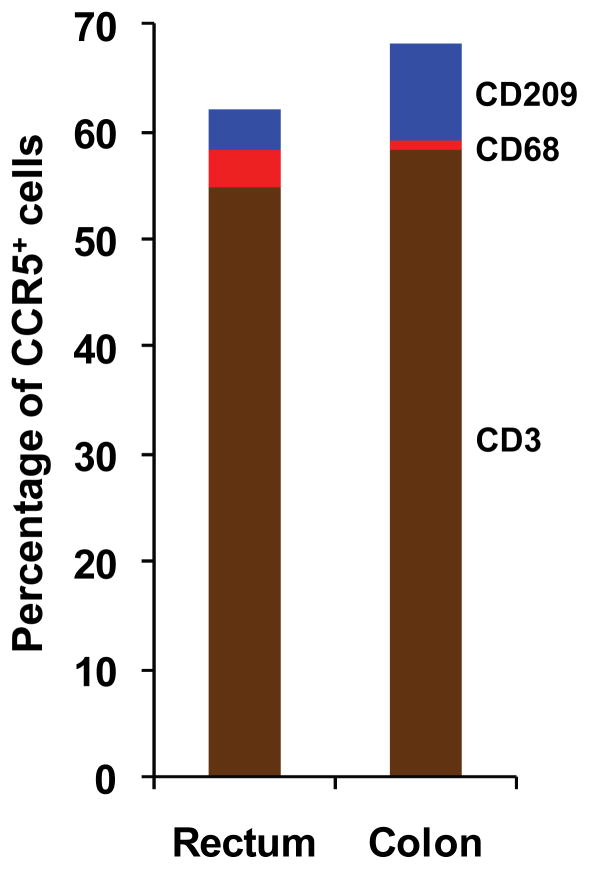
We noted in our initial analysis that some nucleated cells which we could not identify as CD3+ T cells, CD209+ putative DCs or CD68+ macrophages stained positive for CCR5. We therefore also counted the total number of CCR5+ cells in each stained section, irrespective of whether they co-stained for one of our three phenotypic markers. Among the slides co-stained with CD3, a median of 520 total cells/mm2 (mean 494, σ 184) and 459 total cells/mm2 (mean 509, σ 169) tested positive for CCR5 in the rectum and colon, respectively (p unpaired=1.0; p paired=0.81). Among the slides co-stained with either CD209 or CD68, in which the CCR5 stain was amplified by catalyzed signal amplification, a median of 543 total cells/mm2 (mean 604, σ 322) and 693 total cells/mm2 (mean 739, σ 249) tested positive for CCR5 in the rectum and colon, respectively (p unpaired=0.1; p paired=0.37) (data not shown). In the rectum, a median of 54.9% (mean 56.1, σ 9.25) of these CCR5+ cells were CD3+ T cells, 3.69% (mean 4.7, σ 3.81) were CD209+ cells and 3.53% (mean 4.72, σ 4.61) were CD68+ macrophages (Figure 5). In the colon, a median of 58.14% (mean 61.82, σ 8.64) of total CCR5+ cells were CD3+ T cells, 8.93% (mean 11.67, σ 8.96) were CD209+ cells and 1.05% (mean 2.32, σ 4.33) were CD68+ macrophages (Figure 5). The percentage of CCR5+ CD209+ putative DCs among all CCR5+ cells was significantly higher in the colon compared to the rectum (p unpaired=0.001; p paired=0.07), whereas the percentage of CCR5+ CD68+ macrophages among all CCR5+ cells was significantly higher in the rectum compared to the colon (p unpaired=0.0003; p paired=0.09). The percentages of CCR5+ CD3+ T cells among all CCR5+ cells were not significantly different between the rectum and the colon (p unpaired=0.36; p paired=0.22). These results reflect our findings above that CCR5+ macrophages were enriched in the rectum and CCR5+ putative DCs were enriched in the colon. In both anatomical compartments, however, T cells contributed well over half of all CCR5-expressing cells and a far larger share than did DCs and macrophages combined.
Figure 5.
Median percentages of CCR5-expressing CD3+, CD68+ and CD209+ cells among all cells expressing CCR5. Percentages were first calculated separately for each stained tissue section, with subsequent computation of the medians and other descriptive statistics (see text).
Discussion
To our knowledge, this is the first systematic comparison of CCR5 expression on leukocyte subpopulations within the colon versus the distal part of the rectum bordering the anal canal. Intriguingly, we found that macrophages in the rectum express CCR5 quite abundantly. Similarly high CCR5 expression levels have been reported for macrophages in the vagina, a phenotype that correlates with substantial susceptibility to infection with R5-tropic HIV isolates.22 In contrast, low CCR5 expression by macrophages in the colon, as reported by others and confirmed by our study,21–25, 32 correlates with reduced permissiveness to HIV infection.22, 23, 25 Thus, our findings suggest that, in terms of CCR5 expression, macrophages in the human rectum are rather more like vaginal than colonic macrophages, and are therefore likely targets for productive HIV infection. Visually, these rectal macrophages form a dense stromal network of larger CCR5+ cells just beneath the columnar epithelium (Figures 2A and 4A) and should thus be easily accessible to virions penetrating the mucosa. Consequently, considering that the rectum likely bears the highest burden of HIV exposure during anal intercourse, topical microbicide strategies with antiretroviral compounds such as tenofovir need to ensure sufficiently high active drug concentrations in rectal macrophages in addition to T cells. Likewise, our data support that HIV vaccines need to achieve protective immunity in the rectum, which implies that cellular immune responses should also be measured there rather than in surrogate colon tissue. Heretofore, colon was the preferred site for cellular immune assays chiefly because more biopsies can be safely obtained from the colon than the rectum. With the emergence of more sophisticated analysis techniques suitable for lower cell yields, such as single-cell, subnanoliter well arrays and single-cell mass spectrometry cytometers,33, 34 measurements in the rectum have become more feasible and may often be preferable to colon.
Up-regulation of CCR5 on macrophages in the rectum relative to the colon may be understood in the context of other aspects of mucosal immunology in the distal gut. There is a clear link between stromal factors in the colon, in particular TGF-β, and inhibition of CD4/CCR5 up-regulation during monocyte differentiation.23 Using global gene expression analysis in 32 paired colon and rectum samples obtained prior to treatment from participants of a rectal microbicide trial, we have found higher expression of TGF-β antagonistic and macrophage-activating genes in the rectum than in the colon, indicating a more pro-inflammatory environment in the rectum (manuscript in preparation).35 These findings suggest that the TGF-β-driven non-inflammatory state of macrophages in the colon, which likely serves to maintain mucosal homeostasis in the presence of the intestinal microflora,36 is lost toward the external opening of the rectum. We speculate that the reasons for this change are related to preventing intrusion of non-commensal pathogens, the danger of which presumably increases toward the anus.36, 37 These differences in the pathophysiology of the colon and rectum imply that screening for undesirable effects of topical microbicides or vaccination, in particular those that could lead to enhancing the risk of infection, as was observed in the Step Study,30 should include an evaluation of the rectum.
Our data also confirm prior reports that T cells in the colon and rectum of HIV-uninfected men appear similar to each other by phenotypic characterization, including CCR5 expression.29, 38 In the report by McGowan et al., biopsies were obtained from 10 and 30 cm proximal to the pectinate line in the anal canal. Since we found no significant difference in overall and CCR5+ T cell numbers between 4 and ~30 cm in our study, we conclude that the distribution of mucosal T cells and the level of CCR5 expression on T cells is fairly uniform from the descending colon to the anal canal. Both colon and rectum also have been shown to contain T cells expressing the α4β7 integrin,39, 40 which is associated with high susceptibility to HIV infection,41, 42 but no study has systematically evaluated if differences exist in its expression between the two sites. Considering these findings and the strong association between susceptibility to HIV infection and CCR5 expression,11–20, 43 it seems likely that rectal T cells are at least as susceptible to infection as T cells in the colon and small bowel. In vivo models of rectal SIV infection in macaques and ex vivo HIV-1 challenge studies in human explant tissues appear to confirm that, although these studies did not specify the exact anatomical location of the colorectal biopsies that were analyzed.8, 44–49
In our analyses, we could not define all CCR5+ cells as either macrophages, T cells or putative DCs. While it is possible that this was due to failure of our immunofluorescence staining to capture 100% of these three cell types, we believe it is more likely that some other intestinal cell populations express CCR5 as well. Indeed, it has been reported that a fraction of primary intestinal epithelial cells express CCR5 in the jejunum,26 and this could be possible in the colon and rectum as well. In the murine gut, CD4+ CD3− innate lymphoid cells have been described as a critical source of immunoprotective IL-22.50 An equivalent cell type could reside in the human gut and express CCR5. Under certain circumstances, NK cells and apoptotic neutrophils express CCR5,51–53 and even transfer of CCR5 from CCR5+ to CCR5− cells via exosomes has been reported.54 Thus, there are multiple possible explanations for our detection of CCR5+ cells that were neither DCs, macrophages nor T cells. These cells could potentially contribute to HIV disease in the gut.
The most striking finding of this study was that a markedly higher percentage of macrophages expressed CCR5 in the rectum than in the colon. As a consequence, the rectum harbored about three times as many CCR5+ macrophages per mm2 as the colon, with substantial cell numbers present in the superficial stroma of most rectal biopsies, which could heighten susceptibility to HIV infection in the rectum compared to the colon. In terms of prevention of rectal HIV transmission by topical microbicides, this implies that cell-specific studies of intracellular antiretroviral drug concentrations in the rectum should include measurements of macrophages in addition to T cells.55, 56 Experimental approaches with live microbial microbicides will likely need to achieve adequate colonization with the microbicide-producing bacteria in the rectum and not just the sigmoid colon.57 Lastly, for evaluation of immune responses to HIV vaccines and any other immunological studies in the large bowel, our study suggests caution when extrapolating analyses of colon tissues to the rectum.
Supplementary Material
Acknowledgments
This study was supported by NIH grants U01AI068618 (to M.J.M.) and R01HD51455 (to F.H.). We thank the study participants for their time and effort; the HVTN Laboratory Program, SCHARP, and Core staff who contributed to the study implementation and analysis; David Berger and Chris Galloway for enrolling and following study subjects; and Stephen Voght and Allison Mitchell for help with editing of the manuscript.
Footnotes
Conflicts of Interest: No conflicts of interest are declared by the authors.
References
- 1.Hladik F, McElrath MJ. Setting the stage: host invasion by HIV. Nat Rev Immunol. 2008;8:447–457. doi: 10.1038/nri2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gross M, Holte SE, Marmor M, Mwatha A, Koblin BA, Mayer KH. Anal sex among HIV-seronegative women at high risk of HIV exposure. The HIVNET Vaccine Preparedness Study 2 Protocol Team. J Acquir Immune Defic Syndr. 2000;24:393–398. doi: 10.1097/00126334-200008010-00015. [DOI] [PubMed] [Google Scholar]
- 3.Exner TM, Correale J, Carballo-Dieguez A, Salomon L, Morrow KM, Dolezal C, Mayer K. Women’s anal sex practices: implications for formulation and promotion of a rectal microbicide. AIDS Educ Prev. 2008;20:148–159. doi: 10.1521/aeap.2008.20.2.148. [DOI] [PubMed] [Google Scholar]
- 4.Schwandt M, Morris C, Ferguson A, Ngugi E, Moses S. Anal and dry sex in commercial sex work, and relation to risk for sexually transmitted infections and HIV in Meru, Kenya. Sex Transm Infect. 2006;82:392–396. doi: 10.1136/sti.2006.019794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McGowan I, Taylor DJ. Heterosexual anal intercourse has the potential to cause a significant loss of power in vaginal microbicide effectiveness studies. Sex Transm Dis. 2010;37:361–364. [PubMed] [Google Scholar]
- 6.Baggaley RF, White RG, Boily MC. HIV transmission risk through anal intercourse: systematic review, meta-analysis and implications for HIV prevention. Int J Epidemiol. 2010;39:1048–1063. doi: 10.1093/ije/dyq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boily MC, Baggaley RF, Masse B. The role of heterosexual anal intercourse for HIV transmission in developing countries: are we ready to draw conclusions? Sex Transm Infect. 2009;85:408–410. doi: 10.1136/sti.2009.037499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ribeiro Dos Santos P, Rancez M, Pretet JL, Michel-Salzat A, Messent V, Bogdanova A, Couedel-Courteille A, Souil E, Cheynier R, Butor C. Rapid dissemination of SIV follows multisite entry after rectal inoculation. PLoS ONE. 2011;6:e19493. doi: 10.1371/journal.pone.0019493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Louissaint NA, Nimmagadda S, Fuchs EJ, Bakshi RP, Cao YJ, Lee LA, Goldsmith J, Caffo BS, Du Y, King KE, Menendez FA, Torbenson MS, Hendrix CW. Distribution of cell-free and cell-associated HIV surrogates in the colon after simulated receptive anal intercourse in men who have sex with men. J Acquir Immune Defic Syndr. 2012;59:10–17. doi: 10.1097/QAI.0b013e3182373b5e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McKinnon LR, Kaul R. Quality and quantity: mucosal CD4+ T cells and HIV susceptibility. Curr Opin HIV AIDS. 2012;7:195–202. doi: 10.1097/COH.0b013e3283504941. [DOI] [PubMed] [Google Scholar]
- 11.Veazey RS, DeMaria M, Chalifoux LV, Shvetz DE, Pauley DR, Knight HL, Rosenzweig M, Johnson RP, Desrosiers RC, Lackner AA. Gastrointestinal tract as a major site of CD4+ T cell depletion and viral replication in SIV infection. Science. 1998;280:427–431. doi: 10.1126/science.280.5362.427. [DOI] [PubMed] [Google Scholar]
- 12.Smit-McBride Z, Mattapallil JJ, McChesney M, Ferrick D, Dandekar S. Gastrointestinal T lymphocytes retain high potential for cytokine responses but have severe CD4(+) T-cell depletion at all stages of simian immunodeficiency virus infection compared to peripheral lymphocytes. J Virol. 1998;72:6646–6656. doi: 10.1128/jvi.72.8.6646-6656.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mehandru S, Poles MA, Tenner-Racz K, Horowitz A, Hurley A, Hogan C, Boden D, Racz P, Markowitz M. Primary HIV-1 infection is associated with preferential depletion of CD4+ T lymphocytes from effector sites in the gastrointestinal tract. J Exp Med. 2004;200:761–770. doi: 10.1084/jem.20041196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mattapallil JJ, Douek DC, Hill B, Nishimura Y, Martin M, Roederer M. Massive infection and loss of memory CD4+ T cells in multiple tissues during acute SIV infection. Nature. 2005;434:1093–1097. doi: 10.1038/nature03501. [DOI] [PubMed] [Google Scholar]
- 15.Guadalupe M, Reay E, Sankaran S, Prindiville T, Flamm J, McNeil A, Dandekar S. Severe CD4+ T-cell depletion in gut lymphoid tissue during primary human immunodeficiency virus type 1 infection and substantial delay in restoration following highly active antiretroviral therapy. J Virol. 2003;77:11708–11717. doi: 10.1128/JVI.77.21.11708-11717.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brenchley JM, Schacker TW, Ruff LE, Price DA, Taylor JH, Beilman GJ, Nguyen PL, Khoruts A, Larson M, Haase AT, Douek DC. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J Exp Med. 2004;200:749–759. doi: 10.1084/jem.20040874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Q, Duan L, Estes JD, Ma ZM, Rourke T, Wang Y, Reilly C, Carlis J, Miller CJ, Haase AT. Peak SIV replication in resting memory CD4+ T cells depletes gut lamina propria CD4+ T cells. Nature. 2005;434:1148–1152. doi: 10.1038/nature03513. [DOI] [PubMed] [Google Scholar]
- 18.Mehandru S, Poles MA, Tenner-Racz K, Manuelli V, Jean-Pierre P, Lopez P, Shet A, Low A, Mohri H, Boden D, Racz P, Markowitz M. Mechanisms of gastrointestinal CD4+ T-cell depletion during acute and early human immunodeficiency virus type 1 infection. J Virol. 2007;81:599–612. doi: 10.1128/JVI.01739-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poles MA, Elliott J, Taing P, Anton PA, Chen IS. A preponderance of CCR5(+) CXCR4(+) mononuclear cells enhances gastrointestinal mucosal susceptibility to human immunodeficiency virus type 1 infection. J Virol. 2001;75:8390–8399. doi: 10.1128/JVI.75.18.8390-8399.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anton PA, Elliott J, Poles MA, McGowan IM, Matud J, Hultin LE, Grovit-Ferbas K, Mackay CR, Chen ISY, Giorgi JV. Enhanced levels of functional HIV-1 co-receptors on human mucosal T cells demonstrated using intestinal biopsy tissue. Aids. 2000;14:1761–1765. doi: 10.1097/00002030-200008180-00011. [DOI] [PubMed] [Google Scholar]
- 21.Smith PD, Meng G, Sellers MT, Rogers TS, Shaw GM. Biological parameters of HIV-1 infection in primary intestinal lymphocytes and macrophages. J Leukoc Biol. 2000;68:360–365. [PubMed] [Google Scholar]
- 22.Shen R, Richter HE, Clements RH, Novak L, Huff K, Bimczok D, Sankaran-Walters S, Dandekar S, Clapham PR, Smythies LE, Smith PD. Macrophages in vaginal but not intestinal mucosa are monocyte-like and permissive to human immunodeficiency virus type 1 infection. J Virol. 2009;83:3258–3267. doi: 10.1128/JVI.01796-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen R, Meng G, Ochsenbauer C, Clapham PR, Grams J, Novak L, Kappes JC, Smythies LE, Smith PD. Stromal down-regulation of macrophage CD4/CCR5 expression and NF-kappaB activation mediates HIV-1 non-permissiveness in intestinal macrophages. PLoS Pathog. 2011;7:e1002060. doi: 10.1371/journal.ppat.1002060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meng G, Sellers MT, Mosteller-Barnum M, Rogers TS, Shaw GM, Smith PD. Lamina propria lymphocytes, not macrophages, express CCR5 and CXCR4 and are the likely target cell for human immunodeficiency virus type 1 in the intestinal mucosa. J Infect Dis. 2000;182:785–791. doi: 10.1086/315790. [DOI] [PubMed] [Google Scholar]
- 25.Li L, Meng G, Graham MF, Shaw GM, Smith PD. Intestinal macrophages display reduced permissiveness to human immunodeficiency virus 1 and decreased surface CCR5. Gastroenterology. 1999;116:1043–1053. doi: 10.1016/s0016-5085(99)70007-7. [DOI] [PubMed] [Google Scholar]
- 26.Meng G, Wei X, Wu X, Sellers MT, Decker JM, Moldoveanu Z, Orenstein JM, Graham MF, Kappes JC, Mestecky J, Shaw GM, Smith PD. Primary intestinal epithelial cells selectively transfer R5 HIV-1 to CCR5+ cells. Nat Med. 2002;8:150–156. doi: 10.1038/nm0202-150. [DOI] [PubMed] [Google Scholar]
- 27.Shen R, Smythies LE, Clements RH, Novak L, Smith PD. Dendritic cells transmit HIV-1 through human small intestinal mucosa. J Leukoc Biol. 2009;87:663–670. doi: 10.1189/jlb.0909605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gurney KB, Elliott J, Nassanian H, Song C, Soilleux E, McGowan I, Anton PA, Lee B. Binding and transfer of human immunodeficiency virus by DC-SIGN+ cells in human rectal mucosa. J Virol. 2005;79:5762–5773. doi: 10.1128/JVI.79.9.5762-5773.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McGowan I, Elliott J, Cortina G, Tanner K, Siboliban C, Adler A, Cho D, Boscardin WJ, Soto-Torres L, Anton PA. Characterization of baseline intestinal mucosal indices of injury and inflammation in men for use in rectal microbicide trials (HIV Prevention Trials Network-056) J Acquir Immune Defic Syndr. 2007;46:417–425. doi: 10.1097/QAI.0b013e318156ef16. [DOI] [PubMed] [Google Scholar]
- 30.Buchbinder SP, Mehrotra DV, Duerr A, Fitzgerald DW, Mogg R, Li D, Gilbert PB, Lama JR, Marmor M, Del Rio C, McElrath MJ, Casimiro DR, Gottesdiener KM, Chodakewitz JA, Corey L, Robertson MN. Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): a double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet. 2008;372:1881–1893. doi: 10.1016/S0140-6736(08)61591-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McElrath MJ, De Rosa SC, Moodie Z, Dubey S, Kierstead L, Janes H, Defawe OD, Carter DK, Hural J, Akondy R, Buchbinder SP, Robertson MN, Mehrotra DV, Self SG, Corey L, Shiver JW, Casimiro DR. HIV-1 vaccine-induced immunity in the test-of-concept Step Study: a case-cohort analysis. Lancet. 2008;372:1894–1905. doi: 10.1016/S0140-6736(08)61592-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith PD, Meng G, Salazar-Gonzalez JF, Shaw GM. Macrophage HIV-1 infection and the gastrointestinal tract reservoir. J Leukoc Biol. 2003;74:642–649. doi: 10.1189/jlb.0503219. [DOI] [PubMed] [Google Scholar]
- 33.Varadarajan N, Julg B, Yamanaka YJ, Chen H, Ogunniyi AO, McAndrew E, Porter LC, Piechocka-Trocha A, Hill BJ, Douek DC, Pereyra F, Walker BD, Love JC. A high-throughput single-cell analysis of human CD8+ T cell functions reveals discordance for cytokine secretion and cytolysis. J Clin Invest. 2011 doi: 10.1172/JCI58653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bendall SC, Simonds EF, Qiu P, Amir el AD, Krutzik PO, Finck R, Bruggner RV, Melamed R, Trejo A, Ornatsky OI, Balderas RS, Plevritis SK, Sachs K, Pe’er D, Tanner SD, Nolan GP. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 332:687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hladik F, Ballweber L, Dai J, McElrath MJ, McGowan I. Global gene expression analysis reveals a more pro-inflammatory state in the distal than in the proximal rectum. International Microbicides Conference; Sydney. 2012. [Google Scholar]
- 36.Smith PD, Smythies LE, Shen R, Greenwell-Wild T, Gliozzi M, Wahl SM. Intestinal macrophages and response to microbial encroachment. Mucosal Immunol. 2011;4:31–42. doi: 10.1038/mi.2010.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mowat AM, Bain CC. Mucosal macrophages in intestinal homeostasis and inflammation. J Innate Immun. 2011;3:550–564. doi: 10.1159/000329099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fletcher PS, Elliott J, Grivel JC, Margolis L, Anton P, McGowan I, Shattock RJ. Ex vivo culture of human colorectal tissue for the evaluation of candidate microbicides. Aids. 2006;20:1237–1245. doi: 10.1097/01.aids.0000232230.96134.80. [DOI] [PubMed] [Google Scholar]
- 39.Souza HS, Elia CC, Spencer J, MacDonald TT. Expression of lymphocyte-endothelial receptor-ligand pairs, alpha4beta7/MAdCAM-1 and OX40/OX40 ligand in the colon and jejunum of patients with inflammatory bowel disease. Gut. 1999;45:856–863. doi: 10.1136/gut.45.6.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen YB, Kim HT, McDonough S, Odze RD, Yao X, Lazo-Kallanian S, Spitzer TR, Soiffer R, Antin JH, Ritz J. Up-Regulation of alpha4beta7 integrin on peripheral T cell subsets correlates with the development of acute intestinal graft-versus-host disease following allogeneic stem cell transplantation. Biol Blood Marrow Transplant. 2009;15:1066–1076. doi: 10.1016/j.bbmt.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arthos J, Cicala C, Martinelli E, Macleod K, Van Ryk D, Wei D, Xiao Z, Veenstra TD, Conrad TP, Lempicki RA, McLaughlin S, Pascuccio M, Gopaul R, McNally J, Cruz CC, Censoplano N, Chung E, Reitano KN, Kottilil S, Goode DJ, Fauci AS. HIV-1 envelope protein binds to and signals through integrin alpha4beta7, the gut mucosal homing receptor for peripheral T cells. Nat Immunol. 2008;9:301–309. doi: 10.1038/ni1566. [DOI] [PubMed] [Google Scholar]
- 42.Cicala C, Martinelli E, McNally JP, Goode DJ, Gopaul R, Hiatt J, Jelicic K, Kottilil S, Macleod K, O’Shea A, Patel N, Van Ryk D, Wei D, Pascuccio M, Yi L, McKinnon L, Izulla P, Kimani J, Kaul R, Fauci AS, Arthos J. The integrin alpha4beta7 forms a complex with cell-surface CD4 and defines a T-cell subset that is highly susceptible to infection by HIV-1. Proc Natl Acad Sci U S A. 2009;106:20877–20882. doi: 10.1073/pnas.0911796106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grivel JC, Elliott J, Lisco A, Biancotto A, Condack C, Shattock RJ, McGowan I, Margolis L, Anton P. HIV-1 pathogenesis differs in rectosigmoid and tonsillar tissues infected ex vivo with CCR5- and CXCR4-tropic HIV-1. Aids. 2007;21:1263–1272. doi: 10.1097/QAD.0b013e3281864667. [DOI] [PubMed] [Google Scholar]
- 44.Cranage M, Sharpe S, Herrera C, Cope A, Dennis M, Berry N, Ham C, Heeney J, Rezk N, Kashuba A, Anton P, McGowan I, Shattock R. Prevention of SIV rectal transmission and priming of T cell responses in macaques after local pre-exposure application of tenofovir gel. PLoS Med. 2008;5:e157. doi: 10.1371/journal.pmed.0050157. discussion e157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rohan LC, Moncla BJ, Kunjara Na Ayudhya RP, Cost M, Huang Y, Gai F, Billitto N, Lynam JD, Pryke K, Graebing P, Hopkins N, Rooney JF, Friend D, Dezzutti CS. In vitro and ex vivo testing of tenofovir shows it is effective as an HIV-1 microbicide. PLoS One. 2010;5:e9310. doi: 10.1371/journal.pone.0009310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abner SR, Guenthner PC, Guarner J, Hancock KA, Cummins JE, Jr, Fink A, Gilmore GT, Staley C, Ward A, Ali O, Binderow S, Cohen S, Grohskopf LA, Paxton L, Hart CE, Dezzutti CS. A Human Colorectal Explant Culture to Evaluate Topical Microbicides for the Prevention of HIV Infection. J Infect Dis. 2005;192:1545–1556. doi: 10.1086/462424. [DOI] [PubMed] [Google Scholar]
- 47.Herrera C, Cranage M, McGowan I, Anton P, Shattock RJ. Colorectal microbicide design: triple combinations of reverse transcriptase inhibitors are optimal against HIV-1 in tissue explants. Aids. 2011;25:1971–1979. doi: 10.1097/QAD.0b013e32834b3629. [DOI] [PubMed] [Google Scholar]
- 48.Anton PA, Saunders T, Elliott J, Khanukhova E, Dennis R, Adler A, Cortina G, Tanner K, Boscardin J, Cumberland WG, Zhou Y, Ventuneac A, Carballo-Dieguez A, Rabe L, McCormick T, Gabelnick H, Mauck C, McGowan I. First phase 1 double-blind, placebo-controlled, randomized rectal microbicide trial using UC781 gel with a novel index of ex vivo efficacy. PLoS ONE. 2011;6:e23243. doi: 10.1371/journal.pone.0023243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang L, He T, Talal A, Wang G, Frankel SS, Ho DD. In vivo distribution of the human immunodeficiency virus/simian immunodeficiency virus coreceptors: CXCR4, CCR3, and CCR5. J Virol. 1998;72:5035–5045. doi: 10.1128/jvi.72.6.5035-5045.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sonnenberg GF, Monticelli LA, Elloso MM, Fouser LA, Artis D. CD4(+) lymphoid tissue-inducer cells promote innate immunity in the gut. Immunity. 2011;34:122–134. doi: 10.1016/j.immuni.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jiang Y, Zhang Z, Diao Y, Jin X, Shi W, Geng W, Dai D, Zhang M, Han X, Liu J, Wang Y, Shang H. Expression of chemokine receptors on natural killer cells in HIV-infected individuals. Cell Immunol. 2008;251:19–24. doi: 10.1016/j.cellimm.2008.01.010. [DOI] [PubMed] [Google Scholar]
- 52.Bernstein HB, Wang G, Plasterer MC, Zack JA, Ramasastry P, Mumenthaler SM, Kitchen CM. CD4+ NK cells can be productively infected with HIV, leading to downregulation of CD4 expression and changes in function. Virology. 2009;387:59–66. doi: 10.1016/j.virol.2009.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ariel A, Fredman G, Sun YP, Kantarci A, Van Dyke TE, Luster AD, Serhan CN. Apoptotic neutrophils and T cells sequester chemokines during immune response resolution through modulation of CCR5 expression. Nat Immunol. 2006;7:1209–1216. doi: 10.1038/ni1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mack M, Kleinschmidt A, Bruhl H, Klier C, Nelson PJ, Cihak J, Plachy J, Stangassinger M, Erfle V, Schlondorff D. Transfer of the chemokine receptor CCR5 between cells by membrane-derived microparticles: a mechanism for cellular human immunodeficiency virus 1 infection. Nat Med. 2000;6:769–775. doi: 10.1038/77498. [DOI] [PubMed] [Google Scholar]
- 55.Hendrix CW, Cao YJ, Fuchs EJ. Topical microbicides to prevent HIV: clinical drug development challenges. Annu Rev Pharmacol Toxicol. 2009;49:349–375. doi: 10.1146/annurev.pharmtox.48.113006.094906. [DOI] [PubMed] [Google Scholar]
- 56.Patterson KB, Prince HA, Kraft E, Jenkins AJ, Shaheen NJ, Rooney JF, Cohen MS, Kashuba AD. Penetration of tenofovir and emtricitabine in mucosal tissues: implications for prevention of HIV-1 transmission. Science translational medicine. 2011;3:112re114. doi: 10.1126/scitranslmed.3003174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rao S, Hu S, McHugh L, Lueders K, Henry K, Zhao Q, Fekete RA, Kar S, Adhya S, Hamer DH. Toward a live microbial microbicide for HIV: commensal bacteria secreting an HIV fusion inhibitor peptide. Proc Natl Acad Sci U S A. 2005;102:11993–11998. doi: 10.1073/pnas.0504881102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.