Abstract
Despite recent advancements in therapy, melanoma still remains a highly lethal skin cancer. A better understanding of the genetic and epigenetic changes responsible for melanoma formation and progression could result in development of more effective treatments. Advanced melanomas are known to exhibit widespread promoter region CpG island methylation leading to inactivation of key tumor suppressor genes. Meta-analyses of relevant microarray data sets revealed the hematopoietic stem cell regulator gene Latexin (LXN) to be commonly down regulated in approximately 50% of melanomas. The CpG island in the promoter region of LXN was almost universally hypermethylated in melanoma cell lines and tumors and treatment of the cell lines with the demethylating drug, 5-Aza-2-deoxycytidine, resulted in increased LXN expression. In this paper, we demonstrate that exogenous expression of LXN in melanoma cell lines results in a significant inhibition of tumor cell proliferation. In addition, we show that the increased expression of LXN in these lines correlates with reduction in expression levels of stem cell transcription factors OCT4, NANOG, SOX2, KLF4 and MYCN indicating that LXN may exert its tumor suppressive function by altering the stem cell like properties of melanoma cells.
Introduction
Melanoma is the most deadly form of skin cancer. About 76,250 new cases and 9,180 deaths due to melanoma are expected in 2012 in the United States alone (Cancer Facts & Figures 2012, American Cancer Society, http://www.cancer.org). A number of genetic alterations have been identified in melanoma development and progression. Genes regulating the mitogen activated protein kinase (MAPK) pathway are frequently mutated in melanoma, including activating mutations of the BRAF serine/threonine kinase and NRAS which are encountered in 50% and 25% of melanomas, respectively (Davies et al., 2002; Edlundh-Rose et al., 2006; Krauthammer et al., 2012). The CDKN2A locus at 9p21, which encodes two tumor suppressor genes p16INK4A and p14ARF, is altered in the majority of sporadic melanomas and about 40% of familial melanomas (Aitken et al., 1999; Goldstein et al., 2007). Recently, exome sequencing has revealed several novel mutations in genes influencing key melanoma signaling pathways. These mutations are generally rare but nevertheless thought to play an important role in driving the melanomas. Mutations of PPP6C a serine/threonine phosphatase and RAC1, a GTPase were recently reported in 9–12% of melanomas (Hodis et al., 2012; Krauthammer et al., 2012). TRRAP, a component of an important complex possessing histone acetyltransferase activity is mutated in 4% of melanomas (Wei et al., 2011). Mutations of MEK1 and MEK2 kinases were found in 8% of melanomas (Nikolaev et al., 2012). PREX2 a negative regulator of PTEN tumor suppressor gene was found to be altered in 14 % of melanomas (Berger et al., 2012).
Epigenetic silencing has been known play a key role in inactivating tumor suppressors in melanoma and other cancers. Several studies have demonstrated that increased promoter region CpG island hypermethylation invariably occurs in advanced melanomas. A high incidence of promoter methylation of several loci in melanoma has been previously observed including RARB (70%), RASSF1A (55%), PYCARD (50%) and MGMT (34%) (Guan et al., 2003; Hoon et al., 2004; Spugnardi et al., 2003). Using a global screen for promoter hypermethylation in melanoma, we previously identified several genes that are hypermethylated in the vast majority of melanomas, including QPCT and CYP1B1 and LXN, which were methylated in greater than 95% of melanoma tumor samples (Muthusamy et al., 2006).
LXN was originally identified as a neural marker of the lateral neocortex in the developing mammalian brain (Arimatsu, 1994; Hatanaka et al., 1994) and was later described as the human homolog of endogenous inhibitor of rat carboxypeptidase A (Liu et al., 2000). In humans, LXN inhibits the carboxypeptidase (CPA4), a gene known to play a role in development of aggressive prostate cancer phenotypes (Pallares et al., 2005). LXN shows significant homology at the protein level to TIG1, a tumor suppressor silenced by hypermethylation in several types of cancer (Jing et al., 2002; Youssef et al., 2004). Knock out studies in murine models showed LXN to be a modulator of sensory perception, particularly in transmission of pain sensation (Bai et al., 2004; Jin et al., 2006). Liang and colleagues identified the LXN locus as the primary determinant of hematopoetic stem cell (HSC) frequency variation between two inbred mouse strains. Exogenous expression of LXN in the hematopoietic compartment showed it to be a negative regulator of hematopoietic stem cell numbers (Liang et al., 2007). A tumor regulatory role for LXN was suggested where LXN may function as a “tumor progenitor gene” controlling tumor stem cells (Liang and Van Zant, 2008). More recently, LXN was described to be silenced by methylation and function as a tumor suppressor in human gastric cancers and lymphomas (Li et al., 2011; Liu et al., 2012). In this paper, we present evidence that LXN is downregulated in >50% of melanomas by promoter hypermethylation, has a tumor suppressive role, and negatively regulates expression of tumor sustaining stem cell factors.
Results
LXN expression is downregulated in melanoma
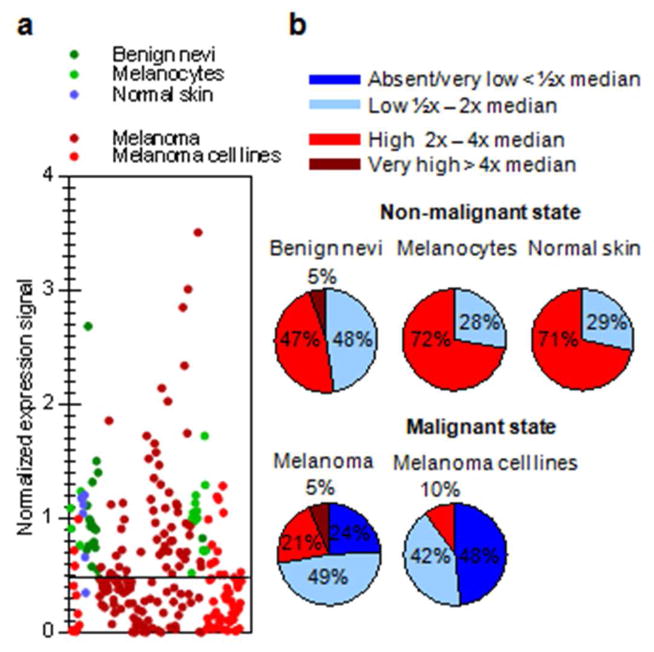
In a previous screen for methylation changes in melanomas we found reduced expression of LXN compared to primary cultured melanocytes which was linked to promoter hypermethylation (Muthusamy et al., 2006). In order to increase the power of our observations and examine LXN status in benign nevi, we performed a meta-analysis for LXN expression in publicly available GEO expression datasets and other published microarray data comprising of a total of: 18 melanocytes, 59 melanoma cell lines, 7 normal skin, 18 benign nevi and 133 melanoma samples GDS1965, (Hoek et al., 2004), GDS 1375, (Talantov et al., 2005), GDS3012, (Muthusamy et al., 2006), (Koga et al., 2009), GSE29377 (Avery-Kiejda et al., 2011). We found that LXN expression was generally lower in melanoma tissue samples and cell lines relative to normal skin, benign nevi and melanocytes (Figure 1a). We used a classification system based on expression compared to median signal intensity of normal controls in each of the experiments, to stratify LXN expression as explained in the methods section. LXN was detectably expressed in all the non-malignant samples included in the study. Relatively high levels of LXN expression was seen in greater than 70% of melanocytes and normal skin samples and in over 50% of benign nevi samples (Figure 1b). In contrast, LXN expression ranged from absent to very low in 48% of the melanoma cell lines and 24% of melanoma tissue samples in the meta-analysis (Figure 1b).
Figure 1. Expression analysis of LXN in human melanoma and melanocytes.
a) Meta-analysis of GEO datasets showing LXN mRNA expression in normal cells – melanocytes, normal skin and benign nevi and malignant cells – melanoma cell lines and tumors. Each dot represents the ratio of expression signal of an individual sample to the median signal of normal samples in that experiment. (b) Stratified levels of LXN expression in normal and malignant states
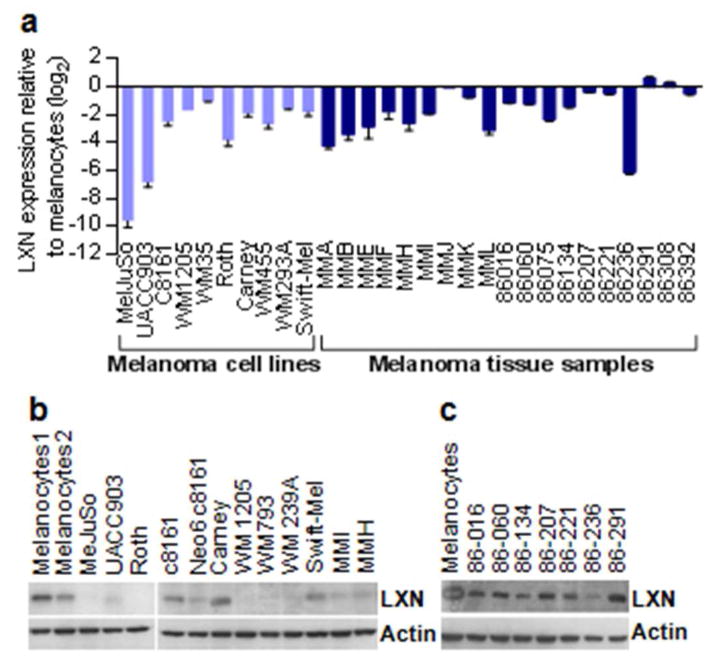
Quantitative PCR for LXN expression performed in a validation set consisting of a panel of melanoma cell lines and microdissected melanoma biopsy samples closely reflected the meta-analysis observations. LXN mRNA levels were decreased in all melanoma cell lines (n= 10) studied, as well as in a majority (74%, n=19) of uncultured melanoma tissue samples compared to primary melanocytes (Figure 2a). Analysis of LXN protein expression levels by western blotting showed absent to low expression of LXN in melanoma cell lines relative to melanocytes, and correlated to mRNA expression levels (Figure 2b). Analysis of protein expression by western blotting in the tumor samples showed slightly reduced levels of LXN compared to melanocytes (Figure 2c). Since immunohistochemistry can resolve expression at a cellular level, we analyzed expression patterns of LXN in melanoma patient samples in The Human Protein Atlas (www.proteinatlas.org) database. Interpretation of the immunohistochemistry data was performed on human melanoma biopsy samples (n=11) that were stained using the validated LXN antibody (HPA 014179, Sigma). A heterogeneous pattern of staining was observed in most of the tumor sections. Nine out of a total of eleven samples showed weak to absent LXN expression and of the two remaining samples one sample showed moderate levels and the other showed strong staining (Supplementary Table 2)
Figure 2. Validation of reduced LXN expression in melanoma.
(a) Quantitiative PCR analysis of LXN expression in melanoma cell lines and tumor tissue samples. (b) Western blot for LXN protein expression in melanoma cell lines (c) Western blot showing LXN protein expression in tumor samples compared to cultured melanocytes
Promoter hypermethylation is frequently encountered in melanoma and is associated with silencing of LXN
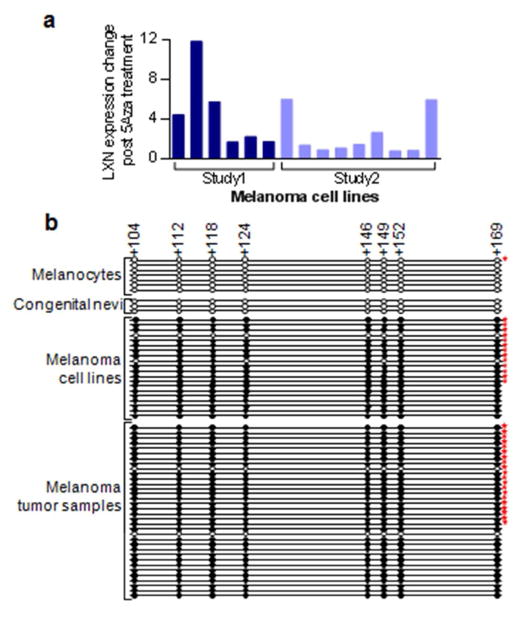
We had previously found that LXN was generally re-expressed in melanoma cell lines upon treatment with the methylation inhibitor 5-aza-2-deoxycytidine (5-aza-dC). This was also observed in another study that included several other melanoma lines (Muthusamy et al., 2006), unpublished data). We evaluated promoter hypermethylation of LXN in blood samples, melanocytes, congenital nevi, melanoma cell lines, and in melanomas. We found that promoter region of LXN was methylated in 95% of the melanoma cell lines and 94% of melanoma tumor samples and was unmethylated in normal melanocytes and in congenital nevi (Figure 3b; Supplementary table 3). To explore the possibility of use of methylated LXN as a tumor biomarker of circulating tumor cells in blood of melanoma patients, we evaluated the LXN promoter methylation status in peripheral blood lymphocyte DNA samples from normal healthy volunteers (n=55), which revealed a complete absence of methylation (Supplementary Figure 1).
Figure 3. Promoter region hypermethylation leads to silencing of LXN in melanoma.
(a) Analysis of microarray data from two previous experiments showing restoration of LXN expression in melanoma cell lines upon treatment with methylation reversing drug 5 Aza 2 deoxycytidine. Study1: light red (Muthusamy et al 2006); Study2: dark red (unpublished data). (b) Sanger bisulfite sequencing of the LXN gene promoter CpG island in melanocytes, melanoma cell lines and tumor samples. CpG positions are indicated by circles in scale to their location in the promoter region. Clear circles indicate absence of methylation, filled circles represent methylated cytosine. The numbers at the top indicate distance from the transcription start site. Stars indicate that the methylation status of these samples were described previously (Muthusamy et al 2006)
LXN has tumor suppressive properties in melanoma
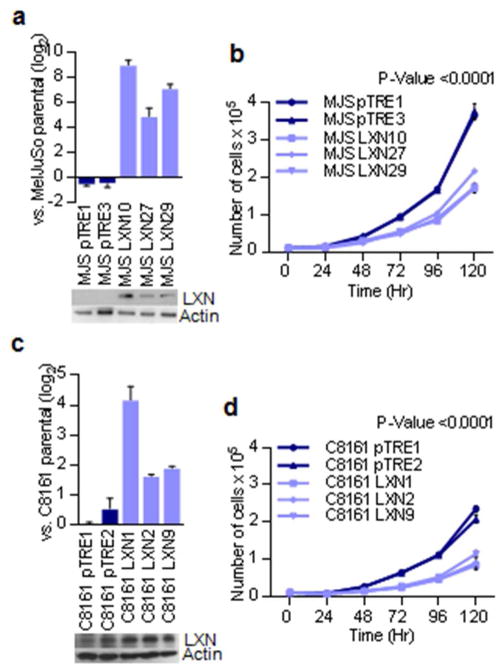
In order to establish the tumor suppressive function of LXN we stably transfected two human melanoma cell lines: 1) MelJuSo, which has no detectable LXN protein expression and 2) C8161, which expresses LXN protein at levels similar to those of melanocytes. In the LXN transfected and selected clones, expression levels of LXN was observed to be generally lower than melanocytes suggesting that high LXN expression was growth suppressive. Further, LXN expression decreased with continued culture of the clonal lines in-vitro eventually reverting to baseline levels of the parental line (data not shown). Cell proliferation assays demonstrated that LXN-transfected stable clones of both MelJuSo and C8161 resulted in slower growth compared to vector transfected controls (Figure 4).
Figure 4. Inhibition of cell proliferation by exogenous expression of LXN.
(a) Quantitative PCR and western blot analysis of LXN expression in the vector and LXN transfected clones of the MelJuSo melanoma cell line. (b) Growth curves showing differences in proliferation of LXN transfected and vector control lines of MelJuSo. (c) Quantitative PCR and western blot analysis of LXN expression in the vector and LXN transfected clones of C8161 melanoma cell line. (d) Growth curves showing difference in proliferation of LXN transfected and vector control lines of C8161. Note that original LXN protein was expressed in the original parental line which is reflected in the vector transfected controls
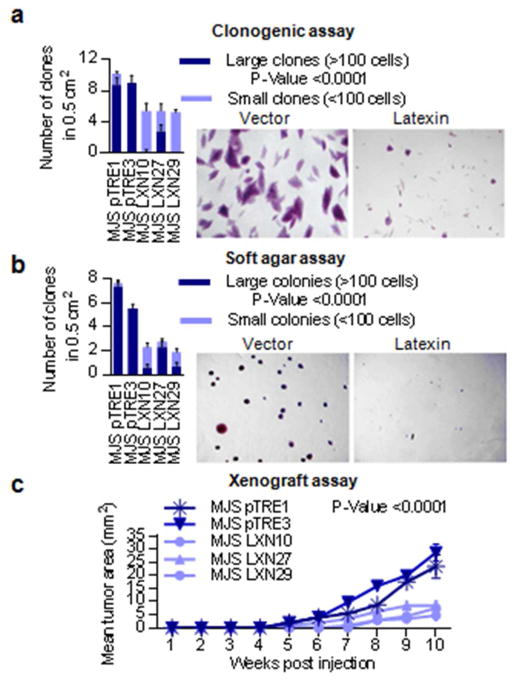
Colony formation assays showed that LXN-expressing MelJuSo clones formed only about half the number of colonies as LXN non-expressing controls. The colonies formed by LXN-expressing cells were generally small and comprised of less than 100 cells, whereas vector control cells typically formed colonies that were 10 times larger (Figure 5a). In order to assess the potential of the transfected cells to grow in an anchorage-independent manner, we performed colony formation assays in soft agar. LXN-expressing MelJuSo cells formed colonies less frequently and were smaller in size compared to vector controls (Figure 5b). In xenograft experiments in immunodeficient mice, significantly smaller tumors were present following injection of LXN expressing clones compared to vector controls. LXN non-expressing vector clones formed tumors earlier than 2 of the 3 LXN clones and grew to form tumors that were on average greater than two fold larger in size (Figure 5c).
Figure 5. Tumor suppressive properties of LXN demonstrated in LXN negative melanoma cell line.
(a) In vitro colony formation assay in LXN transfected MelJuSo compared to vector controls. Large clones comprising >100 cells are represented by dark shading while small clones comprising <100 cells are represented by lighter shading. The results are an average of counts from 10 random squares using a scoring grid, with 5 mm2 squares (b) In vitro colony formation in soft agar by LXN transfected MelJuSo compared to vector controls. Large clones comprising >100 cells are represented by dark shading while small clones comprising <100 cells are represented by lighter shading. The results are an average of counts from 10 random squares using a scoring grid, with 5 mm2 squares (c) In vivo xenograft assay showing reduced formation capability of LXN transfected MelJuSo compared to vector controls. The results depict tumors formed at a total of four injection sites in two mice per condition.
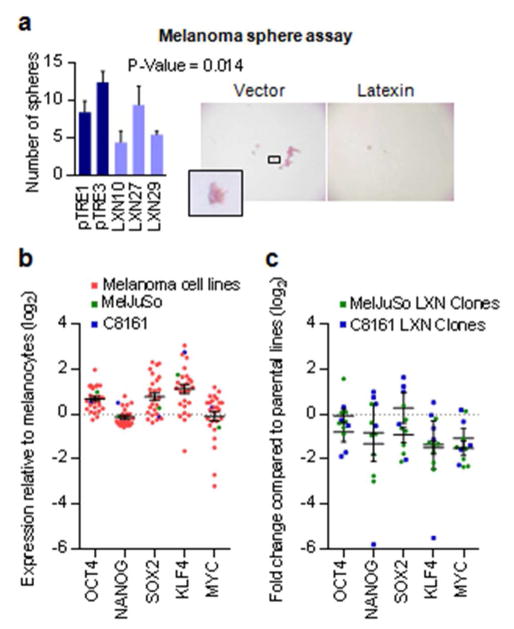
The ability to form non-adherent cellular spheres is thought to be a distinctive attribute of stem cells (Fang et al., 2005; Galli et al., 2004). Under non-adherent culture conditions, we observed a reduction in both number and size of melanoma spheres formed by two out of the three LXN-expressing clones compared to the vector transfected controls. (Figure 6a).
Figure 6. Changes in tumor progenitor properties upon reexpression of LXN.
(a) In vitro melanoma sphere formation assay in non-adherent conditions showing reduced numbers of spheres in LXN-transfected clones compared to vector (pTRE) transfected controls. Data shown is for a seeding of 1000 cells. The results represent an average of two replicate experiments. (b) Expression of stem cell transcription factors in melanoma cell lines compared to melanocytes derived from microarray expression data. (c) Expression of stem cell transcription factors in the LXN expressing clones of MelJuSo and C8161 compared to parental lines.
Regulation of stem cell associated transcription factors by LXN
Analysis of microarray expression data from 27 melanoma cell lines showed that levels of stem cell transcription factors OCT-4, NANOG, SOX2, KLF4, and MYC were generally higher in melanoma cells compared to melanocytes. (Figure 6b) (Koga et al., 2009; Muthusamy et al., 2006). Given the role of Lxn in the negative regulation of hematopoetic stem cells in mice, we sought to determine if tumor suppressor function of LXN is correlated to these stem cell factors in melanoma. Quantitative PCR based expression analysis of LXN transfected stable clonal lines of MelJuSo and C8161 showed that restoration of LXN expression correlated with a general reduction in expression of all the stem cell factors with significant decrease in the expression of KLF4 and MYC transcription factors in both the cell lines compared to their respective vector controls (Figure 6c).
Discussion
LXN was originally identified as a marker of neurons in the lateral cortex region of the developing mammalian brain (Arimatsu, 1994). A diverse range of biological functions has since been attributed to LXN, including modulation of sensory perception (Bai et al., 2004), pain transmission (Jin et al., 2006) regulation of inflammatory responses (Aagaard et al., 2005), inhibition of carboxypeptidase CPA4 (Pallares et al., 2005), and regulation of hematopoetic stem cells (Liang et al., 2007). Lxn regulation of stem cell function raised the possibility of modulation of tumor cells with self renewing capabilities (Liang et al., 2007; Liang and Van Zant, 2008). A tumor suppressor role for LXN has been recently shown in human gastric carcinomas and lymphomas (Li et al., 2011; Liu et al., 2012).
Here we provide evidence that LXN expression is low to absent in the majority of human melanomas and that it has a tumor suppressive function. Robust expression levels in normal cells and frequent loss in melanoma make LXN a potential biomarker for discrimination of melanoma cells from their benign counterparts. Since many benign nevi express melanocytic levels of LXN, its expression can potentially add to increased specificity of multiple marker based approaches that have been described for improved distinction of nevi from melanoma (Kashani-Sabet et al., 2009; Zhang and Li, 2012). Near-universal methylation in tumors compared to absence of methylation in normal cells also make detection of LXN methylation in body fluids a plausible approach for early diagnosis of melanoma, use as a surrogate to estimate tumor burden or for detection of remnant tumor cells post surgery or treatment with drug or immune based therapies. The observation that exogenous expression of LXN in the melanoma cell line C8161, which already expresses LXN at levels seen in melanocytes, still resulted in strong inhibition of proliferation suggests that elevation of LXN protein expression above the steady state level for cancer cells may not be compatible with rapid growth.. This observation raises the possibility of development of cancer therapeutics that can upregulate expression of LXN protein in tumor cells regardless of the status of their default LXN expression. These drugs could potentially include the DNA methyltransferase inhibitors, as we and several others have shown that the promoter region of the LXN gene is commonly hypermethylated in tumor cells compared to their normal counterparts and that 5-Aza-dC can restore expression LXN in tumor cell lines. Phase I trials using Decitabine (5-Aza-dC) have already been carried out in combination with other standard of care melanoma treatments. Clinically objective responses in the form of complete and partial remission have been observed in a small subset of patients receiving such therapy (Gollob et al., 2006; Tawbi et al., 2012).
The term “cancer stem cells” has been proposed for subpopulations of cells that are able to maintain the growth of malignant tumors. Cancer stem cells frequently share cell surface markers with their normal stem cell counterparts, are capable of self-renewal and may give rise to more differentiated progeny (Bonnet and Dick, 1997; Lapidot et al., 1994; Lobo et al., 2007). Lxn was shown to be the primary quantitative trait locus negatively regulating HSC numbers in mice (Liang et al., 2007). Mouse strains expressing low levels of Lxn were found to have significantly more HSCs than their counterparts expressing relatively high levels. Enhancement of Lxn expression by retroviral transfection in bone marrow cells led to a reduction in HSC frequency in mice (Liang et al., 2007). Additional evidence of LXN involvement in stem cell regulation is provided by studies showing that RNAi-mediated depletion of the stem cell factors OCT4 and NANOG, which are required to maintain pluripotency and self-renewal (Chambers et al., 2003; Nichols et al., 1998), leads to a concomitant increase in LXN levels (GSE4189), (Loh et al., 2006). It has recently been demonstrated that transcription factors including OCT4, NANOG, SOX2, KLF4 and MYC are central mediators of stem cell self-renewal and pluripotency (Okita et al., 2007; Takahashi and Yamanaka, 2006; Wernig et al., 2007). Combined expression of these factors has been shown to convert somatic cells into induced pluripotent stem cells (iPS cells) that have a morphology, epigenetic profile, and gene expression signature similar to embryonic stem cells (Okita et al., 2007; Takahashi and Yamanaka, 2006; Wernig et al., 2007). We found elevated levels of OCT4, NANOG, SOX2, and KLF4 in several melanoma cell lines including MelJuSo and C8161 relative to primary melanocytes and that the expression levels of the most oncogenic factors, KLF4 and MYC is associated with exogenous expression of LXN. These findings raise the possibility that LXN might function as a tumor suppressor by regulating cancer progenitor cells with stem-like properties.
Materials and Methods
Blood, tissue specimens, primary cells and cell lines
The melanoma tissue and blood samples were obtained in accordance with the Institutional Review Board approved protocols at the Yale University (New Haven, CT), University of Vermont (Burlington, VT), Memorial Sloan-Kettering Cancer Center (New York, NY) and Dana-Farber Cancer Institute (Boston, MA). Patient consent was not required because the material was de-identified. All of the melanoma samples were derived from lymph nodes or cutaneous metastases. Frozen tumor specimens in OCT were macrodissected to 80% purity of tumor cells guided by examination of a hematoxylin and eosin (H&E) slide by a pathologist (M.B.).
All the cell lines used in the study were grown in DMEM-F12 medium (Invitrogen) supplemented with 5% FBS and non-essential amino acids (Invitrogen). Primary cultured human foreskin melanocytes were grown in Medium 254 supplemented with human melanocyte growth serum (Cascade Biologics). LXN expression constructs were generated by RT-PCR amplification of full-length coding sequence from primary melanocytes and subcloning into the pTRE-tight expression vector (Clontech). The inserts were verified by Sanger sequencing to ensure an absence of reverse transcription-introduced mutations. Stable transfectants were produced by transfection with Lipofectamine (Invitrogen) and selected by growth in media containing 800 micrograms/ml G418 (Invitrogen). Single colonies were ring cloned, expanded, and analyzed for transgene expression using RT-QPCR.
Bisulfite sequencing and quantitative RT-PCR
DNA was isolated from cells and tissues using standard phenol-chloroform extraction. Bisulfite modification was performed as previously described (Herman et al., 1996) using genomic DNA from normal blood, melanocytes, melanoma cell lines and melanoma tissue samples. PCR reactions to interrogate LXN promoter CpG island were carried out using 50 ng of bisulfite modified DNA in a 30 μl volume with a 0.2 μM primers (forward – GGA TGT AGG GAG TTT GGG TT and reverse – TTC CAT TCC RAA TAA ACA ATA AC). PCR products were purified using the Qiaquick Gel Extraction kit (Qiagen) and directly sequenced on the ABI 3100-Avant automated DNA sequencer. Real time quantitative PCR for LXN was performed by using 2.5 μl of 100 fold diluted cDNA template and 0.2 μM LXN specific primers in a 25 μl PCR reaction using JumpStart SYBR green kit (Sigma) according to manufacturer’s instructions with an ABI 7700 (Applied Biosystems, Inc.). Real time quantitative PCR for stem cell associated transcription factors OCT4, NANOG, SOX2, KLF4 and MYC was performed using 2.0 μl of 10 fold diluted cDNA template and 0.2 μM gene specific primers in a 20 μl fast PCR reaction using FastStart Universal SYBR green mix (Roche) in an ABI StepOne Plus Real time PCR system. All the real time reactions including that for LXN and the stem cell associated transcription factors were performed in duplicate, CT values obtained were normalized to GAPDH levels and quantification was performed using the comparative CT method. All primer sequences are provided in Supplementary Table 1.
Western blotting
Western blotting experiments were performed by separation of 20 μg of cell lysate per sample on SDS-PAGE, transfer to Immobilon-P membranes (Millipore), blocking with phosphate buffered saline containing 0.2% Tween 20 and 5% non-fat milk, and incubation with LXN antibodies (N-17: sc-47090), (Santa Cruz Biotechnology). Following washes and incubation with peroxidase-conjugated Donkey anti goat secondary antibodies (Abcam), ECL-Advance detection system (Amersham Biosciences) was employed for chemiluminescent detection. The blots were stripped and re-probed with actin antibodies (AC-40, Sigma) to confirm equal protein loading.
In-vitro proliferation assays, clonogenic assays and colony formation in soft agar
In-vitro growth curve assays were performed as previously described (Muthusamy et al., 2006). Briefly, three replicates of 10,000 cells each of the vector and LXN transfected cells were seeded in 6-well cell culture plates for each time point. The cell were trypsinized, diluted and counted on a flow cytometer (Coulter) at 24h intervals starting at 0h and ending at 120h. Colony formation assays were performed by plating three replicates of 1000 cells of the vector and LXN transfected cells in 6-well cell culture plates and evaluating colony formation at 2 weeks. For the soft agar assay, a base layer of 0.5% agar in growth media was prepared in 6-well tissue culture plates. An upper layer was formed by adding 500 cells into 0.35% agarose in growth media. Samples were plated in duplicate were prepared at the same time to correct for effects on plating efficiency. Colonies were allowed to grow for four weeks within the soft agar. Staining for both colony formation assays and soft agar assays was carried out by adding 0.005% crystal violet in 20% methanol followed by washing with distilled water and destaining with 100% methanol.
Melanoma sphere formation assay
LXN transfected cells and vector controls were seeded at 1000 cells per well of a 24-well Ultra- Low Attachment Costar tissue culture plate (Corning, NY). Melanoma spheres were allowed to form without any disturbance for two weeks and quantified. The cells were also seeded in duplicate in a 24 well tissue culture dish, the cells were allowed to attach and then stained with DAPI to ensure equal seeding of the cells.
Tumor formation in nude mice
For the xenograft assay, two vector control clones and three LXN transfected clones were pelleted, resuspended in cold HBSS and 0.5 × 106 cells in 100 microliters were injected sub-cutaneously into flank skin of nude mice. Two mice were assigned per condition and each mouse was injected twice, one injection to each flank. Tumor dimensions (maximum length and width) were measured every week and the mice were sacrificed at 10 weeks after injection.
Statistics
The results of the proliferation assay, clonogenic assays, melanoma sphere formation assays and xenografting assays were analyzed for statistical significance using the Student’s t-test.
Supplementary Material
Table 1.
RT-QPCR Primer sequences
| Gene | Primer Sequence |
|---|---|
| LXN | F - ATTTAGCCTGGGTTGCCTGT |
| R - TTAGTGCCGTATTGTGGATGC | |
| OCT4 | F - GTGAGAGGCAACCTGGAGAAT |
| R - GTTACAGAACCACACTCGGACC | |
| NANOG | F - AAGAACTCTCCAACATCCTGAAC |
| R - CTGGGGTAGGTAGGTGCTGA | |
| SOX2 | F - GCACACTGCCCCTCTCAC |
| R - ATGCTGTTTCTTACTCTCCTCTTT | |
| KLF4 | F - CGCCGCTCCATTACCAA |
| R - ACAGCCGTCCCAGTCACAG | |
| MYC | F - TCCTCGGATTCTCTGCTCTC |
| R - GATTTCTTCCTCATCTTCTTGTTC |
Table 2.
Expression of LXN in melanoma tissue sections interpreted from The Human Protein Atlas (http://www.proteinatlas.org/) staining with validated antibody – HPA014179
| Melanoma Sample | Absent (−) | Weak (+) | Moderate (++) | Strong (> +++) |
|---|---|---|---|---|
| 1 | 90% | |||
| 2 | 20% | 80% | ||
| 3 | 90% | |||
| 4 | >90% | |||
| 5 | >90% | |||
| 6 | >90% | |||
| 7 | >90% | |||
| 8 | >90% | |||
| 9 | 40% | 60% | ||
| 10 | 20% | 80% | ||
| 11 | 70% | 30% |
Table 3.
List of samples analyzed for promoter hypermethylation of LXN
| Primary cultured melanocytes | Melanocytes1 * |
| Melanocytes c261 | |
| Melanocytes c264 | |
| Melanocytes c293 | |
| Melanocytes c295 | |
| HFSC | |
| HMAP | |
| Congenital nevi | YUOPE |
| YUVATI | |
| YUCLIR | |
| Melanoma cell lines | MelJuSo * |
| UACC903 * | |
| C8161 * | |
| Neo6/C8161 * | |
| WM1205 * | |
| WM455 * | |
| WM1366 * | |
| WM293A * | |
| WM35 * | |
| Roth* | |
| Carney * | |
| WM793 * | |
| Swift-Mel * | |
| YULAC | |
| YURIF | |
| YUSIK | |
| WW165 | |
| MEL501 | |
| YUCAL | |
| YUMAC | |
| Melanoma tumor tissues | 86002 * |
| 86008 * | |
| 86016 * | |
| 86060 * | |
| 86075 * | |
| 86134 * | |
| 86140 * | |
| 86187 * | |
| 86221 * | |
| 86233 * | |
| 86246 * | |
| 86296 * | |
| 86304 * | |
| 86307 * | |
| 86335 * | |
| 86338 * | |
| 86392 * | |
| 86393 * | |
| 86396 * | |
| 86397 * | |
| YURER | |
| YUTUR | |
| YUKIL | |
| YUPAO | |
| YUROL | |
| YUPER | |
| YUHUY | |
| YUBUNE | |
| YUKOLI | |
| YUCHER | |
| YUFIT | |
| YUHOIN | |
| YUKAY | |
| YUMUT |
Gray shading indicates presence of methylation. Light gray indicates partial methylation. Star indicates – methylation previously reported in Muthusamy et al 2006.
Acknowledgments
We would like to thank Dr. Ruth Halaban and Ms. Antonella Bacchiocchi for providing some of the cell lines, melanoma and nevi samples from the Yale SPORE in Skin Cancer Specimen Resource Core.
Grant support: J. Walter Juckett Postdoctoral Fellowship (V.Muthusamy); KO8 CA89124, RO1 CA112054, and a University of Vermont New Research Initiative Grant (M. Bosenberg); and P30 CA22345 and P20 RR16462 (Vermont Cancer Center DNA Analysis Core Facility).
Footnotes
Conflict of interest
The authors state no conflict of interest
References
- Aagaard A, Listwan P, Cowieson N, Huber T, Ravasi T, Wells CA, et al. An inflammatory role for the mammalian carboxypeptidase inhibitor latexin: relationship to cystatins and the tumor suppressor TIG1. Structure. 2005;13:309–17. doi: 10.1016/j.str.2004.12.013. [DOI] [PubMed] [Google Scholar]
- Aitken J, Welch J, Duffy D, Milligan A, Green A, Martin N, et al. CDKN2A variants in a population-based sample of Queensland families with melanoma. Journal of the National Cancer Institute. 1999;91:446–52. doi: 10.1093/jnci/91.5.446. [DOI] [PubMed] [Google Scholar]
- Arimatsu Y. Latexin: a molecular marker for regional specification in the neocortex. Neuroscience research. 1994;20:131–5. doi: 10.1016/0168-0102(94)90030-2. [DOI] [PubMed] [Google Scholar]
- Avery-Kiejda KA, Bowden NA, Croft AJ, Scurr LL, Kairupan CF, Ashton KA, et al. P53 in human melanoma fails to regulate target genes associated with apoptosis and the cell cycle and may contribute to proliferation. BMC cancer. 2011;11:203. doi: 10.1186/1471-2407-11-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai WZ, Ishida M, Arimatsu Y. Chemically defined feedback connections from infragranular layers of sensory association cortices in the rat. Neuroscience. 2004;123:257–67. doi: 10.1016/j.neuroscience.2003.08.056. [DOI] [PubMed] [Google Scholar]
- Berger MF, Hodis E, Heffernan TP, Deribe YL, Lawrence MS, Protopopov A, et al. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature. 2012;485:502–6. doi: 10.1038/nature11071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nature medicine. 1997;3:730–7. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, et al. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–55. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–54. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- Edlundh-Rose E, Egyhazi S, Omholt K, Mansson-Brahme E, Platz A, Hansson J, et al. NRAS and BRAF mutations in melanoma tumours in relation to clinical characteristics: a study based on mutation screening by pyrosequencing. Melanoma research. 2006;16:471–8. doi: 10.1097/01.cmr.0000232300.22032.86. [DOI] [PubMed] [Google Scholar]
- Fang D, Nguyen TK, Leishear K, Finko R, Kulp AN, Hotz S, et al. A tumorigenic subpopulation with stem cell properties in melanomas. Cancer research. 2005;65:9328–37. doi: 10.1158/0008-5472.CAN-05-1343. [DOI] [PubMed] [Google Scholar]
- Galli R, Binda E, Orfanelli U, Cipelletti B, Gritti A, De Vitis S, et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer research. 2004;64:7011–21. doi: 10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
- Goldstein AM, Chan M, Harland M, Hayward NK, Demenais F, Bishop DT, et al. Features associated with germline CDKN2A mutations: a GenoMEL study of melanoma-prone families from three continents. Journal of medical genetics. 2007;44:99–106. doi: 10.1136/jmg.2006.043802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollob JA, Sciambi CJ, Peterson BL, Richmond T, Thoreson M, Moran K, et al. Phase I trial of sequential low-dose 5-aza-2′-deoxycytidine plus high-dose intravenous bolus interleukin-2 in patients with melanoma or renal cell carcinoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2006;12:4619–27. doi: 10.1158/1078-0432.CCR-06-0883. [DOI] [PubMed] [Google Scholar]
- Guan X, Sagara J, Yokoyama T, Koganehira Y, Oguchi M, Saida T, et al. ASC/TMS1, a caspase-1 activating adaptor, is downregulated by aberrant methylation in human melanoma. International journal of cancer Journal international du cancer. 2003;107:202–8. doi: 10.1002/ijc.11376. [DOI] [PubMed] [Google Scholar]
- Hatanaka Y, Uratani Y, Takiguchi-Hayashi K, Omori A, Sato K, Miyamoto M, et al. Intracortical regionality represented by specific transcription for a novel protein, latexin. The European journal of neuroscience. 1994;6:973–82. doi: 10.1111/j.1460-9568.1994.tb00592.x. [DOI] [PubMed] [Google Scholar]
- Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:9821–6. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–63. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoek K, Rimm DL, Williams KR, Zhao H, Ariyan S, Lin A, et al. Expression profiling reveals novel pathways in the transformation of melanocytes to melanomas. Cancer research. 2004;64:5270–82. doi: 10.1158/0008-5472.CAN-04-0731. [DOI] [PubMed] [Google Scholar]
- Hoon DS, Spugnardi M, Kuo C, Huang SK, Morton DL, Taback B. Profiling epigenetic inactivation of tumor suppressor genes in tumors and plasma from cutaneous melanoma patients. Oncogene. 2004;23:4014–22. doi: 10.1038/sj.onc.1207505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin M, Ishida M, Katoh-Fukui Y, Tsuchiya R, Higashinakagawa T, Ikegami S, et al. Reduced pain sensitivity in mice lacking latexin, an inhibitor of metallocarboxypeptidases. Brain research. 2006;1075:117–21. doi: 10.1016/j.brainres.2005.12.099. [DOI] [PubMed] [Google Scholar]
- Jing C, El-Ghany MA, Beesley C, Foster CS, Rudland PS, Smith P, et al. Tazarotene-induced gene 1 (TIG1) expression in prostate carcinomas and its relationship to tumorigenicity. Journal of the National Cancer Institute. 2002;94:482–90. doi: 10.1093/jnci/94.7.482. [DOI] [PubMed] [Google Scholar]
- Kashani-Sabet M, Rangel J, Torabian S, Nosrati M, Simko J, Jablons DM, et al. A multi-marker assay to distinguish malignant melanomas from benign nevi. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:6268–72. doi: 10.1073/pnas.0901185106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga Y, Pelizzola M, Cheng E, Krauthammer M, Sznol M, Ariyan S, et al. Genome-wide screen of promoter methylation identifies novel markers in melanoma. Genome research. 2009;19:1462–70. doi: 10.1101/gr.091447.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauthammer M, Kong Y, Ha BH, Evans P, Bacchiocchi A, McCusker JP, et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nature genetics. 2012;44:1006–14. doi: 10.1038/ng.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645–8. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- Li Y, Basang Z, Ding H, Lu Z, Ning T, Wei H, et al. Latexin expression is downregulated in human gastric carcinomas and exhibits tumor suppressor potential. BMC cancer. 2011;11:121. doi: 10.1186/1471-2407-11-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Jansen M, Aronow B, Geiger H, Van Zant G. The quantitative trait gene latexin influences the size of the hematopoietic stem cell population in mice. Nature genetics. 2007;39:178–88. doi: 10.1038/ng1938. [DOI] [PubMed] [Google Scholar]
- Liang Y, Van Zant G. Aging stem cells, latexin, and longevity. Experimental cell research. 2008;314:1962–72. doi: 10.1016/j.yexcr.2008.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Yu L, Gao J, Fu Q, Zhang J, Zhang P, et al. Cloning, tissue expression pattern and genomic organization of latexin, a human homologue of rat carboxypeptidase A inhibitor. Molecular biology reports. 2000;27:241–6. doi: 10.1023/a:1010971219806. [DOI] [PubMed] [Google Scholar]
- Liu Y, Howard D, Rector K, Swiderski C, Brandon J, Schook L, et al. Latexin is down-regulated in hematopoietic malignancies and restoration of expression inhibits lymphoma growth. PloS one. 2012;7:e44979. doi: 10.1371/journal.pone.0044979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo NA, Shimono Y, Qian D, Clarke MF. The biology of cancer stem cells. Annual review of cell and developmental biology. 2007;23:675–99. doi: 10.1146/annurev.cellbio.22.010305.104154. [DOI] [PubMed] [Google Scholar]
- Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nature genetics. 2006;38:431–40. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- Muthusamy V, Duraisamy S, Bradbury CM, Hobbs C, Curley DP, Nelson B, et al. Epigenetic silencing of novel tumor suppressors in malignant melanoma. Cancer research. 2006;66:11187–93. doi: 10.1158/0008-5472.CAN-06-1274. [DOI] [PubMed] [Google Scholar]
- Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, et al. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–91. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- Nikolaev SI, Rimoldi D, Iseli C, Valsesia A, Robyr D, Gehrig C, et al. Exome sequencing identifies recurrent somatic MAP2K1 and MAP2K2 mutations in melanoma. Nature genetics. 2012;44:133–9. doi: 10.1038/ng.1026. [DOI] [PubMed] [Google Scholar]
- Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–7. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- Pallares I, Bonet R, Garcia-Castellanos R, Ventura S, Aviles FX, Vendrell J, et al. Structure of human carboxypeptidase A4 with its endogenous protein inhibitor, latexin. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:3978–83. doi: 10.1073/pnas.0500678102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spugnardi M, Tommasi S, Dammann R, Pfeifer GP, Hoon DS. Epigenetic inactivation of RAS association domain family protein 1 (RASSF1A) in malignant cutaneous melanoma. Cancer research. 2003;63:1639–43. [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Talantov D, Mazumder A, Yu JX, Briggs T, Jiang Y, Backus J, et al. Novel genes associated with malignant melanoma but not benign melanocytic lesions. Clinical cancer research : an official journal of the American Association for Cancer Research. 2005;11:7234–42. doi: 10.1158/1078-0432.CCR-05-0683. [DOI] [PubMed] [Google Scholar]
- Tawbi HA, Beumer JH, Tarhini AA, Moschos S, Buch SC, Egorin MJ, et al. Safety and efficacy of decitabine in combination with temozolomide in metastatic melanoma: a phase I/II study and pharmacokinetic analysis. Annals of oncology : official journal of the European Society for Medical Oncology/ESMO. 2012 doi: 10.1093/annonc/mds591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Walia V, Lin JC, Teer JK, Prickett TD, Gartner J, et al. Exome sequencing identifies GRIN2A as frequently mutated in melanoma. Nature genetics. 2011;43:442–6. doi: 10.1038/ng.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, et al. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–24. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- Youssef EM, Chen XQ, Higuchi E, Kondo Y, Garcia-Manero G, Lotan R, et al. Hypermethylation and silencing of the putative tumor suppressor Tazarotene-induced gene 1 in human cancers. Cancer research. 2004;64:2411–7. doi: 10.1158/0008-5472.can-03-0164. [DOI] [PubMed] [Google Scholar]
- Zhang G, Li G. Novel multiple markers to distinguish melanoma from dysplastic nevi. PloS one. 2012;7:e45037. doi: 10.1371/journal.pone.0045037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.