Summary
Primary cilium formation is initiated at the distal end of the mother centriole in a highly co-ordinated manner. This requires the capping of the distal end of the mother centriole with a ciliary vesicle and the anchoring of the basal body (mother centriole) to the cell cortex, both of which are mediated by the distal appendages. Here, we show that the distal appendage protein Cep123 (Cep89/CCDC123) is required for the assembly, but not the maintenance, of a primary cilium. In the absence of Cep123 ciliary vesicle formation fails, suggesting that it functions in the early stages of primary ciliogenesis. Consistent with such a role, Cep123 interacts with the centriolar satellite proteins PCM-1, Cep290 and OFD1, all of which play a role in primary ciliogenesis. These interactions are mediated by a domain in the C-terminus of Cep123 (400–783) that overlaps the distal appendage-targeting domain (500–600). Together, the data implicate Cep123 as a new player in the primary ciliogenesis pathway and expand upon the role of the distal appendages in this process.
Key words: Centriolar satellites, Centrosome, Distal appendages, Primary cilium
Introduction
The centrosome performs many functions in the cell including: acting as a microtubule organizing centre (MTOC), establishing cell polarity (Bornens, 2012), contributing to bipolar spindle formation during mitosis and regulating cytokinesis (Khodjakov and Rieder, 2001; Piel et al., 2001). Another important function of the centrosome is to act as a scaffold for the formation of a primary cilium, a sensory organelle, which protrudes from the surface of the cell (Pazour and Witman, 2003). This function is unique to the mother centriole, one of the two centrioles present in the centrosome, which becomes the basal body of the primary cilium. The mother centriole, like the daughter centriole, is formed of nine sets of microtubules that are arranged in a barrel-like shape, but it differs structurally in that it has two sets of appendages (sub-distal and distal appendages) at its distal end (Paintrand et al., 1992; Robbins et al., 1968; Vorobjev and Chentsov, 1982).
The distal appendages play an important, but not yet fully defined, role in primary cilium formation. This process begins with the recruitment and association of vesicles to the distal appendages, to create a ciliary vesicle covering the distal end of the mother centriole (Sorokin, 1962; Sorokin, 1968). Ciliary vesicle formation is dependent upon a cascade of Rab proteins with Rab11 (Knödler et al., 2010; Westlake et al., 2011) and Rab8 being two important components (Nachury et al., 2007; Yoshimura et al., 2007). This cascade is initiated by the Rab11-dependent trafficking of Rabin8, the guanine nucleotide exchange factor (GEF) for Rab8, to a transitory structure known as the pericentrosomal preciliary compartment (PPC) located close to the centrosome (Kim et al., 2010; Knödler et al., 2010; Westlake et al., 2011). Rab11 is not only involved in the trafficking of Rabin8 to the PCC, it also stimulates the GEF activity of Rabin8 (Knödler et al., 2010; Westlake et al., 2011), when it is in its active GTP-bound form. This promotes ciliary membrane formation and entry of Rab8 into the primary cilium. Rabin8 plays a key role in this process and it interacts with many proteins including: the Bardet–Biedl syndrome (BBS) complex (Jin et al., 2010); the exocyst complex through Sec15 (Feng et al., 2012); and the transport protein particle (TRAPP) II complex (Westlake et al., 2011). More recently, Rabin8 has been linked to the distal appendages of the mother centriole by a study showing that it interacts with the distal appendage component Cep164 (Schmidt et al., 2012).
To allow the forming primary cilium to protrude from the surface of the cell the mother centriole has to migrate and attach to the cell cortex, via its distal appendages (Ishikawa et al., 2005), and the ciliary vesicle must fuse with the plasma membrane (Molla-Herman et al., 2010; Sorokin, 1962). Extension of an axoneme, which consists of doublet microtubules, occurs from the distal end of the mother centriole by intraflagellar transport (IFT) (Pazour and Rosenbaum, 2002). This involves the trafficking of IFT components to and from the distal end of the primary cilium by molecular motors.
A large cohort of proteins is required for primary cilium formation (Graser et al., 2007; Kim et al., 2010) and not all of these are directly associated with the centrosome and are transported there in a controlled manner. Centriolar satellites are small electron-dense granules, between 70 to 100 nm in diameter, found in close vicinity to the centrosome (Berns et al., 1977; de-Thé, 1964). Various functions have been ascribed to centriolar satellites including, contributing to bipolar spindle formation during mitosis (Kim and Rhee, 2011; Oshimori et al., 2009), transporting components to the centrosome (Dammermann and Merdes, 2002) and participating in cilium assembly (Graser et al., 2007; Kubo et al., 1999; Nachury et al., 2007; Sedjaï et al., 2010). Pericentriolar material-1 (PCM-1) was the first centriolar satellite protein to be described (Balczon et al., 1994; Kubo et al., 1999), but since its identification the list proteins localizing to centriolar satellites has grown substantially. PCM-1 plays an important role in transporting proteins to the centrosome and its depletion by siRNA-mediated silencing results in decreased centrosomal levels of centrin, pericentrin and ninein (Dammermann and Merdes, 2002). This transport is dependent upon an intact microtubule array and the molecular motor dynein. PCM-1 also interacts with other centriolar satellite proteins including OFD1 (Lopes et al., 2011), Cep290 (Kim et al., 2008) and the BBSome component BBS4 (Kim et al., 2004; Nachury et al., 2007). Recent work has shown that these proteins are dependent upon each other for their proper localization, with the depletion of any one of these proteins causing the disruption of centriolar satellites (Lopes et al., 2011). These centriolar satellite proteins (Ferrante et al., 2006; Graser et al., 2007; Kim et al., 2008; Nachury et al., 2007), and others (Kim et al., 2012; Sedjaï et al., 2010), are also required for primary cilium formation.
In this study, we show that Cep123 interacts with the centriolar satellite proteins PCM-1, OFD1 and Cep290 and is required for primary ciliogenesis. In the absence of Cep123 a ciliary vesicle fails to form at the distal end of the mother centriole, with only small or misshapen vesicles associated with the distal appendages. These results identify Cep123 as a key distal appendage protein whose function is required for primary cilium assembly.
Results
Cep123 is a conserved distal appendage protein
Cep123 was identified as a PLK4-interacting protein in a yeast two-hybrid screen using the kinase domain of PLK4 (residues 1–271) as bait. The precise function of the interaction between Cep123 and PLK4 has not been fully established and will not be discussed in this paper. Four clones corresponded to Cep123 with three clones encoding residues 414–578 and the remaining clone encoding residues 414–444. Three isoforms of Cep123 (Cep89) are described in UniProtKB/Swiss-Prot (Q96ST8), with isoform 1, the longest isoform, predicted to contain two coiled coil domains between residues 234 to 333 and 369 to 719 and a PEST sequence, involved in regulating protein turnover, between residues 112 to 138 (Fig. 1A; supplementary material Fig. S1A). Western blotting of centrosomes purified from KE37 cells with antibodies raised against the N- and C-terminus of the protein indicated that the predominant centrosomal isoform of Cep123 was approximately 100 kDa in molecular weight, which probably corresponds to isoform 1 of Cep123 (supplementary material Fig. S1B). This hypothesis was confirmed by western blotting of soluble and insoluble extracts prepared from Cep123 or control siRNA-treated RPE1 cells with the N- and C-terminal antibodies (supplementary material Fig. S1C).
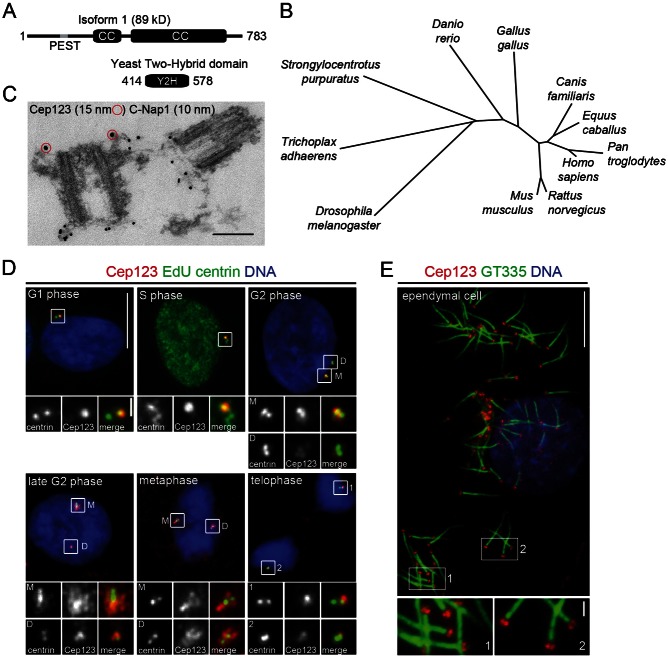
Fig. 1. Cep123 is a conserved protein localizing to the distal appendages of the mother centriole.
(A) A diagram showing the structural domains of Cep123 and the PLK4-interacting fragment identified by yeast two-hybrid screening with a catalytically inactive form of the kinase domain of PLK4 (residues 1–271). Two coiled-coil domains (CC) and a potential PEST sequence (grey line) are present in Cep123. (B) A phylogenetic tree of Cep123 homologs. (C) An electron micrograph of a KE37 cell purified centrosome labelled with anti-Cep123 N-terminal antibody (15 nm gold particles marked with red circles) and C-Nap1 (10 nm gold particles), showing that Cep123 localizes to the distal appendages of the mother centriole. (D) Immunofluorescent images of RPE1 cells labelled with EdU (green) and stained with DAPI (blue) and antibodies against Cep123 (red), centrin (green) and ninein (not shown). Cep123 is associated with the mother centriole (M) throughout the cell cycle and begins to be recruited to the daughter centriole (D) in late G2. (E) Immunofluorescent staining of mouse ependymal cells with antibodies against polyglutamylated tubulin (GT335, green) and Cep123 (red) showed that Cep123 localizes to the base of motile cilia as well as to primary cilia. DNA labelled with DAPI (blue). Scale bars: 10 µm and inset 1 µm.
Phylogenetic analyses indicated that homologs of Cep123 are present in most metazoans (Fig. 1B) including the placazoan T. adhaerens, which is considered to be one of the most primitive metazoans (Srivastava et al., 2008). One notable exception was C. elegans, which possesses centrioles containing only singlet microtubules (O'Connell et al., 2001; O'Toole et al., 2003), where we were unable to identify a Cep123 ortholog.
Cep123 has been shown, by immunofluorescent staining and photoactivatable localization microscopy (PALM), to localize to the distal appendages of the mother centriole (Sillibourne et al., 2011). To confirm this localization, purified KE37 cell centrosomes were stained with antibodies against Cep123 and C-Nap1 or Cep123 alone and thin section EM carried out. This demonstrated conclusively that Cep123 localized to the distal appendages of the mother centriole (Fig. 1C; supplementary material Fig. S1D). Centrosomal proteins often exhibit cell cycle-dependent localization to the organelle, with their levels changing during the cell cycle. Immunofluorescent staining of EdU-labelled RPE1 cells with antibodies against ninein, centrin and Cep123 showed that the localization of Cep123 was indeed cell cycle-dependent with its levels decreasing during mitosis (Fig. 1D). A similar cell cycle-dependent localization has been reported for the distal appendage protein Cep164 (Schmidt et al., 2012). However, in contrast to Cep164, recruitment of Cep123 to the daughter centriole (developing mother centriole) began earlier, in late G2. To evaluate the dynamics of Cep123, live cell imaging and FRAP analysis were carried out on RPE1 cells transiently expressing EGFP-Cep123 and mCherry-centrin-1 (supplementary material Movie 1; Fig. S1E). This revealed that EGFP-Cep123 not only localized to the distal appendages, but also to cytoplasmic particles moving to and from the centrosome. Furthermore, FRAP analysis demonstrated that there was both a mobile and immobile fraction of Cep123 associated with the distal appendages, with the t1/2 measured as being 61.8±40.6 s (supplementary material Fig. S1E).
To investigate if Cep123 is a component of motile cilia, as well as primary cilia, multi-ciliated mouse ependymal cells were stained with antibodies against polyglutamylated tubulin (GT335) and Cep123 (Fig. 1E). Cep123 localized to the base of ependymal cilia confirming that it is a component of both motile and immotile cilia.
Mapping of the distal appendage-targeting domain in Cep123
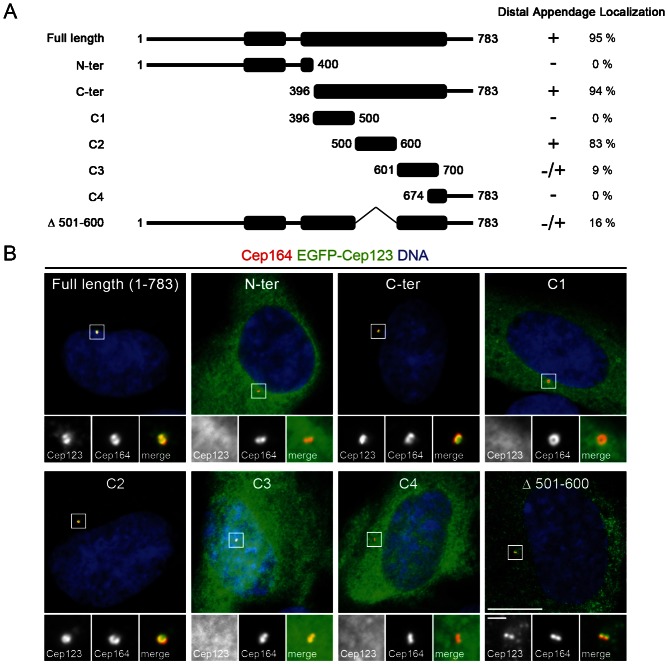
To gain insight into how Cep123 is targeted to the distal appendages, we began by mapping the domains responsible for targeting the protein to these structures. A series of deletion constructs was generated, by fusing fragments of Cep123 to EGFP, and transiently transfected into RPE1 cells (Fig. 2A). Staining of these cells with an antibody against the distal appendage component Cep164 demonstrated that the C-terminal region of Cep123 (residues 396–783) was responsible for targeting Cep123 to the distal appendages, with the C2 construct (residues 500–600) containing the minimal distal appendage-targeting domain (Fig. 2B). Deletion of this domain from Cep123 reduced the efficacy of its targeting to the distal appendages, with only 16% of EGFP-Cep123 Δ501–600 transfected cells exhibiting a distal appendage localization compared with 96% of EGFP-Cep123 transfected cells (Fig. 2A). Thus, while these results indicate that residues 500–600 are not essential for the recruitment of Cep123 to the distal appendages, they do increase the efficiency of its targeting to these structures. It is possible that Cep123 Δ501–600 is able to localize to the distal appendages by self-association with the endogenous protein, as has been noted for many centrosomal proteins (Lukinavičius et al., 2013).
Fig. 2. Mapping of the distal appendage-targeting domain in Cep123.
(A) A diagram showing the EGFP-Cep123 fragments used to identify the distal appendage-targeting domain in Cep123. (B) Immunofluorescent images of RPE1 cells transfected with the EGFP-Cep123 constructs (green) and stained with DAPI (blue) and an antibody against the distal appendage component Cep164 (red). The C2 fragment (residues 500–600) was able to localize to the distal appendages with a similar efficiency as the full length protein. This domain was not essential for the targeting of Cep123 to the distal appendages, although it increased the fidelity of proper localization. Scale bars: 10 µm and inset 1 µm.
Cep123 is required for primary ciliogenesis
During primary ciliogenesis, the distal appendages of the mother centriole are proposed to participate in the anchoring of a ciliary vesicle to the distal end of the centriole and, later on, to anchor the mother centriole (basal body) to the plasma membrane. Cep164, another distal appendage protein, is known to play an important role in primary ciliogenesis (Graser et al., 2007; Schmidt et al., 2012); therefore we assessed whether Cep123 was required for this process by using siRNA to deplete the protein from RPE1 cells.
Depletion of centrosomal proteins has been proposed to trigger a centrosome integrity checkpoint, resulting in p38 kinase-dependent cell cycle arrest (Mikule et al., 2007; Srsen et al., 2006). We first ascertained if Cep123 depletion affected cell cycle progression. Three different sets of siRNA were tested for the efficacy to deplete Cep123, a single duplex, Cep123 siRNA d2, and two pools, Cep123 P1 and Cep123 P2. The latter pool consisted of modified siRNAs designed to reduce off-target effects. As controls, RPE1 cells were also treated with siRNAs directed against PCM-1 and the centriolar linker protein C-Nap1 (Fry et al., 1998), which previous studies have shown to be required for cell cycle progression (Mikule et al., 2007; Srsen et al., 2006). SiRNA-treated RPE1 cells were pulse-labelled with EdU, to detect S phase cells, and stained with an antibody against the cell proliferation marker Ki67 (Fig. 3A). These data showed that, in contradiction to previously published results, neither PCM-1 nor C-Nap1 was required for cell cycle progression, with no significant difference in the number of Ki67- and EdU-positive (S-phase) cells compared with controls (Fig. 3B). However, the siRNAs directed against Cep123 produced variable results, with the number of EdU-positive cells being reduced by approximately 69% (P = 7.3×10−5), 38% (P = 2.2×10−2) and 15% (non-significant) upon treatment with the Cep123 siRNA d2, Cep123 P1 and Cep123 P2 siRNAs, respectively. The number of Ki67-positive cells was also reduced, compared to controls, by 61% (P = 6.4×10−4), 20% (non-significant) and 3% (non-significant) upon treatment with the Cep123 siRNA d2, Cep123 P1 and Cep123 P2 siRNAs, respectively. To ascertain if the stress response pathway was activated in siRNA-treated cells, western blotting was carried out with antibody to T180/Y182 phosphorylated p38 (active p38) (Fig. 3C). Duplicate samples were treated with 500 µM sodium arsenite for 30 minutes to induce and test the robustness of the p38 stress response (Kim et al., 2002). Regardless of the siRNA treatment, the basal level of p38 phosphorylation was similar, while exposure to sodium arsenite induced the stress response and resulted in increased levels of phosphorylated p38. Altogether these results suggest that p38-dependent cell cycle arrest is due to siRNA off-target effects and this is dependent upon the sequence of the siRNA, in agreement with published data (Jackson et al., 2006a; Jackson et al., 2006b).
Fig. 3. Depletion of centrosomal proteins does not necessarily cause cell cycle arrest and activation of the p38 stress response pathway.
(A) Immunofluorescent images of RPE1 cells treated with Cep123, PCM-1, C-Nap1 or a non-targeting (NT) control siRNA, labelled with EdU (green) and stained with DAPI (blue) and an antibody against the cell proliferation marker Ki67 (red). Scale bars: 10 µm and inset 1 µm. (B) Quantification of the number of EdU-positive (S phase) and Ki67-positive (cycling) cells. The data represent the mean of three independent experiments (n = 300) with s.d. shown. Depletion of Cep123 using the Cep123 siRNA d2 caused a significant reduction in the number of both EdU- and Ki67-positive cells (student's t-test, P = 7.3×10−5 and P = 6.4×10−4, respectively), suggesting that the cells had exited the cell cycle. A significant reduction in the number of S-phase cells was also observed upon treatment with the Cep123 P1 siRNA (student's t-test, P = 2.2×10−2). Depletion of PCM-1, C-Nap1 and Cep123 using the Cep123 P2 siRNAs did not cause cell cycle arrest, as there was no significant change in the number of EdU- and Ki67-positive cells compared with the NT control. (C) Western blotting of lysates prepared from siRNA-treated RPE1 cells using antibodies against Cep123 (C-terminal antibody), PCM-1, C-Nap1, p38, phosphorylated p38 and α-tubulin. One set of samples was treated with 500 µM sodium arsenite for 30 minutes to induce a stress response. Regardless of the siRNA treatment used, the basal level of phosphorylated p38 was similar and treatment with sodium arsenite induced a stress response and increased levels of phosphorylated p38.
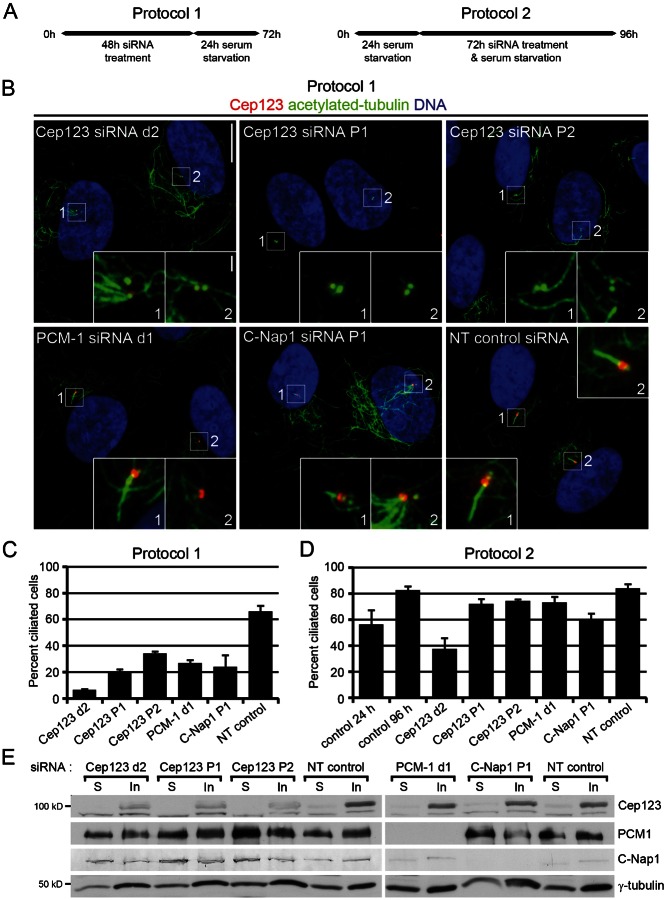
Two different protocols of siRNA treatment were used to establish if Cep123 was required for the formation and/or maintenance of a primary cilium. In the first protocol RPE1 cells were treated with siRNA for 48 hours and deprived of serum for 24 hours to induce primary cilium formation, while in the second protocol the cells were serum-starved for 24 hours before being treated with siRNA for 72 hours in the absence of serum (Fig. 4A). The first protocol assessed whether a protein was required for primary cilium formation, while the second protocol tested if it was necessary for primary cilium maintenance. RPE1 cells were stained with antibodies against acetylated tubulin, to identify primary cilia, and Cep123 to determine the efficacy of Cep123 depletion (Fig. 4B). The results indicated that Cep123 was required for primary cilium formation (Fig. 4C), as a significant reduction in the number of ciliated cells compared with NT controls was observed after its depletion using the first protocol (Cep123 siRNA d2, 90%; Cep123 P1, 69%; Cep123 P2, 48% reduction). However, Cep123 was found to be largely dispensable for primary cilium maintenance (Fig. 4D), as the number of ciliated cells was only marginally decreased after its depletion using the specific pools of siRNA with the second protocol (Cep123 siRNA d2, 56%; Cep123 P1, 14%; Cep123 P2, 12% reduction). Similar results were obtained after PCM-1 depletion, indicating that is also required for the formation, but not the maintenance, of a primary cilium. Depletion of PCM-1 using the first protocol greatly reduced the number of ciliated cells (60% reduction), while only a moderate reduction (13%) was observed using the second protocol (Fig. 4C,D; supplementary material Fig. S2A). Interestingly, C-Nap1 depletion inhibited both primary cilium formation and maintenance, causing a reduction in the number of ciliated cells, using both the first (64% reduction) and the second protocol (30% reduction), compared with NT controls (Fig. 4C,D). The results of these C-Nap1 depletions are consistent with those of a recently published study demonstrating a requirement for C-Nap1 in primary ciliogenesis (Conroy et al., 2012). Western blotting with antibodies against Cep123, PCM-1, and C-Nap1 verified the efficacy of depletion of each centrosomal protein (Fig. 4E). While the siRNAs we used to target Cep123 were equally efficacious in depleting the protein, variation in the penetrance of the knockdown phenotype was observed, with the Cep123 d2 siRNA having the greatest effect upon primary cilium formation. This is probably due to the Cep123 d2 siRNA having off-target effects, such as cell cycle arrest (Fig. 3A,B), which contribute to the reduction in the number of ciliated cells. Nevertheless, depletion of Cep123 with the Cep123 P2 siRNAs significantly reduced primary cilium formation, but did not affect cell cycle progression, thereby supporting a direct role for Cep123 in primary ciliogenesis. We confirmed these results by depleting Cep123 from another cell line, inner medullar collecting duct 3 (IMCD3) cells, and observed a significant reduction in the number of ciliated cells compared with the control (supplementary material Fig. S2B).
Fig. 4. Cep123 is required for primary cilium formation but not maintenance.
(A) A schematic explaining the two protocols used to determine if Cep123, PCM-1 or C-Nap1 is required for the formation (protocol 1) or maintenance (protocol 2) of a primary cilium. (B) Immunofluorescent images of RPE1 cells stained with DAPI to label the DNA (blue) and antibodies against acetylated tubulin (green) and Cep123 (red). Scale bars: 10 µm and inset 1 µm. (C,D) Quantification of the number of ciliated cells, where the results represent the means of three independent experiments (n = 200–450), with the s.d. shown. The depletion of Cep123, PCM-1 or C-Nap1 prevented primary cilium formation, resulting in a significant decrease in the number of ciliated cells (student's t-test: Cep123 siRNA d2, P = 1.8×10−5; Cep123 P1, P = 6.0×10−5; Cep123 P2, P = 2.5×10−4; PCM-1, P = 1.5×10−4; and C-Nap1, P = 1.7×10−3). However, Cep123 and PCM-1 appeared to be dispensable for primary cilium maintenance while C-Nap1 was only partially required (student's t-test: Cep123 siRNA d2, P = 7.9×10−4; Cep123 P1, P = 1.2×10−2; Cep123 P2, P = 7.2×10−3; PCM-1, P = 2.3×10−2; and C-Nap1, P = 2.3×10−3). (E) Western blotting of soluble and insoluble extracts prepared from siRNA-treated RPE1 cells demonstrated the depletion of the appropriate protein.
Centriolar satellite proteins interact with Cep123
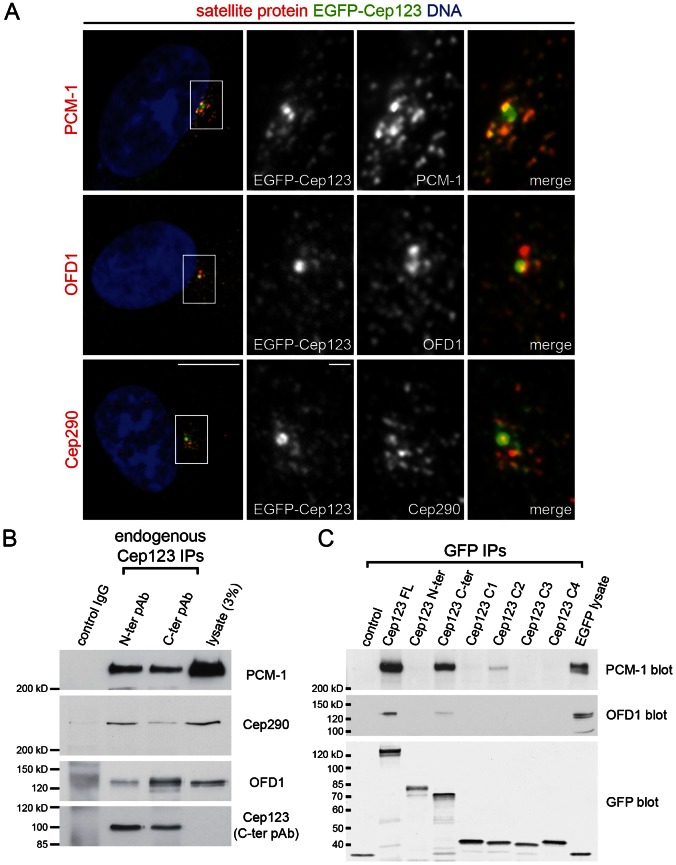
Cep123 localizes to the distal appendages (Sillibourne et al., 2011), but it has also been suggested to localize to centriolar satellites (Jakobsen et al., 2011). To determine if the EGFP-Cep123 particles observed by live imaging of transfected RPE1 cells co-localized with centriolar satellites, RPE1 cells were fixed and stained with antibodies against PCM-1, OFD-1 and Cep290. These centriolar satellite proteins were chosen because they are mutually dependent upon one another for their proper localization to centriolar satellites (Lopes et al., 2011) and are involved in primary cilium formation (Graser et al., 2007; Kim et al., 2008; Nachury et al., 2007; Singla et al., 2010). Co-localization was often observed between PCM-1 satellites and EGFP-Cep123, while fewer OFD1 and Cep290 satellites co-localized with EGFP-Cep123 particles (Fig. 5A). To determine if endogenous Cep123 interacted with any of these centriolar satellite proteins, Cep123 was immunoprecipitated from RPE1 cell lysates using the Cep123 N- and C-terminal antibodies and western blotting carried out (Fig. 5B). PCM-1, OFD1 and Cep290 all co-precipitated with Cep123, suggesting that they interact with it. An interaction between PCM-1 and Cep123 was further supported the co-precipitation of endogenous Cep123 with over-expressed GFP-tagged truncated PCM-1 (residues 1–1468) and full length PCM-1 (supplementary material Fig. S3A). The PCM-1 and OFD1 sites of interaction on Cep123 were identified by transiently transfecting RPE1 cells with EGFP-Cep123 deletion constructs (Fig. 2A) and carrying out IPs with an anti-GFP antibody. This demonstrated that both PCM-1 and OFD1 interact with the C-terminus of Cep123 (Fig. 5C). Furthermore, the distal appendage-targeting domain of Cep123 (residues 500–600) was able to co-precipitate PCM-1, although considerably less than the full length protein (Fig. 5C). While the N-terminus of Cep123 did not interact with centriolar satellite proteins, it might play a role in negatively regulating their interaction with the C-terminus of Cep123, as over-expression of isoform 2 of Cep123 (lacking residues 1–247) increased the frequency of Cep123 particle formation compared with isoform 1 (supplementary material Fig. S3B,C). Furthermore, robust co-localization was observed between particles of isoform 2 of Cep123 and PCM-1 and OFD1. In contrast, only partial co-localization was observed between EGFP-tagged isoform 2 of Cep123 and the centriolar satellite proteins BBS4, Cep290 and FOR20 (Sedjaï et al., 2010). Together these data suggest that Cep123 interacts with centriolar satellite proteins via its C-terminus, with its N-terminus negatively regulating these interactions.
Fig. 5. Centriolar satellite proteins interact with Cep123.
(A) Immunofluorescent images of RPE1 cells transiently transfected with EGFP-Cep123 (green) and stained with DAPI to label the DNA (blue) and antibodies against the centriolar satellite proteins PCM-1, OFD1 and Cep290 (all red). Co-localization between EGFP-Cep123 particles and PCM-1, OFD1 and Cep290 satellites was observed. Scale bars: 10 µm and inset 1 µm. (B) Western blotting of Cep123 immunoprecipitates with antibodies against PCM-1, OFD1 and Cep290 indicated that these satellite proteins form a complex with Cep123. The N- and C-terminal antibodies were used to immunoprecipitate Cep123. (C) EGFP-Cep123 fragments were immunoprecipitated from transfected RPE1 cell lysates using an anti-GFP antibody and western blotting carried out with PCM-1 and OFD1 antibodies. Both of these proteins were co-immunoprecipitated with the C-terminus of Cep123 (residues 396–783).
Cep123 depletion causes defects in ciliary vesicle formation
To understand why Cep123 depletion prevents primary cilium formation, serial thin-section EM was carried out on NT control- and Cep123-siRNA-treated RPE1 cells (Fig. 6A,B). Both the distal and sub-distal appendages of the mother centriole were still present in Cep123-depleted cells, suggesting that the integrity of these structures was not compromised by the absence of Cep123. This was further supported by the staining of siRNA-treated RPE1 cells with anti-Cep164 antibody, followed by fluorescent intensity measurements, which showed that the localization of Cep164 was virtually unperturbed by the loss of Cep123 (supplementary material Fig. S4A,B). A reciprocal experiment demonstrated that Cep123 was still able to localize to the distal appendages in the absence of Cep164 (supplementary material Fig. S4C,D). These results are in agreement with recently published data showing that Cep123 and Cep164 localize to the distal appendages independently of each other (Schmidt et al., 2012). The localization of ninein and γ-tubulin was also unperturbed by the depletion of Cep123 (supplementary material Fig. S4E,F). While the distal appendages still appeared to be intact in Cep123-depleted cells, either only small vesicles or a malformed ciliary vesicle was associated with them (Fig. 6B). These data suggest that in the absence of Cep123 primary ciliogenesis is blocked at the early step of ciliary vesicle formation and anchoring at the distal end of the mother centriole.
Fig. 6. Depletion of Cep123 perturbs ciliary vesicle formation.
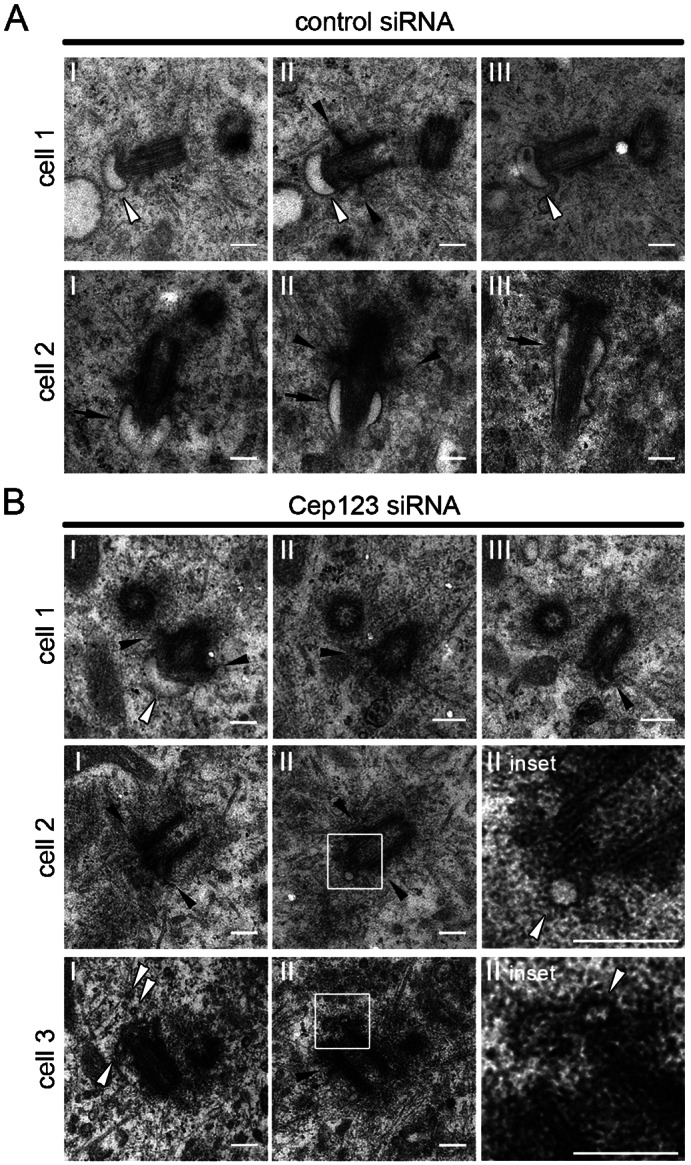
Electron micrographs of RPE1 cells treated with Cep123 or NT control siRNA according to protocol 1. (A) Control cells possessed ciliary vesicles attached to the distal end of the mother centriole (cell 1, white arrowhead) or ciliary pockets (cell 2, black arrows). (B) Cep123-depleted cells had only small vesicles attached to the distal appendages (cells 1 and 2, white arrowheads) or exhibited malformed vesicles surrounded by electron dense material (cell 3, white arrowheads). The black arrowheads mark the sub-distal appendages of the mother centriole. Scale bars: 200 nm.
Discussion
In this paper, we show that Cep123 is a conserved distal appendage protein required for the formation of a ciliary vesicle at the distal end of the mother centriole during primary ciliogenesis. This is apparently linked to the function of the centriolar satellite proteins PCM-1, OFD1 and Cep290, all of which participate in primary ciliogenesis.
Many centrosomal proteins have been implicated in primary ciliogenesis (Graser et al., 2007; Mikule et al., 2007) and it is conceivable that not all of these, given their functional diversity, are directly involved in the process and may play an indirect role. One of these studies (Mikule et al., 2007) proposed that the depletion of centrosomal proteins triggers a p38 kinase-dependent checkpoint, due to structural defects in the centrosome, leading to cell cycle arrest in G0 and the inhibition of primary ciliogenesis. Consequently, these results suggested that it would be difficult to decipher whether a centrosomal protein is directly involved in ciliogenesis or not, as its depletion could trigger the centrosome integrity checkpoint. While the depletion of some centrosomal proteins may cause structural defects in the centrosome and cell cycle arrest, this does not appear to be the case for Cep123. We found that depletion of Cep123, using a pool of modified siRNAs (Cep123 P2), did not cause cell cycle arrest and, furthermore, a combination of serial thin-section EM and immunofluorescent staining with Cep164 antibodies (Fig. 6; supplementary material Fig. S4A) indicated that the structure of the distal appendages had not been compromised by the loss of Cep123. We also found that PCM-1 depletion had no effect upon cell cycle progression, in contradiction to previously published results (Mikule et al., 2007; Srsen et al., 2006), but primary cilium formation was impaired. These results suggest that siRNA off-target effects are responsible for causing cell cycle arrest, in agreement with published results (Jackson et al., 2006a). Under the conditions we used in this paper activation of p38 kinase was not observed after siRNA treatment, arguing against activation of the proposed centrosome integrity checkpoint. Furthermore, a study carried out on RPE1 cells, in which the centrosome was removed either by microsurgery or laser ablation, demonstrated that centrosome loss alone was not sufficient to induce activation of p38 kinase and cell cycle arrest, and that a secondary stress was required (Uetake et al., 2007). Altogether, these data argue that the primary ciliogenesis defect observed upon the loss of Cep123 is due to the protein playing a direct role in the process.
Cep123 has been described to localize to both the distal appendages of the mother centriole (Sillibourne et al., 2011) and to centriolar satellites (Jakobsen et al., 2011). We have shown that the anchoring of Cep123 to the distal appendages occurs via its C-terminus (residues 396–783) with a domain located between residues 500–600 playing a particularly important role in this process. The deletion of this domain from Cep123 decreased the efficacy of targeting of the protein to the distal appendages and when fused to EGFP it was able to localize to the distal appendages with a similar efficiency as full length Cep123. This domain was also able to co-precipitate PCM-1, albeit in smaller amounts than full length or C-terminal Cep123. Such an interaction raises the possibility that PCM-1 is involved in transporting Cep123 to the centrosome, as this has been demonstrated for the PCM-1-interacting proteins centrin, pericentrin and ninein (Dammermann and Merdes, 2002). However, the localization of Cep123 was only partially dependent upon PCM-1, with a moderate reduction in its levels at the distal appendages upon the depletion of PCM-1 (J.E.S. and M.B., unpublished observations).
Several lines of evidence support an interaction between Cep123 and centriolar satellites. Firstly, when Cep123 is over-expressed it associates with cytoplasmic particles that often co-localize with PCM-1-, OFD1- or Cep290-containing centriolar satellites. Secondly, endogenous Cep123 co-precipitates with over-expressed full length PCM-1 and C-terminal truncated PCM-1 (residues 1–1468) and, more importantly, all of the above centriolar satellite proteins co-precipitate with endogenous Cep123. While immunofluorescent staining of cells with an antibody raised against the N-terminus of Cep123 failed to support centriolar satellite localization, one possibility is that the amount of Cep123 present in centriolar satellites is too low to be detected by the N-terminal antibody.
PCM-1 has long been proposed to play a role in the formation of cilia (Kubo et al., 1999) and it interacts with several centriolar satellite proteins, OFD1 (Lopes et al., 2011), Cep290 (Kim et al., 2008), and BBS4 (Kim et al., 2004), involved in primary ciliogenesis. Interestingly, we found that while PCM-1 is required for primary cilium formation, in agreement with previously published results (Graser et al., 2007; Nachury et al., 2007), it is not necessary for primary cilium maintenance. This result, coupled with interaction data, suggests that PCM-1 and Cep123 co-operate during the process of primary ciliogenesis, with Cep123 possibly acting as a receiver of PCM-1-bound cargoes at the distal appendages.
The centriolar satellite protein Cep290 also localizes to the distal ends of centrioles and to the transition zone of the primary cilium, where it is postulated to play a role in regulating the entry of proteins into the ciliary axoneme (Craige et al., 2010; Garcia-Gonzalo et al., 2011; Sang et al., 2011). It is also involved in recruiting Rab8a to the primary cilium and in the absence of Cep290 this GTPase no longer localizes to the ciliary axoneme (Kim et al., 2008). As Cep123-depletion prevents the formation of a ciliary vesicle at the distal end of the mother centriole, it is possible that Cep123 is involved in regulating the recruitment of membranes to the centrosome through its interaction with Cep290.
Together, our data implicate Cep123 as a new player in the process of primary ciliogenesis through its localization to the distal appendages of the mother centriole, its interaction with PCM-1, OFD1 and Cep290 and the failure to assemble a ciliary vesicle in its absence. Further work will be necessary to identify additional Cep123-interacting proteins and determine if they are also involved in primary ciliogenesis.
Materials and Methods
Plasmids and molecular biology techniques
Cep123 sequences were amplified by PCR from the previously described plasmid pEGFP-Cep123 (Sillibourne et al., 2011) using Phusion DNA polymerase (Finnzymes) and cloned into pEGFP (Clontech) or myc-tag vectors. Isoform 3 of Cep123 was amplified from the cDNA clone IRAKp961G0764Q (Source BioScience Lifescience) and cloned into pEGFP-C1 (Clontech). GeneSOEing (Horton, 1993) was used to delete the sequence encoding the distal appendage-targeting domain from Cep123. Two PCRs were setup using the primers Cep123-S01 and Cep123 GS-Rv; and Cep123-AS01 with Cep123 GS-Fw (the sequences of these primers are in supplementary material Table S1). The resulting PCR products were gel purified and used as template to amplify the full length Cep123 sequence using the primers Cep123-S01 and Cep123-AS01. All constructs were verified by sequencing (Applied Biosystems).
Phylogenetic analyses
Cep123 sequences were aligned using ClustalX (Thompson et al., 1997) and a phylogenetic tree created with Phylip (http://evolution.gs.washington.edu/phylip) (Felsenstein, 1989).
Yeast two-hybrid screen
A yeast two-hybrid screen using a catalytically inactive version of the kinase domain of PLK4 (residues 1 to 271 with a K41M mutation) was used as bait to screen a breast cancer cDNA library (Centre of Advanced European Studies and Research [Caesar], Germany).
Antibodies
Rabbit polyclonal antibodies were raised against the N-terminus (amino acids 1–230) (Sillibourne et al., 2011) and the C-terminus (amino acids 674–783) of Cep123. Sequences encoding these fragments of Cep123 were cloned into a 6×HIS-SUMO expression vector, the fusion proteins purified under denaturing conditions (Qiagen) and used for immunization (AgroBio). The N-terminal antibody was affinity purified with maltose-binding protein (MBP) fused to residues 100–200 of Cep123 (Cep123-N2) and the C-terminal antibody with MBP fused to residues 674–783 of Cep123. Immunofluorescent staining was carried out using the Cep123-N2 affinity purified antibody (1:200) and western blotting with affinity-purified Cep123 N- or C-terminal antibodies (both at 1:5000). Primary antibodies were obtained from the following sources: acetylated tubulin mouse monoclonal, 1:4000 (clone 6-11B-1, Sigma); α-tubulin mouse monoclonal (3A2), 1:5000 (L. Lafanechére); Cep290 rabbit polyclonal, 1:1000 (A301-659A, Bethyl Laboratories); C-Nap1 mouse monoclonal 1:250 (BD Pharmingen); FOR20 rat monoclonal antibody, 1:2000 (Olivier Rosnet) (Sedjaï et al., 2010); γ-tubulin mouse monoclonal, 1:2000 (GTU-88, Sigma); GFP mouse monoclonal, 1:2000 (clones 7.1 and 13.1, Roche); GFP rabbit polyclonal (Institut Curie Antibody Production Unit); Ki67 mouse monoclonal, 1:250 (clone B56, BD Pharmingen); myc-tag mouse monoclonal, 1:10000 (9E10, Institut Curie Antibody Production Unit); BBS4 rabbit polyclonal, 1:1000 (Andrew Fry) (Lopes et al., 2011); centrin mouse monoclonal, 1:2000 (20H5, Jeffrey Salisbury) (Salisbury et al., 1988); Cep164 rabbit polyclonal, 1:1000 (Erich Nigg) (Graser et al., 2007); OFD1 rabbit polyclonal, 1:100 (Andrew Fry) (Lopes et al., 2011); PCM-1 rabbit polyclonal, 1:500 (Akiharu Kubo) (Kubo et al., 1999); p38 rabbit polyclonal, 1:1000 (9212, Cell Signaling); phospho-p38 rabbit monoclonal (Thr180/Tyr182), 1:1000 (D3F9, Cell Signaling); polyglutamylated tubulin mouse monoclonal, 1:10000 (GT335, Bernard Eddé) (Wolff et al., 1992); humanized anti-ninein scFv, 1:5 (Sillibourne et al., 2010); and γ-tubulin rabbit polyclonal, 1:500 (Ríos et al., 2004). Anti-mouse, anti-rabbit, and anti-human secondary antibodies labelled with Cy3 or Cy5 were obtained from Jackson Immunoresearch Laboratories Inc. and Alexa 488 labelled antibodies from Invitrogen.
Cell culture
RPE1 cells were cultured in DMEM/F12, IMCD3 cells DMEM and KE37 cells in RPMI1640 (Invitrogen) at 37°C with 5% CO2. All media were supplemented with 10% foetal bovine serum (Biowest Sera), 1% penicillin/streptomycin and 1% L-glutamine (Invitrogen). To induce primary cilium formation RPE1 cells were placed in DMEM/F12 medium containing 0.25% foetal bovine serum.
Transient transfection and siRNA
RPE1 cells were transfected for 24 hours with DNA using XtremeGENE 9 transfection reagent (Roche) according the protocols provided using a 3:1 ratio of transfection reagent to DNA. Small interfering RNAs (siRNAs) were transfected into RPE1 and IMCD3 cells at a final concentration of 10 nM using Dharmafect transfection reagent 4 or 1, respectively. To transfect a single well of a 6 well plate, 2 µL of 10 µM siRNA and 4 µL of transfection reagent were separately diluted with 200 µL of serum-free medium containing 10 mM HEPES pH 7. After incubating the diluted transfection reagent at ambient temperature for 5 minutes the two mixtures were combined and incubated together for a further 20 minutes before being added to cells. Small interfering siRNAs were obtained from Qiagen and Dharmacon and their target sequences can be found in supplementary material Table S2.
Preparation of soluble and insoluble extracts from cells
Cells growing in 60 mm dishes were washed once with PBS and detached using trypsine/EDTA (Invitrogen). The cells were pelleted by centrifugation at 220 g for 3 minutes at 4°C, resuspended in 5 mL of TBS and and pelleted again by centrifugation. The cells were resuspended in 5 mL of hypotonic buffer consisting of 8% sucrose dissolved in 0.1× TBS, pelleted by centrifugation and lysed with 100 µL of lysis buffer (10 mM HEPES pH 7.2, 0.5% NP-40, 0.5 mM MgCl2) containing protease inhibitors. After incubating on ice for 5 minutes, insoluble material was pelleted by centrifugation at 220 g for 5 minutes in a fixed-angle rotor. The soluble material was transferred to a clean Eppendorf tube and precipitated by adding 1.35 mL of −20°C methanol and incubating on ice for 1 hour. Precipitated material was pelleted by centrifugation at 18,000 g for 15 minutes at 4°C, the supernatant removed and the pellet allowed to air dry. The pelleted material, both soluble and insoluble, was resuspended in an equivalent volume of SDS sample buffer.
Immunoprecipitations
RPE1 cells growing in 100 mm dishes were harvested by washing once with PBS and lysing the cells in 500 µL of lysis buffer (50 mM Tris HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100) containing protease inhibitors. The lysates were clarified by centrifugation at 18,000 g for 5 minutes at 4°C, the supernatant transferred to a clean Eppendorf tube containing 10 µL of prewashed magnetic Protein G beads (Invitrogen) and 2 µg of antibody. After incubating the supernatants with the beads for 1 hour at 4°C with end-over-end mixing, the beads were washed five times with 1 mL of lysis buffer and the immunoprecipitates eluted with 15 µL of SDS sample buffer.
Protein fractionation and Western blotting
Sample protein concentration was quantified using the 660 nm protein assay kit in conjunction with the ionic detergent compatibility reagent (Pierce), where necessary. Proteins were fractionated on either 3–8% Tris-acetate gradient (Invitrogen) or Tris-glycine gels and transferred to nitrocellulose using an iBlot (Invitrogen). Membrane blocking was carried out using 5% BSA or 5% non-fat milk in Tris buffered saline (TBS) supplemented with 0.2% Tween 20 (TBST). Primary antibodies were diluted in the same buffer while washing steps were carried out with TBST.
Isolation of centrosomes
Centrosomes were isolated from KE37 cells according to a previously published protocol (Moudjou and Bornens, 1994). Samples were loaded onto 3–8% Tris-acetate gels (Invitrogen) and western blotting carried out as described above.
Edu labelling
Cells were pulsed with 10 µM 5-ethynyl-2′-deoxyuridine (Edu) for 10 minutes at 37°C before being fixed in −20°C methanol. The detection of Edu-labelled cells was carried out according to the manufacturer's instructions (Invitrogen).
Live cell imaging and fluorescence recovery after photobleaching (FRAP)
Transfected cells were plated into 35 mm glass-bottomed dishes (IWAKI) and imaged on a Nikon Ti Eclipse microscope equipped with a spinning disk (Yokogawa) and a HQ2 digital camera (Photometrics). For each fluorescent fusion protein, a 2 µm stack of images was captured, with 2×2 binning and a spacing of 0.2 µm, once every 10 seconds for a total of 10 minutes and maximal intensity projections made. ImageJ was used to create movies from the stacks of maximal intensity projections. FRAP analysis was carried out using the same microscope described above, bleaching for 25 ms and monitoring recovery by capturing one image every 10 sec. Analysis of FRAP images was carried out using EasyFRAP software (Rapsomaniki et al., 2012).
Immunofluorescence staining and imaging
Cells growing on coverslips, were fixed with methanol at −20°C for at least 20 minutes and blocked with antibody blocking buffer (TBS, 1% BSA fraction V and 0.5% Triton X-100). Primary antibodies were diluted in the same buffer and incubated on the cells for 1 hour at ambient temperature. After washing extensively with antibody blocking buffer diluted secondary antibodies were incubated on the cells for 30 minutes at ambient temperature. DNA was labelled with a 0.2 µg/ml solution of 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) (Sigma). After sequentially washing in antibody dilution buffer, TBS and distilled water the coverslips were air-dried and mounted onto glass slides using Mowiol mounting medium. Images were captured on a Leica DMRA2 microscope, fitted with a CoolSNAP camera (Princeton Instruments), using a 100×1.4 N.A. objective lens (Leica) and Metamorph software (Universal Imaging). Processing of images and fluorescence intensity measurements were carried out using either ImageJ or Metamorph software. Briefly, centrosomal fluorescence intensity was calculated by measuring the fluorescence intensity within a circular region placed over the centrosome and subtracting the background fluorescence intensity of a similar sized region adjacent to the centrosome. Figures were assembled using Photoshop (Adobe). Statistical analyses were performed using two-tailed Student's t-test.
Electron microscopy
Approximately 1.5×107 centrosomes were centrifuged onto a 12 mm acid-washed glass coverslip at 10,000 g for 10 minutes at 4°C. Centrosomes were stained without prior fixation with antibodies against either Cep123 diluted in PBS containing 1% BSA (Sigma) for 15 minutes at ambient temperature. Labelling of primary antibodies was carried out with 10 nm or 15 nm gold-conjugated protein A. To double label centrosomes, rabbit antibody/protein A conjugates were fixed with 1% glutaraldehyde for 5 minutes at ambient temperature and washed 5 times with PBS containing 0.1% glycine leaving each wash for 2 minutes. Primary antibody labelling was carried out with a monoclonal antibody against C-Nap1, as described above, followed by labelling with a rabbit anti-mouse bridging antibody and detection with 10 or 15 nm gold-conjugated protein A. All steps were carried out at ambient temperature for 15 minutes. Final fixation was carried out using 2.5% glutaraldehyde/0.1 M cacodylate. Cells for thin serial-section EM were fixed with 2.5% glutaraldehyde and 2% paraformaldehyde in 0.1 M cacodylate pH 7.2 overnight. The samples were processed according the standard techniques, cutting 75 nm thick sections.
Note added in proof
During the preparation of this manuscript a paper by Tanos et al. was published demonstrating that Cep89 (Cep123/CCDC123) is required for primary cilium formation (Tanos et al., 2013).
Supplementary Material
Acknowledgments
We thank Annie Delouvée for help with several experiments, Nathalie Spassky and Jean-Marc Corsi (Ecole Normale et Superieure, Paris) for kindly providing us with samples of mouse ependymal cells, Juliette Azimzadeh for advice on identifying Cep123 homologs. We would also like to thank the following people for providing reagents: Andrew Fry (University of Leicester), Maxence Nachury (University of California San Francisco), Jeffrey Salisbury (Mayo Clinic), Bernard Edde (CRBM, Montpellier), L. Lafanechére (Institut des Neurosciences de Grenoble, Grenoble), Erich Nigg (Biozentrum), Jeremy Reiter (University of California San Francisco), Akiharu Kubo (Kyoto University), Andreas Merdes (Université de Toulouse) and Olivier Rosnet (Université de la Méditerranée, Marseille). We would also like to thank Vincent Fraisier and Lucie Sengmanivong from the Nikon Imaging Centre at the Institut Curie for their help.
Footnotes
Author Contributions: The experiments were conceived by J.E.S. and M.B. and performed by J.E.S. and I.H. The manuscript was written by J.E.S. and M.B. with assistance from T.G.-P., B.G. and P.T.
Competing interests: The authors have no competing interests to declare.
References
- Azimzadeh J., Hergert P., Delouvée A., Euteneuer U., Formstecher E., Khodjakov A., Bornens M. (2009). hPOC5 is a centrin-binding protein required for assembly of full-length centrioles. J. Cell Biol. 185, 101–114 10.1083/jcb.200808082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balczon R., Bao L., Zimmer W. E. (1994). PCM-1, A 228-kD centrosome autoantigen with a distinct cell cycle distribution. J. Cell Biol. 124, 783–793 10.1083/jcb.124.5.783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berns M. W., Rattner J. B., Brenner S., Meredith S. (1977). The role of the centriolar region in animal cell mitosis. A laser microbeam study. J. Cell Biol. 72, 351–367 10.1083/jcb.72.2.351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornens M. (2012). The centrosome in cells and organisms. Science 335, 422–426 10.1126/science.1209037 [DOI] [PubMed] [Google Scholar]
- Conroy P. C., Saladino C., Dantas T. J., Lalor P., Dockery P., Morrison C. G. (2012). C-NAP1 and rootletin restrain DNA damage-induced centriole splitting and facilitate ciliogenesis. Cell cycle 11, 3769–3778 10.4161/cc.21986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craige B., Tsao C. C., Diener D. R., Hou Y., Lechtreck K. F., Rosenbaum J. L., Witman G. B. (2010). CEP290 tethers flagellar transition zone microtubules to the membrane and regulates flagellar protein content. J. Cell Biol. 190, 927–940 10.1083/jcb.201006105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammermann A., Merdes A. (2002). Assembly of centrosomal proteins and microtubule organization depends on PCM-1. J. Cell Biol. 159, 255–266 10.1083/jcb.200204023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de–Thé G. (1964). Cytoplasmic microtubules in different animal cells. J. Cell Biol. 23, 265–275 10.1083/jcb.23.2.265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. (1989). PHYLIP – Phylogeny Inference Package (version 3.2). Cladistics 5, 164–166. [Google Scholar]
- Feng S., Knödler A., Ren J., Zhang J., Zhang X., Hong Y., Huang S., Peränen J., Guo W. (2012). A Rab8 guanine nucleotide exchange factor-effector interaction network regulates primary ciliogenesis. J. Biol. Chem. 287, 15602–15609 10.1074/jbc.M111.333245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrante M. I., Zullo A., Barra A., Bimonte S., Messaddeq N., Studer M., Dollé P., Franco B. (2006). Oral–facial–digital type I protein is required for primary cilia formation and left–right axis specification. Nat. Genet. 38, 112–117 10.1038/ng1684 [DOI] [PubMed] [Google Scholar]
- Fry A. M., Mayor T., Meraldi P., Stierhof Y. D., Tanaka K., Nigg E. A. (1998). C-Nap1, a novel centrosomal coiled-coil protein and candidate substrate of the cell cycle-regulated protein kinase Nek2. J. Cell Biol. 141, 1563–1574 10.1083/jcb.141.7.1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia–Gonzalo F. R., Corbit K. C., Sirerol–Piquer M. S., Ramaswami G., Otto E. A., Noriega T. R., Seol A. D., Robinson J. F., Bennett C. L., Josifova D. J.et al. (2011). A transition zone complex regulates mammalian ciliogenesis and ciliary membrane composition. Nat. Genet. 43, 776–784 10.1038/ng.891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graser S., Stierhof Y. D., Lavoie S. B., Gassner O. S., Lamla S., Le Clech M., Nigg E. A. (2007). Cep164, a novel centriole appendage protein required for primary cilium formation. J. Cell Biol. 179, 321–330 10.1083/jcb.200707181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton R. M. (1993). In vitro recombination and mutagenesis of DNA: SOEing together tailor-made genes. PCR Protocols: Current Methods And Applications (Methods in Molecular Biology, Vol. 15). (ed. White B A.), pp. 251–261 Totowa, NJ: Humana Press. [DOI] [PubMed] [Google Scholar]
- Ishikawa H., Kubo A., Tsukita S., Tsukita S. (2005). Odf2-deficient mother centrioles lack distal/subdistal appendages and the ability to generate primary cilia. Nat. Cell Biol. 7, 517–524 10.1038/ncb1251 [DOI] [PubMed] [Google Scholar]
- Jackson A. L., Burchard J., Leake D., Reynolds A., Schelter J., Guo J., Johnson J. M., Lim L., Karpilow J., Nichols K.et al. (2006a). Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA 12, 1197–1205 10.1261/rna.30706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson A. L., Burchard J., Schelter J., Chau B. N., Cleary M., Lim L., Linsley P. S. (2006b). Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA 12, 1179–1187 10.1261/rna.25706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsen L., Vanselow K., Skogs M., Toyoda Y., Lundberg E., Poser I., Falkenby L. G., Bennetzen M., Westendorf J., Nigg E. A.et al. (2011). Novel asymmetrically localizing components of human centrosomes identified by complementary proteomics methods. EMBO J. 30, 1520–1535 10.1038/emboj.2011.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H., White S. R., Shida T., Schulz S., Aguiar M., Gygi S. P., Bazan J. F., Nachury M. V. (2010). The conserved Bardet–Biedl syndrome proteins assemble a coat that traffics membrane proteins to cilia. Cell 141, 1208–1219 10.1016/j.cell.2010.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov A., Rieder C. L. (2001). Centrosomes enhance the fidelity of cytokinesis in vertebrates and are required for cell cycle progression. J. Cell Biol. 153, 237–242 10.1083/jcb.153.1.237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K., Rhee K. (2011). The pericentriolar satellite protein CEP90 is crucial for integrity of the mitotic spindle pole. J. Cell Sci. 124, 338–347 10.1242/jcs.078329 [DOI] [PubMed] [Google Scholar]
- Kim J. Y., Choi J. A., Kim T. H., Yoo Y. D., Kim J. I., Lee Y. J., Yoo S. Y., Cho C. K., Lee Y. S., Lee S. J. (2002). Involvement of p38 mitogen-activated protein kinase in the cell growth inhibition by sodium arsenite. J. Cell. Physiol. 190, 29–37 10.1002/jcp.10049 [DOI] [PubMed] [Google Scholar]
- Kim J. C., Badano J. L., Sibold S., Esmail M. A., Hill J., Hoskins B. E., Leitch C. C., Venner K., Ansley S. J., Ross A. J.et al. (2004). The Bardet–Biedl protein BBS4 targets cargo to the pericentriolar region and is required for microtubule anchoring and cell cycle progression. Nat. Genet. 36, 462–470 10.1038/ng1352 [DOI] [PubMed] [Google Scholar]
- Kim J., Krishnaswami S. R., Gleeson J. G. (2008). CEP290 interacts with the centriolar satellite component PCM-1 and is required for Rab8 localization to the primary cilium. Hum. Mol. Genet. 17, 3796–3805 10.1093/hmg/ddn277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Lee J. E., Heynen–Genel S., Suyama E., Ono K., Lee K., Ideker T., Aza–Blanc P., Gleeson J. G. (2010). Functional genomic screen for modulators of ciliogenesis and cilium length. Nature 464, 1048–1051 10.1038/nature08895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K., Lee K., Rhee K. (2012). CEP90 is required for the assembly and centrosomal accumulation of centriolar satellites, which is essential for primary cilia formation. PLoS ONE 7, e48196 10.1371/journal.pone.0048196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knödler A., Feng S., Zhang J., Zhang X., Das A., Peränen J., Guo W. (2010). Coordination of Rab8 and Rab11 in primary ciliogenesis. Proc. Natl. Acad. Sci. USA 107, 6346–6351 10.1073/pnas.1002401107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo A., Sasaki H., Yuba–Kubo A., Tsukita S., Shiina N. (1999). Centriolar satellites: molecular characterization, ATP-dependent movement toward centrioles and possible involvement in ciliogenesis. J. Cell Biol. 147, 969–980 10.1083/jcb.147.5.969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes C. A., Prosser S. L., Romio L., Hirst R. A., O'Callaghan C., Woolf A. S., Fry A. M. (2011). Centriolar satellites are assembly points for proteins implicated in human ciliopathies, including oral–facial–digital syndrome 1. J. Cell Sci. 124, 600–612 10.1242/jcs.077156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukinavičius G., Lavogina D., Orpinell M., Umezawa K., Reymond L., Garin N., Gönczy P., Johnsson K. (2013). Selective chemical crosslinking reveals a cep57–cep63–cep152 centrosomal complex. Curr. Biol. 23, 265–270 10.1016/j.cub.2012.12.030 [DOI] [PubMed] [Google Scholar]
- Mikule K., Delaval B., Kaldis P., Jurcyzk A., Hergert P., Doxsey S. (2007). Loss of centrosome integrity induces p38–p53–p21-dependent G1–S arrest. Nat. Cell Biol. 9, 160–170 10.1038/ncb1529 [DOI] [PubMed] [Google Scholar]
- Molla–Herman A., Ghossoub R., Blisnick T., Meunier A., Serres C., Silbermann F., Emmerson C., Romeo K., Bourdoncle P., Schmitt A.et al. (2010). The ciliary pocket: an endocytic membrane domain at the base of primary and motile cilia. J. Cell Sci. 123, 1785–1795 10.1242/jcs.059519 [DOI] [PubMed] [Google Scholar]
- Moudjou M., Bornens M. (1994). Isolation of centrosomes from cultured animal cells. Cell Biology: A Laboratory Handbook (ed. Celis J E.), pp. 595–604 San Diego, CA: Academic Press [Google Scholar]
- Nachury M. V., Loktev A. V., Zhang Q., Westlake C. J., Peränen J., Merdes A., Slusarski D. C., Scheller R. H., Bazan J. F., Sheffield V. C.et al. (2007). A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell 129, 1201–1213 10.1016/j.cell.2007.03.053 [DOI] [PubMed] [Google Scholar]
- O'Connell K. F., Caron C., Kopish K. R., Hurd D. D., Kemphues K. J., Li Y., White J. G. (2001). The C. elegans zyg-1 gene encodes a regulator of centrosome duplication with distinct maternal and paternal roles in the embryo. Cell 105, 547–558 10.1016/S0092-8674(01)00338-5 [DOI] [PubMed] [Google Scholar]
- O'Toole E. T., McDonald K. L., Mäntler J., McIntosh J. R., Hyman A. A., Müller–Reichert T. (2003). Morphologically distinct microtubule ends in the mitotic centrosome of Caenorhabditis elegans. J. Cell Biol. 163, 451–456 10.1083/jcb.200304035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshimori N., Li X., Ohsugi M., Yamamoto T. (2009). Cep72 regulates the localization of key centrosomal proteins and proper bipolar spindle formation. EMBO J. 28, 2066–2076 10.1038/emboj.2009.161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paintrand M., Moudjou M., Delacroix H., Bornens M. (1992). Centrosome organization and centriole architecture: their sensitivity to divalent cations. J. Struct. Biol. 108, 107–128 10.1016/1047-8477(92)90011-X [DOI] [PubMed] [Google Scholar]
- Pazour G. J., Rosenbaum J. L. (2002). Intraflagellar transport and cilia-dependent diseases. Trends Cell Biol. 12, 551–555 10.1016/S0962-8924(02)02410-8 [DOI] [PubMed] [Google Scholar]
- Pazour G. J., Witman G. B. (2003). The vertebrate primary cilium is a sensory organelle. Curr. Opin. Cell Biol. 15, 105–110 10.1016/S0955-0674(02)00012-1 [DOI] [PubMed] [Google Scholar]
- Piel M., Nordberg J., Euteneuer U., Bornens M. (2001). Centrosome-dependent exit of cytokinesis in animal cells. Science 291, 1550–1553 10.1126/science.1057330 [DOI] [PubMed] [Google Scholar]
- Rapsomaniki M. A., Kotsantis P., Symeonidou I. E., Giakoumakis N. N., Taraviras S., Lygerou Z. (2012). easyFRAP: an interactive, easy-to-use tool for qualitative and quantitative analysis of FRAP data. Bioinformatics 28, 1800–1801 10.1093/bioinformatics/bts241 [DOI] [PubMed] [Google Scholar]
- Ríos R. M., Sanchís A., Tassin A. M., Fedriani C., Bornens M. (2004). GMAP-210 recruits gamma-tubulin complexes to cis-Golgi membranes and is required for Golgi ribbon formation. Cell 118, 323–335 10.1016/j.cell.2004.07.012 [DOI] [PubMed] [Google Scholar]
- Robbins E., Jentzsch G., Micali A. (1968). The centriole cycle in synchronized HeLa cells. J. Cell Biol. 36, 329–339 10.1083/jcb.36.2.329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salisbury J. L., Baron A. T., Sanders M. A. (1988). The centrin-based cytoskeleton of Chlamydomonas reinhardtii: distribution in interphase and mitotic cells. J. Cell Biol. 107, 635–641 10.1083/jcb.107.2.635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang L., Miller J. J., Corbit K. C., Giles R. H., Brauer M. J., Otto E. A., Baye L. M., Wen X., Scales S. J., Kwong M.et al. (2011). Mapping the NPHP–JBTS–MKS protein network reveals ciliopathy disease genes and pathways. Cell 145, 513–528 10.1016/j.cell.2011.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt K. N., Kuhns S., Neuner A., Hub B., Zentgraf H., Pereira G. (2012). Cep164 mediates vesicular docking to the mother centriole during early steps of ciliogenesis. J. Cell Biol. 199, 1083–1101 10.1083/jcb.201202126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedjaï F., Acquaviva C., Chevrier V., Chauvin J. P., Coppin E., Aouane A., Coulier F., Tolun A., Pierres M., Birnbaum D.et al. (2010). Control of ciliogenesis by FOR20, a novel centrosome and pericentriolar satellite protein. J. Cell Sci. 123, 2391–2401 10.1242/jcs.065045 [DOI] [PubMed] [Google Scholar]
- Sillibourne J. E., Tack F., Vloemans N., Boeckx A., Thambirajah S., Bonnet P., Ramaekers F. C., Bornens M., Grand–Perret T. (2010). Autophosphorylation of polo-like kinase 4 and its role in centriole duplication. Mol. Biol. Cell 21, 547–561 10.1091/mbc.E09-06-0505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillibourne J. E., Specht C. G., Izeddin I., Hurbain I., Tran P., Triller A., Darzacq X., Dahan M., Bornens M. (2011). Assessing the localization of centrosomal proteins by PALM/STORM nanoscopy. Cytoskeleton (Hoboken) 68, 619–627 10.1002/cm.20536 [DOI] [PubMed] [Google Scholar]
- Singla V., Romaguera–Ros M., Garcia–Verdugo J. M., Reiter J. F. (2010). Ofd1, a human disease gene, regulates the length and distal structure of centrioles. Dev. Cell 18, 410–424 10.1016/j.devcel.2009.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin S. (1962). Centrioles and the formation of rudimentary cilia by fibroblasts and smooth muscle cells. J. Cell Biol. 15, 363–377 10.1083/jcb.15.2.363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin S. P. (1968). Reconstructions of centriole formation and ciliogenesis in mammalian lungs. J. Cell Sci. 3, 207–230. [DOI] [PubMed] [Google Scholar]
- Srivastava M., Begovic E., Chapman J., Putnam N. H., Hellsten U., Kawashima T., Kuo A., Mitros T., Salamov A., Carpenter M. L.et al. (2008). The Trichoplax genome and the nature of placozoans. Nature 454, 955–960 10.1038/nature07191 [DOI] [PubMed] [Google Scholar]
- Srsen V., Gnadt N., Dammermann A., Merdes A. (2006). Inhibition of centrosome protein assembly leads to p53-dependent exit from the cell cycle. J. Cell Biol. 174, 625–630 10.1083/jcb.200606051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanos B. E., Yang H. J., Soni R., Wang W. J., Macaluso F. P., Asara J. M., Tsou M. F. (2013). Centriole distal appendages promote membrane docking, leading to cilia initiation. Genes Dev. 27, 163–168 10.1101/gad.207043.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882 10.1093/nar/25.24.4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetake Y., Loncarek J., Nordberg J. J., English C. N., La Terra S., Khodjakov A., Sluder G. (2007). Cell cycle progression and de novo centriole assembly after centrosomal removal in untransformed human cells. J. Cell Biol. 176, 173–182 10.1083/jcb.200607073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorobjev I. A., Chentsov Y. u. S. (1982). Centrioles in the cell cycle. I. Epithelial cells. J. Cell Biol. 93, 938–949 10.1083/jcb.93.3.938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westlake C. J., Baye L. M., Nachury M. V., Wright K. J., Ervin K. E., Phu L., Chalouni C., Beck J. S., Kirkpatrick D. S., Slusarski D. C.et al. (2011). Primary cilia membrane assembly is initiated by Rab11 and transport protein particle II (TRAPPII) complex-dependent trafficking of Rabin8 to the centrosome. Proc. Natl. Acad. Sci. USA 108, 2759–2764 10.1073/pnas.1018823108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff A., de Néchaud B., Chillet D., Mazarguil H., Desbruyères E., Audebert S., Eddé B., Gros F., Denoulet P. (1992). Distribution of glutamylated alpha and beta-tubulin in mouse tissues using a specific monoclonal antibody, GT335. Eur. J. Cell Biol. 59, 425–432. [PubMed] [Google Scholar]
- Yoshimura S., Egerer J., Fuchs E., Haas A. K., Barr F. A. (2007). Functional dissection of Rab GTPases involved in primary cilium formation. J. Cell Biol. 178, 363–369 10.1083/jcb.200703047 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.