Summary
The basic helix–loop–helix factor Myod initiates skeletal muscle differentiation by directly and sequentially activating sets of muscle differentiation genes, including those encoding muscle contractile proteins. We hypothesize that Pbx homeodomain proteins direct Myod to a subset of its transcriptional targets, in particular fast-twitch muscle differentiation genes, thereby regulating the competence of muscle precursor cells to differentiate. We have previously shown that Pbx proteins bind with Myod on the promoter of the zebrafish fast muscle gene mylpfa and that Pbx proteins are required for Myod to activate mylpfa expression and the fast-twitch muscle-specific differentiation program in zebrafish embryos. Here we have investigated the interactions of Pbx with another muscle fiber-type regulator, Prdm1a, a SET-domain DNA-binding factor that directly represses mylpfa expression and fast muscle differentiation. The prdm1a mutant phenotype, early and increased fast muscle differentiation, is the opposite of the Pbx-null phenotype, delayed and reduced fast muscle differentiation. To determine whether Pbx and Prdm1a have opposing activities on a common set of genes, we used RNA-seq analysis to globally assess gene expression in zebrafish embryos with single- and double-losses-of-function for Pbx and Prdm1a. We find that the levels of expression of certain fast muscle genes are increased or approximately wild type in pbx2/4-MO;prdm1a−/− embryos, suggesting that Pbx activity normally counters the repressive action of Prdm1a for a subset of the fast muscle program. However, other fast muscle genes require Pbx but are not regulated by Prdm1a. Thus, our findings reveal that subsets of the fast muscle program are differentially regulated by Pbx and Prdm1a. Our findings provide an example of how Pbx homeodomain proteins act in a balance with other transcription factors to regulate subsets of a cellular differentiation program.
Key words: Skeletal muscle, Fiber-type differentiation, Pbx, Prdm1, Zebrafish
Introduction
The differentiation of muscle precursor cells into contractile skeletal muscle fibers is necessary for normal muscle development and regeneration. Vertebrate skeletal muscles consist of a range of fiber types, from slow-twitch fibers needed for low-intensity, sustained aerobic activity, to fast-twitch fibers suited for short bursts of intense, anaerobic activity (Wigmore and Evans, 2002). Muscle fiber types are operationally defined by the expression of myosin heavy chain (MyHC) genes (Schiaffino and Reggiani, 1996; Pette and Staron, 2000). In mammals, 11 MyHC genes contribute to skeletal muscle fiber heterogeneity and functional flexibility (Schiaffino and Reggiani, 2011). However, muscle fiber types are distinguished not just by MyHC expression but by the expression of other sarcomeric proteins, most of which also exist as multiple isoforms, as well as by isoforms of intracellular calcium signaling components and levels of metabolic enzymes (Schiaffino and Reggiani, 1996; Pette and Staron, 2000; Schiaffino and Reggiani, 2011). Thus, it is the coordinated expression of fiber-type-specific gene expression programs that gives each fiber type its unique contractile and metabolic properties.
Ultimately, the expression of skeletal muscle differentiation genes, including those encoding the MyHCs and other sarcomeric proteins, is controlled by the family of myogenic basic helix–loop–helix (bHLH) transcription factors: Myf5, Myod, Myf6 and Myog (Buckingham, 2001; Tapscott, 2005). In particular, Myod can directly bind to the promoters and activate the expression of both slow and fast MyHCs (Cao et al., 2010). How do factors like Myod activate their transcriptional targets in a temporally and spatially controlled manner to drive cell-type-specific, or fiber-type-specific, differentiation? Our work has shown that Pbx TALE-class homeodomain proteins function as cofactors for Myod and regulate a subset of Myod target genes, in particular to promote fast-muscle-specific differentiation (Berkes et al., 2004; Maves et al., 2007). We have shown that Pbx proteins bind with Myod on the promoters of a subset of Myod target genes, including Myog and the fast muscle gene mylpfa, and that Pbx proteins are required for Myod to activate myog and mylpfa expression and the fast-muscle-specific differentiation program in zebrafish embryos (Berkes et al., 2004; Maves et al., 2007). Our specific hypothesis is that Pbx homeodomain proteins function as pioneer transcription factors to direct Myod to a subset of its transcriptional targets, in particular fast muscle differentiation genes, thereby regulating the competence of muscle precursor cells to differentiate into a specific fiber type.
Pioneer factors are transcription factors that have the ability to access and bind DNA in repressive chromatin and that bind prior to gene activation and prior to the presence of other transcription factors (Zaret and Carroll, 2011). Pbx proteins have recently been shown to function as pioneer factors in breast cancer cells, where PBX1 binds prior to, and is required for, Estrogen Receptor binding (Magnani et al., 2011). Supporting the role of Pbx proteins as pioneer factors in skeletal muscle differentiation, Pbx proteins can bind silent Myod target gene promoters prior to Myod binding and muscle differentiation (Berkes et al., 2004). Pioneer factors can function in passive roles, in which their presence reduces the number of additional transcription factor binding events needed, or in more active roles, in which they facilitate chromatin opening that is necessary before other factors can bind (Zaret and Carroll, 2011). Whether Pbx proteins act in passive or active roles as pioneer factors is not known.
To address how Pbx proteins function in muscle fiber-type differentiation, here we use zebrafish to examine how Pbx interacts with another fiber-type regulator, Prdm1a. In zebrafish, slow-twitch fibers derive from the adaxial cells, the most medial paraxial mesoderm cells, whereas more lateral somitic cells will give rise to fast-twitch fibers (Devoto et al., 1996). Adaxial cells express the transcriptional repressor Prdm1a/Blimp1, which is both necessary and sufficient for slow MyHC expression and specification of slow twitch fiber identity (Baxendale et al., 2004). In zebrafish, many fast muscle genes are initially transiently co-expressed with slow muscle genes in the adaxial cells before differentiation is completed (Bryson Richardson et al., 2005; Burguière et al., 2011). In prdm1a mutant zebrafish embryos, the expression of smyhc1 and other slow muscle genes is reduced in the adaxial cells, while fast muscle genes show increased and premature expression in the adaxial cells (Roy et al., 2001; Baxendale et al., 2004; von Hofsten et al., 2008; Liew et al., 2008). Pbx and Prdm1a thus have opposite requirements for fast muscle differentiation in zebrafish. Here we combine loss of function of both Pbx proteins and Prdm1a to test the requirements for Pbx in promoting fast muscle differentiation. We use RNA-seq to identify subsets of the fast-muscle program that are either co-regulated by Pbx and Prdm1 or independently regulated by these factors. We find that in embryos lacking both Pbx and Prdm1a, the levels of expression of many fast muscle genes are approximately wild type or are increased relative to wild type, suggesting that, for a subset of the fast muscle program, a major role for Pbx is to counter the repressive action of Prdm1a. However, certain fast muscle genes require Pbx but are not regulated by Prdm1a. Thus, our findings reveal that subsets of the fast muscle program are differentially regulated by Pbx and Prdm1a. Our findings provide an example of how Pbx pioneer factors act in a balance with other transcriptional regulators to direct a specific cellular differentiation pathway.
Results
prdm1a is epistatic to pbx2/4 for fast muscle gene expression in adaxial cells
One advantage of using zebrafish to study Pbx functions in early muscle differentiation is that we have the ability to remove all Pbx function by using morpholinos to knock down maternal and zygotic expression of both pbx2 and pbx4 (pbx2/4), the two pbx genes expressed in young zebrafish embryos (Waskiewicz et al., 2002; Maves et al., 2007). pbx2/4 and prdm1a have opposite requirements for fast muscle gene expression in zebrafish: in the absence of Pbx2/4, fast muscle gene expression in both adaxial cells and lateral cells is reduced and delayed, whereas in the absence of Prdm1a, fast muscle gene expression occurs prematurely and at increased levels in adaxial cells (Maves et al., 2007; Roy et al., 2001; Baxendale et al., 2004; von Hofsten et al., 2008; Liew et al., 2008). We wanted to combine loss of function of both pbx2/4 and prdm1a to test the requirements for Pbx in promoting fast muscle gene expression. We reasoned that if Pbx proteins, as pioneer factors, play a necessary role in assembling factors to drive fast-muscle gene expression, then expression of fast muscle genes should be reduced and delayed in pbx2/4-MO;prdm1a−/− embryos, as in pbx2/4-MO embryos. Alternatively, if, in the adaxial cells, Pbx proteins are primarily needed to overcome the repressive actions of Prdm1a, then expression of fast muscle genes should appear at increased levels in pbx2/4-MO;prdm1a−/− embryos, as in prdm1a−/− embryos. We first confirmed that pbx2/4 and prdm1a are not required for each other's expression (Fig. 1). We then examined the expression of three target genes in pbx2/4-MO;prdm1a−/− double loss-of-function embryos: mylpfa, myhz1.3, and myog (Fig. 2). mylpfa and myhz1.3 encode fast muscle sarcomere proteins whose expression is positively, and, in the case of mylpfa, directly, regulated by Pbx2/4 and directly repressed by Prdm1a (Fig. 2A–D) (Maves et al., 2007; von Hofsten et al., 2008; Liew et al., 2008). myog, which encodes a muscle regulatory transcription factor, is directly and positively regulated by Pbx and shows upregulation in the adaxial cells of prdm1a−/− embryos, indicating that myog is normally repressed by Prdm1a (Fig. 2E,F) (Maves et al., 2007; Maves et al., 2009). In pbx2/4-MO;prdm1a−/− embryos, we find upregulation of mylpfa, myhz1.3, and myog in the adaxial cells relative to the controls (Fig. 2A,C,E), resembling the phenotype of prdm1a−/− embryos. Using real-time PCR (qRT-PCR) to quantitatively examine these markers, we find that mylpfa and myhz1.3 expression are strongly upregulated in pbx2/4-MO;prdm1a−/− embryos relative to both control and pbx2/4-MO embryos, although not quite to the levels seen in prdm1a−/− embryos (Fig. 2B,D). For myog, we find that the level of expression in pbx2/4-MO;prdm1a−/− embryos is close to the control level (Fig. 2F), and, based on the in situ pattern, seems to represent an upregulation of myog in the adaxial cells of pbx2/4-MO;prdm1a−/− embryos but absence of the more lateral Pbx-dependent expression (Fig. 2E). These results show that expression of mylpfa, myhz1.3, and myog within the prdm1a-expressing adaxial cells is upregulated in pbx2/4-MO;prdm1a−/− embryos, resembling the prdm1a−/− phenotype.
Fig. 1. pbx2/4 and prdm1a are not required for each other's expression.
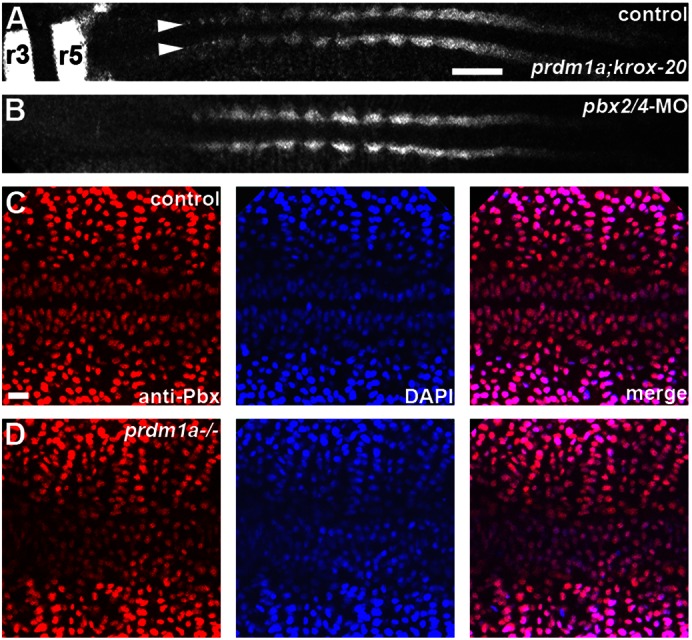
(A,B) RNA in situ expression of prdm1a and krox-20 in (A) control and (B) pbx2/4-MO embryos. prdm1a expression in the adaxial cells along the axis of the embryo is maintained, while krox-20 expression is lost, in pbx2/4-MO embryos. Arrowheads point to adaxial cells, and r3 and r5 indicate krox-20 expression in hindbrain rhombomeres. (C,D) Anti-Pbx (red) and DAPI (blue) staining, along with merged images, in (C) control and (D) prdm1a−/− embryos. Pbx expression is maintained in prdm1a−/− embryos. All embryos are shown in dorsal view, anterior towards the left. Scale bars: (A) 50 µm, (C) 10 µm.
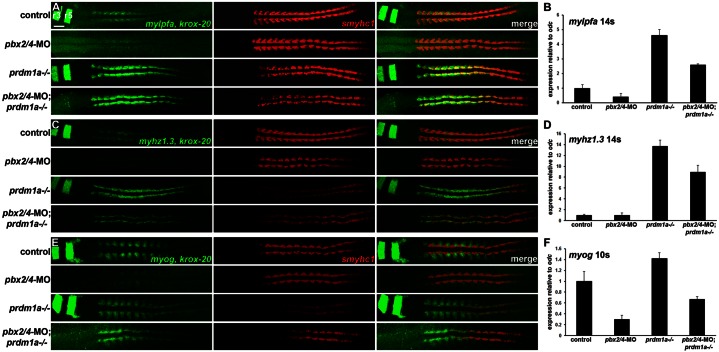
Fig. 2. Expression of Pbx and Prdm1a target genes in pbx2/4-MO;prdm1a−/− embryos.
(A,C,E) RNA in situ expression of (A) mylpfa and krox-20, (C) myhz1.3 and krox-20, and (E) myog and krox-20 in green, smyhc1 in red, and merged images in control, pbx2/4-MO, prdm1a−/−, and pbx2/4-MO;prdm1a−/− embryos. krox-20 expression is included as a control for the pbx2/4-MO knockdown (Maves et al., 2007). r3 and r5 indicate krox-20 expression in hindbrain rhombomeres. smyhc1 is downregulated in prdm1a−/− embryos (Elworthy et al., 2008; von Hofsten et al., 2008). (A,C) embryos are at 14 somites, (E) embryos are at 10 somites. All embryos are shown in dorsal view, anterior towards the left. Somites were counted in all embryos shown to confirm consistent staging. (B,D,F) Graphs of real-time RT-PCR (qRT-PCR) expression levels for (B) mylpfa, (D) myhz1.3, and (F) myog. Expression levels were normalized to odc1 expression. Error bars represent standard deviation. Somite stages (14 s, 10 s) are shown. Scale bar: 50 µm.
To more thoroughly examine the Pbx- and Prdm1a-regulated expression of mylpfa, we examined higher magnification confocal images. In control embryos at 14s, mylpfa is expressed in both lateral cells and in adaxial cells, where its expression coincides with the slow muscle gene smyhc1 (Fig. 3A) (Burguière et al., 2011). In pbx2/4-MO embryos, both adaxial and lateral expression domains are lost (Fig. 3B), showing that Pbx is needed to promote mylpfa expression in both adaxial and lateral cells. In prdm1a−/− embryos, adaxial and lateral expression of mylpfa are present (Fig. 3C). Critically, in pbx2/4-MO;prdm1a−/− embryos, adaxial expression of mylpfa is maintained, as in prdm1−/− embryos, whereas the lateral domain is reduced or absent as in pbx2/4-MO embryos (Fig. 3D). Taken together, the results in Figs 2 and 3 show that prdm1a is epistatic to pbx2/4 for fast muscle gene expression in adaxial cells, revealing that pbx2/4 functions as a genetic suppressor of prdm1a.
Fig. 3. Adaxial cell expression of mylpfa is absent in pbx2/4-MO embryos but present in pbx2/4-MO;prdm1a−/− embryos.
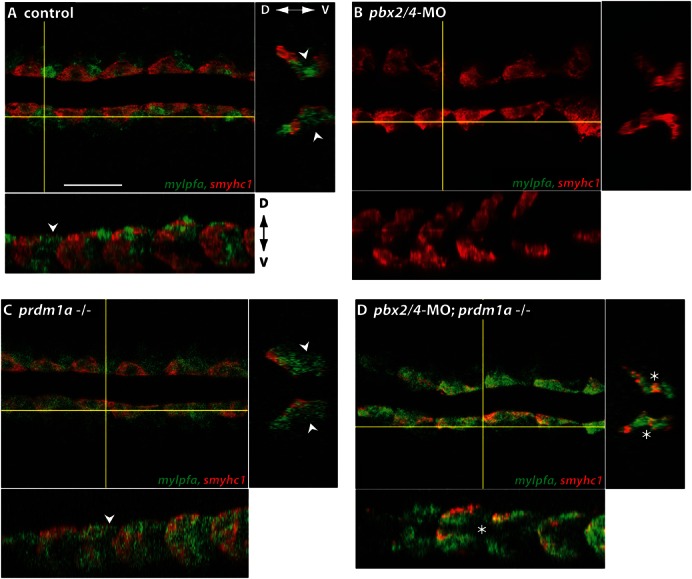
RNA in situ expression of mylpfa (green) and smyhc1 (red) in (A) control, (B) pbx2/4-MO, (C) prdm1a−/−, and (D) pbx2/4-MO;prdm1a−/− embryos. Embryos are 14 somites stage, shown in dorsal view, anterior towards the left. Somite numbers 2–7 are shown for each embryo. Yellow lines indicate where optical sections are taken through the x and y planes (lower and right panels, respectively, each image). Dorsal–ventral (D–V) axes (z axis in xy view) are indicated. Arrowheads point to lateral domain of mylpfa expression, absent or reduced in pbx2/4-MO and pbx2/4-MO;prdm1a−/− (asterisk) embryos. smyhc1-overlapping mylpfa expression in adaxial cells is present in pbx2/4-MO;prdm1a−/− embryos. Scale bar: 50 µm.
RNA-seq approach to identify Pbx2/4- and Prdm1a-regulated genes: comparison with previous expression array studies
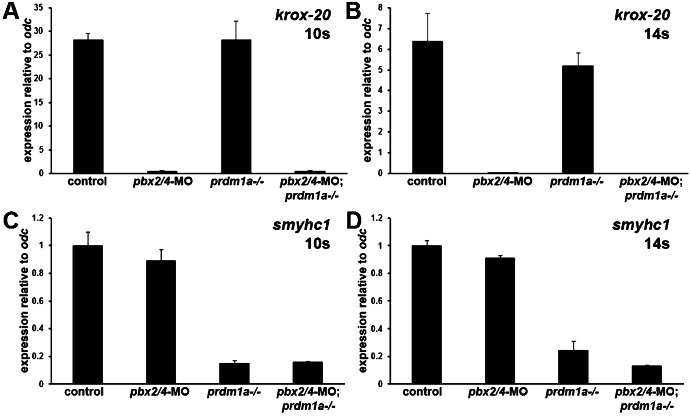
We next wanted to ask whether prdm1a functions epistatically to pbx2/4 broadly across the fast-muscle gene expression program or only at subsets of fast muscle genes. We confirmed that Pbx2/4 and Prdm1a could independently regulate known dependent genes by showing that krox-20 is strongly downregulated in pbx2/4-MO and pbx2/4-MO;prdm1a−/−, but not in prdm1a−/−, embryos, and that smyhc1 is strongly downregulated in prdm1a−/− and pbx2/4-MO;prdm1a−/−, but not in pbx2/4-MO, embryos (Fig. 2A, Fig. 4) (Waskiewicz et al., 2002; Elworthy et al., 2008; von Hofsten et al., 2008). To globally assess which subsets of the fast muscle program are regulated by both Pbx2/4 and Prdm1a versus those that are regulated by either Pbx or Prdm1a, we used high-throughput sequencing of mRNA libraries (RNA-seq). We generated libraries for control prdm1a+/+ embryos, pbx2/4-MO embryos, prdm1a−/− embryos and pbx2/4-MO;prdm1a−/− embryos in triplicate at the 10 somite (s) stage (see Materials and Methods). We chose this stage because it is when early skeletal muscle differentiation initiates in zebrafish and we wanted to assess the earliest muscle requirements for Pbx2/4 and Prdm1a. We also chose this stage in order to compare with previous expression array analyses that we performed on 10 s control and pbx2/4-MO embryos (Maves et al., 2007; Maves et al., 2009). Because we could not reliably sort live prdm1a mutant embryos from their wild-type siblings at 10 s, we developed a technique to genotype RNA samples from individual embryos (see Materials and Methods). We pooled equivalent-genotype RNA samples for our RNA-seq libraries (see Materials and Methods).
Fig. 4. Pbx2/4 and Prdm1a independently regulate genes.
Graphs of qRT-PCR expression levels for (A,B) krox-20 and (C,D) smyhc1. Expression levels were normalized to odc1 expression. Error bars represent standard deviation. Somite stages (10 s, 14 s) are shown.
As an initial assessment of the RNA-seq data, we examined results from pbx2/4-MO and prdm1a−/−, each relative to control. Using the cut-offs of >1.5-fold change and significance of P<0.05, we identified 637 genes that are downregulated in pbx2/4-MO embryos compared to controls (supplementary material Table S1). This group of 637 genes includes the Pbx targets mylpfa, myog, and krox-20 and includes 72/111 (65%) of the genes that we had previously identified with expression arrays as downregulated in pbx2/4-MO 10 s embryos, including 10/12 genes that we had validated by in situ hybridization (supplementary material Table S1) (Maves et al., 2007). Additionally, this group of 637 genes includes 6 genes that we previously identified as downregulated in pbx2/4-MO embryos with expression arrays at 18 s, but not with arrays at 10 s, including mylpfa (supplementary material Table S1) (Maves et al., 2007). Thus, although it is not clear why our RNA-seq analysis did not identify all of the genes previously identified with the expression arrays at 10 s, overall it appears that the RNA-seq analysis is more sensitive than our previous expression array analysis, because we identified genes at 10 s from RNA-seq that the expression arrays only picked up at 18 s. We also identified 142 genes that are upregulated in prdm1a−/− embryos compared to controls (supplementary material Table S2). This group of 142 genes includes the Prdm1a direct targets mylpfa and myhz2 and known prdm1a-regulated genes agr2, myl1, and myhz1.3 (supplementary material Table S2) (von Hofsten et al., 2008; Hernandez-Lagunas et al., 2005; Burguière et al., 2011). Altogether, our initial assessment shows that our RNA-seq analysis has identified Pbx2/4- and Prdm1-dependent genes.
RNA-seq analysis identifies subsets of the fast muscle program that are differentially regulated by Pbx2/4 and Prdm1a
Because Pbx2/4 and Prdm1a have both been shown to broadly regulate the fast muscle gene expression program (Maves et al., 2007; von Hofsten et al., 2008), we used the RNA-seq data to ask if Pbx2/4 and Prdm1a each regulate completely overlapping sets of fast muscle genes. We compared the genes downregulated in pbx2/4-MO embryos (supplementary material Table S1) with the genes upregulated in prdm1a−/− embryos (supplementary material Table S2). At this early muscle differentiation stage, we find 12 genes overlapping between these two groups (Fig. 5A,B). The overlapping genes include fast muscle genes atp2a1, myl1 and mylpfa (Fig. 5B). We then examined the gene lists from supplementary material Tables S1 and S2 and found additional fast muscle genes in either the downregulated-in-pbx2/4-MO set or the upregulated-in-prdm1a−/− set (Fig. 5B). These results show that pbx2/4 and prdm1a regulate overlapping as well as distinct sets of fast muscle differentiation genes.
Fig. 5. pbx2/4 and prdm1a regulate overlapping and distinct subsets of fast muscle differentiation genes, identified through RNA-seq.

(A) Venn diagram identifying the intersection (12 genes) of the set of genes downregulated in pbx2/4-MO embryos (pbx2/4-MO<control, supplementary material Table S1) with the set of genes upregulated in prdm1a−/− embryos (prdm1a−/−>control, supplementary material Table S2). (B) List of the 12 genes identified in A. Also shown are fast muscle genes regulated by Pbx2/4, independently of Prdm1a, and those regulated by Prdm1a, independently of Pbx2/4, as identified from supplementary material Tables S1 and S2. The asterisk refers to fast myosin heavy chain genes that were not identified as Pbx2/4-dependent through RNA-seq, but that are known through additional validation to be Pbx2/4-dependent (see Fig. 2 and Discussion).
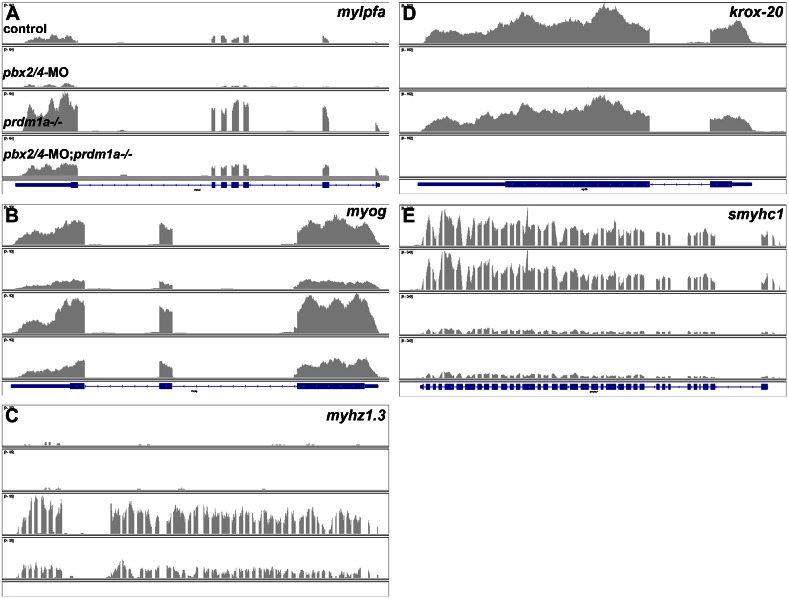
We next examined the RNA-seq results for the target genes analyzed in Fig. 2, using the Integrative Genome Viewer (Robinson et al., 2011) to visualize the data. As expected from our previous in situs and qRT-PCR, mylpfa and myog are downregulated in pbx2/4-MO embryos, upregulated in prdm1a−/− embryos, and remain upregulated in pbx2/4-MO;prdm1a−/− embryos (Fig. 6A,B). For myhz1.3, expression is not detected in control and pbx2/4-MO embryos but is upregulated in prdm1a−/− and pbx2/4-MO;prdm1a−/− embryos (Fig. 6C; see also Discussion). krox-20 and smyhc1 show expected expression patterns (Fig. 6D,E). These results further validate our RNA-seq analysis.
Fig. 6. Visualization and validation of RNA-seq analysis.
Mapping of RNA-seq reads as viewed in the Integrative Genomics Viewer for (A) mylpfa, (B) myog, (C) myhz1.3, (D) krox-20, and (E) smyhc1. Mapped reads are represented by grey histograms. Blue bars at the bottom of each panel represent annotated exons and UTRs (blue boxes) and introns (blue lines with arrowheads for orientation of transcript from 5′ to 3′) for each gene. For C, myhz1.3 is not annotated but the genomic position is confirmed relative to neighboring annotated zebrafish myosin heavy chain genes and mammalian orthologues. For each gene, rows for reads from control, pbx2/4-MO, prdm1a−/−, and pbx2/4-MO;prdm1a−/− are shown, as in A.
To globally determine whether prdm1a functions epistatically to pbx2/4 broadly across the fast-muscle gene expression program, or only at subsets of fast muscle genes, we compared the data sets for genes downregulated in pbx2/4-MO embryos with three different data sets from pbx2/4-MO;prdm1a−/− embryos: 1, genes upregulated relative to control; 2, genes expressed at about control levels; and 3, genes downregulated relative to control. (Fig. 7A–C). The overlap of Set 1 contains only four genes, but two of these are fast muscle myosin genes, myl1 and mylpfa (Fig. 7A; supplementary material Table S3). The overlap of Set 2 contains fast muscle differentiation genes, including atp2a1, a fast muscle SERCA Ca(2+)-ATPase (Wu et al., 1995), ryr1b (Hirata et al., 2007), and also myog (Fig. 7B; supplementary material Table S3). These analyses reveal that many fast muscle genes that are Pbx-dependent are indeed “rescued”, relative to pbx2/4-MO levels, or upregulated in pbx2/4-MO;prdm1a−/− embryos. However, the overlap of Set 3 also contains fast muscle genes, such as atp1a2a, myom1a and srl (Fig. 7C; supplementary material Table S3). Thus, certain fast muscle genes that are Pbx-dependent are still downregulated in pbx2/4-MO;prdm1a−/− embryos. Taken together, our RNA-seq analysis reveals subsets of the fast muscle differentiation program that are differentially regulated by Pbx2/4 and Prdm1a.
Fig. 7. RNA-seq analysis identifies subsets of the fast muscle program that are differentially regulated by Pbx2/4 and Prdm1a.
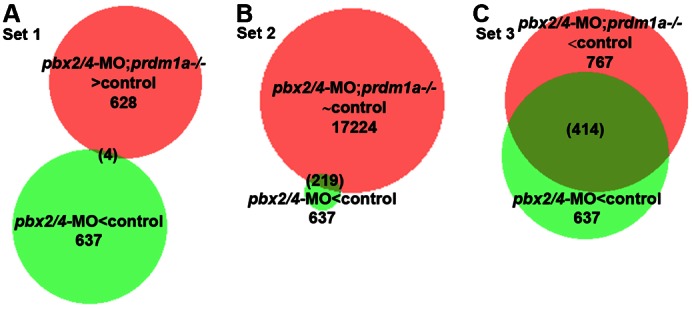
(A–C) Venn diagrams showing overlap of genes whose expression is downregulated in pbx2/4-MO embryos relative to control (green circles) compared to (A) genes that are upregulated in pbx2/4-MO;prdm1−/− embryos, (B) genes that are expressed at about control levels in pbx2/4-MO;prdm1−/− embryos, and (C) genes that are downregulated in pbx2/4-MO;prdm1−/− embryos. The number of genes in each overlap is shown in parentheses; genes are listed in supplementary material Table S3.
Validation of fast muscle genes differentially regulated by Pbx and Prdm1a
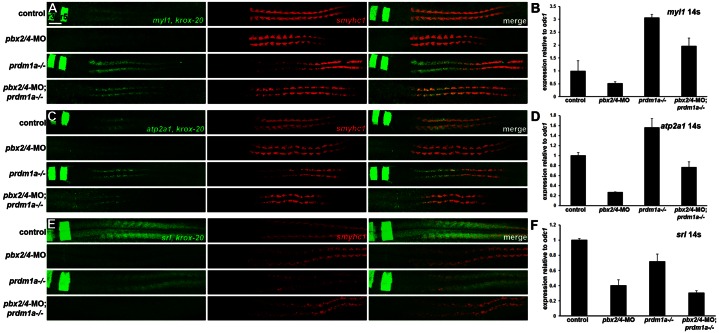
We next validated genes from each of these three sets of loci that are differentially regulated by Pbx and Prdm1a. For Set 1, we already demonstrated that mylpfa is upregulated in pbx2/4-MO;prdm1a−/− embryos (Fig. 2), and we confirmed by both in situ and qRT-PCR that myl1 is downregulated in pbx2/4-MO embryos, upregulated in prdm1a−/− embryos, and upregulated in pbx2/4-MO;prdm1a−/− embryos (Fig. 8A,B). For Set 2, we already demonstrated that myog is expressed at about control levels in pbx2/4-MO;prdm1a−/− embryos (Fig. 2), and we confirmed that atp2a1 is downregulated in pbx2/4-MO embryos, upregulated in prdm1a−/− embryos, and expressed at about control levels in pbx2/4-MO;prdm1a−/− embryos (Fig. 8C,D). Set 3 contains many neural genes, such as krox-20, that are known to be Pbx-dependent (Waskiewicz et al., 2002; Maves et al., 2007; Choe et al., 2009), and we already confirmed that krox-20 is strongly downregulated in pbx2/4-MO and pbx2/4-MO;prdm1a−/− embryos but not affected in prdm1a−/− embryos (Fig. 2, Fig. 4). We have previously shown that atp1a2a is downregulated in pbx2/4-MO embryos (Maves et al., 2007). We also validated that the fast muscle gene srl is downregulated in pbx2/4-MO embryos, not affected in prdm1a−/− embryos, and remains downregulated in pbx2/4-MO;prdm1a−/− embryos (Fig. 8D,E). This analysis demonstrates that certain fast muscle genes are Pbx-dependent but are not repressed by Prdm1a, confirming that subsets of the fast muscle program have different requirements for Pbx and Prdm1a.
Fig. 8. Validation of fast muscle genes differentially regulated by Pbx and Prdm1a.
(A,C,E) RNA in situ expression of (A) myl1 and krox-20, (C) atp2a1 and krox-20, and (E) srl and krox-20 in green, smyhc1 in red, and merged images in control, pbx2/4-MO, prdm1a−/−, and pbx2/4-MO;prdm1a−/− embryos. krox-20 expression is included as a control for the pbx2/4-MO knockdown (Maves et al., 2007). r3 and r5 indicate krox-20 expression in hindbrain rhombomeres. smyhc1 is downregulated in prdm1a−/− embryos (Elworthy et al., 2008; von Hofsten et al., 2008). (A,C) embryos are at 14 somites, (E) embryos are at 18 somites. All embryos are shown in dorsal view, anterior towards the left. Somites were counted in all embryos shown to confirm consistent staging. (B,D,F) Graphs of real-time RT-PCR (qRT-PCR) expression levels for (B) myl1, (D) atp2a1, and (F) srl. Expression levels were normalized to odc1 expression. Error bars represent standard deviation. Somite stage for qRT-PCR (14 s) is shown. Scale bar, 50 µm.
Discussion
The coordinated expression of fiber-type-specific gene expression programs is necessary to give each fiber type its unique contractile properties. Pbx and Prdm1a proteins have previously been shown to have opposite requirements for fast muscle gene expression in zebrafish (Maves et al., 2007; Baxendale et al., 2004; von Hofsten et al., 2008; Liew et al., 2008). Here we combined loss of function of both Pbx and Prdm1a to test the requirements for Pbx in promoting fast muscle differentiation. We used RNA-seq to identify subsets of the fast-muscle program that have different requirements for Pbx and Prdm1a. Our results reveal that there are multiple combinations of Pbx and Prdm1a inputs that regulate fast muscle gene expression (Fig. 9). In lateral cells, fast muscle differentiation genes are positively regulated by Pbx and Myod, likely acting together, as has been shown for mylpfa (Maves et al., 2007) (Fig. 9). In adaxial cells, myl1 is repressed by Prdm1a but not regulated by Pbx, srl is positively regulated by Pbx and Myod but not regulated by Prdm1a, and mylpfa expression appears to be regulated through a balance of Pbx/Myod positive input and Prdm1a repression (Fig. 9 and references therein). Our results thus show that Pbx and Prdm1a transcription factors differentially regulate subsets of the fast skeletal muscle differentiation program in zebrafish.
Fig. 9. Multiple combinations of Pbx and Prdm1a inputs regulate fast muscle gene expression.
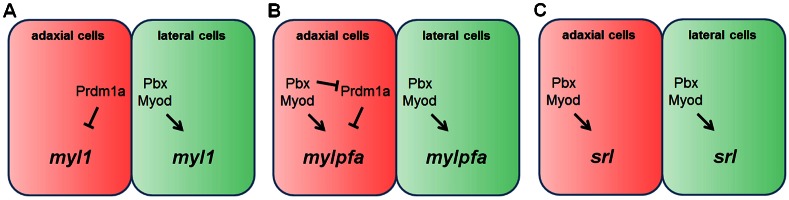
Cartoons illustrating inputs for the expression of three fast muscle genes (A) myl1, (B) mylpfa, and (C) srl. Evidence for inputs comes from A (Burguière et al., 2011; this work), B (Maves et al., 2007; von Hofsten et al., 2008; Liew et al., 2008; this work), and C (Maves et al., 2007; this work). Inputs reflect genetic requirements, although, in the case of mylpfa, direct regulation by Pbx, Myod and Prdm1a has been demonstrated (Maves et al., 2007; von Hofsten et al., 2008; Liew et al., 2008).
Our results support the hypothesis that pioneer factors may function through different mechanisms depending on the target gene (Zaret and Carroll, 2011; Budry et al., 2012). For example, pioneer factors can play passive roles by reducing the number of additional transcription factor binding events needed for gene activation, or more active roles by opening up local chromatin to allow other factors to bind (Zaret and Carroll, 2011). Our findings suggest that, for a subset of fast muscle genes such as mylpfa, Pbx proteins are needed to repress the repressive activity of Prdm1a (Fig. 9B), possibly by inhibiting Prdm1a binding or activity at fast muscle gene promoters. Pbx proteins have been shown to inhibit gene expression through interactions with transcriptional repressors (Saleh et al., 2000; Berghella et al., 2008; Choe et al., 2009), but activating gene expression by repressing a transcriptional repressor may entail a new mechanism for Pbx activity. Pbx proteins could take an active role in inhibiting Prdm1a, while promoting gene expression, by modifying chromatin states at fast muscle gene promoters through Pbx interactions with chromatin factors such as Brg1 or CBP (de la Serna et al., 2005; Choe et al., 2009). Other fast muscle genes, such as srl, are Prdm1a-independent but require Pbx for their activation. Further studies will be needed to understand the mechanisms by which Pbx proteins regulate different subsets of the fast muscle program, in particular how Pbx proteins repress the activity of Prdm1a.
We used an RNA-seq approach in order to globally assess which subsets of the fast muscle program are co-regulated by Pbx2/4 and Prdm1a versus those that are independently regulated. In general, we found that our RNA-seq approach was more sensitive in identifying differentially expressed genes than our previous expression array approach with pbx2/4-MO embryos (Maves et al., 2007). Our analysis at the 10-somite stage allowed us to assess the earliest muscle differentiation requirements for both Pbx2/4 and Prdm1a, but we were not able to assess the entire fast muscle program because many fast muscle genes, such as tnnt, are not activated until later developmental stages (Xu et al., 2000). Some myosin heavy chain genes are expressed early, but we found the fast myosin heavy chain genes to be difficult to analyze by RNA-seq because the sequences of the six genes are so similar that sequencing reads may not get assigned properly. For example, based on our in situ and qRT-PCR analyses, myhz1.3 should have been identified in Fig. 5 as Pbx-dependent and in Fig. 7 in Class 1 (strong expression levels in pbx2/4-MO;prdm1a−/− embryos), but myhz1.3 RNA-seq expression levels were so low in control embryos (Fig. 6C) that myhz1.3 was not identified in the Pbx-dependent set. Another gene that was not identified by our RNA-seq approach is sox6, a known Prdm1a-dependent gene (von Hofsten et al., 2008). However, even though sox6 is expressed within fast muscle precursors and is regulated by Prdm1a and Myod (von Hofsten et al., 2008; Wang et al., 2011), sox6 does not appear to be regulated by Pbx2/4 (L.M., unpublished observations). Even though these examples are caveats to our RNA-seq analysis, they help provide further evidence that Pbx2/4 and Prdm1a regulate shared as well as independent aspects of the fast muscle program.
Why is a balance of factors needed for fiber-type differentiation? Like zebrafish slow muscle precursor cells, embryonic mammalian and avian myocytes and myofibers initially co-express both slow- and fast-type isoforms of many muscle differentiation genes (Narusawa et al., 1987; Sweeney et al., 1989; Lyons et al., 1990; Condon et al., 1990; Sacks et al., 2003; Niro et al., 2010). How this mixed slow/fast phenotype resolves into slow-twitch and multiple types of fast-twitch fibers is not fully understood. Perhaps a balance of factors that activate and repress subsets of fiber-type differentiation programs allows for increased plasticity in generating a variety of fiber types.
One outstanding question is whether Pbx and Prdm1 proteins are required for mammalian muscle differentiation. Mouse cell culture studies support a pioneer factor role for Pbx proteins in mouse skeletal muscle differentiation (Berkes et al., 2004; de la Serna et al., 2005), but direct requirements for Pbx proteins in mammalian skeletal muscle have not yet been tested. A recent analysis of the mouse Prdm1 conditional knock-out in embryonic myogenic cells or myogenic precursor cells revealed normal onset of myogenesis and normal expression of fast and slow muscle differentiation genes in the absence of Prdm1 (Vincent et al., 2012). This study even addressed functional compensation by another Prdm1-like factor by showing that expression of a dominant-negative Prdm1 had no effect on early mouse fast or slow skeletal muscle development (Vincent et al., 2012). If Prdm1 factors are not required in early mammalian skeletal muscle differentiation, it remains a possibility that a different factor with similar activity could act in a balance with Pbx proteins. In future studies it will be important to determine whether Pbx proteins and a Prdm or Prdm-like factor function to regulate mammalian skeletal muscle fiber-type differentiation.
The coordinated expression of contractile gene expression programs is critical not only for skeletal muscle differentiation but also for myocardial and smooth muscle differentiation. Pbx proteins are expressed in myocardial and vascular smooth muscle cells and are needed for proper development of the heart and cardiac outflow tract (Chang et al., 2008; Stankunas et al., 2008; Maves et al., 2009). It is possible that Pbx proteins will act in a balance with other transcription factors to promote subsets of myocardial and smooth muscle differentiation programs.
Materials and Methods
Zebrafish husbandry
All experiments involving live zebrafish (Danio rerio) were carried out in compliance with IACUC guidelines. Zebrafish were raised and staged as previously described (Westerfield, 2000). Time (hpf) refers to hours post-fertilization at 28.5°C. In some cases, embryos were raised for periods at room temperature, about 25°C. The wild-type stock used was AB. The prdm1am805 mutant line has been described and is likely a null allele (Hernandez-Lagunas et al., 2005). prdm1am805 genotyping primers, prdm1adcapsF2 and prdm1adcapsR2 are in Table 1. The genotyping PCR product is 238 base pairs. Digesting with XmnI yields a 204 base pair product from the mutant allele.
Table 1. Primers used.

Morpholino and mRNA injections
Morpholino injections were performed, and working concentrations were determined, as previously described (Maves et al., 2007). Morpholinos were used at the following working concentrations: pbx2-MO2, CCGTTGCCTGTGATGGGCTGCTGCG, 0.25 mg/ml; pbx2-MO3, GCTGCAACATCCTGAGCACTACATT, 0.5 mg/ml; pbx2-MO3 5-base mis-match control, GCTcCAAgATCCTcAcCAgTACATT, 0.5 mg/ml; pbx4-MO1, AATACTTTTGAGCCGAATCTCTCCG, 0.5 mg/ml; pbx4-MO2, CGCCGCAAACCAATGAAAGCGTGTT, 0.5 mg/ml; Standard control-MO, CCTCTTACCTCAGTTACAATTTATA, 1 mg/ml. pbx2-MO2, pbx2-MO3, pbx4-MO1 and pbx4-MO2 were combined to make the pbx2/4-MO combination (3 nl injected). Western analysis confirmed loss of Pbx2 and Pbx4 expression in pbx2/4-MO-treated embryos (Maves et al., 2007). Embryos that are knocked-down for Pbx2 and Pbx4 function show a developmental delay of about 2 somites during somitogenesis stages (Maves et al., 2007). We somite stage-matched sibling control and MO-treated embryos when collecting embryos for staining or for RNA analyses.
RNA in situ hybridization and immunocytochemistry
RNA in situ hybridizations were performed as previously described (Maves et al., 2007; Talbot et al., 2010). The following cDNA probes were used: prdm1a (Hernandez-Lagunas et al., 2005); krox-20 (egr2b-Zebrafish Information Network) (Bradford et al., 2011; Oxtoby and Jowett, 1993); mylpfa (formerly mylz2) (Xu et al., 2000); myhz1.3 (formerly fmyhcX) (von Hofsten et al., 2008); smyhc1 (Bryson-Richardson et al., 2005); myog (Weinberg et al., 1996); atp2a1 (Maves et al., 2007); srl (Maves et al., 2007). Following staining, embryo heads were genotyped for prdm1a as above.
Whole-embryo immunostaining was performed with anti-pan zebrafish Pbx, 1:500 (Pöpperl et al., 2000) as previously described (Maves et al., 2007). For DAPI staining, DAPI was included in the secondary antibody incubation step at a dilution of 1:2000.
Embryos were photographed using a Zeiss Stemi SV6 and Nikon CoolPix 4500 digital camera or imaged using a Zeiss Pascal confocal microscope or Leica TCS SP5 confocal microscope. Images were assembled using Adobe Photoshop.
RNA genotyping of single embryos and quantitative (q) RT-PCR
Embryos were collected from prdm1a+/− parents and injected with either control MO (pbx2-MO3 mismatch or Standard Control MO) or pbx2/4-MO combination. Total RNA from individual embryos was isolated using the RNAqueous Micro Kit (Ambion). RNA samples were genotyped for prdm1a from residual DNA. We then pooled RNA samples from about 10 equivalent-genotype embryos for four classes: control-MO;prdm1+/+; control-MO;prdm1−/−; pbx2/4-MO;prdm1+/+; and pbx2/4-MO;prdm1−/−. These pooled samples were then further cleaned and concentrated using the RNeasy MinElute Kit (Qiagen). 100 ng of pooled RNA samples plus random hexamers were used in a reverse-transcriptase reaction with SuperScriptII Reverse Transcriptase (Invitrogen). Real-time PCRs were performed using SYBR Green (Bio-Rad) and an Applied Biosystems 7900HT System according to the manufacturer's instructions. The relative expression levels were normalized to those of ornithine decarboxylase (odc1) (Maves et al., 2007). For primers, see Table 1.
RNA-seq library preparation and sequencing
Total RNA samples were genotyped and pooled as above for control-MO;prdm1+/+; control-MO;prdm1−/−; pbx2/4-MO;prdm1+/+; and pbx2/4-MO;prdm1−/− embryos at the 10 somite (s) stage from three independent sets of egg collections/injections. An additional set of embryos from each set of injections were processed for in situ hybridization to confirm somite staging as well as loss of krox-20 expression in pbx2/4-MO embryos (see Fig. 2A for example). Library preparation and sequencing was carried out by the FHCRC Genomics Shared Resource. Sequencing libraries were prepared from total RNA using the TruSeq RNA Sample Prep Kit (Illumina) according to the manufacturer's instructions. Library size distributions were validated using an Agilent 2100 Bioanalyzer. Additional library quality control, blending of pooled indexed libraries, and cluster optimization were performed using the QPCR NGS Library Quantization Kit (Agilent Technologies). RNA-seq libraries were pooled (4-plex) and clustered onto a flow cell lane using an Illumina cBot. Sequencing occurred on an Illumina HiSeq 2000 using a single-read, 100 base read length (SR100) sequencing strategy.
RNA-seq data processing and analysis
Image analysis and base calling were performed with Illumina's Real Time Analysis v1.12 software. Files were demultiplexed of indexed reads and generated in FASTQ format using Illumina's CASAVA v1.8. software. Reads were removed that did not pass Illumina's base call quality threshold. Reads were aligned to zebrafish genome build Zv9 release 63, using TopHat 1.4 (Trapnell et al., 2009). SAMtools v0.1.18 (Li et al., 2009) was used to sort and index the TopHat alignments such that they could be visualized in the Integrative Genomics Viewer (Robinson et al., 2011). The gene expression profiles of control-MO;prdm1+/+ versus control-MO;prdm1−/−; pbx2/4-MO;prdm1+/+; and pbx2/4-MO;prdm1−/− embryos were compared using the Bioconductor package edgeR (Robinson et al., 2010). To include reads that mapped to multiple homologs in the zebrafish genome, including the myosin heavy chain genes, reads with multiple hits on the genome were kept, and each hit received a fractional count equal to one over the number of hits. Venn diagrams were generated using BioVenn (Hulsen et al., 2008). Data have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE45532.
Supplementary Material
Acknowledgments
We thank the following colleagues for generously providing reagents: Kristin Artinger, Peter Currie, Zhiyuan Gong, Phil Ingham, and Heike Pöpperl. The Zebrafish International Resource Center (supported by grants RR12546 and RR15402-01 from the NIH) provided cDNA clones. We thank the FHCRC Genomics Shared Resource for excellent technical assistance with the RNA-seq experiments. We are extremely grateful to Cecilia Moens for the use of her zebrafish facility. We also thank Sean Rhodes for zebrafish care, Adam Szofran for technical assistance, and members of the Moens and Tapscott labs for advice. This work was supported by NIH 1R03AR057477 (L.M.), funded in part by the American Recovery and Reinvestment Act, and by NIH R01AR45113 (S.J.T.).
Footnotes
Competing interests: Conceived the experiments: L.M. Performed the experiments: L.M. Analyzed the data: Z.Y., G.H.F., L.M. Provided resources: S.J.T., L.M. Wrote the paper: L.M., with input from all authors.
Competing interests: The authors have no competing interests to declare.
References
- Baxendale S., Davison C., Muxworthy C., Wolff C., Ingham P. W., Roy S. (2004). The B-cell maturation factor Blimp-1 specifies vertebrate slow-twitch muscle fiber identity in response to Hedgehog signaling. Nat. Genet. 36, 88–93 10.1038/ng1280 [DOI] [PubMed] [Google Scholar]
- Berghella L., De Angelis L., De Buysscher T., Mortazavi A., Biressi S., Forcales S. V., Sirabella D., Cossu G., Wold B. J. (2008). A highly conserved molecular switch binds MSY-3 to regulate myogenin repression in postnatal muscle. Genes Dev. 22, 2125–2138 10.1101/gad.468508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkes C. A., Bergstrom D. A., Penn B. H., Seaver K. J., Knoepfler P. S., Tapscott S. J. (2004). Pbx marks genes for activation by MyoD indicating a role for a homeodomain protein in establishing myogenic potential. Mol. Cell 14, 465–477 10.1016/S1097-2765(04)00260-6 [DOI] [PubMed] [Google Scholar]
- Bradford Y., Conlin T., Dunn N., Fashena D., Frazer K., Howe D. G., Knight J., Mani P., Martin R., Moxon S. A.et al. (2011). ZFIN: enhancements and updates to the zebrafish model organism database. Nucleic Acids Res. 39 Suppl. 1, D822–D829 10.1093/nar/gkq1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryson–Richardson R. J., Daggett D. F., Cortes F., Neyt C., Keenan D. G., Currie P. D. (2005). Myosin heavy chain expression in zebrafish and slow muscle composition. Dev. Dyn. 233, 1018–1022 10.1002/dvdy.20380 [DOI] [PubMed] [Google Scholar]
- Buckingham M. (2001). Skeletal muscle formation in vertebrates. Curr. Opin. Genet. Dev. 11, 440–448 10.1016/S0959-437X(00)00215-X [DOI] [PubMed] [Google Scholar]
- Budry L., Balsalobre A., Gauthier Y., Khetchoumian K., L'honoré A., Vallette S., Brue T., Figarella–Branger D., Meij B., Drouin J. (2012). The selector gene Pax7 dictates alternate pituitary cell fates through its pioneer action on chromatin remodeling. Genes Dev. 26, 2299–2310 10.1101/gad.200436.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burguière A. C., Nord H., von Hofsten J. (2011). Alkali-like myosin light chain-1 (myl1) is an early marker for differentiating fast muscle cells in zebrafish. Dev. Dyn. 240, 1856–1863 10.1002/dvdy.22677 [DOI] [PubMed] [Google Scholar]
- Cao Y., Yao Z., Sarkar D., Lawrence M., Sanchez G. J., Parker M. H., MacQuarrie K. L., Davison J., Morgan M. T., Ruzzo W. L.et al. (2010). Genome-wide MyoD binding in skeletal muscle cells: a potential for broad cellular reprogramming. Dev. Cell 18, 662–674 10.1016/j.devcel.2010.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.–P., Stankunas K., Shang C., Kao S.–C., Twu K. Y., Cleary M. L. (2008). Pbx1 functions in distinct regulatory networks to pattern the great arteries and cardiac outflow tract. Development 135, 3577–3586 10.1242/dev.022350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S.–K., Lu P., Nakamura M., Lee J., Sagerström C. G. (2009). Meis cofactors control HDAC and CBP accessibility at Hox-regulated promoters during zebrafish embryogenesis. Dev. Cell 17, 561–567 10.1016/j.devcel.2009.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condon K., Silberstein L., Blau H. M., Thompson W. J. (1990). Development of muscle fiber types in the prenatal rat hindlimb. Dev. Biol. 138, 256–274 10.1016/0012-1606(90)90196-P [DOI] [PubMed] [Google Scholar]
- de la Serna I. L., Ohkawa Y., Berkes C. A., Bergstrom D. A., Dacwag C. S., Tapscott S. J., Imbalzano A. N. (2005). MyoD targets chromatin remodeling complexes to the myogenin locus prior to forming a stable DNA-bound complex. Mol. Cell. Biol. 25, 3997–4009 10.1128/MCB.25.10.3997-4009.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devoto S. H., Melançon E., Eisen J. S., Westerfield M. (1996). Identification of separate slow and fast muscle precursor cells in vivo, prior to somite formation. Development 122, 3371–3380. [DOI] [PubMed] [Google Scholar]
- Elworthy S., Hargrave M., Knight R., Mebus K., Ingham P. W. (2008). Expression of multiple slow myosin heavy chain genes reveals a diversity of zebrafish slow twitch muscle fibres with differing requirements for Hedgehog and Prdm1 activity. Development 135, 2115–2126 10.1242/dev.015719 [DOI] [PubMed] [Google Scholar]
- Hernandez–Lagunas L., Choi I. F., Kaji T., Simpson P., Hershey C., Zhou Y., Zon L., Mercola M., Artinger K. B. (2005). Zebrafish narrowminded disrupts the transcription factor prdm1 and is required for neural crest and sensory neuron specification. Dev. Biol. 278, 347–357 10.1016/j.ydbio.2004.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata H., Watanabe T., Hatakeyama J., Sprague S. M., Saint–Amant L., Nagashima A., Cui W. W., Zhou W., Kuwada J. Y. (2007). Zebrafish relatively relaxed mutants have a ryanodine receptor defect, show slow swimming and provide a model of multi-minicore disease. Development 134, 2771–2781 10.1242/dev.004531 [DOI] [PubMed] [Google Scholar]
- Hulsen T., de Vlieg J., Alkema W. (2008). BioVenn – a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics 9, 488 10.1186/1471-2164-9-488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.; 1000 Genome Project Data Processing Subgroup (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew H. P., Choksi S. P., Wong K. N., Roy S. (2008). Specification of vertebrate slow-twitch muscle fiber fate by the transcriptional regulator Blimp1. Dev. Biol. 324, 226–235 10.1016/j.ydbio.2008.09.020 [DOI] [PubMed] [Google Scholar]
- Lyons G. E., Ontell M., Cox R., Sassoon D., Buckingham M. (1990). The expression of myosin genes in developing skeletal muscle in the mouse embryo. J. Cell Biol. 111, 1465–1476 10.1083/jcb.111.4.1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnani L., Ballantyne E. B., Zhang X., Lupien M. (2011). PBX1 genomic pioneer function drives ERα signaling underlying progression in breast cancer. PLoS Genet. 7, e1002368 10.1371/journal.pgen.1002368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maves L., Waskiewicz A. J., Paul B., Cao Y., Tyler A., Moens C. B., Tapscott S. J. (2007). Pbx homeodomain proteins direct Myod activity to promote fast-muscle differentiation. Development 134, 3371–3382 10.1242/dev.003905 [DOI] [PubMed] [Google Scholar]
- Maves L., Tyler A., Moens C. B., Tapscott S. J. (2009). Pbx acts with Hand2 in early myocardial differentiation. Dev. Biol. 333, 409–418 10.1016/j.ydbio.2009.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narusawa M., Fitzsimons R. B., Izumo S., Nadal–Ginard B., Rubinstein N. A., Kelly A. M. (1987). Slow myosin in developing rat skeletal muscle. J. Cell Biol. 104, 447–459 10.1083/jcb.104.3.447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niro C., Demignon J., Vincent S., Liu Y., Giordani J., Sgarioto N., Favier M., Guillet–Deniau I., Blais A., Maire P. (2010). Six1 and Six4 gene expression is necessary to activate the fast-type muscle gene program in the mouse primary myotome. Dev. Biol. 338, 168–182 10.1016/j.ydbio.2009.11.031 [DOI] [PubMed] [Google Scholar]
- Oxtoby E., Jowett T. (1993). Cloning of the zebrafish krox-20 gene (krx-20) and its expression during hindbrain development. Nucleic Acids Res. 21, 1087–1095 10.1093/nar/21.5.1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pette D., Staron R. S. (2000). Myosin isoforms, muscle fiber types, and transitions. Microsc. Res. Tech. 50, 500–509 [DOI] [PubMed] [Google Scholar]
- Pöpperl H., Rikhof H., Chang H., Haffter P., Kimmel C. B., Moens C. B. (2000). lazarus is a novel pbx gene that globally mediates hox gene function in zebrafish. Mol. Cell 6, 255–267 10.1016/S1097-2765(00)00027-7 [DOI] [PubMed] [Google Scholar]
- Robinson M. D., McCarthy D. J., Smyth G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J. T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E. S., Getz G., Mesirov J. P. (2011). Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S., Wolff C., Ingham P. W. (2001). The u-boot mutation identifies a Hedgehog-regulated myogenic switch for fiber-type diversification in the zebrafish embryo. Genes Dev. 15, 1563–1576 10.1101/gad.195801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacks L. D., Cann G. M., Nikovits W., Jr, Conlon S., Espinoza N. R., Stockdale F. E. (2003). Regulation of myosin expression during myotome formation. Development 130, 3391–3402 10.1242/dev.00541 [DOI] [PubMed] [Google Scholar]
- Saleh M., Rambaldi I., Yang X.–J., Featherstone M. S. (2000). Cell signaling switches HOX-PBX complexes from repressors to activators of transcription mediated by histone deacetylases and histone acetyltransferases. Mol. Cell. Biol. 20, 8623–8633 10.1128/MCB.20.22.8623-8633.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiaffino S., Reggiani C. (1996). Molecular diversity of myofibrillar proteins: gene regulation and functional significance. Physiol. Rev. 76, 371–423. [DOI] [PubMed] [Google Scholar]
- Schiaffino S., Reggiani C. (2011). Fiber types in mammalian skeletal muscles. Physiol. Rev. 91, 1447–1531 10.1152/physrev.00031.2010 [DOI] [PubMed] [Google Scholar]
- Stankunas K., Shang C., Twu K. Y., Kao S.–C., Jenkins N. A., Copeland N. G., Sanyal M., Selleri L., Cleary M. L., Chang C.–P. (2008). Pbx/Meis deficiencies demonstrate multigenetic origins of congenital heart disease. Circ. Res. 103, 702–709 10.1161/CIRCRESAHA.108.175489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney L. J., Kennedy J. M., Zak R., Kokjohn K., Kelley S. W. (1989). Evidence for expression of a common myosin heavy chain phenotype in future fast and slow skeletal muscle during initial stages of avian embryogenesis. Dev. Biol. 133, 361–374 10.1016/0012-1606(89)90040-7 [DOI] [PubMed] [Google Scholar]
- Talbot J. C., Johnson S. L., Kimmel C. B. (2010). hand2 and Dlx genes specify dorsal, intermediate and ventral domains within zebrafish pharyngeal arches. Development 137, 2507–2517 10.1242/dev.049700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapscott S. J. (2005). The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development 132, 2685–2695 10.1242/dev.01874 [DOI] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S. L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent S. D., Mayeuf A., Niro C., Saitou M., Buckingham M. (2012). Non conservation of function for the evolutionarily conserved prdm1 protein in the control of the slow twitch myogenic program in the mouse embryo. Mol. Biol. Evol. 29, 3181–3191 10.1093/molbev/mss125 [DOI] [PubMed] [Google Scholar]
- von Hofsten J., Elworthy S., Gilchrist M. J., Smith J. C., Wardle F. C., Ingham P. W. (2008). Prdm1- and Sox6-mediated transcriptional repression specifies muscle fibre type in the zebrafish embryo. EMBO Rep. 9, 683–689 10.1038/embor.2008.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Ono Y., Tan S. C., Chai R. J., Parkin C., Ingham P. W. (2011). Prdm1a and miR-499 act sequentially to restrict Sox6 activity to the fast-twitch muscle lineage in the zebrafish embryo. Development 138, 4399–4404 10.1242/dev.070516 [DOI] [PubMed] [Google Scholar]
- Waskiewicz A. J., Rikhof H. A., Moens C. B. (2002). Eliminating zebrafish pbx proteins reveals a hindbrain ground state. Dev. Cell 3, 723–733 10.1016/S1534-5807(02)00319-2 [DOI] [PubMed] [Google Scholar]
- Weinberg E. S., Allende M. L., Kelly C. S., Abdelhamid A., Murakami T., Andermann P., Doerre O. G., Grunwald D. J., Riggleman B. (1996). Developmental regulation of zebrafish MyoD in wild-type, no tail and spadetail embryos. Development 122, 271–280. [DOI] [PubMed] [Google Scholar]
- Westerfield M. (2000). The Zebrafish Book. A Guide For The Laboratory Use Of Zebrafish (Danio Rerio), 4th edition. Eugene, OR: University of Oregon Press. [Google Scholar]
- Wigmore P. M., Evans D. J. R. (2002). Molecular and cellular mechanisms involved in the generation of fiber diversity during myogenesis. Int. Rev. Cytol. 216, 175–232 10.1016/S0074-7696(02)16006-2 [DOI] [PubMed] [Google Scholar]
- Wu K.–D., Lee W. S., Wey J., Bungard D., Lytton J. (1995). Localization and quantification of endoplasmic reticulum Ca(2+)-ATPase isoform transcripts. Am. J. Physiol. 269, C775–C784. [DOI] [PubMed] [Google Scholar]
- Xu Y., He J., Wang X., Lim T. M., Gong Z. (2000). Asynchronous activation of 10 muscle-specific protein (MSP) genes during zebrafish somitogenesis. Dev. Dyn. 219, 201–215 [DOI] [PubMed] [Google Scholar]
- Zaret K. S., Carroll J. S. (2011). Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 25, 2227–2241 10.1101/gad.176826.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.