Abstract
When Rhizobium etli CE3 was grown in the presence of Phaseolus vulgaris seed extracts containing anthocyanins, its lipopolysaccharide (LPS) sugar composition was changed in two ways: greatly decreased content of what is normally the terminal residue of the LPS, di-O-methylfucose, and a doubling of the 2-O-methylation of other fucose residues in the LPS O antigen. R. etli strain CE395 was isolated after Tn5 mutagenesis of strain CE3 by screening for mutant colonies that did not change antigenically in the presence of seed extract. The LPS of this strain completely lacked 2-O-methylfucose, regardless of whether anthocyanins were present during growth. The mutant gave only pseudonodules in association with P. vulgaris. Interpretation of this phenotype was complicated by a second LPS defect exhibited by the mutant: its LPS population had only about 50% of the normal amount of O-antigen-containing LPS (LPS I). The latter defect could be suppressed genetically such that the resulting strain (CE395α395) synthesized the normal amount of an LPS I that still lacked 2-O-methylfucose residues. Strain CE395α395 did not elicit pseudonodules but resulted in significantly slower nodule development, fewer nodules, and less nitrogenase activity than lps+ strains. The relative symbiotic deficiency was more severe when seeds were planted and inoculated with bacteria before they germinated. These results support previous conclusions that the relative amount of LPS I on the bacterial surface is crucial in symbiosis, but LPS structural features, such as 2-O-methylation of fucose, also may facilitate symbiotic interactions.
Bacteria that enter into intimate associations with plants and animals, whether pathogenic or mutualistic, respond to signals that are thought to indicate the presence of the host. In the symbiosis between legumes and the bacteria collectively known as rhizobia, one example is the production of Nod factors (lipochitooligosaccharides) by the bacteria in response to flavonoid compounds exuded by the plant (17, 27). Another potential example is the alteration of the lipopolysaccharide (LPS) of Rhizobium etli when grown in the presence of the most potent nod inducers from Phaseolus vulgaris (17), the anthocyanins exuded by germinating seeds (10, 22, 23). These induced LPS alterations have been detected by the lack of recognition of the altered LPS by certain monoclonal antibodies (23). Modifications of the LPS structure in strains of this and other rhizobial species also have been reported to occur during nodule development and in culture in response to low pH and low oxygen, as well as nod inducers (1, 16, 18, 19, 29, 31). However, in no case have precise chemical changes been determined, nor is it known whether these induced LPS modifications are important in the development or functioning of the symbiosis.
Although little is known of the biological roles of specific structural features of the rhizobial LPS, the importance of wild-type LPS in symbiosis is well established. Studies with mutants of a wide variety of rhizobial species have indicated that defects in LPS structure severely impair infection and nodule development on numerous legume hosts (20, 22). Generally, conclusions are based on phenotypes of mutants in which large portions of the LPS molecules are missing. From studies with mutants derived from R. etli CE3, it has been concluded that the LPS O antigen must be present in normal amounts in order for R. etli to infect and incite normal root nodule development on P. vulgaris (22, 24). The cellular basis for this requirement has not been determined, but it is likely that the LPS plays several biological roles that are important in symbiosis and that a different combination of structural features is required in each role (22).
The structure of the LPS of R. etli CE3 grown in tryptone-yeast extract medium has been almost completely determined (Fig. 1) (2, 14, 15, 28). This accomplishment affords the opportunity, not currently possible with other rhizobia, of correlating biological roles with specific structural features of the molecule. One example is a mutant LPS whose N-acetylquinovosamine (QuiNAc) residue is replaced by a QuiNAc precursor. The mutant O-antigen structure is normal otherwise and confers normal sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) mobility and antigenic properties (15a, 24). Even when the mutant O antigen is present in nearly normal amounts, greatly reduced nodule numbers and retarded nodule development result (24). Hence, there may be a specific requirement for QuiNAc or some other structural feature that depends on QuiNAc, as had been suggested by studies of differences in the LPS of another rhizobial strain at different growth stages (8).
FIG. 1.
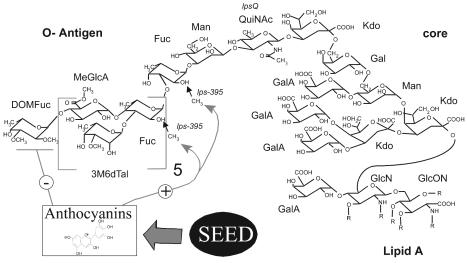
Overview of R. etli LPS I structure and the effects of seed extract. The terminal Fuc has been reported to be either 2,3-di-O-methylated (DOMFuc, as shown above) or tri-O-methylated (11, 14). However, in this study, it was almost exclusively present as DOMFuc. On average, only about one of the other six Fuc residues per LPS molecule is methylated after growth in the absence of seed extract or seed exudate, and the exact distribution of the methylation is unknown. Hence, the residues are depicted as unmethylated, while the small arrows indicate the sites at which the 2-O-methylation occurs (14). Anthocyanins are the components of seed extract or exudate to which the bacteria respond by changing LPS antigenicity (10). In this study it was found that seed extracts induce decreased DOMFuc content and increased 2-O-methylation of Fucs (depicted as the − and + effects, respectively, on the diagram). It is not known which Fuc(s) is up-methylated. Other abbreviations: 3M6dTal, 3-O-methyl-6-deoxytalose; Fuc, fucose; Man, mannose; Gal, galactose; QuiNAc, N-acetylquinovosamine; MeGlcA, methylglucuronate; GalA, galacturonic acid; GlcN, glucosamine; Kdo, 3-deoxy-d-manno-2-octulonic acid; GlcN, glucosamine; GlcON, 2-aminogluconate; R, fatty acyl groups.
In work to be described, changes in LPS sugar composition induced by growth in host anthocyanins were examined. One of the LPS alterations was a doubling in the proportion of fucose (Fuc) that is 2-O-methylated. Mutation lps-395::Tn5 prevented 2-O-methylation of Fuc under inducing and basal conditions. This mutation provided an approach for testing whether this methylation is required in symbiosis.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. For induction of LPS changes with seed extracts, R. etli cultures were grown by gyratory shaking at 200 rpm at 30°C in YEC liquid (0.5% yeast extract, 10 mM CaCl2 [pH 6.0]) (9). For genetic manipulations, R. etli was grown in TY (0.5% tryptone, 0.3% yeast extract, 10 mM CaCl2) solidified with 1.5% agar and supplemented with the appropriate antibiotic (streptomycin [STR], 250 μg/ml; nalidixic acid [NAL], 30 μg/ml; kanamycin [KAN], 30 μg/ml; or tetracycline [TET], 5 μg/ml). Escherichia coli strains were grown at 37°C on solid or liquid Luria-Bertani media supplemented with the appropriate antibiotic (STR, 250 μg/ml; NAL, 30 μg/ml; KAN, 30 μg/ml; TET, 15 μg/ml; or chloramphenicol, 30 μg/ml).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Refer- ence |
|---|---|---|
| E. coli strains | ||
| HB101 | pro leu thi lacY hsd20 endA recA rpsL20 ara-14 galK2 xyl-5 mtl-1 supE44 | 3 |
| MT616 | pro thi endA hsdR supE44 recA-J6 pRK2013Km::Tn9 | 13 |
| R. etli strains | ||
| CE3 | str-1 Lps+ Ndv+ Fix+ | 25 |
| CE3α | CE3 carrying pLPSα | |
| CE395 | CE3 derivative, str-1 lps-395::Tn5 | |
| CE395α | CE395 carrying pLPSα | |
| CE395α395 | CE395 carrying pLPSα395 | |
| CE471 | str-1 lps-395::Tn5, constructed in CE3 by means of pLPSα395 | |
| Plasmids | ||
| pLPSα | pCOS109.11; 30 kb of R. etli CE3 lps region α inserted in pLAFR1 | 7 |
| pLPSα395 | pLPSα in which lps-395::Tn5 replaces wild-type allele | |
| pSUP2021 | Tn5 delivery plasmid; nonreplicating in R. etli | 30 |
Seed extract.
To obtain crude preparations of anthocyanins for induction of LPS modifications, black P. vulgaris (cv. Midnight Black Turtle Soup) (Idaho Seed Bean, Twin Falls, Idaho) seeds were extracted as described previously (9). The seed extract was collected from 720 g of seeds that were thoroughly rinsed in deionized water and surface sterilized with 95% ethanol for 2 min. One hundred twenty grams of each of the treated seeds was added separately to Fernbach flasks containing 250 ml of sterile 0.1% HCl. The flasks were then incubated at 30°C on a shaker at 200 rpm for 16 h. The liquid phase was centrifuged at 700 × g for 20 min at 4°C. The supernatant was filtered over 2 sterile Whatman #2 filter papers. The total volume was adjusted to 900 ml with sterile deionized water. To obtain bacteria with induced LPS changes, 450 ml of this extract was added to a 2.8-liter Fernbach flask with 550 ml of 1.8× YEC (0.91% yeast extract, 18 mM CaCl2), resulting in a final pH of 5.9, and 20 ml of R. etli fully grown in YEC was added. The control cultures were grown at the same time in the same type of flask with 1.0 liters of YEC lacking seed extract. Bacteria were harvested when full growth was indicated by the appearance of a copious cell pellicle on the side of the flask (generally at 24 h).
SDS-PAGE and analysis of LPS antigenicity.
Bacterial cells from 1 ml of fully grown cultures were washed in 1 ml of deionized water two times, pelleted, and extracted in 0.1 ml of SDS-PAGE sample buffer (6) at 100°C for 3 min. Samples of purified LPS or crude phenol-water extracts were prepared by adding 1 mg of the lyophilized material in 1 ml of SDS sample buffer and heating at 100°C for 3 min. After centrifugation to remove insoluble material, the SDS extracts were subjected to electrophoresis in discontinuous SDS-PAGE (6) with 18% (wt/vol) acrylamide resolving gels. LPS in the gels was stained by modification of a previously described periodate-silver method (6). The gels were shaken in six changes of fixative (10% glacial acetic acid, 40% methanol) for at least 1 h. After incubation in 0.7% sodium metaperiodate for 10 min, the gels were rinsed in six changes of glass-distilled water for 35 min, followed by a 10-min incubation in 0.2% silver nitrate and a 1.0-min (no longer) rinse in glass-distilled water, and then developed for 5 to 6 min in Bio-Rad silver stain developer. After SDS-PAGE, some gels were electrotransferred to nitrocellulose before periodate-silver staining of the residual gel contents. The dried nitrocellulose blots were immunostained by using monoclonal antibody (MAb) Jim 28 (a generous gift of N. J. Brewin, Norwich, United Kingdom) and anti-rat immunoglobulin M conjugated with alkaline phosphatase (Sigma Chemical Co.) as previously described (23).
LPS sugar compositions.
LPS was extracted from washed bacterial cell pellets by the hot phenol-water method (4) and purified by Sepharose 4B chromatography (5, 26). After complete hydrolysis of LPS sugar linkages in 2 M trifluoroacetic acid, reduction with NaBD4, and acetylation, alditol acetate derivatives of the LPS were analyzed for neutral and amino sugar compositions by gas chromatography on a 60-m by 0.53-mm by 0.20-μ fused silica capillary SP2330 column (Supelco) (14). The oven temperature was raised 1°C min−1 from 150 to 185°C, 10°C min−1 from 185° to 245°C, and then maintained at 245°C for 35 min. With this protocol, 2-methylfucose was eluted at 25.8 min, and its peak returned completely to baseline before 3-methyl-6-deoxytalose emerged at 26.4 min. 2,3-di-O-methylfucose emerged at 21.7 min, well separated from all other peaks. Acidic sugar compositions were determined on an SPB-1 column (Supelco) as trimethylsilyl-methylglycoside derivatives after methanolysis in methanolic 1 M HCl (14). Identification of tri-O-methylfucose, di-O-methylfucoses (DOMFuc), and 2-O-methylfucose (2MeFuc) was done by comparison of elution times with those of a standard mixture of O-methylated Fucs that varied in position and number of methyl groups, kindly provided by L. Scott Forsberg. Coupled GC and electron impact mass-spectral analyses confirmed the positions of the methyl groups on the 6-deoxyhexose moiety. Where standard deviations are given in the tables that follow, they are based on values from LPS preparations from at least three separately grown cultures. The standard deviations are based on the deviations in the molar ratios before the data were normalized.
Isolation of mutant strain CE395.
This mutant was isolated after Tn5 mutagenesis by immunoblot screening of colonies replicated onto agar amended with seed exudate. The exudate was prepared by aqueous acetone extraction of material exuded into water during germination of P. vulgaris (cv. Midnight Black Turtle Soup) seeds (procedure B in reference 23). After evaporation of the acetone and lyophilization of residual water, the dry material was dissolved in water and added to TY agar plates containing NAL, STR, and KAN, such that the agar contained the exudate from 1.0 seed per ml. Such plates (SE plates) were used within 2 days of preparation and always were protected from light. Colonies on TY agar plates containing NAL, STR, and KAN after Tn5 mutagenesis with pSUP2021 (6) were replicated (using velvet) onto SE plates. After 2 days at 30°C, colonies were lifted onto a dry nitrocellulose sheet (BA85; Schleicher & Schuell) by letting the sheet contact a small area of the plate and, as it became wetted by capillary action, letting it slowly contact the rest of the plate. After being in contact with the colonies for 5 min, the sheet was allowed to dry fully in air and stained with MAb JIM28 by the procedure above used for gel immunoblots. Strain CE395 was a solitary colony that stained darkly among approximately 100 lightly staining colonies.
Genetic suppression.
pLPSα395 was isolated by exploiting in vivo homologous recombination in R. etli strain CE395α that replaced the corresponding wild-type allele on the lps region α in pLPSα with the mutant allele lps-395::Tn5. Such recombinant plasmids were identified by selecting their transfer into E. coli strain HB101. On a TY plate, freshly grown broth cultures of CE395α, HB101, and MT616 were spread together in a 4:1:1 ratio of culture volumes and incubated at 37°C overnight. Bacteria from the plate then were streaked or spread after dilution onto a Luria-Bertani plate containing KAN and TET and incubated at 37°C overnight. Plasmids were isolated from purified colonies that had arisen on this plate. EcoRI digests of each plasmid preparation verified that allele replacement had occurred (absence of the 2.9-kb band from pLPSα and the appearance of a new 8.5-kb band). One such colony (HB101/pLPSα395) was mated with CE395 by means of helper strain MT616 in the same manner as the first mating above. Purified transconjugants from this mating that grew on TY containing STR, NAL, KAN, and TET were deemed to be strain CE395 carrying pLPSα395 and designated CE395α395.
Nodulation tests.
P. vulgaris cv. Midnight Black Turtle Soup seeds disinfected with 5% hypochlorite were pregerminated on water-saturated filter paper, planted, and inoculated in 50-ml serum vials wrapped in aluminum foil containing plant nutrient solidified with 0.7% agar or in plastic growth pouches (Mega International, Minneapolis, Minn.) as previously described (25). Alternatively, disinfected seeds were transferred into the above vials without pregermination, inoculated with bacteria, and put directly into a growth chamber. Regardless of procedure, inoculated plants received 0.25 ml of the respective bacterial culture grown in TY broth. Plants were grown at 27°C with a 14/10-h light/dark cycle. Nitrogenase activity of the acetylene reduction catalyzed by intact nodulated roots was measured by gas chromatography on a Porapak N column (25). Following the acetylene reduction assay, the nodules were stripped off the roots, counted, and weighed. Occupancy of particular strains in nodules was measured by counting CFU on agar plates containing strain-selective antibiotics. The plates were spread with aliquots from the contents of crushed, surface-sterilized nodules suspended in TY broth (26).
Statistical analysis.
The probabilities (P) of the null hypothesis (no difference between data sets) were calculated using unpaired t tests and one-way analysis of variance with Sigma Stat software, version 2.03.
RESULTS
Alterations in wild-type LPS after growth in anthocyanins (seed extract).
As monitored by the extent of binding of MAb JIM28, the LPS alteration in response to anthocyanins is dose dependent (10, 23). LPS was purified from a culture of wild-type strain CE3 (Table 1) that had been exposed to sufficient seed extract so that the synthesized LPS bound very weakly to MAb JIM28 (Fig. 2). The sugar composition of the purified LPS from bacteria grown under these conditions revealed increased content of 2MeFuc, a corresponding decrease in Fuc content, and the near absence of what is normally the terminal sugar of the LPS, DOMFuc (Table 2). The differences in DOMFuc and 2MeFuc after growth in anthocyanins were highly significant (P < 0.001). The differences in Fuc also were significant (P < 0.05). The other values were not significantly different between the two conditions of growth (P ≥ 0.40). Table 2 gives only the compositions of neutral and basic sugars, which were determined as alditol acetates. Acidic sugars Kdo, GlcA, and GalA were determined as trimethylsilyl-methylglycosides; their contents did not differ significantly after growth in anthocyanins (data not shown).
FIG. 2.
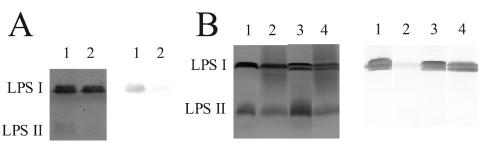
The effect of seed extracts on the antigenicity of LPS. (A) Purified wild-type LPS preparations used to obtain the data in Table 2. LPS was purified after growth in the presence or absence of seed extracts containing anthocyanins, and the LPS was subjected to SDS-PAGE. (B) SDS-PAGE comparison of the LPS in crude phenol-water extracts from strain CE395 and the wild type that had been grown in parallel and, where noted, exposed to the same seed extract. Lanes 1 and 2 in both panels are LPS from wild-type strain CE3. All lanes 3 and 4 are from strain CE395. For all lanes 2 and 4, the bacteria were grown in the presence of seed extract. In each panel the image on the left is of gel lanes stained with silver. (In panel A the gel was stained after being elecroblotted, resulting in barely visible LPS II bands.) On the right in each panel is the blot of these lanes reacted with monoclonal antibody JIM28.
TABLE 2.
Neutral and basic sugar composition of CE3 LPS after growth in seed extractd
| Sugarb | Molar ratioa after growth in:
|
|
|---|---|---|
| YEC | YEC+SEc | |
| DOMFuc | 0.59 ± 0.13 | 0.15 ± 0.06 |
| 2MeFuc | 0.79 ± 0.14 | 1.67 ± 0.08 |
| 3M6dTal | 3.04 ± 0.30 | 2.95 ± 0.28 |
| Fuc | 3.32 ± 0.35 | 2.83 ± 0.20 |
| Man | 1.94 ± 0.40 | 1.74 ± 0.17 |
| Gal | 1.00 ± 0.10 | 1.00 ± 0.10 |
| QuiN | 0.76 ± 0.16 | 0.79 ± 0.15 |
| GlcN | 0.74 ± 0.24 | 0.84 ± 0.34 |
Normalized to 1.00 galactose residues.
Abbreviations are defined in the legend to Fig. 1.
SE, seed extract enriched in anthocyanins.
Results are given ± standard deviation.
Mutant strain CE395.
Strain CE395 was isolated after Tn5 mutagenesis by virtue of its not losing reactivity with MAb JIM28 after growth in the presence of anthocyanins (or crude P. vulgaris seed extract) (Fig. 2B). Figures 2B and 3A illustrate three other properties of this mutant strain. First, the amount of O-antigen-containing LPS (LPS I) relative to a form not carrying the O antigen (LPS II) was about 60% of the wild-type ratio, as determined by integrating the intensities of staining from several gels. Second, the staining of the mutant LPS I by JIM28 in immunoblots was reproducibly more intense than the staining of the wild-type LPS I even in the absence of anthocyanins. The mutant LPS I apparently is a better antigen than the wild type, and this antigenicity is not substantially decreased after growth in anthocyanins (Fig. 2B and 3B). Third, LPS I was split into two almost equally prominent bands. The faster-migrating band comigrated with a very minor band in the wild-type LPS profile.
FIG. 3.
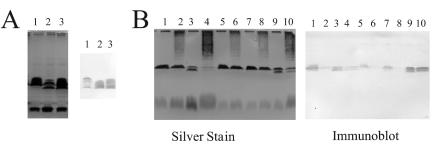
SDS-PAGE and immunoblot analysis showing the effects of recombinant plasmid pLPSα and mutant plasmid pLPSα395. After growth in YEC, with or without seed extract, washed bacteria were processed for SDS-PAGE. (A) Strains CE3 (lanes 1), CE395 (lanes 2), and CE395α395 (lanes 3) grown in YEC without seed extract are compared. (B) Strains carrying pLPSα (CE3α and CE395α) are included; the effects of seed extracts are shown. The analyzed strains were CE3 (lanes 1 and 2), CE395 (lanes 3 and 4), CE3α (lanes 5 and 6), CE395α (lanes 7 and 8), and CE395α395 (lanes 9 and 10). In lanes 2, 4, 6, 8, and 10 of panel B, the strains had been grown in the presence of seed extract, which is responsible for the streaking above LPS I in the silver-stained gel. In each panel the image on the left is of gel lanes stained with silver. (In panel B the gel was stained after being elecroblotted, resulting in relatively less-intense staining of LPS II bands.) On the right in each panel is the blot of these lanes reacted with monoclonal antibody JIM28.
The overall content of O-antigen sugars in the purified LPS of strain CE395 was lower than that of the wild type (Table 3), consistent with the lower content of LPS I revealed by SDS-PAGE. In fact, it is consistent with the mutant LPS population having about 50% of the O-antigen content found in the wild type. In addition, the mutant LPS completely lacked 2MeFuc (Table 3). 2MeFuc could not be detected even by searching for its signature by mass spectral analysis (R. W. Carlson and K. D. Noel, unpublished data). Relative to the other O-antigen sugars, the mean content of Fuc was higher in the mutant, to an extent that roughly correlated with the absence of 2MeFuc (Table 3). Given the lower total O-antigen content in the mutant, the relative proportions of the O-antigen sugars other than 2MeFuc and Fuc were approximately the same in the mutant and the wild type.
TABLE 3.
LPS sugar compositions of 395 and 395α395 grown with or without seed extract
| Sugarb | Molar ratioa found in strain:
|
|||||
|---|---|---|---|---|---|---|
| CE3 | CE3 Ic | CE395 | CE395 I | CE395a395 | CE395a395 I | |
| DOMFuc | 0.59 | 0.15 | 0.43 ± 0.15 | 0.03 | 0.60 ± 0.06 | 0.082 |
| 2MeFuc | 0.79 | 1.67 | NDd | ND | ND | ND |
| 3M6dTal | 3.04 | 2.95 | 1.44 ± 0.42 | 1.14 | 2.77 ± 0.51 | 1.41 |
| Fuc | 3.32 | 2.83 | 1.99 ± 0.48 | 1.71 | 3.97 ± 0.39 | 2.34 |
| Man | 1.94 | 1.74 | 1.28 ± 0.31 | 1.30 | 1.65 ± 0.06 | 1.77 |
| Gal | 1 | 1.00 | 1 ± 0.27 | 1 | 1 ± 0.09 | 1 |
| QuiN | 0.76 | 0.79 | 0.41 ± 0.11 | 0.45 | 0.64 ± 0.13 | 0.31 |
| GlcN | 0.74 | 0.84 | 0.60 ± 0.27 | 0.93 | 0.72 ± 0.37 | 0.31 |
Normalized to 1.00 galactose residues. Results are given ± standard deviation. The values for CE395 I and CE395α395 I were from only one preparation from one culture; hence, averages of the GC runs are shown but not standard deviations. The standard deviations for CE3 and CE3 I are given in Table 2.
Abbreviations are defined in the legend to Fig. 1.
I, cultures grown in the presence of seed extracts.
ND, not detected.
Although the wild type showed an increased relative amount of 2MeFuc in its LPS after growth in seed extract, the mutant LPS still lacked 2MeFuc under this condition (Table 3). The other change in wild-type LPS sugar composition induced by seed extract does appear to occur in mutant CE395; DOMFuc was greatly decreased in the mutant by growth in the presence of seed extract (Table 3).
Suppression of the deficiency in LPS I concentration by harboring lps region α carrying mutation lps-395::Tn5.
Previously reported restriction analysis (24) indicated that the Tn5 insertion of CE395 had occurred in a 2.9-kb EcoRI fragment of the R. etli CE3 lps region α (7). lps region α is operationally defined as the long stretch (at least 18 kb) of lps genes that are contained within the 30-kb insert of recombinant plasmid pLPSα. This plasmid complemented mutation lps-395 such that normal amounts of LPS I were synthesized (Fig. 3). Production of 2MeFuc was also restored in CE395α (strain CE395 carrying pLPSα). In fact, as in the wild type harboring pLPSα (CE3α), the amount of 2MeFuc in CE395α was elevated relative to the content in strain CE3 (Table 4). At the same time, the binding of MAb JIM28 to LPS I was noticeably weaker in strains CE395α and CE3α than in CE3 (Fig. 3). Nevertheless, strains CE395α and CE3α responded to anthocyanin induction; this relatively weak antigenicity was lost after these strains were cultured in the presence of anthocyanins or seed extract (Fig. 3).
TABLE 4.
Effects of pLPSα (lps region α) on 2MeFuc contentsa
| Sugarb | Molar ratioc found in strain:
|
||||
|---|---|---|---|---|---|
| CE3 | CE3αd | CE395 | CE395αd | 395α395 | |
| DOM-Fuc | 0.84 | 0.56 | 0.23 | 0.64 | 0.55 |
| 2MeFuc | 0.94 | 1.46 | ND | 1.46 | ND |
| 3M6dTal | 3.49 | 2.94 | 0.99 | 3.07 | 2.34 |
| Fuc | 4.24 | 3.13 | 1.64 | 3.51 | 3.94 |
| Man | 1.65 | 1.69 | 1.20 | 1.75 | 1.58 |
| Gal | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| QuiN | 0.62 | 0.66 | 0.28 | 0.67 | 0.58 |
| GlcN | 0.37 | 0.39 | 0.38 | 0.37 | 0.39 |
This is from an experiment analyzing phenol-water extracts. Contaminating ribose and glucose contents are not shown. Experiments in which CE3α and CE395α were compared separately with CE3 also showed that these pLPSα-containing strains had at least 50% more 2MeFuc and a corresponding decrease in relative Fuc content.
Abbreviations are defined in the legend to Fig. 1.
Neutral and amino sugars in each strain normalized to 1.00 galactose residues.
α indicates that the strain (CE3 or CE395) carries pLPSα.
Based on a previously reported means of suppressing the deficit in LPS I for strain CE166, it was reasoned that multiple copies of DNA from lps region α outside of the mutant lps gene might suppress the deficiency in the amount of LPS I for CE395. To test this idea, the corresponding wild-type allele on pLPSα was replaced by lps-395::Tn5 through recombination in vivo, and this plasmid (pLPSα395) was mobilized into CE395. For the resulting strain (CE395α395), an approximately normal LPS I/II ratio on SDS-PAGE (Fig. 3) and a normal ratio of core versus O-antigen sugars (Table 3) were restored. However, like CE395, this strain had no detectable 2MeFuc (Table 3). Strain CE395α395 made it possible to separate the biological consequences of the deficiency in LPS I abundance in CE395 from the consequences of other LPS properties conferred by lps-395, including the inability to 2-O-methylate Fuc.
Plasmid pLPSα395 also was used to test the possibility that the LPS I deficiency in CE395 was due to a separate mutation that may have existed in CE395 in addition to lps-395::Tn5. If this were the explanation, then pLPSα395 apparently did not carry this second mutation; otherwise it should not suppress this deficiency. Therefore, pLPSα395 was introduced into CE3 as an agent to reconstruct a mutant that would carry only lps-395::Tn5. This mutant, designated CE471, was identical to CE395 in SDS-PAGE and immunoblot analysis.
Symbiosis.
Regardless of the method of testing nodulation, the mutant strain CE395 elicited pseudonodules that resembled in every way the pseudonodules elicited by R. etli mutants that completely lack O antigen (6, 26). P. vulgaris pseudonodules have a characteristic incomplete nodule development that in past studies has been shown to result from nodulation in the absence of bacterial infection or with bacterial infection that is limited to root hairs before it stops (21, 26, 32). Because there are no bacteroids, pseudonodules lack nitrogenase activity. On the other hand, CE395α395 exhibited only about a 50% deficiency relative to the wild type in total plant nitrogenase activity when inoculated on seedlings that had been pregerminated on filter paper and planted in growth pouches (data not shown). When mixed with the wild type in the inoculum at equal viable cell density, strain CE395α395 was recovered from nodules of plants grown this way at a frequency less than 5% of the wild type's. Therefore, although the difference in symbiotic performance was small when assayed in pouches, it had a measurable impact on the ability to compete for nodule occupancy. A greater deficiency in nodulation and total plant nitrogenase activity was exhibited when symbiosis was assayed by growing the inoculated plants in vials containing agar. The greatest deficiency was observed when seeds were planted without pregermination and inoculated with bacteria at the time they were planted on the agar (Table 5). Fewer, smaller, and less-active nodules resulted from the mutant inoculation. Table 5 presents the results of a representative experiment assayed at 20 days after inoculation. At 17 days after inoculation, the differences between CE395α395 and the wild type were greater, a reflection of the noticeably slower development of the mutant nodules. CE395α behaved like the wild type under each method of testing symbiosis.
TABLE 5.
Nodulation and nitrogenase activity of CE395 and CE395α395i
| Inoculant | ARf plant | Nodules/plant
|
Specific ARg | |
|---|---|---|---|---|
| No. | Mass (mg) | |||
| CE3 | 7.4 ± 1.3a | 11.2 ± 3.9 | 21.9 ± 6.6d | 0.31 ± 0.10 |
| CE3αh | 5.9 ± 1.7 | 8.6 ± 2.4 | 18.3 ± 3.1 | 0.29 ± 0.14 |
| CE395 | 0.00 | 4.5e ± 2.2 | 0.00 | 0.0 ± 0.0 |
| CE395αh | 9.0 ± 2.2a | 13.3 ± 3.5 | 24.8 ± 4.8d | 0.36 ± 0.04 |
| CE395α395 | 2.6 ± 2.2b | 7.5 ± 4.5 | 11.5 ± 6.3c | 0.21 ± 0.10 |
Do not differ from each other (P >0.5).
Differs from a-footnoted values (P <0.01).
Differs from d-footnoted values (P <0.05).
Do not differ from each other (P >0.3).
Pseudonodules.
AR (acetylene reduction) in nanomoles of ethylene produced per minute.
AR per milligram of fresh nodule.
α indicates that the strain carries pLPSα.
Results are given ± standard deviation.
DISCUSSION
The LPS sugar composition after growth of wild-type R. etli in anthocyanins is consistent with the following hypotheses: (i) Anthocyanins somehow inhibit or repress the transfer of DOMFuc to the O-antigen terminus. (ii) At the same time, these seed compounds promote an increase in 2-O-methylation of internal Fuc residues. Both hypotheses are supported by the observed decrease in Fuc content that roughly matches the increase in 2MeFuc. If, instead, the change at the O-antigen terminus were to add a Fuc that was unmethylated or the increase in 2-O-MeFuc were due to the terminal Fuc not having the 3-O-methyl group, the Fuc content would not decrease. Moreover, the phenotype of CE395, including its sugar compositions after growth with and without anthocyanins, strongly supports the idea that the decrease in DOMFuc and the increase in 2MeFuc are two separate effects.
The methods of this study cannot answer the intriguing question of whether the additional methyl group is going to one particular Fuc residue in the O antigen. Indeed, the same question holds with regard to the average of one 2MeFuc per LPS in the wild type grown without anthocyanin. Once the experimental approach to answer this latter question is established with that LPS, which is much easier to obtain in large quantities, the issue should be revisited with regard to the LPS structure of bacteria induced by anthocyanins.
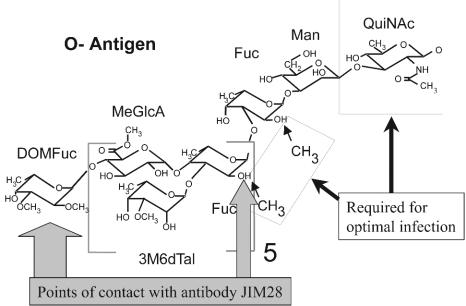
MAb JIM28 has been very useful in tracking the alteration of LPS induced by anthocyanin, low pH, and other environmental conditions (10, 23, 31). Analyses of the LPSs of mutants CE367 (11) and CE395 lead to the following conclusions regarding the epitope of antibody JIM28. (i) As 2-O-methylation of Fuc increases, binding of the antibody decreases. This correlation is illustrated by inspecting the sugar compositions (Table 4) and the immunoblots (Fig. 3) of CE3 and CE395 versus CE3α and CE395α. The greatest antibody-binding affinity is obtained when the O antigen lacks 2MeFuc altogether (as demonstrated by CE395 and CE395α395 [Fig. 1 and 2]). (ii) If DOMFuc is absent and the LPS is changed in no other way (e.g., in mutant CE367), binding of this antibody is eliminated (11). The anthocyanin effect, therefore, reduces binding in at least two ways, by increasing the incidence of 2MeFuc and greatly decreasing the content of the terminal O-antigen residue. These results suggest that the epitope may be at the terminus of the O antigen and that DOMFuc and the 2-hydroxyl portion of a Fuc residue are points of contact with the antibody binding site (Fig. 4). Interestingly, analysis of CE395 LPS after growth in anthocyanin indicates that the enhanced binding conferred by the absence of 2MeFuc overrides the loss of DOMFuc.
FIG. 4.
A model depicting hypotheses that three structural features of the R. etli CE3 O antigen affect symbiotic proficiency and/or binding to antibody JIM28.
As noted in the introduction, it is well established that absence of the O antigen in Lps mutants causes severe breakdowns in the infection process during the development of root nodule symbioses. However, it has not been determined at a cellular-biochemical level why the O antigen is required, nor is there much information regarding any specificity in this structural requirement. 2-O-methylation of Fuc residues, as affected by the lps-395 mutation, provides the second illustration of a specific LPS structural feature whose alteration may affect the symbiosis of R. etli with P. vulgaris. The first such illustration was the absence and/or alteration of the quinovosamine residue (Fig. 1) caused by the lpsQ166 mutation (15a, 24). In all other reported cases of Lps mutants affected in symbiosis, either several residues are missing from the LPS (generally at least the entire O antigen) or the specific structural effect is not known (20, 22). Although the mutants carrying mutation lpsQ166 or lps-395 (CE166 and CE395) are superior in this respect, they have complex LPS phenotypes that preclude simple interpretations of the biochemical causes of the biological effects. Nevertheless, both are subject to one approach that has helped in this regard: genetic suppression.
In wild-type R. etli CE3 bacteria, the LPS molecules that carry O antigen (LPS I) predominate over those that lack O antigen (LPS II) both in planta and ex planta, although the ratio of LPS I to LPS II varies in unexplained ways with culture age and conditions. When compared at a particular growth condition, strains CE166 and CE395 have about 40 and 50%, respectively, of the wild-type amount of LPS I relative to LPS II (24; Fig. 2 and 3 and Tables 3 and 4). This deficiency in LPS I can be suppressed extragenically in each case by introducing multiple copies of DNA from the R. etli CE3 lps region α. Since lpsQ lies outside of lps region α, the extragenic suppression of mutation lpsQ166 was achieved with the wild-type pLPSα, giving the strain designated CE166α. In the case of lps-395, however, it was necessary to introduce the lps-395 mutation on pLPSα to bring about extragenic suppression (in CE395α395) rather than complementation with wild-type alleles of the mutated gene (in CE395α). Each suppressed strain still has the same LPS sugar defect as the unsuppressed mutant: CE166α still lacks QuiN (24), and CE395α395 still lacks 2MeFuc.
Comparing these suppressed and unsuppressed mutants leads to the inference that the amount of LPS I relative to that of LPS II is crucial in the symbiosis. Even though CE166 and CE395 have about half the normal LPS I content, the consequence in symbiosis is the same as having no O antigen (no LPS I) at all. Pseudonodules rather than nodules result, and the rate of appearance of these pseudonodules is much lower than the rate of nodulation elicited by the wild type (i.e., the pseudonodules are relatively dispersed and continue to appear on lower roots, which in wild-type inoculations do not receive nodules). Although infection by CE395 has not been studied by microscopy, infection by CE166 was shown to be blocked at an early point, apparently the same point as that for mutants completely lacking the O antigen (24, 26). Ongoing experiments, in which R. etli is engineered to produce the O antigen of an R. leguminosarum strain in place of its normal O antigen, suggest that overcoming this block does not require a specific O-antigen structure (although, like CE166α and CE395α395, these hybrid strains infect more slowly) (L. Benziger, E. L. Kannenberg, and K. D. Noel, unpublished data). One hypothesis consistent with these results is that R. etli mutants lacking near-normal content of the O antigen are unable to cope with host defenses and/or compounds that become inhibitory to such mutants, and the inhibition becomes insurmountable very early in infection (12).
Having near-normal LPS I content, the suppressed strains do not suffer this block in infection. However, both strains elicit slower nodule development, lower specific nitrogenase activity, and fewer (more dispersed) nodules. Impaired infection is probably the basic defect; infection seems to be slowed from the very start and throughout nodule development. It is tempting to suggest that the symbiotic deficiencies of CE166α and CE395α395 are due to the specific LPS structural features they lack. However, neither of these strains is in all other respects exactly like the wild type. In the case of CE395α395, there is a good possibility that the Tn5 insertion has polar effects on genes that may affect the LPS in ways other than solely 2-O-methylation of Fuc. Nevertheless, its phenotype is consistent with the idea that specific O-antigen structures facilitate infection. This idea is also supported by the R. etli/R. leguminosarum hybrids mentioned above. These hybrids also lead to slower development of (more dispersed) nodules and nitrogenase activity. As one possibility discussed more fully elsewhere (22, 24), these hints of structural specificity in at least one role of the LPS could indicate that R. etli LPS interacts with a plant receptor that somehow promotes infection thread development. More specifically, the phenotypes of CE166α and CE395α395 could be explained if interaction with the hypothetical receptor were enhanced by features conferred by QuiNAc and a 2MeFuc residue (Fig. 4).
Acknowledgments
We thank Russell Carlson for helpful discussions and corroborating data, Scott Forsberg for advice and the gift of a series of methylated Fucs, and Stephanie Rebone and Andrew Zychowicz for research assistance.
This work was funded by grant DE-FG02-98ER20307 from the U.S. Department of Energy.
REFERENCES
- 1.Bhat, U. R., and R. W. Carlson. 1992. Chemical characterization of pH-dependent structural epitopes of lipopolysaccharides from Rhizobium leguminosarum biovar phaseoli. J. Bacteriol. 174:2230-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhat, U. R., L. S. Forsberg, and R. W. Carlson. 1994. Structure of lipid A component of Rhizobium leguminosarum bv. phaseoli lipopolysaccharide. J. Biol. Chem. 269:14402-14410. [PubMed] [Google Scholar]
- 3.Boyer, H. W., and D. Roulland-Dussoix. 1969. A complementation analysis of the restriction and modification of DNA in E. coli. J. Mol. Biol. 41:459-472. [DOI] [PubMed] [Google Scholar]
- 4.Carlson, R. W., S. Kalembassa, D. Turowski, P. Pachori, and K. D. Noel. 1987. Characterization of the lipopolysaccharide from a Rhizobium phaseoli mutant that is defective in infection thread development. J. Bacteriol. 169:4923-4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carlson, R. W., R. E. Sanders, C. Napoli, and P. Albersheim. 1978. Host-symbiont interactions. III. Purification and characterization of Rhizobium lipopolysaccharides. Plant Physiol. 62:912-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cava, J. R., P. M. Elias, D. A. Turowski, and K. D. Noel. 1989. Rhizobium leguminosarum CFN42 genetic regions encoding lipopolysaccharide structures essential for complete nodule development on bean plants. J. Bacteriol. 171:8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cava, J. R., H. Tao, and K. D. Noel. 1990. Mapping of complementation groups within a Rhizobium leguminosarum CFN42 chromosomal region required for lipopolysaccharide synthesis. Mol. Gen. Genet. 221:125-128. [Google Scholar]
- 8.Dazzo, F. B., G. L. Truchet, R. L. Hollingsworth, E. M. Hrabak, H. S. Pankratz, S. Philip-Hollingsworth, J. L. Salzwedel, K. Chapman, L. Appenzeller, A. Squartini, D. Gerhold, and G. Orgambide. 1991. Rhizobium lipopolysaccharide modulates infection thread development in white clover root hairs. J. Bacteriol. 173:5371-5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duelli, D. M. 1999. Plant signals and bacterial components induce modifications in Rhizobium lipopolysaccharide. Ph.D. dissertation. Marquette University, Milwaukee, Wis.
- 10.Duelli, D. M., and K. D. Noel. 1997. Compounds exuded by Phaseolus vulgaris that induce a modification of Rhizobium etli lipopolysaccharide. Mol. Plant-Microbe Interact. 10:903-910. [Google Scholar]
- 11.Duelli, D. M., A. Tobin, J. M. Box, V. S. Kumar Kolli, R. W. Carlson, and K. Dale Noel. 2001. Genetic locus required for antigenic maturation of Rhizobium etli CE3 lipopolysaccharide. J. Bacteriol. 183:6054-6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eisenschenk, L., R. Diebold, J. Perez-Lesher, A. C. Peterson, N. K. Peters, and K. D. Noel. 1994. Inhibition of Rhizobium etli polysaccharide mutants by Phaseolus vulgaris root compounds. Appl. Environ. Microbiol. 60:3315-3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finan, T. M., B. Kunkel, G. F. DeVos, and E. R. Singer. 1986. Second symbiotic megaplasmid in Rhizobium meliloti carrying exopolysaccharide and thiamine synthesis genes. J. Bacteriol. 167:66-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forsberg, L. S., U. R. Bhat, and R. W. Carlson. 2000. Structural characterization of the O-antigenic polysaccharide of the lipopolysaccharide from Rhizobium etli strain CE3. J. Biol. Chem. 275:18851-18863. [DOI] [PubMed] [Google Scholar]
- 15.Forsberg, L. S., and R. W. Carlson. 1998. The structures of the lipopolysaccharides from Rhizobium etli strains CE358 and CE359. The complete structure of the core region of R. etli lipopolysaccharides. J. Biol. Chem. 273:2747-2757. [DOI] [PubMed] [Google Scholar]
- 15a.Forsberg, L. S., K. D. Noel, J. Box, and R. W. Carlson. 2003. Genetic locus and structural characterization of the biochemical defect in the O-antigenic polysaccharide of the symbiotically deficient Rhizobium etli mutant CE166. J. Biol. Chem. 278:51347-51359. [DOI] [PubMed] [Google Scholar]
- 16.Fraysse, N., S. Jabbouri, M. Treilhou, F. Couderc, and V. Poinsot. 2002. Symbiotic conditions induce structural modifications of Sinorhizobium sp. NGR234 surface polysaccharides. Glycobiology 12:741-748. [DOI] [PubMed] [Google Scholar]
- 17.Hungria, M., C. M. Joseph, and D. A. Phillips. 1991. Anthocyanins and flavonols, major nod-gene inducers exuded naturally from seeds of a black-seeded common bean (Phaseolus vulgaris L.). Plant Physiol. 97:751-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kannenberg, E. L., and N. J. Brewin. 1989. Expression of a cell surface antigen from Rhizobium leguminosarum 3841 is regulated by oxygen and pH. J. Bacteriol. 171:4543-4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kannenberg, E. L., E. A. Rathbun, and N. J. Brewin. 1992. Molecular dissection of structure and function in the lipopolysaccharide of Rhizobium leguminosarum strain 3841 using monoclonal antibodies and genetic analysis. Mol. Microbiol. 6:2477-2487. [DOI] [PubMed] [Google Scholar]
- 20.Kannenberg, E. L., B. L. Reuhs, L. S. Forsberg, and R. W. Carlson. 1998. Lipopolysaccharides and K-antigens: their structures, biosynthesis, and functions, p. 119-154. In H. Spaink, A. Kondorosi, and P. Hooykaas (ed.), The Rhizobiaceae: molecular biology of model plant-associated bacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 21.Newman, J. D., B. W. Schultz, and K. D. Noel. 1992. Dissection of nodule development by supplementation of Rhizobium leguminosarum biovar phaseoli purine auxotrophs with 4-aminoimidazole-5-carboxamide riboside. Plant Physiol. 99:401-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Noel, K. D., and D. M. Duelli. 2000. Rhizobium lipopolysaccharide and its role in symbiosis, p. 415-431. In E. W. Triplett (ed.), Prokaryotic nitrogen fixation: a model system for analysis of a biological process. Horizon Scientific Press, Wymondham, United Kingdom.
- 23.Noel, K. D., D. M. Duelli, H. Tao, and N. J. Brewin. 1996. Antigenic change in the lipopolysaccharide of Rhizobium etli CFN42 induced by exudates of Phaseolus vulgaris. Mol. Plant-Microbe Interact. 9:180-186. [Google Scholar]
- 24.Noel, K. D, L. S. Forsberg, and R. W. Carlson. 2000. Varying the abundance of O antigen in Rhizobium etli and its effect on symbiosis with Phaseolus vulgaris. J. Bacteriol. 182:5317-5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Noel, K. D., A. Sanchez, L. Fernandez, J. Leemans, and M. A. Cevallos. 1984. Rhizobium phaseoli symbiotic mutants with transposon Tn5 insertions. J. Bacteriol. 158:148-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Noel, K. D., K. A. VandenBosch, and B. Kulpaca. 1986. Mutations in Rhizobium etli that lead to arrested development of infection threads. J. Bacteriol. 168:1392-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peters, N. K., J. W. Frost, and S. R. Long. 1986. A plant flavone, luteolin, induces expression of Rhizobium meliloti nodulation genes. Science 233:977-980. [DOI] [PubMed] [Google Scholar]
- 28.Que, N. L. S., S. Lin, R. J. Cotter, and C. R. H. Raetz. 2000. Purification and mass spectrometry of six lipid A species from the bacterial endosymbiont Rhizobium etli. J. Biol. Chem. 275:28006-28016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reuhs, B. L., J. S. Kim, A. Badgett, and R. W. Carlson. 1994. Production of cell-associated polysaccharides of Rhizobium fredii USDA205 is modulated by apigenin and host root extract. Mol. Plant-Microbe Interact. 7:240-247. [DOI] [PubMed] [Google Scholar]
- 30.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 31.Tao, H., N. J. Brewin, and K. D. Noel. 1992. Rhizobium leguminosarum CFN42 lipopolysaccharide antigenic changes induced by environmental conditions. J. Bacteriol. 174:2222-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.VandenBosch, K. A., K. D. Noel, Y. Kaneko, and E. H. Newcomb. 1985. Nodule initiation elicited by noninfective mutants of Rhizobium phaseoli. J. Bacteriol. 162:950-959. [DOI] [PMC free article] [PubMed] [Google Scholar]