Abstract
metaXCMS is a software program for the analysis of liquid chromatography/mass spectrometry-based untargeted metabolomic data that is designed to identify differences in metabolic profiles across multiple sample groups (e.g., “healthy” versus “active disease” versus “inactive disease”). By performing second-order (“meta”) analysis, the software facilitates prioritization of interesting metabolite features from large untargeted metabolomic datasets prior to the rate-limiting step of structural identification. Here we provide a detailed step-by-step protocol for going from raw mass spectrometry data to metaXCMS results visualized as Venn diagrams and exported Microsoft Excel spreadsheets. There is no upper limit to the number of sample groups or individual samples that can be compared by the software, and data from most commercial mass spectrometers is supported. The speed of the analysis depends on computational resources and data volume, but will generally be less than one day for most users. metaXCMS is freely available at http://metlin.scripps.edu/metaXCMS/.
INTRODUCTION
Metabolites are the biochemical end products of gene activity and therefore provide a functional readout of cellular phenotype.1–3 Untargeted metabolomics denotes the global and simultaneous profiling of as many metabolites as possible in a search to identify altered pathways that provide a phenotypic signature for the biological system of interest.4–7 The approach has been widely applied to elucidate biomarkers of disease, to discover new therapeutic targets, to assign unknown gene function, and to gain mechanistic insight into physiological processes in plants, yeast, bacteria, and mammals.8–13 Although historically much attention has been dedicated to the analysis of metabolites, until recently most studies focused on a relatively small number of compounds. Developments in high-resolution mass spectrometers, however, now enable the simultaneous detection of thousands of low-concentration species and have largely driven the field of global metabolic profiling over the course of the past ten years.14,15
As with any “omics” technology, the development of metabolomics has relied upon advances in bioinformatic tools that are required for analysis of the complex datasets generated. The analytical technique that has proven to be the most suitable for looking at the largest number of compounds is liquid chromatography/mass spectrometry (LC/MS).14,16 A typical LC/MS analysis of a metabolic extract from a biological tissue or fluid results in the detection of thousands of peaks, each with a unique m/z value and retention time.17,18 The first bioinformatic challenge in LC/MS-based metabolomics was comparing the intensity of each individual peak, known as metabolite features, across all of the samples measured. A complication is that the retention time of a particular metabolite can change slightly from one run to the next due to experimental drift. Deviations in retention time (e.g., from fluctuations in the room temperature, time-dependent changes in the sample, column degradation, etc.) are nonlinear and complicate the feature assignments that are used for correlation between samples.19 In 2005, a metabolomic program was developed called XCMS to identify dysregulated metabolite features between two sample groups by using a novel nonlinear retention-time alignment algorithm that does not require the addition of internal standards.17 XCMS is a freely available and platform-independent R-package that processes, analyzes, and visualizes LC/MS metabolomic data. XCMS is widely used in the field of untargeted metabolomics with over 350 citations of the original paper and more than 45,000 downloads as of 2011.
Although XCMS and other metabolomic programs that have been developed are well suited for the analysis of large sample numbers, the programs are limited in that they only compare two different sample groups directly.20,21 Manual comparisons of multiple sets of XCMS results have been performed, but these studies involve only a small number of sample groups and require additional analysis time.22 metaXCMS was developed to provide a tool for efficient meta-analysis of untargeted metabolomic datasets containing any number of sample groups.23 Meta-analysis can be defined as an approach that compares the results from two or more independently performed studies to identify data points that are unique or shared among all or some of the experimental groups.24 Figure 1 highlights the application of metaXCMS to identify unique and shared metabolite features that are dysregulated between three independent pairwise comparisons. Similar types of meta-analysis tools have been successfully applied in genome-wide association studies to investigate conditions with complex and heterogeneous phenotypes.25–27
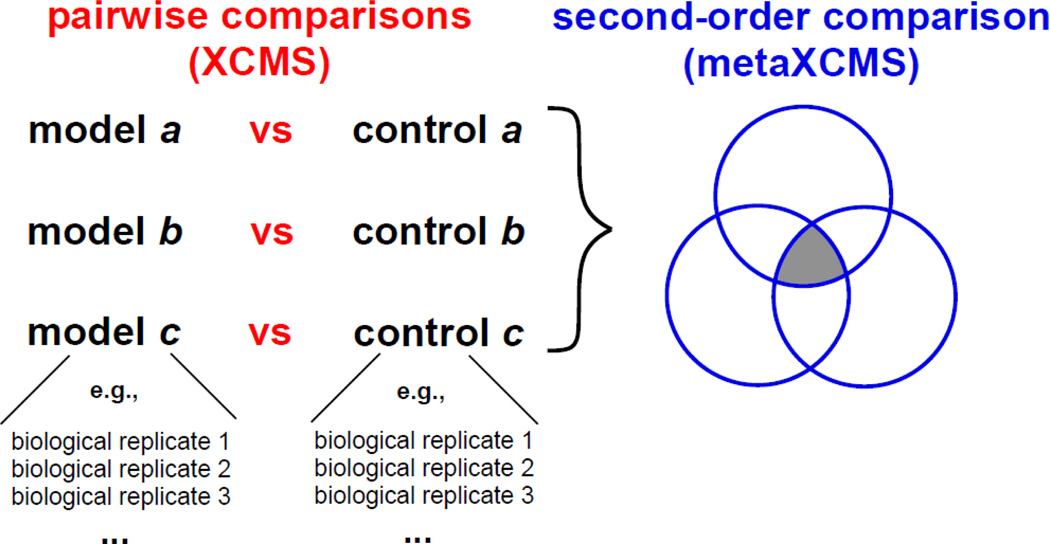
Figure 1.
Introduction of pairwise and second-order comparison. XCMS performs a pairwise comparison of two sample groups with any number of biological replicates. Data from multiple pairwise comparisons is then used by metaXCMS to perform a second-order comparison in which shared and unique differences are identified.
APPLICATIONS
To drive our understanding of chemical physiology, dysregulated metabolites and related cellular pathways need to be specifically correlated with unique biological processes or disease states. Often, however, an untargeted metabolomic analysis results in a significant number of altered metabolite features and it is a major challenge to differentiate molecules causally associated with the phenotype of interest from those that are altered as a downstream effect. Here metaXCMS provides a broadly applicable data-reduction strategy as we recently demonstrated in a study of three different mouse models of pain characterized by unique pathogenic etiology (Figure 2).23 Animals injected with Complete Freund’s Adjuvant were used as an inflammatory model, animals to which noxious heat was acutely applied to the hind paw were used as an acute heat model, and animals intraperitoneally injected with serum from K/BxN mice were used as a pain mode of spontaneous arthritis.28–30 Although the pairwise comparisons of each pain model with its respective control resulted in hundreds of altered metabolite features in total, we suspected that at least some of these molecules may be involved in triggering nociceptive transduction. The second-order analysis of the results with metaXCMS showed that only three of the altered molecules were shared among all of the models. We determined that one of the shared differences was the well-characterized pain mediator histamine, validating the value of the meta-analysis for identifying mechanistically relevant metabolites causally associated with the phenotype of interest. A comparable approach could be applied to any type of disease or stress model.
Figure 2.
Data reduction by meta-analysis. Three pairwise comparisons of different pain models to their respective controls resulted in 22,577 detected metabolite features (model A is animals plantar injected with Complete Freund’s Adjuvant, model B is animals treated with noxious heat, and model C is animals intraperitoneally injected with serum from K/BxN mice, for further details see Tautenhahn et al.23). Next, features with fold changes less than 1.5 and p-values greater than 0.05 were filtered and the remaining 1,825 features were plotted. A second-order comparison by metaXCMS showed that only 3 of these features were commonly shared, one of which was determined to be histamine.
As another example, we analyzed two knockout strains of Halobacterium salinarum. Specifically, knockout strains ΔVNG1816G and ΔVNG2094G were each compared to their parent control strain Δura3. The proteins encoded by VNG1816G and VNG2094G are known to affect glutamic acid metabolism.23,31,32 As expected, results from metaXCMS showed a feature similarly dysregulated in the pairwise comparison of each mutant to its control that was consistent with the accurate mass and retention time of glutamic acid (feature number 88, m/z 148.0606, retention time 5.8 min). The identity of glutamic acid was confirmed by comparing the retention time and MS/MS fragmentation pattern to that of a commercial standard. A truncated version of the XCMS files from each pairwise comparison is available for download as a test dataset at http://metlin.scripps.edu/data/metaXCMS/metaXCMS-testdata.zip. Expected results from performing the protocol described here are provided for comparison within the zip file.
metaXCMS also has broad applicability in the more clinical context of biomarker elucidation. Traditionally, metabolite biomarker discovery has been performed by comparing healthy to disease patients.33 Most disease states of interest, however, are exceedingly complex and highly variable from patient to patient at different stages of progression and severity with potentially different prognoses.34 In addition, there are a number of confounding variables that can be difficult to account for but that are known to influence metabolic profiles such as sex, age, diet, drug regimen, ethnicity, and body mass index.35 Given the relatively good throughput of LC/MS-based metabolomics, it has become readily practical to analyze thousands of human patient samples.10,11 metaXCMS may be applied to compare subgroup populations within these large cohorts to identify metabolic predictors of disease course (Figure 3) and potential risk factors related to other clinical variables. Additionally, metaXCMS analysis of phenotypically stratified subgroup populations similarly has utility in assessing drug efficacy. The comparison of subgroup difference profiles of patients on and off drug (e.g., “low blood pressure on drug” versus “low blood pressure off drug” compared to “high blood pressure on drug” versus “high blood pressure off drug”) will greatly facilitate the identification of variables affecting drug response and potential patients at risk of off-target effects.
Figure 3.
Visualization of theoretical meta-analysis applied to identify biomarkers of disease severity. The left Venn diagram shows shared and unique metabolite features for mild disease, severe disease, and healthy patients. While features in the areas labeled a, b, and c may serve as biomarkers, areas a and c could provide additional markers specific to mild and severe disease respectively. The right Venn diagram shows a second-order visualization of the same comparison that is representative of metaXCMS output when the parameters are set to plot only metabolite features that are unique to disease (i.e., features that are detected in disease but not in healthy samples). The advantage of the second-order visualization is that it is not limited to representing only metabolites unique to a certain sample group. Rather, metabolites that are up- and down-regulated by even small fold changes can be easily represented according to user-defined thresholds. Given that biomarkers may not be metabolites unique to disease samples but instead metabolites that increase by some quantified fold change, second-order visualizations are generally better suited for metabolomic data since they can be used to show up- and down-regulated features (see Venn diagram in Figure 2). Changing the second-order visualization here to include features with smaller fold changes, for example, would result in the display of more features that might represent useful diagnostic markers.
EXPERIMENTAL DESIGN
Although untargeted metabolomics is generally hypothesis generating as opposed to hypothesis driven, it is important to carefully construct an experimental design to assure that the results have value given the significant effort and time that will be required for data analysis. Generally, the rate-limiting step in the untargeted metabolomic workflow is structural identification of metabolites.36 While the untargeted profiling analysis provides the accurate mass of altered features between sample groups, these data must then be searched in metabolite libraries and structurally characterized by comparison of retention time and tandem MS data to that of standard model compounds. Thus, pairwise comparisons that yield hundreds of altered features can be challenging in requiring significant effort and resources for identification. The incorporation of additional physiologically meaningful sample groups into the experimental design, however, can result in a reduced list of interesting features. Importantly, this data reduction by meta-analysis is at the feature level prior to the rate-limiting step of structure determination. metaXCMS therefore has the potential to improve the overall throughput and efficiency of untargeted studies by prioritizing features to be identified that have a high likelihood of being biologically relevant.
Two broadly applicable experimental designs utilizing metaXCMS that have been introduced here include: (i) the comparison of different variations of a disease or stress model to identify shared metabolic alterations related to a mechanistically fundamental response, and (ii) the comparison of phenotypically stratified patient cohorts to deconvolute metabolic responses associated with specific clinical variables and disease heterogeneity. Many other context-dependent applications are also conceivable, but in all cases certain experimental conditions should be followed for best results. First, the samples should be prepared by using the same metabolite-extraction method. Different extraction methods may lead to the removal of different metabolites and thereby introduce artificial differences into the comparison.37 Additionally, because metaXCMS correlates peaks on the basis of m/z values and retention time, all samples being compared should be analyzed by using the same column and chromatographic method. These experimental requirements are limiting in that meta-comparisons of metabolomic analyses from different laboratories are likely to be unreliable. Although such inter-laboratory comparisons have intriguing potential, the protocol described here was not designed for that purpose. It should be noted, however, that meta-analyses from different laboratories should in principle provide the same profile of shared differences despite potential alterations in the retention times of specific compounds across different laboratories.
MATERIALS
Equipment
Hardware requirements
A personal computer with at least 2 GB of RAM. A multi-core processor with at least 2 GB of RAM per core is recommended for the processing of large files/sample groups.
Sufficient hard-drive storage space for raw data files, converted files, and results.
Software requirements
For sample conversion : 32 or 64 bit versions of Windows operating system (XP, Vista, Windows 7).
For XCMS and metaXCMS analysis: any 32 or 64 bit version of Windows, Unix operating system, or Mac OS X (release 10.5 and above) can be used. However, since most 32 bit operating systems cannot allocate more than 2 GB of RAM, 64 bit operating systems are recommended for working with large files/sample groups.
Equipment Setup
Install metaXCMS as described on (http://metlin.scripps.edu/metaxcms/download.php). XCMS will be automatically installed during the installation of metaXCMS.
Download and install ProteoWizard (http://proteowizard.sourceforge.net). CRITICAL: Download the ProteoWizard version that includes vendor reader support.
PROCEDURE
Figure 4 shows an overview of the meta-analysis workflow. The process of file conversion, feature detection and alignment, and second-order analysis is described below step-by-step. For details on data acquisition, see Want et al.38. For more details on result browsing and interpretation, see Smith et al.17 and Tautenhahn et al.23.
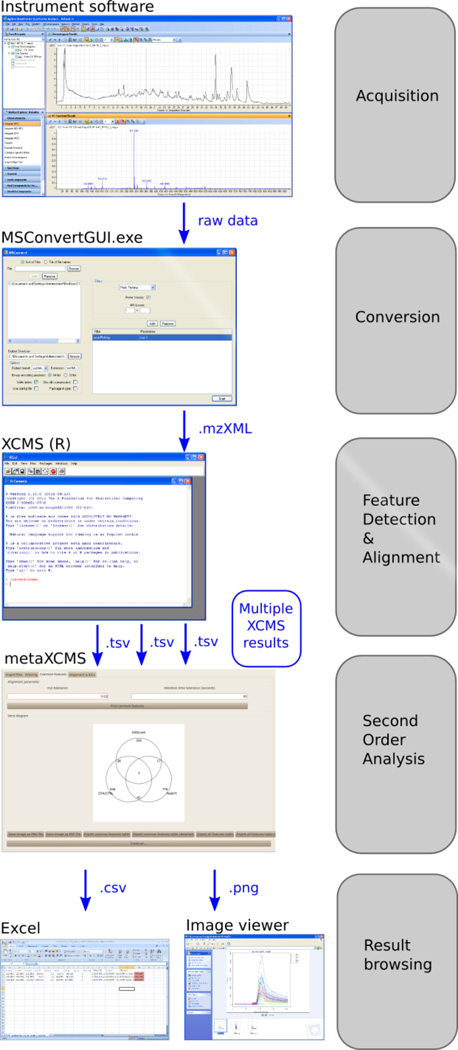
Figure 4.
Overview of the computational workflow. The workflow consists of five stages: acquisition of LC/MS data, conversion of the data to .mzXML files, analysis of the files by XCMS, analysis of XCMS results by metaXCMS, and result browsing and interpretation.
Conversion of vendor-format data files to mzXML
-
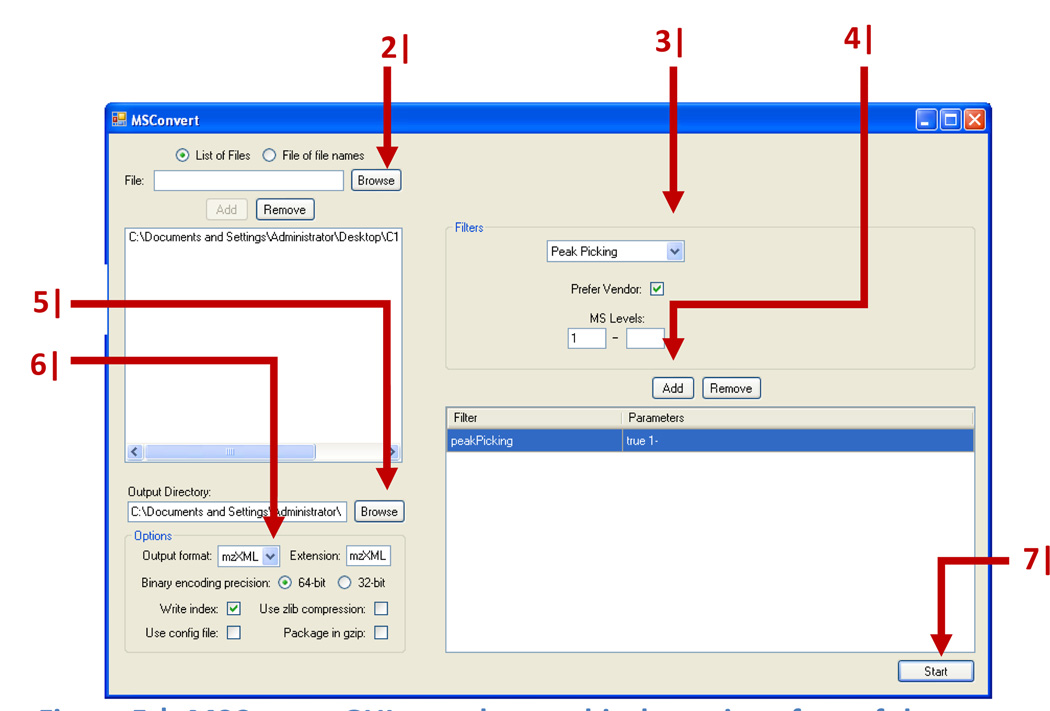
1|
Locate MSConvertGUI.exe in the ProteoWizard folder and run it by double-clicking to call up the graphical user interface as shown in Figure 5.
-
2|
Click ‘Browse’. Select the raw data files to convert. Multiple files can be selected at once.
CRITICAL: Proteowizard currently supports the conversion of Agilent, Applied Biosystems, Bruker, Thermo Fisher, and Waters data files (see http://proteowizard.sourceforge.net/formats.shtml for information about other file formats).
-
3|
Click the filter selection dialog. Select ‘Peak Picking’. Make sure ‘Prefer Vendor’ is activated.
-
4|
Click the ‘Add’ button to add peak picking to the filter list. This will make sure the resulting files are in centroid mode, which is a requirement for the subsequent feature detection.
-
5|
Click ‘Browse’ to select the output directory. Select ‘mzXML’ as output format.
-
6|
Click ‘Start’ to begin file conversion.
Figure 5.
MSConvertGUI.exe, the graphical user interface of the ProteoWizard file converter. The input fields or icons of the software that correspond to specific steps in the protocol are indicated by the step number.
Pairwise Comparison by using XCMS
-
7|
Organize mzXML files in folders. Create a folder for each pairwise comparison. Inside of this folder, create a subfolder for each group. Move all mzXML files that were acquired for the respective sample group into the corresponding folder. For example, make a folder “variationA_vs_controlA” that contains subfolders for each group “variation” and “controlA” where the individual mzXML files are copied.
-
8|
Run R and load the XCMS package.
library(xcms)
-
9|
Set the R working directory to the folder containing the files for the first pairwise comparison, for example setwd(“C:/Data/variationA_vs_controlA”)
CRITICAL: R uses the Unix-style forward slashes / as path separators on Windows operating systems, single backslashes \ do not work.
-
10|
Start the feature detection using the “centWave” method.39
xset <- xcmsSet(method="centWave",ppm =30, peakwidth=c(10,60), prefilter=c(0,0)) If you use a PC with multiple cores, add the argument nSlaves and specify the number of cores (e.g., xset <- xcmsSet(method="centWave", nSlaves=2) for a PC with 2 cores).
-
11|
Perform retention-time correction using the “OBIwarp” method.40
xset1 <- retcor(xset, method="obiwarp", plottype = c("deviation")) The retention time correction curves should be displayed as represented in Figure 6.
-
12|
Group features together across samples.
xset2 <- group(xset1, bw = 5, minfrac = 0.5, mzwid = 0.025)
-
13|
Fill in missing peaks, calculate statistics, generate feature table and extracted ion chromatograms.
xset3 <- fillPeaks(xset2) dr <- diffreport(xset3, filebase="variationA_vs_controlA", eicmax=100)
Use the name of your pairwise comparison for filebase. Output files and folders will be generated with that name.
The parameters used above are optimized for HPLC with ~60min gradient and high-resolution Q-TOF. Suggested parameter settings for most common experimental setups are show in Table 1.
-
14|
Close R session.
q(“no”).
-
15|
Repeat steps 8–14 for each one of the pairwise comparisons.
CRITICAL: Do not rename or move the data folders and do not rename columns in the XCMS result table after XCMS processing. This will make metaXCMS unable to process the XCMS results.
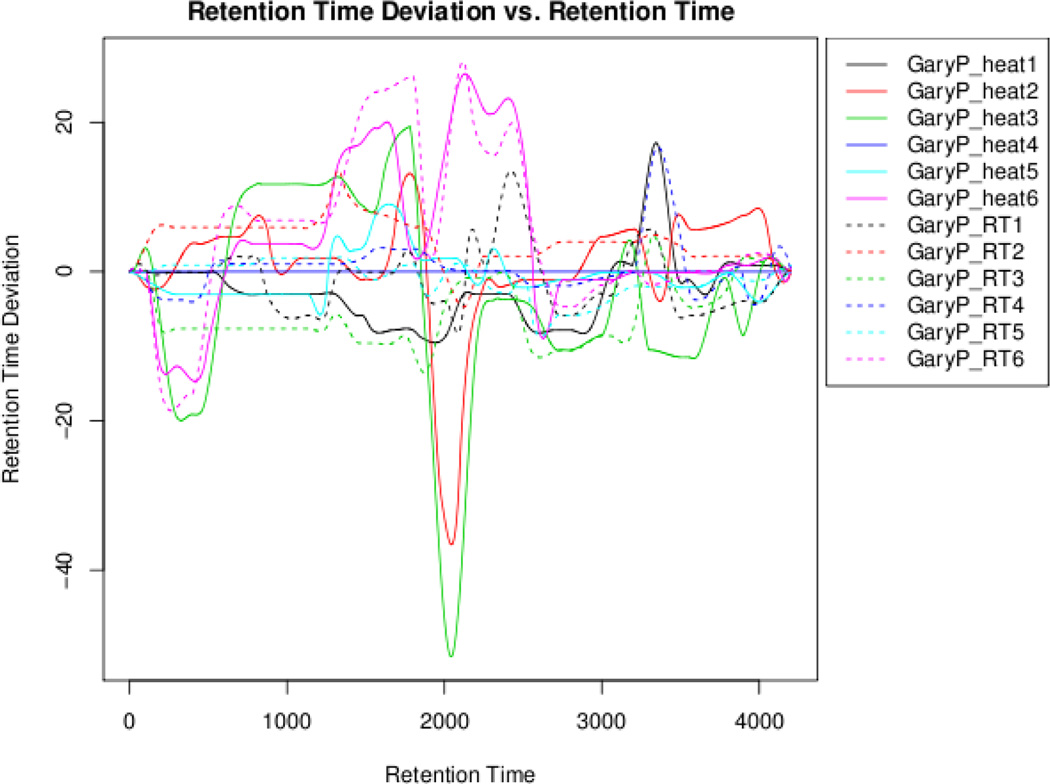
Figure 6.
Retention-time correction curves generated by XCMS. Each colored line represents a different sample processed. Note that the retention-time deviation is different for each sample and that it is not linear.
Table 1.
XCMS Parameter settings
| Experimental setup |
Parameter name and explantation | ||||
|---|---|---|---|---|---|
| ppm | peakwidth | bw | mzwid | prefilter | |
| maximum m/z deviation in consecutive scans in ppm |
chromatographic peak width range in seconds |
measure of retention time tolerance for peak grouping across samples |
width of overlapping m/z slices for grouping peaks across samples |
Intensity prefilter (k,I): features need to have at least k peaks with intensity >= I |
|
| HPLC/QTOF | 30 | c(10,60) | 5 | 0.025 | c(0,0) |
| HPLC/QTOF (high resolution) | 15 | c(10,60) | 5 | 0.015 | c(0,0) |
| HPLC/Orbitrap | 2.5 | c(10,60) | 5 | 0.015 | c(3,5000) |
| UPLC/QTOF | 30 | c(5,20) | 2 | 0.025 | c(0,0) |
| UPLC/QTOF (high resolution) | 15 | c(5,20) | 2 | 0.015 | c(0,0) |
| UPLC/Orbitrap | 2.5 | c(5,20) | 2 | 0.015 | c(3,5000) |
Meta-Analysis by using metaXCMS
-
16|
Run R and load the metaXCMS package (Figure 7).
library(metaXCMS)
-
17|
Click ‘Import XCMS diffreport’. Navigate to the folder that contains the results from one of the pairwise comparisons and open the .tsv file (e.g., variationA_vs_controlA.tsv).
-
18|
Repeat step 17 until all of the results from the pairwise comparisons are loaded.
-
19|
Verify that sample classes have been assigned correctly by clicking on the filename of one of the pairwise comparisons on the left side of the window to select it. All sample names and the automatically assigned sample classes of that comparison are displayed on the right side of the window. If sample classes are assigned incorrectly, double-click and select the correct sample class from the pull-down menu.
-
20|
Verify that the control sample class is assigned correctly. The sample class that is used as a control for each pairwise comparison is shown in the column ‘Control is’. If the assignment is incorrect, double-click and select the correct sample class from the pulldown menu. This will make sure metaXCMS can correctly display and filter up- and down-regulated features.
-
21|
Click ‘Continue’ to view filtering options (Figure 8).
-
22|
Select fold change and p-value thresholds for filtering. Note: p-values are calculated in XCMS by performing a Welch’s t-test for unequal variances.
-
23|
Click ‘Apply filter’. The number of remaining features (green) will be updated.
-
24|
If only up- or down-regulated features should be used from a pairwise comparison, click on ‘all’ and select ‘UP’ or ‘DOWN’ from the pull-down menu. Click ‘Apply filter’.
-
25|
If features from a pairwise comparison should be subtracted from the result (e.g., such as features altered in a sham control) select the “subtract from result” checkbox.
-
26|
Click ‘Continue’.
-
27|
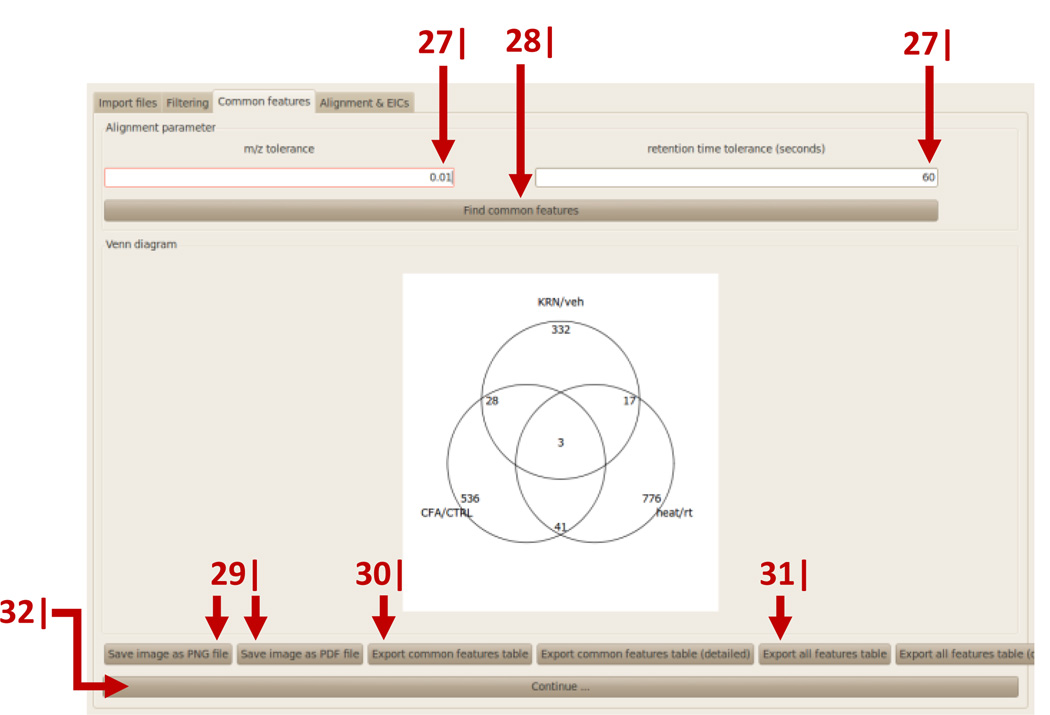
Adjust acceptable m/z and retention-time tolerance. Default values for HPLC/Q-TOF are 0.01 and 60 seconds.
-
28|
Click ‘Find common features’. After the alignment has been calculated, a Venn diagram with the numbers of unique and common features between the pairwise comparisons will be shown (Figure 9).
-
29|
Save the Venn diagram as a .png or .pdf file.
-
30|
To export a table with only the common features, click ‘Export common features table’.
-
31|
To export a table with all features (unique, common, and shared), click ‘Export all features table’.
-
32|
Click ‘Continue’.
-
33|
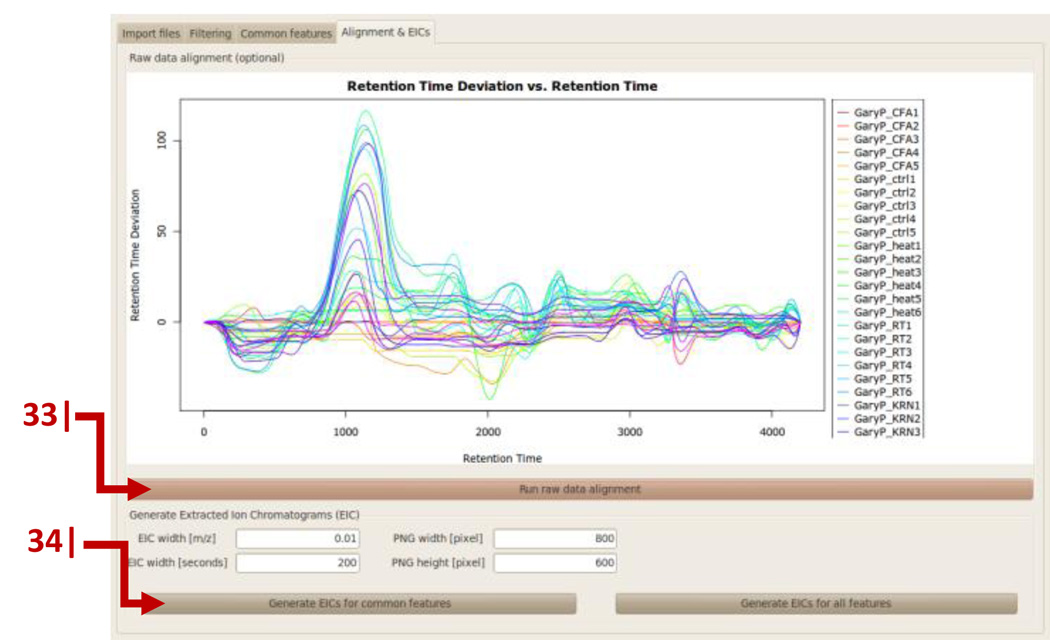
Click ‘Run Raw Data Alignment’. Retention-time correction for all samples will be recalculated (Figure 10).
-
34|
Click ‘Generate EICs for common features’. After an output folder for the extracted ion chromatograms (EIC) has been selected, EICs will be generated for all common features by using the data from all samples.
Figure 7.
Graphical user interface of metaXCMS. The input fields or icons related to the import of XCMS diffreports are indicated by arrows that are numbered according to the protocol step in which they are described.
Figure 8.
Graphical user interface of metaXCMS. Filtering may be performed on the basis of p-value, fold change, and up-/down-regulation. The input fields or icons related to filtering are indicated by arrows that are numbered according to the protocol step in which they are described.
Figure 9.
Graphical user interface of metaXCMS. Features that are uniquely or commonly altered among the pairwise comparisons are displayed as Venn diagrams. The icons related to data visualization and export are indicated by arrows that are numbered according to the protocol step in which they are described.
Figure 10.
Graphical user interface of metaXCMS. Retention-time correction for all samples compared is displayed and EICs are generated. The icons related to retention-time correction and EIC generation are indicated by arrows that are numbered according to the protocol step in which they are described.
TIMING: Depending on the number of samples and the file size, steps 33 and 34 can take up to 1–2 hours each. Also, depending on the number of CPU cores used, XCMS processing (step 10–13) typically takes 15 min - 2 hr per pairwise comparison.
ANTICIPATED RESULTS
metaXCMS will create tables with all features (unique, common, and shared) in CSV format (comma separated values). The files can be opened by Microsoft Excel or Open Office and displayed as spreadsheets. Each row of the spreadsheet will correspond to a feature and list m/z values as well as retention-time values in addition to fold changes and p-values (as originally calculated by XCMS) for each of the pairwise comparisons. EIC’s will be generated for each feature in the table and can be used for visual inspection. It is important to note that metaXCMS does not provide metabolite identifications. To identify interesting features, generally accurate masses are first searched in metabolite databases for making putative metabolite assignments. The putative assignments are subsequently confirmed by additional experiments comparing retention time and tandem MS data to that of model standards.
Table 2.
Troubleshooting table
| Step | Problem | Possible Reason | Solution |
|---|---|---|---|
| 10 | Error in xcmsSet(method = "centWave") : No NetCDF/mzXML/mzData/mzML files were found. | Working directory does not contain any LC/MS raw data files | Make sure the correct folder is selected |
| 10–13, 17–34 | Error: cannot allocate vector of size X Mb | Insufficient RAM | Upgrade RAM, use 64 bit operating system |
| 33 | Cannot find the raw data files for X ? | Raw data files have been deleted or moved | Do not rename or move the data folders and do not rename columns in the XCMS result table after XCMS processing |
| 16 | Error in inDL(x, as.logical(local), as.logical(now), …) : unable to load shared object 'C:/Program Files/[…]/cairoDevice.dll': | GTK+ is not installed | Install GTK+ as described on http://metlin.scripps.edu/metaxcms/download.php |
| 16 | Error in inDL(x, as.logical(local), as.logical(now), …) : unable to load shared object 'C:/Program Files/[…]/RGtk2.dll': | ||
| Any | other | Please describe the problem in the XCMS/metaXCMS user forum http://metlin.scripps.edu/xcms/faq.php |
BOX 1| NUMBER OF SAMPLES PER SAMPLE GROUP.
Currently, there is no consensus in the field with respect to the minimal number of samples that should be included per sample group for an untargeted metabolomic analysis. Similarly, different p-value and fold-change cutoffs are used depending on the biological system under investigation, the methods used for metabolite extraction, and the analytical platform employed. Studies have shown that instrument variability is smaller than biological variability for mammals and suggested that lower-limit fold-change thresholds of 1.5–2.0 be used.41,42 These lower-limit fold-change thresholds from individual pairwise comparisons are likely appropriate thresholds to be used for meta-analysis. Although XCMS and metaXCMS can be used to analyze groups with as few as 2 samples, typically larger sample groups are needed due to intergroup biological variability. It should also be noted that it may be appropriate to apply a statistical correction for multiple comparisons (e.g., a Bonferroni correction) to metaXCMS results depending on experimental design. These additional statistical tests are context-dependent and should be performed manually after metaXCMS analysis when appropriate.
REFERENCES
- 1.Weckwerth W. Unpredictability of metabolism-the key role of metabolomics science in combination with next-generation genome sequencing. Anal Bioanal Chem. 2011;400:1967–1978. doi: 10.1007/s00216-011-4948-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker M. Metabolomics: from small molecules to big ideas. Nat Meth. 2011;8:117–121. [Google Scholar]
- 3.Yanes O, et al. Metabolic oxidation regulates embryonic stem cell differentiation. Nature chemical biology. 2010;6:411–417. doi: 10.1038/nchembio.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wikoff WR, et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A. 2009;106:3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wikoff WR, Pendyala G, Siuzdak G, Fox HS. Metabolomic analysis of the cerebrospinal fluid reveals changes in phospholipase expression in the CNS of SIV-infected macaques. J Clin Invest. 2008;118:2661–2669. doi: 10.1172/JCI34138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vinayavekhin N, Saghatelian A. Untargeted metabolomics. Curr Protoc Mol Biol. 2010;Chapter 30(Unit 30):31–31–24. doi: 10.1002/0471142727.mb3001s90. [DOI] [PubMed] [Google Scholar]
- 7.Wikoff WR, Nagle MA, Kouznetsova VL, Tsigelny IF, Nigam SK. Untargeted Metabolomics Identifies Enterobiome Metabolites and Putative Uremic Toxins as Substrates of Organic Anion Transporter 1 (Oat1) J Proteome Res. 2011 doi: 10.1021/pr200093w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vinayavekhin N, Homan EA, Saghatelian A. Exploring disease through metabolomics. ACS Chem Biol. 2010;5:91–103. doi: 10.1021/cb900271r. [DOI] [PubMed] [Google Scholar]
- 9.McKnight SL. On Getting There from Here. Science. 2010;330:1338–1339. doi: 10.1126/science.1199908. [DOI] [PubMed] [Google Scholar]
- 10.Wang TJ, et al. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011;17:448–453. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Z, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dang L, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olszewski KL, et al. Branched tricarboxylic acid metabolism in Plasmodium falciparum. Nature. 2010;466:774–778. doi: 10.1038/nature09301. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Fernie AR, Trethewey RN, Krotzky AJ, Willmitzer L. Metabolite profiling: from diagnostics to systems biology. Nat Rev Mol Cell Biol. 2004;5:763–769. doi: 10.1038/nrm1451. [DOI] [PubMed] [Google Scholar]
- 15.Lu W, et al. Metabolomic analysis via reversed-phase ion-pairing liquid chromatography coupled to a stand alone orbitrap mass spectrometer. Anal Chem. 2010;82:3212–3221. doi: 10.1021/ac902837x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buscher JM, Czernik D, Ewald JC, Sauer U, Zamboni N. Cross-platform comparison of methods for quantitative metabolomics of primary metabolism. Anal Chem. 2009;81:2135–2143. doi: 10.1021/ac8022857. [DOI] [PubMed] [Google Scholar]
- 17.Smith CA, Want EJ, O'Maille G, Abagyan R, Siuzdak G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem. 2006;78:779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- 18.Yanes O, Tautenhahn R, Patti GJ, Siuzdak G. Expanding coverage of the metabolome for global metabolite profiling. Analytical chemistry. 2011;83:2152–2161. doi: 10.1021/ac102981k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Want EJ, et al. Solvent-dependent metabolite distribution, clustering, and protein extraction for serum profiling with mass spectrometry. Anal Chem. 2006;78:743–752. doi: 10.1021/ac051312t. [DOI] [PubMed] [Google Scholar]
- 20.Lommen A. MetAlign: interface-driven, versatile metabolomics tool for hyphenated full-scan mass spectrometry data preprocessing. Anal Chem. 2009;81:3079–3086. doi: 10.1021/ac900036d. [DOI] [PubMed] [Google Scholar]
- 21.Katajamaa M, Miettinen J, Oresic M. MZmine: toolbox for processing and visualization of mass spectrometry based molecular profile data. Bioinformatics. 2006;22:634–636. doi: 10.1093/bioinformatics/btk039. [DOI] [PubMed] [Google Scholar]
- 22.Böttcher C, et al. The multifunctional enzyme CYP71B15 (PHYTOALEXIN DEFICIENT3) converts cysteine-indole-3-acetonitrile to camalexin in the indole-3-acetonitrile metabolic network of Arabidopsis thaliana. Plant Cell. 2009;21:1830–1845. doi: 10.1105/tpc.109.066670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tautenhahn R, et al. metaXCMS: second-order analysis of untargeted metabolomics data. Anal Chem. 2011;83:696–700. doi: 10.1021/ac102980g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Normand SL. Meta-analysis: formulating, evaluating, combining, and reporting. Stat Med. 1999;18:321–359. doi: 10.1002/(sici)1097-0258(19990215)18:3<321::aid-sim28>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 25.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Bakker PI, et al. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum Mol Genet. 2008;17:R122–R128. doi: 10.1093/hmg/ddn288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sterk P, Hirschman L, Field D, Wooley J. Genomic standards consortium workshop: metagenomics, metadata and metaanalysis (m3) Pac Symp Biocomput. 2010:481–484. [PubMed] [Google Scholar]
- 28.Chu YC, et al. Effect of genetic knockout or pharmacologic inhibition of neuronal nitric oxide synthase on complete Freund's adjuvant-induced persistent pain. Pain. 2005;119:113–123. doi: 10.1016/j.pain.2005.09.024. [DOI] [PubMed] [Google Scholar]
- 29.Bolcskei K, Petho G, Szolcsanyi J. Noxious heat threshold measured with slowly increasing temperatures: novel rat thermal hyperalgesia models. Methods Mol Biol. 2010;617:57–66. doi: 10.1007/978-1-60327-323-7_5. [DOI] [PubMed] [Google Scholar]
- 30.Kyburz D, Corr M. The KRN mouse model of inflammatory arthritis. Springer Semin Immunopathol. 2003;25:79–90. doi: 10.1007/s00281-003-0131-5. [DOI] [PubMed] [Google Scholar]
- 31.Goo YA, et al. Proteomic analysis of an extreme halophilic archaeon, Halobacterium sp. NRC-1. Mol Cell Proteomics. 2003;2:506–524. doi: 10.1074/mcp.M300044-MCP200. [DOI] [PubMed] [Google Scholar]
- 32.Kaur A, et al. A systems view of haloarchaeal strategies to withstand stress from transition metals. Genome Res. 2006;16:841–854. doi: 10.1101/gr.5189606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Branca F, Hanley AB, Pool-Zobel B, Verhagen H. Biomarkers in disease and health. Br J Nutr. 2001;86(Suppl 1):S55–S92. doi: 10.1079/bjn2001339. [DOI] [PubMed] [Google Scholar]
- 34.Cantor RM, Lange K, Sinsheimer JS. Prioritizing GWAS results: A review of statistical methods and recommendations for their application. Am J Hum Genet. 2010;86:6–22. doi: 10.1016/j.ajhg.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crews B, et al. Variability analysis of human plasma and cerebral spinal fluid reveals statistical significance of changes in mass spectrometry-based metabolomics data. Anal Chem. 2009;81:8538–8544. doi: 10.1021/ac9014947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kalisiak J, et al. Identification of a new endogenous metabolite and the characterization of its protein interactions through an immobilization approach. J Am Chem Soc. 2009;131:378–386. doi: 10.1021/ja808172n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yanes O, Tautenhahn R, Patti GJ, Siuzdak G. Expanding coverage of the metabolome for global metabolite profiling. Anal Chem. 2011;83:2152–2161. doi: 10.1021/ac102981k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Want EJ, Nordstrom A, Morita H, Siuzdak G. From exogenous to endogenous: the inevitable imprint of mass spectrometry in metabolomics. J Proteome Res. 2007;6:459–468. doi: 10.1021/pr060505+. [DOI] [PubMed] [Google Scholar]
- 39.Tautenhahn R, Böttcher C, Neumann S. Highly sensitive feature detection for high resolution LC/MS. BMC Bioinformatics. 2008;9:504. doi: 10.1186/1471-2105-9-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prince JT, Marcotte EM. Chromatographic alignment of ESI-LC-MS proteomics data sets by ordered bijective interpolated warping. Anal Chem. 2006;78:6140–6152. doi: 10.1021/ac0605344. [DOI] [PubMed] [Google Scholar]
- 41.Crews B, et al. Variability analysis of human plasma and cerebral spinal fluid reveals statistical significance of changes in mass spectrometry-based metabolomics data. Analytical chemistry. 2009;81:8538–8544. doi: 10.1021/ac9014947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Masson P, Spagou K, Nicholson JK, Want EJ. Technical and biological variation in UPLC-MS-based untargeted metabolic profiling of liver extracts: application in an experimental toxicity study on galactosamine. Anal Chem. 2011;83:1116–1123. doi: 10.1021/ac103011b. [DOI] [PubMed] [Google Scholar]