Abstract
Mutations in VCP cause multisystem degeneration impacting the nervous system, muscle, and/or bone. Patients may present with ALS, Parkinsonism, frontotemporal dementia, myopathy, Paget’s disease or a combination of these. The disease mechanism is unknown. We developed a Drosophila model of VCP mutation-dependent degeneration. The phenotype is reminiscent of PINK1 and parkin mutants, including a pronounced mitochondrial defect. Indeed, VCP interacts genetically with the PINK1/parkin pathway in vivo. Paradoxically, VCP complements PINK1 deficiency but not parkin deficiency. The basis of this paradox is resolved by mechanistic studies in vitro showing that VCP recruitment to damaged mitochondria requires Parkin-mediated ubiquitination of mitochondrial targets. VCP recruitment coincides temporally with mitochondrial fission, and VCP is required for proteasome-dependent degradation of Mitofusins in vitro and in vivo. Further, VCP and its adaptor Npl4/Ufd1 are required for clearance of damaged mitochondria via the PINK1/Parkin pathway, and this is impaired by pathogenic mutations in VCP.
Introduction
Mutations in VCP cause a dominantly inherited, multisystem degenerative disease that affects muscle, bone, and brain. This condition has been called “IBMPFD” to reflect the clinical manifestations of inclusion body myopathy (IBM), frontotemporal dementia (FTD), and Paget’s disease of bone (PDB) in affected families (Watts et al., 2004). More recently the term multisystem proteinopathy (MSP) has been adopted for this disorder to reflect the expanding phenotypic spectrum of VCP-related diseases which include sporadic or familial ALS (Abramzon et al., 2012; Johnson et al., 2010), hereditary spastic paraplegia (de Bot et al., 2012), parkinsonism (Kimonis et al., 2008; Spina et al., 2012), or Parkinson’s disease (Spina et al., 2012). Thus, mutations in a single gene can manifest as any of several, common, age-related degenerative diseases. There does not appear to be genotype-phenotype correlation to account for these different clinical manifestations (Ju and Weihl, 2010a; Mehta et al., 2012). Indeed, the striking pleiotropy associated with VCP mutations is frequently observed within single pedigrees where individuals share not only the same missense mutation but also much genetic background in common. The mechanism whereby mutations in VCP cause disease is unknown, as is the basis for the phenotypic pleiotropy.
VCP is a type II member of the AAA+ (ATPase Associated with diverse cellular Activities) family of proteins (Jentsch and Rumpf, 2007). VCP functions in a plethora of processes, including cell cycle regulation, DNA repair, organelle biogenesis, proteotoxic stress response, endoplasmic reticulum-associated degradation, endolysosomal sorting and autophagosome biogenesis and maturation (Braun et al., 2002; Jentsch and Rumpf, 2007; Ju and Weihl, 2010b; Krick et al., 2010; Rabinovich et al., 2002; Ritz et al., 2011; Tresse et al., 2010; Ye et al., 2001). VCP functions as a “segregase” that extracts ubiquitinated proteins from multimeric complexes or structures for recycling or degradation by the proteasome (Ye et al., 2005). The diversity in VCP activities reflects its ability to interact with a diverse array of adaptor proteins via the N-domain, which in turn enables VCP to interact specifically with a broad array of substrates. The conformation of VCP’s N-domain is regulated allosterically by the status of nucleotide occupancy (ATP vs. ADP) in the nucleotide binding pocket (Tang et al., 2010). Thus, ATP hydrolysis in the D1 domain permits VCP to adopt distinct conformations and interact with distinct subsets of adaptors. All disease-causing mutations in VCP are missense mutations at the interface between the N-domain and the adjacent D1 ATPase domain, which abuts the nucleotide binding pocket in D1. Indeed, crystallographic analysis has confirmed that disease mutations alter the shape of the nucleotide binding pocket, alter nucleotide loading, and impair the normal reordering of the N-domain structure that permits cycling between alternative conformations (Tang et al., 2010). As a consequence, the balance of VCP-adaptor interactions is altered, enhancing the interaction of VCP with some adaptors while diminishing interaction with others (Fernandez-Saiz and Buchberger, 2010).
The central question concerning the pathogenesis of VCP-related disease is which functions of VCP are altered by disease-causing mutations? To address this question in an unbiased way, we generated a Drosophila model that captures VCP mutation-dependent degeneration. Some aspects of this Drosophila model are reminiscent of the phenotypes observed in PINK1 and parkin mutant flies. Indeed, we demonstrate genetic interactions that place VCP downstream of the PINK1/Parkin pathway in vivo. Mechanistic studies in vitro reveal that VCP is recruited to mitochondria in a manner that requires Parkin-dependent ubiquitination of mitochondrial proteins. Moreover, VCP is essential to the regulated degradation of membrane proteins, including Mitofusins, and clearance of damaged mitochondria. Most importantly, these studies reveal that this function of VCP is impaired by pathogenic mutations.
Results
Recapitulation of VCP mutation-dependent phenotypes in motor neurons and muscle in Drosophila
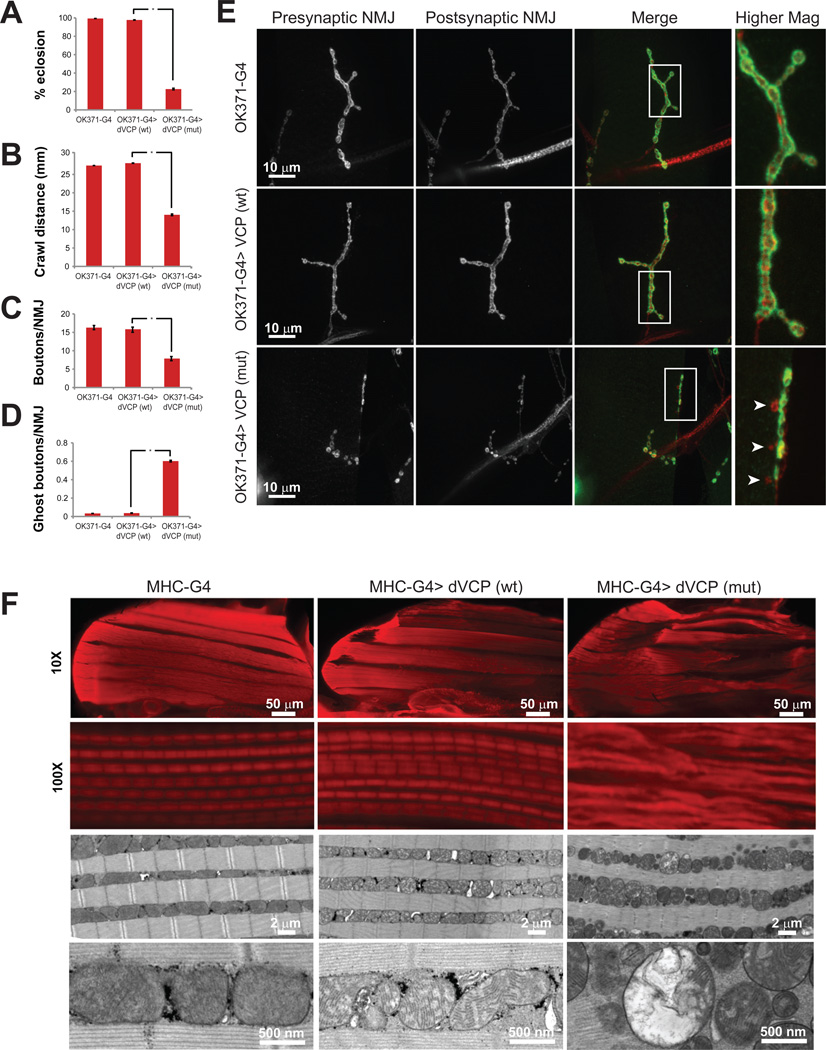
The species Drosophila melanogaster has a single, highly conserved ortholog of human VCP called dVCP. We developed a Drosophila model of VCP-related disease by introducing disease-homologous mutations into dVCP. Expression directed to the eye of these animals resulted in mutation-dependent eye degeneration despite equal levels of transgene expression (Ritson et al., 2010). Expression of wild type dVCP in motor neurons with the driver OK371-GAL4 did not impact fly viability, whereas motor neuron expression of mutant dVCP resulted in substantial pupal lethality (Figure 1A). The few adult escapers expressing mutant dVCP in motor neurons died shortly after eclosion. In 3rd instar larval animals, a mutation-dependent locomotor phenotype was evident, as documented in an assay of larval crawling (Figure 1B). Evaluation of the neuromuscular junction (NMJ) in these animals revealed a striking mutation-dependent morphological phenotype that included reduced numbers of synaptic boutons, an accumulation of ghost boutons, and reduced density of active zones (Figure 1C–E and Supplemental Figure 1). Evaluation of NMJ morphology in rare surviving mutant dVCP adults also revealed morphological defects including an accumulation of synaptic footprints consistent with denervation (Supplemental Figure 2). Consistent with our observations in motor neurons, expression of dVCP in muscle with the driver MHC-GAL4 resulted in mutation-dependent muscle degeneration and a dropped wing phenotype (Figure 1F and data not shown). Many of these flies reached adulthood, which facilitated histological analysis as well as genetic interaction studies. Histological examination of thoracic flight muscle in these flies revealed evidence of pronounced myopathy in flies expressing mutant dVCP, including atrophy of individual muscles and loss of normal sarcomere architecture (Figure 1F). Ultrastructural examination of muscle tissue by transmission electron microscopy (TEM) revealed marked morphological abnormalities in mitochondria with extensive megaconia and pleioconia (Figure 1F). Interestingly, prior phenotypic analysis of flies expressing mutant VCP reported that degeneration was accompanied by reduced cellular ATP levels (Chang et al., 2011). The mechanism of altered ATP levels was not explored in Chang et al. Nevertheless, the relevance of the altered ATP levels was nicely demonstrated since artificial manipulation of ATP levels modified the degenerative phenotype (Chang et al., 2011).
Figure 1. dVCP mutation-dependent motor neuron and muscle phenotypes in Drosophila.
A. Expression of exogenous wild type dVCP in motor neurons with the driver OK371-GAL4 does not impact viability as assessed by rates of eclosion as adults. In contrast, expression of mutant dVCP (R152H) leads to a high rate of pupal lethality. The flies that do eclose die shortly thereafter. Error bars indicate standard error. B. Expression of mutant dVCP (but not wild type) in motor neurons results in a locomotor defect as assessed by monitoring crawling behavior of 3rd instar larvae. Error bars indicate standard error. C. Expression of mutant dVCP (but not wild type) in motor neurons results in abnormal NMJ morphology with reduced total bouton numbers. NMJs at muscle 4 were used for all analyses. Error bars indicate standard error. D. The abnormal NMJs in flies expressing mutant dVCP in motor neurons results in a frequent ‘ghost boutons’ in which presynaptic structure lacks appositional postsynaptic structure. Ghost boutons are very rarely observed in control animals or animals expressing wild type dVCP. Error bars indicate standard error. E. Representative images of NMJs from the genotypes indicated. The arrowheads point out ghost boutons in an animal expressing mutant dVCP. F. Hemithoraces stained with phalloidin from control flies (MHC-GAL4) and flies expressing wild type or mutant dVCP in muscle under control of MHC-GAL4 show a mutation-dependent myopathy at 10X, and a disruption of sarcomere architecture at 100X. TEM revealed profound abnormalities in mitochondrial morphology with numerous swollen mitochondria with disrupted cristae. See also Supplemental Figure 1 and 2 for further analysis.
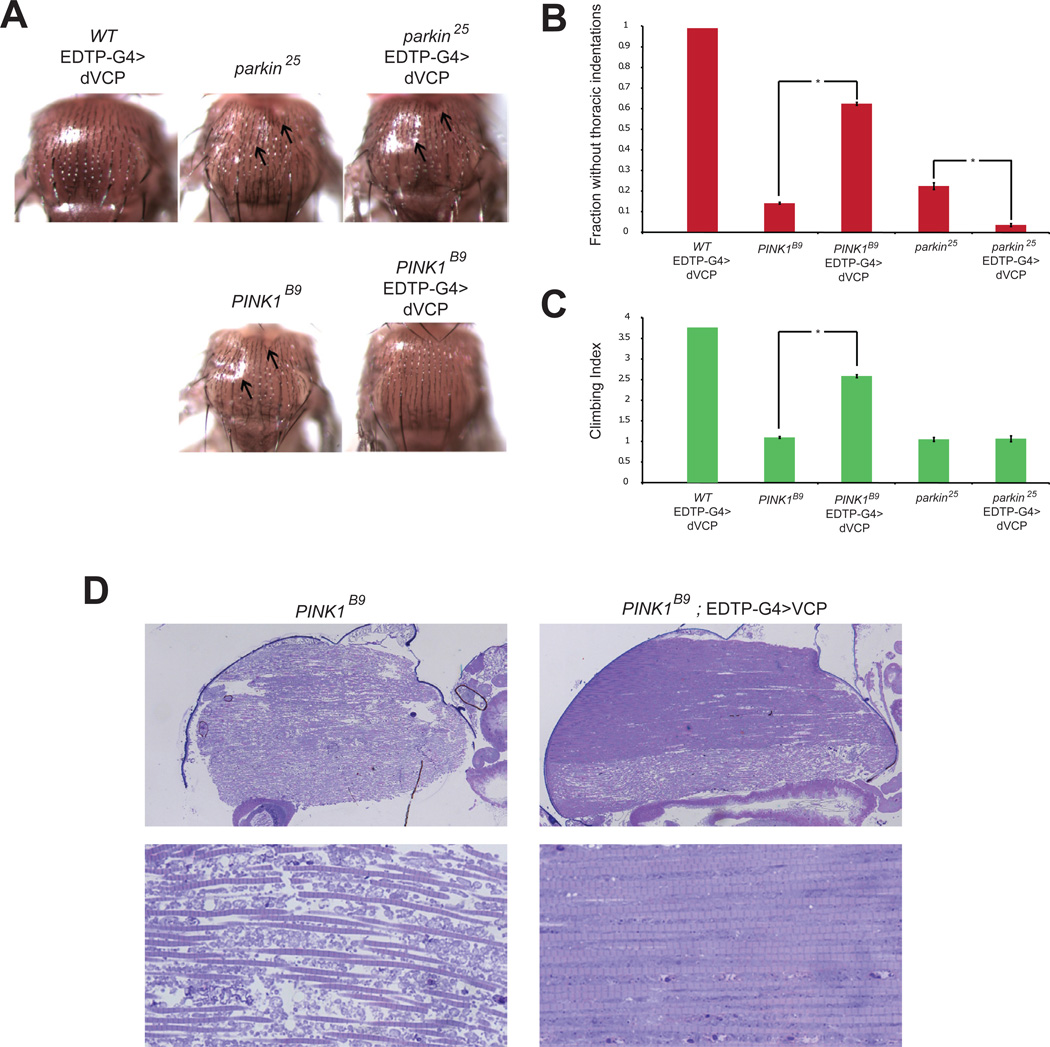
The myopathy and specific mitochondrial abnormalities observed in dVCP-mutant flies are reminiscent of the phenotypes reported in flies null for PINK1 and Parkin (Greene et al., 2003; Poole et al., 2008). Our interest in a possible connection to these genes was heightened by the fact that a subset of patients with VCP mutations present with parkinsonism or Parkinson’s disease (Kimonis et al., 2008; Spina et al., 2012), a clinical phenotype also associated with mutations in PINK1 and Parkin. PINK1 and Parkin participate in a common pathway that regulates mitochondrial dynamics and serve to maintain mitochondrial quality control (Clark et al., 2006; Narendra et al., 2008; Narendra et al., 2010; Park et al., 2006). These observations led us to hypothesize that VCP might be a component of the PINK1/Parkin pathway and contribute to mitochondrial quality control. To test this hypothesis, we performed epistasis studies between VCP, PINK1 and Parkin. We determined that over-expression of VCP rescued the degenerative phenotype associated with PINK1 deficiency, as evidenced by suppression of thoracic indentations (Figure 2A–B) and restoration of normal locomotor function (Figure 2C) in PINK1 null (PINK1B9). Furthermore, histological analysis demonstrated that VCP over-expression rescued the mitochondrial phenotype in PINK1 null flies (Figure 2D). This rescue by VCP is similar to that observed by over-expressing Parkin in PINK1 null flies (Clark et al., 2006; Park et al., 2006). These results indicate that, like Parkin, VCP functions downstream of PINK1 in the mitochondrial quality control pathway. In contrast, VCP did not suppress the degenerative phenotype associated with Parkin deficiency (Figure 2A–C). These data indicate that VCP functions upstream or in concert with Parkin, or alternatively, independent of Parkin in supporting mitochondrial quality control by PINK1. To clarify the relationship between Parkin and VCP and to elucidate the role of VCP in the PINK1/Parkin pathway (especially in mammals) we conducted extensive in vitro studies, as described below.
Figure 2. dVCP overexpression suppresses degeneration associated with PINK1 deficiency but not Parkin deficiency.
A. Micrographs of fly thoraces of the indicated genotypes showing thoracic indentations (arrows) caused by Parkin and PINK1 deficiency. dVCP over-expression rescues this phenotype in PINK1-deficient flies, but not in Parkin-deficient flies. B. Quantitation showing that over-expression of dVCP suppresses thoracic indentations in PINK1-deficient animals but actually enhances this phenotype in Parkin-deficient animals. Error bars indicate standard error. C. Quantitation showing that over-expression of dVCP suppresses the locomotor defect in PINK1-deficient animals but not in Parkin-deficient animals. Error bars indicate standard error. D. Micrographs of sections through fly thoraces showing that over-expression of dVCP suppresses the muscle and mitochondrial phenotypes in PINK1-deficient animals.
VCP re-localizes to mitochondria in response to depolarization
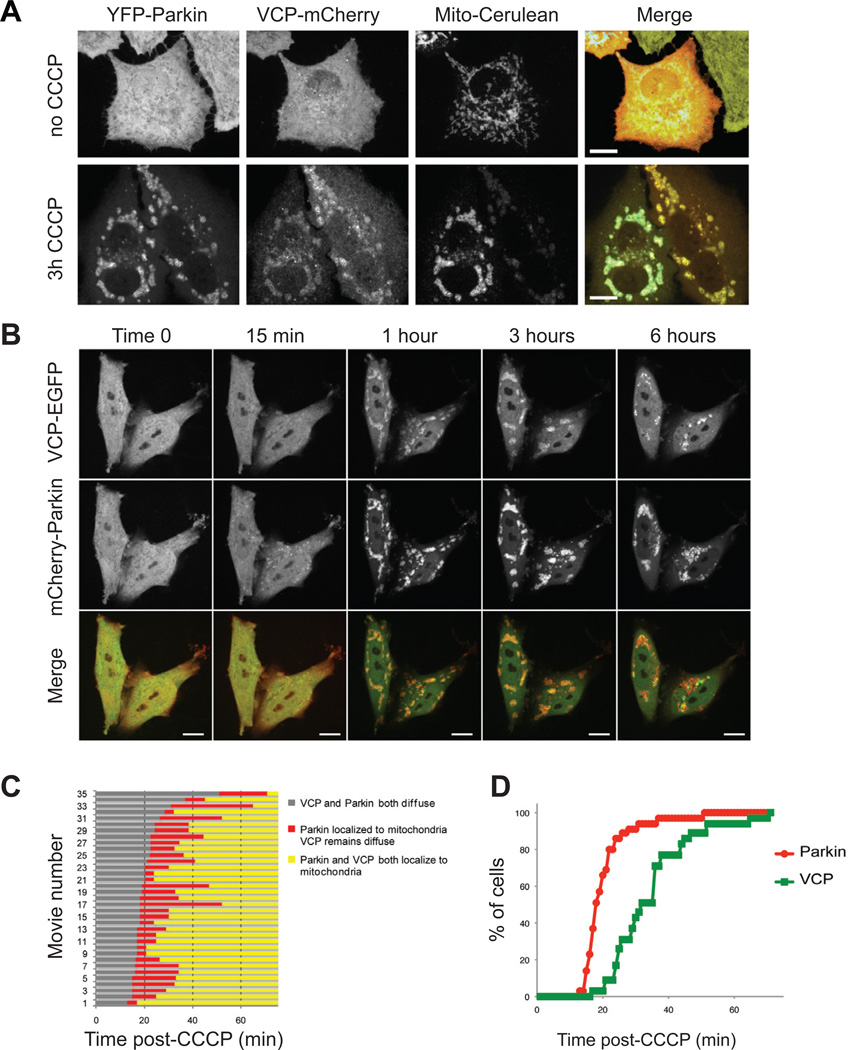
Parkin is an E3 ubiquitin ligase that is rapidly recruited to mitochondria in a PINK1-dependent manner in response to loss of mitochondrial membrane potential (Narendra et al., 2008; Narendra et al., 2010). Parkin recruitment results in ubiquitination of mitochondria, followed by mitochondrial fission, clustering, and subsequent clearance of damaged mitochondria (Matsuda et al.; Narendra et al., 2008; Narendra et al., 2010). To address the hypothesis that VCP participates in the PINK1/Parkin pathway, we first assessed whether VCP is recruited to mitochondria in response to depolarization. We used HeLa cells stably expressing YFP-Parkin (HeLa cells don’t show detectable endogenous Parkin expression, data not shown) and transfected plasmids expressing VCP-mCherry and mito-Cerulean (Cerulean fluorescent protein tagged with a mitochondrial targeting sequence). In untreated cells, Parkin and VCP were both distributed diffusely with no co-localization with mitochondria. However, three hours after treatment with the mitochondrial uncoupling agent carbonyl cyanide mchlorophenyl-hydrazone (CCCP), virtually all of the YFP-Parkin and VCP-mCherry signal co-localized with mito-Cerulean, illustrating recruitment to the mitochondria (Figure 3A).
Figure 3. VCP recruitment to mitochondria follows Parkin.
A. VCP-mCherry and Mito-Cerulean were expressed in YFP-Parkin stable HeLa cells and all 3 proteins were visualized at 0 and 3 hours after addition of CCCP. See also Supplemental Figure 3A for MEFs, Supplemental Figure 4 for Sy5y, Supplemental Figure 5 for C2C12. B. Time lapse imaging of VCP-EGFP and mCherry-Parkin following CCCP treatment in HeLa cells. These still images were extracted from Supplementary Movie 1. See also Supplementary Movie 2. C. Graphic representation of the timing of recruitment of Parkin and VCP to mitochondria in 35 consecutive movies. Each line represents one individual movie of a distinct set of 1–3 cells. D. Graphic representation of the lag time before mitochondrial localization of Parkin and VCP following CCCP treatment. Results represent the sum from all cells captured in 35 consecutive movies.
We also generated mouse embryonic fibroblasts (MEFs) stably expressing mito-Cerulean to enable monitoring of mitochondria. These cells were cotransfected with plasmids expressing mCherry-Parkin and VCP-EGFP, which as expected were distributed diffusely throughout untreated cells. Within three hours of treatment with CCCP, mCherry-Parkin and VCP-EGFP signals were re-localized to mitochondria (Supplemental Figure 3A). Notably, in the absence of exogenous Parkin we observed no recruitment of VCP to mitochondria in response to depolarization, suggesting that VCP recruitment is Parkin-dependent (Supplemental Figure 3B). Entirely consistent with the results in HeLa cells and MEFs, we observed that VCP also was recruited to depolarized mitochondria in neuroblastoma-derived Sy5y cells (Supplemental Figure 4) and myoblast-derived C2C12 cells (Supplemental Figure 5). Thus, VCP is co-recruited to depolarized mitochondria in concert with Parkin in a wide variety of cell types.
VCP recruitment to mitochondria follows Parkin
To characterize mitochondrial recruitment of Parkin and VCP in detail, we performed dynamic imaging in HeLa cells expressing VCP-EGFP and mCherry-Parkin following treatment with CCCP. We consistently observed that recruitment of mCherry-Parkin to mitochondria preceded recruitment of VCP-EGFP (Figure 3B–D and Supplementary Movie 1 and Supplementary Movie 2). Indeed, in an analysis of 35 sequential movies of individual cells we found that Parkin relocalized to mitochondria approximately 20 minutes after CCCP treatment, and that VCP followed Parkin approximately 15 minutes later (Figure 3C, D).
Parkin-dependent recruitment of VCP to depolarized mitochondria in primary neurons
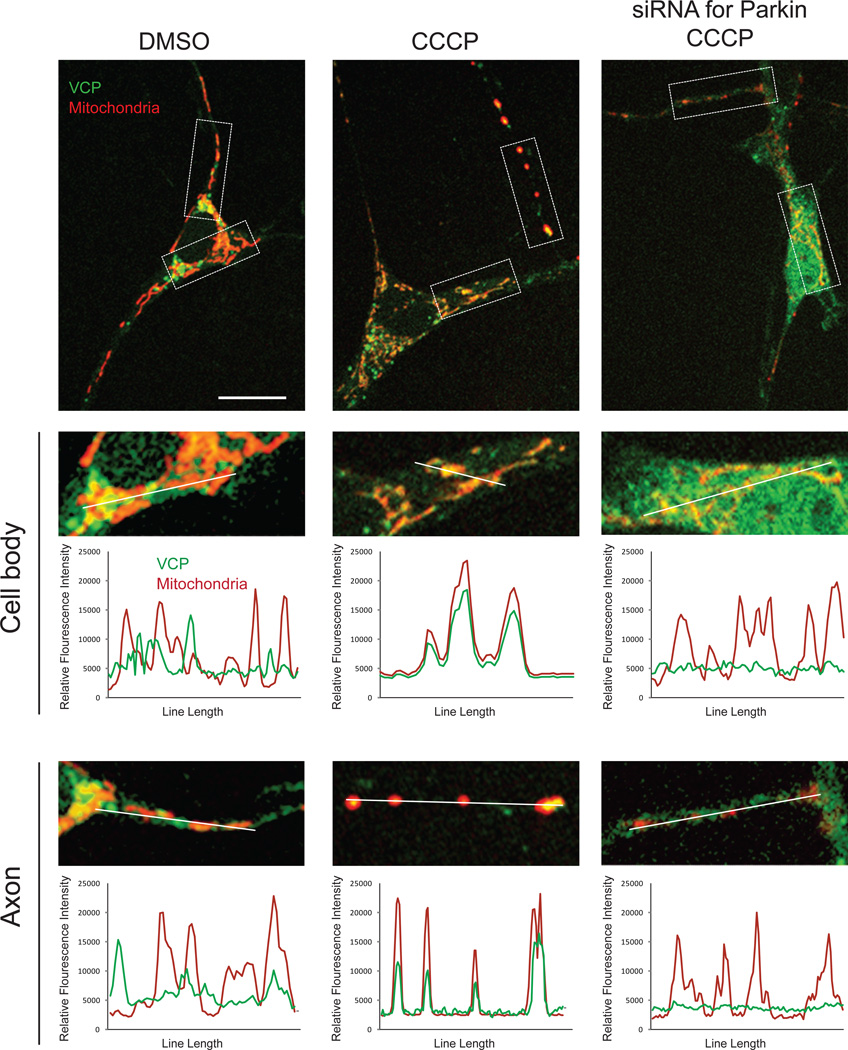
We sought to examine VCP recruitment to mitochondria in a cell type that expresses endogenous Parkin. Entirely consistent with our results in HeLa cells, MEFs, Sy5y cells and C2C12 cells, we determined that VCP relocalizes to mitochondria in primary neurons in response to depolarization with CCCP (Figure 4). Interestingly, this redistribution occurred throughout neuronal cells, including the soma and axonal compartments (Figure 4). Importantly, RNAi-mediated depletion of endogenous Parkin prevented this relocalization of VCP to mitochondria, indicating that VCP recruitment to mitochondria in primary neurons is Parkin-dependent just as it is in MEFs.
Figure 4. VCP recruitment to mitochondria in primary neurons is Parkin-dependent.
Primary neurons were transfected with mito-dsRED and VCP-EGFP and treated with DMSO or CCCP for 48 hrs. VCP-EGFP signal remained diffuse with some accumulation in Golgi in neurons treated with DMSO, but relocalized to mitochondria in neurons treated with CCCP. This relocalization was abolished by knockdown of endogenous Parkin by siRNA.
Mitochondrial ubiquitination by Parkin is essential for VCP recruitment
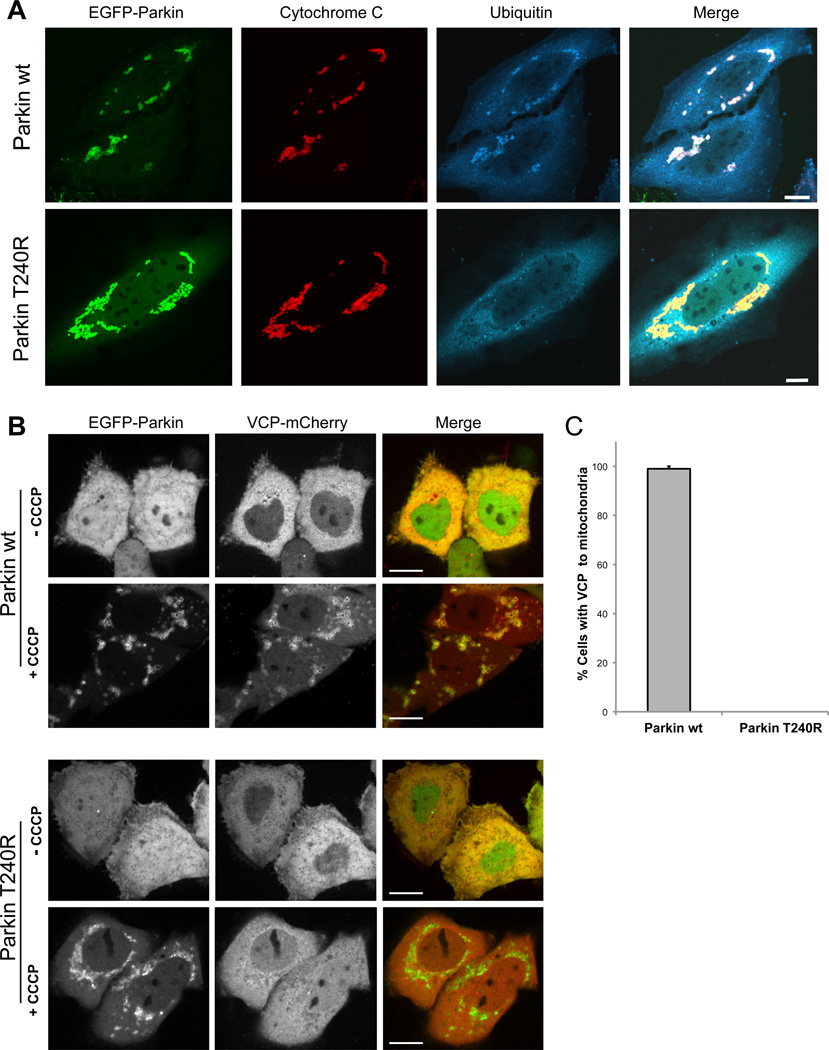
VCP interacts with polyubiquitin chains directly and also indirectly through a broad array of ubiquitin-binding adapter proteins (Dreveny et al., 2004). Given that Parkin is an E3 ubiquitin ligase, we hypothesized that ubiquitination of mitochondria by Parkin is a prerequisite for VCP recruitment. To test this hypothesis, we selected a Parkinson’s disease-associated Parkin mutant that is ubiquitin-ligase-defective due to a missense mutation (T240R) in the first RING domain. Whereas wild type Parkin is recruited to mitochondria and mediates ubiquitination in response to depolarization, Parkin-T240R is recruited to mitochondria but fails to mediate ubiquitination (Lee et al., 2010)(Figure 5A and Supplemental Figure 6). Quantitative analysis revealed that VCP was recruited to mitochondria in all cells expressing wild-type Parkin, but no such VCP recruitment occurred in cells transfected with Parkin-T240R despite the fact that this mutant form of Parkin is itself recruited to mitochondria (Figure 5B, C, and Supplemental Figure 6). We conclude that ubiquitination of mitochondria protein(s) by Parkin is essential to VCP recruitment to mitochondria.
Figure 5. Mitochondrial ubiquitination by Parkin is essential for VCP recruitment.
A. Parkin-dependent mitochondrial ubiquitination. HeLa cells were transfected with EGFP-Parkin wt or Parkin T240R, treated with 10 µM CCCP for 10 h and immunostained for cytochrome C (red, mitochondria) and ubiquitin (blue). Scale bars equal 10 µm. See also Supplemental Figure 6. B. Micrographs of HeLa cells expressing VCP-mCherry and either EGFP-Parkin wt or Parkin T240R. The images are taken during the mitochondrial aggregation stage after CCCP treatment (see Supplemental Figure 6 for the full time course). Scale bars equal 10 µm. C. Quantification of VCP recruitment to mitochondria following CCCP treatment in HeLa cells expressing either Parkin wt or Parkin T240R. Error bars indicate standard error from 3 replicates. 30 cells were counted for each replicate.
VCP contributes to regulation of mitochondrial dynamics
In considering what ubiquitination targets of Parkin might be responsible for recruitment of VCP, we noted a consistent temporal correlation between recruitment of Parkin and VCP and a change in mitochondrial morphology. Specifically, we observed that mitochondria that are fusiform at the time Parkin and VCP are recruited become increasingly fragmented within ∼30 minutes of VCP recruitment (Supplemental Figure 7A and Supplemental Movie 3). This observation is consistent with evidence that PINK1 and Parkin regulate mitochondrial dynamics and interact genetically with some other genes that regulate mitochondrial dynamics in Drosophila (Clark et al., 2006; Deng et al., 2008; Park et al., 2006; Poole et al., 2008). Moreover, it was recently reported that PINK1 and Parkin cooperate to ubiquitinate Mitofusin 1 (Mfn1) in mammalian cells and dMfn in Drosophila (Gegg et al., 2010; Poole et al., 2010; Ziviani et al., 2010). VCP is a ubiquitin-dependent segregase that dissociates ubiquitinated substrates from membrane complexes and makes them accessible to degradation by the proteasome and dominant negative VCP has been shown to stabilize mitochondrial proteins including Mfn (Braun et al., 2002; Rabinovich et al., 2002; Tanaka et al., 2010; Ye et al., 2001). Thus, we hypothesized that VCP works cooperatively with Parkin in response to PINK1 to mediate ubiquitin-dependent degradation of Mfns by the proteasome.
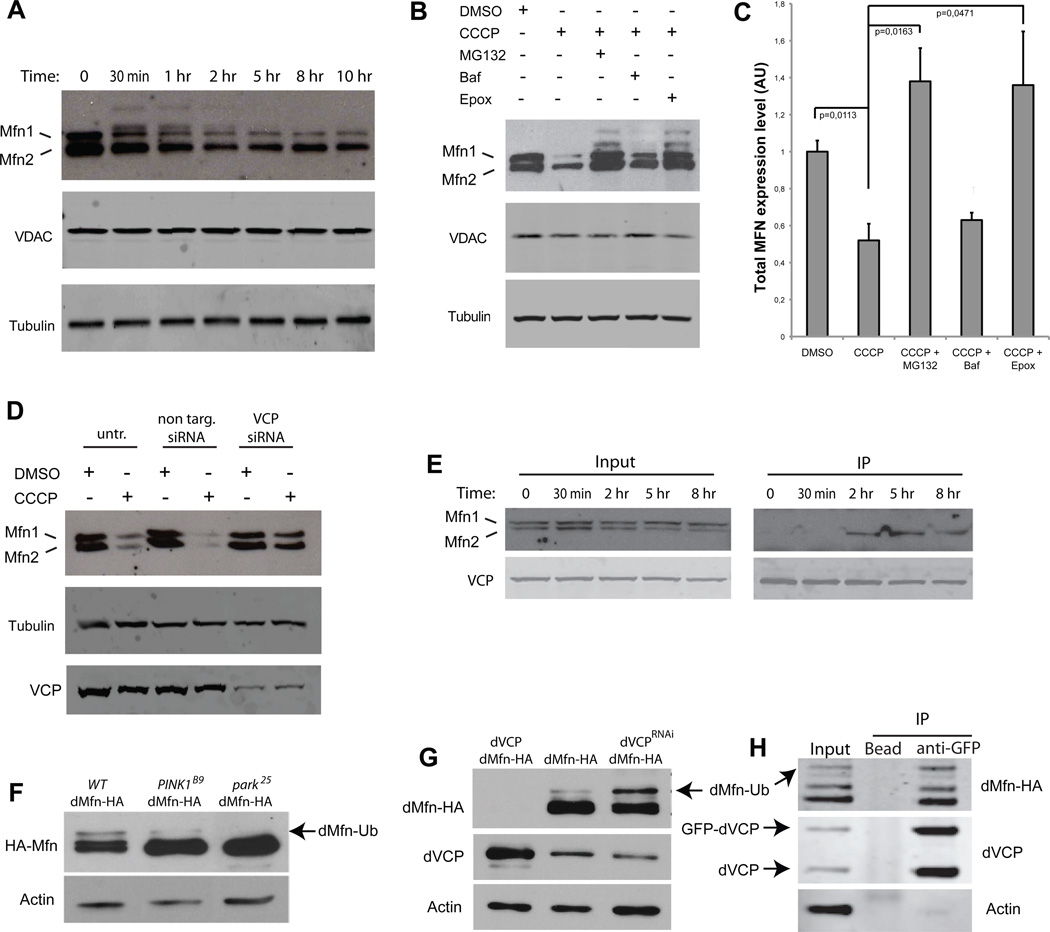
Consistent with this hypothesis, we observed rapid destabilization of Mfns 1 and 2 following CCCP treatment of HeLa cells stably expressing YFP-Parkin (Figure 6A). This degradation occurred prior to degradation of the mitochondria by the autophagic machinery as shown by the levels of VDAC that remain stable until late in the time course (Figure 6A). Moreover, Mfn 1 and 2 degradation is blocked by the proteasome inhibitors MG-132 and epoximicin, but not by the autophagy inhibitor bafilomycin, indicating that degradation of Mfns 1 and 2 is mediated by the proteasome (Figure 6B, C). To test the hypothesis that VCP mediates proteasomal degradation of Mfns 1 and 2, we examined the consequences of siRNA-mediated knockdown of VCP. Whereas non-targeting siRNA has no effect on Mfn 1 and 2 degradation following CCCP treatment, VCP-targeting siRNA blocks Mfn 1 and 2 degradation by the proteasome (Figure 6D). Furthermore, immunoprecipitation shows that VCP interacts with Mfn2 in vitro but only after mitochondrial membrane depolarization (Figure 6E). Thus, we conclude that VCP is essential for proteasome-dependent degradation of Mfns following ubiquitination by the PINK1/Parkin pathway.
Figure 6. Mitofusin degradation by the proteasome is dependent on VCP.
A. Western blots in YFP-Parkin stable HeLa cells against MFN1 and 2, VDAC and tubulin at different time points after CCCP treatment. Ubiquitinated forms of MFNs 1/2 can be observed migrating more slowly. B. Western blots in YFP-Parkin stable HeLa cells against MFN1/2, VDAC, and tubulin. Cells were treated for 12 h with CCCP and either proteasome inhibitors (MG132 or epoxomicin) or the autophagy inhibitor bafilomycin. Ubiquitinated forms of MFN1/2 can be again observed migrating more slowly, particularly with proteasome inhibition. C. Quantification of total MFN expression levels normalized against tubulin in YFP-Parkin stable HeLa cells treated for 12 h with CCCP and either proteasome inhibitors (MG132 or epoxomicin), or the autophagy inhibitor bafilomycin. Error bars indicate standard deviation from triplicates. D. Knockdown of VCP stabilizes MFNs 1 and 2. Western blots in YFP-Parkin stable HeLa cells against MFN1/2, VCP and tubulin. Cells were transfected with non-targeting or VCP-targeting siRNA and treated for 12 h with DMSO or CCCP. E. FLAG IP in HeLa cells cotransfected with YFP-Parkin and VCP-FLAG and treated with CCCP for the indicated times. Immunoprecipitation samples were immunoblotted against MFN1/2 and VCP. Following mitochondrial depolarization VCP interacts with MFN2. F. Total dMfn-HA accumulates in PINK1B9 (lane 2) and Park25 (lane 3) null mutants. Notably, ubiquitinated dMfn is decreased in PINK1B9 null mutants (lane 2) and absent in Park25 null mutants (lane 3). G. Overexpression of dVCP in the compound eye destabilizes dMfn-HA (lane 1), whereas knockdown of endogenous dVCP in the compound eye leads to accumulation of ubiquitinated dMfn-HA (lane 3). H. Immunoprecipitation of HA-dMfn and endogenous (protein-trap) GFP-dVCP from Drosophila brain extract.
To examine the role of VCP in the PINK1/Parkin pathway in vivo we used a transgenic approach to monitor the influence of altered VCP activity on the ubiquitination of the Drosophila mitofusin homolog, dMfn. Specifically, we developed a transgenic line expressing an HA-tagged version of dMfn to permit tissue-specific expression. This approach permitted us to circumvent the lethality associated with reduced VCP activity by selectively knocking-down VCP in a non-essential tissue that is also expressing the tagged version of dMfn. Using this HA-tagged form of dMfn, we find that deficiency in either PINK1 or Parkin results in accumulation of total dMfn (Figure 6F), as previously described for endogenous dMfn (Deng et al., 2008; Ziviani et al., 2010). Despite this accumulation, little ubiquitinated dMfn is detected in PINK1-deficient flies and no ubiquitinated dMfn is detected in Parkin-deficient flies, consistent with the roles of PINK1 and Parkin in mediating dMfn ubiquitination (Figure 6F). Using our system, we found that dVCP levels strongly influence dMfn stability in vivo: over-expression of dVCP eliminates dMfn from detection (Figure 6G, lane 1), whereas RNAi-mediated knockdown of endogenous dVCP leads to accumulation of ubiquitinated dMfn (Figure 6G, lane 3). We also confirmed that dVCP co-immunoprecipitates dMfn in vivo in Drosophila (Figure 6H). These observations are consistent with our hypothesis that dVCP serves to mediate degradation of ubiquitinated dMfn by the proteasome.
VCP knockdown impairs clearance of damaged mitochondria
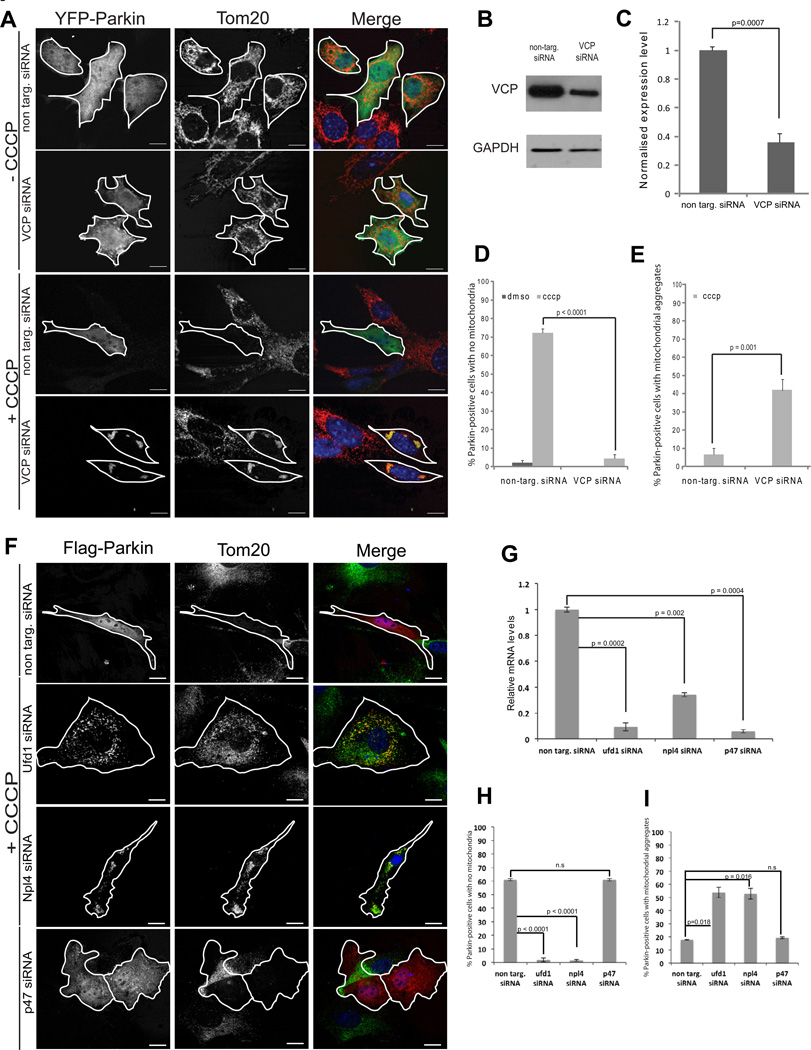
Given that VCP recruitment is dependent on mitochondrial ubiquitination by Parkin, and that abnormal mitochondria accumulate in VCP-mutant Drosophila, we hypothesized that VCP is involved in the process of PINK1/Parkin-dependent clearance of damaged mitochondria. To test this hypothesis, we examined the impact of VCP knockdown on the efficiency of PINK1/Parkin-dependent mitochondrial clearance. We over-expressed YFP-Parkin in MEFs with RNAi-mediated knockdown of VCP, or in control MEFs with non-targeting siRNA, and monitored mitochondrial clearance following CCCP treatment. Twenty-four hours following CCCP treatment, approximately 70% of cells co-transfected with YFP-Parkin and non-targeting siRNA had completely cleared their mitochondria (Figure 7A–D), consistent with previous observations (Narendra et al., 2008). In contrast, cells with YFP-Parkin and VCP-targeting siRNA failed to clear these depolarized mitochondria (Figure 7A–D). We determined that VCP is also essential for Parkin-dependent clearance of depolarized mitochondria in C2C12 myoblast cells (Supplemental Figure 5). Notably, we observed residual, prominent mitochondrial clusters in many cells that failed to clear mitochondrial in response to depolarization (Figure 7A, E, F and I).
Figure 7. VCP and Ufd1/Npl4 are essential for clearance of damaged mitochondria.
A. Immunostaining against TOM20 (red) in MEFs transfected with YFP-Parkin (green) and non-targeting or VCP-targeting siRNA. Cells were treated with DMSO or CCCP for 24 h. B. Immunoblot of VCP and GAPDH in MEFs transfected with non-targeting or VCP-targeting siRNA. C. Quantification of VCP normalized against GAPDH in cells transfected with non-targeting or VCP-targeting siRNA. Error bars indicate standard deviation from triplicates. D. Quantification of cells with mitochondrial clearance. Cells were treated for 24 h with DMSO or CCCP. At least 30 cells were counted for each sample. Errors bars indicate standard error from 3 independent replicates. See also Supplemental Figure 5 for C2C12 cells. E. Quantification of cells with residual aggregates of mitochondria. Cells were treated for 24 h with DMSO or CCCP. F. Immunostaining against TOM20 (red) in MEFs transfected with YFP-Parkin (green) and non-targeting siRNA or siRNA targeting Ufd1, Npl4, or p47. Cells were treated with DMSO or CCCP for 24 h. (Cells treated with DMSO shown in Supplemental Figure 8C). G. Real time PCR quantification of mRNA levels of Ufd1, Npl4, or p47 following treatment with the individual siRNAs. H. Quantification of cells with mitochondrial clearance in the setting of Ufd1, Npl4, or p47 knockdown. I. Quantification of cells with residual aggregates of mitochondria. Cells were treated for 24 h with DMSO or CCCP. At least 30 cells were counted for each sample. Errors bars indicate standard error from 3 independent replicates.
To rule out the possibility that knockdown of VCP merely delays mitochondrial clearance, we carefully monitored the kinetics of YFP-Parkin recruitment, as well as mitochondrial aggregation and clearance, throughout a 24-hour window. In cells without VCP knockdown, YFP-Parkin is recruited to mitochondria within 30 minutes, mitochondria aggregate within 3 hours, and mitochondrial clearance occurs within 10–12 hours. We found that VCP knockdown did not alter the kinetics of YFP-Parkin recruitment or mitochondrial aggregation, but that mitochondrial clearance never occurred (Supplemental Figure 7B–D and data not shown). Thus, VCP is essential for PINK1/Parkin-mediated mitochondrial clearance following depolarization, although mitochondrial aggregation can occur independent of VCP.
The Ufd1/Npl4 complex is recruited to depolarized mitochondria in concert with VCP and is required for clearance of damaged mitochondria
VCP interacts with a variety of adaptor proteins via the N-domain, which enables VCP to serve as a ubiquitin-dependent segregase for a broad array of substrates. In some cases these substrates are targeted for degradation by the proteasome and in this activity VCP often works in concert with the Ufd1/Npl4 complex. We found the Ufd1 and Npl4 are each recruited to depolarized mitochondria in concert with Parkin and VCP (Supplemental Figure 8A–B). The specificity of this adaptor recruitment is confirmed by evidence that the alternative adaptor p47 is not recruited to mitochondria in response to depolarization (Supplemental Figure 8A–B). Further, we show that mitochondrial clearance following depolarization is not only dependent on Parkin and VCP, but also Ufd1 and Npl4, and is not influenced by depletion of p47 (Figure 7F–I and Supplemental Figure 8C).
Mutant VCP impairs clearance of damaged mitochondria
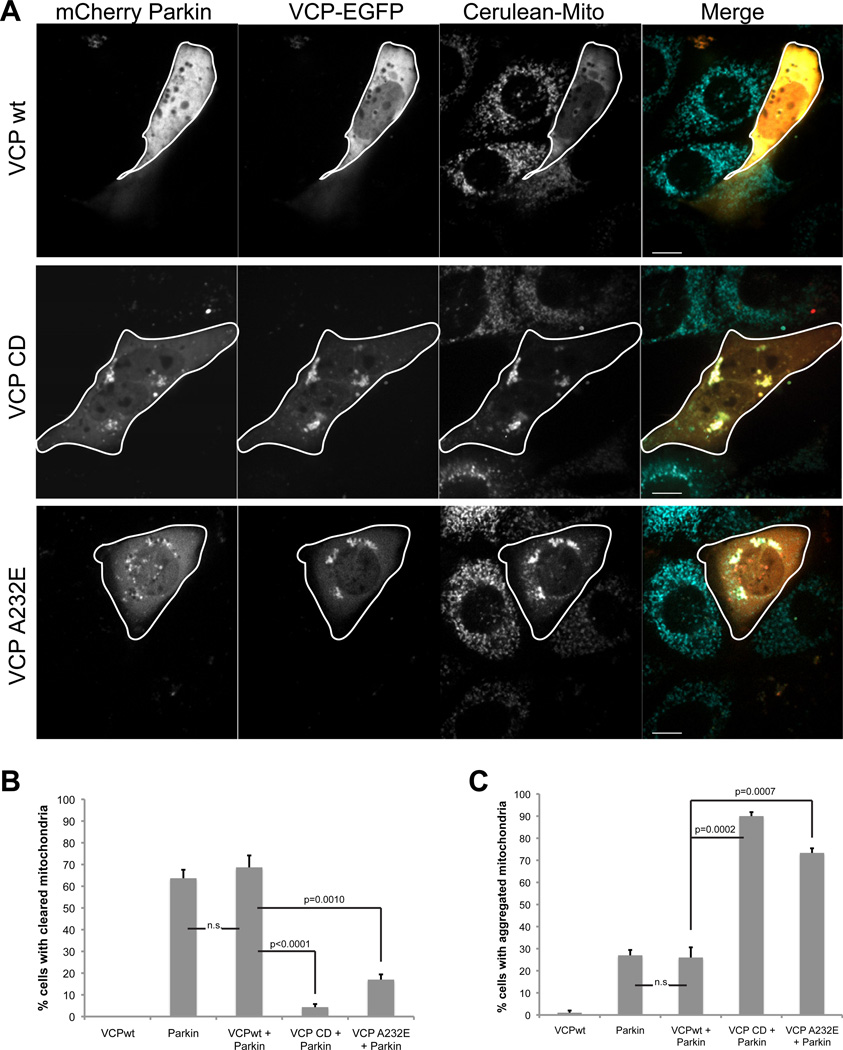
To confirm the involvement of VCP in mitochondrial clearance and investigate the influence of a disease-associated VCP mutation on this phenomenon, we examined the impact of over-expressing catalytically dead (VCP-CD) or disease-associated mutant VCP (VCP-A232E). The VCP-CD mutant was created by introducing mutations that impair both ATPase domains (E305Q/E578Q). VCP-CD functions as a dominant negative when expressed exogenously and has been extensively used to interrogate VCP functions (Dalal et al., 2004). The VCP-A232E missense mutation causes multisystem proteinopathy, a dominantly inherited multisystem degeneration that can present as Parkinson’s disease, frontotemporal dementia, amyotrophic lateral sclerosis, inclusion body myopathy, Paget’s disease of bone, hereditary spastic paraplegia or a combination of these (Guinto et al., 2007; Johnson et al., 2010; Watts et al., 2004).
To assess the impact of these mutations on mitochondrial clearance we co-expressed mCherry-Parkin with either wild type (VCP-wt) or mutant VCP (VCP-CD or VCP-A232E) in mito-Cerulean stable MEFs and quantified mitochondrial clearance in response to depolarization with CCCP. In cells co-transfected with VCP-wt, mitochondria were completely cleared 24 hours post-CCCP in 70% of cells, as expected (Figure 8A–B). In contrast, cells expressing VCP-CD or VCP-A232E failed to clear mitochondria (Figure 8A–B). Instead, we observed mitochondrial aggregates with colocalized Parkin and mutant VCP in most cells (Figure 8A, C). We also examined mitochondrial clearance in C2C12 myoblast cells and determined that cells expressing VCP-CD or VCP-A232E failed to clear mitochondria (Supplemental Figure 5E–G). Thus, VCP is essential to mitochondrial clearance in response to CCCP and a disease-causing mutation in VCP impairs this process.
Figure 8. Overexpression of VCP-CD or A232E inhibits clearance of damaged mitochondria.
A. Imaging VCP-EGFP and mCherry-Parkin in Mito-Cerulean stable MEFs 24h after addition of CCCP. Cells were transfected with VCP-wt, CD or disease mutant A232E -EGFP. Scale bars equal 10µm. B. Quantification of cells with mitochondrial clearance. Cells were prepared as described in A. At least 30 cells were counted for each sample. Errors bars indicate standard error from 3 independent replicates. C. Quantification of cells with residual aggregates of mitochondria. Cells were prepared as described in A. At least 30 cells were counted for each sample. Errors bars indicate standard error from 3 independent replicates. C2C12 cell data are shown in Supplemental Figure 5E–G.
Discussion
VCP is an essential molecular chaperone that contributes to a broad array of cellular activities. The central question concerning the pathogenesis of VCP-related disease is: which functions of VCP are impaired by disease-causing mutations? To address these questions in an unbiased way we generated a Drosophila model that captures VCP mutation-dependent degeneration. We found that these animals have a mitochondrial phenotype resembling that observed in PINK1 and parkin mutant flies (Figure 1). Indeed, this impression was validated by the finding that over-expression of dVCP complements PINK1 and rescues the degeneration and mitochondrial phenotype observed in PINK1 null flies, placing VCP downstream of PINK1 in the mitochondrial quality control pathway (Figure 2). This is similar to prior observations that over-expression of parkin complements PINK1 and rescues PINK1 null flies (Clark et al., 2006; Park et al., 2006). We were intrigued, therefore, when over-expression of dVCP failed to complement parkin. This paradox was resolved by studies in vitro demonstrating that VCP recruitment to mitochondria is Parkin-dependent. Specifically, we showed that VCP recruitment to mitochondria follows Parkin temporally and depends on Parkin-mediated ubiquitination of mitochondrial substrates (Figures 3–5). VCP is required for proteasome-dependent degradation of ubiquitinated Mitofusin-1 and Mitofusin-2 in vitro (Figure 6 and Tanaka et al., 2010) and the Drosophila homolog dMfn in vivo (Figure 6). Furthermore, we showed that VCP and its adaptor complex Ufd1/Npl4 are required for clearance of damaged mitochondria (Figure 7) and that this function is impaired by pathogenic mutations in VCP (Figure 8).
Mutations in VCP cause a dominantly inherited, multisystem degenerative disease that affects muscle, bone, and brain. This condition has been called “IBMPFD” to reflect the clinical manifestations of inclusion body myopathy (IBM), frontotemporal dementia (FTD), and Paget’s disease of bone (PDB) in affected families (Watts et al., 2004). More recently the term multisystem proteinopathy (MSP) has been adopted for this disorder to reflect the expanding phenotypic spectrum of VCP-related diseases. Pathogenic VCP mutations have been identified in more common diseases such as sporadic and familial forms of ALS (Johnson et al., 2010), FTD (Koppers et al., 2012), IBM (Shi et al., 2012), and PDB (Chung et al., 2011). The mechanism whereby mutations in VCP cause disease is incompletely understood. The studies presented here demonstrate not only that VCP functions within the PINK1/Parkin pathway of mitochondrial quality control, but that disease-causing mutations in VCP impair this pathway.
VCP is an essential gene - nullizygous mutations in VCP cause early embryonic lethality in mice (Muller et al., 2007). Thus, the mechanism of VCP-related disease is not likely to be caused by a simple loss-of-function. Indeed, the autosomal dominant inheritance pattern of VCP-related disease and the ability to recapitulate disease by over-expression of mutant forms of VCP in a wild type background in cells or animals (Badadani et al., 2010; Custer et al., 2010; Ritson et al., 2010; Weihl et al., 2007) both argue in favor of a dominant molecular mechanism, resulting in a toxic gain-of-function, dominant negative function or possibly a combination of both.
Structurally, VCP is similar to other type II AAA+ proteins such as NSF and ClpA in that it functions as a homo-hexameric barrel (White and Lauring, 2007). This homohexameric structure suggests the possibility of a dominant-negative effect imposed by disease mutations. Thus, co-mingling of wild type and mutant VCP proteins in the same homohexamer could lead to dominant impairment of wild type VCP function. However, it is clear that loss-of-function alone through a dominant-negative mechanism cannot account for the phenotype observed in Drosophila expressing mutant VCP. If this were the case, co-expression of exogenous wild type dVCP would be expected to suppress the degeneration caused by mutant dVCP, but it does not. Indeed, co-expression of exogenous wild type dVCP enhances the degeneration that accompanies mutant dVCP expression (Chang et al., 2011).
So, how do disease mutations impact VCP function? As stated above, the N-domain of VCP alternates between distinct conformations and this is regulated by the nucleotide occupancy in the D1 domain. These distinct conformation bind to distinct subsets of adaptors that are responsible for different VCP functions. Crystallographic studies of mutant forms of VCP suggest that disease-causing mutations alter nucleotide occupancy in the D1 domain, causing VCP to be locked into a fixed conformation, consequently interfering with interaction of the N-terminal domain with a subset of adaptor proteins (Tang et al., 2010). Thus, disease mutations are predicted to enhance interaction with some adaptors and mitigate interaction with other adaptors, which could amplify some VCP functions and diminish others. Indeed, this precise observation was made when the impact of VCP mutations on several adaptor interactions was examined in vitro (Fernandez-Saiz and Buchberger, 2010).
Consistent with this hypothesis regarding an imbalance in VCP function, we and others have recently shown that disease mutations in VCP do not impair endoplasmic reticulum-associated degradation (Chang et al., 2011; Tresse et al., 2010), but do impair autophagosome maturation (Ju et al., 2009; Tresse et al., 2010), despite the fact that VCP is essential for both of these processes. Pathogenic VCP mutations have also been shown to impair aspects endolysosomal sorting and aspects of myosin assembly (Janiesch et al., 2007; Ritz et al., 2011). The results presented here demonstrate that VCP is essential to mitochondrial quality control by the PINK1/parkin pathway in vitro and in vivo and show that this function must be included in the list of those functions impaired by pathogenic VCP mutations.
Materials and Methods
Cell culture
Mouse embryonic fibroblasts (MEFs), HeLa and C2C12 cells were cultured in DMEM (Hyclone) supplemented with 10% FBS (Hyclone) and GlutaMax-1X (GIBCO). For SH-SY5Y cells, DMEM:F-12 (GIBCO) was used with the same supplements. Drugs were used at the following final concentrations: CCCP at 20 μM, epoxomicin at 100 nM, bafilomycin at 10 μM, and MG132 at 5 μM. Mito-Cerulean stable MEFs were generated by retroviral transfection of a mito-Cerulean plasmid (gift of Fabien Llambi) using Phoenix Eco cells (Orbigen; RVC-10002). The YFP-Parkin HeLa stable line (Narendra et al., 2008) was a gift from Richard Youle.
Plasmids and transfections
HeLa, MEFs and SH-SY5Y cells were transfected using FuGENE6 transfection reagent (Roche; 1988387001) and C2C12 cells were transfected using Lipofectamine 2000 transfection reagent (Invitrogen; 11668-027) following the manufacturer’s instructions respectively. The EGFP-VCP wt and mutant plasmids were previously described (Tresse et al., 2010). VCP-mCherry was constructed by releasing VCP wt from the EGFP-VCP wt plasmid and inserting the gene into the BamHI and HindIII sites of pmCherry-N1 (Clontech). Flag-Parkin was previously described (Lim et al., 2007). EGFP-Parkin wild type and T240R were previously described (Lee et al., 2010). mCherry- and YFP-Parkin plasmids were gifts from Richard Youle. The p47-, npl4- and ufd1-GFP were made by recombination of the Gateway entry clone with pcDNA-DEST47 (Invitrogen) using LR Clonase II Enzyme (Invitrogen, 11791-020) following the manufacturer’s instructions.
The entry vectors (HsCD00042210, HsCD00041106 and HsCD00081676, respectively) were purchased from Dana-Farber/Harvard Cancer Center DNA Resource Core. RNAi knockdown of target genes were performed by transfection of ON-TARGET plus-SMARTpool siRNA (Dharmacon) using Lipofectamin RNAi Max (Invitrogen): non-targeting (1081195), VCP (057592 for mouse cells and 008727 for human cells), p47 (198326), npl4 (199469), ufd1 (011672), parkin (50873).
Neuronal Cell Culture and Transfection
To generate primary neurons, dorsal root ganglia were dissected from postnatal mice (P1) and then dissociated and cultured on glass coverslips coated with poly-D-lysine (Sigma) and Laminin (BD Biosciences). The cultures were maintained at 37°C in Leibovitz’s L-15 medium (phenol red free; Invitrogen) supplemented with 0.6% glucose (Sigma), 2 mM L-glutamine (Sigma), 5 ng/ml nerve growth factor (2.5S; Sigma), 10% fetal bovine serum (Thermo Scientific Hyclone), and 0.5% hydroxypropylmethylcellulose (Methocel; Sigma). Cells were transfected in suspension before plating by electroporation with 75 to 100 mg/ml DNA and 2 mM siRNA by using an Amaxa Nucleofector (SCN program 6; Lonza, Walkersville, MD). Neurons were treated with either 10 mM CCCP (Sigma) in DMSO (Fisher Scientific) or with DMSO alone, supplemented with Z-VAD-FMK (Sigma) for 24 or 48 hours, after which they were washed two times with PBS. The cells were then fixed and observed using a spinning disc confocal Marianas system (Intelligent Imaging Innovations, Denver, CO) configured on a Zeiss Axio Observer. Co-localization was confirmed by line intensity analysis using Slidebook 5.0 (Intelligent Imaging Innovations).
Quantitative Real time PCR
Total RNA was isolated with Trizol (Invitrogen) from MEFS. qPCR performed with TaqMan RNA-to Ct 1 step-kit (Applied Biosystems; 4392938). The primers Mm01243864_g1, Mm01253310_m1, Mm00495912_m1 and 4352339E-1009033 were used to detect the p47, npl4, ufd1 and GAPD levels, respectively. qPCR was performed in an Applied Biosystems 7900HT Fast Real-Time PCR System using the following cycling parameters: 48°C (15 min), 95°C (10 min), and40 cycles of 95°C (15 s), 60°C (1 min). All PCR experiments were conducted in triplicates.
Microscopy
For immunofluorescence, MEFs were plated on chamber slides (Lab-Tek; 154917). Cells were fixed with 4% paraformaldehyde, and then washed twice with PBS and once with NH4Cl (10 mM). The cells were permeabilized with 0.2% saponin and blocked with 10% horse serum and 0.1% saponin. Anti-Tom20 antibody (Santa Cruz Biotechnology; FL-145) was used at a 1:50 dilution. Coverslips were mounted onto microscope slides using ProLong Gold Antifade Reagent with DAPI (Invitrogen; P3691). Cells were observed with a LSM510 (Zeiss) confocal microscope with a 63x objective. Anti-cytochrome C and anti-ubiquitin immunostaining were performed as described in (Lee et al., 2010). For live imaging, MEFs or HeLa cells were plated on glass bottom dishes (MatTek Corporation; P35GC-1.5-10-C) or chambered coverglass (Lab-Tek; 155382). Cells were observed with a Marianas confocal microscope (Leica) with a 63x objective. Movies were collected on a Marianas confocal microscope with 40x objective, with 3-minute intervals during the first 2 hours and 10-minute intervals during the remainder of the time period.
Western blotting
VCP (1:3000; Thermoscientific; MA3-004), tubulin (1:3000; Sigma-Aldrich; T5168), GAPDH (1:500; Sigma-Aldrich; G9545) and VDAC 1–2–3 (1:500; Thermo Scientific; PA1-954A), β-actin (1:1000; Sigma-Aldrich; A5316) and parkin (abcam; 15954) were visualized by the Odyssey system (Li-Cor). MFN1/2 antibody (1:50, Santa Cruz, Z-6) was detected using ECL (34096, Pierce).For western blotting of Drosophila samples the following antibodies were used and detected using ECL or the Odyssey system: HA high Affinity (1:500; Roche; 11867423001), VCP (311–325) (1:3000; Sigma-Aldrich; SAB1100655) and Actin (1:50,000; Chemicon; MAB1501). Western blots were quantified using ImageJ software (NIH).
Statistical analysis
Statistical analysis of mitochondrial clearance and protein quantitation were evaluated by the paired Student’s t-test at p < 0.05 with GraphPad software.
Fly stocks
The EDTP-GAL4, Hsp70-GAL4, GMR-GAL4, and ey-GAL4 drivers were obtained from the Bloomington Drosophila stock center. GFP-dVCP protein trap line (TER94CB04973) was obtained from the Spradling Lab (http://flytrap.med.yale.edu/). The UAS-dMfn-HA transgenic line was a kind gift from Andrea Daga. The UAS-PINK1 transgenic line and PINKB9 mutant were kind gifts from J.K. Chung. The park25 mutants were previously characterized (Greene et al., 2003). UAS-dVCP wt, UAS-dVCP R152H, and UAS-dVCP A229E transgenic flies were previously described (Ritson et al., 2010). MHC-GAL4 and OK371-GAL4 were used to drive expression of dVCP in muscles and motor neurons respectively.
Adult muscle preparation
Adult flies were embedded in a drop of OCT compound (Sakura Finetek; 4583) on a glass slide, frozen with liquid nitrogen and bisected sagitally by a razor blade. After fixation with 4% paraformaldehyde in PBS, hemithoraces were stained by Texas Red-X-Phalloidin (Invitrogen; T7471) according to the manufacturer’s instructions. Samples were mounted in Fluoromount-G mounting medium (SouthernBiotech; 0100-01) and examined by DMIRE2 (Leica) with 10x and 100x objectives for musculature and sarcomere structure, respectively.
Thoracic indentation and pupal lethality phenotypes
To quantitate the frequency of thoracic indentations individual flies were examined 2 to 5 days after eclosion to determine whether there were indentations in the cuticle of the thorax, indicative of flight muscle degeneration. N>90 were examined for PINK1B9 mutants and N>40 were examined for park25 mutants. To monitor viability, total and empty pupal cases were counted (N>200 from 3 independent crosses).
Drosophila neuronal phenotype analysis
Five wandering 3rd instar larvae for each group were collected, washed and placed onto a 3% agarose gel in a 10cm dish. After 5min acclimation, larval crawling behavior was recorded by a digital camera for 30 sec (15fps). Each group was tested three times. Moving distances of each larva were manually measured with ImageJ. Neuromuscular synaptic bouton number (N=18) and active zone density (N=12) were measured as previously described (Nedelsky et al., 2010; Smith and Taylor, 2011) with slight modification. Active zone area and presynaptic area stained by NC82 and anti-HRP antibody were measured with ImageJ and the ratio of active zone area/presynaptic area was calculated. Ghost boutons were counted with the same samples prepared for synaptic bouton number counting. NMJs at muscle 4 were used for all analyses. To check integrity of adult neuromuscular junctions, escapers were dissected and stained with presynaptic anti-HRP antibody (1:200, Jackson Immunoresearch) and postsynaptic anti-discs large antibody (1:50, DSHB) and ventral abdominal muscles were examined.
Coimmunoprecipiation from Drosophila brains
50 brains were dissected from 3rd instar larvae expressing dMFN-HA in motor neurons with a TER94CB04973 allele (TER94CB04973, OK371/+; UAS>dMFN-HA/+). The dissected brains were lysed and immunoprecipitated by anti-GFP agarose beads (Chromotek; ACT-CM-GFA) following the manufacturer’s instruction. Agarose beads were used as a binding control.
Electron microscopy
One-day-old adult thoraces from the appropriate genotypes were fixed in 4% glutaraldehyde in 0.1 M sodium cacodylate buffer pH 7.4 with 5% sucrose and post-fixed in 0.2% osmium tetroxide in 0.1 M sodium cacodylate buffer with 0.3% potassium ferrocyanide for 2 hours. After rinsing in same buffer, the tissue was dehydrated through a series of graded ethanol to propylene oxide, infiltrated and embedded in epoxy resin, and polymerized at 70°C overnight. Semi-thin sections (0.5 mM) were stained with toluidine blue for light microscope examination. Ultrathin sections (70 nm) were cut and stained with Reynolds lead citrate. Examinations were made with a JEOL 1200X transmission electron microscope at 60KV and imaged using an AMT V600 digital camera.
Supplementary Material
Acknowledgements
We thank the Hartwell Center for Bioinformatics and Biotechnology and the Cell and Tissue Imaging Core Facility at St. Jude Children’s Research Hospital. We thank Fabien Llambi and Doug Green for the mito-Cerulean plasmid and Richard Youle for YFP-Parkin stable HeLa cells. Financial support was provided NIH grant NS-054022 to TPY, NIH grant GM086394 to LP and by NIH grant AG031587, a grant for The Robert Packard Foundation for ALS Research at Johns Hopkins and support from ALSAC (American-Lebanese-Syrian Associated Charities) to JPT.
Glossary
Abbreviations list
- ALS
amyotrophic lateral sclerosis
- CCCP
carbonyl cyanide m-chlorophenyl-hydrazone
- CD
catalytically dead
- FTD
frontotemporal dementia
- IBM
inclusion body myopathy
- IBMPFD
inclusion body myopathy with Paget’s disease of bone and frontotemporal dementia
- MEF
mouse embryonic fibroblast
- Mfn1
Mitofusin-1
- MSP
Multisystem proteinopathy
- PDB
Paget’s disease of bone
- PINK1
PTEN-induced putative kinase 1
- TEM
transmission electron microscopy
- VCP
Valosin-containing protein
- VDAC
voltage-dependent anion channel
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abramzon Y, Johnson JO, Scholz SW, Taylor JP, Brunetti M, Calvo A, Mandrioli J, Benatar M, Mora G, Restagno G, et al. Valosin-containing protein (VCP) mutations in sporadic amyotrophic lateral sclerosis. Neurobiology of aging. 2012;33:e2231–e2231. doi: 10.1016/j.neurobiolaging.2012.04.005. 2231 e2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badadani M, Nalbandian A, Watts GD, Vesa J, Kitazawa M, Su H, Tanaja J, Dec E, Wallace DC, Mukherjee J, et al. VCP associated inclusion body myopathy and paget disease of bone knock-in mouse model exhibits tissue pathology typical of human disease. PloS one. 2010;5 doi: 10.1371/journal.pone.0013183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun S, Matuschewski K, Rape M, Thoms S, Jentsch S. Role of the ubiquitin-selective CDC48(UFD1/NPL4 )chaperone (segregase) in ERAD of OLE1 and other substrates. EMBO J. 2002;21:615–621. doi: 10.1093/emboj/21.4.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YC, Hung WT, Chang HC, Wu CL, Chiang AS, Jackson GR, Sang TK. Pathogenic VCP/TER94 alleles are dominant actives and contribute to neurodegeneration by altering cellular ATP level in a Drosophila IBMPFD model. PLoS genetics. 2011;7:e1001288. doi: 10.1371/journal.pgen.1001288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung PY, Beyens G, de Freitas F, Boonen S, Geusens P, Vanhoenacker F, Verbruggen L, Van Offel J, Goemaere S, Zmierczak HG, et al. Indications for a genetic association of a VCP polymorphism with the pathogenesis of sporadic Paget's disease of bone, but not for TNFSF11 (RANKL) and IL-6 polymorphisms. Molecular genetics and metabolism. 2011;103:287–292. doi: 10.1016/j.ymgme.2011.03.021. [DOI] [PubMed] [Google Scholar]
- Clark IE, Dodson MW, Jiang C, Cao JH, Huh JR, Seol JH, Yoo SJ, Hay BA, Guo M. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441:1162–1166. doi: 10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- Custer SK, Neumann M, Lu H, Wright AC, Taylor JP. Transgenic mice expressing mutant forms VCP/p97 recapitulate the full spectrum of IBMPFD including degeneration in muscle, brain and bone. Human molecular genetics. 2010;19:1741–1755. doi: 10.1093/hmg/ddq050. [DOI] [PubMed] [Google Scholar]
- Dalal S, Rosser MF, Cyr DM, Hanson PI. Distinct roles for the AAA ATPases NSF and p97 in the secretory pathway. Mol Biol Cell. 2004;15:637–648. doi: 10.1091/mbc.E03-02-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bot ST, Schelhaas HJ, Kamsteeg EJ, van de Warrenburg BP. Hereditary spastic paraplegia caused by a mutation in the VCP gene. Brain : a journal of neurology. 2012 doi: 10.1093/brain/aws201. [DOI] [PubMed] [Google Scholar]
- Deng H, Dodson MW, Huang H, Guo M. The Parkinson's disease genes pink1 and parkin promote mitochondrial fission and/or inhibit fusion in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:14503–14508. doi: 10.1073/pnas.0803998105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreveny I, Pye VE, Beuron F, Briggs LC, Isaacson RL, Matthews SJ, McKeown C, Yuan X, Zhang X, Freemont PS. p97 and close encounters of every kind: a brief review. Biochemical Society transactions. 2004;32:715–720. doi: 10.1042/BST0320715. [DOI] [PubMed] [Google Scholar]
- Fernandez-Saiz V, Buchberger A. Imbalances in p97 co-factor interactions in human proteinopathy. EMBO reports. 2010;11:479–485. doi: 10.1038/embor.2010.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gegg ME, Cooper JM, Chau KY, Rojo M, Schapira AH, Taanman JW. Mitofusin 1 and mitofusin 2 are ubiquitinated in a PINK1/parkin-dependent manner upon induction of mitophagy. Human molecular genetics. 2010;19:4861–4870. doi: 10.1093/hmg/ddq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene JC, Whitworth AJ, Kuo I, Andrews LA, Feany MB, Pallanck LJ. Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:4078–4083. doi: 10.1073/pnas.0737556100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinto JB, Ritson GP, Taylor JP, Forman MS. Valosin-containing protein and the pathogenesis of frontotemporal dementia associated with inclusion body myopathy. Acta Neuropathol. 2007;114:55–61. doi: 10.1007/s00401-007-0224-7. [DOI] [PubMed] [Google Scholar]
- Janiesch PC, Kim J, Mouysset J, Barikbin R, Lochmuller H, Cassata G, Krause S, Hoppe T. The ubiquitin-selective chaperone CDC-48/p97 links myosin assembly to human myopathy. Nat Cell Biol. 2007;9:379–390. doi: 10.1038/ncb1554. [DOI] [PubMed] [Google Scholar]
- Jentsch S, Rumpf S. Cdc48 (p97): a "molecular gearbox" in the ubiquitin pathway? Trends Biochem Sci. 2007;32:6–11. doi: 10.1016/j.tibs.2006.11.005. [DOI] [PubMed] [Google Scholar]
- Johnson JO, Mandrioli J, Benatar M, Abramzon Y, Van Deerlin VM, Trojanowski JQ, Gibbs JR, Brunetti M, Gronka S, Wuu J, et al. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron. 2010;68:857–864. doi: 10.1016/j.neuron.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju JS, Fuentealba RA, Miller SE, Jackson E, Piwnica-Worms D, Baloh RH, Weihl CC. Valosin-containing protein (VCP) is required for autophagy and is disrupted in VCP disease. The Journal of cell biology. 2009;187:875–888. doi: 10.1083/jcb.200908115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju JS, Weihl CC. Inclusion body myopathy, Paget's disease of the bone and fronto-temporal dementia: a disorder of autophagy. Human molecular genetics. 2010a;19:R38–R45. doi: 10.1093/hmg/ddq157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju JS, Weihl CC. p97/VCP at the intersection of the autophagy and the ubiquitin proteasome system. Autophagy. 2010b;6:283–285. doi: 10.4161/auto.6.2.11063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimonis VE, Mehta SG, Fulchiero EC, Thomasova D, Pasquali M, Boycott K, Neilan EG, Kartashov A, Forman MS, Tucker S, et al. Clinical studies in familial VCP myopathy associated with Paget disease of bone and frontotemporal dementia. American journal of medical genetics Part A. 2008;146A:745–757. doi: 10.1002/ajmg.a.31862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppers M, van Blitterswijk MM, Vlam L, Rowicka PA, van Vught PW, Groen EJ, Spliet WG, Engelen-Lee J, Schelhaas HJ, de Visser M, et al. VCP mutations in familial and sporadic amyotrophic lateral sclerosis. Neurobiology of aging. 2012;33(837):e837–e813. doi: 10.1016/j.neurobiolaging.2011.10.006. [DOI] [PubMed] [Google Scholar]
- Krick R, Bremer S, Welter E, Schlotterhose P, Muehe Y, Eskelinen EL, Thumm M. Cdc48/p97 and Shp1/p47 regulate autophagosome biogenesis in concert with ubiquitin-like Atg8. The Journal of cell biology. 2010;190:965–973. doi: 10.1083/jcb.201002075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Nagano Y, Taylor JP, Lim KL, Yao TP. Disease-causing mutations in parkin impair mitochondrial ubiquitination, aggregation, and HDAC6-dependent mitophagy. The Journal of cell biology. 2010;189:671–679. doi: 10.1083/jcb.201001039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim MK, Kawamura T, Ohsawa Y, Ohtsubo M, Asakawa S, Takayanagi A, Shimizu N. Parkin interacts with LIM Kinase 1 and reduces its cofilin-phosphorylation activity via ubiquitination. Experimental cell research. 2007;313:2858–2874. doi: 10.1016/j.yexcr.2007.04.016. [DOI] [PubMed] [Google Scholar]
- Matsuda N, Sato S, Shiba K, Okatsu K, Saisho K, Gautier CA, Sou YS, Saiki S, Kawajiri S, Sato F, et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. The Journal of cell biology. 2010;189:211–221. doi: 10.1083/jcb.200910140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta SG, Khare M, Ramani R, Watts GD, Simon M, Osann KE, Donkervoort S, Dec E, Nalbandian A, Platt J, et al. Genotype-Phenotype studies of VCP-associated Inclusion Body Myopathy with Paget Disease of Bone and/or Frontotemporal Dementia. Clinical genetics. 2012 doi: 10.1111/cge.12000. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller JM, Deinhardt K, Rosewell I, Warren G, Shima DT. Targeted deletion of p97 (VCP/CDC48) in mouse results in early embryonic lethality. Biochemical and biophysical research communications. 2007;354:459–465. doi: 10.1016/j.bbrc.2006.12.206. [DOI] [PubMed] [Google Scholar]
- Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. The Journal of cell biology. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, Youle RJ. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS biology. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedelsky NB, Pennuto M, Smith RB, Palazzolo I, Moore J, Nie Z, Neale G, Taylor JP. Native functions of the androgen receptor are essential to pathogenesis in a Drosophila model of spinobulbar muscular atrophy. Neuron. 2010;67:936–952. doi: 10.1016/j.neuron.2010.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Lee SB, Lee S, Kim Y, Song S, Kim S, Bae E, Kim J, Shong M, Kim JM, et al. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441:1157–1161. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- Poole AC, Thomas RE, Andrews LA, McBride HM, Whitworth AJ, Pallanck LJ. The PINK1/Parkin pathway regulates mitochondrial morphology. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:1638–1643. doi: 10.1073/pnas.0709336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole AC, Thomas RE, Yu S, Vincow ES, Pallanck L. The mitochondrial fusion-promoting factor mitofusin is a substrate of the PINK1/parkin pathway. PloS one. 2010;5:e10054. doi: 10.1371/journal.pone.0010054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovich E, Kerem A, Frohlich KU, Diamant N, Bar-Nun S. AAA-ATPase p97/Cdc48p, a cytosolic chaperone required for endoplasmic reticulum-associated protein degradation. Mol Cell Biol. 2002;22:626–634. doi: 10.1128/MCB.22.2.626-634.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritson GP, Custer SK, Freibaum BD, Guinto JB, Geffel D, Moore J, Tang W, Winton MJ, Neumann M, Trojanowski JQ, et al. TDP-43 mediates degeneration in a novel Drosophila model of disease caused by mutations in VCP/p97. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2010;30:7729–7739. doi: 10.1523/JNEUROSCI.5894-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritz D, Vuk M, Kirchner P, Bug M, Schutz S, Hayer A, Bremer S, Lusk C, Baloh RH, Lee H, et al. Endolysosomal sorting of ubiquitylated caveolin-1 is regulated by VCP and UBXD1 and impaired by VCP disease mutations. Nat Cell Biol. 2011;13:1116–1123. doi: 10.1038/ncb2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z, Hayashi YK, Mitsuhashi S, Goto K, Kaneda D, Choi YC, Toyoda C, Hieda S, Kamiyama T, Sato H, et al. Characterization of the Asian myopathy patients with VCP mutations. European journal of neurology : the official journal of the European Federation of Neurological Societies. 2012;19:501–509. doi: 10.1111/j.1468-1331.2011.03575.x. [DOI] [PubMed] [Google Scholar]
- Smith R, Taylor JP. Dissection and imaging of active zones in the Drosophila neuromuscular junction. Journal of visualized experiments : JoVE. 2011 doi: 10.3791/2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spina S, Van Laar AD, Murrell JR, Hamilton RL, Kofler JK, Epperson F, Farlow MR, Lopez OL, Quinlan J, Dekosky ST, et al. Phenotypic variability in three families with valosin-containing protein mutation. European journal of neurology : the official journal of the European Federation of Neurological Societies. 2012 doi: 10.1111/j.1468-1331.2012.03831.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka A, Cleland MM, Xu S, Narendra DP, Suen DF, Karbowski M, Youle RJ. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. The Journal of cell biology. 2010;191:1367–1380. doi: 10.1083/jcb.201007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang WK, Li D, Li CC, Esser L, Dai R, Guo L, Xia D. A novel ATP-dependent conformation in p97 N-D1 fragment revealed by crystal structures of disease-related mutants. EMBO J. 2010;29:2217–2229. doi: 10.1038/emboj.2010.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tresse E, Salomons FA, Vesa J, Bott LC, Kimonis V, Yao TP, Dantuma NP, Taylor JP. VCP/p97 is essential for maturation of ubiquitin-containing autophagosomes and this function is impaired by mutations that cause IBMPFD. Autophagy. 2010;6:217–227. doi: 10.4161/auto.6.2.11014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts GD, Wymer J, Kovach MJ, Mehta SG, Mumm S, Darvish D, Pestronk A, Whyte MP, Kimonis VE. Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat Genet. 2004;36:377–381. doi: 10.1038/ng1332. [DOI] [PubMed] [Google Scholar]
- Weihl CC, Miller SE, Hanson PI, Pestronk A. Transgenic expression of inclusion body myopathy associated mutant p97/VCP causes weakness and ubiquitinated protein inclusions in mice. Human molecular genetics. 2007;16:919–928. doi: 10.1093/hmg/ddm037. [DOI] [PubMed] [Google Scholar]
- White SR, Lauring B. AAA+ ATPases: achieving diversity of function with conserved machinery. Traffic. 2007;8:1657–1667. doi: 10.1111/j.1600-0854.2007.00642.x. [DOI] [PubMed] [Google Scholar]
- Ye Y, Meyer HH, Rapoport TA. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature. 2001;414:652–656. doi: 10.1038/414652a. [DOI] [PubMed] [Google Scholar]
- Ye Y, Shibata Y, Kikkert M, van Voorden S, Wiertz E, Rapoport TA. Recruitment of the p97 ATPase and ubiquitin ligases to the site of retrotranslocation at the endoplasmic reticulum membrane. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:14132–14138. doi: 10.1073/pnas.0505006102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziviani E, Tao RN, Whitworth AJ. Drosophila parkin requires PINK1 for mitochondrial translocation and ubiquitinates mitofusin. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:5018–5023. doi: 10.1073/pnas.0913485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.