Abstract
The bacterial decoding region of 16S ribosomal RNA has multiple modified nucleotides. In order to study the role of N4,2′-O-dimethylcytidine (m4Cm), the corresponding phosphoramidite was synthesized utilizing 5′-silyl-2′-ACE chemistry. Using solid-phase synthesis, m4Cm, 5-methylcytidine (m5C), 3-methyluridine (m3U), and 2′-O-methylcytidine (Cm) were site-specifically incorporated into small RNAs representing the decoding regions of different bacterial species. Biophysical studies were then used to provide insight into the stabilizing roles of the modified nucleotides. These studies reveal that methylation of cytidine and uridine has different effects. The same modifications at different positions or sequence contexts within similar RNA constructs also have contrasting roles, such as stabilizing or destabilizing the RNA helix.
Keywords: ribosomal RNA modification, decoding region, N4, 2′-O-dimethylcytidine, 5-methylcytidine, 3-methyluridine
1. Introduction
The ribosome carries out protein synthesis, an essential process for life. The key steps of protein translation include decoding of the mRNA three-nucleotide codons with the correct tRNA anticodons, followed by peptide bond formation.1 The decoding region (helix 44) is a very important functional region of the 16S ribosomal RNA (rRNA), which is involved in translational accuracy (decoding) to synthesize proteins in an error-free manner.1 This region is involved in the selection of initiator tRNA, as well as cognate aminoacyl-tRNAs, according to the mRNA sequence. The decoding region is involved in binding to mRNA, tRNAs, and the large subunit rRNA.2–5 This region is also responsible for translocation.6 In addition, several known antibiotics such as aminoglycosides bind to the decoding region.7
Modified nucleosides are a universal feature of ribosomal RNAs. Over one hundred modified nucleosides have been discovered in RNA,8 which enhance the diverse structural and functional ability of RNA.9 A wide variety of modified nucleosides are present at or near the functional sites of the ribosome.10 The modified nucleotides N4,2′-O-dimethylcytidine (m4Cm), 5-methylcytidine (m5C), and 3-methyluridine (m3U) are found in the bacterial decoding region of 16S rRNA. The modified nucleotide m4Cm is a unique nucleotide only found to date in helix 44.11–16 The modified nucleotides m4Cm at position 1402, m5C at 1407, and m3U at 1498 are found in E. coli 16S rRNA. The modified nucleotides m4Cm and m3U are commonly found at positions 1402 and 1498, respectively, in the decoding region of various other bacterial species; whereas, the nucleotide m5C varies in number and position in different bacterial decoding regions. In Clostridium acetobutylicum, m5C is found at 1409; in Thermus thermophilus, there are three m5C residue at 1400, 1404 and 1407; and in Thermotoga maritime, there is no m5C in the decoding region, but a Cm residue occurs at 1409 (E. coli numbering).14–16
The nucleoside m4Cm was isolated from E. coli and characterized by Nichols and Lane in 1966.17,18 The modified nucleosides m5C and m3U were first isolated by Amos and Korn in 1958 and Hall in 1963.19,20 RsmF and RsmE are specific methyltransferase enzymes that post-transcriptionally methylate pyrimidines to generate m5C1407 and m3U1498, respectively.21,22 The methyltransferase enzymes RsmH and RsmI are responsible for N4-base and 2′-O-sugar methylation of m4Cm1402.23
The fact that modified nucleotides are present in the biologically significant decoding region implies that they have important roles; however, these roles are still not very well understood. A mutational analysis of conserved bases m4Cm1402 and A1500 in 16S rRNA (E. coli) revealed that these nucleotides may be involved in important tertiary interactions.24 Crystal structures of the ribosome show m4Cm and m3U hydrogen bonded with mRNA.4,5 Furthermore, there is a metal ion that bridges m4Cm and G1401 to the codon.5 Another report suggested a connection between the ribosomal modifications and initiator tRNA selection.25 Previous studies provided insight into the effects of functional group modifications on nucleoside structure.26 The next step is to determine the role of modified nucleotides within the context of small rRNA models, which will help to further understand their roles within the complete ribosome.
In order to understand the roles of modified nucleotides within the context of a functionally important model RNA system representing the decoding region of 16S rRNA, they were site-specifically incorporated into the target by using chemical synthesis. A modified nucleotide phosphoramidite of m4Cm was generated and used in solid-phase synthesis, along with Cm, m5C, m3U, and standard nucleotide phosphoramidites, to produce a series of bacterial decoding region model systems (Fig. 1). These analogues were constructed by a mix-and-match approach, using a set of 5′- and 3′-half single-stranded oligonucleotides. The stabilizing roles of the modified nucleotides in the decoding region RNAs were then determined by using UV melting. A related decoding region model system without modifications was used previously for ligand interactions and shown to be a good representation of the native rRNA within the context of complete ribosomes.27–30 The work in this study reveals that the natural pyrimidine modifications have a subtle effect on stability of this functionally important region of the ribosome.
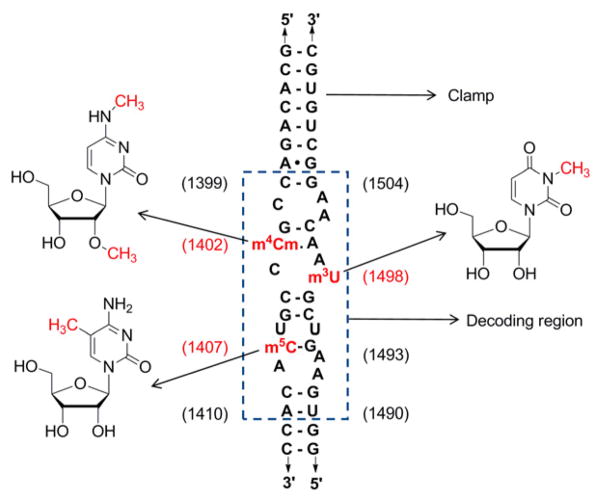
Figure 1.
The decoding region model system is shown with indication of the modified nucleoside identity and positions. Nucleotide numbers are shown in parenthesis (E. coli numbering). The native decoding sequence is boxed, whereas additional nucleotides outside the box are used to stabilize the duplex structure (clamp).
2. Results and Discussion
2.1 Phosphoramidite Synthesis
Chemical synthesis was employed for the site-specific incorporation of modified nucleotides into the small (21–24 nucleotide) rRNA constructs. Among several available approaches, the phosphoramidite method is the most frequently used.31 Synthesis of the modified nucleotide m4Cm phosphoramidite was accomplished using 5′-BzH-2′-ACE protection strategies (Scaringe method).32 This method has several distinct advantages, such as mild deprotection, higher coupling efficiencies, and shorter coupling times for small oligonucleotide constructs. The modified nucleoside m4Cm has methyl groups at the N4 and 2′-O positions; hence, exocyclic amine and 2′-ACE-orthoester protection were not required. Therefore, only two steps were required for synthesis of the m4Cm phosphoramidite (Scheme 1) instead of five steps as needed for the standard 5′-BzH-2′-ACE nucleotides.
Scheme 1.
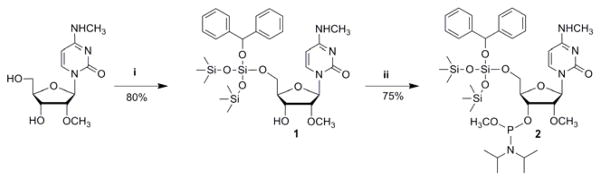
Synthesis of 5′-O-benzhydryloxy-bis(trimethylsilyloxy)silyl-3′-O-(N,N-diisopropylamino)methoxyphosphonyl-N4,2′-O-dimethylcytidine: i) benzhydryloxy-bis(trimethysilyloxy)chlorosilane (BzH-Cl), DMF-CH2Cl2, 0 °C, 3 h; ii) bis(N,N-diisopropylamino)-methoxyphosphine, 1H-tetrazole, CH2Cl2, rt, overnight.
The 5′-BzH protective group was introduced to m4Cm with benzhydryloxy-bis(trimethylsilyloxy)chlorosilane (BzH-Cl) and N,N-diisopropylamine at 0 °C for 3 h to afford compound 1 in 80% yield. The phosphoramidite was obtained by the addition of bis(N,N-diisopropylamino)methoxyphosphine and 1H-tetrazole to a CH2Cl2 solution of compound 1 at room temperature. The final product 5′-O-benzhydryloxy-bis(trimethylsilyloxy)silyl-3′-O-(N,N-diisopropylamino)methoxyphosphonyl-N4,2′-O-dimethylcytidine (2) was obtained in 75% yield from 1. The m4Cm phosphoramidite was synthesized in overall 60% yield in two steps from m4Cm. The Cm phosphoramidite is commercially available.
2.2 Solid-phase Synthesis of Decoding Region RNA
The m4Cm, m5C, m3U, and Cm modified nucleotide phosphoramidites along with standard A, C, G and U phosphoramidites were used to synthesize RNAs containing site-specific modifications by 5′-BzH-2′-ACE solid-phase synthesis on the μmole scale. Nine different RNA strands were synthesized (Supplementary Information, Table S1) by utilizing the 5′-silyl-2′-ACE protocol and deprotected according to literature procedures.33–35 Eleven different decoding region model systems were generated from the single strands (Fig. 2), such that the individual effects of the modified nucleotides could be studied. Nine different decoding region E. coli rRNA constructs were generated, namely unmodified (C-C-U), singly modified (Cm-C-U, m4Cm-C-U, C-m5C-U, and C-C-m3U), doubly modified decoding regions (m4Cm-m5C-U, m4Cm-C-m3U, and C-m5C-m3U), and triply modified (m4Cm-m5C-m3U, wild type, Fig. 1), in which the three symbols indicate the nucleotide composition at positions 1402, 1407 and 1498, respectively. The Cm-C-U decoding region RNA was generated in order to determine the effects of sugar-only modification. The C. acetobutylicum and T. thermophilus decoding region RNAs were synthesized so that the effects of modified nucleotides at different positions or additional locations could be studied.
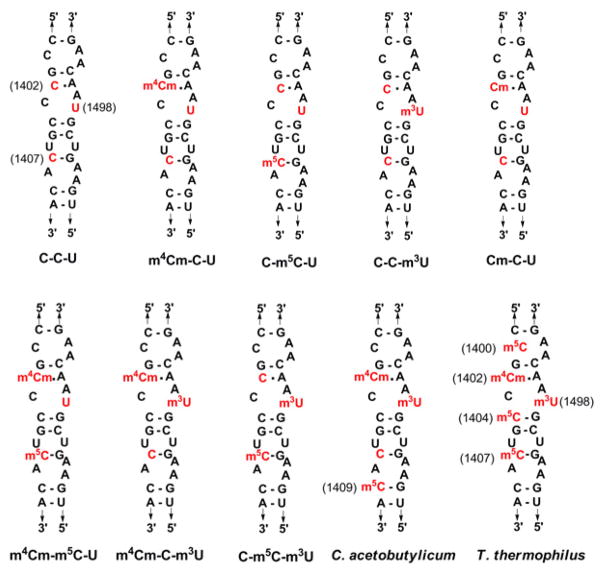
Figure 2.
The decoding region model systems representing unmodified C-C-U, m4Cm-C-U, C-m5C-U, C-C-m3U, Cm-C-U, m4Cm-m5C-U, m4Cm-C-m3U, C-m5C-m3U, Clostridium acetobutylicum, and Thermus thermophilus are shown with E. coli numbering. The modified nucleosides m4Cm and m3U are found at positions 1402 and 1498. The modified nucleotide m5C is found at positions 1407 and 1409 in E. coli and C. acetobutylicum, respectively. There are three m5C residues at positions 1400, 1404, and 1407 in T. thermophilus.
2.3 UV-Melting Studies
Thermal melting studies were carried out on purified single-stranded RNAs in cacodylate buffer (20 mM sodium cacodylate, 1 M NaCl, and 0.5 mM EDTA buffer, pH 7.0). UV-absorbance values of the 5′- and the 3′-half single-stranded decoding region RNA samples were determined at 90 °C, and the strands were mixed in equimolar ratios to obtain the duplex decoding regions. Absorbance versus temperature profiles (UV-melting curves) were obtained for all the RNA constructs at five different concentrations per duplex. The Meltwin 3.5 program was used to derive the thermodynamic parameters (ΔG°37, ΔH°, ΔS°, and Tm).36 Each melting experiment was done in triplicate, and the derived thermodynamic values were averaged. The modified and unmodified decoding region analogues were also studied in KCl buffer (20 mM cacodylic acid, 1 M KCl, and 0.5 mM EDTA, pH 7.0), but no differences were observed. The melting data for the unmodified decoding region were compared to data from all other RNA constructs. The difference of change in free energy (ΔΔG°37) was calculated by Equation 1, in which (A) is the modified construct and (B) is the unmodified decoding region (C-C-U) construct.
| (Eq 1) |
Analysis of the UV-melting data of various decoding region constructs reveals information about the stabilizing roles of the modified nucleotides (Table 1). The duplexes are all relatively stable, with ΔG°37 values ranging from −10.5 to −12.2 kcal/mol. Note that this 16% difference in ΔG° values is significant compared to the smaller error of the experiment (2%).37 Comparison of the UV-melting data (Table 1) for the m4Cm-C-U decoding region with the unmodified decoding region C-C-U gives a ΔΔG°37 value of −1.3 kcal/mol, revealing that m4Cm has a stabilizing role compared to C. In contrast, m5C has a slightly destabilizing role; when the decoding region C-m5C-U is compared with the unmodified decoding region C-C-U, the ΔΔG°37 value is 0.6 kcal/mol. The m4Cm nucleotide is stabilizing, m5C is slightly destabilizing, and the resultant doubly modified decoding region m4Cm-m5C-U has intermediate stability, suggesting that the stabilizing effects are additive.
Table 1.
UV-melting data for decoding region analogues.
| Analogues | ΔG°37 (kcal/mol) | ΔΔG°37 (kcal/mol) | ΔH° (kcal/mol) | ΔS° (cal/K· mol) | Tm(°C) |
|---|---|---|---|---|---|
| E. coli analogues | |||||
| C-C-U | −10.9 ± 0.2 | 0 | −49 ± 5 | −122 ± 15 | 65.6 |
| m4Cm-C-U | −12.2 ± 0.2 | −1.3 | −61 ± 4 | −156 ± 13 | 68.9 |
| C-m5C-U | −10.3 ± 0.2 | 0.6 | −48 ± 5 | −120 ± 17 | 63.6 |
| C-C-m3U | −10.7 ± 0.2 | 0.2 | −51 ± 7 | −129 ± 21 | 63.3 |
| Cm-C-U | −11.5 ± 0.2 | −0.6 | −55 ± 5 | −141 ± 17 | 66.3 |
| m4Cm-m5C-U | −11.4 ± 0.2 | −0.5 | −58 ± 7 | −149 ± 21 | 66.1 |
| m4Cm-C-m3U | −11.3 ± 0.2 | −0.4 | −55 ± 7 | −141 ± 21 | 66.3 |
| C-m5C-m3U | −10.5 ± 0.2 | 0.4 | −47 ± 6 | −119 ± 20 | 65.4 |
| m4Cm-m5C-m3U | −11.3 ± 0.2 | −0.4 | −57 ± 7 | −148 ± 21 | 65.6 |
|
| |||||
| C. acetobutylicum | −11.4 ± 0.2 | −0.5 | −58 ± 7 | −149 ± 21 | 66.1 |
|
| |||||
| T. thermophilus | −11.7 ± 0.2 | −0.8 | −61 ± 6 | −159 ± 20 | 66.3 |
Buffer conditions: 20 mM sodium cacodylate, 1 M NaCl, 0.5 mM Na2EDTA, pH 7.0.
The modified nucleotide m3U has a negligible effect on the decoding region stability in the case of the singly modified construct, C-C-m3U. This result is confirmed by comparing the ΔG°37 values for C-m5C-U and C-m5C-m3U or m4Cm-m5C-U and m4Cm-m5C-m3U, in which m3U has little impact (Table 1). In contrast, the combinations of m3U and/or m5C with m4Cm (m4Cm-C-m3U, m4Cm-m5C-U or m4Cm-m5C-m3U) cause the stabilizing effects of m4Cm to be diminished. In this case, the effects do not appear to be additive, and instead indicate cooperativity of the modifications. These differences are likely the result of unique structural effects of cytidine and uridine methylation. Overall, the three modified nucleotides have distinct, yet at the same time subtle, effects on the decoding region stability.
In order to examine more closely the stabilizing effect of m4Cm, the sugar methylated analogue Cm was analyzed. The ΔG°37 value for Cm-C-U is compared with those for C-C-U and m4Cm-C-U RNAs (Table 1). The ΔΔG°37 value reveals that Cm is stabilizing relative to cytidine, but less stabilizing than dimethylated cytidine, m4Cm. This result shows that the stabilizing effect of m4Cm comes from both sugar and base methylation. The m4C phosphoramidite was not available; therefore, the stabilizing effect of base methylation alone was not examined directly.
Our initial studies were carried out on the E. coli decoding region analogues. It was of interest to consider the effects of m5C at different positions of the decoding region, since this modification is present at different positions, as well as in varying numbers in other organisms. In C. acetobutylicum, m5C is present at position 1409 instead of 1407 as in E. coli. When comparing the stabilities of the two decoding regions, it has been found that there is no noticeable difference (Table 1). In contrast, there is a slight stabilizing effect in the T. thermophilus construct compared to the E. coli decoding region; the former has three m5C nucleotides in its decoding region compared to only one in the latter (Table 1). The two additional m5C nucleotides occur at positions 1400 and 1404 of T. thermophilus (Fig. 2). The modified nucleotide m5C present at position 1407 was shown to have a small destabilizing effect in the E. coli construct. Thus, the additional m5C nucleotides at positions 1400 and 1404 have stabilizing effects to provide overall greater stability to the T. thermophilus analogue compared to the E. coli decoding region. Therefore, as was observed previously with pseudouridine,38 the effects of nucleotide (m5C) modification depend on the locations of those modifications within the RNA construct.
2.4 Circular Dichroism Studies
In order to examine the effects of modified nucleotides on the overall conformation of the bacterial decoding region, circular dichroism (CD) studies of the various decoding region analogues were carried out. The spectrum of the unmodified decoding region RNA (C-C-U) has a peak maximum at 266 nm and peak minimum at 235 nm (Fig. 3A). The spectra for fully modified (m4Cm-m5C-m3U) and singly modified m4Cm decoding region analogues have similar peak maxima and minima. The CD spectra show that all of the analogues have typical A-form RNA conformations.39 Comparison of CD spectra for the three bacterial decoding region analogues (E. coli, C. acetobutylicum and T. thermophilus) reveals that these related RNAs also share similar structural features (Fig. 3B). The spectrum of the T. thermophilus RNA has an additional shoulder between 275 and 285 nm, suggestive of an alternate conformation or unique stacking arrangement with the m5C residues. With the exception of the T. thermophilus analogue, there are no significant effects of modified nucleotides on the overall conformations of the decoding region analogues. These results are consistent with the thermal melting data in which the modified nucleotides have only subtle effects on stability, and the stabilizing and destabilizing effects are compensatory, such that the overall stabilities are changed very little. Similarly, the CD data suggest that modified nucleotides in the decoding region do not change the average conformations of the RNA.
Figure 3.
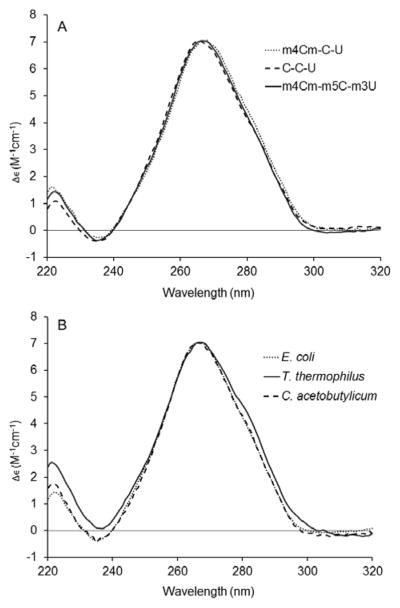
The circular dichroism spectra of the decoding region analogues (panel A: unmodified C-C-U, singly modified m4Cm-C-U, and fully modified m4Cm-m5C-m3U (wild type); panel B: decoding regions from E. coli, C. acetobutylicum and T. thermophilus) are shown. Each curve represents the average of five scans taken at room temperature.
2.5 The Roles of Methylation in Bacterial Decoding Region RNAs
In a crystal structure of E. coli ribosomes,40 m4Cm is stacked between G and C nucleotides. The thermal melting data reveal that m4Cm has a stabilizing role. Hence, methylation likely increases base-stacking interactions with the neighboring nucleotides. In contrast, the modified nucleotide m5C at position 1407 is part of a specific A-site motif in which two key residues, A1492 and A1493, are highly dynamic.41 The modified nucleotide m5C has a slight destabilizing effect, suggesting that its role might involve enhancing the dynamic nature of the A-site motif. The modified nucleotide m3U has only subtle effects on the decoding region stability. Therefore, m3U may have other roles in regulation of ribosome function. The combined modifications appear to have additive effects, such that the overall impact on structure and stability is quite small. Nonetheless, these modifications are known to influence the RNA structure and function within specific locations of the decoding region.
The modified nucleotide m5C is present at different locations within the bacterial decoding regions and also varies in number. Compared to the E. coli decoding region, the T. thermophilus RNA has two additional m5C nucleotides at positions 1400 and 1404, which have slight stabilizing effects. Thus, our results show that the same modified nucleotide at different locations of the decoding region has opposing effects on stability, which was also observed previously with the modified nucleotide pseudouridine.38 These modified nucleotides are located at a functionally significant region of the ribosome, suggesting that they may play a role in fine-tuning the RNA structure and function.10 The fact that m4Cm and m3U are hydrogen bonded with mRNA in ribosome crystal structures,4,5 and m4Cm1402 and A1500 are at the center of the decoding domain involving tertiary interactions with helix 45,24 suggests that the modified nucleotides have roles in binding mRNA and assisting in maintenance of tertiary interactions for proper ribosomal function. Regarding studies with the decoding region analogues from other species, it should be noted that T. thermophilus lives at high temperatures (80 °C), and rRNAs from such thermophiles contain a high number of modifications.14 The m5C nucleotide has an effect on decoding region stability, and therefore may play a role in maintenance of translational fidelity in these organisms.
The knockout of methyltransferase genes (rsmE, rsmF, rsmH and rsmI) that are responsible for modifications in the decoding region leads to growth defects in bacteria. The doubling time of the methyltransferase RsmE knockout strain, which is lacking methylation of uridine to m3U1498, has no significant difference in growth rate compared to wild type; however, mutant cell competition with wild type showed an exponential decrease in the mutant cells as the number of growth cycles increased.22 The doubling time of the methyltransferase RsmF (responsible for m5C1407) knockout strain is increased by 23% compared to wild type in rich media and 48% in minimal media at 42 °C.21 The doubling time of the double knockout strain for loss of methyltransferases RsmH and RsmI, which methylate the N4 and 2′-O-positions of C1402, respectively, is increased by 29% over wild type.23 The luciferase reporter assay illustrated that lack of methylation at C1402 also affected non-AUG initiation and fidelity of translation.23 Our data support these findings in which methylation in the decoding region is important, although not essential, for cell growth, and suggests subtle effects in maintaining key tertiary interactions for proper ribosomal function.
3. Conclusions
In order to site-specifically incorporate modified nucleotides into bacterial decoding region model systems, an N4,2′-O-dimethylcytidine (m4Cm) phosphoramidite was synthesized from the m4Cm nucleoside in 60% overall yield. A series of nine different oligonucleotide strands containing the m4Cm, m5C, m3U, and Cm modified nucleotides were generated by 5′-BzH-2′-ACE solid-phase synthesis on a μmole scale. The various oligonucleotide strands having site-specifically modified nucleotides were employed in the generation of a number of different bacterial decoding region model systems, which enabled biophysical studies to be carried out. The thermal melting and CD studies provide insight into the structural role of the modified nucleotides. The m4Cm stabilizing effect at position 1402 of the decoding region might play a role in stabilizing interactions with helix 45 and mRNA during translation through enhanced base-stacking, as shown in crystal structures. In contrast, the nucleotide m5C at position 1407 is part of the A-site dinucleotide bulge where two adenine residues at positions 1492 and 1493 flip out during tRNA recognition. Therefore, it is not surprising that m5C1407 has a destabilizing effect, which may facilitate dynamics of the decoding region or interactions with key components of the translational machinery. Future experimentation, such as NMR or single-molecule spectroscopy, would be necessary to verify such a mechanism.
These results are consistent with earlier studies showing that modified nucleotides each play unique roles in regulating local RNA stability, structure, or dynamics. Multiple modifications may have additive effects on RNA conformation or stability, and the overall effects may be subtle. Furthermore, the same modified nucleotide at different or multiple locations within the same RNA sequence may have additional stabilizing roles, such as m5C in the T. thermophilus decoding region. The importance of using synthetic approaches combined with biophysical approaches is demonstrated in this study. Future studies such as single molecule analysis on the different RNA constructs may allow more information regarding the individual roles of modified nucleotides to be gleaned. Ligand binding studies may also reveal whether the modified nucleotides influence important interactions with antibiotics in the decoding region.
4. Materials and Methods
All reactions were carried out in anhydrous conditions under argon atmosphere at room temperature, unless noted otherwise. The 1H, 13C, and 31P NMR spectra were recorded on a Varian 400 MHz or Unity 500 MHz spectrometer. ESI mass spectrometry was done on a Waters Micromass Zq instrument. MALDI-TOF spectra was obtained on a Bruker Ultraflex MALDI-TOF mass spectrometer. LC MS was carried out on a Waters LCT Premier XE. Flash column chromatography was carried out on silica gel 60 (240–400 mesh). All NMR solvents were from Cambridge Isotope Laboratories, Inc. Tetrazole was purchased from Glen Research. Benzhydryloxy-bis(trimethylsiloxy)chlorosilane (BzH-Cl) was purchased from Dharmacon Research, Inc. (Lafayette, CO). Methylene chloride (CH2Cl2) was purchased from Fisher and distilled over CaH2. All other chemicals were purchased from Sigma-Aldrich and Fisher and used without further purification. The nucleoside m4Cm was prepared as described previously.26
4.1 Phosphoramidite Synthesis
5′-O-Benzhydryloxy-bis(trimethylsilyloxy)silyl-N4,2′-O-dimethylcytidine (1)
In a round-bottom flask, m4Cm (408 mg, 1.5 mmol, 1 eq.) was dissolved in 5 mL of anhydrous DMF. To prepare solution A, 5 mL of distilled CH2Cl2 and 212 μL of diisopropylamine were added to the solution at 0 °C. Solution B was prepared at 0 °C by adding benzhydryloxy-bis(trimethylsiloxy)chlorosilane (BzH-Cl) (765 mg, 1.8 mmol, 1.2 eq), 5 mL of CH2Cl2, and diisopropylamine (254 μL, 1.8 mmol, 1 eq to BzHCl) to another round-bottom flask. One mL of solution B was added dropwise to solution A, and the mixture was stirred for 10 minutes. Every 15 minutes, 1 mL of solution B was added dropwise to solution A until solution B was empty. Reaction progress was monitored by TLC. The reaction was quenched with 5% NaHCO3 and extracted with CH2Cl2. The organic layer was washed with brine. The organic layer was dried over Na2SO4, filtered, and purified on silica gel using 0.5% triethylamine and 30–60% acetone in a hexane solvent gradient to afford 1 in 80% yield (790 mg). 1H NMR (500 MHz, CDCl3) δ (ppm) 0.01–0.03 (m, 18H), 2.84 (s, 3H), 3.62 (s, 3H), 3.80–4.00 (m, 2H), 4.05–4.17 (m, 3H), 5.13 (d, 1H), 5.90–5.99 (m, 2H), 7.16–7.33 (m, 10H) 7.77 (d, 1H);13C NMR (125 MHz, CDCl3) δ (ppm) 1.61, 1.68, 1.72, 8.83, 28.04, 31.63, 36.70, 46.06, 58.88, 60.99, 61.14, 67.59, 67.79, 83.59, 84.20, 87.85, 94.90, 126.56,126.64, 126.70, 127.26, 127.40, 127.58, 128.35, 128.49, 139.54, 144.20, 156.21, 164.44; HRMS calculated for C30H45N3O8NaSi3 682.2412, found 682.2407.
5′-O-Benzhydryloxy-bis(trimethylsilyloxy)silyl-3′-O-(N,N-diisopropylamino)methoxyphosphonyl-N4,2′-O-dimethylcytidine (2)
Compound 1 (330 mg, 0.5 mmol, 1.0 eq) was added to a round-bottom flask and dissolved in 2 mL of distilled CH2Cl2, and the clear solution was stirred for a few minutes. Bis(N,N-diisopropylamino)methoxyphosphine (400 μL, 367 mg, 1.4 mmol, 2.8 eq) and 1-H-tetrazole (18 mg, 0.25 mmol, 0.5 eq) were added to the clear solution while stirring, and the reaction proceeded for 12 h under argon. The reaction was quenched with 300 μL ethanol. The reaction was dried on rotary evaporator. The crude product was purified using a hexanes: acetone: triethylamine (4:1:0.5%) to hexanes: acetone: triethylamine (2:1:0.5%) solvent gradient to yield 2 as a clear oil in 75% yield (307 mg). 1H NMR (500 MHz, CDCl3) δ (ppm) 0.01–0.03 (m, 18H), 1.00–1.16 (m, 12H), 2.50, 2.52, 2.54, 2.56 (each as a s, 6H), 3.62 (s, 3H), 3.80–4.00 (m, 2H), 4.05–4.17 (m, 3H), 5.13 (d, 1H), 5.90–5.99 (m, 2H), 7.16–7.33 (m, 10H) 7.77 (d, 1H); 13C NMR (125 MHz, CDCl3) δ (ppm) 1.72, 1.74, 1.78, 8.21, 19.24, 19.31, 24.84, 24.89, 24.95, 25.03, 34.68, 43.01, 43.20, 43.25, 43.35, 46.86, 58.80, 83.96, 87.91, 89.03, 126.61, 126.69, 127.41, 128.47, 128.51, 128.57, 139.91, 144.26, 156.25, 164.48; 31P NMR (170 MHz, CDCl3) δ (ppm) 150.74, 151.87; HRMS calculated for C37H61N4O9NaPSi3 843.3382, found 843.3371. (Fig. S1–S4)
4.2 Synthesis of the Decoding Region RNAs
The m4Cm phosphoramidite was sent to Dharmacon Research Inc. (Lafayette, CO) where the oligonucleotide synthesis was carried out. The bacterial decoding region RNAs (Table S1, Supplementary Information) were each synthesized on a 1 μmol scale.
4.3 RNA Deprotection
The removal of protecting groups on the phosphate backbone and exocyclic amines was carried out immediately after synthesis of the oligonucleotide. The methyl protecting groups on the phosphate backbone were removed by using disodium 2-carbamoyl-2-cyanoethylene 1,1-dithiolate (S2Na2) while the RNA was still on the polystyrene support.42 Next, 40% methylamine in water43,44 was used for deprotection of the exocyclic amines. This reaction also removed the acetyl protection of the 2′-bis-acetoxyethyl orthoester to generate 2′-bis-hydroxyethyl orthoester and cleaved the oligonucleotide from the solid support. These steps were done at Dharmacon Research. The RNAs were obtained as the 2′-bis-hydroxyethyl orthoester protected forms. The 2′-orthoester was deprotected with 400 μL of buffer containing 100 mM acetic acid and pH adjusted to 3.8 with TEMED. The RNA was incubated with buffer for 30–35 minutes at 60 °C. The sample was completely dried on a Speedvac evaporator.
4.4 RNA Purification
The dried RNA samples were dissolved in double deionized water (ddH2O). The UV absorbance values were obtained and the concentrations were calculated by using the Beer-Lambert equation (Eq. 2),
| (Eq 2) |
in which ε, C and λ are the extinction coefficient, concentration, and path length, respectively. The extinction coefficients (ε) are 190,400 cm−1M−1 and 242,500 cm−1M−1 for the 5′ and 3′ halves of the decoding region, respectively. Authenticity and purity of RNA samples were confirmed by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) and polyacrylamide gel electrophoresis (PAGE). The oligonucleotides of the decoding region were purified by a using 20% denaturing PAGE (19:1 acrylamide:bisacrylamide) in 0.5 M TBE buffer. Ethanol precipitations were carried out after electroelution. The samples were dialyzed in RNase-free, ddH2O for three days. RNA identities and purities were reconfirmed by MALDI-TOF MS (Fig. S5, Supplementary Information). The incorporation of modified nucleotides was further confirmed by P1 nuclease digestion or NaOH hydrolysis and phosphatase digestion by bovine calf intestinal phosphatase (CIF) and analysis of the resulting nucleosides by either high performance liquid chromatography (HPLC) or liquid chromatography mass spectrometry LCMS. The LCMS spectra of the 5′ half of modified decoding region (5′-GCACAGACCG(m4Cm)CCGU (m5C)ACACC-3′) showed seven different peaks (Fig. S6, Information), corresponding to six nucleosides (C, U, m5C, G, m4Cm and A) and dinucleotide m4CmpC. The dinucleotide peak m4CmpC further confirmed that m4Cm is 2′-O-methylated, which leads to resistance against RNA hydrolysis.
4.5 UV-Melting Experiments
In separate tubes, 2 OD units of purified, desalted RNA strands were dissolved in 100 μL melting buffer (20 mM sodium cacodylate, 1 M NaCl and 0.5 mM Na2EDTA at pH 7.0). Each strand RNA concentration was measured at 90 °C. Equimolar concentrations were calculated for both strands X and Y by using Equation 3.45 Volume VX and VY were mixed together to
| (Eq 3) |
obtain an equimolar ratio of both strands. A dilution series of five different concentrations (8 to 90 μM) of sample were prepared. UV-melting studies of the decoding region RNAs were obtained on an Aviv 14DS UV-visible spectrophotometer equipped with a five-cuvette thermoelectric controller. Three microcuvettes with 1 mm path lengths and volumes 60 μL and two cuvettes with 2 mm path lengths and volumes of 120 μL were employed, and the data was collected at 280 nm every 0.5 °C between 0 and 90 °C at 1 °C/min temperature increments. The exact concentrations of the RNAs were calculated using the UV absorbance (260 nm) at 90 °C. The Meltwin 3.5 program was used to derive the thermodynamic parameters, assuming a two-state-equilibrium for RNA melting.36 Melting experiments were done in triplicate, and the thermodynamic values for each individual experiment were averaged. Reciprocal melting temperature vs. ln(total strand concentration/4) plots gave similar results (Fig. S8).
Single-strand extinction coefficients were calculated as described by Richards46 except that the extinction coefficients for uridine (1.0 × 104 cm−1 M−1 at pH 7.0) and cytidine (7.6 × 103 cm−1 M−1 at pH 7.0) were used instead of 3-methyluridine (1.0 × 104 cm−1 M−1 at pH 7.0)46 and methylated cytidines (7.6 × 103 cm−1 M−1 at pH 7.0)46 for all RNAs because the nearest-neighbor extinction coefficients for methylated nucleotides are unknown, therefore some error might exist for the modified RNAs.
4.6 Circular Dichroism Experiments
The circular dichroism (CD) spectra for all RNAs were obtained on a Chirascan CD spectrometer equipped with a water bath to control the temperature. Equimolar concentrations of the two strands (9.5 μM, 400 μL total volume) were annealed by heating to 90 °C followed by slow cooling to 0 °C. The samples were then diluted to 3.8 °M in 1 mL of buffer. The CD spectra for the RNAs were acquired from 220–320 nm in 1.0 nm steps with a bandwidth of 1.0 nm at 25 °C. The samples were scanned five times and an average was taken. The baseline subtraction and smoothing using the Savitsky-Golay algorithm were performed for the acquired averaged-CD spectra. The ellipticity values, θ, for the 220 to 320 nm wavelength range were converted to Δε Keeping using Equation 4,
| (Eq 4) |
(Eq 4) in which C and l denote the concentration of the sample and path length of the cuvette, respectively.47 The data were plotted as Δε versus the wavelength. Exact RNA concentrations of the CD samples were determined from the UV absorbance using the Beer-Lambert equation. All CD experiments were done in triplicate.
Supplementary Material
Acknowledgments
We thank Drs. J. SantaLucia, A. Feig, B. Ksebati, L. Hryhorczuk, and B. Shay for technical assistance and helpful discussions. This work is supported by the National Institutes of Health (AI061192, AI055496, and GM087596).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ogle JM, Carter AP, Ramakrishnan V. Trends Biochem Sci. 2003;28:259. doi: 10.1016/S0968-0004(03)00066-5. [DOI] [PubMed] [Google Scholar]
- 2.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Science. 2001;292:883. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 3.Ofengand J, Liou R. Biochemistry. 1981;20:552. doi: 10.1021/bi00506a017. [DOI] [PubMed] [Google Scholar]
- 4.Korostelev A, Trakhanov S, Laurberg M, Noller HF. Cell. 2006;126:1065. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 5.Selmer M, Dunham CM, Murphy FV, 4th, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Science. 2006;313:1935. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 6.VanLoock MS, Agrawal RK, Gabashvili IS, Qi L, Frank J, Harvey SC. J Mol Biol. 2000;304:507. doi: 10.1006/jmbi.2000.4213. [DOI] [PubMed] [Google Scholar]
- 7.Moazed D, Noller HF. Nature. 1987;327:389. doi: 10.1038/327389a0. [DOI] [PubMed] [Google Scholar]
- 8.Rozenski J, Crain PF, McCloskey JA. Nucleic Acids Res. 1999;27:196. doi: 10.1093/nar/27.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grosjean H. In: Fine-Tuning of RNA Functions by Modification and Editing. 1. Grosjean H, editor. Springer-Verlag; New York: 2005. p. 1. [Google Scholar]
- 10.Chow CS, Lamichhane TN, Mahto SK. ACS Chem Biol. 2007;2:610. doi: 10.1021/cb7001494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Limbach PA, Crain PF, McCloskey JA. Nucleic Acids Res. 1994;22:2183. doi: 10.1093/nar/22.12.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fellner P, Sanger F. Nature. 1968;219:236. doi: 10.1038/219236a0. [DOI] [PubMed] [Google Scholar]
- 13.McCloskey JA, Rozenski J. Nucleic Acids Res. 2005;33:D135. doi: 10.1093/nar/gki015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guymon R, Pomerantz SC, Crain PF, McCloskey JA. Biochemistry. 2006;45:4888. doi: 10.1021/bi052579p. [DOI] [PubMed] [Google Scholar]
- 15.Emmerechts G, Barbé S, Herdewijn P, Anné J, Rozenski J. Nucleic Acids Res. 2007;35:3494. doi: 10.1093/nar/gkm248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guymon R, Pomerantz SC, Ison JN, Crain PF, McCloskey JA. RNA. 2007;13:396. doi: 10.1261/rna.361607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nichols JL, Lane BG. Biochim Biophys Acta. 1966;119:649. doi: 10.1016/0005-2787(66)90147-x. [DOI] [PubMed] [Google Scholar]
- 18.Nichols JL, Lane BG. Biochim Biophys Acta. 1968;166:605. doi: 10.1016/0005-2787(68)90367-5. [DOI] [PubMed] [Google Scholar]
- 19.Amos H, Korn M. Biochim Biophys Acta. 1958;29:444. doi: 10.1016/0006-3002(58)90214-2. [DOI] [PubMed] [Google Scholar]
- 20.Hall RH. Biochem Biophys Res Comm. 1963;12:361. doi: 10.1016/0006-291x(63)90105-0. [DOI] [PubMed] [Google Scholar]
- 21.Andersen NM, Douthwaite S. J Mol Biol. 2006;359:777. doi: 10.1016/j.jmb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 22.Basturea GN, Rudd KE, Deutscher MP. RNA. 2006;12:426. doi: 10.1261/rna.2283106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kimura S, Suzuki T. Nucleic Acids Res. 2010;38:1341. doi: 10.1093/nar/gkp1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vila-Sanjurjo A, Dahlberg AE. J Mol Biol. 2001;308:457. doi: 10.1006/jmbi.2001.4576. [DOI] [PubMed] [Google Scholar]
- 25.Das G, Thotala DK, Kapoor S, Karunanithi S, Thakur S, Singh NS, Varshney U. EMBO J. 2008;27:840. doi: 10.1038/emboj.2008.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mahto SK, Chow CS. Bioorg Med Chem. 2008;16:8795. doi: 10.1016/j.bmc.2008.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Purohit P, Stern S. Nature. 1994;370:659. doi: 10.1038/370659a0. [DOI] [PubMed] [Google Scholar]
- 28.Stern S, Purohit P. Biochem Cell Biol. 1995;73:899. doi: 10.1139/o95-097. [DOI] [PubMed] [Google Scholar]
- 29.Hobbie SN, Kalapala SK, Akshay S, Bruell C, Schmidt S, Dabow S, Vasella A, Sander P, Böttger EC. Nucleic Acids Res. 2007;35:6086. doi: 10.1093/nar/gkm658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fan-Minogue H, Bedwell DM. RNA. 2008;14:148. doi: 10.1261/rna.805208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chow CS, Mahto SK, Lamichhane TN. ACS Chem Biol. 2008;3:30. doi: 10.1021/cb7002225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scaringe SA, Wincott FE, Caruthers MH. J Am Chem Soc. 1998;120:11820. [Google Scholar]
- 33.Scaringe SA. Methods Enzymol. 2000;317:3. doi: 10.1016/s0076-6879(00)17003-x. [DOI] [PubMed] [Google Scholar]
- 34.Scaringe SA. Methods. 2001;23:206. doi: 10.1006/meth.2000.1132. [DOI] [PubMed] [Google Scholar]
- 35.Hartsel SA, Kitchen DE, Scaringe SA, Marshall WS. Methods Mol Biol. 2005;288:33. doi: 10.1385/1-59259-823-4:033. [DOI] [PubMed] [Google Scholar]
- 36.McDowell JA, Turner DH. Biochemistry. 1996;35:14077. doi: 10.1021/bi9615710. [DOI] [PubMed] [Google Scholar]
- 37.SantaLucia J, Jr, Turner DH. Biopolymers. 1997;44:309. doi: 10.1002/(SICI)1097-0282(1997)44:3<309::AID-BIP8>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 38.Meroueh M, Grohar PG, Qiu J, SantaLucia J, Jr, Scaringe SA, Chow CS. Nucleic Acids Res. 2000;28:2075. doi: 10.1093/nar/28.10.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gray DM, Liu JJ, Ratliff RL, Allen FS. Biopolymers. 1981;20:1337. [Google Scholar]
- 40.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JHD. Science. 2005;310:827. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 41.Ogle JM, Murphy FV, Tarry MJ, Ramakrishnan V. Cell. 2002;111:721. doi: 10.1016/s0092-8674(02)01086-3. [DOI] [PubMed] [Google Scholar]
- 42.Dahl BH, Bjergårde K, Henriksen L, Dahl O. Acta Chem Scand. 1990;44:639. [Google Scholar]
- 43.Reddy MP, Hanna NB, Farooqui F. Tetrahedron Lett. 1994;35:4311. [Google Scholar]
- 44.Wincott F, DiRenzo A, Shaffer C, Grimm S, Tracz D, Workman C, Sweedler D, Gonzalez C, Scaringe S, Usman N. Nucleic Acids Res. 1995;23:2677. doi: 10.1093/nar/23.14.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Santalucia J., Jr . In: Spectrophotometry and Spectrofluorimetry. 2. Gore MG, editor. Oxford University press; Oxford, UK: 2000. p. 329. [Google Scholar]
- 46.Richards EG. Handbook of Biochemistry and Molecular Biology. 3. I CRS Press; Cleaveland, OH, USA: 1975. [Google Scholar]
- 47.Woody RW. Methods Enzymol. 1995;246:34. doi: 10.1016/0076-6879(95)46006-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.