Abstract
Tropomyosin is a ubiquitous actin-binding protein with an extended coiled-coil structure. Tropomyosin-actin interactions are weak and loosely specific, but they potently influence myosin. One such influence is inhibitory, and is due to tropomyosin’s statistically preferred positions on actin that sterically interfere with actin’s strong attachment site for myosin. Contrastingly, tropomyosin’s other influence is activating. It increases myosin’s overall actin affinity ~4-fold. Stoichiometric considerations cause this activating effect to equate to a ~ 47- fold effect of myosin on the actin-affinity of tropomyosin. These positive, mutual, myosin-tropomyosin effects are absent if S. cerevisiae tropomyosin replaces mammalian tropomyosin. To investigate these phenomena, chimeric tropomyosins were generated in which 38 residue muscle tropomyosin segments replaced a natural duplication within S. cerevisiae tropomyosin TPM1. Two such chimeric tropomyosins were sufficiently folded coiled-coils to allow functional study. The two chimeras differed from TPM1, but in opposite ways. Consistent with steric interference, myosin greatly decreased the actin-affinity of chimera 7, which contained muscle tropomyosin residues 228–265. On the other hand, myosin S1 increased by an order of magnitude the actin-affinity of chimera 3, which contained muscle tropomyosin residues 74–111. Similarly, myosin S1-ADP binding to actin was strengthened 2-fold by substitution of chimera 3 tropomyosin for wild type TPM1. Thus, a yeast tropomyosin was induced to mimic the activating behavior of mammalian tropomyosin by inserting a mammalian tropomyosin sequence. The data were not consistent with direct tropomyosin-myosin binding. Rather they suggest an allosteric mechanism, in which myosin and tropomyosin share an effect on the actin filament.
In the absence of Ca2+, muscle tropomyosin attaches to the outer domain of actin, where it blocks substantial portions of the actin site to which myosin otherwise binds strongly to actin (1–3). When Ca2+ binds to the thin filament regulatory protein troponin, tropomyosin shifts position on the filament surface to the outer edge of the inner domain of actin, i.e., to the edge of the myosin binding site. Tropomyosins in general are located in these positions on the actin filament, not limited to tropomyosins that are regulated by troponin (4). Only when myosin S1 is attached to the actin filament does tropomyosin undergoes a further azimuthal shift across more of the actin inner domain. This additional shift fully exposes the myosin binding site on actin (3, 5).
Despite this steric hindrance between the preferred binding sites for tropomyosin and myosin S1 when they attach to actin separately, there is a paradoxical, positive interaction between tropomyosin and myosin. Myosin S1 binds to actin-tropomyosin filaments, or to actin-troponin-tropomyosin filaments, with an affinity 3- to 7-fold higher than to bare actin (6–8). Results with tropomyosin alone are very similar to those with tropomyosin-troponin-Ca2+, indicating that this effect is primarily due to tropomyosin.
This effect has a dramatic consequence that necessarily follows from equilibrium linkage principles. That is, because seven myosin heads can bind to actin for every tropomyosin, the implication is that myosin S1 must increase the actin-affinity of tropomyosin (or of troponin-tropomyosin) by ~ 37- to 77-fold. This very high affinity binding has firm experimental support (9–11). It is an activating effect that is directly opposite of what would be expected based upon steric hindrance. Its importance is supported by other evidence that troponin-tropomyosin enhance, albeit more modestly, force, sliding speed, the rate of ATP product release by myosin, and other aspects of actin-myosin interactions (12–18). The mechanism of these effects remains an open question.
A fruitful, relatively recent approach to investigating tropomyosin’s actions is to investigate and exploit the divergent properties of yeast and mammalian tropomyosins (11, 19–21). In the present work this is pursued for the first time in a ‘host-guest’ manner, so termed in analogy to helical propensity studies in which guest amino acids are inserted into a host helical segment (22). The predominant tropomyosin isoform of S. cerevisiae, TPM1, spans 5 actins and has two properties that lend it to an attempt at such an approach. First, it contains an internal duplication of 38 residues. This duplication is flanked by flexible sites in the heptad repeat of the coiled-coil. Thus, one might be able to replace this one actin segment (i.e., this evolutionary internal duplication) of yeast host tropomyosin with a sequence(s) from foreign guest tropomyosin. Second, yeast tropomyosins have interesting functional differences compared to mammalian tropomyosins, including different spectroscopic effects on actin (23). The most significant dissimilarity is that yeast tropomyosins do not bind more strongly to actin-myosin than to actin (11, 19, 20), unlike all mammalian non-muscle and muscle tropomyosins that have been tested (7–11, 24, 25). (Yeast myosin-yeast tropomyosin effects have not been tested yet in such work, however.) Therefore, in principle one might test whether an important aspect of mammalian tropomyosin function, i.e, the tendency of tropomyosin and myosin to promote each other’s attachment to actin, is produced when yeast tropomyosin hosts inserted guest sequences from mammalian tropomyosin. In practice, utility depends upon such tropomyosins having 3-dimensional structures suitable for functional study, including proper folding and shape.
Following this approach, a panel of several chimeric tropomyosins were generated, by replacement of TPM1 residues 70–107 with various sequences from muscle tropomyosin. Actin binding function was detected for two chimeras with relatively high preservation of folding thermodynamics, but no actin binding was observed for four other chimeras that folded more poorly. As described below, the two functional chimeras had opposite properties. The chimera containing residues mostly from muscle tropomyosin’s 7th quasi-repeat could not bind to actin when myosin was present. In contrast, the chimera containing residues mostly from muscle tropomyosin’s 3rd repeat bound to actin much more strongly when myosin was present. The significance of these findings is discussed.
EXPERIMENTAL PROCEDURES
Tropomyosin mutagenesis
To create the chimeras described in Fig. 1, a 2-stranded synthetic oligonucleotide was created that contained four restriction sites, in sequence: an NcoI site; a BstBI site that is translationally neutral near TPM1 residue 67; a similar AflII site that is translationally neutral near TPM residue 108; a BamHI site. The oligonucleotide was inserted into the NcoI/BamHI sites of expression plasmid pET3d. The 5′-encoding portion of TPM1 was then inserted at the NcoI/BstBI sites, and then the 3′-encoding region was inserted at the AflII/BamHI sites. PCR was performed on the different regions of the α-tropomyosin cDNA targeted for chimera creation, and the PCR fragments successfully ligated into the BstBI/AflII sites of the modified pET3d. Coding sequences were confirmed by automated DNA sequencing (IDT). Each tropomyosin was encoded to contain after expression an N-terminal ala-ser dipeptide, as is necessary for binding of bacterially expressed, unacetylated tropomyosins both from yeast and from striated muscle (20, 26).
FIGURE 1. Design of chimeric host-guest tropomyosins.
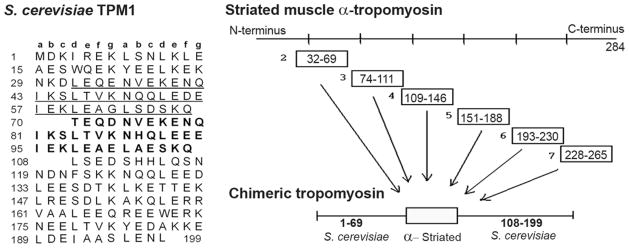
At left, the amino acid sequence of the predominant tropomyosin isoform of S. cerevisiae is shown in its alignment (20) with the heptad repeat (a–g) characteristic of coiled-coils. The heptad phase shifts near the start and finish of the second instance (in bold) of an internal repeat. The first instance of the repeat (underlined) has 31 of 38 residue identity with the second. To create the chimeras, the bold residues were replaced by alternative segments of muscle tropomyosin, as shown at right.
Protein purification
Recombinant tropomyosin, rabbit fast skeletal muscle actin, and rabbit myosin S1 were isolated as previously described (20, 27–29). The concentration of wt TPM1 was determined by quantitative amino acid analysis at the Yale University Keck Center. This agreed within 6% of the result as determined by absorbance, using an extinction coefficient calculated as in Gill and Von Hippel (30). The concentrations of the chimeras were determined by Bradford assay, using the wt TPM1 for the standard. Other purified protein concentrations were determined by absorbance, with myosin S1 A280 = 0.75 ml/mg and actin A290 = 0.62 ml/mg.
Circular dichroism
The circular dichroism of tropomyosin (θ=222 nm) was monitored using a Jasco J-710 spectropolarimeter, recorded as a function of increasing temperature. Conditions were: 50 mM NaH2PO4 (pH 7.0), 100 mM KCl, and 0.20 mg/ml tropomyosin. Fractional unfolding as a function of temperature was assessed as previously (27) to yield values for the enthalpy of a single transition, and the midpoint temperature of that transition. It was assumed that the folded state had ellipticity that varied modestly with temperature, to match the slopes of the lower temperature data. To obtain parameter and parameter error estimates, curve-fitting for this and other experiments in the present study pereformed by nonlinear least squares analysis, using the program SCIENTIST (Micromath).
Binding of tropomyosin to actin filaments
Actin filaments were pelleted by 20 minute centrifugation at 70,000 × g, 25 °C, using a TLA100 rotor in a TL100 ultracentrifuge. Figure 5 conditions were as follows: 10 mM Tris pH 7.5, 1mM dithiothreitol, 120 mM KCl, 5 mM MgCl2, 2.5 μM F-actin, 2.5 μM myosin S1, 0.5 mM glucose, 1unit Hexokinase. Pellets were re-suspended after sedimentation, the actin-bound tropomyosin was visualized by SDS-PAGE (a full experiment within one gel), and the amount of bound tropomyosin measured using GeneTools imaging software from SynGene. Data from at least two independent experiments per tropomyosin were pooled for analysis of combined data sets. Qualitatively similar results were obtained in the presence of 60 mM rather than 120 mM KCl, albeit with tighter actin-tropomyosin affinities that were not measured successfully. Binding curves were analyzed as a linear lattice process (31, 32) dependent on two equilibrium constants: (1) cooperativity parameter y, which is the increase in affinity due to end-to-end overlap of adjacent tropomyosins; (2) the overall Kapp, which is the product of the affinity of tropomyosin for an isolated site on actin and cooperativity parameter, y.
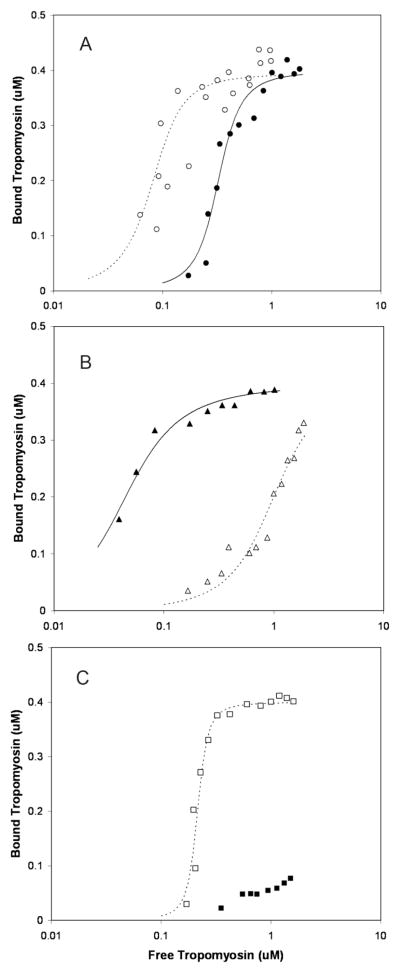
FIGURE 5. Differing effects of myosin S1 on control and chimeric tropomyosin binding to actin.
Increasing concentrations of wt TPM1 (A), chimera 3 (B), or chimera 7 (C) were added to actin filaments in the absence (open symbols) or presence (filled symbols) of myosin S1. Lines are best fit curves with parameters as listed in Table 1. Conditions were as described in Experimental Procedures. Binding was measured by quantitative SDS-PAGE analysis of the amount of tropomyosin present in F-actin pellets.
To assess the effect of S. cerevisiae TPM1 on the binding of 0.3 μM radio-labeled cardiac tropomyosin to 5 μM F-actin, similar procedures were used. However, binding was instead assessed as the difference between total sample radioactivity and supernatant radioactivity, as described (33), with the cardiac tropomyosin labeled on Cys 190.
Binding of Myosin S1 to the Thin Filament
Myosin S1-ADP binding to the thin filament was measured as previously described. Briefly, a Fluoromax Horiba-Jobin-Yvon spectrofluorometer was used to monitor the decrease in fluorescence intensity of cys-374 pyrene-labeled actin, as increasing myosin S1 was added (34, 35). Fluorescence data were analyzed as in previously (8, 36). Intensities were corrected for dilution as myosin S1 was added (8% or less). Conditions: 25 °C, 10 mM Tris-HCl (pH 7.5), 50 mM KCl, 5 mM MgCl2, 2 mM ADP, 1 mM dithiothreitol, 0.2 mg/ml bovine serum albumin, 1 mM glucose, 10 units hexokinase, 200 μM P1,P5-di(adenosine 5′)-pentaphosphate, 1 μM F-actin, 5 μM tropomyosin.
Fluorescence data were averaged from three experiments, and then analyzed by nonlinear least squares fitting to equation 3 from Tobacman and Butters (8). Data for thin filaments with wt or mutant tropomyosins were globally fit, using the assumption that the mutations altered the stability of the active state of the thin filament (thereby changing KT only). The mutations were assumed in this procedure to have no effect on the affinity of myosin for the thin filament active state (Ks), or on tropomyosin’s tendency to move to the M-state position on actin cooperatively (Y). KT is expressed per actin, as in the original derivation (8). In the model of McKillop and Geeves (37) the term KT has a somewhat similar meaning, albeit per tropomyosin.
RESULTS
Design of chimeric tropomyosins
S. cerevisiae tropomyosin isoform TPM1 is well suited as a potential host for a host-guest study of tropomyosin. TPM1 contains an internal duplication of 38 residues, which approximates the number of coiled-coiled residues required to extend fully along one actin monomer on the thin filament, 39.3 residues. Thus, the size of the duplication is a match to function, as it causes TPM1 to stretch along 5 rather than 4 actins (Fig. 1 (20)). This length does not, however, match the n × 7 size required for the characteristic, coiled-coil pattern, in which residues ‘a’ and ‘d’ of successive heptads have hydrophobic side chains that mediate dimerization. A continuous heptad phasing does not accommodate a 38 residue duplication. Instead, in TPM1 the phase shifts at the start and finish of the duplication, in what are termed “stammers”. These can be diagramed as four residue gaps (Fig. 1), or alternatively, as sites where a 3-4-3-3 pattern of hydrophobic residues interrupts the coiled-coil’s characteristic 3-4-3-4 (heptad) pattern. Importantly for the present study, such “stammers are believed to be sites of flexibility in coiled-coils (38).
In the present study this flexibly inserted, natural duplication was swapped out of TPM1, and nearly comparable guest segments of foreign tropomyosin were inserted in its place. The inserted sequences are shown in Fig. 1. To offer the greatest prospect for meaningful results, the guest segments were chosen to retain as much as possible their longitudinal alignment on the actin monomer after insertion into TPM1. Other three-dimensional considerations aside, improper position relative to actin would prevent authentic behavior for the guest segments. In other words, TPM1 residue 70, the start of the duplication, overlies an unknown but particular longitudinal position on an actin monomer. What successive residues in muscle tropomyosin, spaced 39 amino acids apart, overly this same site on actin? Yeast and mammalian tropomyosins lack sequence similarity to each other, and there are no high resolution data of either protein when attached to actin. Therefore, creation of the chimeras required new data on the relative positions, the relative longitudinal alignments, of these two tropomyosins.
To test the relative positions of the ends of muscle tropomyosin and TPM1, we examined whether they could promote each other’s binding to actin. As shown in Fig. 2, TPM1 had a biphasic effect on the binding of a low concentration of 3H-muscle tropomyosin to actin. High concentrations of TPM1 competitively displaced the muscle tropomyosin from actin. More significantly, at lower concentrations (left portion of curve) TPM1 dramatically increased the amount of the muscle tropomyosin that bound to actin. This implies that the ends of muscle tropomyosin and TPM1 bound sufficiently close to the same longitudinal position on actin, that they could cooperatively promote each other’s attachment to the thin filament by hetero-end-to-end polymerization. There remains an alignment imprecision of several residues, because the end-to-end overlaps of the two tropomyosins differ.
FIGURE 2. S. cerevisiae tropomyosin TPM1 cooperatively promotes binding of muscle tropomyosin to actin.
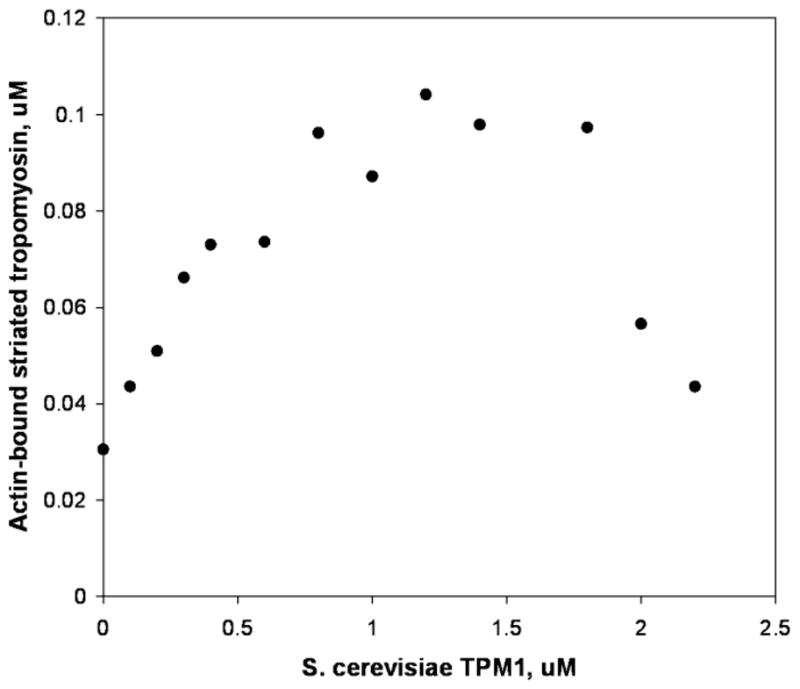
Bovine cardiac tropomyosin was added to F-actin at a concentration (0.3 μM) selected to be near but below the threshold that would produce a sharp increase actin attachment via cooperative tropomyosin end-to-end effects (33). In the experiment shown, a cooperative increase in cardiac tropomyosin actin-binding was produced not by adding more cardiac tropomyosin, but instead by adding S. cerevisiae TPM1 in increasing concentrations. Binding was measured by comparison of pre- vs. post sedimentation radioactivity of cardiac tropomyosin labeled on cys-190 (33).
Muscle tropomyosin has been viewed as comprised of seven loose, quasi-repeats that together result in similar surface charges every 39.3 residues (39, 40). In Fig. 1, the chimeras of the present study are designated 2, 3, 4, 5, 6, and 7, indicating which of these muscle tropomyosin quasi-repeats most closely equaled each guest segment. However, the segments slightly bridged these quasi-repeats, which were not their basis. Rather, the chosen guest inserts were the set of 38 residue regions that (i) began with ‘d’ heptad positions to match TPM1 residue 70, and (ii) best matched the TPM1 70–107 position relative to actin, assuming that the muscle and yeast tropomyosin overlaps are centered at the same longitudinal position. Finally, note in Fig. 1 that end-to-end overlap regions at the C- and N-termini were avoided in creating the chimeras.
Folding and thermal stability of chimeric tropomyosins
As expected for coiled-coils, the chimeric molecules had circular dichroism spectra characteristic of α-helix. Contrary to expectation however, they did not fold equally. Chimeras 5 and 6 had much smaller ellipticity magnitudes at 20 °C (Fig 3A), implying unfolded, non-helical regions. The structural basis for this is unknown, as are the locations of the non-helical portions within these chimeras. However, the decreases in ellipticity matched what would be expected if their NH2-terminal halves, in which the guest segments were inserted, were not helical. The four other chimeras (2, 3, 4, and 7) had ellipticities slightly smaller than that of TPM1. The differences among these four chimeras and wt TPM1 at 20 °C were larger than the 5% imprecision in relative protein concentrations. However, it is unclear whether the weaker signals indicate incomplete folding, or some other altered property.
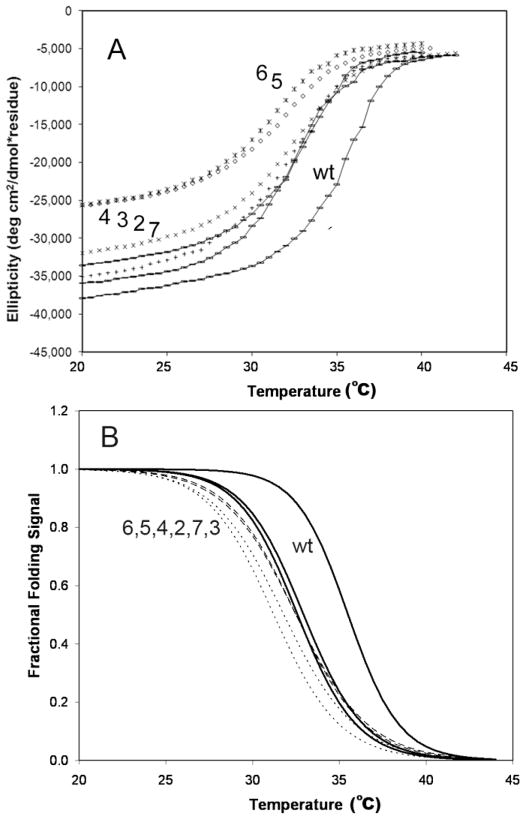
FIGURE 3. Thermal stability of chimeric tropomyosins assessed by circular dichroism.
A, ellipticity at λ= 222 nm of wt TPM1 and chimeric tropomyosins 2, 3, 4, 5, 6, and 7 as a function of temperature. B, Two state thermal melting curves corresponding to best fits the data in Panel A, using parameters listed in Table 1. All of the chimeras had reduced stability. Note that data for chimeras 3 and 7 most resemble data for wt TPM1.
More insight arises from considering these data in normalized form as temperature-induced unfolding transitions (Fig. 3B). TPM1 was more resistant to thermal denaturation than were any of the chimeras, i.e., it had the highest melting temperature (Table 1). All of the chimeras had similar melting temperatures to each other. Notably, chimeras 3 and 7 were the only constructs with melting transitions similarly steep to that of TPM1. Correspondingly, the unfolding enthalpies of the chimeras could be grouped: 128 kcal/mol for wt TPM1, vs. 107–110 kcal/mol for chimeras 3 and 7, vs. 89–97 kcal/mol for chimeras 2, 4, 5, and 6 (Table 1). Thus, the unfolding enthalpies for chimeras 2 and 4 were no greater than those of the two chimeras that clearly did not fold (i.e. chimeras 5 and 6).
Table 1.
Summary of wt Control and Chimeric Tropomyosin Propertiesa
| Tropomyosin | Melting Temperature (°C) | Unfolding Enthalpy, kcal/mol | Tm Affinity for Actin, M−1 × 106 | Tm-Actin Binding Cooperativity Parameter y | Tm Affinity for Acto- myosin S1, M−1 × 106 | Tm-Actomyosin Binding Cooperativity Parameter y | KT x L Actin-S1 Binding Parameter, M−1 × 106 | S1 Affinity for Active State Actin, M−1 × 106 |
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| wt TPM1 | 35.50 ± 0.05 | 128.0 ± 3.5 | 10.3 ± 0.9 | 28 ± 15 | 3.0 ± 0.1 | 78± 27 | 0.21 ± 0.01 | 13.4 |
| Chimera 3 | 32.94 ± 0.04 | 107.3 ± 2.4 | 0.92 ± 0.05 | 19 ± 5 | 20 ± 1 | 13 ± 3 | 0.19 ± 0.01 | 13.4 |
| Chimera 7 | 32.59 ± 0.03 | 109.5 ± 1.8 | 4.6 ± 0.1 | 390 ± 160 | < 0.3 | 0.42 ± 0.01 | 13.4 | |
| Chimera 2 | 32.49 ± 0.06 | 93.5 ± 2.5 | ||||||
| Chimera 4 | 32.49 ± 0.04 | 89.2 ± 1.4 | ||||||
| Chimera 5 | 31.79 ± 0.04 | 90.0 ± 1.6 | ||||||
| Chimera 6 | 31.22 ± 0.04 | 96.8 ± 1.8 | ||||||
Data are presented as mean +/− SEM. Thermal stability results are from Fig. 3 data. Binding to actin (Fig. 5 data) is expressed by an overall Kapp and by a cooperativity parameter (y) indicating the increased affinity for tropomyosin binding to actin when aided by an end-to-end contact (33). The overall affinity of myosin S1 for the thin filament is approximately given by the product of the two rightmost columns in the table (8, 20), with Table values derived from data in Fig. 6. KT × L is the tendency of tropomyosin, when attached to the inner domain of actin, to alter the actin in the same manner that myosin S1alters actin, when myosin strengthens tropomyosin-actin binding.
Overall, the circular dichroism results indicate that TPM1 did not readily accommodate, nor did it equivalently accommodate, replacement of its normal 38 residue repeat by quasi-similar coiled-coil substitutions from muscle tropomyosin. Out of six chimeric molecules, only two, chimeras 3 and 7, retained full folding and large folding enthalpy. Whatever the structural bases for these results, they appear to be functionally significant: chimeras 3 and 7 also were the only chimeras to evidence any actin binding (See below).
Co-sedimentation of control and chimeric tropomyosins with actin and with myosin S1-decorated actin
The chimeric tropomyosins were added to actin in the presence or absence of myosin S1, and thin filaments were collected by ultracentrifugation. Fig. 4 shows SDS-PAGE analyses of representative thin filament pellets. TPM1 bound to actin similarly, with or without myosin S1. More chimera 3 co-sedimented with F-actin in the presence of myosin than co-sedimented with myosin-bare actin. Chimera 7 bound to actin, but bound poorly to actin-S1. In contrast, the figure shows that chimera 7 co-sedimented with actin poorly when myosin S1 was present.
FIGURE 4. SDS-PAGE analysis of control and chimeric tropomyosin co-sedimentation with F-actin.
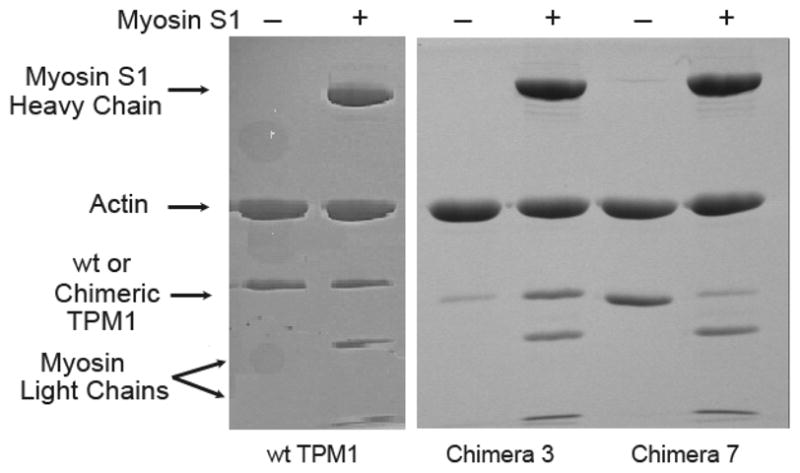
SDS-PAGE lanes show the protein composition of re-suspended F-actin that had been pelleted with various tropomyosins, in each case either without (−) or with (+) myosin S-1. wt TPM1 bound clearly to actin either in the presence or the absence of myosin. Chimera 3 pelleted with actin filaments more effectively when myosin was present. Chimera 7 pelleted with actin filaments more effectively when myosin was absent. Conditions:: 50 mM KCl, 3 mM MgCl2, 50 mM NaH2PO4 (pH 7.0), 0.1 mM dithiothreitol, 9 μM Mg2+ F-actin (51), +/− 8 μM myosin S1, and 4 μM tropomyosin.
None of the other four chimeras (numbers 2, 4, 5, and 6) were detected in actin pellets, in multiple experimental attempts, regardless of the presence of myosin S1. Based on Fig. 3, this finding was attributed to poor coiled-coil formation and stability of these chimeras after insertion of their respective guest inserts.
Effect of the guest sequences on the affinity of tropomyosin for actin and for actin-myosin S1
Figure 5A presents data on the concentration-dependent binding of wt TPM1 to thin filaments. In the absence of myosin, TPM1’s attachment was relatively tight, resulting in an overall Kapp of approximately 107 M−1 (Table 1). Myosin S1 did not increase this affinity, consistent with previous findings (20), and unlike the profound effect observed for mammalian tropomyosins (11, 20). In fact, myosin S1 slightly weakened TPM1-actin binding, a 3-4-fold effect. This agrees closely with findings of Maytum et al (11), using slightly different conditions.
Comparison of Figs. 5A and 5B shows that the guest segment in chimera 3 greatly altered the effect of myosin S1 on tropomyosin-actin attachment. The chimera bound to bare actin with Kapp ~ 106 M−1 (Table 1). Rather than weakening this binding, myosin increased chimera 3’s actin-affinity more than 20-fold. No yeast tropomyosin has ever evidenced this highly characteristic, mammalian tropomyosin-like behavior. The result implies that the guest segment produced a positive, tropomyosin-myosin interaction.
Interestingly, chimera 7 had quite the opposite effect. It attached to bare actin quite readily, but its attachment to myosin S1 decorated-actin was very poor (Fig. 5C). This resembles the behavior of an ultra-short, internally deleted form of TPM1 (41), and is consistent with steric interference between myosin S1 and chimera 7.
Effects of the guest sequences on myosin S1 binding to thin filaments
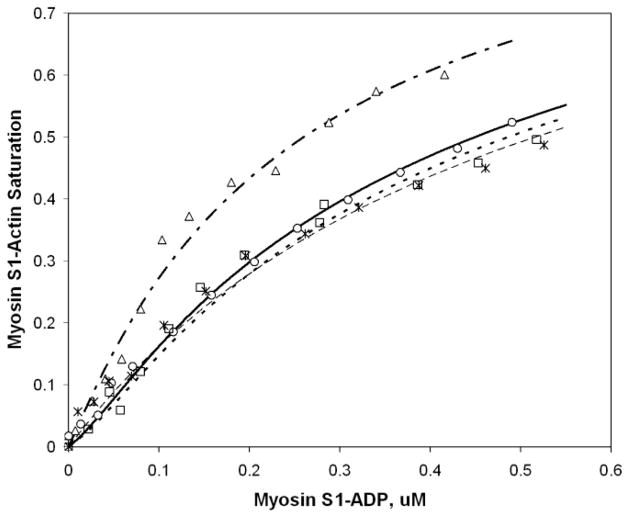
As described above, equilibrium linkage principles require there to be a mutuality to the effects of myosin S1 and tropomyosin on each other’s binding to actin. However, it requires experiment to reveal mechanistically important details. To elucidate these details for chimera 3, the binding of myosin S1 to thin filaments was studied in the presence of ADP (Fig. 6). Myosin S1 bound similarly to actin, to actin with wt TPM1, and to actin with chimera 7. However, the addition of chimera 3 increased the affinity of the myosin 2-fold, a clear difference. Notably, the pattern in the figure is of a general increase in myosin affinity, not limited to the one out of five actins that can be in direct contact with the 38-residue guest segment. The increased affinity is model-independent. For illustration purposes, fitted lines corresponding to one model (20) of myosin binding to the thin filament are shown.
FIGURE 6. Chimera 3 tropomyosin enhances the affinity of myosin S1-ADP for the thin filament.
The data show the effect of myosin S1-ADP on the fluorescence of pyrene-labeled actin, and the effects of the various tropomyosins on this process. Myosin bound similarly to actin (asterisks) and to actin with either wt TPM1(circles) or chimera 7 (squares). In contrast to the lack of effect of wt TPM1 and chimera 7, chimera 3 strengthened myosin binding to the thin filament (triangles). Lines are best fit curves, corresponding to a global fit to Eq. 3 in (8), with Y =100, KT° = 0.17 (20) and fit parameters as shown in Table 1.
DISCUSSION
In the present report, the different properties of mammalian and yeast tropomyosins were examined by creation of six different chimeric tropomyosins, in which part of S. cerevisiae TPM1 was replaced with alternative substitutions. Four of the chimeras were not fully folded and/or had low folding enthalpy, and were non-functional. This suggests an unexpected susceptibility of TPM1 folding thermodynamics to alternative sequence coiled-coil substitutions. More notably, two other chimeras did fold, and they bound to actin with different patterns. Chimeric tropomyosin containing residues mostly from muscle tropomyosin’s 3rd quasi-repeat exhibited positive myosin-tropomyosin cooperativity. That is, unlike findings with wt TPM1, chimera 3 and myosin mutually strengthened each other’s affinity for the thin filament. This reproduced a characteristic function of mammalian tropomyosin, which does not occur for yeast tropomyosin. In contrast, this behavior was not observed for chimeric tropomyosin with inserted residues primarily from the 7th quasi-repeat of muscle tropomyosin. Rather, myosin S1 markedly impaired its attachment to actin. To properly interpret these results, several aspects of tropomyosin structure and function should be considered.
Tropomyosin has a structure with an undeniable overall simplicity - a full length coiled-coil form. Despite this, there is little consensus on how tropomyosin’s structure relates to some important aspects of its function. For example, competing views suggest that tropomyosin’s ability to attach to actin filaments depends upon either of two rather opposing features. In these differing views, actin binding depends primarily on tropomyosin’s tendency either to adopt the specific curved shape of the actin filament (42–44), or to have proper flexibility for attachment to the actin filament (45–48). It may be that flexibility at specific sites is important, rather than global flexibility. In the present study, chimeras 2 and 4 were apparently more flexible (Fig. 3B) at experimental temperatures than were chimeras 3 and 7, yet it was the latter pair that attached to actin.
Similarly, there is no consensus on whether tropomyosin’s actomyosin regulatory actions hinge upon the negative, steric blocking aspect of tropomyosin, or the positive, (presumably) allosteric aspects. Our view has been that both aspects are well supported experimentally, and that both aspects are critical to any understanding of tropomyosin function (8, 20, 36). The findings in the present report are most readily understood by this approach. The behavior of chimera 3 supports the idea that tropomyosin can draw myosin onto the actin filament, allosterically. On the other hand, the behavior of chimera 7 supports the idea that tropomyosin and myosin can interfere with each other’s attachment to actin.
To explain how tropomyosin and myosin promote each other’s binding to actin, we proposed (8, 36) that the changes in actin that accompany myosin binding result in a strengthening of tropomyosin’s otherwise weak association with the actin inner domain. By equilibrium linkage, when tropomyosin is on the actin inner domain it strengthens myosin binding to actin. In this view, the active state of the thin filament is defined by the alterations in actin that accompany strong myosin binding. This proposal, which is supported anew by the present work, provides a framework for understanding the many differences between a fully activated thin filament and bare actin. Tropomyosin shifts cooperatively to the actin inner domain, where it alters actin structure and thus strengthens myosin binding and modulates the rates of steps occurring within the acto-S1 complex.
Finally, previous tropomyosin internal deletion studies (49, 50) suggest that such positive, myosin-tropomyosin effects vary greatly across the seven quasi-repeats that comprise tropomyosin. Deletions of different regions produced dramatically different effects on myosin binding to the thin filament, on myosin cycling, and on the free energy for assembling the thin filament in its active state (49, 50). Quasi-repeats 3, 4, and 5 were particularly and specifically implicated as responsible for positive, myosin-tropomyosin effects. In the present study, too few of the chimeras bound to actin to test these putative distinctions extensively. Furthermore, it is reasonable to assume that the guest segments were affected somewhat by their insertions into the yeast tropomyosin. Nevertheless, it should be noted that the results with chimera 3 and chimera 7 are fully consistent with the prior internal deletion studies. Chimera 3 demonstrated a positive effect on myosin. Chimera 7 demonstrated the opposite.
In summary, the findings of the present study support the view that mammalian tropomyosins have evolved to have opposing effects on myosin. To phrase the matter colloquially, they push and pull. Tropomyosin both pushes myosin away by steric interference with strong myosin binding to actin, and also pulls myosin onto actin by allosteric effects on the thin filament.
Footnotes
This work was supported by NIH grants HL063774 and HL038834.
References
- 1.Lehman W, Vibert P, Uman P, Craig R. Steric-blocking by Tropomyosin Visualized in Relaxed Vertebrate Muscle Thin filaments. J Mol Biol. 1995;251:191–196. doi: 10.1006/jmbi.1995.0425. [DOI] [PubMed] [Google Scholar]
- 2.Xu C, Craig R, Tobacman LS, Horowitz R, Lehman W. Tropomyosin positions in regulated thin filaments revealed by cryoelectron microscopy. Biophys J. 1999;77:985–992. doi: 10.1016/S0006-3495(99)76949-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poole KJ, Lorenz M, Evans G, Rosenbaum G, Pirani A, Craig R, Tobacman LS, Lehman W, Holmes KC. A comparison of muscle thin filament models obtained from electron microscopy reconstructions and low-angle X-ray fibre diagrams from non-overlap muscle. J Struct Biol. 2006;155:273–284. doi: 10.1016/j.jsb.2006.02.020. [DOI] [PubMed] [Google Scholar]
- 4.Lehman W, Hatch V, Korman VL, Rosol M, Thomas LT, Maytum R, Geeves MA, Van Eyk JE, Tobacman LS, Craig R. Tropomyosin and actin isoforms modulate the localization of tropomyosin strands on actin filaments. J Mol Biol. 2000;302:593–606. doi: 10.1006/jmbi.2000.4080. [DOI] [PubMed] [Google Scholar]
- 5.Vibert P, Craig R, Lehman W. Steric-model for activation of muscle thin filaments. J Mol Biol. 1997;266:8–14. doi: 10.1006/jmbi.1996.0800. [DOI] [PubMed] [Google Scholar]
- 6.Geeves MA, Halsall DJ. The dynamics of the interaction between myosin subfragment 1 and pyrene-labelled thin filaments, from rabbit skeletal muscle. Proc R Soc Lond. 1986;B229:85–95. doi: 10.1098/rspb.1986.0076. [DOI] [PubMed] [Google Scholar]
- 7.Williams DL, Greene LE. Comparison of the effects of tropomyosin and troponin-tropomyosin on the binding of myosin subfragment 1 to actin. Biochemistry. 1983;22:2770–2774. doi: 10.1021/bi00280a027. [DOI] [PubMed] [Google Scholar]
- 8.Tobacman LS, Butters CA. A new model of cooperative myosin-thin filament binding. J Biol Chem. 2000;275:27587–27593. doi: 10.1074/jbc.M003648200. [DOI] [PubMed] [Google Scholar]
- 9.Eaton BL. Tropomyosin binding to F-actin induced by myosin heads. Science. 1976;192:1337–1339. doi: 10.1126/science.131972. [DOI] [PubMed] [Google Scholar]
- 10.Cassell M, Tobacman LS. Opposite effects of myosin subfragment 1 on binding of cardiac troponin and tropomyosin to actin. J Biol Chem. 1996;271:12867–12872. doi: 10.1074/jbc.271.22.12867. [DOI] [PubMed] [Google Scholar]
- 11.Maytum R, Konrad M, Lehrer SS, Geeves MA. Regulatory properties of tropomyosin. Effects of length, isoform, and N-terminal sequence. Biochemistry. 2001;40:7334–7341. doi: 10.1021/bi010072i. [DOI] [PubMed] [Google Scholar]
- 12.Homsher E, Lee DM, Morris C, Pavlov D, Tobacman LS. Regulation of force and unloaded sliding speed in single thin filaments: Effects of regulatory proteins and calcium. J Physiol (London) 2000;524.1:233–243. doi: 10.1111/j.1469-7793.2000.00233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heeley DH, Belknap B, White HD. Maximal activation of skeletal muscle thin filaments requires both rigor myosin S1 and calcium. J Biol Chem. 2006;281:668–676. doi: 10.1074/jbc.M505549200. [DOI] [PubMed] [Google Scholar]
- 14.Tobacman LS. Thin filament-mediated regulation of cardiac contraction. Annu Rev Physiol. 1996;58:447–481. doi: 10.1146/annurev.ph.58.030196.002311. [DOI] [PubMed] [Google Scholar]
- 15.Murray JM, Knox MK, Trueblood CE, Weber A. Potentiated state of the tropomyosin actin filament and nucleotide-containing myosin subfragment 1. Biochemistry. 1982;21:906–915. doi: 10.1021/bi00534a015. [DOI] [PubMed] [Google Scholar]
- 16.Fujita H, Kawai M. Temperature effect on isometric tension is mediated by regulatory proteins tropomyosin and troponin in bovine myocardium. J Physiol. 2002;539 (Pt 1):267–276. doi: 10.1113/jphysiol.2001.013220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.VanBuren P, Palmiter KA, Warshaw DM. Tropomyosin directly modulates actomyosin mechanical performance at the level of a single actin filament. Proc Natl Acad Sci U S A. 1999;96:12488–12493. doi: 10.1073/pnas.96.22.12488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.VanBuren P, Work SS, Warshaw DM. Enhanced force generation by smooth muscle myosin in vitro. Proc Natl Acad Sci U S A. 1994;91:202–205. doi: 10.1073/pnas.91.1.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maytum R, Geeves M, Konrad M. Actomyosin regulatory properties of yeast tropomyosin are dependent upon N-terminal modification. Biochemistry. 2000;39:11913–11920. doi: 10.1021/bi000977g. [DOI] [PubMed] [Google Scholar]
- 20.Strand J, Nili M, Homsher E, Tobacman LS. Modulation of Myosin Function by Isoform-Specific Properties of S. Cerevisiae and Muscle Tropomyosins. J Biol Chem. 2001;276:34832–34839. doi: 10.1074/jbc.M104750200. [DOI] [PubMed] [Google Scholar]
- 21.Skoumpla K, Coulton AT, Lehman W, Geeves MA, Mulvihill DP. Acetylation regulates tropomyosin function in the fission yeast Schizosaccharomyces pombe. J Cell Sci. 2007;120:1635–1645. doi: 10.1242/jcs.001115. [DOI] [PubMed] [Google Scholar]
- 22.Sueki M, Lee S, Powers SP, Denton JB, Konishi Y, Scheraga HA. Helix-Coil Stability Constants for the Naturally Occurring Amino Acids in Water. 22 Histidine Parameters from Random Poly[(hydroxybutyl)glutamine-co-L-histidine] Macromolecules. 2010;17:148–155. [Google Scholar]
- 23.Chen W, Wen KK, Sens AE, Rubenstein PA. Differential interaction of cardiac, skeletal muscle, and yeast tropomyosins with fluorescent (pyrene235) yeast actin. Biophys J. 2006;90:1308–1318. doi: 10.1529/biophysj.105.064634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lehrer SS, Golitsina NL, Geeves MA. Actin-tropomyosin activation of myosin subfragment 1 ATPase and thin filament cooperativity. The role of tropomyosin flexibility and end-to-end interactions. Biochemistry. 1997;36:13449–13454. doi: 10.1021/bi971568w. [DOI] [PubMed] [Google Scholar]
- 25.Korman VL, Hatch V, Dixon K, Craig R, Lehman W, Tobacman LS. An actin subdomain 2 mutation that impairs thin filament regulation by troponin and tropomyosin. J Biol Chem. 2000;275:22470–22478. doi: 10.1074/jbc.M002939200. [DOI] [PubMed] [Google Scholar]
- 26.Miller CJ, Bobkova E, Botstein D, Reisler E. Mutational analysis of the role of hydrophobic residues in the 338–348 helix on actin in actomyosin interactions. Biochemistry. 1996;35:3670–3676. doi: 10.1021/bi952645v. [DOI] [PubMed] [Google Scholar]
- 27.Heller M, Tobacman LS, Nili M, Homsher E. Cardiomyopathy mutations that increase calcium-sensitivity and tropomyosin flexibility. J Biol Chem. 2003;278:41742–41748. doi: 10.1074/jbc.M303408200. [DOI] [PubMed] [Google Scholar]
- 28.Spudich JA, Watt S. The regulation of rabbit skeletal muscle contraction. I Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J Biol Chem. 1971;246:4866–4871. [PubMed] [Google Scholar]
- 29.Weeds AG, Taylor RS. Separation of subfragment-1 isoenzymes from rabbit skeletal muscle myosin. Nature. 1975;257:54–56. doi: 10.1038/257054a0. [DOI] [PubMed] [Google Scholar]
- 30.Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 31.McGhee JD, von Hippel PH. Theoretical aspects of DNA-protein interactions: cooperative and non-cooperative binding of large ligands to a one-dimensional lattice. J Mol Biol. 1974;86:469–489. doi: 10.1016/0022-2836(74)90031-x. [DOI] [PubMed] [Google Scholar]
- 32.Adams S, Reisler E. Role of sequence 18–29 on actin in actomyosin interactions. Biochemistry. 1993;32:5051–5056. doi: 10.1021/bi00070a012. [DOI] [PubMed] [Google Scholar]
- 33.Hill LE, Mehegan JP, Butters CA, Tobacman LS. Analysis of troponin-tropomyosin binding to actin. Troponin does not promote interactions between tropomyosin molecules. J Biol Chem. 1992;267:16106–16113. [PubMed] [Google Scholar]
- 34.Kouyama T, Mihashi K. Fluorimetry study of N-(1-pyrenyl)iodoacetamide labelled F-actin. Local structural change of actin protomer both on polymerization and on binding of heavy meromyosin. Eur J Biochem. 1981;114:33–38. [PubMed] [Google Scholar]
- 35.Criddle AH, Geeves MA, Jeffries T. The use of actin labelled with N-(1-pyrenyl)iodoacetamide to study the interaction of actin with myosin subfragments and troponin/tropomyosin. Biochem J. 1985;232:343–349. doi: 10.1042/bj2320343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosol M, Lehman W, Craig R, Landis C, Butters C, Tobacman LS. Three-dimensional reconstruction of thin filaments containing mutant tropomyosin. Biophys J. 2000;78:908–917. doi: 10.1016/S0006-3495(00)76648-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McKillop DFA, Geeves MA. Regulation of the interaction between actin and myosin subfragment 1: evidence for three states of the thin filament. Biophys J. 1993;65:693–701. doi: 10.1016/S0006-3495(93)81110-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brown JH, Cohen C, Parry DAD. Heptad breaks in -helical coiled-coils: stutters and stammers. Proteins: Structure, Function, and Genetics. 1996;26:134–145. doi: 10.1002/(SICI)1097-0134(199610)26:2<134::AID-PROT3>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 39.Phillips GN, Jr, Fillers JP, Cohen C. Tropomyosin crystal structure and muscle regulation. J Mol Biol. 1986;192:111–131. doi: 10.1016/0022-2836(86)90468-7. [DOI] [PubMed] [Google Scholar]
- 40.McLachlan AD, Stewart M. The 14-fold periodicity in -tropomyosin and the interaction with actin. J Mol Biol. 1976;103:271–298. doi: 10.1016/0022-2836(76)90313-2. [DOI] [PubMed] [Google Scholar]
- 41.Maytum R, Hatch V, Konrad M, Lehman W, Geeves MA. Ultra short yeast tropomyosins show novel myosin regulation. J Biol Chem. 2008;283:1902–1910. doi: 10.1074/jbc.M708593200. [DOI] [PubMed] [Google Scholar]
- 42.Holmes KC, Lehman W. Gestalt-binding of tropomyosin to actin filaments. J Muscle Res Cell Motil. 2008;29:213–219. doi: 10.1007/s10974-008-9157-6. [DOI] [PubMed] [Google Scholar]
- 43.Li XE, Lehman W, Fischer S. The relationship between curvature, flexibility and persistence length in the tropomyosin coiled-coil. J Struct Biol. 2010;170:313–318. doi: 10.1016/j.jsb.2010.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sousa D, Cammarato A, Jang K, Graceffa P, Tobacman LS, Li XE, Lehman W. Electron microscopy and persistence length analysis of semi-rigid smooth muscle tropomyosin strands. Biophys J. 2010;99:862–868. doi: 10.1016/j.bpj.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Singh A, Hitchcock-DeGregori SE. Local destabilization of the tropomyosin coiled coil gives the molecular flexibility required for actin binding. Biochemistry. 2003;42:14114–14121. doi: 10.1021/bi0348462. [DOI] [PubMed] [Google Scholar]
- 46.Singh A, Hitchcock-DeGregori SE. Dual requirement for flexibility and specificity for binding of the coiled-coil tropomyosin to its target, actin. Structure. 2006;14:43–50. doi: 10.1016/j.str.2005.09.016. [DOI] [PubMed] [Google Scholar]
- 47.Minakata S, Maeda K, Oda N, Wakabayashi K, Nitanai Y, Maeda Y. Two-crystal structures of tropomyosin C-terminal fragment 176–273: exposure of the hydrophobic core to the solvent destabilizes the tropomyosin molecule. Biophys J. 2008;95:710–719. doi: 10.1529/biophysj.107.126144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nitanai Y, Minakata S, Maeda K, Oda N, Maeda Y. Crystal structures of tropomyosin: flexible coiled-coil. Adv Exp Med Biol. 2007;592:137–151. doi: 10.1007/978-4-431-38453-3_13. [DOI] [PubMed] [Google Scholar]
- 49.Landis CA, Back N, Homsher E, Tobacman LS. Effects of tropomyosin internal deletions on thin filament function. J Biol Chem. 1999;274:31279–31285. doi: 10.1074/jbc.274.44.31279. [DOI] [PubMed] [Google Scholar]
- 50.Hitchcock-DeGregori SE, Song Y, Moraczewska J. Importance of internal regions and the overall length of tropomyosin for actin binding and regulatory function. Biochemistry. 2001;40:2104–2112. doi: 10.1021/bi002421z. [DOI] [PubMed] [Google Scholar]
- 51.Korman VL, Tobacman LS. Mutations in actin subdomain 3 that impair thin filament regulation by troponin and tropomyosin. J Biol Chem. 1999;274:22191–22196. doi: 10.1074/jbc.274.32.22191. [DOI] [PubMed] [Google Scholar]