Abstract
Pediatric cataracts are observed in 1–15 per 10,000 births with 10–25% of cases attributed to genetic causes; autosomal dominant inheritance is the most commonly observed pattern. Since the specific cataract phenotype is not sufficient to predict which gene is mutated, whole exome sequencing (WES) was utilized to concurrently screen all known cataract genes and to examine novel candidate factors for a disease-causing mutation in probands from 23 pedigrees affected with familial dominant cataract. Review of WES data for 36 known cataract genes identified causative mutations in nine pedigrees (39%) in CRYAA, CRYBB1, CRYBB3, CRYGC (2), CRYGD, GJA8 (2), and MIP and an additional likely causative mutation in EYA1; the CRYBB3 mutation represents the first dominant allele in this gene and demonstrates incomplete penetrance. Examination of crystallin genes not yet linked to human disease identified a novel cataract gene, CRYBA2, a member of the βγ-crystallin superfamily. The p.(Val50Met) mutation in CRYBA2 cosegregated with disease phenotype in a four-generation pedigree with autosomal dominant congenital cataracts with incomplete penetrance. Expression studies detected cryba2 transcripts during early lens development in zebrafish, supporting its role in congenital disease. Our data highlight the extreme genetic heterogeneity of dominant cataract as the eleven causative/likely causative mutations affected nine different genes and the majority of mutant alleles were novel. Furthermore, these data suggest that less than half of dominant cataract can be explained by mutations in currently known genes.
Keywords: dominant cataract, whole exome sequencing, CRYBA2, CRYBB3, zebrafish
Introduction
Pediatric cataracts are observed in 1–15 per 10,000 births, with 10–25% of cases attributed to genetic causes (Trumler 2011). Autosomal dominant inheritance is most common for hereditary cases, although autosomal recessive and X-linked patterns are observed (Hejtmancik 2008). Congenital or infantile cataract represents the most severe end of the spectrum with onset within the first year of life and the possibility of permanent blindness due to interference with normal visual development. Genetic studies have identified mutations in numerous genes associated with cataracts, including crystallins, which comprise about half of the known genetic forms of cataract, structural proteins, membrane proteins, and transcription factors (Hejtmancik 2008; Huang and He 2010; Shiels et al. 2010). The exact frequencies of mutations in these identified genes are not known and many cases are still awaiting molecular diagnosis.
Whole exome sequencing has been successfully applied for molecular characterization of a cohort affected with a heterogeneous condition (Aldahmesh et al. 2012; Choi et al. 2012) as well as identification of novel genes (Bamshad et al. 2011). Since the specific cataract phenotype is typically not sufficient to predict which gene is mutated in a family (Hejtmancik 2008), whole exome sequencing may represent an efficient method of screening the known cataract genes in order to identify a disease-causing mutation as well as looking for novel factors involved in this condition. We utilized whole exome sequencing in a population affected with dominant cataract to first provide insight regarding the spectrum/frequency of mutations in known genes and the proportion of hereditary cataract which remains unexplained and second, to perform a comprehensive examination of crystallin genes not previously associated with human disease for mutations in cataract phenotypes.
Methods
Ethics statement
The human study was approved by the Institutional Review Board of the Children’s Hospital of Wisconsin with written informed consent obtained from each participant and/or their legal representative, as appropriate. The studies involving zebrafish embryos were carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Institutional Animal Care and Use Committee at the Medical College of Wisconsin (protocol number AUA00000352).
Subjects
Probands from 23 pedigrees affected with dominant cataract were selected for whole exome sequencing: 18 had cataract diagnosed within the first year of life in at least one member of the family (congenital cataract), 4 were diagnosed between 1 and 5 years of age (juvenile cataract), and one family demonstrated presenile adult-onset (age 40) cataract and retinal detachment (Online Resources 1 and 2). Two pedigrees with cataract microcornea and previously identified mutations in PITX3/FOXE3 were excluded from this study (data not shown). The 23 pedigrees included 19 Caucasian (European and European American), 3 Hispanic, and 1 family with unreported race/ethnicity. Additional family members were available for testing in 21 cases, ranging from 1–12 additional individuals (Online Resource 1).
Whole Exome Sequencing and Data Analysis
Whole exome sequencing was undertaken through Perkin Elmer, Inc (Branford, CT). Patients 1–14 utilized Agilent Sure Select v4 for exome capture while Patients 15–23 utilized v4+UTR. Data was evaluated through the Geospiza GeneSifter Analysis program hosted through Perkin Elmer Bioinformatics using the GATK V2.10 pipeline. The Variant Report was reviewed for the presence of mutations in 36 known cataract genes and 8 additional crystallin genes not yet linked to cataract in humans (Online Resources 2 and 3) while the Gene List was used to verify coverage of the genes of interest. Variants of interest were investigated for presence/absence in NCBI SNP Database (dbSNP) (http://www.ncbi.nlm.nih.gov/projects/SNP/, frequency in the Exome Variant Server (EVS) (http://evs.gs.washington.edu/EVS/), and predicted effect on the protein (Ensembl Variant Effect Predictor: http://www.ensembl.org/info/docs/variation/vep/index.html).
Variant Confirmation
Primers flanking the variants of interest were used to amplify genomic DNA from probands (to confirm variant) and all available family members (to test for cosegregation). PCR products were sequenced in both directions using ABI 3730XL sequencer and protocols (Applied Biosystems/Life Technologies, Carlsbad, CA, USA). Sequences were reviewed manually and using Mutation Surveyor (SoftGenetics, State College, PA).
Mutation Analysis of MAF, FOXE3, and PITX3
MAF, FOXE3 and PITX3 were analyzed by direct DNA sequencing of PCR products in all probands following previously described protocols (Bremond-Gignac et al. 2010; Hansen et al. 2007a).
In situ hybridization in zebrafish embryos
Zebrafish (Danio rerio) were maintained on a 14-hour light/ 10-hour dark cycle. The embryos were obtained by natural spawning and maintained at 28.5°C. Expression analysis of two zebrafish orthologs of human CRYBA2, cryba2a (NM_001002049.1) and cryba2b (NM_001002584.1), was performed by in situ hybridization with 768-nt (cryba2a) and 752-nt (cryba2b) transcript-specific antisense riboprobes and previously described protocols (Sorokina et al. 2011). To generate templates for probe making, the corresponding cryba2a and cryba2b transcripts were amplified using the specific primers, cryba2a forward, GCCAAATCTCTCCCACGACA, and reverse, GAATGGCGACAAGCACACTC, as well as cryba2b forward, GCATTCGCCACTGAATGAGG, and reverse, TGCCTATAGTATTGATACGG, and cloned into pCRII-TOPO vector containing T7 and Sp6 promoters for in vitro RNA synthesis (Invitrogen, Carlsbad, CA).
Results/Discussion
Analysis of thirty-six known genes associated with pediatric cataract
The average mean read depth for the whole exome was 68.5 and on average, 86% of the targeted exome region achieved coverage >10X (Online Resource 2). Analysis of whole exome data for the 36 known cataract genes (Online Resource 3) showed generally good coverage. Three genes (MAF, FOXE3 and PITX3) showed poor coverage via the Agilent Sure Select v4 capture kit with average coding region coverage of 7×; coverage of these three genes was improved via Agilent Sure Select v4+UTR capture kit with average coding region coverage of 53×. MAF, FOXE3 and PITX3 sequencing was completed by Sanger sequencing in all probands. The remaining 33 cataract genes showed good coverage via both v4 (average coding region coverage of 71×) and v4+UTR (average coding region coverage of 64×) capture kits.
Heterozygous causative mutations in known cataract genes were identified in nine families in CRYAA, CRYBB1, CRYBB3, CRYGC (two mutations), CRYGD, GJA8 (two mutations), and MIP, and an additional likely causative mutation was seen in EYA1 (Table 1). Three of the mutations represent new occurrences of previously reported mutations (CRYAA, CRYGC and CRYGD); the remaining seven are novel. None of these mutations were present in dbSNP or the EVS. All nine causative mutations in known genes were identified in families affected with congenital cataract; each of these mutations showed complete cosegregation with the disease phenotype, with incomplete penetrance noted in one family (Table 1). The majority of the identified mutations are consistent with previous reports; two changes are of particular interest, the mutation in CRYBB3 that identifies a novel inheritance pattern for this gene and a likely causative change in EYA1 associated with an isolated cataract phenotype.
Table 1.
Summary of causative and likely causative mutations identified in this study (both known and novel cataract genes).
| Patien t # |
Phenotypea | Race/ ethnicit y |
Mut ated Gene |
Referen ce sequenc e |
DNA effect |
Protein effect |
Cosegregatio n analysis (affected pedigrees) |
Allele freque ncy (contr ols)b |
PolyP hen/ SIFT predic tion |
Previo usly report ed |
|---|---|---|---|---|---|---|---|---|---|---|
| Known cataract genes | ||||||||||
| Patient 2 | Congenital cataract; glaucoma, microcornea | Caucasian (USA) | CRYBB1 | NM_001887.3 | c.286G>T | p.(Val96Phe) | Cosegregates: present in 6 affected, absent in 3 unaffected persons | 0/12,971 | Probably damaging/deleterious | No |
| Patient 3 | Congenital cataract; glaucoma | Caucasian (USA) | GJA8 | NM_005267.4 | c.200A>G | p.(Asp67Gly) | Cosegregates: present in 2 affected persons | 0/13,005 | Probably damaging/ deleterious | No |
| Patient 6 | Congenital nuclear cataract, hyperopia, strabismus | Caucasian (Ashkenazi Jewish, Israel) | CRYGD | NM_006891.3 | c.418C>T | p.(Arg140*) | Cosegregates: present in 3 affected, absent in 1 adult-onset and 2 unaffected persons | 0/12,971 | N/A (premature truncation) | Reviewed in Huang and He, 2010 |
| Patient 7 | Congenital cataract; glaucoma | Caucasian (Italy) | CRYBB3 | NM_004076.3 | c.581T>A | p.(Val194Glu) | Cosegregates with incomplete penetrance: present in 5 affected and 1 unaffected persons | 0/12,999 | Probably damaging/ deleterious | No |
| Patient 9 | Unilateral or bilateral congenital cataract | Caucasi an (USA) | CRYGC | NM_020989.3 | c.157_161 dupGC GGC | p.(Gln55 Valfs*50) | Cosegregates: present in 4 affected, absent in 5 unaffected persons | Not reported | N/A (frame shift) | Reviewed in Huang and He, 2010 |
| Patient 10 | Congenital cataract; strabismus | Caucasi an (USA) | MIP | NM_012064.3 | c.605G>A | p.(Trp2 02*) | Cosegregates: present in 3 affected, absent in 3 unaffected persons | 0/12,178 | N/A (premature truncation) | No |
| Patient 11 | Congenital cataract; pupil defects, glaucoma | Caucasian (USA) | GJA8 | NM_005267.4 | c.226C>T | p.(Arg7 6Cys) | Cosegregates: present in 3 affected, absent in 2 unaffected persons | 0/13,005 | Probably damaging/deleterious | No |
| Patient 15 | Congenital cataract | Unreported (USA) | EYA1 | NM_000503.4 | c.121G>A | p.(Glu4 1Lys) | Not determined: no family members available for testing | 0/12,949 | Possibly damaging/tolerated | No |
| Patient 18 | Congenital cataract; microphthal mia/microcorne a, corneal opacity, glaucoma | Caucasian (USA) | CRYGC | NM_020989.3 | c.417C>G | p.(Tyr139*) | Cosegregates: present in 2 affected, absent in 2 unaffected persons | 0/12,971 | N/A (premature truncation) | No |
| Patient 22 | Congenital cataract, microcornea, macrocephaly; coloboma, glaucoma | Caucasian (Canada) | CRYAA | NM_000394.2 | c.34C>T | p.(Arg1 2Cys) | Cosegregates: present in 3 affected persons | 0/12,961 | Probably damaging/deleterious | Reviewed in Huang and He, 2010 |
| Novel cataract genes | ||||||||||
| Patient 4 | Congenital cataract; myopia, glaucoma | Hispanic (Urugua y) | CRYBA2 | NM_057093.1 | c.148G>A | p.(Val50Met) | Cosegregates with incomplete penetrance: present in 7 affected, 3 obligate carriers; absent in 3 unaffected persons | 0/12,948 | Probably damaging/deleterious | No |
variable features within the family are noted in italics; cataracts, unless specified, are bilateral;
allelle frequency provided for average number of chromosomes covered for the region according to the EVS database.
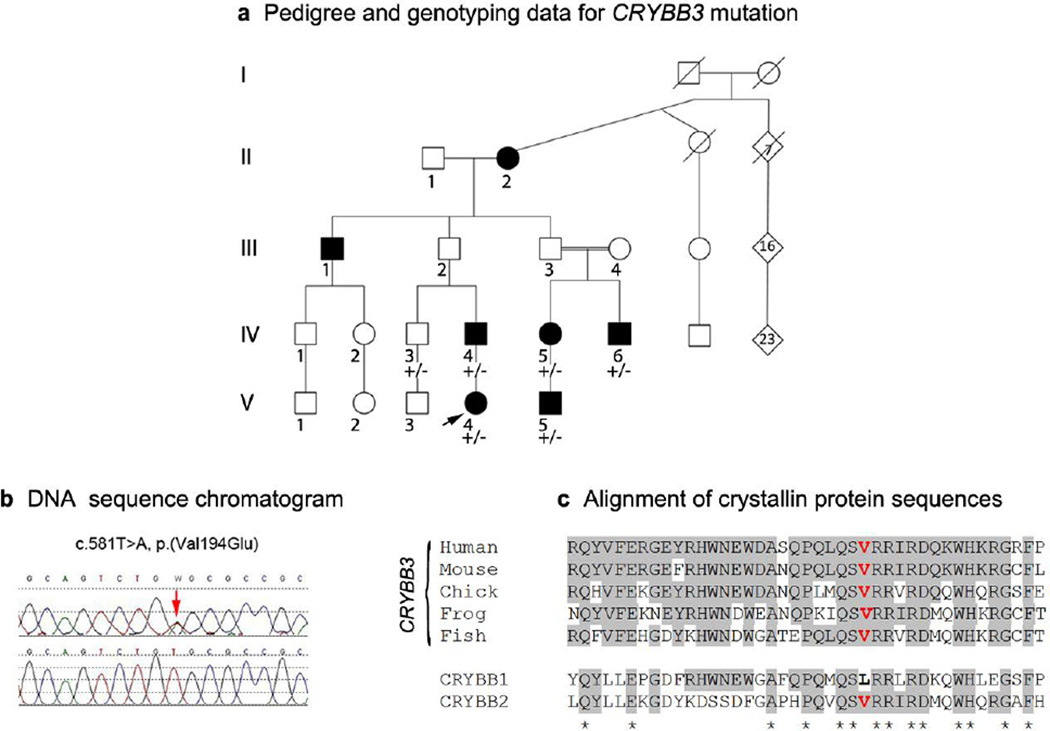
A novel heterozygous missense mutation in CRYBB3, c.581T>A, p.(Val194Glu), was identified in Patient 7 from an extended pedigree affected with bilateral congenital cataract demonstrating dominant inheritance with reduced penetrance (Figure 1). The specific types of cataract varied (posterior polar (IV-4, IV-6), nuclear (IV-5, and V-5), or anterior polar plus cortical (V-4)) and two individuals also had glaucoma (V-4 and V-5); cataracts were diagnosed at birth in four cases and at one year of age in one (IV-5). The two obligate carriers, III-2 and III-3, were last examined by an ophthalmologist at ages 46 and 70, correspondingly, and had no evidence of cataract. The missense mutation is seen in all affected individuals tested (IV-4, IV-5, IV-6, V-4, V-5) as well as an unaffected sibling (IV-3) (last ophthalmological evaluation occurred at age 35 and was reported normal); unfortunately, no other unaffected family members were available for genetic testing (Figure 1). The c.581T>A/ p.(Val194Glu) mutation was concluded to be causative because it cosegregates with affected status in multiple individuals from two branches of the family, is not seen in over 10,000 chromosomes screened by EVS, and results in a nonconservative substitution of a highly conserved amino acid valine within the Greek key motif IV of CRYBB3, which is predicted to be probably damaging/deleterious by PolyPhen/SIFT (accessed through Ensembl Variant Effect Predictor).
Figure 1.
Pedigree, DNA sequence and amino acid conservation associated with CRYBB3 allele. a. Pedigree of family 7 with affected status and genotyping results indicated (+/−: heterozygous CRYBB3 mutation present). The proband is indicated with an arrow; black symbol indicates congenital or juvenile cataract. b. DNA sequence of CRYBB3 showing heterozygous c.581T>A mutation (control sequence on bottom). c. Alignment of protein regions showing conservation of Val194 residue in the CRYBB3 proteins in various species as well as in CRYBB2, but not CRYBB1.
While the previous mutation reported in CRYBB3 was recessive (Riazuddin et al. 2005), all other crystallins are associated with autosomal dominant inheritance and three, CRYAB, CRYAA and CRYBB1 are also associated with both recessive and dominant inheritance, along with GJA8, SIL1, HSF4, and EPHA2 (Huang and He 2010; Kaul et al. 2010). For several genes, there seems to be a clear correlation between the type/location of a mutation and its pattern of inheritance, with nonsense mediated decay (NMD) likely playing a role in some cases (see below). As for CRYBB3, the only previously reported mutation in this gene is the recessive allele c.493G>C, p.(Gly165Arg), located in the loop connecting β-strands 1 and 2 of the Greek-key motif IV (Riazuddin et al. 2005), while the dominant allele reported here, c.581T>A, p.(Val194Glu), occurs within the β-strand 4 of the same motif (Slingsby and Clout 1999). Both changes are nonconservative, with the p.(Val194Glu) mutation causing the hydrophobic amino acid valine to be replaced with hydrophilic glutamic acid, therefore likely resulting in destabilization of the domain structure of the Greek key motif IV and CRYBB3 protein folding. Since this is only the second mutation identified in CRYBB3, additional studies are needed to determine genotype-phenotype correlations and differences in the mechanisms of these mutations. It is also possible that the dominance/recessivity of CRYBB3 alleles can be modified by secondary factors, consistent with the incomplete penetrance of the dominant c.581T>A, p.(Val194Glu) allele reported in this manuscript. Incomplete penetrance has been associated with mutations in several other autosomal dominant cataract genes: GJA8 (He et al. 2011), FTL (Gonzalez-Huerta et al. 2008), GJA3 (Burdon et al. 2004), CRYGC (Ren et al. 2000), and CRYAB (Sacconi et al. 2012).
The nucleotide change in EYA1 observed in Patient 15 results in a nonconservative substitution of the glutamic acid for lysine in the N-terminal region of this regulatory protein. EYA1 mutations are strongly associated with Brachio-Oto-Renal (BOR) syndrome, with no particular genotype-phenotype correlation identified between the type/location of mutation and phenotype severity (Orten et al. 2008). EYA1 mutations in isolated cataract/anterior segment dysgenesis phenotypes are rare and so far documented by only one published report that described two missense changes (Azuma et al. 2000). The c.121G>A, p.(Glu41Lys) substitution observed in Patient 15 with isolated congenital cataract is predicted to be possibly damaging by PolyPhen and is not observed in over 12,000 chromosomes screened by EVS, which suggests that this allele may be causative for the cataract phenotype observed in this patient. Unfortunately, samples from other family members were not available to verify segregation of the c.121G>A, p.(Glu41Lys) allele with the affected phenotype and further confirm (or refute) its pathogenicity.
An additional 14 heterozygous novel or rare (<1% population frequency) variants were identified in 11 cataract genes that did not segregate with the disease phenotype in affected pedigrees and therefore were determined to be non-causative (Online Resource 4); two findings in autosomal dominant genes were of special interest. A rare missense variant in GJA8, c.741T>G, p.(Ile247Met), was seen in Patient 2 (with causative CRYBB1 mutation) but did not cosegregate with the disease phenotype. This allele was initially reported as a disease causing mutation in a small family affected with congenital cataracts (Polyakov et al. 2001), although a subsequent report suggested it was a rare polymorphism (Graw et al. 2009) and it is present in 0.7% of the European American population in the EVS (31/8600 chromosomes); our data provide further evidence that the c.741T>G, p.( Ile247Met) variant in GJA8 is a polymorphism.
The second variant of interest was a novel nonsense allele in CRYGD, c.51T>G, p.(Tyr17*), seen in Patient 23 with adult-onset cataract and not reported in almost 13,000 chromosomes screened in the EVS; the variant did not cosegregate with the disease phenotype, being present in the patient, an affected sibling, and an unaffected sibling but not seen in an affected sibling and their mother. At the same time, four other nonsense mutations in CRYGD (p.(Tyr56*), p.(Tyr134*), p.(Arg140*) and (p.Trp156*)) have been linked to pediatric cataract phenotypes (Huang and He 2010), including an occurrence in Patient 6 from this study. The difference in effects generated by these nonsense mutations is likely to be due to whether or not they are subjected to nonsense mediated decay (NMD) (Holbrook et al. 2004; Khajavi et al. 2006). All previously reported CRYGD dominant nonsense mutations occur later in the gene; three out of four, p.(Tyr134*), p.(Arg140*) and (p.Trp156*), are located the final exon and thus are predicted to escape NMD resulting in truncated protein products with altered structural conformation, which may generate a dominant-negative effect (Hansen et al. 2007b). In contrast, the c.51T>G, p.(Tyr17*), CRYGD variant reported here occurs in the beginning of the second exon and is predicted to be subject to NMD and not expected to yield a protein product; this mutation might result in autosomal recessive disease if it occurred as a homozygous (or compound heterozygous) mutation. Based on this data, the previously reported dominant CRYGD allele, nonsense mutation p.(Tyr56*), may need to be re-examined. While the mutation was reported in a dominant cataract family (Santana et al. 2009), the mutation is located in the second exon more than 55 nucleotides from the final intron and thus would be expected to be subject to NMD, similar to the p.(Tyr17*) variant reported here. NMD alters the patterns of inheritance for premature truncation alleles in many genes with 5’ nonsense mutations (predicted to be subject to NMD) resulting in recessive disease and 3’ nonsense mutations (predicted to escape NMD) leading to dominant disease (Khajavi et al. 2006). In terms of cataract genes, examples include CRYBB1 mutations with dominant changes comprising missense mutations in the Greek key motifs II or IV, a C-terminal extension mutation, and C-terminal truncations that retain ~90% of normal protein sequence but lack part of the Greek key motif IV (all located in the final CRYBB1 exon and predicted to escape NMD); all of these changes are likely to result in production of mutant proteins. In contrast, recessive CRYBB1 changes include an early frameshift mutation that is likely to be subject to NMD, and an initiation codon substitution, both most likely resulting in null alleles (Cohen et al. 2007; Meyer et al. 2009; Wang et al. 2011). Similarly, a nonsense mutation early in the CRYAA gene, p.(Trp9*), is associated with autosomal recessive cataract while most other mutations in this gene result in dominant disease (Graw 2009; Pras et al. 2000). Our analysis of genetic variants highlights the importance of screening all known genes associated with a phenotype and completing cosegregation analysis in the pursuit of a causative mutation in a given pedigree.
Analysis of crystallin genes not yet linked to human disease
Since crystallin mutations comprised the majority of causative alleles (6/9), we proceeded to analyze the sequence of eight crystallin genes not previously linked to cataract in humans: CRYGB, CRYBA2, CRYL1, CRYGN, CRYZ, CRYM, CRYZL1, and CRYBG3 (Online Resource 2).
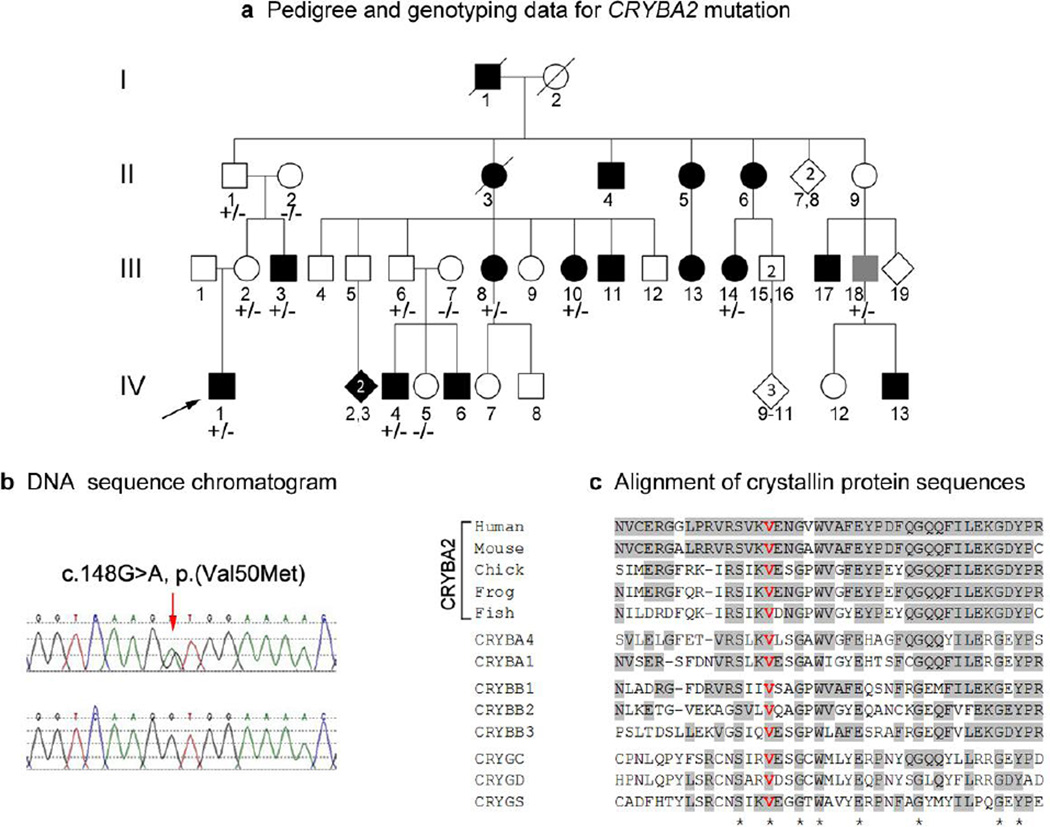
This analysis identified a novel heterozygous missense mutation, c.148G>A, p.(Val50Met), in the CRYBA2 gene in Patient 4 (Table 1, Figure 2). Patient 4 is affected with bilateral multifocal congenital cataract and eccentric pupil. He has a strong family history of autosomal dominant cataracts with incomplete penetrance (Figure 2); glaucoma and/or myopia were also noted in some affected individuals. The cataracts were congenital in all cases, with the exception of individual III-10, whose cataracts were diagnosed at age 9 and individual III-18, whose cataracts were diagnosed at age 34, after the birth of an affected child; it is possible that these individuals may have had mild congenital disease which went undiagnosed due to incomplete evaluation earlier in life. Three unaffected family members who are obligate carriers based on the pedigree were also available for testing. All seven affected individuals tested (III-3, III-8, III-10, III-14, III-18) as well as the obligate carriers (II-1, III-2, and III-6) carry the missense mutation while the unaffected spouses (II-2 and III-7) and an unaffected child (IV-5) do not, providing strong evidence that this mutation is causative of the phenotype in this family. In addition to this, the mutation affects a highly conserved valine residue within the β-strand 4 of the Greek key motif I that is preserved in all βγ- crystallins (Figure 2) and thus is predicted to be probably damaging/deleterious by PolyPhen/SIFT (accessed through Ensembl Variant Effect Predictor) and the mutant allele was not seen in almost 13,000 chromosomes reported by the EVS.
Figure 2.
Pedigree, DNA sequence and amino acid conservation associated with CRYBA2 allele. a. Pedigree of family 4 with affected status and genotyping results indicated (+/−: heterozygous CRYBA2 mutation present, −/−: CRYBA2 mutation absent). The proband is indicated with an arrow; black symbol indicates congenital or juvenile cataract, gray symbol indicates adult-onset cataract. b. DNA sequence of CRYBA2 showing heterozygous c.148G>A mutation (control sequence on bottom). c. Alignment of protein regions showing conservation of Val50 residue in CRYBA2 proteins in various species as well as other beta- and gamma- crystallins.
No other candidates for disease causing mutations were identified in this family (Online Resource 2). Three rare/novel variants in autosomal dominant genes BFSP2 and CRYGS were seen on chromosome 3 in the proband, Patient 4 (Online Resource 4): these three changes did not segregate with the disease phenotype but appeared to travel as a haplotype within the family with the exception of individual IV-3, who had only the second two changes, and individual IV-4, who had only the first (data not shown).
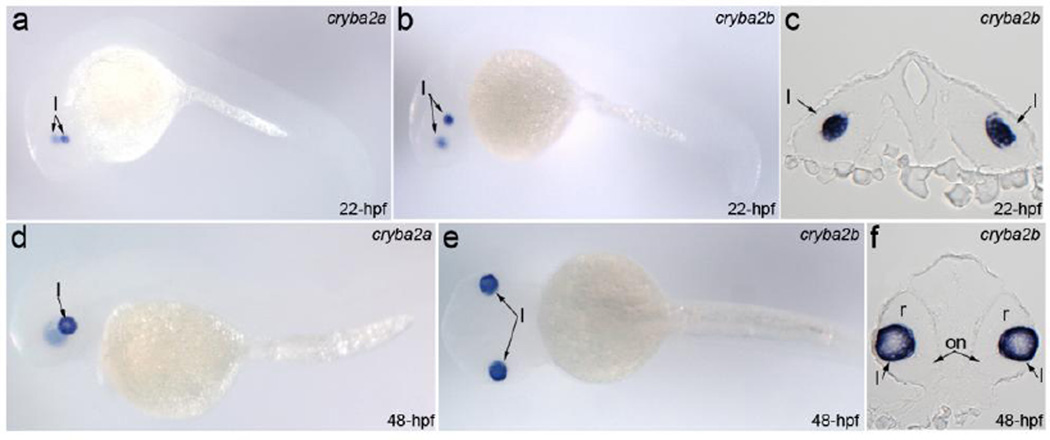
Previous studies demonstrated CRYBA2/Cryba2 expression in adult human lens epithelial and fiber cells (Hawse et al. 2005) as well as in mouse lens epithelial and fiber cells at postnatal days 1, 7, and 21 (Puk et al. 2011). To further evaluate the role of CRYBA2 in lens development, we studied expression of its zebrafish orthologs during embryonic development by in situ hybridization with cryba2a- and cryba2b- specific antisense riboprobes and detected a strong and specific presence of both cryba2 transcripts at early stages of lens development (Figure 3): at 22-hpf, cryba2 orthologs are expressed in the posterior region of the lens mass that delaminates from the head ectoderm (corresponding to lens vesicle formation/separation in 4-week-old human embryos (Smelser 1965) (Figure 3c); at 48-hpf, robust expression is observed in the developing lens fiber cells (Figure 3f). The strong expression of cryba2 transcripts at early stages of lens development provides further support for its role in congenital disease.
Figure 3.
Expression of zebrafish CRYBA2 orthologs, cryba2a and cryba2b, during embryonic development. Expression of cryba2a (a, d) and cryba2b (b, c, e, f) in whole mount embryos (a, b, d, e) and frontal sections of the head at the level of the eyes (c, f) in 22-hpf (a–c) and 48-hpf (d–f) embryos; lateral views (a, b, d) and a dorsal view (e) are shown. Please note strong and specific expression of both genes in the developing lens (l); on- optic nerve, r-retina.
The discovery of a causative mutation in CRYBA2, a member of βγ-crystallin superfamily, provides further evidence of the importance of these proteins to ocular function. Crystallins represent the major structural components of the vertebrate lens and function to increase lens refractory power while preserving its transparency. Mutations in crystallins are widely associated with cataracts in various species (Graw 2009). The βγ-crystallin superfamily is comprised of thirteen members in humans and ten of them have now been shown to cause cataracts in humans when mutated. In mouse, a missense mutation in Cryba2, c.139T>C, p.(Ser47Pro), was associated with small lenses and age-related cataracts at 25 weeks in both heterozygous and homozygous animals; the phenotype was more severe in homozygous mice, but no evidence of congenital cataract was seen in mice with either genotype with histologic analysis performed at postnatal days 1 and 21 (Puk et al. 2011). Interestingly, the disease-causing mutations in both mouse, p.(Ser47Pro), and human, p.(Val50Met), affect invariant amino acids within the Greek key motif I and are separated by only two residues. While it is possible that human eyes are more sensitive to CRYBA2 mutations, other factors potentially contributing to the differences in the age of onset, such as the specific genetic background or differences in lifespan/development between species, cannot be ruled out. Identification of additional human and mouse CRYBA2/cryba2 alleles will allow for better understanding of the phenotypic spectrums associated with mutations in this gene.
No disease-causing mutations were identified in the other seven novel crystallin genes screened (CRYGB, CRYL1, CRYGN, CRYZ, CRYM, CRYZL1, and CRYBG3). One rare variant, c.97C>T, p.(Arg33Trp), was seen in CRYGN in Patient 1 and is predicted to be benign/ deleterious by PolyPhen/SIFT but did not segregate with the disease phenotype; this variant was also seen in 2/8600 EA chromosomes in EVS (0.05% population frequency).
In summary, analysis of twenty-three dominant cataract pedigrees by whole exome sequencing identified causative mutations in known cataract genes (CRYAA, CRYBB1, CRYBB3, CRYGC (2), CRYGD, GJA8 (2), and MIP) in nine families (39%), a novel causative gene mutation in a member of βγ-crystallin superfamily CRYBA2 in one pedigree, and an additional likely causative mutation in EYA1 in one case, thus explaining up to 48% (11/23) of dominant cataract in our study. All mutations were identified in families affected with congenital cataract (11/18); in contrast, no mutations were found in families affected with familial juvenile or adult-onset cataracts (0/5). Our data highlight the extreme genetic heterogeneity of dominant congenital cataract as the eleven causative/likely causative mutations affected nine different genes and the majority of mutant alleles were novel. Furthermore, these data suggest that more than half of hereditary dominant cataract remains to be explained. While it is possible that some of these families may have mutations in known genes in small regions not well-covered by WES or copy number variations affecting these genes, it is likely that the majority are due to novel factors. Ongoing analysis of WES data generated in this study will likely identify novel genetic factors responsible for the cataract seen in the remaining families.
Supplementary Material
Pedigrees of 23 families affected with hereditary cataract and examined in this study. Probands are indicated with an arrow; black symbol indicates congenital or juvenile cataract, gray symbol indicates adult-onset cataract (age of diagnosis indicated under the symbol); individuals included in variant/mutation cosegregation analyses are marked with asterisks; if a causative mutation was identified in the family, the gene is listed in parentheses.
Acknowledgments
The authors gratefully acknowledge the patients and their families for their participation in research studies. This work was supported by the National Institutes of Health awards R01EY015518, R21DC010912 and funds provided by the Children’s Hospital of Wisconsin (EVS), 1UL1RR031973 from the Clinical and Translational Science Award (CTSA) program, and was supported in part by Research to Prevent Blindness, Inc., New York, NY (DPH and DC); the authors are also thankful to Elena A. Sorokina for cloning of zebrafish cryba2 gene sequences and thoughtful discussions.
Footnotes
Ethical standards
All experiments described comply with the current laws of the United States of America.
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Aldahmesh MA, Khan AO, Mohamed JY, Hijazi H, Al-Owain M, Alswaid A, Alkuraya FS. Genomic analysis of pediatric cataract in Saudi Arabia reveals novel candidate disease genes. Genet Med. 2012 doi: 10.1038/gim.2012.86. [DOI] [PubMed] [Google Scholar]
- Azuma N, Hirakiyama A, Inoue T, Asaka A, Yamada M. Mutations of a human homologue of the Drosophila eyes absent gene (EYA1) detected in patients with congenital cataracts and ocular anterior segment anomalies. Hum Mol Genet. 2000;9:363–366. doi: 10.1093/hmg/9.3.363. [DOI] [PubMed] [Google Scholar]
- Bamshad MJ, Ng SB, Bigham AW, Tabor HK, Emond MJ, Nickerson DA, Shendure J. Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet. 2011;12:745–755. doi: 10.1038/nrg3031. [DOI] [PubMed] [Google Scholar]
- Bremond-Gignac D, Bitoun P, Reis LM, Copin H, Murray JC, Semina EV. Identification of dominant FOXE3 and PAX6 mutations in patients with congenital cataract and aniridia. Mol Vis. 2010;16:1705–1711. [PMC free article] [PubMed] [Google Scholar]
- Burdon KP, Wirth MG, Mackey DA, Russell-Eggitt IM, Craig JE, Elder JE, Dickinson JL, Sale MM. A novel mutation in the Connexin 46 gene causes autosomal dominant congenital cataract with incomplete penetrance. J Med Genet. 2004;41:e106. doi: 10.1136/jmg.2004.018333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi BO, Koo SK, Park MH, Rhee H, Yang SJ, Choi KG, Jung SC, Kim HS, Hyun YS, Nakhro K, Lee HJ, Woo HM, Chung KW. Exome sequencing is an efficient tool for genetic screening of Charcot-Marie-Tooth Disease. Hum Mutat. 2012;33:1610–1615. doi: 10.1002/humu.22143. [DOI] [PubMed] [Google Scholar]
- Cohen D, Bar-Yosef U, Levy J, Gradstein L, Belfair N, Ofir R, Joshua S, Lifshitz T, Carmi R, Birk OS. Homozygous CRYBB1 deletion mutation underlies autosomal recessive congenital cataract. Invest Ophthalmol Vis Sci. 2007;48:2208–2213. doi: 10.1167/iovs.06-1019. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Huerta L, Ramirez-Sanchez V, Rivera-Vega M, Messina-Baas O, Cuevas-Covarrubias S. A family with hereditary hyperferritinaemia cataract syndrome: evidence of incomplete penetrance and clinical heterogeneity. Br J Haematol. 2008;143:596–598. doi: 10.1111/j.1365-2141.2008.07345.x. [DOI] [PubMed] [Google Scholar]
- Graw J. Genetics of crystallins: cataract and beyond. Exp Eye Res. 2009;88:173–189. doi: 10.1016/j.exer.2008.10.011. [DOI] [PubMed] [Google Scholar]
- Graw J, Schmidt W, Minogue PJ, Rodriguez J, Tong JJ, Klopp N, Illig T, Ebihara L, Berthoud VM, Beyer EC. The GJA8 allele encoding CX50I247M is a rare polymorphism, not a cataract-causing mutation. Mol Vis. 2009;15:1881–1885. [PMC free article] [PubMed] [Google Scholar]
- Hansen L, Eiberg H, Rosenberg T. Novel MAF mutation in a family with congenital cataract-microcornea syndrome. Mol Vis. 2007a;13:2019–2022. [PubMed] [Google Scholar]
- Hansen L, Yao W, Eiberg H, Kjaer KW, Baggesen K, Hejtmancik JF, Rosenberg T. Genetic heterogeneity in microcornea-cataract: five novel mutations in CRYAA, CRYGD, and GJA8. Invest Ophthalmol Vis Sci. 2007b;48:3937–3944. doi: 10.1167/iovs.07-0013. [DOI] [PubMed] [Google Scholar]
- Hawse JR, DeAmicis-Tress C, Cowell TL, Kantorow M. Identification of global gene expression differences between human lens epithelial and cortical fiber cells reveals specific genes and their associated pathways important for specialized lens cell functions. Mol Vis. 2005;11:274–283. [PMC free article] [PubMed] [Google Scholar]
- He W, Li X, Chen J, Xu L, Zhang F, Dai Q, Cui H, Wang DM, Yu J, Hu S, Lu S. Genetic linkage analyses and Cx50 mutation detection in a large multiplex Chinese family with hereditary nuclear cataract. Ophthalmic Genet. 2011;32:48–53. doi: 10.3109/13816810.2010.535886. [DOI] [PubMed] [Google Scholar]
- Hejtmancik JF. Congenital cataracts and their molecular genetics. Semin Cell Dev Biol. 2008;19:134–149. doi: 10.1016/j.semcdb.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbrook JA, Neu-Yilik G, Hentze MW, Kulozik AE. Nonsense-mediated decay approaches the clinic. Nat Genet. 2004;36:801–808. doi: 10.1038/ng1403. [DOI] [PubMed] [Google Scholar]
- Huang B, He W. Molecular characteristics of inherited congenital cataracts. Eur J Med Genet. 2010;53:347–357. doi: 10.1016/j.ejmg.2010.07.001. [DOI] [PubMed] [Google Scholar]
- Kaul H, Riazuddin SA, Shahid M, Kousar S, Butt NH, Zafar AU, Khan SN, Husnain T, Akram J, Hejtmancik JF, Riazuddin S. Autosomal recessive congenital cataract linked to EPHA2 in a consanguineous Pakistani family. Mol Vis. 2010;16:511–517. [PMC free article] [PubMed] [Google Scholar]
- Khajavi M, Inoue K, Lupski JR. Nonsense-mediated mRNA decay modulates clinical outcome of genetic disease. Eur J Hum Genet. 2006;14:1074–1081. doi: 10.1038/sj.ejhg.5201649. [DOI] [PubMed] [Google Scholar]
- Meyer E, Rahman F, Owens J, Pasha S, Morgan NV, Trembath RC, Stone EM, Moore AT, Maher ER. Initiation codon mutation in betaB1-crystallin (CRYBB1) associated with autosomal recessive nuclear pulverulent cataract. Mol Vis. 2009;15:1014–1019. [PMC free article] [PubMed] [Google Scholar]
- Orten DJ, Fischer SM, Sorensen JL, Radhakrishna U, Cremers CW, Marres HA, Van Camp G, Welch KO, Smith RJ, Kimberling WJ. Branchio-oto-renal syndrome (BOR): novel mutations in the EYA1 gene, and a review of the mutational genetics of BOR. Hum Mutat. 2008;29:537–544. doi: 10.1002/humu.20691. [DOI] [PubMed] [Google Scholar]
- Polyakov AV, Shagina IA, Khlebnikova OV, Evgrafov OV. Mutation in the connexin 50 gene (GJA8) in a Russian family with zonular pulverulent cataract. Clin Genet. 2001;60:476–478. doi: 10.1034/j.1399-0004.2001.600614.x. [DOI] [PubMed] [Google Scholar]
- Pras E, Frydman M, Levy-Nissenbaum E, Bakhan T, Raz J, Assia EI, Goldman B, Pras E. A nonsense mutation (W9X) in CRYAA causes autosomal recessive cataract in an inbred Jewish Persian family. Invest Ophthalmol Vis Sci. 2000;41:3511–3515. [PubMed] [Google Scholar]
- Puk O, Ahmad N, Wagner S, Hrabe de Angelis M, Graw J. First mutation in the betaA2-crystallin encoding gene is associated with small lenses and age-related cataracts. Invest Ophthalmol Vis Sci. 2011;52:2571–2576. doi: 10.1167/iovs.10-6443. [DOI] [PubMed] [Google Scholar]
- Ren Z, Li A, Shastry BS, Padma T, Ayyagari R, Scott MH, Parks MM, Kaiser-Kupfer MI, Hejtmancik JF. A 5-base insertion in the gammaC-crystallin gene is associated with autosomal dominant variable zonular pulverulent cataract. Hum Genet. 2000;106:531–537. doi: 10.1007/s004390000289. [DOI] [PubMed] [Google Scholar]
- Riazuddin SA, Yasmeen A, Yao W, Sergeev YV, Zhang Q, Zulfiqar F, Riaz A, Riazuddin S, Hejtmancik JF. Mutations in betaB3-crystallin associated with autosomal recessive cataract in two Pakistani families. Invest Ophthalmol Vis Sci. 2005;46:2100–2106. doi: 10.1167/iovs.04-1481. [DOI] [PubMed] [Google Scholar]
- Sacconi S, Feasson L, Antoine JC, Pecheux C, Bernard R, Cobo AM, Casarin A, Salviati L, Desnuelle C, Urtizberea A. A novel CRYAB mutation resulting in multisystemic disease. Neuromuscul Disord. 2012;22:66–72. doi: 10.1016/j.nmd.2011.07.004. [DOI] [PubMed] [Google Scholar]
- Santana A, Waiswol M, Arcieri ES, Cabral de Vasconcellos JP, Barbosa de Melo M. Mutation analysis of CRYAA, CRYGC, and CRYGD associated with autosomal dominant congenital cataract in Brazilian families. Mol Vis. 2009;15:793–800. [PMC free article] [PubMed] [Google Scholar]
- Shiels A, Bennett TM, Hejtmancik JF. Cat-Map: putting cataract on the map. Mol Vis. 2010;16:2007–2015. [PMC free article] [PubMed] [Google Scholar]
- Slingsby C, Clout NJ. Structure of the crystallins. Eye (Lond) 1999;13((Pt 3b)):395–402. doi: 10.1038/eye.1999.113. [DOI] [PubMed] [Google Scholar]
- Smelser GK. Embryology and Morphology of the Lens. Invest Ophthalmol. 1965;4:398–410. [PubMed] [Google Scholar]
- Sorokina EA, Muheisen S, Mlodik N, Semina EV. MIP/Aquaporin 0 represents a direct transcriptional target of PITX3 in the developing lens. PLoS One. 2011;6:e21122. doi: 10.1371/journal.pone.0021122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trumler AA. Evaluation of pediatric cataracts and systemic disorders. Curr Opin Ophthalmol. 2011;22:365–379. doi: 10.1097/ICU.0b013e32834994dc. [DOI] [PubMed] [Google Scholar]
- Wang KJ, Wang S, Cao NQ, Yan YB, Zhu SQ. A novel mutation in CRYBB1 associated with congenital cataract-microcornea syndrome: the p.Ser129Arg mutation destabilizes the betaB1/betaA3-crystallin heteromer but not the betaB1-crystallin homomer. Hum Mutat. 2011;32:E2050–E2060. doi: 10.1002/humu.21436. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Pedigrees of 23 families affected with hereditary cataract and examined in this study. Probands are indicated with an arrow; black symbol indicates congenital or juvenile cataract, gray symbol indicates adult-onset cataract (age of diagnosis indicated under the symbol); individuals included in variant/mutation cosegregation analyses are marked with asterisks; if a causative mutation was identified in the family, the gene is listed in parentheses.