Abstract
Introduction
The tumor suppressor gene HIC1 (Hypermethylated in Cancer 1) which encodes a transcriptional repressor with multiple partners and multiple targets is epigenetically silenced but not mutated in tumors. HIC1 has broad biological roles during normal development and is implicated in many canonical processes of cancer such as control of cell growth, cell survival upon genotoxic stress, cell migration and motility.
Areas covered
The HIC1 literature herein discussed include its discovery as a candidate tumour suppressor gene hypermethylated or deleted in many human tumours, animals models establishing it as tumour suppressor gene, its role as a sequence specific transcriptional repressor recruiting several chromatin regulatory complexes, its cognate target genes and its functional roles in normal tissues. Finally, this review will discuss how its loss of function contributes to the early steps in tumorigenesis.
Expert opinion
Given HIC1’s ability to direct repressive complexes to sequence specific binding sites associated with its target genes, its loss results in specific changes in the transcriptional program of the cell. An understanding of this program through identification of HIC1’s target genes and their involvement in feedback loops and cell process regulation will yield the ability to leverage this knowledge for therapeutic translation.
Keywords: Epigenetics, tumour suppressor genes, transcriptional repression, HIC1
1-Introduction
1.1 Tumor Suppressor Gene Inactivation
Tumor suppressor genes (TSG) normally function to control cellular proliferation. The loss of TSG function is one of the major mechanisms of tumor formation and is contrasted with the gain-of-function of oncogenes, which similarly confer growth advantages upon pre-neoplastic cells. Given that there are two alleles of each gene, it normally requires two genetic events to inactivate a TSG. Knudson originally proposed the two hit hypothesis to explain the inactivation of one of the most common TSGs, the Retinoblastoma gene (Rb) [1]. At the time, these two hits were thought to require deletion or mutation. In the past two decades, a third process has been implicated in the silencing of tumor suppressor genes, epigenetic inactivation. These epigenetic alterations, so called because the genetic sequence of the DNA remains unaltered, involve post-translational modifications of the histone tail, altered chromatin conformation, transcription factor exclusion and DNA methylation. Increasingly, these mechanisms are becoming better understood, making possible an era of therapeutic intervention designed to reverse epigenetic TSG silencing.
1.2 Chromosome arm 17p deletions in cancer
The search for tumor suppressor genes has historically been driven by studies of high frequency chromosomal losses in tumor tissues. The short arm of chromosome 17 comprises a region that is commonly reduced to homozygosity in many human cancers including those of the breast, lung, liver, colon, kidney and brain [2–8]. This chromosomal aberration is particularly frequent in neural tumors. Childhood brain tumors that exhibit this chromosomal loss include medulloblastoma, ependymoma, and astrocytoma [9–17]. In medulloblastoma, the most common malignant brain tumor of childhood, early studies indicated that 2/3 of tumors exhibit LOH (Loss of Heterozygosity) converging at the 17p13.3 locus [6]. Notably, this region is telomeric to p53 and its deletion is distinct from p53 inactivating events [13, 15, 18-209-17] as has also been shown in high grade glioma [11, 14]. Subsequent evaluations of medulloblastoma clinical samples by single nucleotide polymorphism chips have demonstrated that chromosome 17p LOH events are found in 25%, 42% and 63% of sonic hedgehog, group 3 and group 4 tumors respectively whereas 17p LOH in the context of isochrome 17q is confined to subgroups 3 and 4 [21–23]. Therefore, an important question in tumorigenesis has been “what gene(s) is/are affected by this common deletional event and by extension, what are the functional/biological consequences of this event ?”
1.3 Identification of HIC1
The search for relevant tumor suppressor genes in tumors with a distal 17p deletion was narrowed down to the minimal region of deletion in close proximity to the VNTR (Variable Number of Tandem Repeats) marker YNZ22/D17S5/D17S30 (17p13.3) [9, 10, 13, 24–26].
HIC1 (Hypermethylated In Cancer-1) is one of the genes found in this 17p13.3 region. Increasing its potential significance, HIC1 resides completely within a CpG island that is frequently hypermethylated in tumors, a feature associated with the transcriptional silencing of tumor suppressor genes [27]. HIC1’s status as a tumor suppressor gene results from its location in a commonly deleted area, its propensity toward epigenetic inactivation, the spontaneous formation of tumors in a knockout murine model and the fact that the enforced re-expression of HIC1 in tumor cell lines significantly decreases their clonogenic survival [27]. The regulatory region upstream of the HIC1 sequence contains a p53 binding site and the transcription of HIC1 is activated by wild type p53 [27–29]. As detailed below, HIC1 is involved in an elaborate regulatory relationship with p53 that involves SIRT1 via post-translational modifications. Its relationship with p53 is compelling given the prominence of p53 alterations in human cancers. HIC1 is also linked to another prominent tumor suppressive pathway, Rb. When phosphorylated, Rb releases E2F1, a transcription factor whose target genes mediate transit of the G1/S checkpoint. E2F1 is also a positive regulator of HIC1, potentially as part of a negative feedback loop to promote orderly cell cycle progression [30]. E2F1 also takes part in cellular responses to DNA damage and has been found to be a necessary component of HIC1 upregulation in that context [31, 32].
2-HIC1 inactivation in tumorigenesis
The presence of HIC1 methylation has been demonstrated in various tumor types including renal tumors, pancreatic carcinoma, prostate cancer, gastric cancer, colorectal carcinoma, myeloid leukemia, non-small cell lung cancer (NSCLC), hepatocellular carcinoma, breast cancer, astrocytoma, ependymoma and medulloblastoma [3–6, 16, 17, 35–40]. Consistent with an epigenetically determined tumor phenotype, methylated HIC1 expression can be de-methylated and re-activated by 5-azacytidine resulting in decreased cellular proliferation, cell cycle arrest, decreased tumor aggressiveness and increased apoptosis in head and neck squamous cell and pancreatic carcinomas [36, 40, 41]. Many studies support the postulate that epigenetic inactivation of HIC1 is a progression event in oncogenesis. For example, methylation of HIC1 increases across the spectrum from normal liver tissues to pre-cancerous liver conditions to hepatocellular carcinoma [35]. In colon cancer, DNA in the region of HIC1 exhibits increasing methylation density from normal colonic mucosa to pre-cancerous colonic polyps to colon cancer [2]. Notably, among colorectal cancer patients, those arising in the context of ulcerative colitis exhibit the highest levels of HIC1 methylation [37]. Finally, HIC1 was found to be methylated with a significantly greater incidence in both tumorous and non-tumorous lung tissues from smokers compared to non-smokers [34].
It should be noted that although HIC1 is frequently hypermethylated in leukemias, some leukemias have a reduction in HIC1 expression that is not correlated with HIC1 promoter hypermethylation. This strongly suggests the existence of other inhibitory mechanisms such as direct transcriptional repression [32].
HIC1 inactivation has been shown to correlate with a more aggressive phenotype and poorer survival in multiple tumor types. For example, LOH of 17p13.3 has been associated with the higher grades of astrocytoma, namely anaplastic astrocytoma and glioblastoma multiforme [14]. In a study of non-small cell lung carcinoma, decreased HIC1 expression in tumors, in comparison to paired normal lung tissue, predicted a poorer survival (80% versus 46% 5 year survival; p=0.034). Suppression of HIC1 expression and its prognostic implications were found to be invariable by clinical stage, indicating that it is an early event in tumor development [6]. In renal cell carcinoma, HIC1 hypermethylation correlates with an increased risk of disease recurrence [42]. Likewise, HIC1 hypermethylation is an independent predictor of poor survival in laryngeal squamous cell carcinoma [43]. The authors have found LOH and DNA methylation to be silencing events involving the region containing the HIC1 gene in medulloblastoma and correlated HIC1 methylation with decreased overall survival [16].
3-Animal models
Both the cloning of the HIC1 gene from a CpG island that is commonly subject to DNA methylation in neoplastic tissues and its association with malignant progression gave impetus to the quest to identify the functional role played by HIC1 in normal tissues that is lost in tumorigenesis. The first Hic1 murine knockout model was created in the Baylin laboratory at Johns Hopkins and showed that Hic1 plays a prominent role in neural and somatic development. Mice with homozygous loss of Hic1 are marked by perinatal death, small size, acrania, exencephaly, craniofacial abnormalities, limb defects and omphalocele [44]. Heterozygous knockouts did not have these defects. These abnormalities are very similar to those found in the Miller-Dieker syndrome (MDS), a contiguous-gene syndrome marked by deletion of multiple genes in the 17p13.3/D17S5 region including HIC1. MDS consists of a constellation of brain malformation (lissencephaly), mental retardation, craniofacial dysmorphology, defects of the limbs and digits and omphalocele.
The knock-out mouse model also provided evidence for HIC1’s tumor suppressor function. The heterozygous mice develop various tumors beginning after 70 weeks and involving 16.4% of mice by 90 weeks compared to 0% for wild-type litter mates. Interestingly, the observed tumors followed a sex determined pattern with male mice developing epithelial cancers while female mice exhibited a predominance of sarcomas and lymphomas. In all of these tumors, the remaining wild-type Hic1 allele had undergone epigenetic silencing marked by DNA methylation[45].
Given the delayed time course of tumor formation allowing for the second epigenetic hit to occur, it was logical to inquire whether Hic1 silencing could cooperate with other known tumorigenic pathways to hasten tumor formation. A double heterozygous model incorporating p53 knockout (Hic1−/+p53−/+) demonstrated, when both events occurred on the same chromosome, an increased incidence of osteosarcoma formation. When the deletions occurred on opposite chromosomes, breast and ovarian carcinomas and metastatic osteosarcomas occurred exhibiting epigenetic inactivation of the wild type Hic1 allele. This model showed the importance of cooperation between genetic deletion and epigenetic inactivation in tumorigenesis [46]. Another double heterozygote cross, between the medulloblastoma forming Ptch1−/+ model and Hic1−/+, resulted in an unchanged latency of tumor onset but a four-fold increase in tumor incidence compared to the Ptch1−/+ mouse. In this model, the remaining wild type allele of Hic1 was again found to be methylated and transcriptionally silent. Notably, tumors arising from the Ptch1−/+ model also demonstrate epigenetic silencing of Hic1 [47].47
Consistent with the observation that HIC1 is methylated in intestinal polyps, it was shown that loss of a single Hic1 allele resulted in crypt hyperplasia and GI tract neoplasia in the Hic1/Apc double heterozygous model. When the Hic1+/− mouse was crossed with the polyp forming mutant Apc murine line (Apc +/Δ716), the resulting double heterozygous mice displayed a further increase in polyp formation. These polyps, with absent Hic1 expression, also featured deregulated WNT signaling with nuclear Beta-catenin and upregulated Sox9 and Sirt1, target genes of Hic1 mediated transcriptional repression that play regulatory roles in the WNT pathway [48].
4-HIC1 target genes
The positional cloning of HIC1 close to the D17S5 microsatellite region and the sequencing of the corresponding 11.0Kbp region identified an open reading frame containing two well-defined functional domains: five canonical C-terminal C2H2 Krüppel-like Zinc fingers motifs involved in sequence-specific DNA-binding and an N-terminal BTB/POZ (bric à brac, Broad-Complex, tramtrack/Pox viruses and Zinc fingers) domain. The BTB/POZ domain is a functional domain which at that time had just been identified in actin-binding proteins such as the Drosophila Kelch and Poxviruses proteins and in a subset of Zinc fingers transcription factors [27, 49–51].49 These two functional domains, as well as the C-terminal region are highly homologous to the chicken γFBB-B protein characterized as a transcriptional repressor of the γF-crystallin gene [52]. These results strongly suggested that HIC1 was a sequence-specific transcriptional repressor belonging to the BTB/POZ and C2H2 zinc fingers family [27, 53].
HIC1 contains a cluster of four C2H2 Zinc fingers (ZF2-5) which are separated by a canonical 7–8 amino acids H/C link found in Krüppel-like Zinc fingers strongly suggesting that they are involved in sequence-specific DNA-binding. By contrast, ZF1, albeit also phylogenetically conserved, is more distant and is therefore unlikely to contribute to DNA-binding. Instead, this ZF could be involved in protein-protein interactions [54]. As a first step toward the identification of HIC1 direct target genes, we have determined the HIC1-specific DNA binding sequence using random oligomers and a GST-HIC1 5ZF-C-Term fusion protein in multiple rounds of SAAB (Selection and Amplification of Binding sites) [55]. These studies identified the sequence 5′ C/GNGC/GGGCAC/ACC 3′ as the optimal in vitro HIC1 binding site 55. Functional assays and mutational analyses highlighted a GGCA core motif bound by ZF3 and ZF4 [55].
The BTB/POZ domain is a dimerization and protein-protein interaction domain [49, 51]. Indeed, crystal structures of the BTB/POZ domains of two sequence-specific transcriptional repressors (BCL6 and PLZF) have revealed a tightly intertwined dimer with an extensive hydrophobic interface [56–58]. In addition, the BTB/POZ domain forms dimer-dimer interactions in the crystal opening the possibility for higher order protein complexes. This might explain why in transient transfection assays, BTB/POZ proteins invariably are observed in nuclear dots structures [59–61]. Moreover, this punctuated nuclear localization has been observed for several endogenous BTB/POZ proteins, including HIC1 [62]. Therefore, HIC1, as is the case with many BTB/POZ transcriptional repressors must exist as a branched transcription factor with two or more C-terminal DNA-binding modules. Consistent with this fact, we have demonstrated that the full length HIC1 protein binds poorly to a single HIC1 responsive element (HiRE) in electrophoretic mobility shift assays (EMSA) performed with reticulocyte lysates or with nuclear extracts of transfected cells, due to an inhibitory effect mediated by the BTB/POZ domain, as demonstrated earlier [49, 55, 63]. Conversely, the full-length HIC1 proteins bind cooperatively to a probe containing multiple HIC1 consensus binding sites (5xHIRE) [55]. Our studies together with several others suggest a model in which BTB/POZ proteins could simultaneously bind several, not necessarily optimal, distant binding sites thereby linking cis enhancers of silencers and their promoters in a single regulatory unit. This architectural role has been better demonstated for the Drosophila Bab proteins which bind several sites in the large bab locus [64]or for the vertebrate PLZF protein which can form a DNA loop bringing together multiple distant PLZF responsive elements in the HoxD gene [65].
Despite this wealth of information concerning the in vitro binding properties of HIC1, only 12 direct target genes have been validated so far. The following six direct target genes were identified through educated guesses and/or bioinformatics analyses of gene promoters for the presence of HIC1 binding sites: SIRT1, FGF-BP1, ΔNP73, CCND1, P57KIP2 and P21Waf1.
4.1 SIRT1
The first bona fide HIC1 target gene identified was SIRT1, a member of the Sirtuin family of NAD+-dependent deacetylases. Involved in epigenetic regulation through histone deacetylation but also in post-translational modifications of a still growing list of non-histone substrates, notably transcriptional regulators, SIRT is implicated in numerous essential biological processes including aging, metabolism and response to stress. Several animal models clearly demonstrated that loss of HIC1 and P53 synergized in tumorigenesis [45, 46]. To decipher how HIC1 and P53 cooperate, these authors analyzed the role of HIC1 in the P53-dependant DNA damage response [66]. Hic1 knock-out murine embryonic fibroblasts (Hic1−/− MEFs) were more resistant than wild-type MEFS to etoposide treatment (80μM for 16 hours) induced apoptosis. Conversely ectopic expression of HIC1 in MCF7 cells through adenoviral infection resulted in massive apoptosis 2 days after drug removal whereas cells infected with a control adenoviral vector were resistant [66]. This effect was strictly p53-dependent since it was lost in cells transfected with shP53. SIRT-1 mRNA and protein expression levels were significantly increased in Hic1−/− MEFs. This effect is mediated by a cluster of two adjacent HIC1 binding sites (HiRE) in the same orientation (−1116 and −1039) located 5′ to the SIRT1 promoter [66]. Chromatin immunoprecipitation (ChIP) assays in normal human WI38 fibroblasts demonstrated binding of endogenous HIC1 and SIRT1 proteins on these two sites leading to the conclusion that a complex containing HIC1-SIRT1 directly represses transcription of SIRT1 [66]. Several studies have confirmed and extended these results. First, we synchronized the cell cycles of normal human WI38 fibroblasts by serum starvation to obtain mid-G1 cells and observed a slight increase in SIRT1 mRNA levels in mid-G1 WI38 cells as compared to quiescent cells. Strikingly, ChIP experiments clearly demonstrated the binding of HIC1 on the two SIRT1 promoter HiRE in mid-G1 but not in quiescent cells [67]. The reappraisal of the SIRT1 promoter sequence and the identification of another proximal HiRE helped to solve this conundrum. Indeed, ChIP experiments revealed a switch of HIC1 binding from the previously described distal (−1116 and −1039) HiREs to this newly identified proximal site when growing cells are serum-starved for 72 hours [67]. Notably, this proximal site is perfectly conserved in the human, murine and rat genomes by contrast with the distal sites. Recently, this proximal HiRE has been implicated in the suppression of SIRT1 transcription, mediated in skeletal muscle cells by the pro-inflammatory cytokine IFN-γ through the direct binding of a HIC1-CTIIA (Class II transactivator) [68].68
Therefore, SIRT1 is a direct target gene of HIC1 in normal cells subjected to various stress and/or metabolic conditions through a complex interplay between two functionally different clusters of HIRE and different types of HIC1-corepressor complexes.
4.2 FGF-BP1
The repression of the gene coding for Fibroblast Growth factor-binding protein 1 (FGF-BP1) by TGFβ during smooth cell muscle differentiation in vitro is strictly dependent on a HIC1 binding site in its promoter region [69]. FGF-BP1 is essential for the activation of FGFs since it allows their release from the extracellular matrix where FGFs are tightly bound to heparin sulfate proteoglycans and hence inactive. Besides a role in embryogenesis and wound healing through blood vessel formation, FGF-BP1 is also essential for tumor angiogenesis. FGF-BP1 is an angiogenic switch in human cancers and is thus up-regulated in several tumors [70].
4.3 ΔNP73
ΔNP73 is a N-terminally truncated isoform of P73, a member of the P53 family. Since it lacks the N-terminal transactivation domain,ΔNP73 is a potent dominant-negative inhibitor of P53 and P73 and has potent anti-apoptotic and oncogenic properties. ΔNP73 is up-regulated in many tumors including breast, ovarian, prostate, lung, and esophagus [71]. Its expression is inversely correlated with HIC1 expression in normal and tumor tissues. HIC1 directly regulates ΔNP73 via a phylogenetically conserved HiRE in its 5′ untranslated region [72].
4.4 CCND1
HIC1 is a direct target gene of P53 [27–29]and E2F1 [30]which strongly suggests a direct relationship between HIC1 and cell-cycle regulation. In silico analyses of genes encoding key regulators of the cell cycle identified potential HiRE in various Cyclin genes, notably in the cyclin D1 promoter region. Cyclin D1 is essential for the G1 to S phase transition in mammalian cells. Cyclin D1 binds to and activates its associated cyclin-dependent kinases (CDK4 and CDK6) to phosphorylate Rb. Phosphorylated Rb is unable to bind E2F1 which can then transactivate its S phase target genes. To address the potential regulation of Cyclin D1 by HIC1, we synchronized the cell cycle of normal human WI38 fibroblasts by serum starvation to obtain mid-G1 cells. Chromatin immunoprecipitation (ChIP) experiments clearly demonstrated the binding of HIC1 and two co-repressor complexes to the Cyclin D1 HiRE in quiescent cells and its absence in Mid-G1 [67]. Cyclin D1 expression is mainly controlled by post-transcriptional mechanisms. However, the lack of HIC1 repressive complexes on the Cyclin D1 promoter nicely correlated with its increase of expression during Mid-G1. Together, these results identified Cyclin D1 as a new direct HIC1 target gene.
4.5 P57KIP2
P57KIP2 (CDKN1C) is also a direct target gene of a transcription factor that we have isolated as a potential HIC1 partner in a yeast two-hybrid screen (Van Rechem; unpublished results). In the same experimental setting of cell cycle synchronized human fibroblasts, we demonstrated that HIC1 repressive complexes are bound on the P57Kip2 HiRE in quiescent cells but not in Mid-G1 cells [67]. Thus, the direct HIC1-mediated repression of the CDK inhibitor, P57KIP2, and of the CDK activator, Cyclin D1, might at first sight seem contradictory but could allow for fine-tuned regulation of cell cycle progression. In fact, P57KIP2 is a positive or negative regulator of G1 phase progression; its expression is high during Go and G1 phases and decreases during progression from G1 to S. In addition, at low levels, P57KIP2 binds to the CDK-cyclin heterodimers and promotes their assembly whereas at high levels it abrogates CDK activity.
4.6 P21CIP/Waf1
P21(CIP/WAF1) is, as is P57KIP2, a member of the CIP/KIP family of cyclin-dependent kinase inhibitors. P21CIP/Waf1 is a direct target gene of P53 which is activated by stabilized P53 in response to repairable DNA-damage to reversibly arrest the cell cycle, thus allowing safe completion of the DNA repair process [73]. Recently, using ectopic expression of HIC1 in MDA-MB231 breast cancer cells as well as HIC1 knock-down and ChIP experiments in BJ fibroblasts, we identified P21(CIP/WAF1) as a new direct target gene of HIC1 [74]. These results add a new layer of complexity to the P53-HIC1-SIRT1 regulatory network, modulating the cellular response to genotoxic stress. Indeed, P53 activates the transcription of P21(CIP/WAF1), SIRT1 and HIC1 whereas HIC1 directly represses transcription of SIRT1 and P21(CIP/WAF1) [27, 73–74]. While the P53-mediated activation of the P21(CIP/WAF1) cell cycle inhibitor is necessary early in the DNA damage response to immediately block the cell cycle progression, its repression by HIC1 could occur later, allowing the cell cycle to resume in cases of reparable damage or trigger apoptosis in case of non repairable damage [27, 74].
The other HIC1 direct target genes came from four independent gene profiling experiments using forced re-expression of HIC1 in tumour cell lines deficient for HIC1; the osteosarcoma U2OS [75], the medulloblastoma D425 [47], the breast cancer MCF-7 [76]and the PC3 prostatic [77]cell lines. These target genes include ATOH1, CXCR7, ephrinA1, EphA2, ADRB2 and Sox9. By contrast, SIRT1 is not repressed but is up-regulated (1.5 fold) both in U2OS and D425 cells infected by Adenoviral vectors expressing HIC1, presumably reflecting a stress response of these cells to the infection [47, 75].
4.7 ATOH1/MATH1
In the Ptch1−/+Hic1+/− double heterozygotes model, Hic1 inactivation results in the increased incidence of medulloblastomas. These authors therefore investigated the potential role of HIC1 as a transcription factor essential for normal cerebellar development. Briefly, the cerebellum is composed of two main cell layers which proliferate and differentiate during early post-natal development [78]. The external granule cell layer (EGL) is composed of immature proliferative cells driven by the Sonic hedgehog (Shh) pathway. At day 7 of development, these cells begin to differentiate and migrate to form the internal granule cell layer (IGL) composed of non-proliferating, differentiated granule cells [79]. By immunohistochemistry, Hic1 expression was absent in the outer EGL, weakly detectable in cells lining the inner EGL and strongly expressed in the mature granule cells of the IGL [47]. This expression pattern inversely mirrored that of ATOH1/MATH1 a pro-neuronal transcription factor essential for normal cerebellar development whose deregulation contributes to medulloblastoma formation [80] Re-expression through adenoviral expression in the HIC-1-deficient (through promoter hypermethylation) D425 medulloblastoma cell line and ChIP experiments in normal murine cerebellum at selected times of post-natal development clearly demonstrated that ATOH1 is a direct target gene of HIC1 through binding to tandem HiRE sites in a distal 3′ enhancer [47] as well as to HiREs in the 5′ promoter [81].
4.8 CXCR7/RDC1
CXCR7, also known as RDC1, is a G-protein coupled seven-span transmembrane receptor (GPCR). CXCR7 is involved in normal development and in tumorigenesis [82]. Two CXCR7 knock-out models in mice have defined a role in cardiac development. In addition, CXCR7 is highly expressed in many tumor types and in activated, tumor-associated endothelial cells. The chemokine CXCL12/SDF-1 has been shown to bind with high affinity to the CXCR4 and CXCR7 receptors. In contrast with CXCR4, CXCR7 fails to activate heterotrimeric G proteins and is thus considered to be a non-signalling scavenger receptor counter-acting CXCR4 function. CXCR7 has been recently shown to affect cellular signalling networks through heterodimerization with CXCR4. These CXCR4-CXCR7 heterodimeric complexes and the ligand binding to CXCR7 result in preferential activation of β-arrestin linked signalling pathways instead of canonical G protein pathways, to enhance cell migration through MAP kinase [83–84]. Therefore, mounting evidence implicates the CXCR7-CXCR4 and CXCL12/SDF-1 axis in tumorigenesis. In particular, CXCR7 is important for prostate and breast cancer metastasis [85–86]. CXCR7 has been shown to be a direct HIC1 target gene, in part through a phylogenetically conserved HiRE located close to the transcription start site, in various types of transformed cells re-expressing HIC1 as well as in normal fibroblasts [67, 75–77, 81]. CXCR4 was an absent call in our U2OS gene profiling experiments and has not been described as a HIC1 direct target genes in other studies. By contrast, SDF1b appears to be a potential HIC1 target gene, either direct or indirect, as its expression was down-regulated (7-fold) in MCF-7 cells over expressing HIC1 [76]. Clearly, the regulation of the CXCR4-CXCR7-SDF-1 axis by HIC1 deserves further attention as a potential therapeutic opportunity in many cancer types.
4.9 ephrinA1 and EphA2
These two target genes have been isolated in two independent studies, in MCF-7 and U2OS cells overexpressing HIC1, respectively, but will be discussed together since they are functionally linked; ephrin-A1 is the cell-bound ligand of the Tyrosine Receptor (RTK) EphA2 [76-, 87–88]. These results are in good agreement with the mutually exclusive expression of EphA2 and ephrinA1 observed in a panel of 28 breast cancer cell lines including MCF-7 and MDA-MB-231 [89]. Indeed, EphA2 is expressed in cells with mesenchymal characteristics such as MDA-MB-231 whereas ephrinA1 expression is restricted to cancer cell lines which have retained epithelial cell markers [89]. A similar inverse correlation of expression has been recently described in metastatic ductal carcinoma samples whereas normal breast and in situ ductal carcinomas expressed both EphA2 and ephrin-A1 proteins [90]. The impact of the EphA and ephrinA1 signalling on cell behaviour is complex and highly context-dependent, sometimes with opposite effects [91]. Nevertheless, a widely recognized function of this complex signalling is the regulation of cell adhesion, positioning and migration [92]. In line with these observations, the over-expression of HIC1 in MDA-MB-231 strongly inhibits their anchorage-dependent and anchorage-independent growth as well as their migration and invasion properties [88]. These effects could at least partly rely on the direct transcriptional repression of EphA2 by HIC1, as demonstrated by various biological and functional assays [88]. Similarly, ephrin-A1 is a direct HIC1 target gene in normal WI38 human fibroblasts and in MCF-7 cells over-expressing HIC1 [76]. In addition, misexpression of ephrinA1 was observed in Hic1 −/− e14.5 mouse embryos as compared to wild-type littermates [76] consistent with the roles of Ephrins and Hic1 in normal development [45, 93]. As a whole, HIC1 directly regulates EphA2 and ephrinA1 expression in normal mammary cells. The early epigenetic alteration of HIC1 coupled with additional genetic or epigenetic mutational events might then yield distinct populations of transformed cells expressing either the tyrosine kinase receptor or its ligand. This could ultimately contribute to intra-tumoral heterogeneity.
4.10 Sox9
This HMG (high-mobility group) transcription factor is down-regulated in U2OS cells overexpressing HIC1 [75]. In the gastrointestinal (GI) tract, Sox9 is a target of the Wnt pathway and is essential to repress genes involved in intestinal development, notably in colonic crypts. Since HIC1 is epigenetically silenced in human colon cancers, the gastrointestinal tract of Hic1+/− heterozygous mice, which spontaneously develop tumors, has been analysed by immunohistochemistry. These experiments demonstrated that loss of a single Hic1 allele can promote crypt hyperplasia and neoplasia of the GI tract [48]. Furthermore, HIC1 can cooperate with APC (Adenomatous polyposis coli), another gene mutated in gastrointestinal tract cancers. Indeed, mice double heterozygote for Hic1+/−, and for APC+/Δ716, an APC mutant encoding a truncated protein, develop increased numbers of polyps throughout the GI tract [48]. In these polyps, the lack of HIC1 expression nicely correlated with a concomitant up-regulation of Sox9 and SIRT1. ChIP assays confirmed that Sox9 is a direct transcriptional target of HIC1 [48].
4.11 ADRB2
Given the direct impact of HIC1 on the transcription of several, membrane receptors (CXCR7, EphA2) and a ligand (ephrinA1) involved in migration properties of mammary epithelial cells, we recently focused on the β-2 adrenergic receptor (ADRB2), another cell membrane receptor significantly repressed in U2OS cells infected by Ad-HIC1 [75, 94]. ADRB2 is a GPCR whose activation by the stress hormones adrenaline/noradrenaline stimulates migration and invasion in in vitro cellular models and promotes angiogenesis, tumor growth and metastasis in vivo [95]. Through various biochemical and functional assays in normal fibroblasts endogenously expressing HIC1 or in the HIC1-deficient metastatic breast cancer cell MDA-MB-231 infected with a retrovirus expressing HIC1, we established that ADRB2 is a new direct target gene of HIC1 [94]. In addition, we demonstrated by qRT-PCR that ADRB2 but not the related ADRB1 and ADRB3 receptors was strongly expressed in these cells, making them a suitable experimental model to study the impact of ADRB2 and HIC1 dysregulation on invasion and migration properties. Indeed, overexpression of HIC1 inhibited an ADRB2-mediated boost of migration and invasion of MDA-MB-231 cells both in basal conditions and upon ADRB2 activation by pretreatment of the cells with isoproterenol, a synthetic cathecholamine targeting and activating β subtype adrenergic receptors [94]. siRNA inhibition of HIC1 in hTERT-HMEC (immortalized normal mammary epithelial cells) led to an increase in their migration properties [88]. However, despite the expression of ADRB2 in these cells, this migratory effect continued to be delayed upon isoproterenol treatment. Therefore, ADRB2 up-regulation per se could not favor cell motility in normal cells whereas it can in cells like MDA-MB-231 which have undergone an epithelial-mesenchymal transition (EMT). In agreement with the multistep process of breast cancer progression, HIC1 inactivation in early tumorigenesis and the resulting dysregulation of direct target genes could contribute to deregulation of growth, stress/survival and migration pathways (e.g. CXCR7, SIRT1, EphA2 and ephrinA1). It could also contribute to late steps such as metastasis of EMT-prone cells, via ADRB2 activation.
4.12 Indirect transcriptional effects through transcription factor sequestration
Besides these direct DNA-dependent transcriptional effects, HIC1 could also regulate the expression of target genes through indirect effects implicating protein-protein interactions with other transcription factors. For example, HIC1 can antagonize TCF/β-catenin mediated transcription in Wnt-stimulated cells through direct multi-domain interactions with the HMG box transcription factor TCF4 [94]. HIC1 recruits TCF/β-catenin onto discrete nuclear structures called “HIC1 bodies”. Depending on HIC1 protein levels, TCF-4 and β-catenin are thus prevented from direct association with the promoters of TCF target genes. This mechanism is not unique in the BTB/POZ family of transcriptional repressors as exemplified by the repression of AP-1 function by the BCL6 (B cell lymphoma-6) transcriptional repressor [96]. As for HIC1, it would be interesting to define if this indirect regulation through “sequestration” in HIC1 bodies might affect additional transcription factors since a yeast two-hybrid screen experiment has identified numerous transcription factors as potential interacting partners of HIC1 (our unpublished results).
5-HIC1, a multifaceted transcriptional repressor
The structural organization of HIC1, an N-terminal BTB/POZ domain and C-terminal Zinc Fingers domains separated by a central region, is shared by many BTB/POZ transcriptional repressors, including the well known BCL6 and PLZF [57].
To demonstrate that HIC1 was indeed another BTB/POZ transcriptional repressor, we first used the GAL4 repression assay in which various fragments of a protein are cloned in frame with the GAL4-DNA-binding domain and tested in transient transfection assays with a Luciferase reporter gene driven by GAL4 responsive elements. These experiments demonstrated that both the HIC1 BTB/POZ domain and central region were autonomous transcriptional repression domains as previously demonstrated for BCL6 and PLZF [57, 61, 97–100].
However, in striking contrast with these two examples, the HIC1 BTB/POZ domain is insensitive to Class I and Class II HDAC inhibitors such as Trichostatin A (TSA) [97]. The repression mechanisms brought about by the BTB/POZ domain are not fully deciphered even though two studies have clearly demonstrated an interaction between the isolated HIC1 BTB/POZ domain and the NAD+ dependent Class III deacetylase SIRT1 [54, 66].
The central region is also an autonomous repression domain but sensitive to TSA indicating that this region is able to interact with HDAC-containing repression complexes [61, 98]. The central region is not phylogenetically conserved between HIC1 and its paralog located on chromosome 22, HRG22 [60], or even in the evolution of HIC1 proteins from zebrafish to human, with the notable exception of five small conserved peptide motifs [101]. The function of two of them has been elucidated; the GLDLSKK and the MKHEP motif have been implicated in the interaction with two components of HDAC-containing repression complexes, CtBP and MTA1, respectively [61, 67, 99].
The GLDLSKK motif identified through CLUSTAL alignments of HIC1 proteins is highly similar to the canonical CtBP-interacting domain (CID) containing a PxDLSxK/R motif found in most proteins interacting with the related CtBP1 and CtBP2 corepressors [101, 102]. Indeed, crystallographic studies have defined a PLDLS-binding cleft lined with several hydrophobic residues that is implicated in the interaction with a large majority of DNA-binding transcriptional repressors interacting with CtBPs [102]. Various functional assays have unambiguously demonstrated that HIC1 interacts with CtBP1 and CtBP2 [61]. Furthermore, these analyses have highlighted the central Leucine residue (Leucine 225 in the GLDL225SKK motif of HIC1) as the sole invariant residue essential for the interaction of transcription factors with CtBPs [98]. In crystal structures, CtBP proteins exist as dimers whose formation is favoured by NADH over NAD+ and CtBP is thus considered to be a redox sensor linking cellular metabolism to transcriptional repression [102]. In particular, hypoxia has been shown to enhanced CtBP-mediated repression of the E-cadherin gene promoter, a key regulator of EMT through its increased interaction with the ZEB transcriptional repressor [102]. The same group reported that the glycolytic inhibitor 2-deoxy-D-glucose (2DG) activates the transcription of SIRT1, an NAD+ dependent deacetylase, through increased NADH levels and the resulting decrease of HIC1-CtBP interaction with the SIRT1 promoter [67, 98, 104].
The MKHEP motif has been identified as a potential consensus site for SUMOylation, a versatile modification of numerous nuclear proteins [105]. SUMO (small ubiquitin-related modifier) is an 11 KDa polypeptide structurally related to ubiquitin and covalently conjugated in target proteins to Lysine residues in the consensus ψ-Lys-x-Glu (ψKxE), where ψ is a large hydrophobic residue and x any amino-acid. In contrast to ubiquitination, which usually marks proteins for rapid degradation, SUMOylation is involved in the regulation of DNA-binding activity, transcriptional activity, nuclear sublocalization and assembly of multiprotein complexes [106, 107]. We have demonstrated that HIC1 is SUMOylated on the conserved K314R lysine residue [99]. In addition the K314R point mutation which inhibits HIC1 SUMOylation does not influence HIC1 subnuclear localization in punctuate nuclear dots. However, this K314R mutation, as well as the E316A mutation of the other mandatorily conserved residue in the ψKxE motif, severely impaired the transcriptional repression potential of HIC1 [99]. SUMOylated HIC1 proteins were detected by sequential ChIP analyses on promoters of HIC1 direct target genes, in agreement with the widely accepted model that SUMOylation participates in the assembly of multiprotein complexes [67, 105]. However, the identity of these transcriptional partners dependent upon HIC1 SUMOylation remained elusive.
To answer this question and more generally to better define transcriptional repression mechanisms brought about by HIC1, we conducted yeast two hybrid screen experiments using the two autonomous repression domains of HIC1, the BTB/POZ domain and the central region, as bait to screen a human mammary gland cDNA library. In close agreement with our previous work, the complete coding sequences of CtBP1 and CtBP2 were among the interacting clones isolated [61]. Two clones corresponded to a small region of the specific C-terminal region of MTA1 (Metastasis-associated protein 1), an integral subunit of the NuRD (Nucleosome Remodeling and histone Deacetylase) complex which is one of the two major types of complexes that modify DNA accessibility to co-factors [108]. NuRD complexes are the only chromatin complexes known so far which associate two enzymatic activities - histone deacetylation and chromatin remodelling through ATPase. The NuRD complexes are highly heterogeneous, strongly suggesting that they could target various promoters with different outcomes linked to transcriptional control, mainly though not exclusively resulting in repression [109]. The NuRD shared core proteins include the histone deacetylases HDAC1/HDAC2, the histone chaperones RBBP7/RbAp46 and RBBP4/RbAP48 and the p66α and p68β proteins. The interchangeable, mutually exclusive proteins are the Mi2α or Mi-2β ATPases, the Methyl-CpG binding proteins MBD2 or MBD3 and a protein of the MTA family [110]. This family comprises three distinct members, the related MTA1 and MTA2 genes and the more distant MTA3 gene, each also encoding truncated isoforms [111]. The full-length proteins share a common bi-modular structural organization, an N-terminal moiety consisting of four individual functional domains implicated in interactions with the NuRD core components together with an unstructured and more divergent C-terminal end implicated in interactions with various co-factors or sequence-specific transcription factors. We have validated through functional assays the interaction between HIC1 and MTA1. More importantly, we have demonstrated that this interaction is regulated by two competitive post-translational modifications of HIC1 at lysine 314, promotion by SUMOylation and inhibition by acetylation [67]. Thus, whereas the recruitment of the CtBP corepressor to HIC1-target genes is regulated by hypoxia, and more generally by cellular metabolism, the recruitment of NuRD complexes to HIC1 target genes is controlled by post-translational modifications of HIC1 lysine 314. It is therefore important to identify the external cues and signalling pathways controlling these latter modifications [67].
Another interacting partner isolated in the yeast two-hybrid screen with the HIC1 bait is ARID1A (A-T rich Interaction domain)/BAF250A (Brg1/Brm associated factor), one of the two mutually exclusive ARID1 containing subunits of SWI/SNF ATP-dependent chromatin remodelling complexes [112, 113]. SWI/SNF complexes correspond to a small series of related complexes of variable composition associating either of the two related but distinct core ATPases BRG1 and BRM to about ten non-catalytic subunits [112]. These non-catalytic subunits, referred to as ARID or BAFs, contain various DNA-binding and protein-binding motifs which modulate the targeting and activity of the ATPases [114]. These distinct subsets of SWI/SNF complexes have clearly distinct functions, since anti-proliferative complexes contain ARID1A and pro-proliferative complexes contain ARID1B [114]. Recently, BAF250B and BAF250A have been characterized as E3 ubiquitin ligases targeting Histone H2B but the functional link between this new ubiquitination pathway and chromatin remodelling is still not yet fully understood [115]. Finally, next generation sequencing of whole exomes has described frequent mutations of SWI/SNF components, especially of ARID1A, in breast, liver, gastric, esophagus, bladder and gynecological carcinomas [116]. Together, these analyses have not only confirmed the wide role of SWI/SNF complexes in cancers but also have established ARID1A as another candidate tumor suppressor gene.
The last interacting partner that we have recently validated is hPCL3/PHF19 [117], one of the three human homologs of the Drosophila Polycomb-like protein (PCL) that is found in Polycomb PRC2 repression complexes [81, 118–120]. Polycomb complexes first discovered in Drosophila are global epigenetic transcriptional regulators of cell fate decisions in all metazoans, playing important roles in normal development, stem cell renewal and cancer. They are organized in multiprotein complexes containing variable chromatin-modifying Polycomb group (PcG) proteins. In mammals, the best characterized complexes are the Polycomb Repressive Complexes 1 and 2 (PRC1 and PRC2). PRC2 complex core components include SUZ12, EED and EZH2, a histone methyltransferase which catalyzes the di-methylation and trimethylation of lysine 27 of histone 3 (H3K27me3) thereby generating an epigenetic repressive mark bound by the Polycomb (Pc) protein of PRC1 complexes [119, 120]. PRC2 is associated with accessory components that are essential to modulate its recruitment to specific loci, notably CpG islands, such as the H3K4me3 demethylase Jarid1b (KDM5b) and/or its repressive activity as with proteins of the Polycomb like (PCL) family. In line with the significant expansion of PcG genes during evolution, three orthologs of the unique Drosophila Polycomb-like gene have been characterized in the human genome, hPCL1/PHF1 (human Polycomb-like 1/PHD finger protein 1), hPCL2/MTF2 and hPCL3/PHF19 (for a review, [81, 118]). These three genes are differentially expressed suggesting that their expression pattern could comprise potential regulatory mechanisms of PcG target genes. These PCL proteins have not been implicated in the formation and the stability of PRC2 complex in contrast with its two other core components EED and SUZ12 but are essential for high levels of H3K27 trimethylation as well as for the cell-specific targeting of PRC2 to specific loci [121, 122]. PHF1, hPCL2 and hPCL3 are highly similar and share an N-terminal module consisting of three well-defined functional domains, namely a TUDOR domain and two adjacent PHD (Plant HomeoDomain) fingers [122]. In contrast with the PHD1 finger, the conserved PHD2 finger of hPCL3 has been implicated in its interaction with EZH2 118. Several recent studies have unraveled specific binding of the TUDOR domain of all human Polycomb-like proteins to trim ethylated lysine 36 of Histone H3, H3K36me3 [123–126]., an epigenetic mark mainly associated with transcribed genes [127]. Despite some unresolved apparent discrepancies between the different studies, the following model begins to emerge. The TUDOR domain of hPCL proteins recognizes H3K36me3 epigenetic marks and allows targeting and intrusion of PRC2 into active chromatin regions. Next, the two demethylases associated with PRC2, KDM5a/Jarid1b and PHF19, KDM2b/NO66 erase the activating H3K4me3 and H3K36me3 epigenetic marks [123–126]. In addition, this demethylation activates the catalytic activity of PRC2, thereby favoring the establishment of transcriptional silencing [128].
With the repression domains (BTB/POZ and CR regions) of HIC1 as prey, we have characterized the N-terminal half of the full-length hPCL3 isoform (amino acids 32–361) as interacting bait. Transient transfection assays and co-immunoprecipitation experiments have confirmed that HIC1 interacts with hPCL3 or PHF1 and with PRC2 core components. This interaction relies both on the HIC1 BTB/POZ domain and on the hPCL3 TUDOR and PHD2 domains which also interact with H3K36me3 [124, 125] and EZH2 respectively [81, 118]. As a functional consequence, hPCL3 promotes the formation of a ternary complex between HIC1 and PRC2. ChIP-qPCR experiments have detected EZH2 and H3K27 trimethylation on a subset of HIC1 target genes, namely ATOH1, EFNA1 and CXCR7 [81]. Moreover, extended HIC1 inactivation by RNA interference resulted in a significant decrease of EZH2 recruitment and H3K27me3 levels on the ATOH1 enhancer and promoter in BJ-TERT fibroblasts. Finally, ChIP analyses of post-natal murine cerebella at P5 and P21 clearly demonstrated that HIC1 and Polycomb complexes are functionally linked on ATOH1 during normal mouse cerebellar development in vivo [81].
Besides their well-established role as epigenetic regulators maintaining the repressed status of numerous genes involved in cell growth and development, recent data have emphasized a potential role of Polycomb group proteins in the DNA-damage response [129]. Besides PRC1 components involved in histone ubiquitination, PRC2 core components and the epigenetic H3K27me3 mark are found at double-strand breaks (DSBs). In addition, PHF1 is rapidly but transiently recruited to DSBs immediately after irradiation and physically interacts with several proteins involved in the DNA damage response: Ku70/Ku80, RAD50, SMC1, DHX9 and P53 [130]. PHF1 was recently shown to control P53 stability and activity in normal and DNA-damage conditions by interfering with MDM2-mediated ubiquitination and degradation of P53 [131]. Clearly, these new provocative results strongly suggest that the human Polycomb-like proteins have not yet divulged all their secrets regarding their connection with transcription factors such as P53 and HIC1 during the DNA-damage response.
6- HIC1 SIRT1 interaction: epigenetic regulation and/or Posttranslational modifications
Beginning with the search for a candidate tumor suppressor gene on 17p in the context of wild-type P53 and its positional cloning [27], HIC1 has been closely linked to P53 and hence to one of its essential regulators, the deacetylase SIRT1. The HIC1-P53-SIRT1 feed-back loop has gradually emerged from the phenotypic characterization of sophisticated animal models, notably the murine double Hic1+/− P53+/− heterozygote in cis or in trans [46], from in vitro molecular biology studies [66, 67] and from analyses of human tumors [132]. Clearly, SIRT1 interacts with HIC1 to repress transcription, including its own. However the underlying mechanisms are still not yet fully deciphered, particularly the exact relative contribution of epigenetic silencing mechanisms versus transcriptional regulation through SIRT1-mediated post-translational modifications of HIC1. Indeed, SIRT1 is a double-edge sword. SIRT1 is a Class III NAD+ dependent histone deacetylase participating in the creation of the “histone code”, the complex cross-talk of post-translation modifications of histone tails, which regulates gene expression by modulating the compaction status of chromatin. In that context, SIRT1 preferentially deacetylates lysines 9 and 14 of Histone H3 as well as lysine 16 of Histone H4. However, it is becoming increasingly clear that SIRT1 also regulates many biological processes such as longevity, aging, metabolism and response to various stresses by deacetylating non-histone substrates including P53, E2F1, BCL6 and Foxo1 [133]. We have shown that SIRT1 deacetylates HIC1, notably at lysine 314 which is competitively targeted for both acetylation by CBP/P300 and for SUMOylation [54, 99]. Such a competitive acetylation or SUMOylation of the same lysine has been detected in several proteins [134]. This is a simplified version of a multisite modification mechanism whereby a neighbouring phosphorylation synergizes with the SUMOylation process, as first demonstrated for the MEF2D transcription factor and its so-called SUMOylation-Acetylation Switch (SAS) motif [135] or Phospho-SUMOyl switch motif (PDSM) [134–136]. In that case a complex between two deacetylases belonging to two different classes: the Class III NAD+-dependent SIRT1 and a Class II TSA-sensitive HDAC, such as HDAC4, HDAC5 or HDAC7 is responsible for the acetylation/SUMOylation switch [137–139]. SIRT1 is involved in deacetylation of the lysine residue which allows its subsequent SUMOylation by the Class II HDAC. This activity is independent of the C-terminal catalytic deacetylase domain but relies on the regulatory N-terminal region which interacts with the E2 conjugating enzyme Ubc9 and has thus been named E3 ligase-like or SUMO-facilitase activity [137–139].
In HIC1, the MK314HEP motif is not a PDSM motif but its acetylation/SUMOylation switch also depends on a SIRT1/HDAC4 complex [99]. On the one hand, HIC1 interacts with HDAC4 [99] and with Ubc9, but only in the presence of HDAC4 [140]. On the other hand, HIC1 interacts with SIRT1 through its BTB/POZ domain [66] and its last four Zinc fingers, which are also involved in sequence-specific DNA-binding [54]. Conversely, with a series of SIRT1 deletion mutants, we demonstrated that the amino-acids 610–677 of SIRT1 are required for the interaction with HIC1 [54]. Notably, this region contains the ESA (Essential for SIRT1 deacetylase Activity) domain which increases the interaction between the deacetylase and its substrates, e.g. Ac-P53 and native Ac-Histone H3 [141]. Interestingly, this 610–677 region of SIRT1 also contains two CK2 consensus sites, which are phosphorylated upon DNA-damage to increase SIRT1 substrate binding affinity [142]. We also demonstrated that TBCA, a CK2 inhibitor, severely impaired the HIC1/SIRT1 interaction [54]. HIC1 collaborates with SIRT1 to regulate the P53-dependent apoptotic DNA-damage response in part through the direct repression of the SIRT1 promoter itself; but the epigenetic status of this promoter in normal and apoptotic DNA-damaged conditions has not been reported [66]. Recently, we have shown that the SUMOylation of HIC1 is increased via an ATM-dependent pathway upon repairable (1 hour exposure or apoptotic (16 hours of exposure) double stranded DNA-damage induced by Etoposide [140]. In accordance with our previous results, we again observed an increased interaction between endogenous HIC1 and MTA1 proteins upon Etoposide treatment of normal human fibroblasts [140]. Collectively, our results strongly suggest that HIC1 could be activated upon DNA-damage by its SIRT1-mediated deacetylation followed by its subsequent SUMOylation to favour the recruitment of MTA1-containing NURD complexes on target genes including, among others which remained to be identified, SIRT1 itself [54, 67, 99, 140].
7-Conclusion
18 years of HIC1 research since the gene was first cloned in the Baylin laboratory has established the biological importance of HIC1 in various essential physiological roles ranging from normal development to cell growth control and survival upon various genotoxic or metabolic stresses. HIC1 is a multi-faceted transcriptional repressor interacting with four major repression and chromatin remodelling complexes. These interactions, at least for the NuRD and CtBP complexes, are regulated by post-translational modifications allowing fine-tuned repression of gene-specific pathways depending on the external cues. So far, only a limited number of HIC1 direct target genes have been validated, but all of them confirmed the developmental regulation and tumor suppressor functions of HIC1. In addition to transcription factors involved in developmental processes and cell cycle regulators, HIC1 also represses transcription of various RTK and GPCR membrane receptors involved in proliferation and migration/invasion properties. Several animal models (double heterozygotes) have highlighted the collaboration between HIC1 and its direct target genes to accelerate and/or modify tumor formation. The next breakthroughs will come as a result of genome-wide ChIP-seq analyses of HIC1 direct target genes yielding a wide coverage and unbiased genome-wide analyses of HIC1 binding sites and therefore providing further insight in regulatory networks implicating HIC1.
8-Expert Opinion
The translational power of epigenetics lies in the fact that epigenetic determinants of gene transcription are actively maintained and therefore reversible [143]. Epigenetically targeted translational strategies are beginning to be tested in clinical trials and these efforts are accelerating as a greater understanding of, and appreciation for, the role of epigenetic gene regulation in cancer is realized.
Initial attempts to impact the epigenetic programming of cancer cells focused on DNA methylation. The DNA methyltransferases DNMT1, 2, 3a and b are variously responsible for transferring a methyl group to the 5′ position of cytosine residues in CpG dinucleotides in different contexts. DNMT inhibitors have been sought to prevent the maintenance of DNA methylation yielding demethylation with cell division, thereby causing the re-expression of silenced genes. The most mature DNMT inhibitors are the nucleoside analogues 5-aza-2′-deoxycytabine (Decitabine) and 5-azacytadine (Vidaza) which have been extensively tested in leukemia and myelodysplastic syndromes, achieving FDA approval for the latter indication. The small molecules hydralazine and procainamide, both approved for cardiovascular conditions, also exert competitive inhibition of DNMT1 and are being explored in cancer trials [144]. One drawback to this approach is its lack of precision; global hypomethylation may also act to accelerate tumor progression pathways via reactivation of tumor promoting genes [145].
With the discovery of the importance of the histone code and the recognition of the association between histone deacetylation and repression of gene transcription, histone deacetylase inhibitors (HDACis) were developed. In general, the net effect of HDACis is to re-express epigenetically silenced genes. A number of HDACis have been developed with varying specificities for Class I, II and IV histone deacetylates. Among those in early phase clinical trials, the best studied include short chain fatty acids (the anti-epileptic drug valproic acid, sodium phenylbutyrate, Pivanex) and the hydroxamic acids (trichostatin A, suberoylanilide hydroxamic acid (a.k.a. Vorinostat, SAHA), Panobinostat, CHR-3996, SB939, Givinostat, etc.). HDACis are now being widely tested in combination with radiation and cytotoxic chemotherapy for hematologic malignancies and solid tumors (for review see Nebbioso, et al. [143]). Another group of endogenous histone deacetylases are the sirtuins, responsible for 2 important histone tail modifications- the acetylation status of H4K16 and H3K9 -deacetylation of which results in heterochromatic formation and gene silencing. The sirtuins also act on non-histone proteins and Sirt1’s de-acetylation of p53 leading to its inactivation has positioned it as a tumor promoter. Of course, Sirt1 is also a HIC1 target gene and loss of HIC1 increases levels of Sirt1 as well as its activity upon p53 [66]. Therefore, Sirt1 inhibition is one example of a translational opportunity stemming from an understanding of HIC1’s function. Another refinement has been the linkage between the methylation state of lysine and arginine residues in the histone tail and gene transcriptional activity yielding a class of drugs effecting histone de-methylation. The multiple potential sites of methylation coupled with their ability to be mono-, di-, or tri- methylated yields an enormous complexity of potential methylation states which, if understood along with the molecules responsible for their creation, could address the main weakness of epigenetic therapies, specificity. One modification associated with the repression of transcription is methylation of the 27th lysine of histone H3 (H3K27), carried out by EZH2 as part of the polycomb repressive complex 2 (PRC2). As noted above, HIC1 plays an important role in recruiting PRC2 to its target genes. Importantly, EZH2 gain of function mutations are found in B cell lymphomas causing increased H3K27 trimethylation via enhanced PRC2 activity [146]. Two groups have reported EZH2 inhibitors that show remarkable specificity causing epigenetic reprogramming and apoptosis primarily in mutated lymphoma cells but also in some wildtype lymphoma cells, demonstrating the potential power of targeted inhibition of epigenetic mechanisms [147–149].
Epigenetic reprogramming resulting from the loss of HIC1 is not a global unfocused event. Rather, given HIC1’s ability to direct the repressive PRC2 machinery to sequence specific binding sites associated with its target gene promoters, its loss results in specific changes in the transcriptional program of the cell. An understanding of this program through identification of HIC1’s target genes and their interrelationships in feedback loops and cell process regulation will yield the ability to leverage knowledge about the consequences of HIC1 loss for therapeutic translation.
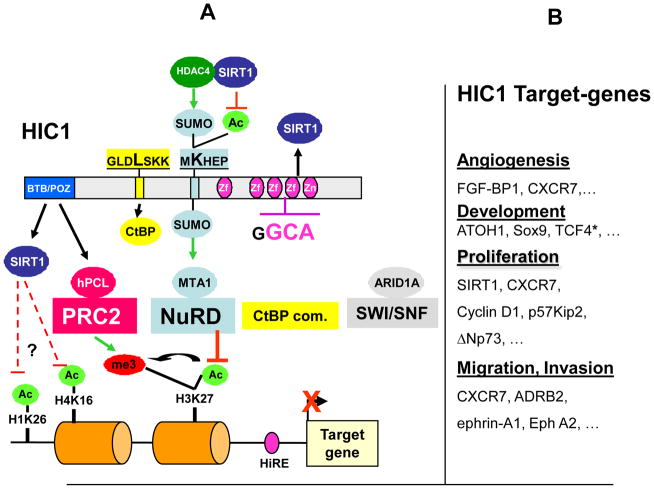
Figure 1. HIC1, its associated transcriptional co-repressors and their target genes.
A) HIC1 can recruit to its target genes four major chromatin repression and remodelling complexes: CtBP com., NuRD, Polycomb PRC2 and SWI/SNF.
From top to bottom: Linear representation of the HIC1 protein with its functional domains, the BTB/POZ and the central region which are autonomous repression domains as well as the four C-Terminal Zinc fingers which are involved in sequence-specific binding to a HIC1 responsive element (HiRE). HIC1 Lysine 314 in the MKHEP conserved motif can be competitively modified by Acetylation or SUMOylation, this latter modification favouring the interaction with MTA1. The direct interacting partners of HIC1 characterized by yeast two hybrid screen, hPCL3, MTA1 and ARID 1A are shown as ovals with the respective complexes to which they belong shown as boxes. SIRT1 interacts with the BTB/POZ and zinc finger domains of HIC1. This interaction can be implicated in the post-translational modification of HIC1 in conjunction with HDAC4 as a SUMO E3 ligase-like or in epigenetic modification of the promoter regions of HIC1 target genes.
B) Summary of HIC1 target genes. HIC1 directly represses genes involved in angiogenesis, embryonic development, proliferation and in cell migration and invasion properties.* Unique to the transcription factor TCF4, HIC1 interacts with the transcription factor TCF4 and “sequesters” it away from its direct target genes into “HIC1 nuclear bodies” thereby indirectly suppressing the transcriptional response induced by Wnt signalling.
Article highlights.
HIC1 is a tumor suppressor gene located in 17p13.3 which is frequently epigenetically silenced or deleted in many types of cancers.
HIC1 is implicated in various physiological processes ranging from normal development to control of cell growth to survival after genotoxic and metabolic stresses.
HIC1 is involved in complex regulatory loops with P53 and the deacetylase SIRT1.
HIC1 is a multifaceted, sequence-specific transcriptional repressor that interacts with several major repression and chromatin remodelling complexes.
To date, only a few HIC1 direct target genes have been validated: they include transcription factors involved in developmental processes, cell cycle regulators, and various RTK and GPCR membrane receptors.
Footnotes
Declaration of interest
The authors state no conflict of interest. This research is supported by the following grants: BRR NIH/NINDS grant 1K08NS051477 and a grant from the Thomas Family Foundation, DL grants from AICR (Association for International Cancer Research) from ARC (Association pour la recherché contre le Cancer, Villejuif, France and from Ligue Nationale contre le Cancer (Comité du Septentrion, Lille, France).
Bibliography
Papers of special note have been highlighted as either of importance (*) or of considerable importance (**) to readers.
- 1.Knudson AG., Jr Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci U S A. 1971;68:820–3. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Makos M, Nelkin BD, Lerman MI, et al. Distinct hypermethylation patterns occur at altered chromosome loci in human lung and colon cancer. Proc Natl Acad Sci U S A. 1992;89:1929–33. doi: 10.1073/pnas.89.5.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Makos M, Nelkin BD, Chazin VR, et al. DNA hypermethylation is associated with 17p allelic loss in neural tumors. Cancer Res. 1993;53:2715–8. [PubMed] [Google Scholar]
- 4.Makos M, Nelkin BD, Reiter RE, et al. Regional DNA hypermethylation at D17S5 precedes 17p structural changes in the progression of renal tumors. Cancer Res. 1993;53:2719–22. [PubMed] [Google Scholar]
- 5.Fujii H, Biel MA, Zhou W, et al. Methylation of the HIC-1 candidate tumor suppressor gene in human breast cancer. Oncogene. 1998;16:2159–64. doi: 10.1038/sj.onc.1201976. [DOI] [PubMed] [Google Scholar]
- 6.Hayashi M, Tokuchi Y, Hashimoto T, et al. Reduced HIC-1 gene expression in non-small cell lung cancer and its clinical significance. Anticancer Res. 2001;21:535–40. [PubMed] [Google Scholar]
- 7.Crawley JJ, Furge KA. Identification of frequent cytogenetic aberrations in hepatocellular carcinoma using gene-expression microarray data. Genome Biol. 2002;3:RESEARCH0075. doi: 10.1186/gb-2002-3-12-research0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weber-Mangal S, Sinn HP, Popp S, et al. Breast cancer in young women (< or = 35 years): Genomic aberrations detected by comparative genomic hybridization. Int J Cancer. 2003;107:583–92. doi: 10.1002/ijc.11460. [DOI] [PubMed] [Google Scholar]
- 9.Cogen PH, Daneshvar L, Metzger AK, Edwards MS. Deletion mapping of the medulloblastoma locus on chromosome 17p. Genomics. 1990;8:279–85. doi: 10.1016/0888-7543(90)90283-z. [DOI] [PubMed] [Google Scholar]
- 10.Cogen PH, Daneshvar L, Metzger AK, et al. Involvement of multiple chromosome 17p loci in medulloblastoma tumorigenesis. Am J Hum Genet. 1992;50:584–9. [PMC free article] [PubMed] [Google Scholar]
- 11.Saxena A, Clark WC, Robertson JT, et al. Evidence for the involvement of a potential second tumor suppressor gene on chromosome 17 distinct from p53 in malignant astrocytomas. Cancer Res. 1992;52:6716–21. [PubMed] [Google Scholar]
- 12.von Haken MS, White EC, Daneshvar-Shyesther L, et al. Molecular genetic analysis of chromosome arm 17p and chromosome arm 22q DNA sequences in sporadic pediatric ependymomas. Genes, chromosomes & cancer. 1996;17:37–44. doi: 10.1002/(SICI)1098-2264(199609)17:1<37::AID-GCC6>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 13.Steichen-Gersdorf E, Baumgartner M, Kreczy A, et al. Deletion mapping on chromosome 17p in medulloblastoma. Br J Cancer. 1997;76:1284–7. doi: 10.1038/bjc.1997.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chattopadhyay P, Rathore A, Mathur M, et al. Loss of heterozygosity of a locus on 17p13. 3, independent of p53, is associated with higher grades of astrocytic tumours. Oncogene. 1997;15:871–4. doi: 10.1038/sj.onc.1201238. [DOI] [PubMed] [Google Scholar]
- 15.Aldosari N, Rasheed BK, McLendon RE, et al. Characterization of chromosome 17 abnormalities in medulloblastomas. Acta neuropathologica. 2000;99:345–51. doi: 10.1007/s004010051134. [DOI] [PubMed] [Google Scholar]
- 16.Rood BR, Zhang H, Weitman DM, Cogen PH. Hypermethylation of HIC-1 and 17p allelic loss in medulloblastoma. Cancer Res. 2002;62:3794–7. [PubMed] [Google Scholar]
- 17.Waha A, Koch A, Hartmann W, et al. Analysis of HIC-1 methylation and transcription in human ependymomas. Int J Cancer. 2004;110:542–9. doi: 10.1002/ijc.20165. [DOI] [PubMed] [Google Scholar]
- 18.Adesina AM, Nalbantoglu J, Cavenee WK. p53 gene mutation and mdm2 gene amplification are uncommon in medulloblastoma. Cancer Res. 1994;54:5649–51. [PubMed] [Google Scholar]
- 19.Badiali M, Iolascon A, Loda M, et al. p53 gene mutations in medulloblastoma. Immunohistochemistry, gel shift analysis, and sequencing. Diagn Mol Pathol. 1993;2:23–8. [PubMed] [Google Scholar]
- 20.Biegel JA, Burk CD, Barr FG, Emanuel BS. Evidence for a 17p tumor related locus distinct from p53 in pediatric primitive neuroectodermal tumors. Cancer Res. 1992;52:3391–5. [PubMed] [Google Scholar]
- 21.Kool M, Korshunov A, Remke M, et al. Molecular subgroups of medulloblastoma: an international meta-analysis of transcriptome, genetic aberrations, and clinical data of WNT, SHH, Group 3, and Group 4 medulloblastomas. Acta neuropathologica. 2012;123:473–84. doi: 10.1007/s00401-012-0958-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Northcott PA, Jones DT, Kool M, et al. Medulloblastomics: the end of the beginning. Nat Rev Cancer. 2012;12:818–34. doi: 10.1038/nrc3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taylor MD, Northcott PA, Korshunov A, et al. Molecular subgroups of medulloblastoma: the current consensus. Acta neuropathologica. 2012;123:465–72. doi: 10.1007/s00401-011-0922-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McDonald JD, Daneshvar L, Willert JR, et al. Physical mapping of chromosome 17p13. 3 in the region of a putative tumor suppressor gene important in medulloblastoma. Genomics. 1994;23:229–32. doi: 10.1006/geno.1994.1481. [DOI] [PubMed] [Google Scholar]
- 25.Hoff C, Seranski P, Mollenhauer J, et al. Physical and transcriptional mapping of the 17p13. 3 region that is frequently deleted in human cancer. Genomics. 2000;70:26–33. doi: 10.1006/geno.2000.6353. [DOI] [PubMed] [Google Scholar]
- 26.Schultz DC, Vanderveer L, Berman DB, et al. Identification of two candidate tumor suppressor genes on chromosome 17p13. 3. Cancer Res. 1996;56:1997–2002. [PubMed] [Google Scholar]
- 27**.Wales MM, Biel MA, el Deiry W, et al. p53 activates expression of HIC-1, a new candidate tumour suppressor gene on 17p13.3. Nat Med. 1995;1:570–7. doi: 10.1038/nm0695-570. This paper describes the positional cloning and identification of HIC1as a BTB/POZ and zinc finger transcription factor. [DOI] [PubMed] [Google Scholar]
- 28.Guerardel C, Deltour S, Pinte S, et al. Identification in the human candidate tumor suppressor gene HIC-1 of a new major alternative TATA-less promoter positively regulated by p53. J Biol Chem. 2001;276:3078–89. doi: 10.1074/jbc.M008690200. [DOI] [PubMed] [Google Scholar]
- 29.Britschgi C, Rizzi M, Grob TJ, et al. Identification of the p53 family-responsive element in the promoter region of the tumor suppressor gene hypermethylated in cancer 1. Oncogene. 2006;25:2030–9. doi: 10.1038/sj.onc.1209240. [DOI] [PubMed] [Google Scholar]
- 30.Jenal M, Trinh E, Britschgi C, et al. The tumor suppressor gene hypermethylated in cancer 1 is transcriptionally regulated by E2F1. Mol Cancer Res. 2009;7:916–22. doi: 10.1158/1541-7786.MCR-08-0359. [DOI] [PubMed] [Google Scholar]
- 31.Dehennaut V, Leprince D. Implication of HIC1 (Hypermethylated In Cancer 1) in the DNA damage response. Bull Cancer. 2009;96:E66–72. doi: 10.1684/bdc.2009.0959. [DOI] [PubMed] [Google Scholar]
- 32.Jenal M, Britschgi C, Fey MF, Tschan MP. Inactivation of the hypermethylated in cancer 1 tumour suppressor--not just a question of promoter hypermethylation? Swiss medical weekly. 2010;140:w13106. doi: 10.4414/smw.2010.13106. [DOI] [PubMed] [Google Scholar]
- 33.Issa JP, Zehnbauer BA, Kaufmann SH, et al. HIC1 hypermethylation is a late event in hematopoietic neoplasms. Cancer Res. 1997;57:1678–81. [PubMed] [Google Scholar]
- 34.Eguchi K, Kanai Y, Kobayashi K, Hirohashi S. DNA hypermethylation at the D17S5 locus in non-small cell lung cancers: its association with smoking history. Cancer Res. 1997;57:4913–5. [PubMed] [Google Scholar]
- 35.Kanai Y, Hui AM, Sun L, et al. DNA hypermethylation at the D17S5 locus and reduced HIC-1 mRNA expression are associated with hepatocarcinogenesis. Hepatology. 1999;29:703–9. doi: 10.1002/hep.510290338. [DOI] [PubMed] [Google Scholar]
- 36.Waha A, Waha A, Koch A, et al. Epigenetic silencing of the HIC-1 gene in human medulloblastomas. J Neuropathol Exp Neurol. 2003;62:1192–201. doi: 10.1093/jnen/62.11.1192. [DOI] [PubMed] [Google Scholar]
- 37.Abouzeid HE, Kassem AM, Abdel Wahab AH, et al. Promoter hypermethylation of RASSF1A, MGMT, and HIC-1 genes in benign and malignant colorectal tumors. Tumour Biol. 2011;32:845–52. doi: 10.1007/s13277-011-0156-7. [DOI] [PubMed] [Google Scholar]
- 38.Yu J, Liu P, Cui X, et al. Identification of novel subregions of LOH in gastric cancer and analysis of the HIC1 and TOB1 tumor suppressor genes in these subregions. Molecules and cells. 2011;32:47–55. doi: 10.1007/s10059-011-2316-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kilinc D, Ozdemir O, Ozdemir S, et al. Alterations in promoter methylation status of tumor suppressor HIC1, SFRP2, and DAPK1 genes in prostate carcinomas. DNA and cell biology. 2012;31:826–32. doi: 10.1089/dna.2011.1431. [DOI] [PubMed] [Google Scholar]
- 40.Zhao G, Qin Q, Zhang J, et al. Hypermethylation of HIC1 Promoter and Aberrant Expression of HIC1/SIRT1 Might Contribute to the Carcinogenesis of Pancreatic Cancer. Annals of surgical oncology. 2012 May 3; doi: 10.1245/s10434-012-2364-9. [DOI] [PubMed] [Google Scholar]
- 41.Brieger J, Pongsapich W, Mann SA, et al. Demethylation treatment restores hic1 expression and impairs aggressiveness of head and neck squamous cell carcinoma. Oral oncology. 2010;46:678–83. doi: 10.1016/j.oraloncology.2010.06.016. [DOI] [PubMed] [Google Scholar]
- 42.Eggers H, Steffens S, Grosshennig A, et al. Prognostic and diagnostic relevance of hypermethylated in cancer 1 (HIC1) CpG island methylation in renal cell carcinoma. International journal of oncology. 2012;40:1650–8. doi: 10.3892/ijo.2012.1367. [DOI] [PubMed] [Google Scholar]
- 43.Stephen JK, Chen KM, Shah V, et al. DNA hypermethylation markers of poor outcome in laryngeal cancer. Clinical epigenetics. 2010;1:61–9. doi: 10.1007/s13148-010-0005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carter MG, Johns MA, Zeng X, et al. Mice deficient in the candidate tumor suppressor gene Hic1 exhibit developmental defects of structures affected in the Miller-Dieker syndrome. Hum Mol Genet. 2000;9:413–9. doi: 10.1093/hmg/9.3.413. [DOI] [PubMed] [Google Scholar]
- 45**.Chen WY, Zeng X, Carter MG, et al. Heterozygous disruption of Hic1 predisposes mice to a gender-dependent spectrum of malignant tumors. Nat Genet. 2003;33:197–202. doi: 10.1038/ng1077. This research demonstrates that HIC1 is a bona fide tumour suppressor gene. [DOI] [PubMed] [Google Scholar]
- 46**.Chen W, Cooper TK, Zahnow CA, et al. Epigenetic and genetic loss of Hic1 function accentuates the role of p53 in tumorigenesis. Cancer Cell. 2004;6:387–98. doi: 10.1016/j.ccr.2004.08.030. This research establishes a functional cooperation between HIC1 and P53 in animal models. [DOI] [PubMed] [Google Scholar]
- 47.Briggs KJ, Corcoran-Schwartz IM, Zhang W, et al. Cooperation between the Hic1 andPtch1 tumor suppressors in medulloblastoma. Genes Dev. 2008;22:770–85. doi: 10.1101/gad.1640908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mohammad HP, Zhang W, Prevas HS, et al. Loss of a single Hic1 allele accelerates polyp formation in Apc(Delta716) mice. Oncogene. 2011;30:2659–69. doi: 10.1038/onc.2010.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bardwell VJ, Treisman R. The POZ domain: a conserved protein-protein interaction motif. Genes Dev. 1994;8:1664–77. doi: 10.1101/gad.8.14.1664. [DOI] [PubMed] [Google Scholar]
- 50.Zollman S, Godt D, Prive GG, et al. The BTB domain, found primarily in zinc finger proteins, defines an evolutionarily conserved family that includes several developmentally regulated genes in Drosophila. Proc Natl Acad Sci U S A. 1994;91:10717–21. doi: 10.1073/pnas.91.22.10717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Albagli O, Dhordain P, Deweindt C, et al. The BTB/POZ domain: a new protein-protein interaction motif common to DNA- and actin-binding proteins. Cell Growth Differ. 1995;6:1193–8. [PubMed] [Google Scholar]
- 52.Deltour S, Guerardel C, Stehelin D, Leprince D. The carboxy-terminal end of the candidate tumor suppressor gene HIC-1 is phylogenetically conserved. Biochim Biophys Acta. 1998;1443:230–2. doi: 10.1016/s0167-4781(98)00219-x. [DOI] [PubMed] [Google Scholar]
- 53.Fleuriel C, Touka M, Boulay G, et al. HIC1 (Hypermethylated in Cancer 1) epigenetic silencing in tumors. Int J Biochem Cell Biol. 2009;41:26–33. doi: 10.1016/j.biocel.2008.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dehennaut V, Loison I, Pinte S, Leprince D. Molecular dissection of the interaction between HIC1 and SIRT1. Biochem Biophys Res Commun. 2012;421:384–8. doi: 10.1016/j.bbrc.2012.04.026. [DOI] [PubMed] [Google Scholar]
- 55*.Pinte S, Stankovic-Valentin N, Deltour S, et al. The tumor suppressor gene HIC1 (hypermethylated in cancer 1) is a sequence-specific transcriptional repressor: definition of its consensus binding sequence and analysis of its DNA binding and repressive properties. J Biol Chem. 2004;279:38313–24. doi: 10.1074/jbc.M401610200. This work identifies the DNA binding site of HIC1, critical for the identification of HIC1 target genes. [DOI] [PubMed] [Google Scholar]
- 56.Ahmad KF, Engel CK, Prive GG. Crystal structure of the BTB domain from PLZF. Proc Natl Acad Sci U S A. 1998;95:12123–8. doi: 10.1073/pnas.95.21.12123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ahmad KF, Melnick A, Lax S, et al. Mechanism of SMRT corepressor recruitment by the BCL6 BTB domain. Mol Cell. 2003;12:1551–64. doi: 10.1016/s1097-2765(03)00454-4. [DOI] [PubMed] [Google Scholar]
- 58.Li X, Peng H, Schultz DC, et al. Structure-function studies of the BTB/POZ transcriptional repression domain from the promyelocytic leukemia zinc finger oncoprotein. Cancer Res. 1999;59:5275–82. [PubMed] [Google Scholar]
- 59.Dhordain P, Albagli O, Ansieau S, et al. The BTB/POZ domain targets the LAZ3/BCL6 oncoprotein to nuclear dots and mediates homomerisation in vivo. Oncogene. 1995;11:2689–97. [PubMed] [Google Scholar]
- 60.Deltour S, Pinte S, Guerardel C, Leprince D. Characterization of HRG22, a human homologue of the putative tumor suppressor gene HIC1. Biochem Biophys Res Commun. 2001;287:427–34. doi: 10.1006/bbrc.2001.5624. [DOI] [PubMed] [Google Scholar]
- 61.Deltour S, Pinte S, Guerardel C, et al. The human candidate tumor suppressor gene HIC1 recruits CtBP through a degenerate GLDLSKK motif. Mol Cell Biol. 2002;22:4890–901. doi: 10.1128/MCB.22.13.4890-4901.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Valenta T, Lukas J, Doubravska L, et al. HIC1 attenuates Wnt signaling by recruitment of TCF-4 and beta-catenin to the nuclear bodies. Embo J. 2006;25:2326–37. doi: 10.1038/sj.emboj.7601147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li JY, English MA, Ball HJ, et al. Sequence-specific DNA binding and transcriptional regulation by the promyelocytic leukemia zinc finger protein. J Biol Chem. 1997;272:22447–55. doi: 10.1074/jbc.272.36.22447. [DOI] [PubMed] [Google Scholar]
- 64.Lours C, Bardot O, Godt D, et al. The Drosophila melanogaster BTB proteins bric a brac bind DNA through a composite DNA binding domain containing a pipsqueak and an AT-Hook motif. Nucleic acids research. 2003;31:5389–98. doi: 10.1093/nar/gkg724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Barna M, Merghoub T, Costoya JA, et al. Plzf mediates transcriptional repression of HoxD gene expression through chromatin remodeling. Dev Cell. 2002;3:499–510. doi: 10.1016/s1534-5807(02)00289-7. [DOI] [PubMed] [Google Scholar]
- 66**.Chen WY, Wang DH, Yen RC, et al. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell. 2005;123:437–48. doi: 10.1016/j.cell.2005.08.011. This research deciphers a regulatory loop between HIC1, P53 and the deacetylase SIRT1. [DOI] [PubMed] [Google Scholar]
- 67*.Van Rechem C, Boulay G, Pinte S, et al. Differential regulation of HIC1 target genes by CtBP and NuRD, via an acetylation/SUMOylation switch, in quiescent versus proliferating cells. Mol Cell Biol. 2010;30:4045–59. doi: 10.1128/MCB.00582-09. This research demonstrates that competitive post translational modifications of HIC1 lysine regulate its interaction with the NuRD repressive complex. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li P, Zhao Y, Wu X, et al. Interferon gamma (IFN-gamma) disrupts energy expenditure and metabolic homeostasis by suppressing SIRT1 transcription. Nucleic acids research. 2012;40:1609–20. doi: 10.1093/nar/gkr984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Briones VR, Chen S, Riegel AT, Lechleider RJ. Mechanism of fibroblast growth factor-binding protein 1 repression by TGF-beta. Biochem Biophys Res Commun. 2006;345:595–601. doi: 10.1016/j.bbrc.2006.04.052. [DOI] [PubMed] [Google Scholar]
- 70.Tassi E, McDonnell K, Gibby KA, et al. Impact of fibroblast growth factor-binding protein-1 expression on angiogenesis and wound healing. Am J Pathol. 2011;179:2220–32. doi: 10.1016/j.ajpath.2011.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bailey SG, Cragg MS, Townsend PA. Family friction as DeltaNp73 antagonises p73 and p53. Int J Biochem Cell Biol. 2011;43:482–6. doi: 10.1016/j.biocel.2010.12.022. [DOI] [PubMed] [Google Scholar]
- 72.Vilgelm AE, Hong SM, Washington MK, et al. Characterization of DeltaNp73 expression and regulation in gastric and esophageal tumors. Oncogene. 2010;29:5861–8. doi: 10.1038/onc.2010.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.el-Deiry WS, Tokino T, Velculescu VE, et al. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–25. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 74.Dehennaut V, Loison I, Boulay G, et al. Identification of p21 (CIP1/WAF1) as a direct target gene of HIC1 (Hypermethylated In Cancer 1) Biochem Biophys Res Commun. 2013;430:49–53. doi: 10.1016/j.bbrc.2012.11.045. [DOI] [PubMed] [Google Scholar]
- 75*.Van Rechem C, Rood BR, Touka M, et al. Scavenger Chemokine (CXC Motif) Receptor 7 (CXCR7) Is a Direct Target Gene of HIC1 (Hypermethylated in Cancer 1) J Biol Chem. 2009;284:20927–35. doi: 10.1074/jbc.M109.022350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76*.Zhang W, Zeng X, Briggs KJ, et al. A potential tumor suppressor role for Hic1 in breast cancer through transcriptional repression of ephrin-A1. Oncogene. 2010;29:2467–76. doi: 10.1038/onc.2010.12. These two papers contain the first lists of HIC1 target genes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zheng J, Wang J, Sun X, et al. HIC1 modulates prostate cancer progression by epigenetic modification. Clin Cancer Res. 2013 Jan 22; doi: 10.1158/1078-0432.CCR-12-2888. [DOI] [PubMed] [Google Scholar]
- 78.Roussel MF, Hatten ME. Cerebellum development and medulloblastoma. Current topics in developmental biology. 2011;94:235–82. doi: 10.1016/B978-0-12-380916-2.00008-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Briggs KJ, Eberhart CG, Watkins DN. Just say no to ATOH: how HIC1 methylation might predispose medulloblastoma to lineage addiction. Cancer Res. 2008;68:8654–6. doi: 10.1158/0008-5472.CAN-08-1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ayrault O, Zhao H, Zindy F, et al. Atoh1 inhibits neuronal differentiation and collaborates with Gli1 to generate medulloblastoma-initiating cells. Cancer Res. 2010;70:5618–27. doi: 10.1158/0008-5472.CAN-09-3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Boulay G, Dubuissez M, Van Rechem C, et al. Hypermethylated in cancer 1 (HIC1) recruits polycomb repressive complex 2 (PRC2) to a subset of its target genes through interaction with human polycomb-like (hPCL) proteins. J Biol Chem. 2012;287:10509–24. doi: 10.1074/jbc.M111.320234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Singh AK, Arya RK, Trivedi AK, et al. Chemokine receptor trio: CXCR3, CXCR4 and CXCR7 crosstalk via CXCL11 and CXCL12. Cytokine & growth factor reviews. 2012 Sep 15; doi: 10.1016/j.cytogfr.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Decaillot FM, Kazmi MA, Lin Y, et al. CXCR7/CXCR4 heterodimer constitutively recruits beta-arrestin to enhance cell migration. J Biol Chem. 2011;286:32188–97. doi: 10.1074/jbc.M111.277038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rajagopal S, Kim J, Ahn S, et al. Beta-arrestin- but not G protein-mediated signaling by the “decoy” receptor CXCR7. Proc Natl Acad Sci U S A. 2010;107:628–32. doi: 10.1073/pnas.0912852107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang J, Shiozawa Y, Wang J, et al. The role of CXCR7/RDC1 as a chemokine receptor for CXCL12/SDF-1 in prostate cancer. J Biol Chem. 2008;283:4283–94. doi: 10.1074/jbc.M707465200. [DOI] [PubMed] [Google Scholar]
- 86.Hernandez L, Magalhaes MA, Coniglio SJ, et al. Opposing roles of CXCR4 and CXCR7 in breast cancer metastasis. Breast Cancer Res. 2011;13:R128. doi: 10.1186/bcr3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Brantley-Sieders DM. Clinical relevance of Ephs and ephrins in cancer: lessons from breast, colorectal, and lung cancer profiling. Seminars in cell & developmental biology. 2012;23:102–8. doi: 10.1016/j.semcdb.2011.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Foveau B, Boulay G, Pinte S, et al. Receptor Tyrosyne Kinase Epha2 Is a Direct Target-Gene of Hic1 (Hypermethylated in Cancer 1) J Biol Chem. 2012;287:5366–78. doi: 10.1074/jbc.M111.329466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Macrae M, Neve RM, Rodriguez-Viciana P, et al. A conditional feedback loop regulates Ras activity through EphA2. Cancer Cell. 2005;8:111–8. doi: 10.1016/j.ccr.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 90.Brantley-Sieders DM, Jiang A, Sarma K, et al. Eph/ephrin profiling in human breast cancer reveals significant associations between expression level and clinical outcome. PloS one. 2011;6:e24426. doi: 10.1371/journal.pone.0024426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Miao H, Wang B. EphA receptor signaling--complexity and emerging themes. Seminars in cell & developmental biology. 2012;23:16–25. doi: 10.1016/j.semcdb.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pasquale EB. Eph receptors and ephrins in cancer: bidirectional signalling and beyond. Nat Rev Cancer. 2010;10:165–80. doi: 10.1038/nrc2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pasquale EB. Eph-ephrin bidirectional signaling in physiology and disease. Cell. 2008;133:38–52. doi: 10.1016/j.cell.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 94.Boulay G, Malaquin N, Loison I, et al. Loss of hypermethylated In Cancer 1 (HIC1) in breast cancer cells contributes to stress induced migration and invasion through beta-2 adrenergic receptor (ADRB2) misregulation. J Biol Chem. 2012;287:5379–89. doi: 10.1074/jbc.M111.304287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Perez-Sayans M, Somoza-Martin JM, Barros-Angueira F, et al. Beta-adrenergic receptors in cancer: therapeutic implications. Oncology research. 2010;19:45–54. doi: 10.3727/096504010x12828372551867. [DOI] [PubMed] [Google Scholar]
- 96.Vasanwala FH, Kusam S, Toney LM, Dent AL. Repression of AP-1 function: a mechanism for the regulation of Blimp-1 expression and B lymphocyte differentiation by the B cell lymphoma-6 protooncogene. J Immunol. 2002;169:1922–9. doi: 10.4049/jimmunol.169.4.1922. [DOI] [PubMed] [Google Scholar]
- 97.Deltour S, Guerardel C, Leprince D. Recruitment of SMRT/N-CoR-mSin3A-HDAC-repressing complexes is not a general mechanism for BTB/POZ transcriptional repressors: the case of HIC-1 and gammaFBP-B. Proc Natl Acad Sci U S A. 1999;21(96):14831–6. doi: 10.1073/pnas.96.26.14831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Stankovic-Valentin N, Verger A, Deltour-Balerdi S, et al. A L225A substitution in the human tumour suppressor HIC1 abolishes its interaction with the corepressor CtBP. Febs J. 2006;273:2879–90. doi: 10.1111/j.1742-4658.2006.05301.x. [DOI] [PubMed] [Google Scholar]
- 99.Stankovic-Valentin N, Deltour S, Seeler J, et al. An acetylation/deacetylation-SUMOylation switch through a phylogenetically conserved psiKXEP motif in the tumor suppressor HIC1 regulates transcriptional repression activity. Mol Cell Biol. 2007;27:2661–75. doi: 10.1128/MCB.01098-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Parekh S, Polo JM, Shaknovich R, et al. BCL6 programs lymphoma cells for survival and differentiation through distinct biochemical mechanisms. Blood. 2007;110:2067–74. doi: 10.1182/blood-2007-01-069575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bertrand S, Pinte S, Stankovic-Valentin N, et al. Identification and developmental expression of the zebrafish orthologue of the tumor suppressor gene HIC1. Biochim Biophys Acta. 2004;1678:57–66. doi: 10.1016/j.bbaexp.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 102.Chinnadurai G. Transcriptional regulation by C-terminal binding proteins. Int J Biochem Cell Biol. 2007;39:1593–607. doi: 10.1016/j.biocel.2007.01.025. [DOI] [PubMed] [Google Scholar]
- 103.Zhang Q, Wang SY, Nottke AC, et al. Redox sensor CtBP mediates hypoxia-induced tumor cell migration. Proc Natl Acad Sci U S A. 2006;103:9029–33. doi: 10.1073/pnas.0603269103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhang Q, Wang SY, Fleuriel C, et al. Metabolic regulation of SIRT1 transcription via a HIC1:CtBP corepressor complex. Proc Natl Acad Sci U S A. 2007;104:829–33. doi: 10.1073/pnas.0610590104. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 105.Hay RT. SUMO: a history of modification. Mol Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 106.Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol. 2007;8:947–56. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- 107.Gareau JR, Lima CD. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat Rev Mol Cell Biol. 2010;11:861–71. doi: 10.1038/nrm3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Denslow SA, Wade PA. The human Mi-2/NuRD complex and gene regulation. Oncogene. 2007;26:5433–8. doi: 10.1038/sj.onc.1210611. [DOI] [PubMed] [Google Scholar]
- 109.Miccio A, Wang Y, Hong W, et al. NuRD mediates activating and repressive functions of GATA-1 and FOG-1 during blood development. Embo J. 2010;29:442–56. doi: 10.1038/emboj.2009.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ramirez J, Hagman J. The Mi-2/NuRD complex: a critical epigenetic regulator of hematopoietic development, differentiation and cancer. Epigenetics. 2009;4:532–6. doi: 10.4161/epi.4.8.10108. [DOI] [PubMed] [Google Scholar]
- 111.Manavathi B, Kumar R. Metastasis tumor antigens, an emerging family of multifaceted master coregulators. J Biol Chem. 2007;282:1529–33. doi: 10.1074/jbc.R600029200. [DOI] [PubMed] [Google Scholar]
- 112.Wang X, Nagl NG, Wilsker D, et al. Two related ARID family proteins are alternative subunits of human SWI/SNF complexes. The Biochemical journal. 2004;383:319–25. doi: 10.1042/BJ20040524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Van Rechem C, Boulay G, Leprince D. HIC1 interacts with a specific subunit of SWI/SNF complexes, ARID1A/BAF250A. Biochem Biophys Res Commun. 2009;385:586–90. doi: 10.1016/j.bbrc.2009.05.115. [DOI] [PubMed] [Google Scholar]
- 114.Nagl NG, Jr, Wang X, Patsialou A, et al. Distinct mammalian SWI/SNF chromatin remodeling complexes with opposing roles in cell-cycle control. Embo J. 2007;26:752–63. doi: 10.1038/sj.emboj.7601541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Li XS, Trojer P, Matsumura T, et al. Mammalian SWI/SNF--a subunit BAF250/ARID1 is an E3 ubiquitin ligase that targets histone H2B. Mol Cell Biol. 2010;30:1673–88. doi: 10.1128/MCB.00540-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bourdeaut F, Bieche I. Chromatin remodeling defects and cancer: the SWI/SNF example. Bull Cancer. 2012;99:1133–40. doi: 10.1684/bdc.2012.1664. [DOI] [PubMed] [Google Scholar]
- 117.Wang S, Robertson GP, Zhu J. A novel human homologue of Drosophila polycomblike gene is up-regulated in multiple cancers. Gene. 2004;343:69–78. doi: 10.1016/j.gene.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 118.Boulay G, Rosnoblet C, Guerardel C, et al. Functional characterization of human Polycomb-like 3 isoforms identifies them as components of distinct EZH2 protein complexes. The Biochemical journal. 2011;434:333–42. doi: 10.1042/BJ20100944. [DOI] [PubMed] [Google Scholar]
- 119.Morey L, Helin K. Polycomb group protein-mediated repression of transcription. Trends Biochem Sci. 2010;35:323–32. doi: 10.1016/j.tibs.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 120.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–9. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Nekrasov M, Klymenko T, Fraterman S, et al. Pcl-PRC2 is needed to generate high levels of H3-K27 trimethylation at Polycomb target genes. Embo J. 2007;26:4078–88. doi: 10.1038/sj.emboj.7601837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hunkapiller J, Shen Y, Diaz A, et al. Polycomb-like 3 promotes polycomb repressive complex 2 binding to CpG islands and embryonic stem cell self-renewal. PLoS Genet. 2012;8:e1002576. doi: 10.1371/journal.pgen.1002576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Cai L, Rothbart SB, Lu R, et al. An H3K36 Methylation-Engaging Tudor Motif of Polycomb-like Proteins Mediates PRC2 Complex Targeting. Mol Cell. 2013;49:1–12. doi: 10.1016/j.molcel.2012.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Brien GL, Gambero G, O’Connell DJ, et al. Polycomb PHF19 binds H3K36me3 and recruits PRC2 and demethylase NO66 to embryonic stem cell genes during differentiation. Nature structural & molecular biology. 2012;19:1273–81. doi: 10.1038/nsmb.2449. [DOI] [PubMed] [Google Scholar]
- 125.Ballare C, Lange M, Lapinaite A, et al. Phf19 links methylated Lys36 of histone H3 to regulation of Polycomb activity. Nature structural & molecular biology. 2012;19:1257–65. doi: 10.1038/nsmb.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Musselman CA, Avvakumov N, Watanabe R, et al. Molecular basis for H3K36me3 recognition by the Tudor domain of PHF1. Nature structural & molecular biology. 2012;19:1266–72. doi: 10.1038/nsmb.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Chantalat S, Depaux A, Hery P, et al. Histone H3 trimethylation at lysine 36 is associated with constitutive and facultative heterochromatin. Genome Res. 2011;21:1426–37. doi: 10.1101/gr.118091.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Schmitges FW, Prusty AB, Faty M, et al. Histone methylation by PRC2 is inhibited by active chromatin marks. Mol Cell. 2011;42:330–41. doi: 10.1016/j.molcel.2011.03.025. [DOI] [PubMed] [Google Scholar]
- 129.Vissers JH, van Lohuizen M, Citterio E. The emerging role of Polycomb repressors in the response to DNA damage. J Cell Sci. 2012;125:3939–48. doi: 10.1242/jcs.107375. [DOI] [PubMed] [Google Scholar]
- 130.Hong Z, Jiang J, Lan L, et al. A polycomb group protein, PHF1, is involved in the response to DNA double-strand breaks in human cell. Nucleic acids research. 2008;36:2939–47. doi: 10.1093/nar/gkn146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yang Y, Wang C, Zhang P, et al. Polycomb Group Protein PHF1 Regulates p53-dependent Cell Growth Arrest and Apoptosis. J Biol Chem. 2013;288:529–39. doi: 10.1074/jbc.M111.338996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Tseng RC, Lee CC, Hsu HS, et al. Distinct HIC1-SIRT1-p53 loop deregulation in lung squamous carcinoma and adenocarcinoma patients. Neoplasia. 2009;11:763–70. doi: 10.1593/neo.09470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Liu T, Liu PY, Marshall GM. The critical role of the class III histone deacetylase SIRT1 in cancer. Cancer Res. 2009;69:1702–5. doi: 10.1158/0008-5472.CAN-08-3365. [DOI] [PubMed] [Google Scholar]
- 134.Yang XJ, Gregoire S. A recurrent phospho-sumoyl switch in transcriptional repression and beyond. Mol Cell. 2006;23:779–86. doi: 10.1016/j.molcel.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 135.Shalizi A, Gaudilliere B, Yuan Z, et al. A calcium-regulated MEF2 sumoylation switch controls postsynaptic differentiation. Science. 2006;311:1012–7. doi: 10.1126/science.1122513. [DOI] [PubMed] [Google Scholar]
- 136.Hietakangas V, Anckar J, Blomster HA, et al. PDSM, a motif for phosphorylation-dependent SUMO modification. Proc Natl Acad Sci U S A. 2006;103:45–50. doi: 10.1073/pnas.0503698102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gregoire S, Yang XJ. Association with class IIa histone deacetylases upregulates the sumoylation of MEF2 transcription factors. Mol Cell Biol. 2005;25:2273–87. doi: 10.1128/MCB.25.6.2273-2287.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zhao X, Sternsdorf T, Bolger TA, et al. Regulation of MEF2 by histone deacetylase 4- and SIRT1 deacetylase-mediated lysine modifications. Mol Cell Biol. 2005;25:8456–64. doi: 10.1128/MCB.25.19.8456-8464.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Gao C, Ho CC, Reineke E, et al. Histone deacetylase 7 promotes PML sumoylation and is essential for PML nuclear body formation. Mol Cell Biol. 2008;28:5658–67. doi: 10.1128/MCB.00874-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140**.Dehennaut V, Loison I, Dubuissez M, et al. DNA double-strand breaks lead to activation of Hyperrmethylated in Cancer 1 (HIC1) gene by SUMOylation to regulate DNA repair. J Biol Chem. 2013 Feb 15; doi: 10.1074/jbc.M112.421610. This research demonstrates that HIC1 plays a key role in the DNA-damage response (DDR) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kang H, Suh JY, Jung YS, et al. Peptide switch is essential for Sirt1 deacetylase activity. Mol Cell. 2011;44:203–13. doi: 10.1016/j.molcel.2011.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kang H, Jung JW, Kim MK, Chung JH. CK2 is the regulator of SIRT1 substrate-binding affinity, deacetylase activity and cellular response to DNA-damage. PloS one. 2009;4:e6611. doi: 10.1371/journal.pone.0006611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Nebbioso A, Carafa V, Benedetti R, Altucci L. Trials with ‘epigenetic’ drugs: an update. Molecular oncology. 2012;6:657–82. doi: 10.1016/j.molonc.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Amatori S, Bagaloni I, Donati B, Fanelli M. DNA demethylating antineoplastic strategies: a comparative point of view. Genes & cancer. 2010;1:197–209. doi: 10.1177/1947601910365081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Veeck J, Esteller M. Breast cancer epigenetics: from DNA methylation to microRNAs. Journal of mammary gland biology and neoplasia. 2010;15:5–17. doi: 10.1007/s10911-010-9165-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Yap DB, Chu J, Berg T, et al. Somatic mutations at EZH2 Y641 act dominantly through a mechanism of selectively altered PRC2 catalytic activity, to increase H3K27 trimethylation. Blood. 2011;117:2451–9. doi: 10.1182/blood-2010-11-321208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Knutson SK, Wigle TJ, Warholic NM, et al. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nature chemical biology. 2012;8:890–6. doi: 10.1038/nchembio.1084. [DOI] [PubMed] [Google Scholar]
- 148.Melnick A. Epigenetic therapy leaps ahead with specific targeting of EZH2. Cancer Cell. 2012;22:569–70. doi: 10.1016/j.ccr.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.McCabe MT, Ott HM, Ganji G, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492:108–12. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]