Abstract
Humans express at least seven alcohol dehydrogenase (ADH) isoforms that are encoded by ADH gene cluster (ADH7–ADH1C–ADH1B–ADH1A–ADH6–ADH4–ADH5) at chromosome 4. ADHs are key catabolic enzymes for retinol and ethanol. The functional ADH variants (mostly rare) have been implicated in alcoholism risk. In addition to catalyzing the oxidation of retinol and ethanol, ADHs may be involved in the metabolic pathways of several neurotransmitters that are implicated in the neurobiology of neuropsychiatric disorders. In the present study, we comprehensively examined the associations between common ADH variants [minor allele frequency (MAF) >0.05] and 11 neuropsychiatric and neurological disorders. A total of 50,063 subjects in 25 independent cohorts were analyzed. The entire ADH gene cluster was imputed across these 25 cohorts using the same reference panels. Association analyses were conducted, adjusting for multiple comparisons. We found 28 and 15 single nucleotide polymorphisms (SNPs), respectively, that were significantly associated with schizophrenia in African-Americans and autism in European-Americans after correction by false discovery rate (FDR) (q <0.05); and 19 and 6 SNPs, respectively, that were significantly associated with these two disorders after region-wide correction by SNPSpD (8.9 × 10−5 ≤ p ≤ 0.0003 and 2.4 × 10−5 ≤ p ≤ 0.0003, respectively). No variants were significantly associated with the other nine neuropsychiatric disorders, including alcohol dependence. We concluded that common ADH variants conferred risk for both schizophrenia in African-Americans and autism in European-Americans.
Introduction
Humans express at least seven alcohol dehydrogenase (ADH) isoforms, each with slightly different properties (Luo et al. 2008). ADHs are expressed predominantly in the liver, the upper digestive tract (from mouth to stomach), and kidney, and partly in the brain (Yoshida et al. 1998). Particularly, because ADHs are key catabolic enzymes for ethanol, ADH variants have been implicated in the risk for alcohol dependence by previous studies [reviewed by (Luo et al. 2006)]. However, in addition to catalyzing the oxidation of retinol and ethanol, ADHs may be involved in the metabolic pathways of several neurotransmitters including serotonin, epinephrine, norepinephrine, and dopamine (Holmes 1994; Svensson et al. 1999). The functions of ADHs in the metabolism of these monoamines suggest their potential roles in the etiology of other neuropsychiatric disorders.
ADH isoforms are encoded by ADH7–ADH1C–ADH1B–ADH1A–ADH6–ADH4–ADH5 gene cluster at chromosome 4. It has been widely reported by candidate gene studies that at least four functional ADH gene variants, i.e., rs1229984 (ADH2*2; Arg48His), rs2066702 (ADH2*3; Arg370Cys), rs1693482 (ADH3*2; Arg272Gln), and rs698 (ADH3*2; Ile350Va), significantly affect the risk for alcohol dependence [reviewed by (Luo et al. 2006)]. These variants are rare in most populations, e.g., in Europeans (minor allele frequency (MAF)rs2066702 = 0.000 and MAFrs1229984 = 0.008) and Africans (MAFrs1229984 = 0.000, MAFrs1693482 = 0.052, and MAFrs698 = 0.042). In one of our previous studies, we also found that the rare variant constellation across the entire ADH cluster was associated with alcohol dependence in European-Americans, European-Australians, and African-Americans (Zuo et al. 2013b). So far, numerous genome-wide association studies (GWASs) of alcohol dependence using common variants as markers have also been performed; however, only one GWAS identified one common ADH variant (rs1789891; MAF = 0.192) that was associated with alcohol dependence at the genome-wide significance level (p = 1.3 × 10−8; OR = 1.46; α = 5 × 10−8) (Frank et al. 2012). This leads to a hypothesis that common ADH variants might be associated with other diseases rather than alcohol dependence only. For example, one candidate gene study reported that common variants at ADH7 were associated with Parkinson’s disease (Buervenich et al. 2000). To further test this hypothesis, in the present study, we comprehensively examined the associations between common ADH variants (MAF >0.05 in both cases and controls) and 11 neuropsychiatric and neurological disorders including schizophrenia, autism, attention deficit hyperactivity disorder (ADHD), alcoholism, major depression, bipolar disorder, Alzheimer’s disease, amyotrophic lateral sclerosis (ALS), early onset stroke, ischemic stroke, and Parkinson’s disease in subjects of European or African descent.
Materials and methods
Subjects
A total of 50,063 subjects in 25 independent cohorts with 11 different neuropsychiatric and neurological disorders were analyzed. They included case–control and family-based samples, genotyped on Illumina, Affymetrix, or PERLEGEN microarray platforms. All subjects gave informed consent. Diagnoses, ethnicities, study designs, sample sizes, and dataset names for these cohorts are shown in Table 1. More detailed demographics data of these cohorts were published previously (Stefansson et al. 2009; Anney et al. 2010; Zuo et al. 2011, 2012, 2013a, b).
Table 1.
Associations between ADH gene cluster and different neuropsychiatric or neurological disorders
| Most sig. SNP | Location | Affected
|
Unaffected
|
Minimal p value | SNP # (total) | SNP # (p < 0.05) | SNP # (p < α) | SNP # (q < 0.05) | Ethnicity | Human disease | Dataset | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | MAF | N | MAF | ||||||||||
| rs1789916 | Intron 3 of ADH1C | 1,195 | 0.114 | 954 | 0.158 | 8.9 × 10−5 | 632 | 50 | 19 | 28 | AA (CC) | Schizophrenia | GAIN |
| rs284781 | Between ADH1C and ADH7 | 1,351 | 0.068 | 1378 | 0.090 | 0.0029 | 630 | 7 | 0 | 0 | EA (CC) | Schizophrenia | GAIN |
| rs6811453 | 3′UTR of ADH1A | 1,437 | 0.334 | 1347 | 0.289 | 0.0108 | 554 | 25 | 0 | 0 | EA (CC) | Schizophrenia | MGS_nonGAIN |
| rs6532803 | Between ADH4 and ADH6 | 98 | 0.136 | 20 | 0.625 | 0.0049 | 459 | 43 | 0 | 0 | AA (CC) | Schizophrenia | MGS_nonGAIN |
| rs62323588 | Between ADH4 and ADH5 | 1,331 | 0.107 | 2745 | 0.116 | 2.4 × 10−5 | 921 | 141 | 6 | 15 | EA (Fam) | Autism | AGP |
| rs10516441 | Between ADH1C and ADH7 | 924 | 0.062 | 1833 | 0.051 | 0.0048 | 1028 | 7 | 0 | 0 | CA (Fam) | ADHD | IMAGE |
| rs904092 | 5′ flanking to ADH1A | 681 | 0.181 | 508 | 0.243 | 0.0005 | 916 | 26 | 0 | 0 | AA (CC) | Alcoholism | SAGE + COGA |
| rs1583977 | Between ADH1C and ADH7 | 1,409 | 0.113 | 1518 | 0.093 | 0.0010 | 965 | 13 | 0 | 0 | EA (CC) | Alcoholism | SAGE + COGA |
| rs1442486 | 5′UTR of ADH7 | 1,645 | 0.424 | 3928 | 0.446 | 0.0019 | 940 | 14 | 0 | 0 | EAu (Fam) | Alcoholism | OZ-ALC |
| rs1154415 | Intron 4 of ADH5 | 1,754 | 0.408 | 1800 | 0.446 | 0.0030 | 1027 | 120 | 0 | 0 | CA (CC) | Major depression | PRSC |
| rs4147547 | Intron 7 of ADH6 | 368 | 0.346 | 1034 | 0.234 | 0.0046 | 623 | 64 | 0 | 0 | EA (CC) | Bipolar disorder | BDO + GRU |
| rs1693435 | Between ADH1B and ADH1C | 653 | 0.048 | 1034 | 0.105 | 0.0118 | 620 | 1 | 0 | 0 | EA (CC) | Bipolar disorder | BARD + GRU |
| rs2851012 | 3′UTR of ADH7 | 141 | 0.158 | 671 | 0.244 | 0.0230 | 606 | 4 | 0 | 0 | AA (CC) | Bipolar disorder | BARD + GRU |
| rs10014818 | 3′UTR of ADH6 | 2,320 | 0.084 | 2244 | 0.082 | 0.0048 | 959 | 19 | 0 | 0 | CA (Fam) | Alzheimer’s disease | LOAD × 4 |
| rs1229981 | 5′UTR of ADH1B | 806 | 0.132 | 782 | 0.087 | 0.0118 | 629 | 12 | 0 | 0 | EA (CC) | Alzheimer’s disease | GenADA |
| rs1154455 | Intron 8 of ADH7 | 261 | 0.587 | 246 | 0.388 | 0.0017 | 893 | 61 | 0 | 0 | CA (CC) | ALS | GRU |
| rs1154479 | 5′UTR of ADH7 | 372 | 0.381 | 430 | 0.292 | 0.0012 | 969 | 264 | 0 | 0 | EA (CC) | Early Onset stroke | GEOS × 3 |
| rs1497378 | Between ADH1A and ADH6 | 309 | 0.139 | 290 | 0.207 | 0.0013 | 880 | 45 | 0 | 0 | AA (CC) | Early onset stroke | GEOS × 3 |
| rs2298754 | Intron 8 of ADH1C | 219 | 0.135 | 266 | 0.070 | 0.0050 | 916 | 26 | 0 | 0 | CA (CC) | Ischemic stroke | ISGS |
| rs4699701 | Intron 4 of ADH5 | 2,000 | 0.094 | 1,986 | 0.076 | 0.0043 | 959 | 85 | 0 | 0 | CA (CC) | Parkinson’s disease | NGRC |
| rs2718682 | 5′ flanking to ADH7 | 900 | 0.294 | 867 | 0.334 | 0.0113 | 933 | 7 | 0 | 0 | CA (CC) | Parkinson’s disease | PDRD + GRU |
| rs67631357 | Between ADH1B and ADH1C | 677 | 0.273 | 538 | 0.216 | 0.0016 | 953 | 32 | 0 | 0 | CA (CC) | Parkinson’s disease | pd_v3_550v3 |
| rs2584462 | Between ADH1C and ADH7 | 263 | 0.091 | 263 | 0.238 | 0.0264 | 944 | 5 | 0 | 0 | CA (CC) | Parkinson’s disease | pd_v3_300v1 |
| rs7655945 | 5′UTR of ADH6 | 263 | 0.276 | 263 | 0.210 | 0.0702 | 771 | 0 | 0 | 0 | CA (CC) | Parkinson’s disease | pd_v3_250 s |
| rs146919815 | 5′UTR of ADH7 | 940 | 0.122 | 801 | 0.167 | 0.0381 | 947 | 10 | 0 | 0 | CA (CC) | Parkinson’s disease | lng_coriell_pd_v3 |
Only the most significant risk markers are listed. The significance level (α) is corrected for the numbers of effective genetic markers (calculated by SNPSpD) [GenADA: Li et al. 2008; Filippini et al. 2009; AGP: The AGP Consortium 2007, 2010a, b]
N sample size, MAF minor allele frequency, CC case–control sample, Fam family sample, EA European-American, AA African-American, EAu European-Australian, CA Caucasian, ADHD attention deficit hyperactivity disorder, ALS amyotrophic lateral sclerosis
The African-American schizophrenia cohort came from the GAIN dataset (dbGaP access number: phs000021.v3.p2), including 1,195 cases with schizophrenia and 954 controls. The subjects were genotyped on AFFYMETRIX AFFY_6.0 platform. All subjects were at least 18 years old. The cases included 746 males (41.9 ± 10.8 years) and 449 females (43.0 ± 9.8 years); and the controls included 362 males (46.2 ± 13.7 years) and 592 females (45.0 ± 12.9 years). Affected subjects met lifetime DSM-IV criteria for schizophrenia (American Psychiatric Association 1994). Cases were excluded if they had worse than mild mental retardation, or if their psychotic illness was judged to be secondary to substance use or a neurological disorder. Controls were excluded if they did not deny all of the following psychosis screening questions: treatment for or diagnosis of schizophrenia or schizoaffective disorder; treatment for or diagnosis of bipolar disorder or manic-depression; treatment for or diagnosis of psychotic symptoms such as auditory hallucinations or persecutory delusions.
The Autism cohort came from the AGP dataset (dbGaP access number: phs000267.v1.p1). A total of 1,366 families (trios) contained 4,075 European-American subjects including 1,330 probands with autism. The probands consisted of 1,121 males (7.2 ± 3.2 years) and 209 females (7.1 ± 3.0 years). Affected subjects were diagnosed using the Autism Diagnostic Interview-Revised (ADI-R) and Autism Diagnostic Observation Schedule (ADOS) instruments, and met DSM-IV criteria for autism (American Psychiatric Association 1994). Cases with known karyotypic abnormalities, fragile X mutations, or other genetic disorders were excluded. The subjects were genotyped on ILLUMINA_Human_1 M platform.
Imputation
To make the genetic marker sets highly consistent across the different samples, we imputed the missing single nucleotide polymorphisms (SNPs) across the entire ADH gene cluster (Chr4: 100204900-100631900) in all samples of 25 cohorts using the same reference panels (i.e., 1,000 genome project and HapMap 3 panels). We used the programs IMPUTE2 (Howie et al. 2009) and BEAGLE (Browning and Browning 2009) for imputation, with the reference CEU panel for the samples of European descent and the reference YRI panel for the samples of African descent. We maximized the success rate and accuracy of imputation and minimized the false-positives during the imputation process. Only the genotypes that were consistently imputed from the two independent reference panels (i.e., 1,000 genome project and HapMap 3 panels) and the genotypes that were consistently imputed by both IMPUTE2 and BEAGLE were selected for analysis. The uncertainty rate of inference for missing genotypes was controlled at <1 %. Furthermore, only the SNPs that had similar MAFs (with frequency difference <2 % within the same ethnicity) in the healthy controls across different cohorts and HapMap database were selected for analysis. After this strict selection, we were highly confident with the quality of these imputed genotype data. Checking the imputed genotypes in all of our four family-based cohorts, we did not find any one individual with more than 0.1 % Mendelian inconsistency (considering all SNPs tested) or any one SNP with more than 0.1 % Mendelian inconsistency (considering all individuals tested).
Data cleaning
We stringently cleaned the phenotype data [detailed previously (Zuo et al. 2012)] and then the imputed genotype data. Subjects with poor genotypic data, allele discordance, sample relatedness, a mismatch between self-identified and genetically inferred ethnicity, or a missing genotype call rate ≥2 % across all SNPs were filtered out. Furthermore, we filtered out the monomorphic SNPs and the SNPs with allele discordance, Mendelian errors (in family samples), an overall missing genotype call rate ≥2 %, MAFs <0.05 in either cases or controls, or Hardy–Weinberg Equilibrium (HWE) (p <10−4) within controls. We also filtered out the SNPs with MAF differences ≥2 % or missing rate differences ≥2 % between two samples that had the same phenotype and microarray platform. The cleaned sample sizes and cleaned SNP numbers are shown in Table 1.
Association test
For case–control cohorts, the allele frequencies were compared between cases and controls using logistic regression analysis as implemented in the program PLINK (Purcell et al. 2007). Diagnosis served as the dependent variable, alleles served as the independent variables, and ancestry proportions (to control for admixture effects) (Zuo et al. 2012), sex, and age served as covariates. The ancestry proportions for each individual were estimated using the program STRUCTURE (Pritchard et al. 2000). For those non-alcoholism cohorts, alcohol drinking behavior, if available, was also included as a covariate. Furthermore, for family cohorts, we used DFAM as implemented in PLINK to test associations (as effective as the program FBAT). The −log(p) value distribution is shown in Fig. 1. The MAFs and minimal p values of the most significant risk SNPs are shown in Table 1. The statistically significant risk SNPs associated with diseases (p <α) are shown in Table 2. Finally, we did bioinformatic analysis of these significant risk SNPs to explore their potential functions using the UCSC Genome Browser including ENCODE data (http://genome.ucsc.edu).
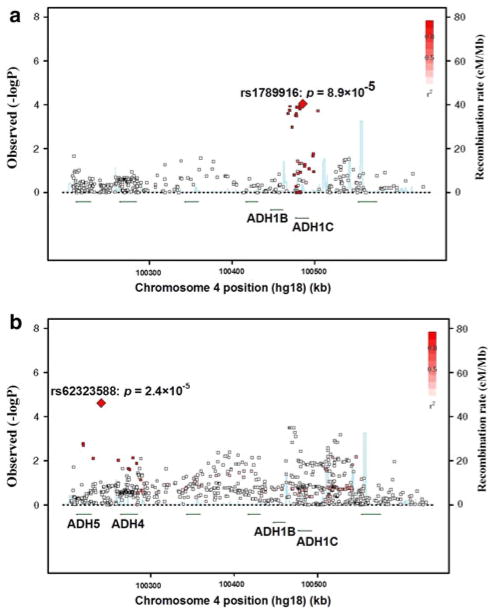
Fig. 1.
Regional association plots in ADH cluster [left Y-axis corresponds to −log(p) value; right Y-axis corresponds to recombination rates; X-axis corresponds to genomic positions; quantitative color gradient corresponds to r2; red squares represent peak SNPs. a Regional association plot in African-American GAIN schizophrenia sample; b regional association plot in European-American autism sample]
Table 2.
Significant risk SNPs associated with schizophrenia and autism (p <α)
| Disease | SNP | Position | Gene | Location | OR | p value | q value | Bioinformatics |
|---|---|---|---|---|---|---|---|---|
| Schizophrenia | rs35348915 | 100467780 | Intergenic | Between ADH1B and 1C | 1.47 | 0.0003 | 0.008 | |
| Schizophrenia | rs71612682 | 100467929 | Intergenic | Between ADH1B and 1C | 1.47 | 0.0003 | 0.008 | CpG |
| Schizophrenia | rs1789892 | 100469681 | Intergenic | Between ADH1B and 1C | 1.47 | 0.0002 | 0.008 | TFBS |
| Schizophrenia | rs36070167 | 100469884 | Intergenic | Between ADH1B and 1C | 1.49 | 0.0001 | 0.008 | |
| Schizophrenia | rs2866152 | 100470090 | Intergenic | Between ADH1B and 1C | 1.48 | 0.0001 | 0.008 | TFBS |
| Schizophrenia | rs1612735 | 100477030 | ADH1C | Intron 8 | 1.40 | 0.0003 | 0.009 | TFBS |
| Schizophrenia | rs1442480 | 100477704 | ADH1C | Intron 8 | 1.41 | 0.0003 | 0.008 | TFBS/CNV |
| Schizophrenia | rs1789900 | 100477740 | ADH1C | Intron 8 | 1.41 | 0.0003 | 0.008 | TFBS/CNV |
| Schizophrenia | rs1442481 | 100478070 | ADH1C | Intron 8 | 1.40 | 0.0003 | 0.009 | Conserved |
| Schizophrenia | rs1789901 | 100478392 | ADH1C | Intron 8 | 1.42 | 0.0001 | 0.008 | TFBS |
| Schizophrenia | rs1662060 | 100478864 | ADH1C | Intron 8 | 1.43 | 0.0001 | 0.008 | TFBS |
| Schizophrenia | rs1789904 | 100481431 | ADH1C | Intron 6 | 1.44 | 0.0001 | 0.008 | TFBS |
| Schizophrenia | rs1789906 | 100482242 | ADH1C | Intron 6 | 1.43 | 0.0001 | 0.008 | TFBS |
| Schizophrenia | rs904094 | 100482275 | ADH1C | Intron 6 | 1.43 | 0.0001 | 0.008 | |
| Schizophrenia | rs1789912 | 100482965 | ADH1C | Intron 6 | 1.43 | 0.0001 | 0.008 | TFBS/CpG/Conserved |
| Schizophrenia | rs1789916 | 100485719 | ADH1C | Intron 3 | 1.45 | 8.9 × 10−5 | 0.008 | TFBS/CpG/CNV |
| Schizophrenia | rs1693427 | 100485850 | ADH1C | Intron 3 | 1.46 | 8.9 × 10−5 | 0.008 | TFBS |
| Schizophrenia | rs1693428 | 100485954 | ADH1C | Intron 3 | 1.45 | 0.0001 | 0.008 | TFBS |
| Schizophrenia | rs1154434 | 100504035 | Intergenic | 11 kb to 5′ of ADH1C | 1.52 | 0.0002 | 0.008 | TFBS |
| Autism | rs62323588 | 100240894 | Intergenic | Between ADH5 and 4 | 1.50 | 2.4 × 10−5 | 0.012 | Long RNA |
| Autism | rs2213041 | 100466374 | Intergenic | Between ADH1B & 1C | 1.33 | 0.0003 | 0.026 | TFBS |
| Autism | rs1789889 | 100466888 | Intergenic | Between ADH1B and 1C | 1.33 | 0.0003 | 0.026 | TFBS |
| Autism | rs2226898 | 100467950 | Intergenic | Between ADH1B and 1C | 1.33 | 0.0003 | 0.026 | TFBS |
| Autism | rs1789890 | 100468907 | Intergenic | Between ADH1B and 1C | 1.33 | 0.0003 | 0.026 | TFBS |
| Autism | rs1229863 | 100471409 | Intergenic | Between ADH1B and 1C | 1.33 | 0.0003 | 0.026 | Conserved |
Conserved located at a species-conserved element, TFBS located at a transcription factor binding site, CpG located at a methylated CpG island, CNV located at a copy number variant (CNV), long RNA located at a long RNA transcript (>200 bases)
Correction for multiple testing
The experiment-wide significance level (α) was corrected for the number of effective markers that were calculated from the entire marker set by the program SNPSpD. SNPSpD is based on an adjusted Bonferroni correction method (Li and Ji 2005). The linkage disequilibrium (LD) structures were highly similar across different phenotype groups within the same ethnicity. Approximately, 100 effective SNPs captured most information of all common SNPs across the entire ADH gene cluster both in subjects of European and African descents. Thus, the corrected significance level (α) was set at 0.0005. The numbers of risk SNPs that were nominally (p <0.05) or significantly (p <α) associated with phenotypes are shown in Table 1. Finally, q value for each SNP was estimated from p values within each phenotype group using the R package QVALUE (Storey and Tibshirani 2003). The numbers of risk SNPs with q <0.05 and the q values for the significant risk SNPs are shown in Tables 1 and 2, respectively.
Results
Among a total of 632 common SNPs in African-American GAIN samples, 50 SNPs were nominally associated with schizophrenia (p <0.05), 28 of which were significantly associated with schizophrenia after false discovery rate (FDR) correction (q <0.05). With region-wide correction for multiple testing by SNPSpD, 19 SNPs were significantly associated with schizophrenia (8.9 × 10−5 ≤ p ≤ 0.0003). These 19 SNPs were in high LD with one another (D′ = 1). Among a total of 921 common SNPs in European-Americans, 141 SNPs were nominally associated with autism (p <0.05), 15 of which survived FDR correction (q <0.05), and 6 of which survived region-wide SNPSpD correction (2.4 × 10−5 ≤ p ≤ 0.0003) (Tables 1, 2). These six SNPs were in high LD with one another (D′ >0.9). After further corrected by the number of cohorts examined, these associations still remained suggestively significant. In addition, as introduced above, a recent GWAS identified a common variant (rs1789891 between ADH1B and ADH1C) that was significantly associated with alcohol dependence in the subjects of German descent (Frank et al. 2012). Interestingly, this SNP was suggestively associated with autism (p = 0.0015) in the present study, but not with alcohol dependence (p >0.05).
Bioinformatic analysis showed that most of the significant risk SNPs (p <α; Table 2) were located at transcription factor binding sites (TFBS). Three SNPs, i.e., rs1442481 and rs1789912 at ADH1C and rs1229863 between ADH1B and ADH1C, were located at species-conserved elements. Three SNPs, i.e., rs71612682 between ADH1B and ADH1C and rs1789916 and rs1789912 at ADH1C, were located at methylated CpG islands. rs1789900 and rs1442480 at ADH1C were located at a 60-bp-long copy number variant (CNV: A_16_ P16787293), and rs1789916 at ADH1C was located at another 60-bp-long CNV (A_16_P36841645). In addition, rs62323588 between ADH5 and ADH4 was located at a long RNA transcript (>200 bases).
Among a total of 916 common SNPs in African-Americans, 26 SNPs were nominally associated with alcohol dependence (p <0.05), some of which were suggestively associated with alcohol dependence at a non-significant trend level. The most significant one was rs904092 at 5′ flanking region of ADH1A (p = 0.00053), and the second most significant one was rs2066702 (Arg370Cys; ADH2*3) at exon 9 of ADH1B (p = 0.0015; f = 0.142 in cases and 0.193 in controls). However, no SNPs survived either FDR or SNPSpD correction (Table 1). Similarly, although some SNPs were nominally associated with other neuropsychiatric and neurological disorders (p <0.05), no SNPs survived either FDR or SNPSpD correction (Table 1).
Discussion
The principal finding of the current study was that common ADH variants were significantly associated with the risk for schizophrenia and autism, but not other neuropsychiatric disorders, including alcohol dependence. There is growing evidence that schizophrenia and autism share genetic risk variants including SNPs and CNVs (McCarthy et al. 2009; Sebat et al. 2009; Owen et al. 2011). The present study provided new evidence in support of this shared risk.
The location of the ADH variants within the ADH gene cluster may have functional significance. All of the 19 significant risk SNPs for schizophrenia and five of the six significant risk SNPs for autism were located within or flanking ADH1C (i.e., in 5′ flanking region of ADH1C or between ADH1C and ADH1B) (Table 2). These risk SNPs may have potential biological functions based on the bioinformatic analyses. It has been known that the lower functioning γγADH enzyme (mainly) (encoded by ADH1C) and ββADH enzyme (partially) (encoded by ADH1B) inhibit the turnover of 5-HIAL to 5-HTOL and increase 5-HIAA levels (Svensson et al. 1999). 5-HIAA is an important metabolite of serotonin. Alterations in 5-HIAA levels variably associated with schizophrenia (Wieselgren and Lindstrom 1998) and autism (Adamsen et al. 2011) have been interpreted as providing evidence of disturbances in serotonergic neurotransmission associated with these disorders (Cook and Leventhal 1996; Abi-Dargham et al. 1997; Chugani 2004). Thus, it is conceivable that ADH1B and ADH1C are involved in serotonergic dysfunction associated with these disorders. In addition, we noted that one significant risk SNP (rs62323588) for autism was located between ADH4 and ADH5 (Table 2). It has been known that the increased ππADH enzyme (encoded by ADH4) activity could lead to a very high turnover of norepinephrine aldehydes (Holmes 1994), and norepinephrine has been reported to be involved in the development of autism (Leboyer et al. 1992). These functional links may be supported, at least partially, by our current finding of the association between rs62323588 and autism.
It is also worth noting that the two top-ranked common ADH variants, i.e., rs904092 and rs2066702, that were suggestively associated with alcohol dependence in African–Americans at a trend level, are located in the 5′ flanking region of ADH1A and within ADH1B, respectively. The functional rs2066702 (ADH2*3) reduces the activity of ββADH enzyme in the oxidation of ethanol, and thus may affect risk for alcohol dependence (Thomasson et al. 1995). ADH1A encodes ααADH enzyme that has similar properties to ββADH and γγADH and contributes to the oxidization of ethanol. Thus, ADH1A is also a reasonable candidate gene for alcohol dependence (Zuo et al. 2009). In view of the apparent biological functions of these ADHs, the trend-level associations between these variants and alcohol dependence may reflect the smaller effects of common variants than rare variants. Future studies with larger samples are warranted to examine whether the associations between common ADH variants and alcohol dependence can really reach a significant level.
In conclusion, human diseases may be caused by a constellation of rare variants (Dickson et al. 2010), common variants, or both. Our studies, including a previous work (Zuo et al. 2013b) and the present one, suggest that rare ADH variants are associated with alcohol dependence; common ADH variants were suggestively associated with alcohol dependence, but significantly associated with schizophrenia and autism. These findings may support a hypothesis that rare and common ADH variants play different roles in the ADH properties. The rare ADH variants (e.g., those four functional variants introduced above) may influence the ADH functions that are related to the ethanol metabolism, and may thus be implicated in risk for alcoholism; however, the common ADH variants are more likely to affect the ADH activity that is related to the monoamines’ metabolic pathways, and may thus be implicated in risk for schizophrenia, autism, and possible, more other, neuropsychiatric disorders.
Acknowledgments
We thank for Dr. Daniel Winstead’s helpful comments. This work was supported in part by National Institute on Drug Abuse (NIDA) Grants K01 DA029643 and R01DA016750, National Institute on Alcohol Abuse and Alcoholism (NIAAA) grants R01 AA016015 and R21 AA020319 and the National Alliance for Research on Schizophrenia and Depression (NARSAD) Award 17616 (L.Z.). We thank NIH GWAS Data Repository, the Contributing Investigator(s) who contributed the phenotype and genotype data from his/her original study (e.g., Drs. Bierut, Edenberg, Heath, Singleton, Hardy, Foroud, Myers, Gejman, Faraone, Sonuga-Barke, Sullivan, Nurnberger, Devlin, Monaco, etc.), and the primary funding organization that supported the contributing study. Funding and other supports for phenotype and genotype data were provided through the National Institutes of Health (NIH) Genes, Environment and Health Initiative (GEI) (U01HG004422, U01HG004436 and U01HG00 4438); the GENEVA Coordinating Center (U01HG004446); the NIAAA (U10AA008401, R01AA013320, P60AA011998); the NIDA (R01DA013423); the National Cancer Institute (P01 CA089392); the Division of Neuroscience, the NIA National Institute of Neurological Disorders and Stroke (NINDS); the NINDS Human Genetics Resource Center DNA and Cell Line Repository; the NIH contract “High throughput genotyping for studying the genetic contributions to human disease” (HHSN268200782096C); the Center for Inherited Disease Research (CIDR); a Cooperative Agreement with the Division of Adult and Community Health, Centers for Disease Control and Prevention; the NIH Office of Research on Women’s Health (ORWH) (R01NS45012); the Department of Veterans Affairs; the University of Maryland General Clinical Research Center (M01RR165001), the National Center for Research Resources, NIH; the National Institute of Mental Health (K01MH086621, R01MH059160, R01MH59565, R01MH59566, R01MH59571, R01 MH59586, R01MH59587, R01MH59588, R01MH60870, R01MH 60879, R01MH61675, R01MH62873, R01MH081803, R01MH67 257, R01MH81800, U01MH46276, U01MH46282, U01MH46289, U01MH46318, U01MH79469, U01MH79470 and R01MH67257); the NIMH Genetics Initiative for Bipolar Disorder; the Genetic Association Information Network (GAIN); the Genetic Consortium for Late Onset Alzheimer’s Disease; the Autism Genome Project, the MARC: Risk Mechanisms in Alcoholism and Comorbidity; the Molecular Genetics of Schizophrenia Collaboration; the Medical Research Council (G0601030) and the Wellcome Trust (075491/Z/04), University of Oxford; the Netherlands Scientific Organization (904-61-090, 904-61-193, 480-04-004, 400-05-717, NWO Genomics, SPI 56-464-1419) the Centre for Neurogenomics and Cognitive Research (CNCR-VU); Netherlands Study of Depression and Anxiety (NESDA) and the Netherlands Twin Register (NTR); and the European Union (EU/WLRT-2001-01254), ZonMW (geestkracht program, 10-000-1002). Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, was provided by the Genetic Consortium for Late Onset Alzheimer’s Disease, the GENEVA Coordinating Center (U01 HG004446), and the National Center for Biotechnology Information. Genotyping was performed at the Johns Hopkins University Center for Inherited Disease Research, and GlaxoSmithKline, R&D Limited. The datasets used for the analyses described in this manuscript were obtained from dbGaP at http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gap. The dbGaP accession numbers include phs000125.v1.p1, phs000021. v3.p2, phs000021.v3.p2, phs000167.v1.p1, phs000167.v1.p1, phs00 0267.v1.p1, phs000016.v2.p2, phs000092.v1.p1, phs000092.v1.p1, phs000181.v1.p1, phs000020.v2.p1, phs000017.v3.p1, phs000017. v3.p1, phs000017.v3.p1, phs000168.v1.p1, phs000219.v1.p1, phs000 101.v3.p1, phs000292.v1.p1, phs000292.v1.p1, phs000102.v1.p1, phs000196.v2.p1, phs000126.v1.p1, phs000089.v3.p2, phs000089. v3.p2, phs000089.v3.p2 and phs000089.v3.p2.
Footnotes
Conflict of interest There is no conflict of interest to declare.
Contributor Information
Lingjun Zuo, Email: Lingjun.Zuo@yale.edu, Department of Psychiatry, Yale University School of Medicine, New Haven CT 06516, USA.
Kesheng Wang, Department of Biostatistics and Epidemiology, College of Public Health, East Tennessee State University, Johnson City, TN, USA.
Xiang-Yang Zhang, Menninger Department of Psychiatry and Behavioral Sciences, Baylor College of Medicine, Houston, TX, USA.
Xinghua Pan, Department of Genetics, Yale University School of Medicine, New Haven, CT, USA.
Guilin Wang, Yale Center for Genome Analysis, Yale University School of Medicine, Orange, CT, USA.
Yunlong Tan, Biological Psychiatry Research Center, Beijing Huilongguan Hospital, Beijing, China.
Chunlong Zhong, Department of Neurology, Renji Hospital, Shanghai Jiaotong University, Shanghai, China.
John H. Krystal, Department of Psychiatry, Yale University School of Medicine, New Haven CT 06516, USA
Matthew State, Department of Psychiatry, Yale University School of Medicine, New Haven CT 06516, USA.
Heping Zhang, Department of Biostatistics, Yale University School of Epidemiology and Public Health, New Haven, CT, USA.
Xingguang Luo, Email: xingguang.luo@yale.edu, Department of Psychiatry, Yale University School of Medicine, New Haven CT 06516, USA.
References
- Abi-Dargham A, Laruelle M, Aghajanian GK, Charney D, Krystal J. The role of serotonin in the pathophysiology and treatment of schizophrenia. J Neuropsychiatry Clin Neurosci. 1997;9:1–17. doi: 10.1176/jnp.9.1.1. [DOI] [PubMed] [Google Scholar]
- Adamsen D, Meili D, Blau N, Thony B, Ramaekers V. Autism associated with low 5-hydroxyindoleace acid in CSF and the heterozygous SLC6A4 gene Gly56Ala plus 5-HTTLPR L/L promoter variants. Mol Genet Metab. 2011;102:368–373. doi: 10.1016/j.ymgme.2010.11.162. [DOI] [PubMed] [Google Scholar]
- AGP: The AGP Consortium. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 2007;39(3):319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AGP: The AGP Consortium. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466(7204):368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AGP: The AGP Consortium. A genome-wide scan for common alleles affecting risk for autism. Hum Mol Genet. 2010;19(20):4072–4082. doi: 10.1093/hmg/ddq307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Psychiatric Association. Diagnostic and statistical manual of mental disorders. 4. American Psychiatric Press; Washington DC: 1994. [Google Scholar]
- Anney R, Klei L, Pinto D, Regan R, Conroy J, Magalhaes TR, Correia C, Abrahams BS, Sykes N, Pagnamenta AT, Almeida J, Bacchelli E, Bailey AJ, Baird G, Battaglia A, Berney T, Bolshakova N, Bolte S, Bolton PF, Bourgeron T, Brennan S, Brian J, Carson AR, Casallo G, Casey J, Chu SH, Cochrane L, Corsello C, Crawford EL, Crossett A, Dawson G, de Jonge M, Delorme R, Drmic I, Duketis E, Duque F, Estes A, Farrar P, Fernandez BA, Folstein SE, Fombonne E, Freitag CM, Gilbert J, Gillberg C, Glessner JT, Goldberg J, Green J, Guter SJ, Hakonarson H, Heron EA, Hill M, Holt R, Howe JL, Hughes G, Hus V, Igliozzi R, Kim C, Klauck SM, Kolevzon A, Korvatska O, Kustanovich V, Lajonchere CM, Lamb JA, Laskawiec M, Leboyer M, Le Couteur A, Leventhal BL, Lionel AC, Liu XQ, Lord C, Lotspeich L, Lund SC, Maestrini E, Mahoney W, Mantoulan C, Marshall CR, McConachie H, McDougle CJ, McGrath J, McMahon WM, Melhem NM, Merikangas A, Migita O, Minshew NJ, Mirza GK, Munson J, Nelson SF, Noakes C, Noor A, Nygren G, Oliveira G, Papanikolaou K, Parr JR, Parrini B, Paton T, Pickles A, Piven J, Posey DJ, Poustka A, Poustka F, Prasad A, Ragoussis J, Renshaw K, Rickaby J, Roberts W, Roeder K, Roge B, Rutter ML, Bierut LJ, Rice JP, Salt J, Sansom K, Sato D, Segurado R, Senman L, Shah N, Sheffield VC, Soorya L, Sousa I, Stoppioni V, Strawbridge C, Tancredi R, Tansey K, Thiruvahindrapduram B, Thompson AP, Thomson S, Tryfon A, Tsiantis J, Van Engeland H, Vincent JB, Volkmar F, Wallace S, Wang K, Wang Z, Wassink TH, Wing K, Wittemeyer K, Wood S, Yaspan BL, Zurawiecki D, Zwaigenbaum L, Betancur C, Buxbaum JD, Cantor RM, Cook EH, Coon H, Cuccaro ML, Gallagher L, Geschwind DH, Gill M, Haines JL, Miller J, Monaco AP, Nurnberger JI, Jr, Paterson AD, Pericak-Vance MA, Schellenberg GD, Scherer SW, Sutcliffe JS, Szatmari P, Vicente AM, Vieland VJ, Wijsman EM, Devlin B, Ennis S, Hallmayer J. A genome-wide scan for common alleles affecting risk for autism. Hum Mol Genet. 2010;19:4072–4082. doi: 10.1093/hmg/ddq307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning BL, Browning SR. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet. 2009;84:210–223. doi: 10.1016/j.ajhg.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buervenich S, Sydow O, Carmine A, Zhang Z, Anvret M, Olson L. Alcohol dehydrogenase alleles in Parkinson’s disease. Mov Disord. 2000;15:813–818. doi: 10.1002/1531-8257(200009)15:5<813::aid-mds1008>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- Chugani DC. Serotonin in autism and pediatric epilepsies. Ment Retard Dev Disabil Res Rev. 2004;10:112–116. doi: 10.1002/mrdd.20021. [DOI] [PubMed] [Google Scholar]
- Cook EH, Leventhal BL. The serotonin system in autism. Curr Opin Pediatr. 1996;8:348–354. doi: 10.1097/00008480-199608000-00008. [DOI] [PubMed] [Google Scholar]
- Dickson SP, Wang K, Krantz I, Hakonarson H, Goldstein DB. Rare variants create synthetic genome-wide associations. PLoS Biol. 2010;8:e1000294. doi: 10.1371/journal.pbio.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippini N, Rao A, Wetten S, Gibson RA, Borrie M, Guzman D, Kertesz A, Loy-English I, Williams J, Nichols T, Whitcher B, Matthews PM. Anatomically-distinct genetic associations of APOE epsilon4 allele load with regional cortical atrophy in Alzheimer’s disease. Neuroimage. 2009;44(3):724–728. doi: 10.1016/j.neuroimage.2008.10.003. [DOI] [PubMed] [Google Scholar]
- Frank J, Cichon S, Treutlein J, Ridinger M, Mattheisen M, Hoffmann P, Herms S, Wodarz N, Soyka M, Zill P, Maier W, Mossner R, Gaebel W, Dahmen N, Scherbaum N, Schmal C, Steffens M, Lucae S, Ising M, Muller-Myhsok B, Nothen MM, Mann K, Kiefer F, Rietschel M. Genome-wide significant association between alcohol dependence and a variant in the ADH gene cluster. Addict Biol. 2012;17:171–180. doi: 10.1111/j.1369-1600.2011.00395.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes RS. Alcohol dehydrogenases: a family of isozymes with differential functions. Alcohol Alcohol Suppl. 1994;2:127–130. [PubMed] [Google Scholar]
- Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leboyer M, Bouvard MP, Launay JM, Tabuteau F, Waller D, Dugas M, Kerdelhue B, Lensing P, Panksepp J. Brief report: a double-blind study of naltrexone in infantile autism. J Autism Dev Disord. 1992;22:309–319. doi: 10.1007/BF01058158. [DOI] [PubMed] [Google Scholar]
- Li J, Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity. 2005;95:221–227. doi: 10.1038/sj.hdy.6800717. [DOI] [PubMed] [Google Scholar]
- Li H, Wetten S, Li L, St Jean PL, Upmanyu R, Surh L, Hosford D, Barnes MR, Briley JD, Borrie M, Coletta N, Delisle R, Dhalla D, Ehm MG, Feldman HH, Fornazzari L, Gauthier S, Goodgame N, Guzman D, Hammond S, Hollingworth P, Hsiung GY, Johnson J, Kelly DD, Keren R, Kertesz A, King KS, Lovestone S, Loy-English I, Matthews PM, Owen MJ, Plumpton M, Pryse-Phillips W, Prinjha RK, Richardson JC, Saunders A, Slater AJ, St George-Hyslop PH, Stinnett SW, Swartz JE, Taylor RL, Wherrett J, Williams J, Yarnall DP, Gibson RA, Irizarry MC, Middleton LT, Roses AD. Candidate single-nucleotide polymorphisms from a genomewide association study of Alzheimer disease. Arch Neurol. 2008;65(1):45–53. doi: 10.1001/archneurol.2007.3. [DOI] [PubMed] [Google Scholar]
- Luo X, Kranzler HR, Zuo L, Wang S, Schork NJ, Gelernter J. Diplotype trend regression analysis of the ADH gene cluster and the ALDH2 gene: multiple significant associations with alcohol dependence. Am J Hum Genet. 2006;78:973–987. doi: 10.1086/504113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Kranzler HR, Zuo L, Zhang H, Wang S, Gelernter J. ADH7 variation modulates extraversion and conscientiousness in substance-dependent subjects. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:179–186. doi: 10.1002/ajmg.b.30589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy SE, Makarov V, Kirov G, Addington AM, McClellan J, Yoon S, Perkins DO, Dickel DE, Kusenda M, Krastoshevsky O, Krause V, Kumar RA, Grozeva D, Malhotra D, Walsh T, Zackai EH, Kaplan P, Ganesh J, Krantz ID, Spinner NB, Roccanova P, Bhandari A, Pavon K, Lakshmi B, Leotta A, Kendall J, Lee YH, Vacic V, Gary S, Iakoucheva LM, Crow TJ, Christian SL, Lieberman JA, Stroup TS, Lehtimaki T, Puura K, Haldeman-Englert C, Pearl J, Goodell M, Willour VL, Derosse P, Steele J, Kassem L, Wolff J, Chitkara N, McMahon FJ, Malhotra AK, Potash JB, Schulze TG, Nothen MM, Cichon S, Rietschel M, Leibenluft E, Kustanovich V, Lajonchere CM, Sutcliffe JS, Skuse D, Gill M, Gallagher L, Mendell NR, Craddock N, Owen MJ, O’Donovan MC, Shaikh TH, Susser E, Delisi LE, Sullivan PF, Deutsch CK, Rapoport J, Levy DL, King MC, Sebat J. Microduplications of 16p11.2 are associated with schizophrenia. Nat Genet. 2009;41:1223–1227. doi: 10.1038/ng.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen MJ, O’Donovan MC, Thapar A, Craddock N. Neuro-developmental hypothesis of schizophrenia. Br J Psychiatry. 2011;198:173–175. doi: 10.1192/bjp.bp.110.084384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, Levy DL, McCarthy SE. Rare structural variants in schizophrenia: one disorder, multiple mutations; one mutation, multiple disorders. Trends Genet. 2009;25:528–535. doi: 10.1016/j.tig.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson H, Ophoff RA, Steinberg S, Andreassen OA, Cichon S, Rujescu D, Werge T, Pietilainen OP, Mors O, Mortensen PB, Sigurdsson E, Gustafsson O, Nyegaard M, Tuulio-Henriksson A, Ingason A, Hansen T, Suvisaari J, Lonnqvist J, Paunio T, Borglum AD, Hartmann A, Fink-Jensen A, Nordentoft M, Hougaard D, Norgaard-Pedersen B, Bottcher Y, Olesen J, Breuer R, Moller HJ, Giegling I, Rasmussen HB, Timm S, Mattheisen M, Bitter I, Rethelyi JM, Magnusdottir BB, Sigmundsson T, Olason P, Masson G, Gulcher JR, Haraldsson M, Fossdal R, Thorgeirsson TE, Thorsteinsdottir U, Ruggeri M, Tosato S, Franke B, Strengman E, Kiemeney LA, Melle I, Djurovic S, Abramova L, Kaleda V, Sanjuan J, de Frutos R, Bramon E, Vassos E, Fraser G, Ettinger U, Picchioni M, Walker N, Toulopoulou T, Need AC, Ge D, Yoon JL, Shianna KV, Freimer NB, Cantor RM, Murray R, Kong A, Golimbet V, Carracedo A, Arango C, Costas J, Jonsson EG, Terenius L, Agartz I, Petursson H, Nothen MM, Rietschel M, Matthews PM, Muglia P, Peltonen L, St Clair D, Goldstein DB, Stefansson K, Collier DA. Common variants conferring risk of schizophrenia. Nature. 2009;460:744–747. doi: 10.1038/nature08186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genome-wide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson S, Some M, Lundsjo A, Helander A, Cronholm T, Hoog JO. Activities of human alcohol dehydrogenases in the metabolic pathways of ethanol and serotonin. Eur J Biochem/FEBS. 1999;262:324–329. doi: 10.1046/j.1432-1327.1999.00351.x. [DOI] [PubMed] [Google Scholar]
- Thomasson HR, Beard JD, Li TK. ADH2 gene polymorphisms are determinants of alcohol pharmacokinetics. Alcohol Clin Exp Res. 1995;19:1494–1499. doi: 10.1111/j.1530-0277.1995.tb01013.x. [DOI] [PubMed] [Google Scholar]
- Wieselgren IM, Lindstrom LH. CSF levels of HVA and 5-HIAA in drug-free schizophrenic patients and healthy controls: a prospective study focused on their predictive value for outcome in schizophrenia. Psychiatry Res. 1998;81:101–110. doi: 10.1016/s0165-1781(98)00090-0. [DOI] [PubMed] [Google Scholar]
- Yoshida A, Rzhetsky A, Hsu LC, Chang C. Human aldehyde dehydrogenase gene family. Eur J Biochem/FEBS. 1998;251:549–557. doi: 10.1046/j.1432-1327.1998.2510549.x. [DOI] [PubMed] [Google Scholar]
- Zuo L, Gelernter J, Kranzler HR, Stein MB, Zhang H, Wei F, Sen S, Poling J, Luo X. ADH1A variation predisposes to personality traits and substance dependence. Am J Med Genet B Neuropsychiatr Genet. 2009;153B:376–386. doi: 10.1002/ajmg.b.30990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo L, Zhang CK, Wang F, Li CS, Zhao H, Lu L, Zhang XY, Lu L, Zhang H, Zhang F, Krystal JH, Luo X. A novel, functional and replicable risk gene region for alcohol dependence identified by genome-wide association study. PLoS One. 2011;6:e26726. doi: 10.1371/journal.pone.0026726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo L, Gelernter J, Zhang CK, Zhao H, Lu L, Kranzler HR, Malison RT, Li CR, Wang F, Zhang XY, Deng HW, Krystal JH, Zhang F, Luo X. Genome-wide association study of alcohol dependence implicates KIAA0040 on chromosome 1q. Neuro-psychopharmacology. 2012;37:557–566. doi: 10.1038/npp.2011.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo L, Wang F, Zhang XY, Li CSR, Lu L, Ye L, Zhang H, Krystal JH, Deng HW, Luo X. Genome-wide significant association signals in IPO11-HTR1A region specific for alcohol and nicotine co-dependence. Alcohol Clin Exp Res. 2013a doi: 10.1111/acer.12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo L, Zhang H, Malison RT, Li CSR, Zhang XY, Wang F, Lu L, Lu L, Wang X, Krystal JH, Zhang F, Deng H, Luo X. Alcohol Alcohol. Vol. 48. Oxford: Oxfordshire; 2013b. Rare ADH variant constellations are specific for alcohol dependence; pp. 9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]