Abstract
The Notch signaling system features a growing number of modulators that include extracellular proteins that bind to the Notch ectodomain. Collagens are a complex, heterogeneous family of secreted proteins that serve both structural and signaling functions, most prominently through binding to integrins and DDR. The shared widespread tissue distribution of Notch and collagen prompted us to investigate the effects of collagen on Notch signaling. In a cell co-culture signaling assay, we found that type IV collagen inhibited Notch signaling in H460 and A7R5 cell lines. Moreover, Notch-stimulated expression of mature smooth muscle genes SMA, MHC, SM22, and calponin, which define the physiologic phenotype of normal vascular smooth muscle, was inhibited by type IV collagen in A7R5 cells. Cloned promoters of three of these genes were also inhibited by exposure to collagen. Collagen-dependent repression of Notch signaling required an RBP-jK site within the SM22 promoter. Moreover, repression by collagen required extracellular stimulation of the Notch signaling pathway. Type IV collagen bound to both Notch3 and Jagged1 proteins in purified protein binding assays. In addition, type I collagen also inhibited Notch signaling and bound to Notch and Jagged. We conclude that type IV and type I collagen repress canonical Notch signaling to alter expression of Notch target genes.
Keywords: collagen, Notch, smooth muscle, inhibition
1. Introduction
Notch signaling is an evolutionarily conserved pathway that plays an essential role in early development and frequently participates in adaptive responses to disease. Loss of Notch results in early embryonic lethality that is accompanied by failure to develop functional vasculature (Domenga et al., 2004, Gale et al., 2004, High et al., 2007, Iso et al., 2003, Krebs et al., 2000, Limbourg et al., 2005, McCright et al., 2001, Uyttendaele et al., 2001, Xue Y, 1999). Postnatal inhibition of Notch signaling results in dysfunctional angiogenesis, and, therefore, Notch inhibition has been proposed as a treatment strategy for cancer (Li et al., 2007, Noguera-Troise et al., 2006, Ridgway et al., 2006). Graded regulation of the level of Notch signaling also plays a significant role in development. Incremental losses of Notch1 and Notch2 alleles in melanocytes result in progressive whitening of the hair proportional to the number of null alleles (Schouwey et al., 2007). These observations indicate that quantitative regulation of Notch signaling may play a role in modulating phenotype.
All Notch receptors contain a large array of EGF-like repeats. Canonical Notch signaling requires interactions between a small subset of these EGF-like segments of Notch with the EGF-like repeats of ligands, Jagged and Delta (Cordle et al., 2008, Joutel et al., 2004, Shimizu et al., 1999). The significance of the large number of additional Notch EGF-like repeats, which have been conserved between all species, remains undetermined, but these protein domains may participate in modulatory interactions with extracellular proteins that fine tune Notch signaling. Indeed, numerous extracellular Notch enhancers and Notch inhibitors have now been described (D’Souza et al., 2010, Wang, 2011). Many of the heretofore described modulators contain EGF-like repeats, implicating EGF-like to EGF-like interactions as a potential protein heterodimerization interface.
Collagens are among the most common extracellular proteins of the human body and are expressed during development and in all postnatal tissues. Like Notch, collagens are composed of numerous subtypes which are dynamically regulated and also participate in disease pathogenesis (Myllyharju and Kivirikko, 2004). In further analogy to Notch, the collagens are large, complex molecules composed of repetitive biochemical units that are extensively posttranslationally modified; these proteins perform not only structural functions but also stimulate cell signaling events through integrins (Barczyk et al., 2010) and DDR proteins (Shrivastava et al., 1997, Vogel et al., 1997). Here, we perform the first investigation of the effects of collagen on Notch signaling.
2. Results
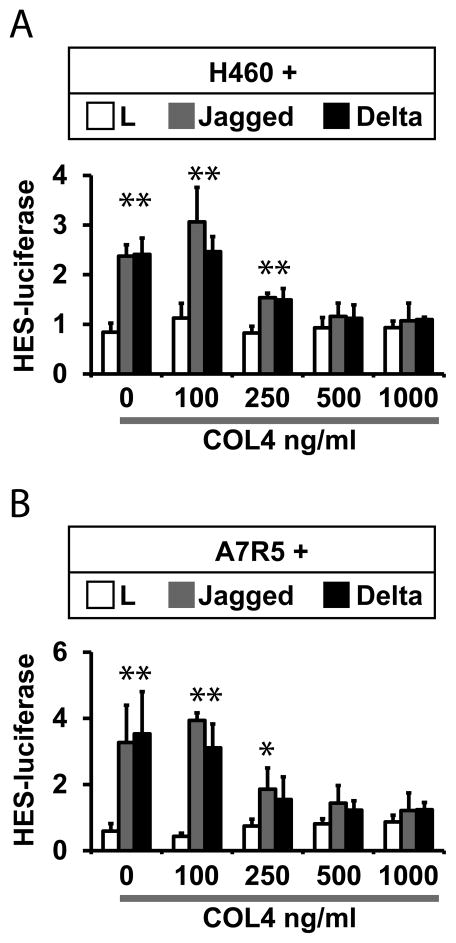
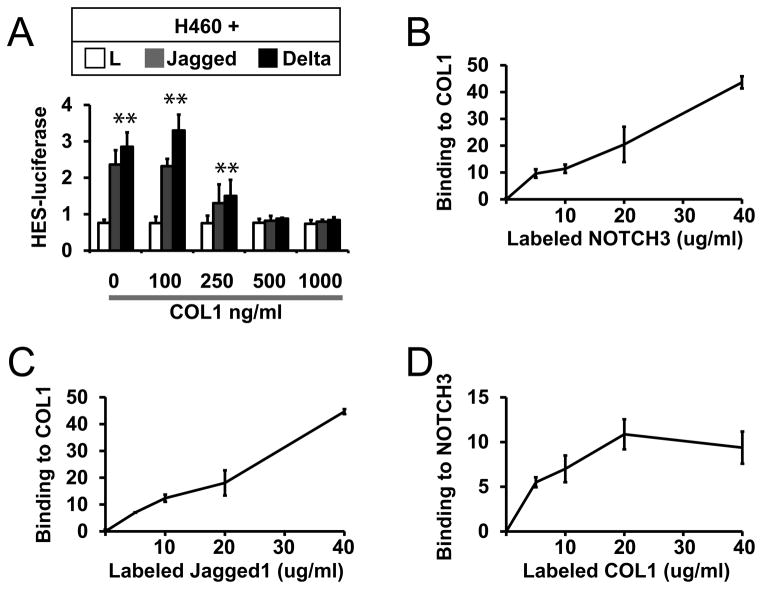
A coculture system was used to measure the effects of collagen on canonical transcellular Notch signaling (Meng et al., 2009, Meng et al., 2010, Meng et al., 2012). Notch-expressing cell lines were transfected with a HES-luciferase reporter; transfected cells were then cocultured with Notch ligand-expressing fibroblast cell lines to stimulate signaling, which was quantified by measuring luciferase activity. Parallel cocultures were performed in the presence of increasing amounts of collagen added to the culture media (Figure 1). Concentrations of 500 ng/ml or higher of type IV collagen completely blocked Jagged and Delta-mediated activation of HES-luciferase. The inhibitory function of type IV collagen was reproduced in Notch-expressing human H460 and the rat A7R5 cell lines.
Figure 1.
Collagen inhibition of Notch pathway activation. Quantitation of Notch signaling was determined by measuring activity of a HES1-Luciferase reporter transfected into Notch receptor expressing cells. Signaling was activated by coculture with mouse fibroblasts (L cells) stably transfected with empty vector control, Jagged1, or Delta-like1. Notch receptor expressing cells H460 (A) or A7R5 (B) were co-transfected with HES1-Luciferase and a plasmid consititutively expressing Renilla luciferase. Cells were co-cultured with ligand expressing cells for 24 hours, and culture media was then supplemented with type IV collagen (concentrations as indicated) before lysates were analyzed for the ratio of firefly/Renilla luciferase. All groups included three replicates. Experiments were performed at least three times and produced the same results. * denotes significant differences between control and Notch ligand-stimulated cultures (p<0.05).
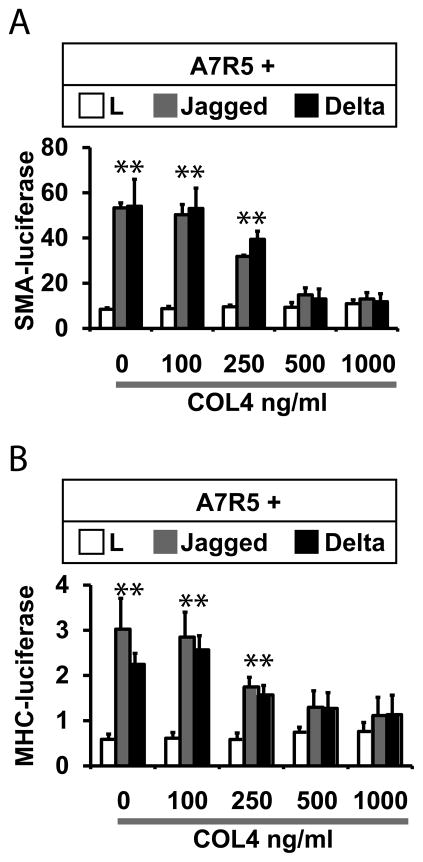
To determine the effects of collagen on vascular marker gene expression, we tested the effects of the protein on Notch dependent expression of mature smooth muscle genes. Notch signaling has been shown to increase mRNA levels of mature smooth muscle genes encoding SM22, smooth muscle actin, MHC, and calponin (Noseda et al., 2006, Tang et al., 2010, Tang et al., 2008, Doi et al., 2006). Moreover, the cloned proximal promoters of SM22 (Doi et al., 2006), SMA (Tang et al., 2010, Tang et al., 2008), and MHC (Meng et al., 2012) have been shown to respond to Notch ligands. To test the effects of collagen on Notch in smooth muscle-derived A7R5 cells, we again used coculture assays. A7R5 cells were transfected with cloned smooth muscle promoter sequences fused to a luciferase reporter gene. Transfected cells were then stimulated with control or Notch ligand-expressing cells in the presence in increasing concentrations of type IV collagen, and luciferase expression was used to quantify promoter activity. As seen in Figure 2, type IV collagen efficiently repressed Notch activation of the cognate promoters for SMA and MHC genes at concentrations of 500 ng/mL and above.
Figure 2.
Collagen blocks Notch regulation of smooth muscle promoters. Experiments were performed as in Fig 1 to determine the effects of type IV collagen on SMA and MHC promoters. A7R5 cells were cotransfected with either SMA-Luciferase (A) or MHC-Luciferase (B) plasmids and Renilla luciferase (as a transfection control). Cocultures were treated with increasing concentrations of type IV collagen. Smooth muscle promoter activation was determined using the ratio of firefly/Renilla luciferase activity. Collagen effectively inhibited expression of both promoter constructs. All groups included three replicates. * denotes significant differences between control and Notch ligand-stimulated cultures (p<0.05).
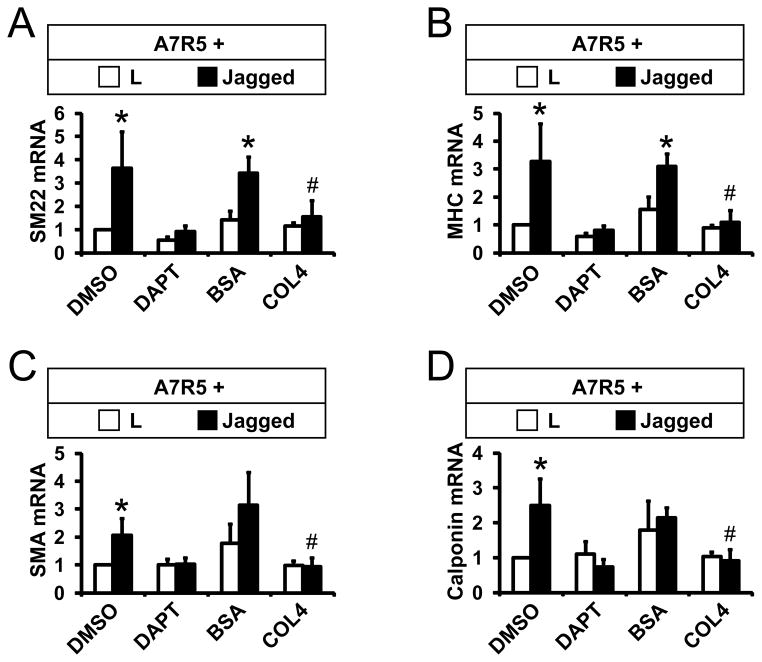
Figure 3 shows experiments to test the effects of collagen on endogenous Notch-regulated genes. Jagged-stimulated expression of four endogenous target smooth muscle genes (SM22, MHC, SMA, and calponin) was inhibited by DAPT, a gamma-secretase inhibitor, suggesting that these genes are coordinately regulated through canonical Notch signaling. Application of type IV collagen to A7R5 cells also inhibited Notch ligand-stimulated expression of the same four endogenous smooth muscle target genes. In sum, these findings demonstrate that type IV collagen blocks coordinated Notch activation of mature smooth muscle targets by repressing their cognate promoters.
Figure 3.
Collagen blocks Jagged-dependent expression of mature smooth muscle genes. (AD) A7R5 cells were cocultured with L or Jagged cells. Some cultures were treated with DMSO (vehicle) or 10uM DAPT, which inhibits Notch signaling. For other groups, after one day, BSA or type IV collagen (500 ng/mL) was added to co-cultures. The next day mRNA was harvested, and quantitative RT-PCR was conducted using rat-specific primers to measure production of smooth muscle genes from A7R5 cells; values were normalized to rat 18S RNA levels. All groups included three replicates. All experiments were performed at least three times with the same results. * denotes significant differences between control and Jagged-stimulated cultures; # denotes significant differences between BSA and collagen treated cultures (p<0.05).
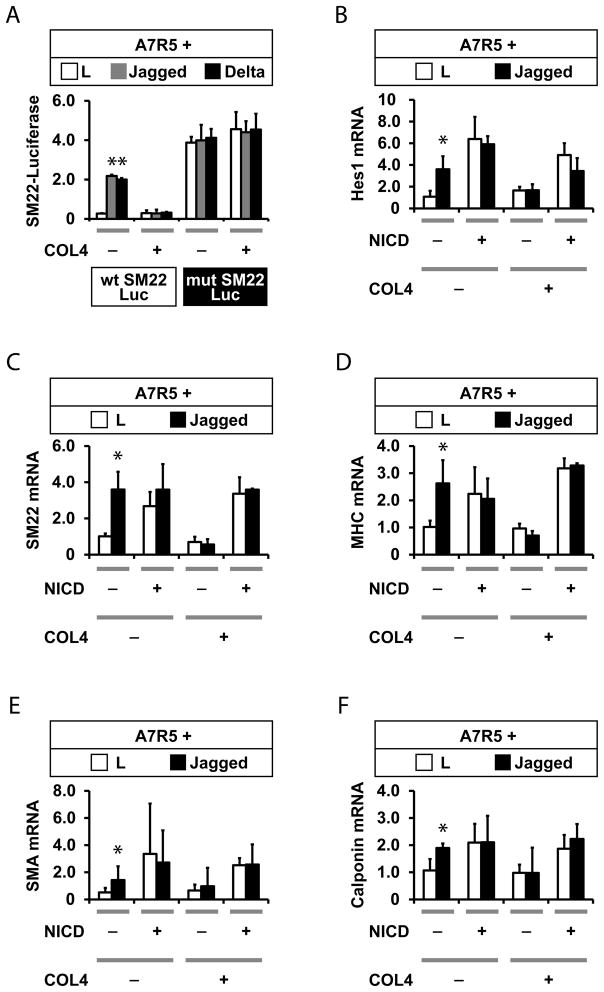
To further define the mechanism of collagen regulation of Notch signaling, we compared the effect of type IV collagen on wildtype and mutant SM22 promoters; the latter contains a mutant RBJ-Jkappa site, which mediates canonical Notch responses (Meng et al., 2012). Mutations in this site ablates Notch-regulated inhibition of the promoter, rendering the construct constitutively active, even in the presence of Notch ligands. Unlike the wildtype sequence, the mutant SM22 promoter was not inhibited by collagen in co-culture experiments, suggesting that collagen inhibits canonical RBP-jK mediated Notch signaling (Figure 4A).
Figure 4.
Collagen inhibits canonical Notch signaling. (A) Effects of collagen on the SM22 promoter require its RBP-jK site, the target of canonical Notch signaling. A7R5 cells were transfected with a proximal SM22 promoter luciferase reporter construct prior to co-culturing with control or Notch ligand expressing cells. Cocultures were treated with increasing concentrations of type IV collagen. Smooth muscle promoter activation was determined using the ratio of firefly/Renilla luciferase activity. Collagen effectively inhibited expression of the wild type SM22 construct, but the mutant SM22-luciferase exhibited constitutive activation that was not affected by extracellular collagen. (B–F) A7R5 cells were transfected NICD from NOTCH3 to constitutively activate the Notch pathway in the absence of extracellular stimulation. Transfected cells were treated with type IV collagen, which did not repress NICD-stimulated reporter expression. Target gene regulation was assessed by quantitative RT-PCR of genes indicated in the Y-axis. RNA content was normalized to 18S control rRNA levels in each group. All groups included three replicates. * denotes significant differences between control and Jagged-stimulated cultures (p<0.05).
Collagen could interfere with Notch signaling by affecting either extracellular Notch or by triggering events that repress intracellular Notch function. To distinguish between these possibilities, we tested the effect of collagen on Notch signaling stimulated by expression of the Notch intracellular domain (NICD), which activates the canonical Notch pathway by bypassing extracellular signaling events (Figures 4B–F). NICD stimulated the expression of target genes Hes1, SM22, MHC, SMA, and calponin; as anticipated, presentation of Jagged cells did not further induce gene expression, demonstrating that NICD-activated expression was independent of extracellular ligand presentation. Presentation of extracellular type IV collagen failed to reduce activation of any of the five target genes in NICD transfected cells (Figures 4B–4F). Because collagen failed to affect intracellular (NICD-stimulate) Notch activation, we conclude that collagen inhibits Notch via an extracellular mechanism.
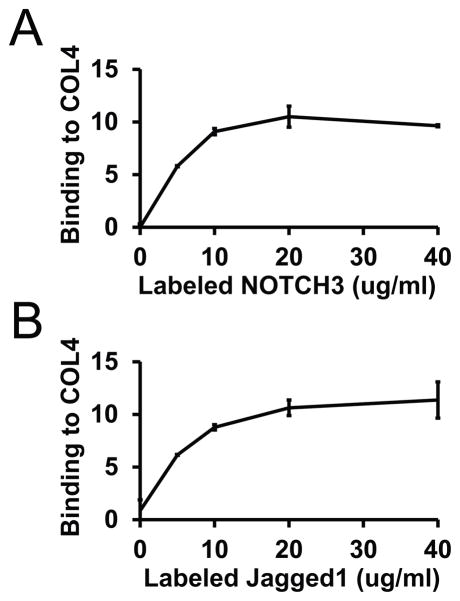
Since extracellular Notch modulators alter signaling through molecular interactions with Notch ectodomains or ligands (D’Souza et al., 2010, Wang, 2011), we tested whether collagen could bind to Notch extracellular signaling components. Indeed, in solid state binding assays (Figure 5A), type IV collagen interacted with NOTCH3 ectodomain. In addition, type IV collagen bound to the Notch ligand Jagged (Figure 5B), likely the most important ligand in vascular smooth muscle cells.
Figure 5.
Interactions between type IV collagen, Notch, and Jagged. Type IV collagen was adsorbed to plastic wells, and wells were then blocked with BSA. Labeled, purified NOTCH3 (A) or Jagged (B) was added to wells at indicated concentrations to allow protein binding. After washing, bound proteins were quantified. NOTCH3 and Jagged did not bind to BSA coated wells. Labeled Fc was used as a control probe; results represent NOTCH3 or Jagged bound probe after subtraction of the Fc signal.
Finally, we tested whether other types of collagen could modulate Notch signaling. In addition to type IV collagen, in normal vascular smooth muscle and in the NOTCH3-related vascular disease CADASIL, type I collagen is highly expressed (Dong et al., 2012). Extracellular presentation of soluble type I collagen, the major fiber forming collagen, inhibited the activation of Notch signaling in NOTCH3-positive H460 cells (Figure 6A). Like type IV collagen, type I collagen formed molecular complexes with both NOTCH3 ectodomain and Jagged (Figures 6B–6D).
Figure 6.
Type I collagen effects on Notch signaling and binding to Notch protein. (A) H460 cells were transfected and co-cultured for Notch signaling assays as in Figure 1. Co-cultures were treated with indicated concentrations of type I collagen. Type I collagen inhibited Notch signaling in a dose-dependent fashion (p<0.05 for control versus Notch ligand stimulation). Binding assays of type I collagen to NOTCH3 (B) and Jagged (C) were performed as in Figure 5. (D) Labeled type I collagen bound to immobilized NOTCH3 in a similar assay, using Fc binding to the target as a negative control.
3. Discussion
Collagen is ubiquitous and plays a role in both normal physiology and in pathology (Myllyharju and Kivirikko, 2004). Aside from its important structural role, collagen also modulates signaling systems including integrins (Barczyk et al., 2010) and DDR (Shrivastava et al., 1997, Vogel et al., 1997). Our studies suggest a novel function of extracellular collagen: inhibition of Notch.
We exemplify the effects of collagen on Notch signaling by studying functional collagen/Notch interactions on mature smooth muscle gene expression. Significant evidence suggests that a key element of vascular pathology involves transition of smooth muscle cells into a pathological, proliferative cell, which features repressed levels of markers such as SM22, MHC, calponin, and SMA (Deaton et al., 2009, Duband et al., 1993, Gabbiani et al., 1984, Hungerford et al., 1996, Miano et al., 1994). At the anatomic level, vessel pathology also frequently features fibrosis with the deposition of large amounts of collagens of multiple types (Dong et al., 2012, Roggendorf et al., 1988). As such, our studies suggest a molecular mechanism that could link collagen deposition in arteries to the promotion or propagation of smooth muscle dedifferentiation.
What is the molecular mechanism of collagen actions on Notch? Our results show that type IV collagens inhibit canonical Notch signaling that involves RBP-jK signaling. Since collagens are only able to inhibit Notch signaling that is stimulated by extracellular ligands, it is likely that collagens directly interfere with extracellular Notch signaling. This is supported by protein binding studies, in which collagen binds to both Notch and Notch ligands. Interestingly, however, collagen did not interfere with Notch-Jagged interactions, suggesting a non-competitive mechanism of inhibition.
Type I and IV collagen, applied in concentrations that affect Notch signaling, did not grossly affect cell morphology or cytoskeletal architecture (Supplemental Figure 1). Moreover, over the time courses relevant to our Notch assays, we observed no collagen-related differences in cellular migration (Supplemental Figure 2). Thus, we there is no evidence that collagens affects Notch indirectly by changing cell morphology or by altering cell motility.
These findings shed new light on regulators of Notch. This is the first study that demonstrates a role for collagen molecules in Notch signaling. Since collagens are ubiquitous proteins that are dynamically expressed and modified, our studies could substantially expand the complexity of Notch regulation in vivo. Moreover, since numerous additional proteins contain collagen-like domains, future studies should be performed to test whether proteins containing these domains can modulate Notch signaling as well; one such protein, von Willebrand factor, has been shown to block Notch and to also deposit in large quantities in cerebral small vessel disease.
Both type I and type IV collagen (fibrillary and network forming collagens, respectively) inhibit Notch. The two proteins differ substantially in quaternary structure and diverge in their non-collagenous domains. These distinctions suggest that the conserved repetitive collagen domain may mediate interactions with Notch. If this is true, then other collagen molecules, of which 28 have been identified so far, may also modulate Notch signaling. Examination of the Notch modulating activities of other types of collagen may help elucidate the structural basis of their actions on Notch signaling. Moreover, the effects of degradation of collagen by a wide variety of collagenases (eg. numerous matrix metalloproteinases) remain to be explored.
In conclusion, we demonstrate that two abundant forms of collagen inhibit canonical Notch signaling. Since collagen is widely expressed and modulated in the adult, additional studies that probe the role of collagen on Notch mediated developmental and pathological processes are warranted.
4. Materials and Methods
Cell culture and luciferase reporter assays
L fibroblasts (control and Notch ligand producing cell lines; (Hicks et al., 2000)), and A7R5 were propagated in Dulbecco’s modified Eagle’s medium (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen). H460 cells were grown in RPMI 1640 with 10% fetal bovine serum (Invitrogen). Notch receptor expressing cells (H460 or A7R5) were transfected with HES-luciferase and Renilla luciferase reporter for Notch assays which was performed as previously described (Meng et al., 2009, Meng et al., 2010). Other reporters for smooth muscle promoters (SMA, MHC and SM22 proximal regulatory regions fused to firefly luciferase) have been described before (Meng et al., 2012). Purified collagens (Sigma) were dissolved in 500 mM acetic acid and then dialyzed extensively against PBS (pH 7.4); the proteins were added directly to the culture medium.
Solid phase binding assays
Recombinant proteins human Notch-3-Fc (first 11 EGF repeats) and rat Jagged 1-Fc were purchased from R&D Systems. As a negative control, we used Fc protein alone. Purified human collagens were prepared as above and adsorbed to plates in neutral PBS at 5 ug/ml. Solid phase assays were performed as described (Meng et al., 2009, Meng et al., 2010). In short, we labeled purified proteins with Alexa 700 succinimide and then removed free label by gel filtration chromatography. Protein-coated plates were blocked with 0.5 % BSA in Tris-buffered saline (50 mM Tris, 150 mM NaCl) with 2 mM CaCl2 for 1 h at room temperature. Subsequently, the labeled proteins were incubated with protein-coated ELISA plates in Tris-buffered saline with 2 mM CaCl2 overnight at 4 °C. Afterwards, plates were rapidly washed with TBS with 2 mM calcium three times at room temperature. The total probe adhering to wells were quantified using a LiCor flatbed infrared scanner. We subtracted the signal from Fc binding to plates, which was negligible. None of the probes bound to BSA coated plates.
Quantitative reverse transcriptase PCR assay
For mRNA expression assays, we co-cultured A7R5 cells (three wells per group) with L cells expressing Jagged ligands or L cells without ligand as a control (Hicks et al., 2000). After overnight co-culture, the media was changed to serum-free DMEM supplemented with BSA or collagen for another 24 hours. We then prepared total RNA from each well. The RNA was reverse transcribed and quantified by real time PCR using rat 18S RNA as a control to assess target gene regulation by Notch activation; the primer sequences and protocol have been described before (Meng et al., 2012). In some experiments, A7R5 cells were first transfected with Notch3 NICD cDNA 24 hours prior to co-culture.
Statistical analysis
Results are displayed with standard deviations. All luciferase and quantitative PCR studies were done with groups of at least three. T-tests were applied with statistically significant differences considered for p<0.05.
Supplementary Material
Acknowledgments
The National Institutes of Health and NINDS supported this work (NS052681, NS054724, and NS062816). The Department of Veterans Affairs also provided funding for these studies (5I01BX000375).
List of Abbreviations and Acronyms
- EGF
epidermal growth factor
- DDR
discoidin domain receptor
- HES
hairy enhancer of split
- SMA
smooth muscle actin
- MHC
myosin heavy chain
- DAPT
N-[N-(3,5-Difluorophenacetyl)-L-alanyl]-S-phenylglycine t-butyl ester
- RNA
ribonucleic acid
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barczyk M, Carracedo S, Gullberg D. Integrins. Cell Tissue Res. 2010;339:269–280. doi: 10.1007/s00441-009-0834-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordle J, Johnson S, Tay JZ, Roversi P, Wilkin MB, de Madrid BH, Shimizu H, Jensen S, Whiteman P, Jin B, Redfield C, Baron M, Lea SM, Handford PA. A conserved face of the Jagged/Serrate DSL domain is involved in Notch trans-activation and cis-inhibition. Nat Struct Mol Biol. 2008;15:849–857. doi: 10.1038/nsmb.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza B, Meloty-Kapella L, Weinmaster G. Canonical and non-canonical Notch ligands. Curr Top Dev Biol. 2010;92:73–129. doi: 10.1016/S0070-2153(10)92003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deaton RA, Gan Q, Owens GK. Sp1-dependent activation of KLF4 is required for PDGF-BB-induced phenotypic modulation of smooth muscle. Am J Physiol Heart Circ Physiol. 2009;296:H1027–1037. doi: 10.1152/ajpheart.01230.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi H, Iso T, Sato H, Yamazaki M, Matsui H, Tanaka T, Manabe I, Arai M, Nagai R, Kurabayashi M. Jagged1-selective notch signaling induces smooth muscle differentiation via a RBP-Jkappa-dependent pathway. J Biol Chem. 2006;281:28555–28564. doi: 10.1074/jbc.M602749200. [DOI] [PubMed] [Google Scholar]
- Domenga V, Fardoux P, Lacombe P, Monet M, Maciazek J, Krebs LT, Klonjkowski B, Berrou E, Mericskay M, Li Z, Tournier-Lasserve E, Gridley T, Joutel A. Notch3 is required for arterial identity and maturation of vascular smooth muscle cells. Genes Dev. 2004;18:2730–2735. doi: 10.1101/gad.308904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Blaivas M, Wang MM. Bidirectional encroachment of collagen into the tunica media in cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy. Brain Res. 2012;1456:64–71. doi: 10.1016/j.brainres.2012.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duband JL, Gimona M, Scatena M, Sartore S, Small JV. Calponin and SM 22 as differentiation markers of smooth muscle: spatiotemporal distribution during avian embryonic development. Differentiation. 1993;55:1–11. doi: 10.1111/j.1432-0436.1993.tb00027.x. [DOI] [PubMed] [Google Scholar]
- Gabbiani G, Kocher O, Bloom WS, Vandekerckhove J, Weber K. Actin expression in smooth muscle cells of rat aortic intimal thickening, human atheromatous plaque, and cultured rat aortic media. J Clin Invest. 1984;73:148–152. doi: 10.1172/JCI111185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale NW, Dominguez MG, Noguera I, Pan L, Hughes V, Valenzuela DM, Murphy AJ, Adams NC, Lin HC, Holash J, Thurston G, Yancopoulos GD. Haploinsufficiency of delta-like 4 ligand results in embryonic lethality due to major defects in arterial and vascular development. Proc Natl Acad Sci U S A. 2004;101:15949–15954. doi: 10.1073/pnas.0407290101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks C, Johnston SH, diSibio G, Collazo A, Vogt TF, Weinmaster G. Fringe differentially modulates Jagged1 and Delta1 signalling through Notch1 and Notch2. Nat Cell Biol. 2000;2:515–520. doi: 10.1038/35019553. [DOI] [PubMed] [Google Scholar]
- High FA, Zhang M, Proweller A, Tu L, Parmacek MS, Pear WS, Epstein JA. An essential role for Notch in neural crest during cardiovascular development and smooth muscle differentiation. J Clin Invest. 2007;117:353–363. doi: 10.1172/JCI30070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hungerford JE, Owens GK, Argraves WS, Little CD. Development of the aortic vessel wall as defined by vascular smooth muscle and extracellular matrix markers. Dev Biol. 1996;178:375–392. doi: 10.1006/dbio.1996.0225. [DOI] [PubMed] [Google Scholar]
- Iso T, Hamamori Y, Kedes L. Notch signaling in vascular development. Arterioscler Thromb Vasc Biol. 2003;23:543–553. doi: 10.1161/01.ATV.0000060892.81529.8F. [DOI] [PubMed] [Google Scholar]
- Joutel A, Monet M, Domenga V, Riant F, Tournier-Lasserve E. Pathogenic mutations associated with cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy differently affect Jagged1 binding and Notch3 activity via the RBP/JK signaling Pathway. Am J Hum Genet. 2004;74:338–347. doi: 10.1086/381506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs LT, Xue Y, Norton CR, Shutter JR, Maguire M, Sundberg JP, Gallahan D, Closson V, Kitajewski J, Callahan R, Smith GH, Stark KL, Gridley T. Notch signaling is essential for vascular morphogenesis in mice. Genes Dev. 2000;14:1343–1352. [PMC free article] [PubMed] [Google Scholar]
- Li JL, Sainson RC, Shi W, Leek R, Harrington LS, Preusser M, Biswas S, Turley H, Heikamp E, Hainfellner JA, Harris AL. Delta-like 4 Notch ligand regulates tumor angiogenesis, improves tumor vascular function, and promotes tumor growth in vivo. Cancer Res. 2007;67:11244–11253. doi: 10.1158/0008-5472.CAN-07-0969. [DOI] [PubMed] [Google Scholar]
- Limbourg FP, Takeshita K, Radtke F, Bronson RT, Chin MT, Liao JK. Essential role of endothelial Notch1 in angiogenesis. Circulation. 2005;111:1826–1832. doi: 10.1161/01.CIR.0000160870.93058.DD. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCright B, Gao X, Shen L, Lozier J, Lan Y, Maguire M, Herzlinger D, Weinmaster G, Jiang R, Gridley T. Defects in development of the kidney, heart and eye vasculature in mice homozygous for a hypomorphic Notch2 mutation. Development. 2001;128:491–502. doi: 10.1242/dev.128.4.491. [DOI] [PubMed] [Google Scholar]
- Meng H, Zhang X, Hankenson KD, Wang MM. Thrombospondin 2 potentiates notch3/jagged1 signaling. J Biol Chem. 2009;284:7866–7874. doi: 10.1074/jbc.M803650200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng H, Zhang X, Lee SJ, Strickland DK, Lawrence DA, Wang MM. Low density lipoprotein receptor-related protein-1 (LRP1) regulates thrombospondin-2 (TSP2) enhancement of Notch3 signaling. J Biol Chem. 2010;285:23047–23055. doi: 10.1074/jbc.M110.144634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng H, Zhang X, Yu G, Lee SJ, Chen YE, Prudovsky I, Wang MM. Biochemical Characterization and Cellular Effects of CADASIL Mutants of NOTCH3. PLoS One. 2012;7:e44964. doi: 10.1371/journal.pone.0044964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miano JM, Cserjesi P, Ligon KL, Periasamy M, Olson EN. Smooth muscle myosin heavy chain exclusively marks the smooth muscle lineage during mouse embryogenesis. Circ Res. 1994;75:803–812. doi: 10.1161/01.res.75.5.803. [DOI] [PubMed] [Google Scholar]
- Myllyharju J, Kivirikko KI. Collagens, modifying enzymes and their mutations in humans, flies and worms. Trends Genet. 2004;20:33–43. doi: 10.1016/j.tig.2003.11.004. [DOI] [PubMed] [Google Scholar]
- Noguera-Troise I, Daly C, Papadopoulos NJ, Coetzee S, Boland P, Gale NW, Lin HC, Yancopoulos GD, Thurston G. Blockade of Dll4 inhibits tumour growth by promoting non-productive angiogenesis. Nature. 2006;444:1032–1037. doi: 10.1038/nature05355. [DOI] [PubMed] [Google Scholar]
- Noseda M, Fu Y, Niessen K, Wong F, Chang L, McLean G, Karsan A. Smooth Muscle alpha-actin is a direct target of Notch/CSL. Circ Res. 2006;98:1468–1470. doi: 10.1161/01.RES.0000229683.81357.26. [DOI] [PubMed] [Google Scholar]
- Ridgway J, Zhang G, Wu Y, Stawicki S, Liang WC, Chanthery Y, Kowalski J, Watts RJ, Callahan C, Kasman I, Singh M, Chien M, Tan C, Hongo JA, de Sauvage F, Plowman G, Yan M. Inhibition of Dll4 signalling inhibits tumour growth by deregulating angiogenesis. Nature. 2006;444:1083–1087. doi: 10.1038/nature05313. [DOI] [PubMed] [Google Scholar]
- Roggendorf W, Opitz H, Schuppan D. Altered expression of collagen type VI in brain vessels of patients with chronic hypertension. A comparison with the distribution of collagen IV and procollagen III. Acta Neuropathol. 1988;77:55–60. doi: 10.1007/BF00688243. [DOI] [PubMed] [Google Scholar]
- Schouwey K, Delmas V, Larue L, Zimber-Strobl U, Strobl LJ, Radtke F, Beermann F. Notch1 and Notch2 receptors influence progressive hair graying in a dose-dependent manner. Dev Dyn. 2007;236:282–289. doi: 10.1002/dvdy.21000. [DOI] [PubMed] [Google Scholar]
- Shimizu K, Chiba S, Kumano K, Hosoya N, Takahashi T, Kanda Y, Hamada Y, Yazaki Y, Hirai H. Mouse jagged1 physically interacts with notch2 and other notch receptors. Assessment by quantitative methods. J Biol Chem. 1999;274:32961–32969. doi: 10.1074/jbc.274.46.32961. [DOI] [PubMed] [Google Scholar]
- Shrivastava A, Radziejewski C, Campbell E, Kovac L, McGlynn M, Ryan TE, Davis S, Goldfarb MP, Glass DJ, Lemke G, Yancopoulos GD. An orphan receptor tyrosine kinase family whose members serve as nonintegrin collagen receptors. Mol Cell. 1997;1:25–34. doi: 10.1016/s1097-2765(00)80004-0. [DOI] [PubMed] [Google Scholar]
- Tang Y, Urs S, Boucher J, Bernaiche T, Venkatesh D, Spicer DB, Vary CP, Liaw L. Notch and transforming growth factor-beta (TGFbeta) signaling pathways cooperatively regulate vascular smooth muscle cell differentiation. J Biol Chem. 2010;285:17556–17563. doi: 10.1074/jbc.M109.076414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Urs S, Liaw L. Hairy-related transcription factors inhibit Notch-induced smooth muscle alpha-actin expression by interfering with Notch intracellular domain/CBF-1 complex interaction with the CBF-1-binding site. Circ Res. 2008;102:661–668. doi: 10.1161/CIRCRESAHA.107.165134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyttendaele H, Ho J, Rossant J, Kitajewski J. Vascular patterning defects associated with expression of activated Notch4 in embryonic endothelium. Proc Natl Acad Sci U S A. 2001;98:5643–5648. doi: 10.1073/pnas.091584598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel W, Gish GD, Alves F, Pawson T. The discoidin domain receptor tyrosine kinases are activated by collagen. Mol Cell. 1997;1:13–23. doi: 10.1016/s1097-2765(00)80003-9. [DOI] [PubMed] [Google Scholar]
- Wang MM. Notch signaling and Notch signaling modifiers. Int J Biochem Cell Biol. 2011;43:1550–1562. doi: 10.1016/j.biocel.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue YGX, Lindsell CE, Norton CR, Chang B, Hicks C, Gendron-Maguire M, Rand EB, Weinmaster G, Gridley T. Embryonic lethality and vascular defects in mice lacking the Notch ligand Jagged1. Hum Mol Genet. 1999;1999:723–730. doi: 10.1093/hmg/8.5.723. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.