Abstract
Considerable progress has been made recently toward understanding the processes of mitochondrial DNA (mtDNA) damage and repair. However, a paucity of information still exists regarding the physiological effects of persistent mtDNA damage. This is due, in part, to experimental difficulties associated with targeting mtDNA for damage, while sparing nuclear DNA. Here, we characterize two systems designed for targeted mtDNA damage based on the inducible (Tet-ON) mitochondrial expression of the bacterial enzyme, exonuclease III, and the human enzyme, uracil-N-glyosylase containing the Y147A mutation. In both systems, damage was accompanied by degradation of mtDNA, which was detectable by six hours after induction of mutant uracil-N-glycosylase and by twelve hours after induction of exoIII. Unexpectedly, increases in the steady-state levels of single-strand lesions, which led to degradation, were small in absolute terms indicating that both abasic sites and single-strand gaps may be poorly tolerated in mtDNA. mtDNA degradation was accompanied by the loss of expression of mtDNA-encoded COX2. After withdrawal of the inducer, recovery from mtDNA depletion occurred faster in the system expressing exonuclease III, but in both systems reduced mtDNA levels persisted longer than 144h after doxycycline withdrawal. mtDNA degradation was followed by reduction and loss of respiration, decreased membrane potential, reduced cell viability, reduced intrinsic reactive oxygen species production, slowed proliferation, and changes in mitochondrial morphology (fragmentation of the mitochondrial network, rounding and “foaming” of the mitochondria). The mutagenic effects of abasic sites in mtDNA were low, which indicates that damaged mtDNA molecules may be degraded if not rapidly repaired. This study establishes, for the first time, that mtDNA degradation can be a direct and immediate consequence of persistent mtDNA damage and that increased ROS production is not an invariant consequence of mtDNA damage.
Keywords: mtDNA damage, mtDNA degradation, mtDNA repair, exoIII, UNG1 Y147A, Reactive Oxygen Species
1. Introduction
Among the organelles found in the mammalian cell, mitochondria and the nucleus are unique in that they serve as repositories for genetic information in the form of DNA. The human mitochondrial genome is a 16,569 bp circular DNA molecule, which encodes 13 polypeptide subunits of the electron transport chain, as well as 22 tRNAs and 2 rRNAs, which are required for the translation of these subunits on mitochondrial ribosomes using a genetic code distinct from that used to encode nuclear genes [1]. Therefore, mitochondrial DNA (mtDNA) integrity is critical for the proper function of the electron transport chain and ATP production through respiration.
Apart from supplying the bulk of ATP for vitally important cellular processes in most (but not all [2]) cell types [3], mitochondria serve as a major cellular source of reactive oxygen species [4, 5], play a pivotal role in the regulation of apoptosis [6, 7], and are involved in numerous other cellular functions [8]. Therefore, it comes as no surprise that mutations in mtDNA have been associated with various human pathologies, such as mitochondrial diseases [9-11], diabetes [12-14], cancer [15-18] and neurodegenerative disorders [19], just to name a few.
Moreover, it has been established that not only mtDNA mutations, but, also, a reduction in mtDNA copy number can be either pathogenic [20-22], or can negatively affect the normal function of the cell in other ways. For instance, depletion of mtDNA can cause a defect in glucose utilization [23]. Also, mtDNA copy number was strongly correlated with adipocyte lipogenesis [24]. Furthermore, mtDNA depletion induces radioresistance [25]. Finally, cells that lack mtDNA (ρ° cells) have been found to resist apoptosis [26, 27]. Surprisingly, mutation and depletion of mtDNA also were associated with increased susceptibility to apoptosis [28].
While significant progress has been achieved in understanding the effects of mutation and copy number alterations of mtDNA on cellular physiology, the effects of mtDNA damage remain poorly understood. Available evidence suggests that both acute mtDNA damage [29-31] and an imbalance in mtDNA repair [32, 33] may be associated with both apoptosis and mtDNA degradation [34] in different cell types. This uncertainty is further confounded by the fact that precise targeting of mtDNA by damaging agents remains challenging and that it is difficult to control for the contribution of incompletely understood off-target effects. Indeed, externally applied Reactive Oxygen Species (ROS), such as pure H2O2, or H2O2 generated extracellularly by enzymatic reactions, such as xanthine oxidase [35] or glucose oxidase [36], have the capacity to inflict damage upon both nuclear DNA (nDNA) and mtDNA. Even though mtDNA is preferentially targeted by ROS for strand breaks [37], it can be argued that nDNA damage is more consequential because of the high redundancy of the mitochondrial genome, and that it is nDNA damage, rather than mtDNA damage, that is responsible for the physiological effects following the exposure to ROS . Apart from indiscriminately targeting all cellular DNA, extracellular ROS may interfere with normal cellular ROS signaling, thus adding another layer of complexity. Intracellular ROS generation with the help of redox cyclers such as alloxan (in the cytosol) or menadione (in mitochondria) also is associated with off-target effects [38]. Similarly, alkylating agents target both nDNA and mtDNA (and appear to preferentially damage nDNA [39]). Moreover, while the repair of alkylation damage to mtDNA is well documented [40, 41], the observed kinetics of the repair is faster than can be explained by alkylation-mediated destabilization of the glycosidic bond [42]. The current dearth of knowledge on the molecular identity of the initiating glycosylase(s), also complicates the interpretation of the outcomes of experiments with alkylating agents.
The challenges associated with the interpretation of the outcomes of experiments, which employ conventional DNA damaging agents/protocols and aimed at the clarification of the effect of mtDNA damage on cellular function highlight the need for novel, targeted approaches for mtDNA damage with minimal off-target effects. Targeted mtDNA damage can be achieved by expressing DNA-damaging enzymes in the mitochondrial compartment, or by suppressing the activity of mitochondria-specific DNA-repair enzyme(s) thus slowing repair and increasing the steady-state level of mtDNA damage.
Here, we report that mitochondrial targeting of two different enzymes, each of which can damage mtDNA by a unique mechanism, results in mtDNA depletion, morphological abnormalities in mitochondria, suppressed respiration and slowed proliferation in HeLa cells.
2. Materials and Methods
2.1. Cells and DNA constructs. HeLa cells and their derivatives were propagated in Dulbecco’s Modified Eagle Medium (DMEM) containing 10% Fetal Bovine Serum, 50 μg/ml gentamycin, 50 μg/ml uridine, and 1 mM sodium pyruvate in a humidified atmosphere containing 5% CO2 at 37°C. For inducible expression, HeLa cells were modified by introducing a Tet-On Advanced transactivator with retrovirus rv2641 [43]. Cloning of the Escherichia coli exonuclease III (exoIII) gene has been described previously [44]. The wild type (WT) human uracil DNA glycosylase 1 (UNG1) gene was cloned by RT-PCR from total RNA isolated from HeLa cells. The first 231bp encoding the matrix targeting sequence (MTS) were removed from the WT UNG1 gene by PCR and replaced with the MTS from human ornithine transcarbamylase, followed by a myc-tag. The Y147A mutation was introduced into UNG1 by overlap extension PCR [45] using primers UNGriF (gcgaattcgccaccatgggcgtcttctgccttgg), UNGxbaR (gctctagactcacagctccttccagtcaa), UNGy147aF (gggacaggatccagcccatggacctaatca), and UNGy147aR (tgattaggtccatgggctggatcctgtccc).
2.2. Production of lentiviral supernatants and infection of target cells. Lentivirus-containing supernatants were produced by CaPO4-mediated transfection of the HEK293FT cell line, using established protocols [46]. Gag, Pol and Env functions for lentiviral constructs were provided in trans by cotransfecting the vector plasmid with two helper plasmids, psPAX2 and pMD2.G. Target cells were infected with lentiviruses in 35-mm dishes at 20% confluence by incubating them overnight with corresponding supernatant in the presence of 10 μg/mL polybrene (Sigma-Aldrich Corp., St. Louis, MO). The next day, the supernatant was removed and cells were allowed to recover for 24h in DMEM, after which cells were trypsinized, transferred into 145-mm dishes, and puromycin selection (4 μg/mL) was applied for 6 days.
2.3. Western blotting. Protein extracts from treated and control cells were prepared using lysis solution containing 10 mM Tris-HCl, 1% SDS, 1× EDTA-free protease inhibitor cocktail (Roche, Indianapolis, IN). Protein concentrations were measured using the BCA assay (Pierce, Rockford, IL, USA). Proteins were separated by PAAG electrophoresis and transferred to PVDF membranes, blocked and incubated with primary and secondary antibodies using standard techniques [47]. Blots were developed with SuperSignal West Pico and exposed to CL-Xposure film (both Pierce). Primary antibodies were α-myc tag (Cell Signaling), α-HSP60 (mitochondrial, BD Biosciences), α-cytochrome oxidase subunit 2 (AbCam).
2.4. Cellular respiration. in whole attached cells was measured with the help of an XF-24 extracellular flux analyzer (Seahorse Biosciences, Billerica, MA, U.S.A) according to the manufacturer’s recommendations and expressed as pMol/min/μg protein. ATP-linked respiration was determined with the help of oligomycin (OLIG, 5 μM), maximal respiration was induced with Carbonyl cyanide m-chlorophenyl hydrazone (CCCP, 1 μM), and non-mitochondrial respiration was determined after injection of rotenone and antimycin A (R+A, 1 μM each).
2.5. Mitochondrial membrane potential. was measured with TMRM. Briefly, cells were plated in 12-well plates, allowed to attach overnight and then were incubated with 200 nM TMRM in DMEM for 10 min at 37°C in an atmosphere of 5% CO2. After treatment, cells were trypsinized and immediately subjected to flow cytometry on a BD FACS Aria with excitation at 561 nm and an emission bandpass filter 582/15. Measurements were performed on three biological replicas. Membrane potential was expressed as arbitrary fluorescence units.
2.6. Growth curves. were generated by plating 20,000 cells/well in a series of 6-well plates, and allowing attachment overnight in the presence or absence of 4 μg/ml doxycycline, an initial cell count (Ci) was determined by trypsinization and counting of cells with a Coulter counter using one plate per series. The remaining plates were grown for another 5 days with or without 4 μg/ml doxycycline, and cell counts were determined daily using a single 6-well plate per series as described above. Doubling time was calculated according to the formula Td=T/log2(Ci/CT), where Td is doubling time and T is time in hours between initial (Ci) and final (CT) cell counts.
2.7. Mitochondrial ROS. were measured with MitoSOX Red (Invitrogen, Carlsbad, CA). For experiments which involved no longer than 24h induction, cells transduced with exoIII, wtUNG1 or mUNG1 constructs (Figure 1) were grown in 12-well plates with or without inductor (doxycycline), after which they were loaded with 5 μM MitoSOX in DMEM for 10 min at 37°C in an atmosphere of 5% CO2. After treatment, cells were trypsinized and immediately subjected to flow cytometry on a BD FACS Aria with excitation at 561 nm and an emission bandpass filter 582/15. Measurements were performed on three biological replicates. The amount of ROS produced was expressed as percent of uninduced control. For induction longer than 24h cells were grown and induced in 100-mm dishes, and plated in 12-well plates 24h prior to assay.
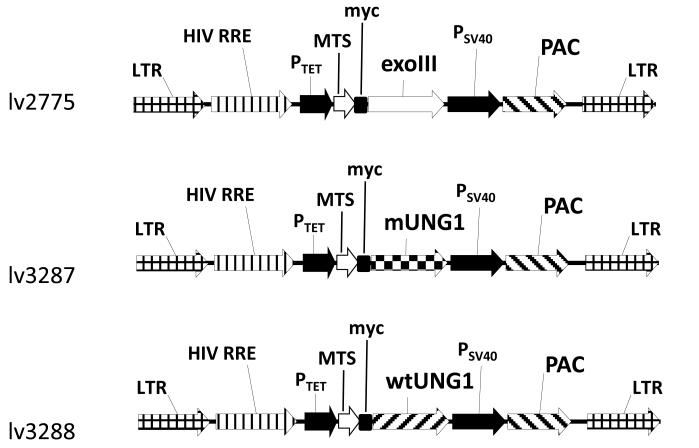
Figure 1. Vector maps.
Abbreviations: exoIII, Escherichia coli exonuclease III gene; HIV RRE, human immunodeficiency virus rev response element; LTR, lentiviral long terminal repeat; MTS, mitochondrial matrix targeting sequence of human ornithine transcarbamylase [72]; mUNG1, mutant human UNG1 Y147A gene; myc, myc tag epitope; PAC, puromycin resistance gene; PSV40, SV40 promoter; PTet, doxycycline-regulated promoter; wtUNG1, wild type human UNG1 gene; wPRE, woodchuck hepatitis virus posttranscriptional regulatory element.
2.8. Microscopy. For phase contrast imaging, cells were plated into 35 mM glass bottom MaTek dishes, allowed to attach overnight, and imaged with a Nikon TE200U microscope (10× objective). For mitochondria visualization, cells were plated as above, loaded with 120 nM MitoTracker Red for 10 min at 37°C in an atmosphere of 5% CO2, after which mitochondria were imaged with a Nikon A1R confocal microscope using a 60× water immersion objective.
2.9. Quantitative Southern Blotting. Quantitative Southern Blotting under alkaline (denaturing) conditions (QSBA) and Quantitative Southern Blotting under non-denaturing conditions (QSBN) conditions were performed essentially as described earlier [34]. Briefly, cells were lysed in a solution containing 10 mM Tris, pH7.5, 1 mM EDTA, 0.5% SDS and 300 μg/ml proteinase K, and incubated in this solution overnight at 37°C with shaking. The next day, the solution was adjusted to 1M NaCl with 5M stock solution in water, and DNA was extracted twice with chloroform:isoamyl alcohol (24:1). Then, DNA was precipitated with 2 volumes of 100% EtOH, collected by centrifugation, washed with 70% EtOH, briefly dried, and dissolved in H2O overnight at 4°C. The next day, DNA was digested with restriction enzyme (1.5 unit/μl) in the presence of RNAseA (2μg/ml) overnight at 37°C. Digested DNA was precipitated with 2 volumes of EtOH, dissolved in TE buffer overnight at 4°C, and quantitated by fluorometry. After DNA quantitation, a master mix was prepared containing 10 μg of total DNA in 45 μl of TE buffer. 20 μl aliquots of this master mix then were withdrawn and subjected to either QSBN (gel run in TBE buffer) or QSBA (electrophoresis employing an alkaline loading buffer (10mM EDTA, 25% Ficoll, 0.25% Bromcresol purple and 500mM NaOH) and a running buffer containing 23 mM NaOH and 1 mM EDTA). After blotting, the membrane was cut at the level of the 9 kb band of the lambda/HindIII marker. The upper portion then was hybridized with the mtDNA probe (detects 16,569 bp fragment), and the lower portion was hybridized with the 18S rDNA probe (detects 5,102 bp fragment). In a given experiment, QSBN and QSBA membranes were hybridized in a single roll bottle with either mtDNA probe or nDNA probe to ensure uniformity of hybridization conditions. After hybridization, membranes were exposed to an imaging screen to determine band intensity. The number of pixels per band was determined by encompassing bands with identical rectangular regions of interest and subtracting the background.
2.10. mtDNA damage. Hela/2641 cells transduced with either exoIII or mUNG1 constructs were grown either with or without 4 μg/ml doxycycline for 6h, after which cells were lysed, total DNA was isolated and subjected to QSBA and QSBN. To determine mtDNA single-strand lesion frequency (BF), intensities of mtDNA bands on QSBA and QSBN were normalized using intensities of nDNA bands (since nDNA is not expected to be damaged by mitochondrially targeted enzymes). After normalization, BF was determined using the Poisson expression (BF = −ln (QSBA/QSBN)), where QSBA and QSBN are adjusted intensities of mtDNA bands on QSBA and QSBN, respectively. Since QSBA, but not QSBN, detects single-strand lesions, QSBA/QSBN ratio is indicative of the fraction of mtDNA molecules free of single-strand lesions. BF was expressed as the number of breaks per 16,569 bp (the size of mtDNA molecule).
2.11. The percent of mtDNA remaining (a measure of mtDNA degradation) was determined using QSBN. First, the ratios of intensities of mtDNA to nDNA bands for uninduced and induced cells were determined as a surrogate for mtDNA copy number. Then, an average ratio in uninduced cells was determined from the three biological replicas and assigned a value of 100%. Finally, the percent of remaining mtDNA was expressed as a fraction of this value.
2.12. mtDNA mutagenesis. mtDNA mutation loads were determined by a PCR-cloning-sequencing approach. Total DNA was extracted with the help of a Blood and Tissue kit (Qiagen, Valencia, CA) from uninduced cells and from cells that were induced for 48h and then allowed to grow without induction for 240h to restore mtDNA levels. To ensure representativeness of the results, three different ~1 kb regions of mtDNA were amplified with primers hMT2198-3251F1 (AAAGCGTTCAAGCTCAACACCCAC) plus hMT2198-3251R1 (TACCGGGCTCTGCCATCTTAACAA), hMT7113-8210F2 (ACCCTAGACCAAACCTACGCCAAA) plus hMT7113-8210R2, and hMT12534-13612F3 (ACACTGAGCCACAACCCAAACAAC) plus hMT12534-13612R3 (TTCGAGTGCTATAGGCGCTTGTCA). Amplification of mtDNA was performed in triplicate 100 μl reactions per each independent experimental condition. Each reaction contained 50 μl of 2× Phusion high fidelity PCR master mix (Thermo Fisher Scientific Inc., Waltham, MA), 300 ng of total DNA, 1 μM of each primer, and the balance of water. PCR parameters were: initial denaturation for 1 min at 98°C, and then 25 cycles of denaturation for 10 sec at 98°C, annealing for 5 sec at 55°C, and extension for 30 sec at 55°C. PCR product was directly purified from the reaction mixture using a PCR purification kit (Qiagen, Valencia, CA) and ligated to an EcoRV-digested plasmid, pBluescriptII SK+ , overnight at 25°C. The ligated mixture was introduced into E. coli cells, allowed to incubate for 1h at 37°C and then 25% of the total mixture was diluted 1:4 and 100μl aliquots of this dilution were plated on LB agar plates containing 200 μg/ml ampicillin, 40 μg/ml X-gal and 1mM IPTG. Typically, no more than 10% of electroporated cells were plated, which ensured that no duplicates were sequenced. White colonies were picked, inoculated into LB medium in 96-well deep well plates, grown overnight with shaking at 300 rpm at 37°C, overnight cultures were mixed with equal volume of 50% glycerol in 96-well plates, frozen, and submitted for Sanger sequencing to Functional Biosciences (Madison, WI). The resulting chromatograms were aligned with human mtDNA, with the help of the SeqMan II program.
2.13. Statistical analyses. Pairwise comparisons were made using unpaired two-tailed T-test assuming unequal variance.
3. Results
3.1. Experimental system
In this study, for targeted damage of mtDNA, we utilized inducible mitochondrial expression of either E. coli exoIII or the Y147A mutant of the human UNG1 gene. The former protein comes from a prokaryotic host, where it serves as a major apurinic/apyrimidinic (AP) endonuclease, which also exhibits a 3′-repair diesterase, 3′-->5′ exonuclease, 3′-phosphomonoesterase and ribonuclease activities [48]. Previously, we demonstrated that mitochondrial expression of this bacterial DNA repair enzyme sensitizes cells to oxidative damage [44]. UNG1 is a DNA repair glycosylase that normally excises uracil from DNA. However, mutation Y147A in the active site of this enzyme has been shown to extend the specificity of this enzyme to include naturally occurring thymine [49]. Excision of thymine from DNA generates a lesion in the form of an AP site, which is mutagenic and cytotoxic in E. coli [49]. To achieve regulated expression of these enzymes in recipient cells, three lentiviral constructs were generated using a previously developed expression system [43]. lv2775 encodes exoIII fused to the MTS of the human ornithine transcarbamylase and a myc tag (further referred to as the exoIII construct), and lv3287 and lv3288 encode Y147A mutant and WT UNG1 genes, respectively fused to the same MTS and a myc tag (further referred to as mUNG1 and wtUNG1 constructs, Figure 1).
3.2. Mitochondrial expression of either mUNG1 or exoIII results in rapid loss of mtDNA
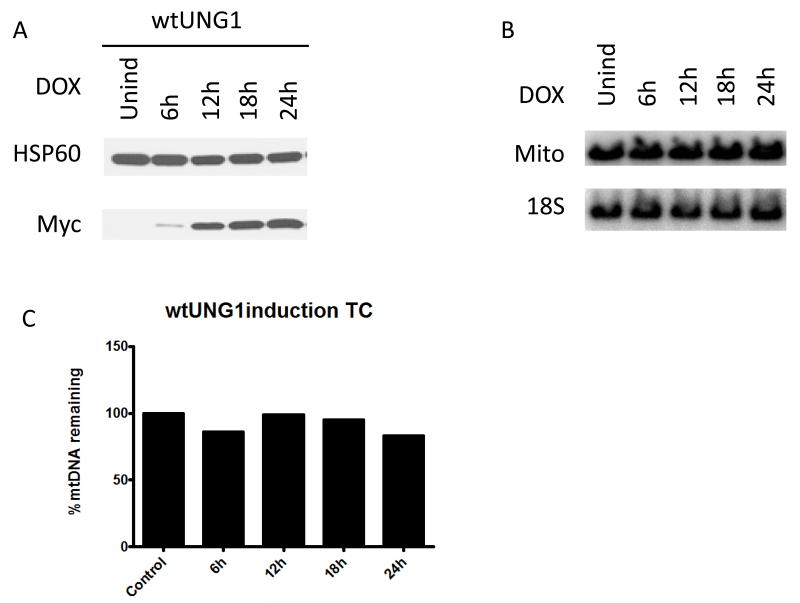
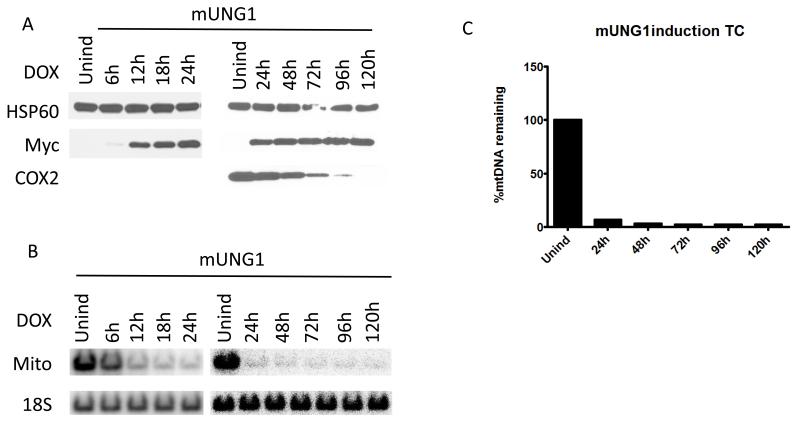
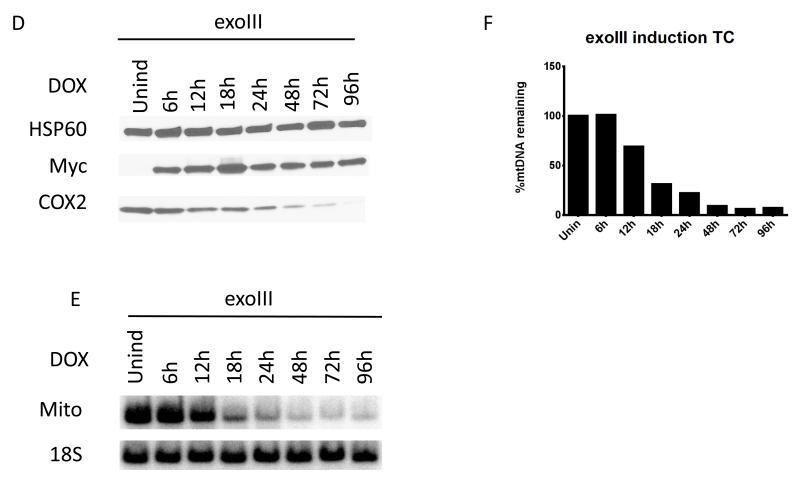
UNG1 is normally found in mitochondria [50]. Therefore we used overexpression of the wtUNG1 enzyme in the mitochondrial compartment to control for non-specific effects of induction. Upon induction of the cells infected with wtUNG1construct, recombinant protein was detectable in whole cell lysates by 6h after induction (Figure 2 A). However, this had no effect on the mtDNA content in the induced cells (Figure 2 B and C). In contrast, induction of the mUNG1, which proceeded with similar kinetics (Figure 3 A), led to a rapid loss of mtDNA (Figure 3 B and C). This loss was substantial at the earliest time point (6h) at which expression of the mUNG1was detectable in the whole cell lysates. We were unable to detect mUNG1 expression in whole cell lysates at 3h after induction (results not shown). While the kinetics of the exoIII and mUNG1 induction were similar, the loss of mtDNA upon exoIII induction was not as steep and trailed detectable protein expression by approximately 6h (Figure 3 D-F). Importantly, the loss of mtDNA in both cases was followed by a reduction in the cellular levels of the mtDNA-encoded subunit 2 of cytochrome oxidase (COX2, part of the mitochondrial respiratory complex IV, see Figure 3). The delay between the loss of mtDNA and reduction in cellular COX 2 content can be explained by the fact that both the COX2 transcripts and the protein need to be degraded and/or diluted by proliferation of the cells before steady-state levels of this protein in cells decreases.
Figure 2. Overexpression of the wtUNG1 in mitochondria has no effect on mtDNA content.
The time course (TC) of the wtUNG1 (A) induction visualized by Western blotting for the myc tag, mtDNA content in induced cells visualized with QSBN (B), and quantitation of these changes as ratio of intensities of mtDNA/nDNA bands (C). HSP60, mitochondrial heat shock protein 60 kDa; myc, myc tag; Mito, mtDNA; 18S, 18S (nuclear) DNA.
Figure 3. Targeting of both mUNG1 and exoIII to mitochondria results in the rapid loss of mtDNA.
The time course of mUNG1 (A) and exoIII (D) induction, corresponding changes in mtDNA content in induced cells visualized with QSBN (B and E, respectively), and quantitation of these changes (C and F, respectively). Quantitative data from separate QSBNs corresponding to 0-24h and 24-120h time courses were combined in a single graph in C. HSP60, mitochondrial heat shock protein 60 kDa; myc, myc tag; COX2, mitochondrial complex IV (cytochrome oxidase) subunit 2 encoded by mtDNA; Mito, mtDNA; 18S, 18S (nuclear) DNA.
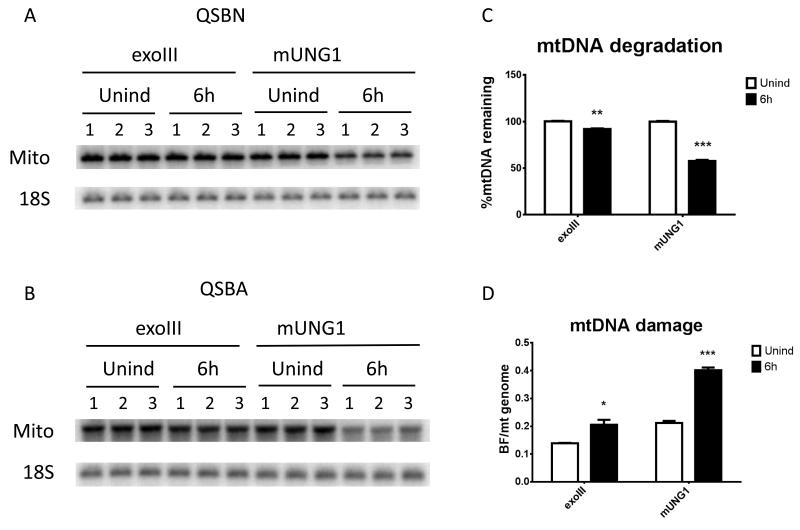
3.3. mtDNA degradation is associated with increased frequency of single-strand lesions
One important question pertinent to damage-mediated mtDNA degradation is what steady-state levels of lesions in mtDNA can induce degradation. To address this, exoIII or mUNG1were either induced or not for 6h, and steady-state levels of mtDNA lesions were determined using a combination of QSBA and QSBN as described in the Materials and Methods. The 6h time point was chosen because preliminary experiments (Figure 3) suggested that at this time point cells expressing exoIII will just begin degrading their mtDNA, while cells expressing mUNG1will lose more than 40% of their mtDNA, and both cell lines will be on their way to degrade more mtDNA, i.e. the steady-state level of mtDNA lesions in cells expressing each enzyme will favor degradation. Indeed, at 6h after induction cells expressing exoIII and mUNG1have lost virtually no and 43% of mtDNA, respectively despite the fact that exoIII was expressed at a higher level at this time point (Figure 4). Concomitantly, a statistically significant increase in the frequency of single-strand lesions was observed in both types of cells (Figure 4).
Figure 4. mtDNA degradation is associated with increased frequency of single-strand lesions.
Cells transduced with exoIII or mUNG1 constructs were either induced or not for 6h, total DNA was extracted, and subjected to QSBN (A) and QSBA (B). Unind, uninduced; 6h, cells induced for 6h; Mito, mtDNA band; 18S, 18S rDNA (nuclear) band. C, mtDNA degradation in cells expressing either exoIII or mUNG1 constructs with and without induction for 6h. D, Frequency of mitochondrial single-strand lesions in induced (6h) or uninduced (Unind) cells transduced with either exoIII or mUNG1 constructs. Error bars, SEM. *, P<0.05; **, P<0.01; ***, P<0.001, unpaired T-rest assuming unequal variance with Holm-Sidak correction for multiple comparisons.
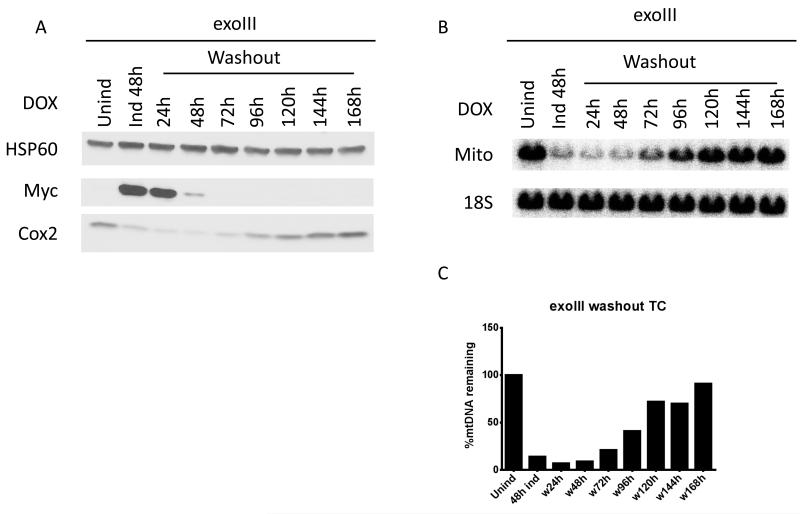
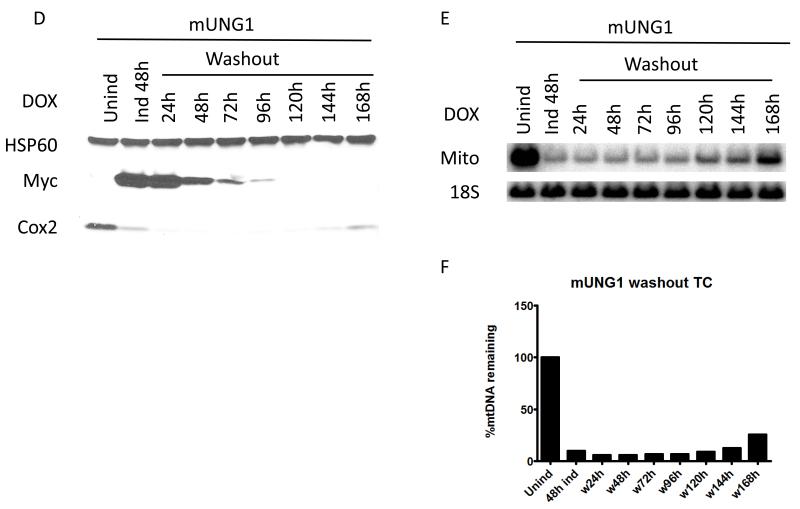
3.4. Recovery of the mtDNA copy number is delayed after the removal of the inducer
To study the recovery of mtDNA levels after degradation induced by mitochondrial expression of exoIII and mUNG1, cells transduced with corresponding constructs were first induced for 48h, and then were grown in medium without inducer for up to 168h. In order for mtDNA copy number to be restored, cellular levels of either exoIII or mUNG1 had to drop below those that induce degradation at rates higher than the mtDNA replication rate. In the absence of inducer (i.e. in the absence of de novo synthesis of mtDNA-damaging proteins), this is achieved by turning over both message and protein for exoIII or mUNG1 and by diluting the remaining proteins through cell proliferation. Upon doxycycline withdrawal, the levels of exoIII in whole cell lysates dropped below detection levels by 72h. In contrast, mUNG1 persisted in cells 48h longer (Figure 5 A and D). Since proliferation of cells expressing these two proteins occurred at similar rates (see below), this observation indicates higher stability of either mUNG1 message or the protein, or both. The kinetics of the recovery of mtDNA-encoded COX2 protein paralleled that of mtDNA recovery regardless of the protein used to induce depletion (Figure 5). However, this recovery occurred faster in cells transduced with the exoIII construct. This was facilitated by both the faster loss of exoIII protein and by a longer delay between the disappearance of the damaging enzyme and the initiation of recovery (24h for exoIII vs. 48h for mUNG1). Importantly, recovery of mtDNA levels in cells transduced with mUNG1 was incomplete even after 168h of doxycycline withdrawal.
Figure 5. Recovery of the mtDNA copy number after removal of the inducer.
Cells transduced with either exoIII or mUNG1 constructs were first induced with 4 μg/ml doxycycline for 48h, and then grown without inducer. A and D, changes in protein levels. B,C,E and F, changes in mtDNA content. Labels are as in the Figure 3.
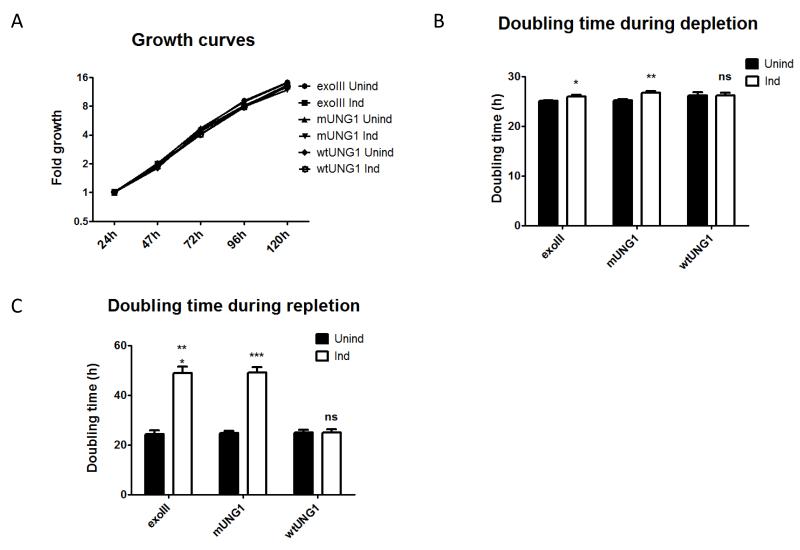
3.5. mtDNA depletion reduces the rate of cell proliferation
During the first 120h of depletion, the growth rate of induced cells was similar to that of uninduced cells for all three lentiviral constructs, as evidenced by the growth curves (Figure 6A). Nevertheless, doubling times in cells expressing exoIII and mUNG1, but not wtUNG1, were statistically greater than in the corresponding uninduced cells, although the difference was less than 10% (Figure 6B). When cells, which were either induced or not for 120h, were allowed to recover in medium without inducer, the difference in doubling times between induced and uninduced cells became dramatic. Cells, which were induced for the expression of exoIII or mUNG1 were proliferating at half the rate of their uninduced counterparts. Importantly, the doubling time in cells transduced with a construct for wtUNG1 remained the same, regardless of prior history of induction, suggesting that the difference in growth rates is attributable to mtDNA depletion (Figure 6C).
Figure 6. mtDNA depletion reduces the rate of cell proliferation.
Growth curve during depletion (A) and doubling time during depletion (B) and repletion (C) in cells transduced with exoIII, wtUNG1, or mUNG1 constructs. A, cells were plated at 20,000/well in 6-well plates, and grown with or without 4 μg/ml doxycycline for up to 5 days. Cell counts were determined daily by trypsinization. B, doubling time is average over 96h of depletion; C, doubling time is average over 120h repletion after 120h depletion. Error bars, SEM. *, P<0.05; **, P<0.01; ***, P<0.001; ns, not significant.
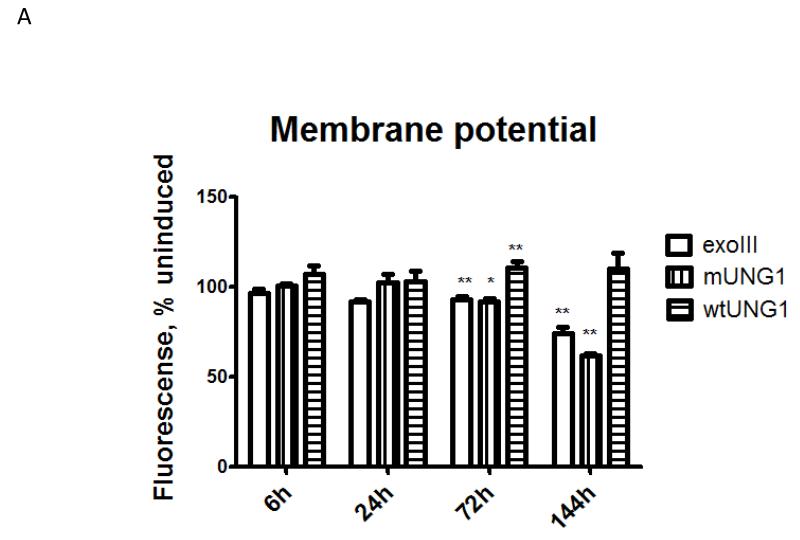
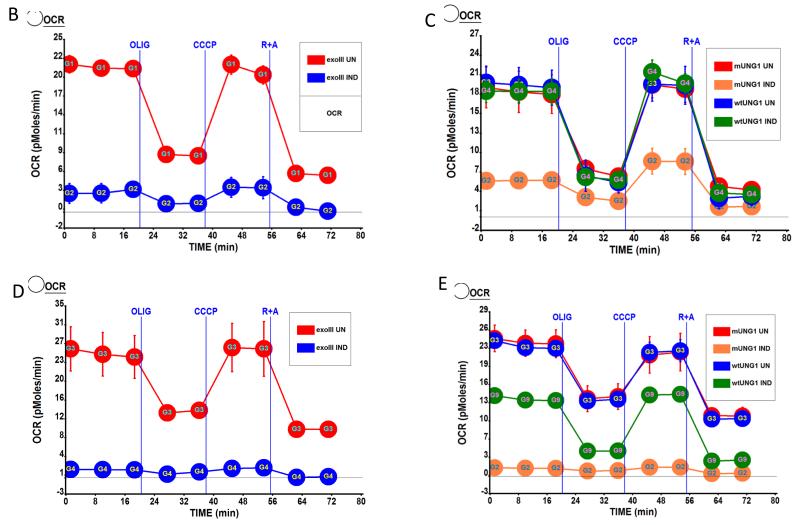
3.6. Prolonged mtDNA depletion reduces respiration and membrane potential
mtDNA encodes at least one of the subunits of each of the three respiratory complexes involved in the generation of mitochondrial membrane potential. Therefore, it is reasonable to expect that mtDNA depletion may result in the reduction of mitochondrial membrane potential. Nevertheless, the relationship between mitochondrial membrane potential and mtDNA-encoded subunits of the respiratory chain is not straightforward, as it has been reported that cells devoid of mtDNA (ρ° cells) may still maintain either residual [51] or full [52] membrane potential. In our experimental paradigm, induction of mtDNA – damaging enzymes did not affect membrane potential for at least 24h after induction (Figure 7 A), which is consistent with only a slight drop in mtDNA-encoded COX2 protein at 24h after induction (Figure 3 A and D). However, mitochondrial membrane potential was significantly reduced in cells expressing exoIII and mUNG1, but not in cells expressing wtUNG1, at 72h and 144h after induction (Figure 7 A), which is consistent with Western blotting, which shows a reduction of COX2 expression at these time points (Figure 3 and results not shown). Also, mitochondrial expression of both mtDNA-damaging enzymes, but not wtUNG1 led to a reduction in both baseline and CCCP-induced maximal respiration at 72h and 120h after induction (Figure 7 B-E).
Figure 7. Effect of mtDNA depletion on mitochondrial membrane potential and respiration.
A, mitochondrial membrane potential in cells transduced with three different constructs and induced or not for indicated periods of time. Error bars, SEM. Significant changes as compared to uninduced control are designated as follows: *, P<0.05; **, P<0.01; n≥3. B-E, respiratory profiles of the cells transduced with three different constructs and induced or not for 72h (B and C) or 120h (D and E) as measured by XF-24 analyzer. Oxygen consumption expressed in pMoles O2/min/μg protein. UN, uninduced; IND, induced; Olig, ATP synthase (complex V) inhibitor oligomycin, 5 μM; CCCP, uncoupler carbonyl cyanide m-chlorophenyl hydrazine 1 μM; R+A, rotenone (complex I inhibitor) plus antimycin A (complex III inhibitor), 1 μM each.
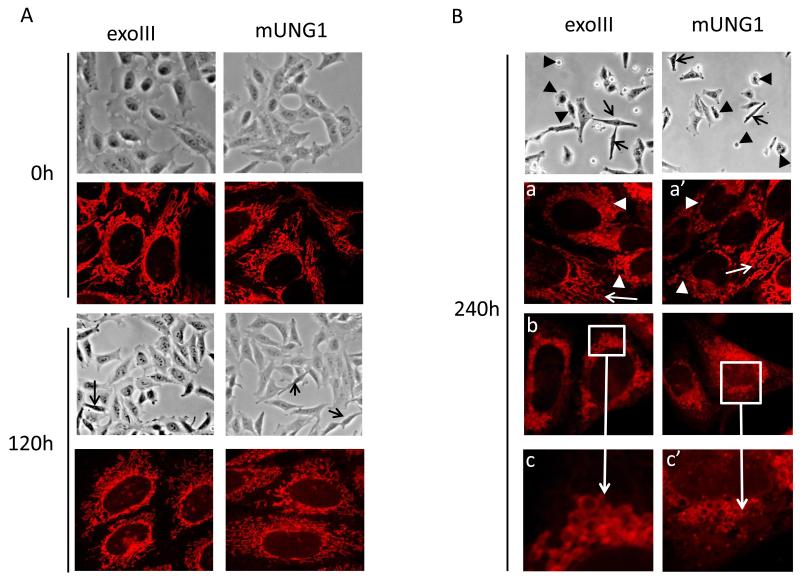
3.7. Effect of mtDNA depletion on mitochondrial morphology and ROS production
In some instances, complete loss of mtDNA may lead to alterations in mitochondrial morphology [53, 54]. To determine whether mtDNA damage and associated depletion may lead to similar consequences, we employed a combination of phase contrast microscopy to evaluate changes in overall cell health, and laser scanning confocal microscopy to evaluate corresponding alterations in mitochondrial morphology over the time course of induction of exoIII and mUNG1. Even though induction led to a rapid loss of mtDNA, no substantial changes in either cellular or mitochondrial morphology were observed for the first 4 days of induction. By 120h after induction, cells expressing both constructs remained viable, although the proportion of elongated cells with smaller, retracted lamellipodia increased (Figure 8A, black arrows). Correspondingly, an accumulation of cells with fragmented (shorter) mitochondria was observed. At 240h after induction, cultures of cells expressing either mtDNA damaging protein displayed substantial number of floating dead cells and cells displaying membrane blebbing, while elongated cells with retracted lamellipodia persisted (Figure 8B, black arrows and black arrowheads, respectively). While cells displaying normal or nearly normal mitochondrial morphology were present in substantial numbers (Figure 8B, panels a and a’, white arrows), often neighbouring cells with completely fragmented and rounded mitochondria were observed (Figure 8B, panels a and a’, white arrowheads). Overall, the proportion of cells with fragmented mitochondria, as well as cells in which mitochondria failed to take up the dye and, instead, the cytosol was stained uniformly, was substantially increased at this time point. Finally, the 240h time point was characterized by the presence of cells with “foamy” mitochondria (Figure 8B, panels b and b’, magnified in c and c’). While the exact mechanism leading to the change in mitochondrial morphology, which accompanies mtDNA loss remains to be identified, shortening and rounding up of mitochondria appears to be a general phenomenon associated with mtDNA loss [55-58].
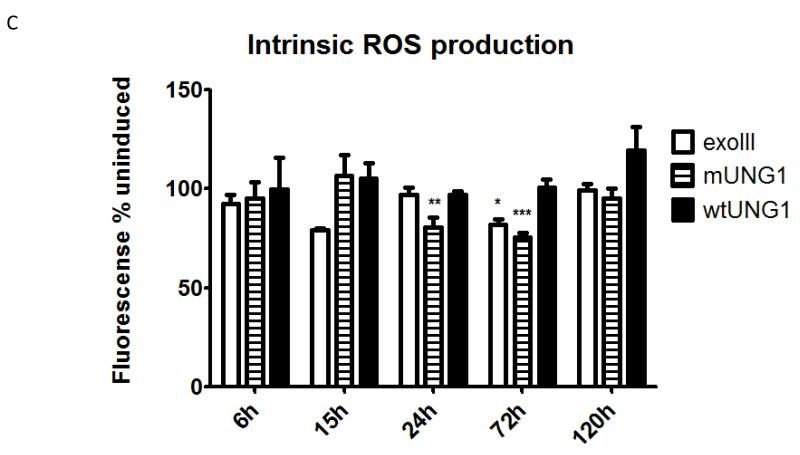
Figure 8. Effect of mtDNA depletion on cellular and mitochondrial morphology, and on the production of ROS.
A and B, cellular and mitochondrial morphology in uninduced cells and in cells induced for various periods of time. Black arrows at 120h, elongated cells with retracted lamellipodia. Black arrowheads, floating cells and cells undergoing membrane blebbing. White arrows, normal elongated mitochondria. White arrowheads, fragmented and rounded mitochondria. White boxes in b and b’ are areas with “foamy” mitochondria magnified in c and c’. C, effect of mtDNA damage and depletion on ROS production. MitoSOX fluorescence intensities of induced cells expressed as percent of the corresponding uninduced controls. Error bars, SEM. Significant changes as compared to uninduced control are designated as follows: *, P<0.05; **, P<0.01; n≥3.
Recently, it was reported that SSB in mtDNA may lead to increased ROS production [59]. In our experimental system, mUNG1 is expected to generate mtDNA damage in the form of abasic sites followed by SSBs, while exoIII may generate both SSBs and single-strand gaps. Therefore, we evaluated ROS production in induced and uninduced cells. Unexpectedly, mtDNA damage and depletion up to 24h after induction did not result in alterations in ROS production (Figure 8C and results not shown). However, at 24h after induction, cells expressing mUNG1 and at 72h after induction, cells expressing either exoIII or mUNG1, but not cells expressing wtUNG1, showed a decrease in ROS production (Figure 8 C). Temporarily, changes in ROS production by induced and uninduced cells better correlated with changes in membrane potential (Figure 7A), than with mtDNA loss. As expected, the cells responded to treatment with 100 μM menadione or with 5 μM rotenone by increasing ROS production (results not shown). However, this increased ROS production also was attenuated in cells transduced with exoIII or mUNG1 and induced for 120h or longer (not shown).
3.8. Persistent mtDNA damage has low mutagenicity
A quick and dramatic reduction in mtDNA copy number indicates a fairly uniform accessibility of the majority of mtDNA molecules to damaging enzymes. In our previous study, we proposed that mtDNA degradation is induced when mtDNA repair machinery is overwhelmed with damage [34]. Many DNA polymerases are able to bypass an abasic site by inserting predominantly (but not exclusively) an A across from the lesion [60]. However, yeast DNA polymerase Rev1 inserts predominantly a C [61, 62], DNA polymerase | (pol |) prefers a G [63]. Another study reported that pol η preferentially inserts T and A, pol | inserts T, G, and A, and pol κ inserts C and A across from the abasic site [61]. Mitochondrial pol γ, in vitro, conforms to the A-rule (inserts predominantly an A) [64]. So far, in vivo adherence of pol γ to this rule has not been tested. Therefore, we induced cells transduced with mUNG1 for 48h to inflict damage, and then grew these cells without induction for 240h to allow for a slow turnover and dilution of the damaging enzyme by cell proliferation followed by restoration of mtDNA levels. After that, the mtDNA mutation loads were determined in induced and uninduced cells by PCR-cloning-sequencing using high-fidelity Phusion DNA polymerase. The baseline mtDNA mutation load in uninduced cells was found to be 0.37 mutations/mtDNA molecule, similar to that reported earlier [34]. In cells, which underwent induction and recovery, this load increased 2.5 fold, to 0.94 mutations/mtDNA molecule. However, this increase was not statistically significant (P=0.4444, two-tailed paired t-test assuming unequal variance, n=3), a finding consistent with previous observations in vivo [65]. Mutation load in uninduced cells transduced with the exoIII construct was 0.56 mutations/mtDNA molecule. Surprisingly, no mutations were detected among 94,810 bp sequenced from these cells after inducion-recovery (mutation load below 0.17 mutations/mtDNA molecule). However, in this case, too, the increase was not statistically significant (P=0.1861), two-tailed paired t-test assuming unequal variance, n=3).
4. Discussion
While the effects of total loss of mtDNA (ρ° phenotype) on cellular physiology are relatively well described, the effects and consequences of persistent mtDNA damage remain poorly understood. This is due, in part, to difficulties associated with targeting damage to mtDNA, while sparing nuclear DNA. In previous studies, this specificity has been achieved with the expression of mitochondrially targeted restriction endonucleases. The reported effect was, invariably, either a shift from heteroplasmy to a mtDNA species, which is not recognized by the restriction endonuclease [66, 67], or a total loss of mtDNA if such species do not exist [58]. However, double-strand breaks are rarely induced in mtDNA under physiological conditions. The predominant types of physiological mtDNA damage are to bases, the sugar phosphate backbone, and single-strand breaks, which can be either directly induced in mtDNA, or can arise in the course of cellular processing of base or sugar phosphate backbone damage.
Recently, Lauritzen et al. used regulated expression of the mitochondrially targeted mUNG1 to induce abasic sites in the mitochondrial genome in mouse brain neurons in vivo [65]. However, the kinetics and extent of mtDNA loss in those studies could not be fully characterized [68], due in part to the intrinsically low temporal resolution of the in vivo systems and in part to the fact that the brain represents a complex mixture of neuronal (expressing mUNG1) and glial (not expressing mUNG1) cells [69], whose separation was not achieved in that study.
In the present study, we employed regulated mitochondrial targeting of two unrelated enzymes, bacterial exoIII and the Y147A mutant of the UNG1, to study the effects of persistent mtDNA damage on cellular physiology. mtDNA damage induced a rapid loss of mtDNA with 43% of the mtDNA lost by 6h after induction of mUNG1. In contrast, mtDNA levels were reduced by only 31% after 12h of induction in cells expressing exoIII. This slower kinetics for mtDNA loss in cells expressing exoIII may reflect differences in the mechanism of induction of mtDNA damage. While mUNG1 can attack virtually any accessible region of mtDNA and induce damage by cleaving off thymines thus creating abasic sites, exoIII can only act at the sites of existing damage (SSB or abasic sites), or by antagonizing mitochondrial pol γ at SSBs thus creating single-strand gaps. Therefore, mUNG1 may be more efficient at depleting mtDNA, due to the higher availability and number of targets. In both cases, initiation of mtDNA double-strand breaks (and, therefore, degradation [34]) can be mediated either by closely apposed single-strand lesions on complementary DNA strands, or by stalled DNA or RNA polymerase.
With both exoIII and mUNG1 the rate of mtDNA loss was faster than what would be expected to occur through a damage-induced mtDNA replication block followed by a dilution of the remaining mtDNA molecules in the process of cell proliferation. Indeed, the doubling time in cells expressing exoIII and mUNG1 is at least 24h (Figure 6B). However, less than 25% and 5% of mtDNA remained in cells after 24h induction of exoIII and mUNG1, respectively (Figure 3). Therefore, mtDNA degradation is responsible, at least in part, for the reduction in cellular mtDNA copy number following induction of exoIII and mUNG1.
At 6h after induction, mtDNA degradation was under way in cells expressing exoIII or mUNG1. This indicates that steady-state levels of mtDNA lesions were higher than those, which can be tolerated without degradation. Indeed, single-strand lesion frequency was increased in both types of cells. However, the low magnitude of this increase (0.07 and 0.19 lesions per mtDNA molecule for exoIII and mUNG1, respectively) indicates that the lesions induced in our experimental system are poorly tolerated if not rapidly repaired, and efficiently channel mtDNA for degradation.
After the removal of the inducer, exoIII and mUNG1 persisted in cells for extended periods of time delaying the restoration of mtDNA levels. In the first five days, the damage and loss of mtDNA had a small, but statistically significant effect on cell proliferation. However, when cells were allowed to recover 120h without doxycycline, the doubling time increased twofold. This can be explained by both depletion of the mtDNA - encoded polypeptides and by increased cell death during this period.
A recent report employed downregulation of EXOG to induce persistent SSB in mtDNA. That study suggested that a fairly modest persistent mtDNA damage may lead to a substantial increase in the production of ROS and apoptosis in HeLa cells [59]. In the present report, mtDNA damage and degradation had no effect on ROS production for up to 72h, after which ROS production, mitochondrial membrane potential, and cellular respiration were all significantly decreased. Increased cell death was observed in both studies. However, in our experimental system, it became substantial much later than the latest time point examined in the previous report. Further studies are needed to establish the reason for this discrepancy. Nevertheless, several differences stand out. First, in our system damage occurred earlier during treatment. Moreover, the chemical nature of the lesions is different in these two systems. Finally, EXOG, like other components of the mtDNA repair machinery [70], may play a role in more than one cellular process. Reported examples of “eclipsed” subcellular distribution of proteins [71] leave open the possibility that a fraction of the EXOG may still be localized to another cellular compartment, e.g. the nucleus. Finally, the eventual reduction in ROS production in our experimental system was likely caused by reduced expression of mtDNA-encoded subunits of the respiratory chain, which led to an overall reduction in the cellular abundance of respiratory complexes.
While mtDNA damage and degradation in this study were associated with both reduced viability and alterations in mitochondrial morphology, the substantial delay between the loss of the bulk of mtDNA and the onset of these events suggests that they are not an immediate consequence of mtDNA damage, but rather are secondary to the loss of expression of mtDNA encoded genes.
DNA repair is an error-prone process. We have reported previously that acute oxidative damage to mtDNA is associated with both repair and degradation of damaged mtDNA molecules, while mutagenic consequences appear to be minimal [34]. Here, we report that in a very different experimental paradigm of mtDNA damage and repair using mUNG1 the mutagenic outcome is still modest (2.5-fold increase over the baseline), considering that mtDNA was subjected to persistent damage over at least 9 days. The magnitude of damage varied over this period in parallel with the changing cellular content of mutagenic enzymes thus providing “optimization” of the damage. This low mutagenicity has at least three possible explanations. First, pol γ in vivo may strictly adhere to the A-rule. Therefore, abasic site bypass may not lead to a mutation because a correct residue (A) is inserted opposite an abasic site generated by the cleavage of a T. This notion is consistent with observations in vitro [64]. The second explanation is that regardless of how strictly the A-rule is observed in mitochondria, pol γ fails to efficiently extend the nascent DNA strand beyond the lesion, and terminates either before the abasic site, or at it. The inability of the pol γ to synthesize DNA beyond the lesion may result in the degradation of the damaged mtDNA molecule. This explanation is also supported by observations in vitro [64]. The third possibility is that once mUNG1 gains access to the mtDNA molecule, it inflicts so much damage (because of the abundance of the substrate), that the affected mtDNA molecule degrades. These three mechanisms are not mutually exclusive, and may operate concomitantly.
In contrast to mUNG1, lesions induced in mtDNA by exoIII (single-strand gaps) are not promutagenic. This may explain our observation that mtDNA mutagenesis is not increased upon induction-recovery of cells transduced with exoIII construct.
In general, despite substantial differences in the experimental systems, our findings with mUNG1 are in a very good agreement with those of Lauritzen et al. [65, 68]. Differences in the kinetics and amplitude of changes notwithstanding, the loss of mtDNA, cell death, the lack of mtDNA mutagenesis, reduced respiration, and the reduction in expression of mtDNA-encoded subunits were observed in both studies.
Conclusions
Our data from two different enzymatic systems indicate that persistent damage can lead to mtDNA degradation in the absence of increased ROS production. Persistence of both abasic sites and single-strand gaps in mtDNA is poorly tolerated, and a small increase in the steady-state levels of these lesions channels mtDNA to degradation. In spite of persistent and, as judged by degradation, substantial mtDNA damage, mutagenicity of abasic sites in mtDNA vivo appears to be low. The changes in the membrane potential, mitochondrial morphology, doubling time and cell viability are secondary to the reduced expression of the mtDNA-encoded polypeptides as a result of mtDNA damage and degradation. Overall, regulated mitochondrial expression of mUNG1 and exoIII provides a unique system for targeted mtDNA damage, while sparing nDNA, and enables precise elucidation of the physiological effects of persistent mtDNA damage and of mtDNA depletion. Although not shown here explicitly, persistent mtDNA damage may provide a novel approach to generating mtDNA-less (ρ°) cells.
Two enzymatic systems for regulated and targeted mtDNA damage are characterized
mtDNA damage is inflicted by mitochondrial targeting of either exoIII or mUNG1 mutant
mtDNA damage is immediately followed by degradation
mtDNA damage by mUNG1 and exoIII has low mutagenicity
Observed physiological changes are secondary to reduced expression of mitochondrial polypeptides
Acknowledgments
The authors acknowledge excellent technical assistance by Victoriya Pastukh, Luanne Oliveira, and Larysa Yuzefovich. This study was supported by the National Institutes of Health grants ES03456, PO1 HL66299 and RR031286/OD010944. The purchase and operation of the Nikon A1R confocal system was supported by NIH grant S10RR027535. The funding sources had no role in the study design, in the collection, analysis and interpretation of data, in the writing of the report, or in the decision to submit the article for publication.
Abbreviations
- CCCP
carbonyl cyanide m-chlorophenyl hydrazine
- COX2
cytochrome oxidase subunit 2
- exoIII
Escherichia coli exonuclease III
- HSP60
mitochondrial heat shock protein, 60 kDa
- mtDNA
mitochondrial DNA
- MTS
matrix targeting sequence
- mUNG1
mutant Y147A human uracil DNA glycosylase 1
- nDNA
nuclear DNA
- OLIGO
oligomycin
- pol
DNA polymerase
- QSBN
Quantitative Southern Blotting under the non-denaturing conditions
- R+A
rotenone and antimycin A
- ROS
reactive oxygen species
- SSB
single strand breaks
- wtUNG1
wild type human uracil DNA glycosylase 1
- WT
wild type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Contributor Information
Inna N. Shokolenko, Department of Cell Biology and Neuroscience, University of South Alabama, Mobile, AL (USA) 36688. Tel (251) 460-6772, Fax (251) 460-6771 ishokolenko@southalabama.edu.
Glenn L. Wilson, Department of Cell Biology and Neuroscience, University of South Alabama, Mobile, AL (USA) 36688. Tel (251) 460-6765, Fax (251) 460-6771 gwilson@usouthal.edu
Mikhail F. Alexeyev, Department of Cell Biology and Neuroscience, University of South Alabama, Mobile, AL (USA) 36688..
References
- [1].Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- [2].Noll T, Wissemann P, Mertens S, Krutzfeldt A, Spahr R, Piper HM. Hypoxia tolerance of coronary endothelial cells. Adv Exp Med Biol. 1990;277:467–476. doi: 10.1007/978-1-4684-8181-5_52. [DOI] [PubMed] [Google Scholar]
- [3].Zu XL, Guppy M. Cancer metabolism: facts, fantasy, and fiction. Biochemical and biophysical research communications. 2004;313:459–465. doi: 10.1016/j.bbrc.2003.11.136. [DOI] [PubMed] [Google Scholar]
- [4].Alexeyev MF. Is there more to aging than mitochondrial DNA and reactive oxygen species? FEBS J. 2009;276:5768–5787. doi: 10.1111/j.1742-4658.2009.07269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Murphy MP. How mitochondria produce reactive oxygen species. The Biochemical journal. 2009;417:1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Carlsen O. Root complex and root canal system: a correlation analysis using one-rooted mandibular second molars. Scandinavian journal of dental research. 1990;98:273–285. doi: 10.1111/j.1600-0722.1990.tb00973.x. [DOI] [PubMed] [Google Scholar]
- [7].Wang C, Youle RJ. The role of mitochondria in apoptosis*. Annu Rev Genet. 2009;43:95–118. doi: 10.1146/annurev-genet-102108-134850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Wallace DC. A Mitochondrial Paradigm of Metabolic and Degenerative Diseases, Aging, and Cancer: A Dawn for Evolutionary Medicine. Annu Rev Genet. 2005 doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ylikallio E, Suomalainen A. Mechanisms of mitochondrial diseases. Ann Med. 2011 doi: 10.3109/07853890.2011.598547. [DOI] [PubMed] [Google Scholar]
- [10].Schapira AH. Mitochondrial diseases. Lancet. 2012 doi: 10.1016/S0140-6736(11)61305-6. [DOI] [PubMed] [Google Scholar]
- [11].Zheng J, Ji Y, Guan MX. Mitochondrial tRNA mutations associated with deafness. Mitochondrion. 2012 doi: 10.1016/j.mito.2012.04.001. [DOI] [PubMed] [Google Scholar]
- [12].Maassen JA, Janssen GM, t Hart LM. Molecular mechanisms of mitochondrial diabetes (MIDD) Ann Med. 2005;37:213–221. doi: 10.1080/07853890510007188. [DOI] [PubMed] [Google Scholar]
- [13].Bannwarth S, Abbassi M, Valero R, Fragaki K, Dubois N, Vialettes B, Paquis-Flucklinger V. A Novel Unstable Mutation in Mitochondrial DNA Responsible for Maternally Inherited Diabetes and Deafness. Diabetes Care. 2011 doi: 10.2337/dc11-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Supale S, Li N, Brun T, Maechler P. Mitochondrial dysfunction in pancreatic beta cells. Trends Endocrinol Metab. 2012 doi: 10.1016/j.tem.2012.06.002. [DOI] [PubMed] [Google Scholar]
- [15].Kurelac I, Mackay A, Lambros MB, Di Cesare E, Cenacchi G, Ceccarelli C, Morra I, Melcarne A, Morandi L, Calabrese FM, Attimonelli M, Tallini G, Reis-Filho JS, Gasparre G. Somatic complex I disruptive mitochondrial DNA mutations are modifiers of tumourigenesis that correlate with low genomic instability in pituitary adenomas. Hum Mol Genet. 2012 doi: 10.1093/hmg/dds422. [DOI] [PubMed] [Google Scholar]
- [16].Larman TC, Depalma SR, Hadjipanayis AG, Protopopov A, Zhang J, Gabriel SB, Chin L, Seidman CE, Kucherlapati R, Seidman JG. Spectrum of somatic mitochondrial mutations in five cancers. Proc Natl Acad Sci U S A. 2012;109:14087–14091. doi: 10.1073/pnas.1211502109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Wallace DC. Mitochondria and cancer. Nat Rev Cancer. 2012;12:685–698. doi: 10.1038/nrc3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Yu M. Somatic mitochondrial DNA mutations in human cancers. Adv Clin Chem. 2012;57:99–138. doi: 10.1016/b978-0-12-394384-2.00004-8. [DOI] [PubMed] [Google Scholar]
- [19].Milone M. Mitochondria, diabetes, and Alzheimer’s disease. Diabetes. 2012;61:991–992. doi: 10.2337/db12-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Clay Montier LL, Deng JJ, Bai Y. Number matters: control of mammalian mitochondrial DNA copy number. J Genet Genomics. 2009;36:125–131. doi: 10.1016/S1673-8527(08)60099-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Rotig A, Poulton J. Genetic causes of mitochondrial DNA depletion in humans. Biochimica et biophysica acta. 2009;1792:1103–1108. doi: 10.1016/j.bbadis.2009.06.009. [DOI] [PubMed] [Google Scholar]
- [22].Arnaudo E, Dalakas M, Shanske S, Moraes CT, DiMauro S, Schon EA. Depletion of muscle mitochondrial DNA in AIDS patients with zidovudine-induced myopathy. Lancet. 1991;337:508–510. doi: 10.1016/0140-6736(91)91294-5. [DOI] [PubMed] [Google Scholar]
- [23].Park SY, Choi GH, Choi HI, Ryu J, Jung CY, Lee W. Depletion of mitochondrial DNA causes impaired glucose utilization and insulin resistance in L6 GLUT4myc myocytes. J Biol Chem. 2005;280:9855–9864. doi: 10.1074/jbc.M409399200. [DOI] [PubMed] [Google Scholar]
- [24].Kaaman M, Sparks LM, van Harmelen V, Smith SR, Sjolin E, Dahlman I, Arner P. Strong association between mitochondrial DNA copy number and lipogenesis in human white adipose tissue. Diabetologia. 2007;50:2526–2533. doi: 10.1007/s00125-007-0818-6. [DOI] [PubMed] [Google Scholar]
- [25].Cloos CR, Daniels DH, Kalen A, Matthews K, Du J, Goswami PC, Cullen JJ. Mitochondrial DNA depletion induces radioresistance by suppressing G2 checkpoint activation in human pancreatic cancer cells. Radiat Res. 2009;171:581–587. doi: 10.1667/RR1395.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kim JY, Kim YH, Chang I, Kim S, Pak YK, Oh BH, Yagita H, Jung YK, Oh YJ, Lee MS. Resistance of mitochondrial DNA-deficient cells to TRAIL: role of Bax in TRAIL-induced apoptosis. Oncogene. 2002;21:3139–3148. doi: 10.1038/sj.onc.1205406. [DOI] [PubMed] [Google Scholar]
- [27].Park SY, Chang I, Kim JY, Kang SW, Park SH, Singh K, Lee MS. Resistance of mitochondrial DNA-depleted cells against cell death: role of mitochondrial superoxide dismutase. J Biol Chem. 2004;279:7512–7520. doi: 10.1074/jbc.M307677200. [DOI] [PubMed] [Google Scholar]
- [28].Liu CY, Lee CF, Hong CH, Wei YH. Mitochondrial DNA mutation and depletion increase the susceptibility of human cells to apoptosis. Annals of the New York Academy of Sciences. 2004;1011:133–145. doi: 10.1007/978-3-662-41088-2_14. [DOI] [PubMed] [Google Scholar]
- [29].Ruchko M, Gorodnya O, LeDoux SP, Alexeyev MF, Al-Mehdi AB, Gillespie MN. Mitochondrial DNA damage triggers mitochondrial dysfunction and apoptosis in oxidant-challenged lung endothelial cells. American journal of physiology. 2005;288:L530–535. doi: 10.1152/ajplung.00255.2004. [DOI] [PubMed] [Google Scholar]
- [30].Alexeyev MF, Ledoux SP, Wilson GL. Mitochondrial DNA and aging. Clin Sci (Lond) 2004;107:355–364. doi: 10.1042/CS20040148. [DOI] [PubMed] [Google Scholar]
- [31].Rachek LI, Grishko VI, Alexeyev MF, Pastukh VV, LeDoux SP, Wilson GL. Endonuclease III and endonuclease VIII conditionally targeted into mitochondria enhance mitochondrial DNA repair and cell survival following oxidative stress. Nucleic acids research. 2004;32:3240–3247. doi: 10.1093/nar/gkh648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Fishel ML, Seo YR, Smith ML, Kelley MR. Imbalancing the DNA base excision repair pathway in the mitochondria; targeting and overexpressing N-methylpurine DNA glycosylase in mitochondria leads to enhanced cell killing. Cancer research. 2003;63:608–615. [PubMed] [Google Scholar]
- [33].Rinne M, Caldwell D, Kelley MR. Transient adenoviral N-methylpurine DNA glycosylase overexpression imparts chemotherapeutic sensitivity to human breast cancer cells. Molecular cancer therapeutics. 2004;3:955–967. [PubMed] [Google Scholar]
- [34].Shokolenko I, Venediktova N, Bochkareva A, Wilson GL, Alexeyev MF. Oxidative stress induces degradation of mitochondrial DNA. Nucleic acids research. 2009;37:2539–2548. doi: 10.1093/nar/gkp100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Grishko V, Solomon M, Wilson GL, LeDoux SP, Gillespie MN. Oxygen radical-induced mitochondrial DNA damage and repair in pulmonary vascular endothelial cell phenotypes. American journal of physiology. 2001;280:L1300–1308. doi: 10.1152/ajplung.2001.280.6.L1300. [DOI] [PubMed] [Google Scholar]
- [36].Salazar JJ, Van Houten B. Preferential mitochondrial DNA injury caused by glucose oxidase as a steady generator of hydrogen peroxide in human fibroblasts. Mutation research. 1997;385:139–149. doi: 10.1016/s0921-8777(97)00047-5. [DOI] [PubMed] [Google Scholar]
- [37].Yakes FM, Van Houten B. Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc Natl Acad Sci U S A. 1997;94:514–519. doi: 10.1073/pnas.94.2.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Kruglov AG, Subbotina KB, Saris NE. Redox-cycling compounds can cause the permeabilization of mitochondrial membranes by mechanisms other than ROS production. Free radical biology & medicine. 2007 doi: 10.1016/j.freeradbiomed.2007.10.049. [DOI] [PubMed] [Google Scholar]
- [39].Furda AM, Marrangoni AM, Lokshin A, Van Houten B. Oxidants and not alkylating agents induce rapid mtDNA loss and mitochondrial dysfunction. DNA repair. 2012;11:684–692. doi: 10.1016/j.dnarep.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Pettepher CC, LeDoux SP, Bohr VA, Wilson GL. Repair of alkali-labile sites within the mitochondrial DNA of RINr 38 cells after exposure to the nitrosourea streptozotocin. J Biol Chem. 1991;266:3113–3117. [PubMed] [Google Scholar]
- [41].Ledoux SP, Shen CC, Grishko VI, Fields PA, Gard AL, Wilson GL. Glial cell-specific differences in response to alkylation damage. Glia. 1998;24:304–312. [PubMed] [Google Scholar]
- [42].Beranek DT. Distribution of methyl and ethyl adducts following alkylation with monofunctional alkylating agents. Mutation research. 1990;231:11–30. doi: 10.1016/0027-5107(90)90173-2. [DOI] [PubMed] [Google Scholar]
- [43].Alexeyev MF, Fayzulin R, Shokolenko IN, Pastukh V. A retro-lentiviral system for doxycycline-inducible gene expression and gene knockdown in cells with limited proliferative capacity. Mol Biol Rep. 2010;37:1987–1991. doi: 10.1007/s11033-009-9647-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Shokolenko IN, Alexeyev MF, Robertson FM, LeDoux SP, Wilson GL. The expression of Exonuclease III from E. coli in mitochondria of breast cancer cells diminishes mitochondrial DNA repair capacity and cell survival after oxidative stress. DNA repair. 2003;2:471–482. doi: 10.1016/s1568-7864(03)00019-3. [DOI] [PubMed] [Google Scholar]
- [45].Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- [46].Zufferey R, Nagy D, Mandel RJ, Naldini L, Trono D. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat Biotechnol. 1997;15:871–875. doi: 10.1038/nbt0997-871. [DOI] [PubMed] [Google Scholar]
- [47].Sambrook J, Russel DW. A laboratory manual. Cold Spring Harbor Laboratory Press; New York: 2001. Molecular Cloning. [Google Scholar]
- [48].Mol CD, Kuo CF, Thayer MM, Cunningham RP, Tainer JA. Structure and function of the multifunctional DNA-repair enzyme exonuclease III. Nature. 1995;374:381–386. doi: 10.1038/374381a0. [DOI] [PubMed] [Google Scholar]
- [49].Kavli B, Slupphaug G, Mol CD, Arvai AS, Peterson SB, Tainer JA, Krokan HE. Excision of cytosine and thymine from DNA by mutants of human uracil-DNA glycosylase. Embo J. 1996;15:3442–3447. [PMC free article] [PubMed] [Google Scholar]
- [50].Anderson CT, Friedberg EC. The presence of nuclear and mitochondrial uracil-DNA glycosylase in extracts of human KB cells. Nucleic acids research. 1980;8:875–888. [PMC free article] [PubMed] [Google Scholar]
- [51].Appleby RD, Porteous WK, Hughes G, James AM, Shannon D, Wei YH, Murphy MP. Quantitation and origin of the mitochondrial membrane potential in human cells lacking mitochondrial DNA. Eur J Biochem. 1999;262:108–116. doi: 10.1046/j.1432-1327.1999.00350.x. [DOI] [PubMed] [Google Scholar]
- [52].Marchetti P, Susin SA, Decaudin D, Gamen S, Castedo M, Hirsch T, Zamzami N, Naval J, Senik A, Kroemer G. Apoptosis-associated derangement of mitochondrial function in cells lacking mitochondrial DNA. Cancer research. 1996;56:2033–2038. [PubMed] [Google Scholar]
- [53].Kukat A, Kukat C, Brocher J, Schafer I, Krohne G, Trounce IA, Villani G, Seibel P. Generation of {rho}0 cells utilizing a mitochondrially targeted restriction endonuclease and comparative analyses. Nucleic acids research. 2008 doi: 10.1093/nar/gkn124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Holmuhamedov E, Jahangir A, Bienengraeber M, Lewis LD, Terzic A. Deletion of mtDNA disrupts mitochondrial function and structure, but not biogenesis. Mitochondrion. 2003;3:13–19. doi: 10.1016/S1567-7249(03)00053-9. [DOI] [PubMed] [Google Scholar]
- [55].Contamine V, Picard M. Maintenance and integrity of the mitochondrial genome: a plethora of nuclear genes in the budding yeast. Microbiology and molecular biology reviews : MMBR. 2000;64:281–315. doi: 10.1128/mmbr.64.2.281-315.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Margineantu DH, Gregory Cox W, Sundell L, Sherwood SW, Beechem JM, Capaldi RA. Cell cycle dependent morphology changes and associated mitochondrial DNA redistribution in mitochondria of human cell lines. Mitochondrion. 2002;1:425–435. doi: 10.1016/s1567-7249(02)00006-5. [DOI] [PubMed] [Google Scholar]
- [57].Qian W, Van Houten B. Alterations in bioenergetics due to changes in mitochondrial DNA copy number. Methods. 2010;51:452–457. doi: 10.1016/j.ymeth.2010.03.006. [DOI] [PubMed] [Google Scholar]
- [58].Kukat A, Kukat C, Brocher J, Schafer I, Krohne G, Trounce IA, Villani G, Seibel P. Generation of rho0 cells utilizing a mitochondrially targeted restriction endonuclease and comparative analyses. Nucleic acids research. 2008;36:e44. doi: 10.1093/nar/gkn124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Tann AW, Boldogh I, Meiss G, Qian W, Van Houten B, Mitra S, Szczesny B. Apoptosis induced by persistent single-strand breaks in the mitochondrial genome: Critical role of EXOG (5′ EXO/Endonuclease) in their repair. J Biol Chem. 2011;286:31975–31983. doi: 10.1074/jbc.M110.215715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Huang H, Greenberg MM. Synthesis and analysis of oligonucleotides containing abasic site analogues. J Org Chem. 2008;73:2695–2703. doi: 10.1021/jo702614p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Choi JY, Lim S, Kim EJ, Jo A, Guengerich FP. Translesion synthesis across abasic lesions by human B-family and Y-family DNA polymerases alpha, delta, eta, iota, kappa, and REV1. J Mol Biol. 2010;404:34–44. doi: 10.1016/j.jmb.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. DNA synthesis across an abasic lesion by yeast REV1 DNA polymerase. J Mol Biol. 2011;406:18–28. doi: 10.1016/j.jmb.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. DNA synthesis across an abasic lesion by human DNA polymerase iota. Structure (London, England : 1993) 2009;17:530–537. doi: 10.1016/j.str.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Pinz KG, Shibutani S, Bogenhagen DF. Action of mitochondrial DNA polymerase gamma at sites of base loss or oxidative damage. J Biol Chem. 1995;270:9202–9206. doi: 10.1074/jbc.270.16.9202. [DOI] [PubMed] [Google Scholar]
- [65].Lauritzen KH, Moldestad O, Eide L, Carlsen H, Nesse G, Storm JF, Mansuy IM, Bergersen LH, Klungland A. Mitochondrial DNA toxicity in forebrain neurons causes apoptosis, neurodegeneration, and impaired behavior. Mol Cell Biol. 2010;30:1357–1367. doi: 10.1128/MCB.01149-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Alexeyev MF, Venediktova N, Pastukh V, Shokolenko I, Bonilla G, Wilson GL. Selective elimination of mutant mitochondrial genomes as therapeutic strategy for the treatment of NARP and MILS syndromes. Gene Ther. 2008;15:516–523. doi: 10.1038/gt.2008.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Bayona-Bafaluy MP, Blits B, Battersby BJ, Shoubridge EA, Moraes CT. Rapid directional shift of mitochondrial DNA heteroplasmy in animal tissues by a mitochondrially targeted restriction endonuclease. Proc Natl Acad Sci U S A. 2005;102:14392–14397. doi: 10.1073/pnas.0502896102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Lauritzen KH, Cheng C, Wiksen H, Bergersen LH, Klungland A. Mitochondrial DNA toxicity compromises mitochondrial dynamics and induces hippocampal antioxidant defenses. DNA repair. 2011;10:639–653. doi: 10.1016/j.dnarep.2011.04.011. [DOI] [PubMed] [Google Scholar]
- [69].Hilgetag CC, Barbas H. Are there ten times more glia than neurons in the brain? Brain Struct Funct. 2009;213:365–366. doi: 10.1007/s00429-009-0202-z. [DOI] [PubMed] [Google Scholar]
- [70].Kelley MR, Georgiadis MM, Fishel ML. APE1/Ref-1 role in redox signaling: translational applications of targeting the redox function of the DNA repair/redox protein APE1/Ref-1. Current molecular pharmacology. 2012;5:36–53. doi: 10.2174/1874467211205010036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Regev-Rudzki N, Pines O. Eclipsed distribution: a phenomenon of dual targeting of protein and its significance. Bioessays. 2007;29:772–782. doi: 10.1002/bies.20609. [DOI] [PubMed] [Google Scholar]
- [72].Pastukh V, Shokolenko IN, Wilson GL, Alexeyev MF. Mutations in the passenger polypeptide can affect its partitioning between mitochondria and cytoplasm : Mutations can impair the mitochondrial import of DsRed. Mol Biol Rep. 2008;35:215–223. doi: 10.1007/s11033-007-9073-7. [DOI] [PMC free article] [PubMed] [Google Scholar]