Abstract
In the present study, a collection of 187 Enterococcus food isolates mainly originating from European cheeses were studied for the phenotypic and genotypic assessment of tetracycline (TC) resistance. A total of 45 isolates (24%) encompassing the species Enterococcus faecalis (n = 33), E. durans (n = 7), E. faecium (n = 3), E. casseliflavus (n = 1), and E. gallinarum (n = 1) displayed phenotypic resistance to TC with MIC ranges of 16 to 256 μg/ml. Eight of these strains exhibited multiresistance to TC, erythromycin, and chloramphenicol. By PCR detection, TC resistance could be linked to the presence of the tet(M) (n = 43), tet(L) (n = 16), and tet(S) (n = 1) genes. In 15 isolates, including all of those for which the MIC was 256 μg/ml, both tet(M) and tet(L) were found. Furthermore, all tet(M)-containing enterococci also harbored a member of the Tn916-Tn1545 conjugative transposon family, of which 12 erythromycin-resistant isolates also contained the erm(B) gene. Filter mating experiments revealed that 10 E. faecalis isolates, 3 E. durans isolates, and 1 E. faecium isolate could transfer either tet(M), tet(L), or both of these genes to E. faecalis recipient strain JH2-2. In most cases in which only tet(M) was transferred, no detectable plasmids were acquired by JH2-2 but instead all transconjugants contained a member of the Tn916-Tn1545 family. Sequencing analysis of PCR amplicons and evolutionary modeling showed that a subset of the transferable tet(M) genes belonged to four sequence homology groups (SHGs) showing an internal homology of ≥99.6%. Two of these SHGs contained tet(M) mosaic structures previously found in Tn916 elements and on Lactobacillus and Neisseria plasmids, respectively, whereas the other two SHGs probably represent new phylogenetic lineages of this gene.
As lactic acid bacteria, enterococci are natural inhabitants of the gastrointestinal systems of mammals, but they are also known to occur in soil and fecally polluted surface waters and on plants and vegetables (26, 31, 35). Because of their high prevalence in the gastrointestinal tracts of many food animals, it is often unavoidable that these organisms enter the human food chain via contamination of raw milk or raw meat. For many years, the presence of enterococci in foods has been highly controversial. On the one hand, Enterococcus strains can harbor specific biochemical traits that are essential in the manufacturing of various fermented milk products, and some strains are technologically exploited as functional starters or probiotics (12, 17). On the other hand, enterococci have also been implicated in the spoilage of processed meats (14, 51) and include strains that have been recognized as emerging human pathogens mostly in nosocomial but also in community-acquired infections (for a review, see reference 29).
Triggered by the apparent duality between their beneficial and harmful properties, a lot of research has focused on the potential role of food enterococci as reservoirs and/or vehicles of antibiotic resistance (AR) and virulence factors (12, 24, 25) and on the possible interaction between AR and virulence (28). During the antibiotic era, an increasing number of food enterococci have developed resistance to various therapeutic antibacterial agents, including vancomycin (18, 40, 52), gentamicin (11), and streptogramines (42). From studies in which patterns of AR to a broader range of therapeutic agents were determined, it appears that tetracycline (TC) resistance (Tcr) is one of the most common phenotypes of acquired AR in food isolates of the genus Enterococcus (34, 45). Although the broad-spectrum clinical use of TCs is declining, new applications of minocycline and glycylcycline in human therapy have been identified (7). Moreover, this group of agents is still used in veterinary and aquaculture settings and some TCs are still in use as animal growth promoters in several countries outside Europe (7).
In enterococci, two major groups of Tcr (tet) genes have been recognized (7). The first group confers resistance by ribosomal protection (RP) and includes the genes tet(M), tet(O), and tet(S), which have been detected in Enterococcus spp. A second group mediates energy-dependent efflux of TC from cells and is represented in enterococci by the tet(K) and tet(L) genes. A sixth gene, tet(U), encodes low-level resistance in Enterococcus faecium through an unknown mechanism (39). So far, the great majority of studies dealing with the distribution of tet genes in Enterococcus spp. have focused on human or animal isolates. In contrast, data on the occurrence of tet genes in Enterococcus isolates obtained from food and food products are still relatively scarce. Teuber and coworkers (45, 46) reported the presence of tet(M) in six Enterococcus isolates from cheese and salami, some of which were located both on the chromosome and on a large plasmid. In one isolate, E. faecalis FO1, the tet(M) gene was located on a Tn916-like transposon named TnFO1 (33).
Of the six types of tet genes detected in veterinary or clinical enterococci, only tet(M) has been reported in Enterococcus isolates from food. To obtain further insight into the prevalence and diversity of tet genes among food enterococci, the present study pursued the phenotypic and genotypic assessment of Tcr among a collection of taxonomically well-characterized Enterococcus isolates originating mainly from cheeses but also from other food sources. In addition, a subset of tet genes belonging to the tet(M) group that could be transferred in vitro by filter conjugation to E. faecalis recipient strain JH2-2 was further characterized by partial sequencing to examine their degree of nucleotide polymorphism and their sequence similarities to previously determined tet(M) genes.
MATERIALS AND METHODS
Bacterial isolates.
The 187 Enterococcus isolates included in this study were obtained from the BCCM/LMG Bacteria Collection, Ghent University, Ghent, Belgium (http://www.belspo.be/bccm/db/bacteria_search.htm), and all originated from food sources. The majority of the isolates (n = 139) were isolated from different types of European cheeses (mostly from Italy, Ireland, and Greece) that were obtained in the course of European Union research project FAIR-CT97-3078 (Enterococci in Food Fermentations: Functional and Safety Aspects). More information on these isolates can be found in the catalogue of enterococci of the FAIR-E collection (50). All of the enterococci included in this study were previously identified to the species level by protein profiling. Isolates were preserved with the Microbank bead storage system (Pro-LAB Diagnostics, Wirral, United Kingdom) at −80°C and routinely grown on MRS agar (CM359; Oxoid, Basingstoke, United Kingdom) at 30 or 37°C under aerobic conditions.
Determination of phenotypic resistance.
First, all 187 isolates were screened for the Tcr phenotype by an agar dilution method. Cultures grown overnight were inoculated on MRS plates containing TC at 25 μg/ml (T-3383; Sigma, St. Louis, Mo.), and growth was scored visually after 24 to 72 h. All isolates that displayed the Tcr phenotype after agar dilution were subjected to MIC testing in the range of 4 to 512 μg of TC per ml based on the broth microdilution method with cation-adjusted Mueller-Hinton II broth (212322; Becton Dickinson, Cockeysville, Md.) as recommended by the National Committee for Clinical Laboratory Standards (NCCLS; 30). Following a 24-h incubation, the MIC was visually read from a microtiter plate as the concentration at which ≥80% inhibition of growth occurred. Each batch of MIC determinations included the control strain E. faecalis LMG 8222 (ATCC 29212).
The presence of additional phenotypic resistances was determined by the agar disk diffusion method as previously described (30). Antibiotic disks (Oxoid) containing TC (30 μg), doxycycline (30 μg), minocycline (30 μg), ampicillin (10 μg), rifampin (30 μg), vancomycin (30 μg), erythromycin (ER, 15 μg), and chloramphenicol (CM; 30 μg) were used. Inhibition zones were interpreted in accordance with the NCCLS guideline tables (30).
Detection of AR and int genes.
Total genomic DNA was prepared by using a protocol based on the method of Pitcher and coworkers (36). Isolation of plasmid DNA was based on conventional alkaline lysis (2).
For all detection assays, a common PCR core mixture (total volume, 50 μl) was used that consisted of 1× PCR buffer (Applied Biosystems, Warrington, United Kingdom), deoxynucleoside triphosphates (dATP, dCTP, dGTP, and dTTP; Applied Biosystems) at a concentration of 200 μM each, 1 U of AmpliTaq DNA polymerase (Applied Biosystems), and 20 pmol of each primer (Sigma-Genosys Ltd., Cambridgeshire, United Kingdom). A 50-ng portion of intact total DNA was used as the PCR template. All genes were detected with previously described primers and PCR control strains. In a first PCR assay, tet genes encoding RP proteins were detected with the primers DI and DII (15) and primers Ribo2-FW and Ribo2-RV (1). Isolates harboring an RP protein-type tet gene were further subjected to PCR detection of tet(M), tet(O), tet(Q), tet(S), tet(T), and tet(W) with primers DI and TetM-R, primers TetO-FW1 and TetO-RV1, primers TetQ-FW and TetQ-RV, primers TetS-FW and TetS-RV, primers TetT-FW and TetT-RV, and primers TetW-FW and TetW-RV (1, 15), respectively. The presence of the efflux genes tet(K) and tet(L) was determined with primers TetK-FW1 and TetK-RV1 and primers TetL-FW3 and TetL-RV3, respectively (15). The presence of the erm(B) gene was investigated with primers ErmB-FW and ErmB-RV (15). The occurrence of conjugative transposons of the Tn916-Tn1545 family was determined with primers Int-FW and Int-RV targeting the transposon integrase (int) gene (10, 15). All PCR amplifications were performed in a GeneAmp 9600 PCR system (Perkin-Elmer) with the temperature program previously described for each primer set (1, 15). PCR amplicons were checked electrophoretically on 1% agarose and visualized by ethidium bromide fluorescence.
Filter matings.
All strains harboring one or more tet genes were included in filter matings with E. faecalis strain JH2-2 (21) on the basis of the protocol described by Perreten and coworkers (33). In short, 1 ml of a culture of the donor or recipient strain grown overnight in brain heart infusion (BHI) broth (Difco) at 37°C was added to 5 ml of fresh BHI broth and further incubated for 4 h. Equal volumes (1 ml) of donor and recipient cultures were mixed and filtered through a sterile membrane filter with a pore size of 0.45 μm (MF-Millipore membrane filter HAWP 2500; Millipore, Bedford, Mass.) contained in a Swinnex filter holder (SX00 02500; Millipore). Subsequently, filters were gently rinsed once with 2 ml of a sterile peptone-physiological saline (PPS) solution (8.5 g of NaCl per liter, 1g of neutralized bacteriological peptone [Oxoid L34] per liter). Filters were incubated on BHI agar (Difco) for 24 h at 37°C, and after mating, cells were washed from the filter with 2 ml of PPS. Finally, serial dilutions in PPS of the mixed suspension and of the donor and recipient strains were plated on BHI agar supplemented with 10 μg of TC per ml, 50 μg of rifampin per ml, and 100 μg of fusidic acid per ml (triple selective medium). The TC-susceptible JH2-2 recipient strain and the potential donor strains are resistant and susceptible, respectively, to the latter two antibiotics at the indicated concentrations. Following incubation at 37°C for 24 to 72 h, plates were checked for the absence (donor and recipient plates) or presence (mating mixture plate) of growth and up to 10 colonies were picked from the latter plate and inoculated into BHI broth supplemented with 10 μg of TC per ml. Upon growth, potential transconjugant cultures were purified on BHI agar and stored with the Microbank bead storage system at −80°C. Phenotypic and genotypic properties of possible transconjugants were determined by disk diffusion susceptibility testing, determination of the MIC of TC, (GTG)5-PCR DNA fingerprinting (16), plasmid profiling, and PCR-based detection of tet genes and other resistance elements as described above.
Sequencing of PCR amplicons.
A subset of PCR amplicons of tet(M) genes detected in JH2-2 transconjugants were purified and sequenced with primers DI, DII, and TetM-R (15). As a sequencing control, the sequence of the tet(M) gene of strain E. faecalis FO1 (EMBL accession no. X92947) (35) was partially redetermined. Sequencing was performed with a BigDye Terminator (version 2) Ready Reaction cycle sequencing kit (Applied Biosystems) on an ABI Prism 3100 Genetic Analyzer (Applied Biosystems). Reference sequences of tet(M) were retrieved from the EMBL database (http://www.ebi.ac.uk) and compared with the new sequences by using BioNumerics version 3.5 software (Applied Maths, St.-Martens Latem, Belgium). Additional sequence similarity searches were performed with the EMBL Fasta program.
Nucleotide sequence accession numbers.
The partial sequences of the tet(M) genes described in this paper have been submitted to the EMBL database under accession numbers AJ585076 to AJ585084.
RESULTS AND DISCUSSION
Prevalence of Tcr among food enterococci.
In order to increase our understanding of the molecular ecology and population biology of Tcr determinants and their hosts, surveillance and characterization of Tcr should not only be restricted to pathogenic bacteria but should also include organisms from other environmental niches, such as food-associated and commensal bacteria (43; http://www.tufts.edu/med/apua/ROAR/roarhome.htm). In this regard, enterococci constitute an interesting group as they are commensals of humans and animals that occur and grow in a variety of foods and have also been implicated in nosocomial infections. In the present survey, a collection of 187 enterococcal isolates, mostly from cheese but also from other food sources (Table 1), were studied for the presence of Tcr, of which 45 isolates (24%) displayed the Tcr phenotype on the basis of agar dilution testing with a breakpoint concentration of 25 μg of TC per ml. The majority of the Tcr isolates belonged to the species E. faecalis (n = 33) and E. durans (n = 7), whereas only a few Tcr isolates of E. faecium (3 out of 55) were found (Table 1). Of the remaining species, only E. casseliflavus and E. gallinarum both yielded one Tcr isolate, but clearly more isolates should be included in order to assess the prevalence of the Tcr phenotype in these taxa. Most of the Tcr Enterococcus isolates originated from cheese (n = 34), followed by a few Tcr isolates from milk, fermented milks, and fish, among other food sources (Table 1). The relative prevalence of Tcr enterococci encountered in this study (24%) falls within the range of a previous estimate of 20 to 97% Tcr among enterococci from animals and their meat (45). However, it should be stressed that the prevalence of Tcr among enterococcal isolates from food can vary significantly, depending on the type of food product (5), geographical origin (37), and taxonomic identity (13). The recovery of only a small minority of Tcr E. faecium strains in our study (5%) may be due to the fact that most of the isolates originated from cheese. Considerably higher percentages of Tcr E. faecium isolates (28 to 74%) can be expected when enterococci from retail poultry are investigated (5, 37). Some of the isolates used in our study were also previously included for an E-test analysis (13), in which it was found that E. faecium displayed fewer resistance phenotypes than E. faecalis among cheese isolates.
TABLE 1.
Prevalence of the Tcr phenotype in Enterococcus isolates from food
| Type(s) of food | No. of Tcr isolates/total (% above arbitrary threshold)
|
||||||
|---|---|---|---|---|---|---|---|
| E. casseliflavus | E. durans | E. faecalis | E. faecium | E. gallinarum | Other speciesa | Total | |
| Cheese | 1/2 | 6/21 (29) | 24/65 (37) | 2/39 (5) | 1/5 | 0/7 | 34/139 (24) |
| Milk | 0/1 | 0/1 | 3/9 | 0/3 | 0/1 | 3/15 (20) | |
| Meat and meat products | 1/4 | 0/6 | 1/10 | ||||
| Fish | 1/1 | 2/5 | 3/6 | ||||
| Vegetables | 0/1 | 0/3 | 0/1 | 0/5 | |||
| Fermented milk | 0/1 | 2/2 | 2/3 | ||||
| Others | 1/4 | 1/4 | 0/1 | 2/9 | |||
| Total | 1/3 | 7/24 (29) | 33/90 (37) | 3/55 (5) | 1/5 | 0/10 | 45/187 (24) |
Isolates of E. hirae (n = 7), E. malodoratus (n = 2), and E. flavescens (n = 1).
For all 45 Tcr Enterococcus isolates, the minimum TC MIC was 16 μg/ml, which is the cutoff level proposed by the NCCLS (30) for the classification of enterococci as Tcr. For E. faecalis strains, MICs ranged from 16 to 256 μg/ml, whereas a somewhat lower range of 32 to 128 μg/ml was found for the Tcr E. durans and E. faecium strains (Table 2). Susceptibilities to additional antibiotics were assessed by disk diffusion testing by the NCCLS method with Mueller-Hinton agar (30), which was compared with the previously reported agar overlay method using MRS agar (20). In terms of interpretive zone reading, the data sets generated by both methods were compatible, with the exception of inhibition zones for ER, which were occasionally smaller on MRS agar (results not shown). The Tcr phenotype was uniformly confirmed by disk diffusion testing, and all Tcr isolates also displayed resistance to other TCs, i.e., doxycycline and minocycline, but were susceptible to ampicillin, rifampin, and vancomycin (data not shown). In addition, about one-third of the Tcr E. faecalis isolates were coresistant to ER (12 of 33 strains) and/or CM (10 of 33 strains) (Table 2), of which 8 displayed multiple resistance to TC, ER, and CM. Teuber and coworkers (45) indicated that resistance to TC, ER, CM, and gentamicin was a common multiple drug resistance phenotype displayed by Enterococcus isolates from various types of cheeses.
TABLE 2.
Phenotypic and genotypic resistance properties of Tcr food enterococcia
| Species | No. of isolates | TC MIC (μg/ml) | Additional resistanceb
|
tet(M) | tet(L) | tet(S) | Tn916-Tn1545 | erm(B) | Transfer to JH2-2 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ER + CM | ER | CM | |||||||||
| E. casseliflavus | 1 | 128 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| E. durans | 7 | 32-128 | 0 | 1 | 0 | 7 | 1 | 0 | 7 | 1 | 3 |
| E. faecalis | 33 | 16-256 | 8 | 2 | 2 | 32 | 13 | 1 | 30 | 10 | 10 |
| E. faecium | 3 | 32-128 | 0 | 1 | 0 | 3 | 2 | 0 | 3 | 1 | 1 |
| E. gallinarum | 1 | 64 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 |
| Total | 45 | 8 | 4 | 2 | 43 | 16 | 1 | 41 | 12 | 14 | |
Unless noted otherwise, each value is the number of isolates with the property indicated.
As determined by disk diffusion.
Prevalence of tet genes and other resistance elements.
Various studies on the antibiotic susceptibilities of human enterococci obtained in the early 1950s indicate that a significant pool of Tcr determinants was already prevalent in clinical isolates during the first years of therapeutic use of TC (3, 23, 48). In the present study, the genotypic basis of the Tcr phenotype found in food isolates of Enterococcus spp. was investigated by PCR-based detection of eight tet genes, all of which, except tet(T), tet(Q), and tet(W), have been previously reported in this genus (7). As shown in Table 2, the tet(M) gene was found in all but two Tcr Enterococcus isolates and one E. faecalis strain contained tet(S) whereas none of the other RP genes tested for [tet(O), tet(Q), tet(T), or tet(W)] were detected. Of the efflux genes tet(K) and tet(L), only the latter was detected in 16 Tcr isolates mainly belonging to the species E. faecalis and in 1 Tcr E. durans isolate. The sole Tcr E. casseliflavus isolate encountered in this study did not contain any of the above-mentioned tet genes. For this isolate, originating from smear-ripened raw milk cheese, the MIC was 128 μg/ml, which indicates that it contains an unknown mechanism conferring high-level Tcr. The finding that all Tcr isolates harbored an RP tet gene is concordant with the minocycline resistance phenotype found by disk diffusion testing, which is mediated only by RP mechanisms in enterococci (7). A similar situation was found during a major survey of 229 enterococcal isolates collected in 10 hospitals in France, where tet(M) and tet(L) were the dominant Tcr determinants, followed by tet(S), whereas tet(O) was found in only 1 isolate (6). It should be kept in mind that the tet gene distribution reported in this study is based on a relatively limited set of isolates, which highlights the need for regular monitoring of additional sets of biologically and geographically diverse strains in order to obtain a broader view of the prevalence of these genes in food-associated enterococci. Except for one strain, all of the strains in our study that were found to contain tet(L) also possessed the tet(M) gene. It is noteworthy that the six E. faecalis strains for which the MIC of TC was the highest (256 μg/ml) contained both tet(M) and tet(L), whereas for the sole strain that harbored only tet(L) and no RP gene, the MIC was the lowest observed in this study (16 μg/ml). This observation suggests that a higher level of Tcr is reached when the organism contains both an active-efflux mechanism and an RP mechanism. Such a reinforcement effect has previously also been reported for methicillin-resistant Staphylococcus aureus (49), for isolates containing either tet(M) or tet(K) the TC MIC for 90% of the strains tested was 64 μg/ml whereas for those that contained both of these tet genes the TC MIC for 90% of the strains tested was 256 μg/ml.
In many enterococci and streptococci of clinical or food origin, drug resistance genes occur more frequently on conjugative transposons than on plasmids. Also in this study, the majority of the tet(M)-containing isolates (41 of 43 isolates) were positive by PCR for the integrase element int, indicating that they contain a member of the broad-host-range Tn916-Tn1545 conjugative transposon family (Table 2). By definition, all members of this family carry the tet(M) gene, sometimes joined by additional determinants, e.g., those encoding resistance to ER and kanamycin in the case of Tn1545 (8). In our study, the 10 E. faecalis isolates and single isolates of E. durans and E. faecium that were Err all contained erm(B), which is considered to be the most widespread macrolide resistance gene among enterococci from food animals (22) or foods (45). Furthermore, all of these erm(B)-containing strains were also positive for the detection of a Tn916-Tn1545 element. In this context, it should be mentioned that erm(B) genes in enterococci can also occur on other mobile elements, such as conjugative multiresistance plasmids (47) or members of the Tn917 family (41).
Conjugal transfer of Tcr.
The historical spread of Tcr into and among enterococcal populations, including those from food environments, has been strongly associated with the fact that many tet genes are located on mobile genetic elements (45). On the basis of the finding that most of the Tcr isolates in the present study also carried a member of the Tn916-Tn1545 family (Table 2) or harbored one or more plasmids (data not shown), we investigated the mobility of the detected tet genes in filter mating experiments with TC-susceptible recipient strain E. faecalis JH2-2 (21). Initially, potential transconjugant colonies were obtained from 18 out of 45 tested Tcr donor strains representing the species E. faecalis, E. durans, and E. faecium. However, after phenotypic and genotypic confirmation, transconjugants of four E. faecalis isolates were found to be susceptible to rifampin and/or displayed (GTG)5-PCR fingerprints highly similar, if not identical, to those of the respective donors. As a result, the number of successful matings with strain JH2-2 was reduced to 14 (31% of analyzed donors), all of which were obtained from donor strains originating from Italian or Irish cheeses (n = 12), milk (n = 1), or shellfish (n = 1) (Table 3). Overall, transfer frequencies obtained during filter matings were in the range of 10−6 to 10−8 per recipient.
TABLE 3.
Phenotypic and genotypic resistance properties of Enterococcus donors and resulting JH2-2 transconjugants from filter mating experiments
| Donor strain (FAIR-collection no.) | Biologic origin | Geographic origin (yr of isolation) | TC MIC (μg/ml)
|
Additional resistance(s)
|
tet gene(s)
|
Tn916-Tn1545
|
erm(B)
|
No. of plasmids transferred (size[s] of plasmid[s] [kb]) | tet(M) SHG | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Donor | Trans- conjugant | Donor | Trans- conjugant | Donor | Trans- conjugant | Donor | Trans- conjugant | Donor | Trans- conjugant | |||||
| E. durans | ||||||||||||||
| LMG 20930 (FAIR-E 387) | Casera cheese | Italy (1980) | 128 | 64 | tet(M) | tet(M) | + | + | − | − | Not detected | |||
| LMG 20931 (FAIR-E 388) | Casera cheese | Italy (1980) | 128 | 64 | tet(M) | tet(M) | + | + | − | − | Not detected | |||
| LMG 20932 (FAIR-E 389) | Valtellina cheese | Italy (1981) | 128 | 64 | tet(M) | tet(M) | + | + | − | − | Not detected | III | ||
| E. faecalis | ||||||||||||||
| LMG 20647 (FAIR-E 42) | Shellfish | Italy (1997) | 64 | 64 | ER | tet(M) | tet(M) | + | + | − | − | Not detected | I | |
| LMG 20678 (FAIR-E 74) | Montasio cheese | Italy (1997) | 32 | 128 | ER | tet(M) | tet(M) | + | + | − | − | Not detected | V | |
| LMG 20693 (FAIR-E 90) | Goat cheese | Italy (1997) | 32 | 64 | ER | tet(M) | tet(M) | + | + | − | − | Not detected | V | |
| LMG 20786 (FAIR-E 224) | Cheddar cheese | Ireland (1994) | 64 | 64 | ER + CM | CM | tet(M), tet(L) | tet(M) | + | + | + | − | Not detected | IV |
| LMG 20790 (FAIR-E 228) | Cheddar cheese | Ireland (1994) | 64 | 64 | ER | ER | tet(M) | tet(M) | + | + | + | + | Not detected | IV |
| LMG 20791 (FAIR-E 229) | Cheddar cheese | Ireland (1994) | 256 | 32 | ER + CM | CM | tet(M), tet(L) | tet(L) | + | − | − | − | 1 (>20) | |
| LMG 20798 (FAIR-E 236) | Smear-ripened cheese | Ireland (1995) | 256 | 64 | CM | CM | tet(M), tet(L) | tet(M), tet(L) | + | + | − | − | 2 (>20) | IV |
| LMG 20799 (FAIR-E 237) | Cheddar cheese | Ireland (1995) | 32 | 64 | ER | tet(M), tet(L) | tet(M) | + | + | − | − | 3 (4.8, 5, >20) | ||
| LMG 20831 (FAIR-E 275) | Milk | Italy (1997) | 32 | 32-64 | tet(M) | tet(M) | + | + | − | − | 5 (4.8, 5, 9.8, 20, >20) | III | ||
| LMG 20843 (FAIR-E 287) | Fontina cheese | Italy (1997) | 128 | 64 | ER | tet(M) | tet(M) | + | + | − | − | 2 (5, 9.8) | III | |
| E. faecium LMG 20927 (FAIR-E 384) | Grana cheese | Italy (1976) | 128 | 32-128 | ER | ER | tet(M), tet(L) | tet(L)/tet(M), tet(L)a | + | + | + | + | 3 (4.7, 5.3, >20)a | |
Transconjugants of LMG 20927 acquired either tet(L) or both tet(M) and tet(L). Plasmid profiles of these transconjugants differed in the number of acquired plasmids.
For transconjugants obtained from 10 E. faecalis donors, the TC MIC was equal to or lower than that for the corresponding donor, except for isolates LMG 20678, LMG 20693, LMG 20799, and LMG 20831, for which the MICs were 1 or 2 doubling concentrations higher (Table 3). For transconjugants obtained from three E. durans isolates, the MICs were consistently lower than those for the donors, whereas for a set of transconjugants obtained from E. faecium LMG 20927, the MICs did not exceed 128 μg/ml. Because of the limited number of Tcr donors included, it is not clear to what extent these differences in MICs may reflect increased or decreased levels of tet gene expression. For matings with E. faecalis donors, Tcr in JH2-2 transconjugants was linked to acquisition of the tet(M) gene (n = 8), the tet(L) gene (n = 1), or both of these genes (n = 1). All transconjugants from E. durans acquired tet(M) and, in contrast, transconjugants from E. faecium LMG 20927 harbored either tet(M) or both tet(M) and tet(L), which explains the relatively broad MIC range found for this mating (Table 3). One E. faecalis donor and one E. faecium donor (LMG 20790 and LMG 20927, respectively), both carrying an erm(B) gene, could transfer this gene to recipient strain JH2-2, whereas three other E. faecalis donors cotransferred Cmr (Table 3). To our knowledge, conjugal transfer of TC, ER, and/or CM resistance by food enterococci has previously only been reported by Teuber and colleagues (46) for isolates from cheese and meat products. These authors reported that Tcr acquisition by JH2-2 was due to transfer of the tet(M) gene, which in some cases was located both on the chromosome and on a large plasmid (46). Our study provides the first indication that also tet(L), sometimes in combination with tet(M), can be transferred in vitro by food isolates of E. faecalis and E. faecium. For three donors, transfer of tet(L) was associated with the acquisition of one or more large plasmids (>20 kb). In most other cases (8 out of 11 matings), in which only tet(M) was transferred to recipient JH2-2, no plasmid transfer was detected (data not shown) but all transconjugants contained a representative of the Tn916-Tn1545 family (Table 3). Members of this family have been found in more than 50 different bacterial species, most of which are of clinical importance (8). Our data thus seem to reinforce the evidence that many enterococci, regardless of having a clinical background or a food origin, use conjugative transposition as an important mechanism for the dissemination of tet(M) and possibly other resistance genes.
Genetic diversity of tet(M) genes from JH2-2 transconjugants.
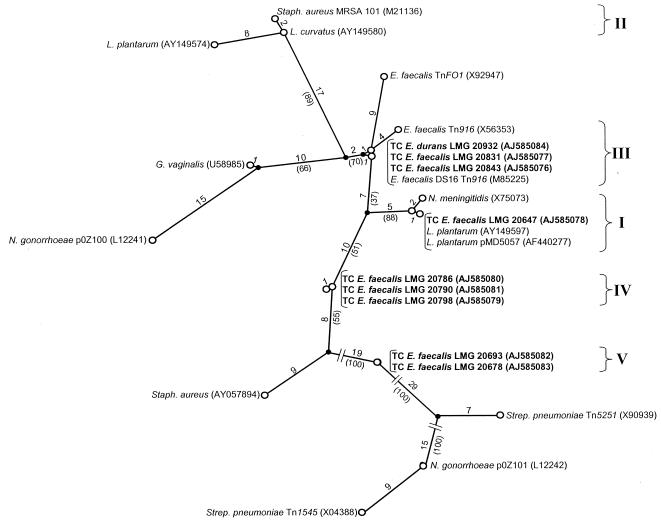
The finding that the same types of tet genes, such as tet(M), occur in veterinary, clinical, and food enterococci raises the question of the extent to which genetic transfer and/or recombination events have played a role in the global distribution of such genes. On the basis of gene sequence alignments (19, 32) or high-resolution gene restriction analysis (10), several authors have shown that the tet(M) gene exhibits several mosaic structures, possibly as a result of a series of homologous recombinations. To examine if any of these previously observed mosaic structures also occur in transferable tet(M) genes of food enterococci, a subset of tet(M) genes that could be transferred by nine donor isolates to recipient strain JH2-2 was further analyzed by sequencing of the DI-TetM-R PCR amplicons obtained from the corresponding transconjugants. Although it can be expected that sequence analysis of the complete tet(M) gene will result in a more robust evolutionary model, analysis of amplicon sequences that encompass 74% of the 1,920-bp coding sequence already provides a reliable indication of the phylogenetic diversity among enterococcal tet(M) genes. Global alignment and evolutionary modeling by maximum-parsimony analysis allowed us to group these sequences into four different sequence homology groups (SHGs) compared with reference tet(M) sequences retrieved from the EMBL database (Fig. 1). The delineation of these SHGs was based on an internal sequence homology level of ≥99.6%, which generally corresponded to zero to six nucleotide substitutions. Concordant with a previously proposed nomenclature in which two tet(M) SHGs (I and II) were defined by maximum-parsimony analysis (15), the newly obtained Enterococcus tet(M) sequences were assigned to SHGs I, III, IV, and V, whereas no new members of SHG II were found. In addition to the five currently defined SHGs, at least seven other tet(M) structures have previously been recognized, some of which may represent additional SHGs (Fig. 1).
FIG. 1.
Unrooted maximum-parsimony tree of multiple aligned partial tet(M) sequences of enterococcal JH2-2 transconjugants and reference tet(M) sequences retrieved from the EMBL database. SHGs showing ≥99.6% internal sequence homology are indicated by roman numerals I to V. The number of base conversions over the tree is indicated along the phylogenetic distance lines, and bootstrap percentages for analysis of 100 replicates are in parentheses. If available, the designation of the tet(M)-carrying strain, plasmid, or transposon is indicated, followed by the EMBL accession number in parentheses. Newly obtained tet(M) sequences are in boldface.
SHG I comprised one new tet(M) sequence of E. faecalis LMG 20647, which was joined by tet(M) genes previously found on plasmids in Neisseria meningitidis (accession no. X75073) and only recently also in lactobacilli (9, 15). Although SHG I contained tet(M) sequences originating from different gram-positive and gram-negative genera, all of these sequences were very highly related, showing only zero to three nucleotide variations. Transfer of tet(M) from isolate LMG 20647 to recipient JH2-2 was not accompanied by the acquisition of one or more detectable plasmids, suggesting that this transfer may be mediated by a conjugative transposon element of the Tn916-Tn1545 family, as indicated by the presence of an int gene in the resulting transconjugants. SHG III included tet(M) genes from E. faecalis LMG 20831 and LMG 20843 and E. durans LMG 20932 and also comprised the prototype mosaic structure of the tet(M) gene located within conjugative transposon Tn916 (4, 44). The partial tet(M) gene of the three Italian Enterococcus isolates displayed only zero to two nucleotide differences from each other or from the tet(M) sequence of Tn916-carrying E. faecalis strain DS16 (accession no. M85225). Together with the fact that these isolates indeed contained a member of the Tn916-Tn1545 family (Table 3), the high sequence homology with reference sequences of Tn916 seems to indicate that in these three isolates the tet(M) gene is integrated into a transposon of the latter type. It is noteworthy that the tet(M) gene of Tn916-like transposon TnFO1 (32) was closely linked to members of SHG III (9 to 13 nucleotide variations, mostly including unknown bases). The remaining two SHGs, IV and V, comprised the tet(M) genes of three Irish and two Italian cheese isolates of E. faecalis, respectively, and exhibited extremely high internal sequence homology (zero to one nucleotide substitution). Both SHGs did not include any other previously determined tet(M) sequences, and none of them were closely joined by known tet(M) sequences after similarity searches with the EMBL Fasta program, indicating that these sequences may represent two new phylogenetic lineages of tet(M). Like isolate LMG 20647, the two isolates of SHG V harbored a Tn916-Tn1545 element but seemingly did not transfer any plasmids to JH2-2. Possibly, these tet(M)-containing isolates harbor Tn916-like structures previously found in Enterococcus spp. such as Tn5381 (38), the tet(M) gene sequences of which are not available or even contain currently undescribed members of this conjugative-transposon family. Taken together, the main conclusion from our sequencing data is that transferable tet(M) genes carried by food enterococci belong to several different lineages, which may explain why this gene has been widely disseminated among at least 18 gram-positive and 8 gram-negative hosts (7).
Conclusions.
Collectively, the data presented in this study demonstrate that Tcr in food enterococci mainly originating from European cheeses is conferred basically by the same types of tet genes [tet(M), tet(L), and tet(S)] as those previously found among veterinary or clinical Enterococcus isolates. Moreover, the finding that a significant proportion of Tcr isolates exhibited coresistance to ER and/or CM reinforces the suggestion that the selection of Tcr genotypes may provide a suitable molecular basis for the further selection of multiple resistances (27). In vitro conjugation experiments showed that a considerable fraction of the tet genes in food enterococci is located on active mobile genetic elements including known and, potentially, undescribed members of the tet(M)-containing Tn916-Tn1545 family. The high evolutionary diversity of tet(M) structures, as evidenced by the delineation of several SHGs, should provide the basis for further molecular studies. Next to physical linkage of tet and other resistance genes to enterococcal plasmids or chromosomal elements by Southern blotting, detailed molecular dissection of up- and downstream regions of the targeted int gene of Tn916/Tn1545 and full sequence analysis of tet gene-containing self-transmissible plasmids need to be emphasized in follow-up studies. In order to establish the broader ecological and clinical relevance of Tcr enterococci occurring along the food chain, extensive assessment of the in vitro and in vivo host range spectra of these tet gene carriers is needed. It is expected that these new insights will contribute to the definition of well-argued safety requirements for the approved use of Enterococcus strains as food starter cultures or as human or animal probiotics, an issue that is currently under debate in Europe (http://www.europa.eu.int/comm/food/fs/sc/scan/index_en.html) and the United States (http://www.fao.org/es/ESN/Probio/probio.htm).
Acknowledgments
This work was supported by the Fund for Scientific Research—Flanders (Belgium) (F.W.O.-Vlaanderen; contract G.0309.01). G.H. is a postdoctoral fellow of the Fund for Scientific Research—Flanders (Belgium) (F.W.O.-Vlaanderen).
We thank R. Coopman and M. Cnockaert for excellent technical assistance with the sequencing work and G. Dasen (Institut für Lebensmittelwissenschaft, Zürich, Switzerland) for the gift of E. faecalis strain FO1.
REFERENCES
- 1.Aminov, R. I., N. Garrigues-Jeanjean, and R. I. Mackie. 2001. Molecular ecology of tetracycline resistance: development and validation of primers for detection of tetracycline resistance genes encoding ribosomal protection proteins. Appl. Environ. Microbiol. 67:22-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, D. G., and L. L. McKay. 1983. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl. Environ. Microbiol. 46:549-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atkinson, B. A., A. Abu-Al-Jaibat, and D. J. LeBlanc. 1997. Antibiotic resistance among enterococci isolated from clinical specimens between 1953 and 1954. Antimicrob. Agents Chemother. 41:1598-1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burdett, V. 1990. Nucleotide sequence of the. tet(M) gene of Tn916. Nucleic Acids Res. 18:6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Butaye, P., K. Van Damme, L. A. Devriese, L. Van Damme, M. Baele, S. Lauwers, and F. Haesebrouck. 2000. In vitro susceptibility of Enterococcus faecium isolated from food to growth-promoting and therapeutic antibiotics. Int. J. Food Microbiol. 54:181-187. [DOI] [PubMed] [Google Scholar]
- 6.Charpentier, E., G. Gerbaud, and P. Courvalin. 1994. Presence of the Listeria tetracycline resistance gene tet(S) in Enterococcus faecalis. Antimicrob. Agents Chemother. 38:2330-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chopra, I., and M. Roberts. 2001. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65:232-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clewell, D. B., S. E. Flannagan, and D. D. Jaworski. 1995. Unconstrained bacterial promiscuity: the Tn916-Tn1545 family of conjugative transposons. Trends Microbiol. 3:229-236. [DOI] [PubMed] [Google Scholar]
- 9.Danielsen, M. 2002. Characterization of the tetracycline resistance plasmid pMD5057 from Lactobacillus plantarum 5057 reveals a composite structure. Plasmid 48:98-103. [DOI] [PubMed] [Google Scholar]
- 10.Doherty, N., K. Trzcinski, P. Pickerill, P. Zawadzki, and C. Dowson. 2000. Genetic diversity of the tet(M) gene in tetracycline-resistant clonal lineages of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 44:2979-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donabedian, S. M., L. A. Thal, E. Hershberger, M. B. Perri, J. W. Chow, P. Bartlett, R. Jones, K. Joyce, S. Rossiter, K. Gay, J. Johnson, C. Mackinson, E. Debess, J. Madden, F. Angulo, and M. J. Zervos. 2003. Molecular characterization of gentamicin-resistant enterococci in the United States: evidence of spread from animals to humans through food. J. Clin. Microbiol. 41:1109-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franz, C. M. A. P., W. H. Holzapfel, and M. Stiles. 1999. Enterococci at the crossroads of food safety? Int. J. Food Microbiol. 47:1-24. [DOI] [PubMed] [Google Scholar]
- 13.Franz, C. M. A. P., A. B. Muscholl-Silberhorn, N. M. K. Yousif, M. Vancanneyt, J. Swings, and W. H. Holzapfel. 2001. Incidence of virulence factors and antibiotic resistance among enterococci isolated from food. Appl. Environ. Microbiol. 67:4385-4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franz, C. M. A. P., and A. von Holy. 1996. Thermotolerance of meat spoilage lactic acid bacteria and their inactivation in vacuum-packaged Vienna sausages. Int. J. Food Microbiol. 29:59-73. [DOI] [PubMed] [Google Scholar]
- 15.Gevers, D., M. Danielsen, G. Huys, and J. Swings. 2003. Molecular characterization of tet(M) genes in Lactobacillus isolates from different types of fermented dry sausage. Appl. Environ. Microbiol. 69:1270-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gevers, D., G. Huys, and J. Swings. 2001. Applicability of rep-PCR fingerprinting for identification of Lactobacillus species. FEMS Microbiol. Lett. 205:31-36. [DOI] [PubMed] [Google Scholar]
- 17.Giraffa, G., D. Carminati, and E. Neviani. 1997. Enterococci isolated from dairy products: a review of risks and potential technological use. J. Food Microbiol. 60:732-737. [DOI] [PubMed] [Google Scholar]
- 18.Giraffa, G., A. M. Olivari, and E. Neviani. 2000. Isolation of vancomycin-resistant Enterococcus faecium from Italian cheeses. Food Microbiol. 17:671-677. [Google Scholar]
- 19.Huang, R., D. M. Gascoyne-Binzi, P. M. Hawkey, M. Yu, J. Heritage, and A. Eley. 1997. Molecular evolution of the tet(M) gene in Gardnerella vaginalis. J. Antimicrob. Chemother. 40:561-565. [DOI] [PubMed] [Google Scholar]
- 20.Huys, G., K. D'Haene, and J. Swings. 2002. Influence of the culture medium on antibiotic susceptibility testing of food-associated lactic acid bacteria with the agar overlay disc diffusion method. Lett. Appl. Microbiol. 34:402-406. [DOI] [PubMed] [Google Scholar]
- 21.Jacob, A. E., and S. J. Hobbs. 1974. Conjugal transfer of plasmid-borne multiple antibiotic resistance in Streptococcus faecalis var. zymogenes. J. Bacteriol. 117:360-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jensen, L. B., N. Fridmodt-Moller, and F. M. Aarestrup. 1999. Presence of erm gene classes in gram-positive bacteria of animal and human origin in Denmark. FEMS Microbiol. Lett. 170:151-158. [DOI] [PubMed] [Google Scholar]
- 23.Jones, W. F., Jr., and M. Finland. 1957. Susceptibility of Enterococcus to eleven antibiotics in vitro. Am. J. Clin. Pathol. 27:467-481. [DOI] [PubMed] [Google Scholar]
- 24.Klare, I., C. Konstabel, D. Badstübner, G. Werner, and W. Witte. 2003. Occurrence and spread of antibiotic resistances in Enterococcus faecium. Int. J. Food Microbiol. 88:269-290. [DOI] [PubMed] [Google Scholar]
- 25.Klein, G. 2003. Taxonomy, ecology and antibiotic resistance of enterococci from food and the gastro-intestinal tract. Int. J. Food Microbiol. 88:123-131. [DOI] [PubMed] [Google Scholar]
- 26.Leclerc, H., L. A. Devriese, and D. A. A. Mossel. 1996. Taxonomical changes in intestinal (faecal) enterococci and streptococci: consequences on their use as indicators of faecal contamination in drinking water. J. Appl. Bacteriol. 81:459-466. [DOI] [PubMed] [Google Scholar]
- 27.Levy, S. B. 1992. The antibiotic paradox: how miracle drugs are destroying the miracle. Plenum Press, New York, N.Y.
- 28.Martinez, J. L., and F. Baquero. 2002. Interactions among strategies associated with bacterial infection: pathogenicity, epidemicity, and antibiotic resistance. Clin. Microbiol. Rev. 15:647-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morrison, D., N. Woodford, and B. Cookson. 1997. Enterococci as emerging pathogens of humans. J. Appl. Bacteriol. Symp. Suppl. 83:89S-99S. [PubMed] [Google Scholar]
- 30.National Committee for Clinical Laboratory Standards. 2003. Performance standards for antimicrobial susceptibility testing. Thirteenth informational supplement. M100-S13. National Committee for Clinical Laboratory Standards, Villanova, Pa.
- 31.Niemi, R. M., S. I. Niemelä, D. H. Bamfort, J. Hantula, T. Hyvärinen, T. Forsten, and A. Raateland. 1993. Presumptive fecal streptococci in environmental samples characterized by one-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Appl. Environ. Microbiol. 59:2190-2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oggioni, M. R., C. G. Dowson, J. M. Smith, R. Provvedi, and G. Pozzi. 1996. The tetracycline resistance gene tet(M) exhibits mosaic structure. Plasmid 35:156-163. [DOI] [PubMed] [Google Scholar]
- 33.Perreten, V., B. Kollöfel, and M. Teuber. 1997. Conjugal transfer of the Tn916-like transposon TnFO1 from Enterococcus faecalis isolated from cheese to other gram-positive bacteria. Syst. Appl. Microbiol. 20:27-38. [Google Scholar]
- 34.Peters, J., K. Mac, H. Wichmann-Schauer, G. Klein, and L. Ellerbroek. 2003. Species distribution and antibiotic resistance patterns of enterococci isolated from food of animal origin in Germany. Int. J. Food Microbiol. 88:311-314. [DOI] [PubMed] [Google Scholar]
- 35.Petersen, A., J. S. Andersen, T. Kaewmak, T. Somsiri, and A. Dalsgaard. 2002. Impact of integrated fish farming on antimicrobial resistance in a pond environment. Appl. Environ. Microbiol. 68:6036-6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pitcher, D. G., N. A. Saunders, and R. J. Owen. 1989. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett. Appl. Microbiol. 8:151-156. [Google Scholar]
- 37.Quednau, M., S. Ahrné, A. C. Petersson, and G. Molin. 1998. Antibiotic-resistant strains of Enterococcus isolated from Swedish and Danish chicken and pork. J. Appl. Microbiol. 84:1163-1170. [DOI] [PubMed] [Google Scholar]
- 38.Rice, L. B., S. H. Marshall, and L. L. Carias. 1992. Tn5381, a conjugative transposon identifiable as a circular form in Enterococcus faecalis. J. Bacteriol. 174:7308-7315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ridenhour, M. B., H. M. Fletcher, J. E. Mortensen, and L. Daneo-Moore. 1996. A novel tetracycline-resistant determinant, tet(U), is encoded on the plasmid pKQ10 in Enterococcus faecium. Plasmid 35:71-80. [DOI] [PubMed] [Google Scholar]
- 40.Robredo, B., K. V. Singh, F. Baquero, B. Murray, and C. Torres. 2000. Vancomycin-resistant enterococci isolated from animals and food. Int. J. Food Microbiol. 54:197-204. [DOI] [PubMed] [Google Scholar]
- 41.Rollins, L. D., L. N. Lee, and D. J. LeBlanc. 1985. Evidence for a disseminated erythromycin resistance determinant mediated by Tn917-like sequences among group D streptococci isolated from pigs, chickens, and humans. Antimicrob. Agents Chemother. 27:439-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simjee, S., D. G. White, J. Meng, D. D. Wagner, S. Qaiyumi, S. Zhao, J. R. Hayes, and P. F. McDermott. 2002. Prevalence of streptogramin resistance genes among Enterococcus isolates recovered from retail meats in the greater Washington DC area. J. Antimicrob. Chemother. 50:877-882. [DOI] [PubMed] [Google Scholar]
- 43.Smith, D. L., A. D. Harris, J. A. Johnson, E. K. Silbergeld, and J. G. Morris, Jr. 2002. Animal antibiotic use has an early but important impact on the emergence of antibiotic resistance in human commensal bacteria. Proc. Natl. Acad. Sci. USA 99:6434-6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Su, Y. A., P. He, and D. B. Clewell. 1992. Characterization of the tet(M) determinant of Tn916: evidence for regulation by transcription attenuation. Antimicrob. Agents Chemother. 36:769-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Teuber, M., L. Meile, and F. Schwarz. 1999. Acquired antibiotic resistance in lactic acid bacteria from food. Antonie van Leeuwenhoek 76:115-137. [PubMed] [Google Scholar]
- 46.Teuber, M., V. Perreten, and F. Wirsching. 1996. Antibiotikumresistente Bakterien: eine neue Dimension in der Lebensmittelmikrobiologie. Lebensmittel-Technol. 29:182-199. [Google Scholar]
- 47.Teuber, M., F. Schwarz, and V. Perreten. 2003. Molecular structure and evolution of the conjugative multiresistance plasmid pRE25 of Enterococcus faecalis isolated from a raw-fermented sausage. Int. J. Food Microbiol. 88:325-329. [DOI] [PubMed] [Google Scholar]
- 48.Toala, P., A. McDonald, C. Wilcox, and M. Finland. 1970. Comparison of antibiotic susceptibility of group D streptococcus strains isolated at Boston City Hospital in 1953-54 and 1968-69, p. 479-484. Antimicrob. Agents Chemother. 1969. [PubMed]
- 49.Trzcinski, K., B. S. Cooper, W. Hryniewicz, and C. G. Dowson. 2000. Expression of resistance to tetracyclines in strains of methicillin-resistant Staphylococcus aureus. J. Antimicrob. Chemother. 45:763-770. [DOI] [PubMed] [Google Scholar]
- 50.Vancanneyt, M., K. Kersters, and J. Swings. 1999. Catalogue of enterococci of the FAIR-E collection. BCCM/LMG Bacteria Collection, Ghent, Belgium.
- 51.von Holy, A., T. E. Cloete, and W. H. Holzapfel. 1991. Quantification and characterization of microbial populations associated with spoiled, vacuum-packaged Vienna sausages. Food Microbiol. 8:95-104. [DOI] [PubMed] [Google Scholar]
- 52.Wegener, H. C., M. Madsen, N. Nielsen, and F. M. Aarestrup. 1997. Isolation of vancomycin resistant Enterococcus faecium from food. Int. J. Food Microbiol. 35:57-66. [DOI] [PubMed] [Google Scholar]