Abstract
Estrogen hormone (E2) is involved in the physiology and pathology of many tissues. E2 information is conveyed by the transcription factors estrogen receptors (ER) α and β that mediate a complex array of nuclear and non-nuclear events. The interaction of ER with specific DNA sequences, estrogen responsive elements (EREs), constitutes one critical nuclear signaling pathway. In addition, E2-ER regulates transcription through interactions with transfactors bound to their cognate regulatory elements on DNA, hence the ERE-independent signaling pathway. However, the relative importance of the ERE-independent pathway in E2-ERβ signaling is unclear. To address this issue, we engineered an ERE binding defective ERβ mutant (ERβEBD) by changing critical residues in the DNA binding domain required for ERE binding. Biochemical and functional studies revealed that ERβEBD signaled exclusively through the ERE independent pathway. Using adenovirus infected ER-negative cancer cell models, we found that although E2-ERβEBD regulated the expression of a number of genes identified by microarrays, it was ineffective to alter cellular proliferation, motility and death in contrast to E2-ERβ. Our results indicate that genomic responses from the ERE-independent pathway to E2-ERβ are not sufficient to alter cellular phenotype. These findings suggest that the ERE-dependent pathway is a required signaling route for E2-ERβ to induce cellular responses.
Keywords: Estrogen, ERβ, ERE-Dependent, ERE-Independent, Gene Expression
INTRODUCTION
Estrogen hormones, particularly the main circulating estrogen, 17β-estradiol (E2), play important roles in the regulation of many tissue functions (Deroo and Korach 2006; Huang, et al. 2005a). E2 is also involved in the initiation and development of target tissue malignancies. The E2 information is primarily conveyed by estrogen receptor (ER) α and β (Deroo and Korach 2006; Huang et al. 2005a). ERs are members of the conserved superfamily of hormone receptors and are ligand-activated transcription factors. The effects of E2-ER are exerted through a complex array of convergent and divergent signaling pathways that mediate genomic events involved in the regulation of mitogenesis, motogenesis and apoptosis (Hall, et al. 2001; Huang et al. 2005a; Nilsson, et al. 2001). The interaction of E2-ER with specific DNA sequences, estrogen responsive elements (EREs), constitutes one primary genomic signaling pathway (Hall et al. 2001; Huang et al. 2005a; Nilsson et al. 2001). The ERE-bound ER recruits an ensemble of multi-subunit complexes responsible for the alteration of local chromatin structure and the interaction with the basal transcription machinery. The integrated effects of these complexes regulate transcription. This type of E2-ER mediated signaling is referred to as the ERE-dependent signaling pathway (Hall et al. 2001; Huang et al. 2005a; Nilsson et al. 2001).
In addition, the E2-ER complex regulates the expression of E2 responsive genes through functional interactions with transcription factors bound to their cognate regulatory elements on DNA (Kushner, et al. 2000; Safe 2001). In this DNA-dependent and ERE-independent signaling pathway, transcriptional responses are dependent upon ER-subtype, promoter- and cell-context (Kushner et al. 2000; Safe 2001). Despite the large body of experimental evidence, the relative importance of the ERE-independent pathway in physiology, and its contribution to pathophysiology, of E2-ERβ signaling is unclear. We envisioned that a selective regulation of the ERE-independent genes would allow us to begin to address this issue. To accomplish this, we generated an ERE binding defective ERβ mutant (ERβEBD) that renders the receptor nonfunctional at the ERE-dependent pathway, while conserving the regulatory potential at the ERE-independent pathway. We used ER-negative cells as experimental models, with which exogenously introduced ERs were shown to regulate the expression of responsive genes (Kian Tee, et al. 2004; Licznar, et al. 2003; Moggs, et al. 2005; Monroe, et al. 2005; Stossi, et al. 2004) and to induce phenotypic changes (Garcia, et al. 1992; Jiang and Jordan 1992; Lazennec, et al. 2001; Lazennec and Katzenellenbogen 1999; Licznar et al. 2003; Zajchowski, et al. 1993). We found in adenovirus-infected cells that genomic responses induced by ERβEBD in response to a physiological level of E2 are insufficient to alter cellular proliferation, death or motility in contrast to E2-ERβ. These results imply that the ERE-dependent pathway is the required signaling route mediated by the E2-ERβ complex.
MATERIALS AND METHODS
Generation of ERβ DNA binding defective mutant (ERβEBD)
The human ERβ cDNAs that encodes the 530 amino-acid long ERβ was described previously (Yi, et al. 2002a). This ERβ cDNA also contains sequences encoding an amino-terminal Flag epitope (Yi et al. 2002a). For the engineering of an ERE binding defective ERβ (ERβEBD), we utilized an overlapping PCR with the ERβ cDNA as the template and primers that contain amino acid substitutions to replace glutamic acid and glycine at position 167 and 168, respectively, with alanine residues in the first zinc finger of the DNA binding domain (DBD) of the receptor.
Restriction and DNA modifying enzymes were obtained from New England Bio-Labs (Beverly, MA) and Invitrogen Corp. (Carlsbad, CA).
Cell culture
Culturing of MDA-MB-231 and HeLa cells was described previously (Yi et al. 2002a). U-2 OS cells derived from osteosarcoma were purchased from ATCC (Manassas, VA). U-2 OS cells were grown with in McCoy’s 5α medium supplemented with 10% fetal bovine serum (FBS, Invitrogen). In all experiments, medium was changed every third day.
Transient transfections
Transient transfections for the simulated ERE-dependent and -independent pathways were accomplished as described previously (Huang, et al. 2004; Yi et al. 2002a). Transfected cells were treated without or with 10−9 M 17β-estradiol (E2), 10−7 M 4-hydroxyl-tamoxifen (4-OHT) (Sigma-Aldrich), 10−7 M Imperial Chemical Industries 182,780 (ICI, Tocris Inc., Ballwin, MO), and 10−8 M diarylpropionitrile (DPN, Tocris) for 24h or 40h to assess the effects of ligands on ER-mediated transcriptional responses from the ERE-dependent or -independent signaling, respectively.
Generation of a recombinant adenovirus bearing an ER cDNA
Recombinant adenovirus bearing none, the cDNA of Flag-ERβ or Flag-ERβEBD were produced by using the AdEasy-XL Adenoviral System (Stratagene, La Jolla, CA) as described previously (Huang, et al. 2005b). The purified viruses were titered using an Adeno-X Rapid Titer Kit (BD Biosciences, Palo Alto, CA) to determine multiplicity of infection (MOI).
Western blot (WB), Electrophoretic mobility shift assay (EMSA), and Immunocytochemistry (ICC)
Transfected or infected cells in a time-dependent manner were processed for WB, EMSA and ICC as described previously (Muyan, et al. 2001; Yi et al. 2002a). For WB, proteins were probed with horseradish peroxidase conjugated monoclonal Flag antibody (M2-HRP, Sigma-Aldrich) using the ECL-Plus Western Blotting kit (Amersham-Pharmacia). Images were captured by PhosphorImager (Molecular Dynamics, Sunnyvale, CA). For ICC, we used an ERβ-specific antibody (Zymed Laboratories, San Fransisco, CA) followed by a fluorescein conjugated secondary antibody (Santa Cruz Biotechology).
In situ E2 binding and ERE competition assays
To assess the functionality of ERβ species in transfected cells, we used the in situ E2 binding assay and the in situ ERE competition assays as described previously (Huang et al. 2005b).
Chromatin Immunoprecipitation Assay (ChIP)
ChIP assay was performed using Flag-M2 antibody conjugated agarose beads (Sigma-Aldrich) as described previously (Huang et al. 2005b). The generation of a 366 bp PCR fragment indicates the specificity of PCR reactions.
Endogenous Gene Expression
MDA-MB-231 cells (100,000 cells/well) plated in 6-well tissue culture plates in phenol red-free DMEM containing 10% CD-FBS for 24h were infected with recombinant adenoviruses without or with 10−9 M E2 for 48h to assess the effects of ER on the expression of the TFF1, C3, MMP1 and RARA genes.
Cells were also infected with recombinant adenoviruses in the absence of E2 and maintained for 48h, the time at which the synthesis of ERs reaches comparable levels (Fig. 1). Infected cells were then treated with 10−9 M E2 for 6, 12 or 24h to confirm the identities of genes determined by microarrays using quantitative PCR (qPCR). At the termination, cells were collected and subjected to total RNA extraction using the RNeasy Mini Kit (Qiagen, Valencia, CA) for qPCR, which we used custom TaqMan Low-Density Arrays with proprietary primer and probe sequences (Applied Biosystems, Foster City, CA). All qPCRs were carried out at the Functional Genomic Center of the University of Rochester, NY. The expression of the ACTB (actin β) and GAPDH (Glyceraldehyde-3-phosphate dehydrogenase) genes was used as controls. The real-time RT-PCR amplifications were accomplished using an ABI Prism 7900HT Sequence Detection System with a TaqMan Low Density Array Upgrade (Applied Biosystems). Relative Quantification Analysis was performed using the Comparative CT Method (Livak and Schmittgen 2001).
Fig. 1.
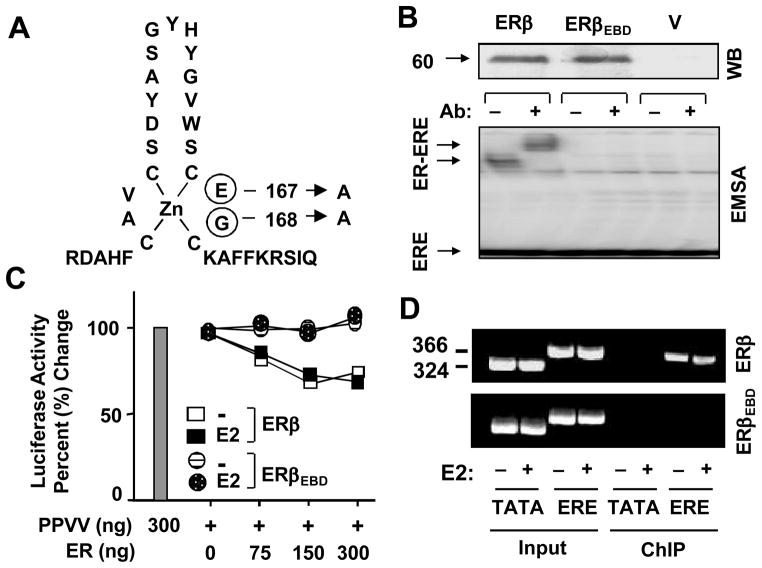
Generation of an ERE binding defective ERβ (ERβEBD). (A) ERβEBD was engineered by changing glutamic acid and glycine at positions 167 and 168 of the first zinc-finger of the DNA binding domain of the ERβ, respectively, to alanine residues. (B) The synthesis of ERβ species. MDA-MB-231 cells were transiently transfected with an expression vector bearing none (V), the ERβ or ERβEBD cDNA. Cell extracts (10 μg) were subjected to western blotting (WB) using a horseradish peroxidase conjugated monoclonal Flag antibody. MW in KDa is indicated. Cell extracts (20 μg) of transfected cells were also subjected to electrophoretic mobility shift assay (EMSA) without (−) or with (+) a Flag antibody (Ab). ERE denotes unbound and ER-ERE indicates ER bound radiolabeled ERE. Representative result from three independent experiments of WB and EMSA are shown. (C) The in situ ERE competition assay. MDA-MB-231 cells were transiently transfected with a reporter vector bearing one ERE upstream of the TATA box promoter driving the expression of Firefly luciferase cDNA and the expression plasmid for PPVV, together with various concentrations of expression vector bearing the ERβ or ERβEBD cDNA. Cells were also co-transfected with a reporter plasmid bearing Renilla luciferase cDNA to monitor transfection efficiency. Cells were then treated without (−E2) or with (+E2) 10−9 M E2 for 24h. The relative luciferase activity was presented as percentage (%) change compared to control (PPVV alone in the absence of E2), which was set to 100%. Shown is the mean of three independent experiments performed in duplicate. SEM, which was less than 15% of the mean, is not shown for simplicity. (D) Chromatin immunoprecipitation (ChIP) assay. MDA-MB-231 cells were transiently transfected with an expression vector bearing the ERβ or ERβEBD cDNA together with a reporter bearing none (TATA) or one ERE (ERE) TATA box promoter. Cells were treated without (−) with (+) 10−7 M E2 for 1h prior to ChIP using the Flag antibody-conjugated agarose beads. Sizes of DNA fragments in base-pair are indicated. A representative image from three independent experiments is shown.
Cell proliferation
MDA-MB-231 cells (5,000 cells/well) plated in 24-well tissue culture plates in phenol red-free DMEM containing 10% CD-FBS for 24h were infected with recombinant adenoviruses in the absence or presence of 10−9 M E2 for different durations of time. Cells were collected and counted using a hemacytometer (Hausser Scientific, Horsham, PA).
Additionally we used a colorimetric proliferation assay, MTT assay (Mosmann 1983). Cells, plated and infected with adenoviruses as described for hemacytometric cell counting, were incubated with 200 μl of phenol red-free DMEM with 10% CD-FBS that contains 60 μM of MTT (Invitrogen) for 2h. The spent medium was removed and 200 μl of dimethyl sulfoxide (DMSO) were added to each well to dissolve the MTT formazan. The absorbance was measured by a Microplate Spectrophotometer, SpectraMax Plus (Molecular Devices, Sunnyvale, CA) to estimate cell number.
For U-2 OS cell proliferation, cells (5,000 cells/well) were plated in 24-well tissue culture plates, pre-coated with poly-L-lysine (Sigma-Aldrich), in McCoy’s α medium containing 10% FBS for 24h. Cells were subsequently incubated with McCoy’s α medium containing 10% CD-FBS for an additional 24h. Cells were then infected with recombinant adenoviruses in the absence or presence of 10−9 M E2 for different durations of time. We used Ad5-ERβ at 40 MOI. At this concentration, the recombinant adenovirus synthesizes a concentration of ERβ that is dependent upon E2 for function. Ad5-ERβEBD was used at 50 MOI, which produced comparable levels of receptor to that of ERβ. At the termination of an experiment, cells were subjected to cell counting and MTT assays.
Cell cycle analysis
MDA-MB-231 cells (50,000 cells/well) in 6-well tissue culture plates were infected with recombinant adenoviruses in the absence or presence of 10−9 M E2 for different durations. Cells were collected and pelleted. Pelleted cells were resuspended in ethanol (70%) to fix and permeabilize at 4C overnight. Cells were subsequently incubated with 1 mg/ml of RNase A (Sigma-Aldrich) for 30 min followed by 20 μg/ml of propidium iodide (PI) (Sigma-Aldrich) for 10 min. Cells were then subjected to a fluorescence-activated cell sorting (FACS) using EPICS Elite (Coulter Corp., Miami, FL).
Caspase 3/7 assay
MDA-MB-231 cells, (12,500 cells/well) plated onto poly-L-Lysine coated 96-well tissue culture black plates with clear bottom (BD Biosciences, Franklin Lakes, NJ) in phenol red-free DMEM containing 10% CD-FBS for 24h were infected with recombinant adenoviruses, in the absence or presence of 10−9 M E2, for different lengths of time. Cells were then subjected to Apo-ONE Homogeneous Caspase-3/7 Assay (Promega, Madison, WI) according to the manufacturer’s protocol. The fluorescence was then measured by a Spectrophotometer.
Annexin V assay
To study apoptosis by examining the loss of cell membrane asymmetry as an indicator of middle stages of apoptosis, we used the Vybrant Apoptosis Assay Kit (Invitrogen). This assay is based on the specific recognition of phosphatidyl-serine (PS) by FITC conjugated annexin V. MDA-MB-231 cells (100,000 cells/well) were infected with recombinant adenoviruses in the absence or presence of 10−9 M E2 for different lengths of time. Cells were collected and subjected to Annexin V assay according to the instruction of the manufacturer prior to FACS analysis.
TUNEL assay
To study late stages of apoptosis by examining the fragmentation of genomic DNA (Korsmeyer 1999), MDA-MB-231 cells (25,000 cells/well) plated in poly-L-Lysine coated 48-well tissue culture plates in phenol red-free DMEM containing 10% CD-FBS for 24h were infected with recombinant adenoviruses in the absence or presence of 10−9 M E2 for different lengths of time. Cells were then subjected to a Terminal dUTP Nick-End Labeling (TUNEL) assay utilizing the DeadEnd Flurometric TUNEL System (Promega) according to the manufacturer’s protocol. DAPI (Vector Labs) was used to stain cell nuclei. Stained cells were imaged under a microscope with corresponding filters.
Wound-healing assay
MDA-MB-231 cells (200,000 cells/well in 12-well tissue culture plates) were infected with recombinant adenoviruses in the absence or presence of 10−9 M E2 for 48h for confluence. A wound was then created using a one ml pipette tip. The gap closure was photographed at every 24h. Due to irregular shape of the edges of a wound, five randomly selected cross edges were used to obtain a mean gap measure for wound-healing.
Invasion assay
We used BD Matrigel Invasion Chambers (BD Biosciences, San Diego, CA) for the invasion assay. MDA-MB-231 cells (100,000 cells/well) in 6-well tissue culture plates were infected with recombinant adenoviruses in the absence or presence of 10−9 M E2 for 48h. Cells were then trypsinized and counted. The same number (25,000 cells/chamber) of cells was seeded on the upper section of the chamber, which contained phenol red-free DMEM without or with 10−9 M E2. The lower section of the chamber contained phenol red-free DMEM supplemented with 10% CD-FBS and 30 μg/ml fibronectin in the absence or presence of 10−9 M E2. After 24h incubation, cells on the upper section of the membrane were removed by cotton swab. Cells on the bottom of chamber membrane were stained with the Diff-Quik Stain Set (Dade Behring, Newark, DE), dried and mounted onto a glass slide. Images were captured and stained cells were counted from images.
Microarray Analysis
To examine the effects of E2 on endogenous gene expression mediated by ERs, MDA-MB-231 cells were infected with recombinant adenoviruses in the absence of E2 for 48h. Infected cells were then treated with 10−9 M E2 for 6h. This duration of E2 treatment was expected to induce significant changes in the level of immediate/early gene expression, as observed with the responses to E2 in ERα synthesizing breast cancer cell lines (Frasor, et al. 2003). At the termination, cells were subjected to RNeasy Mini kit (Qiagen) for total RNA extraction. Processing of RNA for microarray analysis was carried out at the Functional Genomic Center of the University of Rochester, NY. The cDNA synthesis and subsequent fragmentation and biotinylation of cDNA fragments were carried out using the Ovation kit (NuGEN, San Carlos, CA) according to the manufacturer’s procedure. The biotinylated cDNA fragments were then used for hybridization with microarrays. We used Affymetrix HG-U133 Plus 2.0 arrays. Arrays were scanned with the GeneChip Scanner 3000 7G. GeneChip Operating Software (GCOS, Affymetrix) was used for initial processing of the scanner data, including generation of cel files. Array normalization for the Affymetrix signal method (GCOS) involves multiplying raw signals by a scaling factor such that the trimmed mean (excluding highest and lowest 2%) of all expression scores is 500 arbitrary units for every array.
Experimental sets for microarrays were replicated six independent times executed on different days. Although we also conducted microarray data analysis based on probe sets that relied on earlier genome and transcriptome annotation (Dai, et al. 2005; Gautier, et al. 2004; Harbig, et al. 2005) and posted at http://dbb.urmc.rochester.edu/labs/muyan/ArrayAddendum.htm, we present here data analysis using reorganized and updated probe sets (Dai, et al. 2005; Gautier, et al. 2004; Harbig, et al. 2005) based on the up-to-date genome, cDNA/EST clustering and single nucleotide polymorphism information through web-based custom GeneChip library files (Chip Definition Files or CDFs, http://arrayanalysis.mbni.med.umich.edu) that increase accuracy and reduce false discovery rate (Dai, et al. 2007). Following UniGene transformation, data sets were subjected to N-statistic test (Klebanov, et al. 2006) in conjunction with the step-down Westfall-Young procedure (Westfall and Young 1993) controlling the family-wise error rate (FWER) at a level of 0.05, which was reported here. MIAME (Minimum Information About a Microarray Experiment)-compliant microarray data for the six independent replicate studies have been submitted to the Gene Expression Omnibus (GEO) database (GEO 2004) with an accession number GSE9761.
Statistical Analysis
Results were presented as the mean ± SEM of, at least, three independent experiments. Student’s t test was employed for comparison of the means between two groups wherein P < 0.05 was considered significant.
RESULTS
The functional characterization of the ERE binding defective ERβ (ERβEBD)
ERs recognize an ERE as a dimer mediated by a dimerization interface located in the ligand binding domain (LBD) and a weak interaction surface in the DNA binding domain (DBD) (Parker 1998). The DBD of ERα contains two zinc finger-like modules that fold to form a single functional domain. Each DBD of the ERα dimer makes analogous contacts with one of the inverted motifs of ERE that results in a rotationally symmetrical structure (Luisi, et al. 1994). Distinct residues in a region of the first zinc-finger module of DBD, the P-box, particularly glutamic acid and glycine at positions 203 and 204, respectively, determine the DNA binding specificity critical for sequence discrimination (Green, et al. 1988). Residues in the second zinc-finger-like module, the D-box, are involved in the discrimination of half-site spacing through a protein-protein interaction between two ER monomers (Green et al. 1988). The human ERα and ERβ share a 97% amino-acid identity in their DBDs with identical residues at the P- and D-boxes (Mosselman, et al. 1996). This structural homology is reflected in the abilities of ERs to bind various ERE sequences with a similar specificity and affinity by interacting with the same nucleotides (Yi, et al. 2002b).
Studies showed that the regulation of E2 responsive genes through ERE-independent signaling involves a functional interaction between ERs and a transcription factor bound to its cognate response element on DNA through regions that also encompass the DBD of ERs (Kushner et al. 2000; Safe 2001). Changing of glutamic acid and glycine at positions 207 and 208 of the mouse ERα, and at positions 167 and 168 of human ERβ, respectively, at the P-box of the first zinc finger of the DBD hinders the ER-ERE interactions, while conserving the capacity to regulate transcription from the ERE-independent signaling pathway (Bjornstrom and Sjoberg 2002; Jakacka, et al. 2001). Based on these observations, we engineered an ERE binding defective ERβ variant (ERβEBD) bearing alanine residues at positions 167 and 168 to exclusively regulate the DNA-dependent and ERE-independent pathway (Fig. 1A).
The initial biochemical characterization of ERβEBD was carried out in transiently transfected ER-negative cell models. Cell extracts, shown for MDA-MB-231 cells, were subjected to Western blot (WB) and electrophoretic mobility shift assay (EMSA). The detection of receptor proteins by WB using a Flag antibody (M2) directed to the Flag epitope at the amino-termini of the receptors indicated that receptors were synthesized at similar levels (Fig. 1B, WB). EMSA, using a 32P-end labeled DNA fragment bearing the consensus ERE, revealed that despite the comparable level of synthesis, ERβ, but not ERβEBD, interacted in vitro with ERE (Fig. 1B, EMSA).
To ensure that ERβEBD is indeed defective in binding to ERE in situ, we used in situ ERE competition and chromatin immunoprecipitation (ChIP) assays. The in situ ERE competition assay is based on the ability of ER to compete for ERE binding with a designer activator, designated as PPVV, which potently and constitutively induces transcription from an ERE-driven reporter construct (Huang et al. 2005b). Interference of activator-mediated transcription by unliganded or liganded ERs is then taken as an indication of ER-ERE interaction. The reporter TATA box plasmid bearing none (TATA) or one ERE (ERE) was co-transfected with an expression vector encoding the PPVV cDNA into cell models, shown for MDA-MB-231 cells, in the absence or presence of varying amounts of an expression vector bearing an ER cDNA without or with a physiological concentration (10−9 M) of E2 for 24h. The normalized luciferase activity mediated by PPVV alone in the absence of E2 was set to 100%. Alterations in the reporter enzyme activity as a result of a co-transfected ER cDNA without or with E2 are depicted as percentage (%) change compared to the activity induced by PPVV alone. Previously we (Huang et al. 2005b) and others (Hall and McDonnell 1999) showed that ERβ, in contrast to ERα, interacts with ERE in situ independent of E2. Consistent with this finding, our results here also reveal that transfection with increasing concentrations of the expression vector bearing the ERβ, but not ERβEBD, cDNA gradually decreased the luciferase activity induced by PPVV independent of E2 (Fig. 1C), without altering responses from the reporter plasmid bearing the TATA box promoter (data not shown).
A ChIP assay was also employed to corroborate our finding (Fig. 1D). The expression vectors were co-transfected with the reporter TATA box promoter vector bearing none (TATA) or one ERE (ERE) into MDA-MB-231 cells. Cells were treated without or with a saturating concentration (10−7 M) of E2 for 1h, and processed for ChIP. We found that ERβ, but not ERβEBD or the parent vector (data not shown), produced a PCR product from cells co-transfected with the reporter vector bearing ERE whether or not cells were treated with E2 as observed with the in situ ERE competition assay (Fig. 1C). Thus, these results collectively suggest that ERβEBD does not interact with ERE in situ as well.
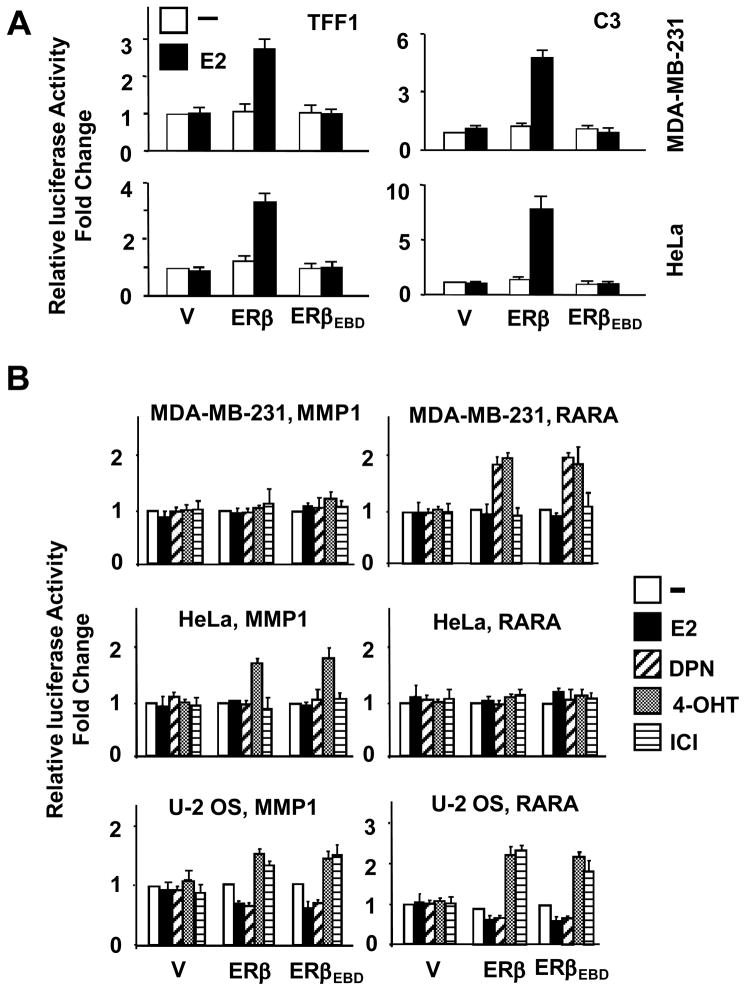
To ensure that ERβEBD is functional only in the ERE-independent pathway, the expression vectors were transiently transfected into ER-negative MDA-MB-231, HeLa or U-2 OS cells. Cells were also co-transfected with a reporter vector containing a promoter driving the expression of the firefly luciferase cDNA as the reporter enzyme. Promoters were derived from the estrogen responsive TFF1 (Trefoil factor 1, pS2) and C3 (Complement component 3) genes that contain ERE sequences, hence modeling the ERE-dependent signaling pathway (Huang et al. 2004; Yi et al. 2002a). Normalized activity from each reporter construct was compared to the parent expression vector (V) in the absence of E2, with the latter value set to one. Results showed that the ERβEBD had no effect on luciferase activity in the absence or presence of 10−9 M E2 from either the TFF1 or C3 gene promoter in all cell lines tested, shown MDA-MB-231 and HeLa cells (Fig. 2A). In contrast, ERβ increased the reporter enzyme activity in response to E2 from both promoters in all cell lines.
Fig. 2.
Transcriptional responses to ERβs from heterologous reporter systems. (A) Responses from the ERE-dependent signaling pathway. MDA-MB-231 or HeLa cells were transiently transfected with an expression vector bearing none (V), ERβ or ERβEBD cDNA together with a reporter vector bearing the TFF1 or C3 gene promoter in the absence or presence of 10−9 M E2 for 24h. Normalized luciferase values represented as fold changes compared with the luciferase values obtained with control vector in absence of E2, which was set to 1. Shown are the mean ± SEM of three independent experiments performed in duplicate. (B) Transactivation ability of ERβEBD in simulated ERE-independent signaling pathways. MDA-MB-231, HeLa, or U-2 OS cells were transiently transfected with an expression vector bearing ERβ or ERβEBD cDNA together with reporter vector bearing the MMP1 gene or the RARA gene proximal promoter. Cells were treated without or with 10−9 M E2, 10−8 DPN, 10−7 M 4-OHT or ICI for 40h. Normalized luciferase values are represented as fold change compared with the luciferase values obtained without ligand. Results are the mean ± SEM of three independent experiments performed in triplicate.
To confirm that the transregulatory function of ERβEBD is restricted to the ERE-independent signaling pathway, we used a reporter construct that bears the proximal MMP1 (Matrix metallopeptidase 1, Collagenase) or RARA (Retinoic acid receptor α) gene promoter (Huang et al. 2004). The functional interaction of ERs with the Jun/Fos family of proteins bound to an activator protein-1 (AP1) element in the proximal promoter of the MMP1 gene provides the basis for the responsiveness to ERs in a ligand- and cell-type dependent manner in experimental systems (Kushner et al. 2000; Paech, et al. 1997; Webb, et al. 1995; Webb, et al. 1999). Similarly, the interaction of ER with specificity protein-1 (SP1) bound to GC boxes of the proximal promoter is critical for the ligand-mediated regulation of the RARA gene (Rishi, et al. 1995; Safe 2001; Sun, et al. 1998). Transiently transfected MDA-MB-231, HeLa or U-2 OS cells were incubated without or with 10−9 M E2, 10−8 M 2,3-bis(4-hydroxyphenyl)-propionitrile (DPN), 10−7 M 4-hydroxytamoxifen (4-OHT), and 10−7 M Imperial Chemical Industries 182,780 (ICI). DPN is an ERβ subtype-specific estrogenic compound (Harrington, et al. 2003). Results revealed that while the parent vector had no effect on transcription in the absence or presence of ligands in all cell lines tested, ERβ and ERβEBD mediated the effects of ligands on the reporter enzyme activity similarly independent of promoter and cell type (Fig. 2B). In MDA-MB-231 cells, only DNP and 4-OHT enhanced both ERβ- and ERβEBD-induced responses from the RARA promoter construct, while E2 or ICI had no effect on the reporter enzyme activity mediated by either ER. In contrast to MDA-MB-231 cells, ERs expressed in HeLa cells enhanced the reporter enzyme activity from the MMP1 promoter in response to only 4-OHT. In this cell lines, ERβ or ERβEBD had no effect on transcriptional responses from the RARA gene promoter in the absence or presence of ligands. In U2- OS cells, on the other hand, E2 and DPN repressed reporter enzyme activity mediated by ERβ and ERβEBD from both the MMP1 and the RARA promoter constructs, whereas 4-OHT and ICI augmented responses to both receptors.
While the transcriptional responses mediated by ER species varied depending upon the nature of ligand, promoter and cell context, our findings clearly indicate that ERβEBD mirrored the effect of ERβ on the reporter enzyme activity from reporter constructs emulating the ERE-independent signaling pathway. Our results suggest that ERβEBD retains its transregulatory function in the ERE-independent pathway, despite the fact that it does not bind to ERE and, consequently, is nonfunctional in the ERE-dependent signaling pathway.
Regulation of endogenous gene expression by E2-ERβs
The experimental reporter systems we used here contain minimal promoter sequences responsive to E2-ER signaling. While transient transfection into mammalian cells has been a valuable tool for the understanding of action mechanisms of transfactors, nucleosome deposition onto non-replicating reporter vector displays an incompletely organized nucleosome array in contrast to the replicative cellular chromatin (Archer, et al. 1992; Lee and Archer 1994; Pennie, et al. 1995). This results in a more accessible chromatin of the transfected DNA that affects basal and transfactor-regulated transcription compared to responses observed with cellular chromatin (Archer et al. 1992; Lee and Archer 1994; Pennie et al. 1995). Moreover, the temporal regulation of endogenous E2 target gene expression is the result of integrated effects of transcription factors that bind to cognate responsive elements within the regulatory regions. The combinatorial effects of transfactors and ER ultimately determine the magnitude and/or direction of the endogenous gene expression (Nunez, et al. 1989; Vyhlidal, et al. 2000). Consistent with heterologous expression systems, as we demonstrated here (Fig. 2), the presence of an ERE in the TFF1 gene promoter provides the endogenous responsiveness to E2-ER signaling (Nunez et al. 1989). The regulation of the RARA gene expression is, on the other hand, attributed to ER-SP1 interactions in experimental systems (Rishi, et al. 1995; Safe 2001; Sun, et al. 1998), as we also showed here (Fig. 2) and previously (Li, et al. 2004). However, a recent genomics approach identified a long-distance regulatory module that contains a functional ERE responsible for the regulation of the RARA gene expression in breast cancer cell models expressing endogenous ER (Laganiere, et al. 2005).
To ensure that ERβEBD can indeed discriminately regulate endogenous gene expression, we used MDA-MB-231 cells as a model within which exogenously expressed ERs were shown to induce gene expressions that affect phenotypic characteristics (Garcia et al. 1992; Jiang and Jordan 1992; Lazennec et al. 2001; Lazennec and Katzenellenbogen 1999; Licznar et al. 2003; Moggs et al. 2005; Zajchowski et al. 1993). We also used recombinant adenoviruses for an efficient gene delivery (Huang et al. 2005b).
Recent studies showed that augmented levels of unliganded ERα lead to aberrant gene expression and an altered phenotype through mechanisms that are distinct from those mediated by E2 (Fowler, et al. 2004; Fowler, et al. 2006). To circumvent this potential problem, we used various concentrations of the recombinant adenovirus bearing ERβ cDNA (Ad5-ERβ) to obtain an optimal MOI that leads to a level of receptor synthesis that requires E2 to regulate gene expression and cellular growth. We found that MDA-MB-231 cells infected with Ad5-ERβ at 600 MOI synthesize a concentration of ERβ that is strictly dependent upon E2 for function (data not shown). The recombinant adenovirus expressing ERβEBD cDNA (ERβEBD) was used at 900 MOI producing comparable levels of receptor to that of ERβ (see also Fig. 3B). We therefore used 900 MOI of the parent recombinant adenovirus (Ad5), 600 MOI of Ad5-ERβ, which was brought to 900 MOI by supplementing with 300 MOI of Ad5, and 900 MOI of ERβEBD in subsequent experiments.
Fig. 3.
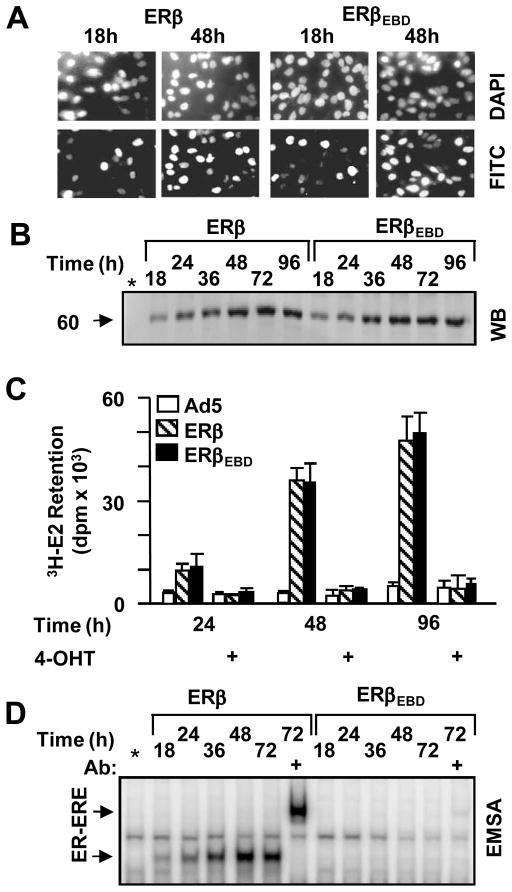
Functional ER synthesis in infected model cells. (A) MDA-MB-231 cells were infected with the parent recombinant adenovirus (Ad5) at MOI 900, a recombinant adenovirus bearing ERβ cDNA (ERβ) at 600 MOI together with 300 MOI Ad5 to equalize the total adenovirus titer, or 900 MOI for virus bearing ERβEBD cDNA. Intracellular localization of receptor proteins was examined by immunocytochemistry. Infected cells as a function of time (shown at 18 and 48h post-infection) were probed with an ERβ specific antibody followed by a fluorescein-conjugated secondary antibody (FITC). DAPI was used to stain nuclei. (B) Cell extracts (10 μg) of infected cells at indicated times were subjected to WB using the horseradish peroxidase conjugated monoclonal Flag antibody. Molecular weight in KDa is indicated. * indicates extracts of the parent adenovirus infected cells at 96h. The image is a representative of three independent experiments. (C) Cell extracts (20 μg) of infected cells for the indicated times were subjected EMSA in the absence (−) or presence (+) of the anti-flag antibody. * denotes extracts of the parent adenovirus infected cells at 72h. ER-ERE indicates the radiolabeled ERE bound ERs. Unbound radiolabelled ERE is not shown for simplicity. A representative image from three independent experiments is shown. (D) In situ E2 binding assay. Infected MDA-MB-231 cells, maintained for various durations of time as indicated, were incubated in medium containing 10−7 M of 3H-E2 for 1h. Radioactivity retained in cells was quantified. That 3H-E2 was specifically retained by ERs in cells was assessed by the co-incubation of cells with 10−6 M 4-OHT. The graph represents the mean ± SEM of three independent experiments performed in duplicate.
The synthesis of functional receptor protein was examined by immunocytochemistry (ICC), WB, E2 binding and EMSA assays in a time dependent manner in infected MDA-MB-231 cells. ICC revealed that receptor proteins became detectable in the nuclei of infected cells as a function of time such that at 48h post-infection nearly all cells showed staining for ERβ and ERβEBD (Fig. 3A). This was a reflection of the level of ER synthesis as assessed by WB, which showed that ERβ and ERβEBD were synthesized comparably in a time dependent manner (Fig. 3B). The in situ E2 binding assay further showed that 3H-E2 was similarly retained in cells that synthesize ERβ and ERβEBD but not the parent recombinant adenovirus (Fig. 3C). The co-incubation of cells with 4-OHT prevented the retention of radiolabeled E2. This indicates that 3H-E2 was specifically retained by ERs in cells. These results demonstrate that MDA-MB-231 cells infected with recombinant adenoviruses synthesize comparable amounts of receptor proteins capable of binding to E2. Moreover, EMSA further revealed that ERβEBD did not interact with ERE, while ERβ effectively bound to ERE (Fig. 3D).
Based on these observations, we anticipated that ERβEBD would not regulate endogenous gene expression mediated by the ERE-dependent pathway. Our studies using qPCR revealed that ERβ only in the presence of E2 effectively induced the expression of estrogen responsive genes mediated by ER-ERE interactions, exemplified by the TFF1 and C3 genes, while ERβEBD had no effect on the expression of these genes (Fig. 4A). We also observed that E2-ERβ, but not E2-ERβEBD, activated the endogenous RARA gene expression. This is consistent with the finding that the regulation of the RARA gene expression is dependent upon ER-ERE interactions (Laganiere et al. 2005), which is clearly in contrast to the response from the reporter system we observed here and reported previously (Rishi, et al. 1995; Safe 2001; Sun, et al. 1998). On the other hand, both ERs in the presence of E2 repressed the expression of the MMP1 gene that models an ERE-independent signaling-regulated gene.
Fig. 4.
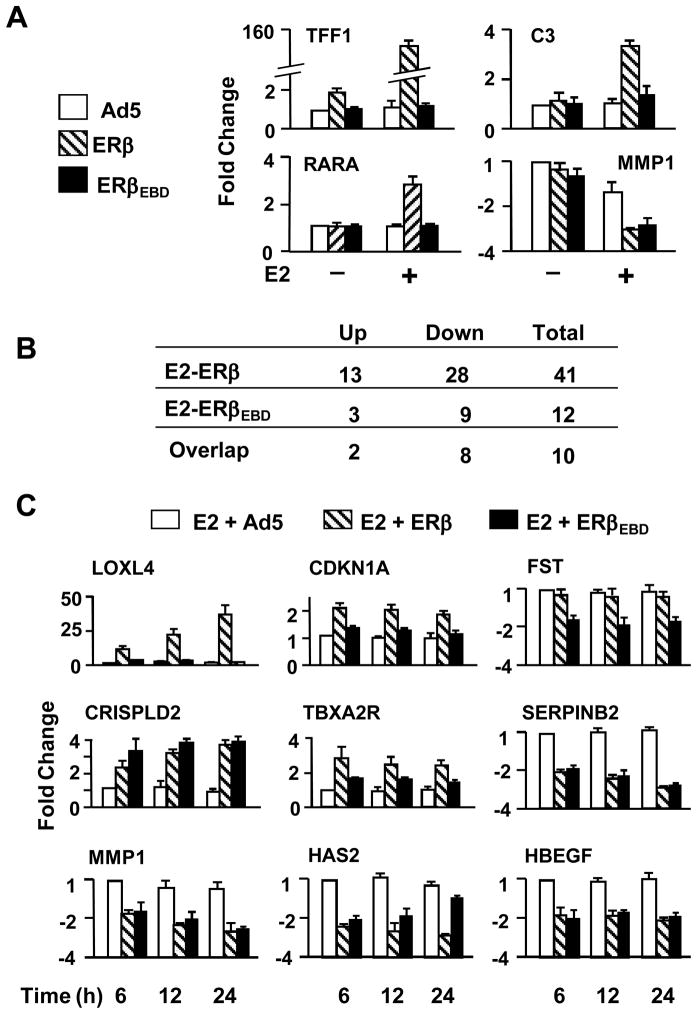
Effects of ERs on endogenous gene expression. (A) MDA-MB-231 cells were infected with recombinant adenoviruses in the absence (−) or presence (+) of 10−9 M E2 for 48h. Total RNA from infected cells was subjected to qPCR for the expression of the TFF1, C3, MMP1 and RARA genes. (B) Summary of genes identified with microarrays. MDA-MB-231 cells were infected with recombinant adenoviruses in the absence of E2 for 48h. Cells were then treated with 10−9 M E2 for 6h. Total RNA was subjected to arrays. Results are the mean of six independent determinations. (C) Verification of subset of genes identified with arrays. Infected MDA-MB-231 cells were maintained in the absence of E2 for 48h. Cells were then treated without or with 10−9 M E2 for 6, 12 or 24h. Total RNA was subjected to qPCR. Results, which are the mean ± SEM of four independent determinations in quadruplicate, are depicted as fold change compared with the level of gene expression in cells infected with the parent recombinant adenovirus (Ad5) in the absence of E2 at 6h of E2 treatment. The expression of the LOXL4, CDKN1A, FST, CRISPLD2, TBXA2R, SERPINB2, MMP1, HAS2, HBEGF genes are shown.
To further verify the functionality of ERβEBD and to preliminarily identify genes mediated by the ERE-independent pathway, we used a global gene expression profiling approach. MDA-MB-231 cells were infected with recombinant adenoviruses in the absence of E2 for 48h. At this time of post-infection, nearly all cells synthesized receptor proteins at levels that were maximal and comparable (Fig. 3). Cells were then treated with 10−9 M E2 for 6h, which is expected to induce significant changes in the level of transcription of immediate/early estrogen responsive genes (Frasor et al. 2003).
Expression profiling revealed that the genes regulated by ERβ and ERβEBD are involved in a broad range of function including metabolism, signal transduction, proliferation, apoptosis, and motility (Tables 1 & 2). As summarized in Fig. 4B, E2-ERβ significantly altered the expression of 41 genes, whereas E2-ERβEBD regulated the expression of 12 genes, 10 of which were also modified by E2-ERβ. The majority of the identified genes were suppressed by both E2-ERβ and E2-ERβEBD. E2-ERβ enhanced the expression of 12 responsive genes, whereas E2-ERβEBD augmented the expression of three genes.
Table 1.
Genes mediated by E2-ERβ
| Gene Name | Symbol | Fold Change | Process |
|---|---|---|---|
| Serpin peptidase inhibitor, clade B (ovalbumin), member 2 | SERPINB2 | −3.70 | Anti-apoptosis |
| Matrix metallopeptidase 1 (interstitial collagenase) | MMP1 | −3.57 | Collagen catabolism, Proteolysis |
| Endothelial differentiation, sphingolipid G-protein-coupled receptor, 1 | EDG1 | −2.33 | Signal transduction |
| Hyaluronan synthase 2 | HAS2 | −2.33 | Matrix formation |
| Gap junction protein, β2, 26kDa (connexin 26) | GJB2 | −2.27 | Cell-cell signaling |
| Matrix metallopeptidase 3 (stromelysin 1, progelatinase) | MMP3 | −2.17 | Collagen catabolism, proteolysis |
| Hypothetical protein LOC285628 | LOC285628 | −2.13 | Unknown |
| Plasminogen activator, urokinase receptor | PLAUR | −1.75 | Cell motility, Signaling, Chemotaxis |
| Rho GTPase activating protein 18 | ARHGAP18 | −1.75 | Signal transduction |
| Transforming growth factor, beta 2 | TGFB2 | −1.76 | Proliferation, apoptosis, migration |
| CDNA FLJ25677 fis, clone TST04054 | --- | −1.69 | Unknown |
| Heparin-binding EGF-like growth factor | HBEGF | −1.64 | Signal transduction |
| Adenosine A2b receptor | ADORA2B | −1.59 | Signal transduction |
| KIAA1609 protein | KIAA1609 | −1.56 | Unknown |
| Dickkopf homolog 1 (Xenopus laevis) | DKK1 | −1.56 | Signal Transduction |
| Forkhead box Q1 | FOXQ1 | −1.49 | Transcription |
| Adhesion molecule with Ig-like domain 2 | AMIGO2 | −1.47 | Cell adhesion |
| Dual specificity phosphatase 5 | DUSP5 | −1.45 | Protein dephosphorylation |
| G protein-coupled receptor | GPRC5A | −1.43 | Signal transduction |
| CD44 antigen | CD44 | −1.35 | Cell adhesion |
| Interleukin 18 (interferon-gamma-inducing factor) | IL18 | −1.35 | Signal transduction |
| Connective tissue growth factor | CTGF | −1.32 | Signal transduction |
| Follicular lymphoma variant translocation 1 | FVT1 | −1.27 | Metabolism |
| Baculoviral IAP repeat-containing 2 | BIRC2 | −1.27 | Apoptosis |
| Immediate early response 2 | IER2 | −1.25 | Mitogenesis |
| Myeloid-associated differentiation marker | MYADM | −1.22 | Transport vehicle biogenesis |
| UDP-glucose ceramide glucosyltransferase | UGCG | −1.22 | Glycosphingolipid biosynthesis |
| Fucosyltransferase 1 | FUT1 | −1.20 | Glycosphingolipid biosynthesis |
| Ectodermal-neural cortex | ENC1 | 1.42 | Organ development, apoptosis |
| Kruppel-like factor 10 | KLF10 | 1.68 | Cell Proliferation, Cell-cell Signaling |
| Olfactomedin-like 3 | OLFML3 | 1.69 | Matrix-related functions |
| Cyclin-dependent kinase inhibitor 1A (p21, Cip1) | CDKN1A | 1.85 | Cell cycle |
| Hypothetical protein LOC283551 | LOC283551 | 1.90 | Unknown |
| 3′-phosphoadenosine 5′-phosphosulfate synthase 2 | PAPSS2 | 1.92 | Nucleic acid metabolism |
| Pim-1 oncogene | PIM1 | 2.41 | Cell proliferation, apoptosis |
| Cdk5 and Abl enzyme substrate 2 | CABLES2 | 2.44 | Cell division |
| Transcribed locus | --- | 2.58 | Unknown |
| Cysteine-rich secretory protein LCCL domain containing 2 | CRISPLD2 | 2.80 | Signal transduction |
| Thromboxane A2 Receptor | TBXA2R | 3.16 | Signal transduction |
| SLIT and NTRK-like family, member 4 | SLITRK4 | 5.65 | Axonogenesis |
| Lysyl oxidase-like 4 | LOXL4 | 24.99 | Ion binding |
Table 2.
Genes mediated by E2-ERβEBD
| Gene Name | Gene Symbol | Fold Change | Process |
|---|---|---|---|
| Serpin peptidase inhibitor, clade B (ovalbumin), member 2 | SERPINB2 | −4.76 | Anti-apoptosis |
| Matrix metallopeptidase 1 (interstitial collagenase) | MMP1 | −3.70 | Collagen catabolism, proteolysis |
| Matrix metallopeptidase 3 (stromelysin 1, progelatinase) | MMP3 | −2.56 | Collagen catabolism, proteolysis |
| Hyaluronan synthase 2 | HAS2 | −2.13 | Matrix formation |
| Follistatin | FST | −2.08 | Signal transduction |
| Heparin-binding EGF-like growth factor | HBEGF | −1.72 | Signal transduction |
| Interleukin 18 (interferon-gamma-inducing factor) | IL18 | −1.59 | Signal transduction |
| Dickkopf homolog 1 (Xenopus laevis) | DKK1 | −1.49 | Signal transduction |
| G protein-coupled receptor | GPRC5A | −1.35 | Signal transduction |
| Thromboxane A2 receptor | TBXA2R | 1.73 | Signal transduction |
| PDZ and LIM domain 1 (elfin) | PDLIM1 | 1.98 | Signal transduction, Cyoskelatal reorganization |
| Cysteine-rich secretory protein LCCL domain containing 2 | CRISPLD2 | 4.04 | Signal transduction |
We further verified the expression of a subset of the identified genes with qPCR. Infected MDA-MB-231 cells were maintained for 48h in the absence of E2 (Fig. 4C). Cells were then treated without or with 10−9 M E2 for 6, 12 and 24h. Total RNA was subjected to qPCR. As observed with microarrays, only ERβ in the presence of E2 induced the expression of the CDKN1A (Cyclin dependent kinase inhibitor, WAF1 p21, CIP1) and LOX4 (Lysyl oxidase-like 4) genes. The expression of the FST (Follistatin) gene was indeed repressed by only E2-ERβEBD.
E2-ERβ and E2-ERβEBD activated the CRISPLD2 (Cysteine-rich secretory protein LCCL domain containing 2) and TBXA2R (Thromboxane A2 Receptor) gene expressions. On the other hand, both receptors repressed the expression of the MMP1, SERPINB2 (Serpin peptidase inhibitor, clade member 2), HAS2 (Hyaluronan synthase 2) and HBEGF (Heparin-binding EGF like growth factor) genes when E2 is present.
Thus, ERβEBD is capable of modulating the expression of endogenous genes in response to E2.
Differential effects of E2-ERβ and E2-ERβEBD on cell growth
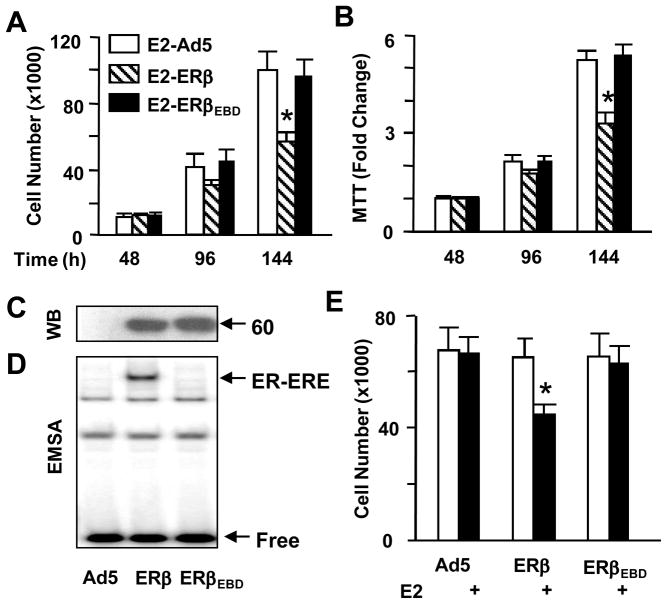
To examine whether the genomic responses mediated by E2-ERβEBD participate in the regulation of cellular growth, we used proliferation assays. Infected MDA-MB-231 cells were grown for various durations of time in the absence or presence of 10−9 M E2. Cells were then subjected to cell counting (Fig. 5A) and MTT assay (Fig. 5B). Results revealed that ERβ or ERβEBD had no effect on cellular growth in the absence of E2 (data not shown). On the other hand, E2 treatment of the infected cells synthesizing ERβ, but not ERβEBD, decreased the cell number as a function of time. These findings indicate that the E2-ERβEBD signaling has no discernable effect on cell proliferation.
Fig. 5.
Effects of ERβs on cell growth. (A & B) Infected MDA-MB-231 cells were maintained in the absence (data not shown) or presence of 10−9 M E2. At 48, 96 and 144h post-infection, cells were subjected to cell counting (A), or (B) MTT assay. The graphs represent the mean ± SEM of three independent experiments performed in duplicate. (C & D) U-2 OS cells were infected with Ad5 (50 MOI), Ad-5ERβ (40 MOI supplemented with 10 MOI of Ad5) or Ad5-ERβEBD (50 MOI) for 48h. Cell extracts were subjected to WB (C) and EMSA (D). Molecular weight in KDa is shown. Free (unbound) and the ER-bound ERE are indicated. Representative images from three independent experiments are shown. (E) Infected U-2 OS cells were maintained in the absence or presence of 10−9 M E2. At 144h post-infection, cells were subjected to cell counting. The mean ± SEM indicates three independent experiments performed in duplicate. * denotes a significant effect compared to the parent adenovirus (Ad5).
Moreover, despite the comparable synthesis as assessed by WB (Fig 5C) and the in situ E2 binding assay (data not shown), the ERE-binding defective ERβEBD (Fig. 5D) had no effect on cellular growth in the absence or presence of E2 in infected U-2 OS cells in contrast to E2-ERβ, which effectively suppressed the proliferation (Fig. 5E), as observed with MDA-MB-231 cells.
These results suggest that the ERE-independent pathway does not play a critical role in E2-ERβ signaling to mediate cellular growth.
Effects of E2-ERβ and E2-ERβEBD on cell cycle
The absence of an effect of E2-ERβEBD on cellular growth predicts that the ERE-independent pathway mediated by E2-ERβ is also insufficient to alter cell cycle distribution. To address this prediction, MDA-MB-231 cells were infected with recombinant adenoviruses in the absence or presence of 10−9 M E2 for various durations of time. Cells were then subjected to FACS. Kinetic analysis of histograms (Supplemental Data, Fig. 1A), summarized as the percentile of cells in G1 phase (Fig. 6A), revealed that ERβ, only in the presence of E2, increased the number of cells in G1 phase, and decreased the cell number accumulated in S and G2 phases (data not shown). On the other hand, ERβEBD had little effect on cell cycle distribution whether or not cells were treated with E2. Thus, it appears that E2 signaling conveyed by ERβ through the ERE-independent signaling pathway is insufficient to alter cell cycle distribution.
Fig. 6.
Effects of ERs on cell cycle, death and motility. (A) To examine the effects of ERs on cell cycle distribution, MDA-MB-231 cells were infected with recombinant adenoviruses in the absence (data not shown) or presence of 10−9 M E2 for 24, 48, or 96h. Cells were then subjected to FACS. Results showing the percent of cells in G1 phase as a function of time are the mean ± SEM of four independent experiments. (B) Infected MDA-MB-231 cells in the absence or presence of 10−9 M E2 as a function of time as indicated were subjected to a Caspase 3/7 assay. The fluorescence was measured by fluorometer. Results are the mean ± SEM of three independent experiments. (C & D) To examine the effects of ERs on cell death, infected MDA-MB-231 cells at indicated times were subjected to (C) an Annexin V assay using FACS or (D) a TUNEL assay. Results, which are the mean ± SEM of three independent experiments, are summarized as the number cells bound to Annexin V or as the number cells that incorporated FITC conjugated dUTP into the fragmented DNA (TUNEL). (E) To assess the role of the ERE-independent signaling pathway in cellular motility, infected MDA-MB-231 cells were incubated in the absence or presence of 10−9 M E2 for 48h to allow cells to reach near 100% confluence. A wound was then created and images were captured at 0h and at every 24h thereafter. Results, which are the mean ± SEM of three independent experiments performed in duplicate, are summarized as the wound closure at 96h relative to 0h. (F) To examine the effects of ERs on cellular invasiveness, MDA-MB-231 cells were infected with recombinant adenoviruses in the absence or presence of 10−9 M E2 for 48h. Cells were collected and counted. The same number of cells from each experimental group was then seeded on the upper section of the invasion chamber. After 24h incubation, cells with invasive capabilities on the bottom of chamber membrane were counted. Results, which represent the mean ± SEM of three independent experiments performed in duplicate, are percent change compared to cells infected with the parent adenovirus in the absence of E2, which is set to 100%. * indicates a significant effect compared to the parent adenovirus (Ad5).
Since the effects of ERβ on the ERE-independent signaling are also dependent on the nature of ligand, we wanted to examine whether ER agonists or antagonists differentially affect cellular growth mediated by ERβEBD. Infected MDA-MB-231 cells were treated without or with 10−9 M E2, 10−8 M DPN, 10−7 M 4-OHT or ICI (data not shown) for 48h. Cells were also co-treated with 10−9 M E2 and 10−7 M 4-OHT (or ICI, data not shown), or 10−8 M DPN together with 10−7 M 4-OHT (or ICI, data not shown) for 48h. These concentrations of ligands are optimally effective to alter transcription in response to ERβ (Fig. 2 & data not shown). Cells were then subjected to FACS (Supplemental Data, Fig. 1B). Treatment of infected cells synthesizing ERβ with DPN repressed the cell number accumulated in G1 phase comparable to that observed with the E2 treatment, while the compound had no effect on the growth of cells infected with Ad5 or Ad5-ERβEBD. Furthermore, the ER antagonist 4-OHT (or ICI, data not shown) alone did not affect cell cycle distribution in cells synthesizing either ERβ species, but it effectively blocked the effect of E2 or DPN on G1 phase mediated solely by ERβ. The effects of ligand-ERb on cell cycle distribution were mirrored in cellular growth, which was suppressed only by agonists (Supplemental Data, Fig. 1C).
Regulation of apoptosis by E2-ERβs
Since cellular growth encompasses cell proliferation and death, the inability of E2-ERβEBD to affect cell growth suggests that the ERE-independent pathway participates minimally in inducing apoptosis, which is a complex, multiple-step event that culminates in cell death (Korsmeyer 1999). To address this issue, MDA-MB-231 cells were infected in the absence or presence of 10−9 M E2 for different lengths of time. Cells were then subjected to Caspase 3/7, annexin V and TUNEL assays, which assess early-, mid- and late-stages of apoptosis, respectively.
We found that ERβEBD had no effect on the activity of caspase 3/7 whether or not cells were treated with E2, while ERβ in the presence of E2 increased the enzyme activities compared to cells infected with Ad5 (Fig. 6B).
Similar results were obtained with annexin V staining of infected cells (Fig. 6C & Supplemental Data, Fig. 2A). The number of apoptotic cells in a population of cells infected with a recombinant adenovirus bearing a receptor cDNA was compared with the number of apoptotic cells infected with the parent adenovirus in the absence of E2 as a function time. We observed that the unliganded or E2 bound ERβEBD had no effect on the cell population stained with annexin V, while ERβ in response to E2 increased the number of stained cells.
We corroborated our results with a TUNEL assay by incorporating FITC-conjugated nucleotides into the fragmented genomic DNA (Gorczyca, et al. 1998). Infected MDA-MB-231 cells were incubated without or with 10−9 M E2 for various durations of time. Cells were then subjected to the TUNEL assay. Results, depicted as number of cells stained with TUNEL (Supplemental Data, Fig. 2B) and summarized as a function of time (Fig. 6D), showed that E2-ERβ induced the fragmentation of genomic DNA of the infected cells. In contrast, ERβEBD had no discernable effect.
Collectively, these results indicate that ERβ but not ERβEBD induced apoptosis in infected cells treated with E2. These results suggest that the ERE-dependent pathway is the critical E2-ERβ signaling for the induction of apoptosis.
Differential effects of ERβ and ERβEBD on cell migration and invasion
Studies showed that ER-positive breast cancer cell models and ER-negative cell lines expressing ectopically introduced ER cDNA are less motile and invasive than the parent cells (Garcia, et al. 1997; Lazennec et al. 2001).
To examine whether the ERE-independent pathway participates in the anti-motogenic effect of E2-ERβ signaling, we used wound-healing and invasion assays. MDA-MB-231 cells infected with recombinant adenoviruses in the absence or presence of 10−9 M E2 were grown for 48h to allow cells to reach near confluence. A wound was then created and the rate of wound closure was assessed. Results showed that while ERβEBD had little effect on wound closure in the absence or presence of E2, ERβ without E2 delayed the wound healing which was further delayed by the presence of E2 (Fig. 6E & Supplemental Data, Fig. 2C).
We also utilized an invasion assay that assesses the capacity of cells to migrate through a reconstituted basement membrane to independently verify our findings. MDA-MB-231 cells were infected with recombinant adenoviruses in the absence or presence of 10−9 M E2 for 48h. Cells were then collected, and an equal number of cells was seeded on the reconstituted basement membrane of invasion chambers in the absence or presence of E2. After 24h incubation, cells on the bottom of the chamber membrane were stained and imaged (Supplemental Data, Fig. 2D). Quantitative analysis of images revealed that ERβ only in response to E2, but not E2-ERβEBD, reduced invasive cell number (Fig. 6F).
These results collectively suggest that the ERE-independent pathway mediated by E2-ERβ signaling plays a minimal role in cellular motility.
DISCUSSION
The integrated effects of the ERE-dependent and ERE-independent nuclear E2-ER signaling are critical for the homeodynamic regulation of estrogen responsive tissue functions. However, the relative importance of each pathway in physiology and pathophysiology of E2 signaling is unclear. Using an ERE binding mutant of ERβ (ERβEBD) that acts primarily through the ERE-independent pathway, we show here that genomic responses to E2-ERβEBD are not sufficient to alter phenotypic characteristics of model cells.
The elements of the nuclear action of ERs involve a multi-step regulation in which ERs are converted from an inactive form to a transcriptionally active state mediated by a conformational change upon E2 binding (Parker 1998). The interaction of E2-ER with an ERE constitutes the initial step in the ERE-dependent signaling in which gene expression is cyclically regulated (Metivier, et al. 2003). In the ERE-independent pathway, E2-ER functionally interacts with transcription factors bound to their cognate regulatory elements on DNA to modulate gene expression (Kushner et al. 2000; Safe 2001). This interaction involves the stabilization of transfactor binding to DNA and/or recruitment of coregulators to the complex (Kushner et al. 2000; Safe 2001). Estrogen regulation of the MMP1 and the IGF1 (insulin like growth factor-1) genes is, for example, characterized by a functional interaction between the members of the AP1 family proteins and ER (Kushner et al. 2000; Umayahara, et al. 1994). The modulation of the FOS (cellular oncogene fos) and the PGR (progesterone receptor) gene expressions, on the other hand, are dependent upon an interaction between ER and SP1 (Duan, et al. 1998; Schultz, et al. 2003). Similarly, ER interactions with the signal transducers and activators of transcription (STATs) family proteins provides a mechanism for the estrogen responsiveness of the CSN2 (β-casein) gene (Bjornstrom, et al. 2001).
In the ERE-independent signaling, ER establishes interactions with transfactors through regions that also encompass the DNA binding domain (DBD), while the integrated effects of the amino- and carboxyl-termini are responsible for the gene expression (Bjornstrom and Sjoberg 2002; Cheung, et al. 2005; Kushner et al. 2000; Safe 2001). Studies also demonstrated that the prevention of ER-ERE interactions by changing critical residues in the DBD of the mouse ERα and the human ERβ prevent the transcription from the ERE-dependent pathway, while conserving the regulatory capacity of the receptors in simulated ERE-independent pathways (Bjornstrom and Sjoberg 2004; Jakacka et al. 2001). Consistent with these findings, we also show here that ERβEBD, which had no effect on gene expression from the ERE-dependent pathway, mimics the effect of ERβ on transcriptional responses from simulated ERE-independent signaling pathways in a ligand, promoter, and cell context-dependent manner. Although underlying mechanisms are unclear, distinct conformational changes induced by a specific ligand (Nettles, et al. 2007) could alter the extent of interaction with a transfactor specific to each gene, thereby altering transcription output. Additionally, cellular differences in the concentration of transfactors and/or co-regulatory proteins (McKenna, et al. 1999) could contribute to differential effects of ligand-ERs on responses from promoter constructs.
Extending these findings, our microarray and qPCR studies further indicate that ERβEBD in the presence of E2 regulates the expression of a number of genes, the majority of which (10 out of 12) are similarly modulated by E2-ERβ in infected MDA-MB-231 cells (Table 1 and 2). Although the nature of the regulatory elements critical for the E2 responsiveness of the identified genes are unknown and currently being investigated, these genes are also targets for various cytokines and growth factors that utilize transfactors involved in cross-talk with ERs. Growth factor-mediated expression of the MMP1 and MMP3 genes, for example, involves the AP1 proteins (Aho, et al. 1997; Buttice, et al. 1996). Similarly, while the basal expression of the HAS2 gene is primarily controlled by the SP family of proteins (Monslow, et al. 2006), the responsiveness of the gene to endocrine and paracrine signaling is mediated by STAT and the NFkB transcription factors (Pasonen-Seppanen, et al. 2003; Saavalainen, et al. 2005). The protein products of these E2-ERβEBD responsive genes play critical roles in cell growth, death and migration. Matrix metalloproteases (MMPs), for example, are a large family of endopeptidases responsible for the tissue remodeling and degradation of the extracellular matrix (ECM) that includes collagens, elastins, gelatin, laminin and matrix glycoproteins (Duffy, et al. 2000). MMP1 is involved in the degradation of collagen, while MMP3 participates in the degradation of many ECM substrates including glycoproteins, laminin, fibronectin and collagen (Duffy et al. 2000). Increased MMP1 and MMP3 gene expressions are associated with advanced stages of breast cancer and are involved in the development of metastasis (Duffy et al. 2000). Similarly, aberrant synthesis of hyaluronan (HA), the major glycosaminoglycan found in the ECM, is implicated in a diverse range of ECM-mediated processes including cellular proliferation, adhesion and migration (Gotte and Yip 2006). HA is synthesized by the hyaluronan synthase family of transmembrane glycosyltransferases that are encoded by the corresponding HAS1, -2, -3a, and -3b genes (Gotte and Yip 2006). Preferential and high expression of the HAS2 gene appears to be critical for invasiveness of breast cancer cells, including MDA-MB-231 cells (Gotte and Yip 2006). Studies also indicate that aberrant expression of the SERPINB2 (Andreasen, et al. 1997), HBEGF (Ito, et al. 2001), DKK1 (Forget, et al. 2007), GPRC5A (Nagahata, et al. 2005) and TBXA2R (Watkins, et al. 2005) genes alters the proliferation and motility of breast cancer cells.
Considering the ability of ERβEBD to alter gene expression, the absence of an effect of ERβEBD on phenotypic characteristics of model cells is surprising. Studies indicate that the suppression of HAS2 (Li, et al. 2007; Udabage, et al. 2005) or MMP1 (Wyatt, et al. 2005) synthesis by a silencing RNA approach decreases growth and invasiveness of MDA-MB-231 cells. One explanation is that the extent of transcriptional alterations mediated by E2-ERβEBD and subsequent changes in RNA levels are not sufficient to modify the corresponding protein concentrations in cells, in contrast to silencing RNA approaches. This is unlikely, since the magnitude of E2-ERβ-mediated transcriptional responses was comparable to those modulated by E2-ERβEBD (Fig. 3 and Table 1 & 2), while only E2-ERβ induced alterations in cellular phenotypes. An alternative explanation is that the integrated regulation of genes involved in a signaling cascade through both ERE-dependent and ERE-independent pathways is crucial for the manifestation of phenotypic alterations in response to E2-ER signaling. Interaction of CD44 with HA, for example, induces signaling events that promote anchorage-independent cell growth and migration, thereby increasing metastatic spread (Gotte and Yip 2006). Importantly, post-transcriptionally-mediated repression of HAS2 leads to a concomitant decrease in its cell surface receptor CD44 transcript (Udabage et al. 2005). A similar mechanism is also suggested for the members of MMP proteins (Wyatt et al. 2005). Thus, the concomitant regulation of the HAS2 and the CD44 gene expression, or genes involved in the MMP signaling cascade, by E2-ERβ could be the underlying reason for the efficacy of the receptor to induce phenotypic alterations, in contrast to E2-ERβEBD that decreased only the HAS2 gene expression without an effect on cellular phenotype.
Transcript profiling of infected MDA-MB-231 cells synthesizing an ERE binding defective mouse ERα indicated that E2 alters the expression of a number of genes involved in various cellular functions (Glidewell-Kenney, et al. 2005). However, of the 29 identified genes, the expression of only the HAS2 and SERPINB2 genes coincides with the altered gene expression reported here. Since transcriptional responses from ERE-independent signaling pathways in experimental systems are also dependent upon ER-subtype, a discordant gene regulation could be critical for differences in subtype-mediated cellular responses.
Augmented synthesis of ERα, as observed in atypical hyperplasia and in situ carcinoma of breast, causes an E2-independent, tamoxifen-insensitive proliferation through aberrant gene expressions (Fowler et al. 2004; Fowler et al. 2006). Our studies presented here were designed to assess the role of the ERE-independent signaling pathway mediated by ERβ to induce cellular responses at concentrations that strictly depend upon E2 for function. Circumvention of E2-dependency by heightened ERβ levels could contribute to the characteristics of target tissue malignancies through a selective expansion, or altered expression, of target genes mediated by the ERE-independent signaling pathway.
In summary, our results show that genomic responses mediated by the ERE binding defective ERβ mutant in response to E2 are insufficient to alter the phenotypic characteristics of model cell lines. These results imply that the ERE-dependent pathway is necessary for E2-ERβ to regulate growth of the responsive cells. Studies that aim to target specifically ERE-bearing genes through the use of designer transcription factors are underway to address this issue.
Supplementary Material
Acknowledgments
We thank Michelle Zanche and Andrew Cardillo of the Functional Genomic Center at the University of Rochester for the guidance, assistance and execution of GeneArrays and qPCRs. We are also grateful to Dr. Peter Keng, Michael Strong and Brian Warsop of The Cell Separation and Flow Cytometry Facility at the University of Rochester for the supervision and assistance for FACS studies. We thank Christine Brower of the Biostatistics and Computational Biology Department of the University of Rochester for the assistance in microarray analysis. This work was supported by an NIH grant, CA113682 (MM), grants from the Susan G. Komen Foundation (MM) and the University of Rochester Clinical and Translational Science Institute (MM).
Footnotes
The authors declare no conflict of interest.
References
- Aho S, Rouda S, Kennedy SH, Qin H, Tan EM. Regulation of human interstitial collagenase (matrix metalloproteinase-1) promoter activity by fibroblast growth factor. Eur J Biochem. 1997;247:503–510. doi: 10.1111/j.1432-1033.1997.00503.x. [DOI] [PubMed] [Google Scholar]
- Andreasen PA, Kjoller L, Christensen L, Duffy MJ. The urokinase-type plasminogen activator system in cancer metastasis: a review. Int J Cancer. 1997;72:1–22. doi: 10.1002/(sici)1097-0215(19970703)72:1<1::aid-ijc1>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Archer TK, Lefebvre P, Wolford RG, Hager GL. Transcription factor loading on the MMTV promoter: a bimodal mechanism for promoter activation. Science. 1992;255:1573–1576. doi: 10.1126/science.1347958. [DOI] [PubMed] [Google Scholar]
- Bjornstrom L, Kilic E, Norman M, Parker MG, Sjoberg M. Cross-talk between Stat5b and estrogen receptor-α and -β in mammary epithelial cells. J Mol Endocrinol. 2001;27:93–106. doi: 10.1677/jme.0.0270093. [DOI] [PubMed] [Google Scholar]
- Bjornstrom L, Sjoberg M. Mutations in the estrogen receptor DNA-binding domain discriminate between the classical mechanism of action and cross-talk with Stat5b and activating protein 1 (AP-1) J Biol Chem. 2002;277:48479–48483. doi: 10.1074/jbc.C200570200. [DOI] [PubMed] [Google Scholar]
- Bjornstrom L, Sjoberg M. Estrogen receptor-dependent activation of AP-1 via non-genomic signalling. Nucl Recept. 2004;2:3. doi: 10.1186/1478-1336-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttice G, Duterque-Coquillaud M, Basuyaux JP, Carrere S, Kurkinen M, Stehelin D. Erg, an Ets-family member, differentially regulates human collagenase1 (MMP1) and stromelysin1 (MMP3) gene expression by physically interacting with the Fos/Jun complex. Oncogene. 1996;13:2297–2306. [PubMed] [Google Scholar]
- Cheung E, Acevedo ML, Cole PA, Kraus WL. Altered pharmacology and distinct coactivator usage for estrogen receptor-dependent transcription through activating protein-1. Proc Natl Acad Sci U S A. 2005;102:559–564. doi: 10.1073/pnas.0407113102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, Bunney WE, Myers RM, Speed TP, Akil H, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33:e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai M, Wang P, Jakupovic E, Watson SJ, Meng F. Web-Based GeneChip Analysis System for Large-Scale Collaborative Projects. Bioinformatics. 2007;23:2185–2187. doi: 10.1093/bioinformatics/btm297. [DOI] [PubMed] [Google Scholar]
- Deroo BJ, Korach KS. Estrogen receptors and human disease. J Clin Invest. 2006;116:561–570. doi: 10.1172/JCI27987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan R, Porter W, Safe S. Estrogen-induced c-fos protooncogene expression in MCF-7 human breast cancer cells: role of estrogen receptor Sp1 complex formation. Endocrinology. 1998;139:1981–1990. doi: 10.1210/endo.139.4.5870. [DOI] [PubMed] [Google Scholar]
- Duffy MJ, Maguire TM, Hill A, McDermott E, O’Higgins N. Metalloproteinases: role in breast carcinogenesis, invasion and metastasis. Breast Cancer Res. 2000;2:252–257. doi: 10.1186/bcr65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forget MA, Turcotte S, Beauseigle D, Godin-Ethier J, Pelletier S, Martin J, Tanguay S, Lapointe R. The Wnt pathway regulator DKK1 is preferentially expressed in hormone-resistant breast tumours and in some common cancer types. Br J Cancer. 2007;96:646– 653. doi: 10.1038/sj.bjc.6603579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler AM, Solodin N, Preisler-Mashek MT, Zhang P, Lee AV, Alarid ET. Increases in estrogen receptor-α concentration in breast cancer cells promote serine 118/104/106-independent AF-1 transactivation and growth in the absence of estrogen. FASEB J. 2004;18:81–93. doi: 10.1096/fj.03-0038com. [DOI] [PubMed] [Google Scholar]
- Fowler AM, Solodin NM, Valley CC, Alarid ET. Altered target gene regulation controlled by estrogen receptor-α concentration. Mol Endocrinol. 2006;20:291–301. doi: 10.1210/me.2005-0288. [DOI] [PubMed] [Google Scholar]
- Frasor J, Danes JM, Komm B, Chang KC, Lyttle CR, Katzenellenbogen BS. Profiling of estrogen up- and down-regulated gene expression in human breast cancer cells: insights into gene networks and pathways underlying estrogenic control of proliferation and cell phenotype. Endocrinology. 2003;144:4562–4574. doi: 10.1210/en.2003-0567. [DOI] [PubMed] [Google Scholar]
- Garcia M, Derocq D, Freiss G, Rochefort H. Activation of estrogen receptor transfected into a receptor-negative breast cancer cell line decreases the metastatic and invasive potential of the cells. Proc Natl Acad Sci U S A. 1992;89:11538–11542. doi: 10.1073/pnas.89.23.11538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia M, Derocq D, Platet N, Bonnet S, Brouillet JP, Touitou I, Rochefort H. Both estradiol and tamoxifen decrease proliferation and invasiveness of cancer cells transfected with a mutated estrogen receptor. J Steroid Biochem Mol Biol. 1997;61:11–17. doi: 10.1016/s0960-0760(96)00255-5. [DOI] [PubMed] [Google Scholar]
- Gautier L, Moller M, Friis-Hansen L, Knudsen S. Alternative mapping of probes to genes for Affymetrix chips. BMC Bioinformatics. 2004;5:111. doi: 10.1186/1471-2105-5-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glidewell-Kenney C, Weiss J, Lee EJ, Pillai S, Ishikawa T, Ariazi EA, Jameson JL. ERE-independent ERα target genes differentially expressed in human breast tumors. Mol Cell Endocrinol. 2005;245:53–59. doi: 10.1016/j.mce.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Gorczyca W, Melamed MR, Darzynkiewicz Z. Analysis of apoptosis by flow cytometry. Methods Mol Biol. 1998;91:217–238. doi: 10.1385/0-89603-354-6:217. [DOI] [PubMed] [Google Scholar]
- Gotte M, Yip GW. Heparanase, hyaluronan, and CD44 in cancers: a breast carcinoma perspective. Cancer Res. 2006;66:10233–10237. doi: 10.1158/0008-5472.CAN-06-1464. [DOI] [PubMed] [Google Scholar]
- Green S, Kumar V, Theulaz I, Wahli W, Chambon P. The N-terminal DNA-binding ‘zinc finger’ of the oestrogen and glucocorticoid receptors determines target gene specificity. EMBO J. 1988;7:3037–3044. doi: 10.1002/j.1460-2075.1988.tb03168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JM, Couse JF, Korach KS. The multifaceted mechanisms of estradiol and estrogen receptor signaling. J Biol Chem. 2001;276:36869–36872. doi: 10.1074/jbc.R100029200. [DOI] [PubMed] [Google Scholar]
- Hall JM, McDonnell DP. The estrogen receptor β-isoform (ERβ) of the human estrogen receptor modulates ERα transcriptional activity and is a key regulator of the cellular response to estrogens and antiestrogens. Endocrinology. 1999;140:5566–5578. doi: 10.1210/endo.140.12.7179. [DOI] [PubMed] [Google Scholar]
- Harbig J, Sprinkle R, Enkemann SA. A sequence-based identification of the genes detected by probesets on the Affymetrix U133 plus 2.0 array. Nucleic Acids Res. 2005;33:e31. doi: 10.1093/nar/gni027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington WR, Sheng S, Barnett DH, Petz LN, Katzenellenbogen JA, Katzenellenbogen BS. Activities of estrogen receptor α- and β-selective ligands at diverse estrogen responsive gene sites mediating transactivation or transrepression. Mol Cell Endocrinol. 2003;206:13–22. doi: 10.1016/s0303-7207(03)00255-7. [DOI] [PubMed] [Google Scholar]
- Huang J, Li X, Hilf R, Bambara RA, Muyan M. Molecular basis of therapeutic strategies for breast cancer. Curr Drug Targets Immune Endocr Metabol Disord. 2005a;5:379–396. doi: 10.2174/156800805774912944. [DOI] [PubMed] [Google Scholar]
- Huang J, Li X, Maguire CA, Hilf R, Bambara RA, Muyan M. Binding of estrogen receptor β to estrogen response element in situ is independent of estradiol and impaired by its amino terminus. Mol Endocrinol. 2005b;19:2696–2712. doi: 10.1210/me.2005-0120. [DOI] [PubMed] [Google Scholar]
- Huang J, Li X, Yi P, Hilf R, Bambara RA, Muyan M. Targeting estrogen responsive elements (EREs): design of potent transactivators for ERE-containing genes. Mol Cell Endocrinol. 2004;218:65–78. doi: 10.1016/j.mce.2003.12.005. [DOI] [PubMed] [Google Scholar]
- Ito Y, Takeda T, Higashiyama S, Noguchi S, Matsuura N. Expression of heparin-binding epidermal growth factor-like growth factor in breast carcinoma. Breast Cancer Res Treat. 2001;67:81–85. doi: 10.1023/a:1010667108371. [DOI] [PubMed] [Google Scholar]
- Jakacka M, Ito M, Weiss J, Chien PY, Gehm BD, Jameson JL. Estrogen receptor binding to DNA is not required for its activity through the nonclassical AP1 pathway. J Biol Chem. 2001;276:13615–13621. doi: 10.1074/jbc.M008384200. [DOI] [PubMed] [Google Scholar]
- Jiang SY, Jordan VC. Growth regulation of estrogen receptor-negative breast cancer cells transfected with complementary DNAs for estrogen receptor. J Natl Cancer Inst. 1992;84:580–591. doi: 10.1093/jnci/84.8.580. [DOI] [PubMed] [Google Scholar]
- Kian Tee M, Rogatsky I, Tzagarakis-Foster C, Cvoro A, An J, Christy RJ, Yamamoto KR, Leitman DC. Estradiol and selective estrogen receptor modulators differentially regulate target genes with estrogen receptors α and β. Mol Biol Cell. 2004;15:1262–1272. doi: 10.1091/mbc.E03-06-0360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebanov L, Gordon A, Xiao Y, Land H, Yakovlev A. A permutation test motivated by microarray data analysis. Compatrative statistical data analaysis. 2006;50:3619–3628. [Google Scholar]
- Korsmeyer SJ. Programmed cell death and the regulation of homeostasis. Harvey Lect. 1999;95:21–41. [PubMed] [Google Scholar]
- Kushner PJ, Agard DA, Greene GL, Scanlan TS, Shiau AK, Uht RM, Webb P. Estrogen receptor pathways to AP-1. J Steroid Biochem Mol Biol. 2000;74:311–317. doi: 10.1016/s0960-0760(00)00108-4. [DOI] [PubMed] [Google Scholar]
- Laganiere J, Deblois G, Giguere V. Functional genomics identifies a mechanism for estrogen activation of the retinoic acid receptor α1 gene in breast cancer cells. Mol Endocrinol. 2005;19:1584–1592. doi: 10.1210/me.2005-0040. [DOI] [PubMed] [Google Scholar]
- Lazennec G, Bresson D, Lucas A, Chauveau C, Vignon F. ERβ inhibits proliferation and invasion of breast cancer cells. Endocrinology. 2001;142:4120–4130. doi: 10.1210/endo.142.9.8395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazennec G, Katzenellenbogen BS. Expression of human estrogen receptor using an efficient adenoviral gene delivery system is able to restore hormone-dependent features to estrogen receptor-negative breast carcinoma cells. Mol Cell Endocrinol. 1999;149:93–105. doi: 10.1016/s0303-7207(98)00254-8. [DOI] [PubMed] [Google Scholar]
- Lee HL, Archer TK. Nucleosome-mediated disruption of transcription factor-chromatin initiation complexes at the mouse mammary tumor virus long terminal repeat in vivo. Mol Cell Biol. 1994;14:32–41. doi: 10.1128/mcb.14.1.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Huang J, Yi P, Bambara RA, Hilf R, Muyan M. Single-chain estrogen receptors (ERs) reveal that the ERα/β heterodimer emulates functions of the ERα dimer in genomic estrogen signaling pathways. Mol Cell Biol. 2004;24:7681–7694. doi: 10.1128/MCB.24.17.7681-7694.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Li L, Brown TJ, Heldin P. Silencing of hyaluronan synthase 2 suppresses the malignant phenotype of invasive breast cancer cells. Int J Cancer. 2007;120:2557–2567. doi: 10.1002/ijc.22550. [DOI] [PubMed] [Google Scholar]
- Licznar A, Caporali S, Lucas A, Weisz A, Vignon F, Lazennec G. Identification of genes involved in growth inhibition of breast cancer cells transduced with estrogen receptor. FEBS Lett. 2003;553:445–450. doi: 10.1016/s0014-5793(03)01090-1. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Luisi BF, Schwabe JW, Freedman LP. The steroid/nuclear receptors: from three-dimensional structure to complex function. Vitam Horm. 1994;49:1–47. doi: 10.1016/s0083-6729(08)61145-0. [DOI] [PubMed] [Google Scholar]
- McKenna NJ, Lanz RB, O’Malley BW. Nuclear receptor coregulators: cellular and molecular biology. Endocrine Rev. 1999;20:321–344. doi: 10.1210/edrv.20.3.0366. [DOI] [PubMed] [Google Scholar]
- Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, Gannon F. Estrogen receptor-α directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–763. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- Moggs JG, Murphy TC, Lim FL, Moore DJ, Stuckey R, Antrobus K, Kimber I, Orphanides G. Anti-proliferative effect of estrogen in breast cancer cells that re-express ERα is mediated by aberrant regulation of cell cycle genes. J Mol Endocrinol. 2005;34:535–551. doi: 10.1677/jme.1.01677. [DOI] [PubMed] [Google Scholar]
- Monroe DG, Secreto FJ, Subramaniam M, Getz BJ, Khosla S, Spelsberg TC. Estrogen receptor α and β heterodimers exert unique effects on estrogen- and tamoxifen-dependent gene expression in human U2OS osteosarcoma cells. Mol Endocrinol. 2005;19:1555–1568. doi: 10.1210/me.2004-0381. [DOI] [PubMed] [Google Scholar]
- Monslow J, Williams JD, Fraser DJ, Michael DR, Foka P, Kift-Morgan AP, Luo DD, Fielding CA, Craig KJ, Topley N, et al. Sp1 and Sp3 mediate constitutive transcription of the human hyaluronan synthase 2 gene. J Biol Chem. 2006;281:18043–18050. doi: 10.1074/jbc.M510467200. [DOI] [PubMed] [Google Scholar]
- Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- Mosselman S, Polman J, Dijkema R. ERβ: identification and characterization of a novel human estrogen receptor. FEBS Letters. 1996;392:49–53. doi: 10.1016/0014-5793(96)00782-x. [DOI] [PubMed] [Google Scholar]
- Muyan M, Yi P, Sathya G, Willmert LJ, Driscoll MD, Hilf R, Bambara RA. Fusion estrogen receptor proteins: toward the development of receptor-based agonists and antagonists. Mol Cell Endocrinol. 2001;182:249–263. doi: 10.1016/s0303-7207(01)00493-2. [DOI] [PubMed] [Google Scholar]
- Nagahata T, Sato T, Tomura A, Onda M, Nishikawa K, Emi M. Identification of RAI3 as a therapeutic target for breast cancer. Endocr Relat Cancer. 2005;12:65–73. doi: 10.1677/erc.1.00890. [DOI] [PubMed] [Google Scholar]
- Nettles KW, Bruning JB, Gil G, O’Neill EE, Nowak J, Guo Y, Kim Y, DeSombre ER, Dilis R, Hanson RN, et al. Structural plasticity in the oestrogen receptor ligand-binding domain. EMBO Rep. 2007;8:563–568. doi: 10.1038/sj.embor.7400963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson S, Makela S, Treuter E, Tujague M, Thomsen J, Andersson G, Enmark E, Pettersson K, Warner M, Gustafsson JA. Mechanisms of estrogen action. Physiol Rev. 2001;81:1535– 1565. doi: 10.1152/physrev.2001.81.4.1535. [DOI] [PubMed] [Google Scholar]
- Nunez AM, Berry M, Imler JL, Chambon P. The 5′ flanking region of the pS2 gene contains a complex enhancer region responsive to oestrogens, epidermal growth factor, a tumour promoter (TPA), the c-Ha-ras oncoprotein and the c-jun protein. EMBO J. 1989;8:823– 829. doi: 10.1002/j.1460-2075.1989.tb03443.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paech K, Webb P, Kuiper GG, Nilsson S, Gustafsson J, Kushner PJ, Scanlan TS. Differential ligand activation of estrogen receptors ERα and ERβ at AP1 sites. Science. 1997;277:1508–1510. doi: 10.1126/science.277.5331.1508. [DOI] [PubMed] [Google Scholar]
- Parker MG. Transcriptional activation by oestrogen receptors. Biochem Soc Symp. 1998;63:45–50. [PubMed] [Google Scholar]
- Pasonen-Seppanen S, Karvinen S, Torronen K, Hyttinen JM, Jokela T, Lammi MJ, Tammi MI, Tammi R. EGF upregulates, whereas TGF-β downregulates, the hyaluronan synthases Has2 and Has3 in organotypic keratinocyte cultures: correlations with epidermal proliferation and differentiation. J Invest Dermatol. 2003;120:1038–1044. doi: 10.1046/j.1523-1747.2003.12249.x. [DOI] [PubMed] [Google Scholar]
- Pennie WD, Hager GL, Smith CL. Nucleoprotein structure influences the response of the mouse mammary tumor virus promoter to activation of the cyclic AMP signalling pathway. Mol Cell Biol. 1995;15:2125–2134. doi: 10.1128/mcb.15.4.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rishi AK, Shao ZM, Baumann RG, Li XS, Sheikh MS, Kimura S, Bashirelahi N, Fontana JA. Estradiol regulation of the human retinoic acid receptor α gene in human breast carcinoma cells is mediated via an imperfect half-palindromic estrogen response element and Sp1 motifs. Cancer Research. 1995;55:4999–5006. [PubMed] [Google Scholar]
- Saavalainen K, Pasonen-Seppanen S, Dunlop TW, Tammi R, Tammi MI, Carlberg C. The human hyaluronan synthase 2 gene is a primary retinoic acid and epidermal growth factor responding gene. J Biol Chem. 2005;280:14636–14644. doi: 10.1074/jbc.M500206200. [DOI] [PubMed] [Google Scholar]
- Safe S. Transcriptional activation of genes by 17β-estradiol through estrogen receptor-Sp1 interactions. Vitam Horm. 2001;62:231–252. doi: 10.1016/s0083-6729(01)62006-5. [DOI] [PubMed] [Google Scholar]
- Schultz JR, Petz LN, Nardulli AM. Estrogen receptor α and Sp1 regulate progesterone receptor gene expression. Mol Cell Endocrinol. 2003;201:165–175. doi: 10.1016/s0303-7207(02)00415-x. [DOI] [PubMed] [Google Scholar]
- Stossi F, Barnett DH, Frasor J, Komm B, Lyttle CR, Katzenellenbogen BS. Transcriptional profiling of estrogen-regulated gene expression via estrogen receptor (ER) α or ERβ in human osteosarcoma cells: distinct and common target genes for these receptors. Endocrinology. 2004;145:3473–3486. doi: 10.1210/en.2003-1682. [DOI] [PubMed] [Google Scholar]
- Sun G, Porter W, Safe S. Estrogen-induced retinoic acid receptor α 1 gene expression: role of estrogen receptor-Sp1 complex. Mol Endocrinol. 1998;12:882–890. doi: 10.1210/mend.12.6.0125. [DOI] [PubMed] [Google Scholar]
- Udabage L, Brownlee GR, Waltham M, Blick T, Walker EC, Heldin P, Nilsson SK, Thompson EW, Brown TJ. Antisense-mediated suppression of hyaluronan synthase 2 inhibits the tumorigenesis and progression of breast cancer. Cancer Res. 2005;65:6139–6150. doi: 10.1158/0008-5472.CAN-04-1622. [DOI] [PubMed] [Google Scholar]
- Umayahara Y, Kawamori R, Watada H, Imano E, Iwama N, Morishima T, Yamasaki Y, Kajimoto Y, Kamada T. Estrogen regulation of the insulin-like growth factor I gene transcription involves an AP-1 enhancer. Journal of Biological Chemistry. 1994;269:16433–16442. [PubMed] [Google Scholar]
- Vyhlidal C, Samudio I, Kladde MP, Safe S. Transcriptional activation of transforming growth factor α by estradiol: requirement for both a GC-rich site and an estrogen response element half-site. J Mol Endocrinol. 2000;24:329–338. doi: 10.1677/jme.0.0240329. [DOI] [PubMed] [Google Scholar]
- Watkins G, Douglas-Jones A, Mansel RE, Jiang WG. Expression of thromboxane synthase, TBXAS1 and the thromboxane A2 receptor, TBXA2R, in human breast cancer. Int Semin Surg Oncol. 2005;2:23. doi: 10.1186/1477-7800-2-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb P, Lopez GN, Uht RM, Kushner PJ. Tamoxifen activation of the estrogen receptor/AP-1 pathway: potential origin for the cell-specific estrogen-like effects of antiestrogens. Mol Endocrinol. 1995;9:443–456. doi: 10.1210/mend.9.4.7659088. [DOI] [PubMed] [Google Scholar]
- Webb P, Nguyen P, Valentine C, Lopez GN, Kwok GR, McInerney E, Katzenellenbogen BS, Enmark E, Gustafsson JA, Nilsson S, et al. The estrogen receptor enhances AP-1 activity by two distinct mechanisms with different requirements for receptor transactivation functions. Mol Endocrinol. 1999;13:1672–1685. doi: 10.1210/mend.13.10.0357. [DOI] [PubMed] [Google Scholar]
- Westfall PH, Young S. Resampling-based multiple testing. New York: Wiley; 1993. [Google Scholar]
- Wyatt CA, Geoghegan JC, Brinckerhoff CE. Short hairpin RNA-mediated inhibition of matrix metalloproteinase-1 in MDA-231 cells: effects on matrix destruction and tumor growth. Cancer Res. 2005;65:11101–11108. doi: 10.1158/0008-5472.CAN-05-2446. [DOI] [PubMed] [Google Scholar]
- Yi P, Bhagat S, Hilf R, Bambara RA, Muyan M. Differences in the abilities of estrogen receptors to integrate activation functions are critical for subtype-specific transcriptional responses. Mol Endocrinol. 2002a;16:1810–1827. doi: 10.1210/me.2001-0323. [DOI] [PubMed] [Google Scholar]
- Yi P, Driscoll MD, Huang J, Bhagat S, Hilf R, Bambara RA, Muyan M. The effects of estrogen-responsive element- and ligand-induced structural changes on the recruitment of cofactors and transcriptional responses by ERα and ERβ. Mol Endocrinol. 2002b;16:674–693. doi: 10.1210/mend.16.4.0810. [DOI] [PubMed] [Google Scholar]
- Zajchowski DA, Sager R, Webster L. Estrogen inhibits the growth of estrogen receptor-negative, but not estrogen receptor-positive, human mammary epithelial cells expressing a recombinant estrogen receptor. Cancer Res. 1993;53:5004–5011. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.