Abstract
We have cloned, sequenced, and expressed the gene for a unique ATP- and NADPH-dependent carboxylic acid reductase (CAR) from a Nocardia species that reduces carboxylic acids to their corresponding aldehydes. Recombinant CAR containing an N-terminal histidine affinity tag had Km values for benzoate, ATP, and NADPH that were similar to those for natural CAR, and recombinant CAR reduced benzoic, vanillic, and ferulic acids to their corresponding aldehydes. car is the first example of a new gene family encoding oxidoreductases with remote acyl adenylation and reductase sites.
Aromatic, aliphatic, and alicyclic aldehydes and alcohols are useful intermediates in the chemical, pharmaceutical, and food industries. Chemical methods for carboxylic acid reductions are limited, and they usually require prior derivatization and product deblocking with reactants containing competing functional groups. Biocatalytic reductions of carboxylic acids are attractive because the substrates are water soluble, blocking chemistry is not necessary, reductions are enantiospecific, and the scope of the reaction is very broad (24, 32).
Although microbial reductions of carboxylic acids, usually producing the acids' corresponding aldehydes or alcohols, have been observed with whole-cell reactions of bacteria and fungi (3, 4, 6, 8, 20, 22, 24, 25, 30, 36-38, 40), enzymatic reductions of carboxylic acids are relatively new and unexploited biocatalytic reactions of great potential value in organic synthesis (12).
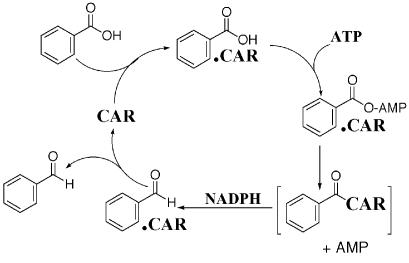
Aldehyde oxidoreductases, also known as carboxylic acid reductases (CAR), require ATP, Mg2+, and NADPH as cofactors (16, 17, 18, 21, 24). The reduction is a stepwise process involving initial binding of both ATP and the carboxylic acid to the enzyme in order to form mixed 5′-adenylic acid-carbonyl anhydride intermediates (9, 15, 25, 27, 39) that are subsequently reduced by hydride delivery from NADPH to form aldehyde products (16, 25) (Fig. 1). Aromatic CARs have been purified to homogeneity only from Neurospora (17) and Nocardia (21, 24) species. Although N-terminal and internal amino acid sequences were reported for our Nocardia enzyme (24), sequences have never been determined for any gene coding for CARs. CAR from Nocardia sp. strain NRRL 5646 has an extremely wide substrate range, and it enantiospecifically reduces carboxylic acids (8, 24, 32). We report here the cloning and expression of the first CAR gene, car, and the use of cloned enzyme in vitro and in vivo to reduce carboxylic acid substrates.
FIG. 1.
The reduction cycle of Nocardia sp. strain NRRL 5646 CAR. Brackets enclose a putative, covalently linked functional CAR-carbonyl complex.
Strains, plasmids, media, and growth.
Bacteria and plasmids used in this study are listed in Table 1. Nocardia sp. strain NRRL 5646 (14) was grown at 30°C in Luria-Bertani (LB) medium containing 0.05% Tween 80 (vol/vol [liquid medium only]). With Escherichia coli as the recombinant host for pHAT-based vectors, cells were grown at 37°C on solid or in liquid LB medium. Ampicillin (100 μg/ml) was incorporated into LB medium to select for recombinants, and isopropyl-β-d-thiogalactopyranoside (IPTG) (1 mM) and/or 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (80 μg/ml) were included for recombinant identification.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant propertiesa | Source or reference |
|---|---|---|
| Strains | ||
| Nocardia sp. strain NRRL 5646 | Wild type | 24 |
| E. coli JM 109 | RecA−, recombinant vector host strain | Promega |
| E. coli BL21 (DE3) | Inducible T7 RNA polymerase, Ampr | Stratagene |
| E. coli BL21-CodonPlus (DE3)-RP | Has argU and proL tRNA genes to provide tRNA for rare codons found in many GC-rich bacteria | Stratagene |
| Plasmids | ||
| pGEM-T easy | T/A PCR cloning vector, Ampr | Promega |
| pHAT10 | Cloning vector for in-frame addition of HAT tag (MTMITPSLKDHLIHNVH KEEHAHAHNKIDDDDKVDGS) to the N terminus of CAR, pLac for expression, Ampr | Clontech |
| pHAT-305 | pHAT-10 with car insert | This study |
| pHAT-DHFR | Positive control expression vector with dihydrofolate reductase gene tagged with HAT at the N terminus | Clontech |
Ampr, ampicillin resistance.
Molecular biology techniques.
DNA manipulations were performed by standard protocols (33). Restriction enzymes, T4 DNA ligase, and shrimp alkaline phosphatase were from New England Biolabs (Beverly, Mass.); the pGEM-T easy vector kit was from Promega (Madison, Wis.); polyclonal rabbit anti-histidine-affinity-tag (HAT) antibody, pHAT10 vector, and Talon resin were from Clontech (Palo Alto, Calif.); goat anti-rabbit immunoglobulin G-conjugated alkaline phosphatase and Immun-Star chemiluminescent substrate kit were from Bio-Rad (Hercules, Calif.); and Qiaprep Spin Miniprep kit and Qiaquick kit were from Qiagen, Inc. (Chatsworth, Calif.). All other chemicals were from Sigma (St. Louis, Mo.) unless otherwise specified.
Nocardia sp. strain NRRL 5646 chromosomal DNA (gDNA) was purified as described previously by Pelicic et al. (29), with modifications. Briefly, ampicillin (0.2 mg/ml) and glycine (1.5%, vol/vol) were added into 100-ml stationary-phase cultures 2 h before harvest. Cells (wet weight, 1.5 g) were resuspended in 5 ml of 25% sucrose in 50 mM Tris-HCl (pH 8.0) containing 50 mM EDTA and 12 mg of lysozyme per ml and incubated at 37°C with shaking at 50 rpm for 1.5 h. Next, 3 ml of 100 mM Tris-HCl (pH 8.0) (containing 1% sodium dodecyl sulfate [SDS] and 700 μg of proteinase K per ml) was added, and the sample was incubated at 55°C for 4 h. Then, 45 μl of RNase (500 μg/ml) was added, and the lysate was incubated with shaking at 50 rpm and 37°C for 1 h and then extracted with phenol-chloroform-isoamyl alcohol (25:24:1 [vol/vol/vol]; Invitrogen), followed by ethanol precipitation, yielding 90 μg of gDNA. Recombinant plasmids from E. coli were purified with a Qiaprep Spin Miniprep kit, and Qiaquick kits were used for PCR cleanup and gel extractions of vector constructs. PCR cloning amplification was done either with Platinum TaqDNA polymerase or Platinum Pfx DNA polymerase (Invitrogen).
Oligonucleotides (Table 2) corresponding to N-terminal and internal amino acid sequences from purified CAR (24) were constructed. Degeneracy was minimized by taking advantage of reported Nocardia codon preferences (11). A typical 50-μl reaction in 1× PCR buffer contained 500 ng of Nocardia DNA, 5 mM Mg2+, a 500 μM concentration of each deoxynucleotide triphosphate, a 0.5 μM concentration of each primer, 1% dimethyl sulfoxide (vol/vol), and 3.5 U of TaqDNA polymerase. Reaction mixtures were subjected to the following cycles: 1 cycle at 94°C for 4 min; 30 cycles at 94°C for 45 s, 56°C for 45 s, and 72°C for 2 min; and finally 1 cycle at 72°C for 10 min. PCR products were separated on a 1% agarose gel. The desired band was excised, extracted, and ligated into pGEM-T; it was then transformed by heat shock into E. coli JM109. Following selection and identification of clones on LB-X-Gal-ampicillin plates, colonies were used to inoculate to LB-ampicillin broth and incubated overnight. Following culture harvest and a plasmid miniprep procedure, recombinant plasmid was sequenced in both directions with sequencing primers (Table 2).
TABLE 2.
Oligonucleotides used in this study
| Oligo- nucleotide | Sequence (5′-3′)a | Source or reference |
|---|---|---|
| Cloning primers | ||
| CA-1 | GTSGATTCACCSGATGAG | This study |
| CA-2 | CCSGATGARCGSCTACAG | This study |
| CA-3 | TGSGCSACSGTSACGAAC | This study |
| CA-4 | SACGAAYTCSCCCTGSGAC | This study |
| CA-5 | GGTCGGGATCAATCTCAACTACATG | This study |
| CA-6 | CCTGCTCATCTTCTGCAAACAACTG | This study |
| carF | CGCGGATCCGCAGTGGATTCACCG GATGAG | This study |
| carR | CGGGGTACCCCTGATATCCGTCAG AGCAGCTG | This study |
| Sequencing primers | ||
| T7 | TAATACGACTCACTATAGGG | |
| SP6 | CATACGATTTAGGTGACACTATAG | Sigma-Genosys |
| M13 reverse | CAGGAAACAGCTATGAC | Sigma-Genosys |
| Scar-1 | CTCGACCTGGCCGATATCCAC | This study |
| Scar-2 | GAGGACGGCTTCTACAAGAC | This study |
| Scar-3 | GACGCGCACTTCACCGACCTG | This study |
| Scar-4 | GTCGACCTGATCGTCCATCC | This study |
| Sacr-5 | GACCTACGACGTGCTCAATC | This study |
| Scar-6 | CGTACGACGATGGCATCTC | This study |
| Scar-7 | GTGGATATCGGCCAGGTCGAG | This study |
| He-32 | GGTGGCAGGATGGAATCGG | This study |
| He-33 | CGTCGATTCGCGATTCCCTG | This study |
Restriction cleavage sites are underlined. R represents A or G, Y represents C or T, and S represents G or C.
Inverse PCR was used to obtain the entire car sequence. Nocardia gDNA (1 μg) was completely digested with 20 U of SalI or Acc65I and was then diluted fivefold, ligated, and used as the template for inverse PCR. PCR primers CA-5 (forward) and CA-6 (reverse) were used for inverse PCR with TaqDNA polymerase for a total of 30 cycles with the following cycling pattern: 94°C for 45 s, 57°C for 45 s, and 72°C for 2 min. The amplified PCR product was cloned in pGEM-T and transformed into E. coli JM109. Plasmid preparations from independent clones were sequenced in both directions. The resulting sequence combined with the above part of Nocardia car gave a 4.6-kb sequence containing the entire Nocardia car gene (with Acc65I-digested and subsequently religated gDNA used as the template). A sequence of 2.5 kb upstream from car was obtained with SalI-digested and subsequently religated gDNA used as the PCR template.
Nocardia car was generated by PCR using the primers carF and carR with Nocardia gDNA used as the template, which resulted in a BamHI site and a KpnI site, respectively, at the 5′ and 3′ ends of car. PCR was performed using Platinum Pfx DNA polymerase for a total of 30 cycles as follows: 94°C for 18 s, 59°C for 30 s, and 68°C for 4 min. PCR products digested with BamHI and KpnI were separated on a 1% agarose gel, purified, and then subcloned into pHAT10 to form pHAT-305.
Open reading frames were identified with the National Center for Biotechnology Information (NCBI) orf finder (http://www.ncbi.nlm.nih.gov). Derived amino acid sequences were compared to those of database proteins by the NCBI BlastP program (2). Protein sequence relationships were examined by the NCBI CD program. The Sanger Centre TBLASTN program was used to locate and analyze the Mycobacterium bovis and Mycobacterium leprae sequences that were homologous with car and the surrounding regions (http://www.sanger.ac.uk). CAR homologue sequences were compared by using the Clustal W multiple-alignment program (35) with default settings available at the Baylor College of Medicine search launcher (http://dot.imgen.bcm.tmc.edu). Alignments were prepared for examination or presentation by shading with the Boxshade program at the Swiss Institute for ExperimentalCancer Research (http://www.ch.embnet.org/software/BOX_form.html).
A 100-ml culture of E. coli [BL21(DE3) or BL21-CodonPlus(DE3)-RP] harboring pHAT-305 was grown overnight in LB-ampicillin medium, diluted 20-fold in fresh medium, and then incubated at 170 rpm in a rotary shaker at 37°C to an optical density at 600 nm of 0.6, which was followed by the addition of 1 mM IPTG and further incubation for 4.5 h. The cells were harvested by centrifugation and stored at −65°C.
Enzyme assays, gel analysis, and HAT-CAR purification methods.
The standard reaction mixture contained 1 mM ATP, 0.15 mM NADPH, 5 mM sodium benzoate, 10 mM MgCl2, and enzyme in 0.05 M Tris buffer (pH 7.5) containing 1 mM EDTA, 1 mM dithiothreitol, and 10% glycerol (vol/vol), all in a final volume of 1.4 ml. The reference cuvette contained all components except enzyme and benzoate, and the background control cuvette contained all components except benzoate. The background control was subtracted from the complete reaction to give the true enzyme activity. Reactions were initiated by adding enzyme and were monitored at 340 nm at 25°C. Enzyme kinetics were examined as previously described (10). One unit of the enzyme catalyzed the reduction of 1 μmol of benzoate to benzaldehyde · min−1 under standard assay conditions.
Protein was measured by the Bradford protein microassay (5) with bovine serum albumin used as the standard. Gel analysis of proteins was carried out with SDS-10% polyacrylamide gel electrophoresis (PAGE) (23).
E. coli CodonPlus cells (wet weight, 4.3 g) transformed with pHAT-305 were suspended in 26 ml of 0.05 M K2HPO4 (pH 7.5) buffer containing 0.3 M NaCl, 10% (vol/vol) glycerol, 0.2 mM phenylmethylsulfonyl fluoride, and 3 mM β-mercaptoethanol, and the cells were disrupted twice by a French press cell at 12,000 lb/in2. Cell debris was removed by centrifugation for 60 min at 25,000 × g and 4°C. The resulting supernatant (27 ml) was referred to as cell extract and used for HAT-CAR purification. Cell extract (24 ml) was loaded onto a 6-ml bed volume column of Talon resin (cobalt-complexed resin made by Clontech that specifically binds the HAT tag) equilibrated with 0.05 M K2HPO4 buffer (pH 7.5) containing 0.3 M NaCl and 10% (vol/vol) glycerol and then washed with the same buffer. The HAT-CAR was eluted sequentially by 16-ml portions of 5, 7.5, 10, and 20 mM concentrations of imidazole in the same buffer. Active fractions were pooled and concentrated by ultrafiltration in an Amicon concentrator (PM-10 membrane) and then diluted with 100 ml of 50 mM Tris buffer (pH 7.5) containing 1 mM EDTA, 1 mM dithiothreitol, and 10% glycerol. This preparation was loaded onto a DEAE Sepharose column (dimensions, 1.5 by 20 cm; bed volume, 24 ml) equilibrated with the preceding Tris buffer. The column was then washed with 30 ml of this buffer and eluted with a 0 to 0.5 M NaCl linear gradient (total, 100 ml). Active 2-ml fractions (fractions 29 to 34) were combined for analysis (Table 3).
TABLE 3.
Purification of recombinant HAT-CAR from E. coli CodonPlus carrying pHAT-305
| Step | Total protein (mg) | Total activity (U)a | Sp act (U/mg) | Yield (%) | Purifi- cation (n-fold) |
|---|---|---|---|---|---|
| Crude extract | 600 | 5.21 | 0.009 | 100 | 1 |
| Talon matrix | 69.1 | 4.57 | 0.066 | 87.7 | 7.3 |
| DEAE Sepharose | 40 | 4.43 | 0.11 | 85 | 12.2 |
One unit of enzyme catalyzed the reduction of 1 μmol of benzoate to benzaldehyde per min at 25°C.
Western blot samples were subjected to SDS-PAGE and transferred to a polyvinylidene difluoride membrane. The membrane was incubated with 2% fat-free milk in Tris-buffered saline, polyclonal anti-HAT antibody (diluted 1:20,000), and finally polyclonal goat anti-rabbit immunoglobulin G conjugated to alkaline phosphatase (diluted 1:20,000), which was used with the Bio-Rad Immuno-Star chemoluminescent substrate. Tagged proteins were identified with photographic film. E. coli JM 109 carrying a vector expressing HAT-tagged dihydrofolate reductase (DHFR; Clontech) was used as a positive control, and E. coli CodonPlus carrying the pHAT10 vector was used as a negative control.
Large-scale in vitro enzyme reactions were carried out in 50 ml of 50 mM Tris-HCl buffer (pH 7.5) containing 0.1 mmol of substrate, 12.5 mg of NADPH, 55 mg of ATP, 101 mg of MgCl2, 33.6 mg of glucose-6-phosphate, 3 U of glucose-6-phosphate dehydrogenase, and 1 mg of purified HAT-CAR (0.1 U). Reaction mixtures were incubated at 30°C with gentle shaking at 50 rpm for 24 h. In vivo reactions were conducted with 100-ml cultures of E. coli CodonPlus carrying pHAT-305. Cultures were induced by treatment with 1 mM IPTG for 4 h before receiving 1 mg of benzoic acid, vanillic acid, or ferulic acid per ml. Samples (∼2 ml) were removed at intervals, the pH was adjusted to pH 2.0 with 6 N HCl, and samples were extracted with 1 ml of ethyl acetate; centrifugation for 2 min at 1,000 × g followed. Organic phases were removed and used for silica gel GF254 thin-layer chromatography. A solvent system of hexanes-ethyl acetate-formic acid (12:6:0.03) was used for benzoic acid (Rf, 0.7) and benzaldehyde (Rf, 0.4). Hexanes-ethyl acetate-formic acid (18:6:0.03) was used for vanillic acid (Rf, 0.33) and vanillin (Rf, 0.5). Hexanes-ethyl acetate-formic acid (12:6:0.035) was used for ferulic acid (Rf, 0.2), coniferyl aldehyde (Rf, 0.3), and coniferyl alcohol (Rf, 0.15). Developed thin-layer chromatography plates were examined under 254-nm-wavelength UV light before being sprayed with 0.5% phosphomolybdic acid in 95% ethanol and warming with a heat gun to visualize spots. Reduction products were isolated by adjustment of incubation mixtures to pH 2.0 with 6 N HCl and extracting them each three times with half-volumes of ethyl acetate. After removal of solvent by rotary evaporation, reduction products were purified by preparative thin-layer chromatography for comparison with authentic standards, and yields were estimated by weighing dried extractions.
Sequence of Nocardia car.
Degenerate primers (two forward [CA-1 and CA-2] and two reverse [CA-3 and CA-4]) were used to amplify sequences between the known N-terminal and internal amino acid sequences (24). PCR products were cloned into pGEM-T, and 1.6 kb of sequence was determined. Gene sequence-specific primers (CA-5 and CA-6) based on this identified fragment were synthesized for inverse PCR to clone the entire Nocardia car gene. The sequence derived from two inverse PCR experiments and the above-obtained sequence gave a total of 6.9 kb of data, which included the entire Nocardia car gene and its flanking regions. The DNA sequence and the deduced amino acid sequence of Nocardia car have been deposited in the GenBank database (accession number AY495697). Nocardia car consists of 3,525 bp, corresponding to a protein with a length of 1,173 amino acid residues with a calculated molecular mass of 128.3 kDa and a pI of 4.74. The N-terminal amino acid sequence of purified Nocardia CAR exactly matched the deduced amino acid sequence of the N terminus, with Ala as the first amino acid (24). The assignment of ATG as the start codon is supported by analysis of the 5′ flank region: 6 bp upstream from the start codon ATG lies a conserved Streptomyces ribosomal binding site (GGGAGG) (28, 34).
The deduced amino acid sequence of Nocardia CAR was 60% identical to the putative acyl-coenzyme A synthase-substrate-coenzyme A ligase fadD9 of Mycobacterium tuberculosis and M. bovis and was 57% identical with its M. leprae homologue. This finding suggests that the mycobacterial proteins function in carboxylic acid reduction, but this supposition remains to be tested. Three open reading frames were found adjacent to car, all coding for putative membrane proteins (data not shown). BLAST analysis showed that CAR contains two major domains and a possible phosphopantetheine attachment site. The N-terminal domain (amino acids 90 to 544) has high homology with AMP-binding proteins. The C-terminal domain has high homology with NADPH binding proteins. We suggest that if a 4′-phosphopantetheine prosthetic group exists in active CAR, it may act as a “swinging arm” for transferring acyl-AMP intermediates to the C-terminal reductase domain. This arrangement of the CAR protein would reflect a sequential catalytic mechanism wherein the N-terminal domain catalyzes substrate activation by formation of an initial acyl-AMP intermediate, while the C-terminal portion then catalyzes the reduction of acyl-AMP by NADPH to finish a catalytic cycle (Fig. 1). We are attempting to establish the existence of a possible 4′-phosphopantetheine prosthetic group for the catalytic process as with aminoadipic acid reductase (13).
Heterologous expression of car.
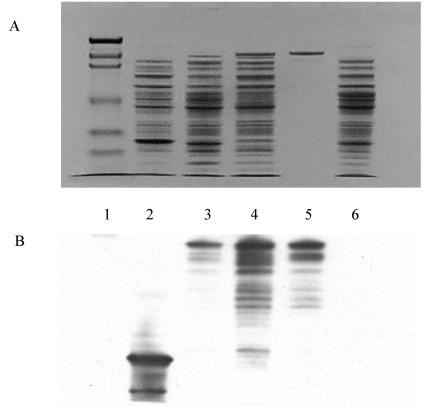
After Pfx DNA polymerase amplification, car was cloned in frame into pHAT10 to form the expression vector pHAT-305. Lysate from E. coli BL21(DE3) and CodonPlus cells carrying pHAT-305 had moderate CAR activity (0.003 and 0.009 U/mg, respectively) compared to that of Nocardia wild-type cells (0.03 U/mg of protein) (24), although SDS-PAGE showed that recombinant CAR is overexpressed in CodonPlus cells. This enzyme activity definitively established that we cloned active CAR. When these cultures were examined by SDS-PAGE, the Coomassie blue-stained band with an apparent molecular mass of 132.4 kDa was confirmed to be HAT-CAR by Western blot analysis (Fig. 2). The DHFR-positive control and empty pHAT10 negative control showed the absence of a 132.4-kDa band by SDS-PAGE and Western blot analysis.
FIG. 2.
SDS-PAGE and Western blot analysis of Nocardia CAR expression in E. coli carrying pHAT10-based vectors. Samples were taken from the lysates of E. coli cells carrying different vectors and were separated in duplicate by SDS-10% PAGE and either stained with 0.1% Coomassie blue R-250 (A) or subjected to Western blotting using a HAT-specific antibody (B). Lane 1, molecular mass markers for myosin (209 kDa), β-galactosidase (124 kDa), bovine serum albumin (80 kDa), ovalbumin (49.1 kDa), carbonic anhydrase (34.8 kDa), soybean trypsin inhibitor (21.5 kDa), lysozyme (20.6 kDa), and aprotinin (7.1 kDa). Lane 2, E. coli CodonPlus cells carrying pHAT-DHFR. Lane 3, E. coli BL21(DE3) cells carrying pHAT-305 (induced); lane 4, E. coli CodonPlus cells carrying pHAT-305 (induced). Lane 5, purified HAT-CAR. Lane 6, E. coli CodonPlus cells carrying pHAT10.
The HAT-CAR protein from E. coli was purified to SDS-PAGE homogeneity by Talon and DEAE chromatography (85% recovery). Recombinant CAR bound weakly to the Talon affinity matrix, being eluted from columns by 10 mM imidazole rather than the 100 mM usually required for HAT-tagged proteins. Minor impurities occurring after the affinity step were removed by DEAE Sepharose column chromatography. Trace impurities not detected by SDS-PAGE were detected by Western blot analysis (Fig. 2). These impurities were HAT tag-containing proteins that are likely hydrolyzed fragments of HAT-CAR cleaved by metal proteases. Metal protease inhibitors were not used to prevent protease cleavage during cell disruption because they would be incompatible with Talon matrix chromatography. The purified HAT-CAR showed a specific activity of 0.11 μmol · min−1 · mg−1 of protein, which was less than that of CAR purified from Nocardia (5.89 U/mg of protein) (24). Km values for benzoate, ATP, and NADPH were 852 ± 82, 69 ± 6.6, and 57 ± 3.6 μM, respectively (10). These are similar to the native enzyme Km values. The Vmax was 0.135 ± 0.004 μmol · min−1 · mg−1 of protein, which is less than that of the natural enzyme (0.902 ± 0.04 μmol · min−1 · mg−1 of protein) (24).
In vitro, nonoptimized biotransformation reactions showed that pure HAT-CAR reduced several carboxylic acids to their corresponding aldehydes. In 24-h in vitro reactions, 10.2 mg of benzaldehyde (82% yield), 7.5 mg of vanillin (48% yield), and 4 mg of coniferyl aldehyde (23% yield) were obtained by preparative layer chromatography. In vivo biotransformations of the same substrates showed that benzoate was rapidly converted to benzyl alcohol in 2 h, while vanillin and coniferyl aldehyde observed at 2 h were converted to their corresponding alcohols in 24 h.
Conclusions.
CAR differs from coniferyl aldehyde dehydrogenase, which uses NAD+ to oxidize aldehydes to acids, does not use ATP, and has no homology with CAR (1). ATP-dependent CAR catalyzes the energetically unfavorable reduction of acids to aldehydes by using ATP for energy to drive the reaction. It can also catalyze the oxidation of aldehyde to acid without ATP, but the cofactor for CAR is NADP(H) instead of NAD(H). The size of car (3.5 kb) is much larger than that of aldehyde dehydrogenase genes (1.5 kb) (1). CAR also differs from fatty acid reductases in luminescent bacteria, which contain three polypeptide components (31).
CAR was only moderately expressed in E. coli BL21(DE3) carrying pHAT-305. This low expression might be due to codon bias that can cause early termination and slow transcription since the G+C content of the sequence is 66%, and expression of pHAT-305 improved about threefold when it was carried out in E. coli CodonPlus cells. The HAT-CAR protein bands from CodonPlus cells were more clearly seen on SDS-PAGE than was extract from normal BL21 (DE3) cells (Fig. 2).
We speculate that two forms of the enzyme exist in E. coli. One form (holo-CAR) is active, while the other (apo-CAR) is inactive. Conversion of apo-CAR to catalytically active holo-CAR might occur by posttranslational modification, e.g., phosphopantetheinylation at a position such as Ser688 to function as a swinging arm (13). In holo-CAR, the SH of the phosphopantetheine prosthetic group would react with acyl-AMP to form an acyl-S-pantotheine-CAR intermediate (shown as C6H5-CO-CAR in brackets in Fig. 1). The C-terminal reductase domain would finish the catalytic cycle by delivering hydride from NADPH to the acyl-S-pantotheine-CAR intermediate, freeing an aldehyde product. This may explain the different specific activity observations between the wild-type and recombinant enzyme, in which the majority of the latter enzyme may be in the form of apo-CAR.
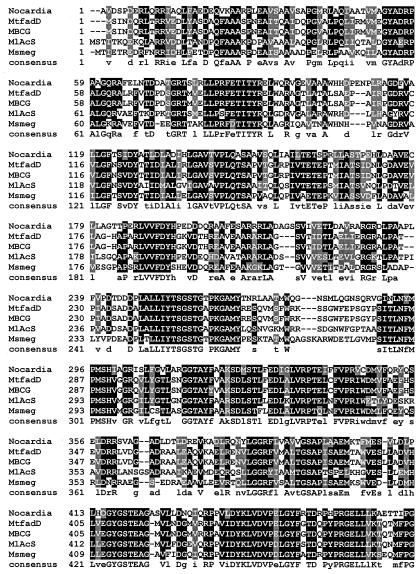
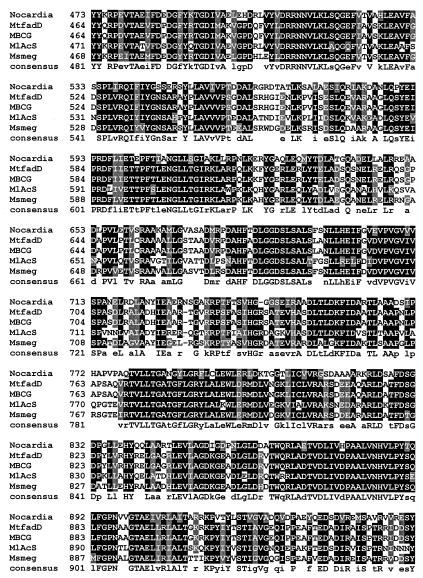
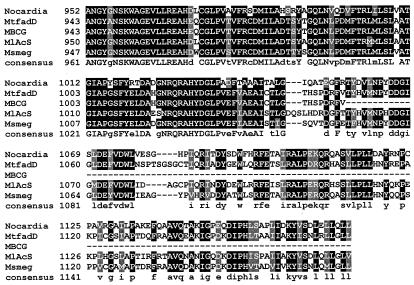
α-Aminoadipate reductase is well studied, and motifs responsible for adenylation of α-aminoadipate, reduction, NADPH binding, and attachment of a phosphopantetheinyl group have been identified previously (7, 19). While traditional BLAST analysis does not reveal the expected common motifs in the N-terminal portion of CAR, they do appear in the C-terminal portion. As with α-aminoadipate reductase, a phosphopantotheine attachment site, domain J, is clearly present in CAR (LGGxSxxA), as are the reduction domain (GYxxSKW) and the NADP binding domain (GxxGxLG). These motifs are fully conserved in the Mycobacterium CAR homologues (Fig. 3). Whether benzoate induction (21, 24) increases CAR expression or increases the conversion of an inactive form of the enzyme to an active form by a posttranslational modification remains to be established, as does the possible involvement of a Lys5-like protein (13) catalyzing posttranslational 4′-phosphopantetheinylation of apo-CAR.
FIG. 3.
Alignment of the deduced amino acid sequence of Nocardia CAR with a representative sample of putative homologous molecules from other organisms. Identical amino acids are highlighted in black, and similar amino acids are highlighted in gray. The Clustal W program was used to align the above sequences, and Boxshade (setting, 0.7) was used to determine the degree of residue shading. Accession numbers for the other protein sequences above are as follows: MtfadD, M. tuberculosis (Z77724); MlAcS, M. leprae (NP_301424); Msmeg, Mycobacterium smegmatis (Contig 3313); MBCG, M. bovis BCG (unnamed hypothetical protein at bases 2,885,319 to 2,888,822).
Biotransformation reactions using IPTG-induced whole E. coli CodonPlus cells carrying pHAT-305 were simple to conduct and resulted in the smooth conversion of carboxylic acids to aldehydes—and subsequently to alcohols. With whole cells, expensive cofactors are unnecessary (26), rendering the biocatalytic reaction more practical at large scales. Reduction of aldehydes formed by CAR to alcohols by an endogenous E. coli alcohol dehydrogenase similar to that observed in Nocardia (26) is relatively slow. Biochemical engineering approaches with the recombinant organism might be exploited to diminish this unwanted side reaction.
The unique car sequence for the CAR enzyme may be used to produce easily grown recombinant E. coli cultures for direct use in whole-cell biocatalytic conversions of natural or synthetic carboxylic acids (24, 32). Alternatively, this gene sequence or its homologues may be incorporated into the genomes of multiply recombinant strains through pathway engineering to be used in combinatorial biocatalytic syntheses of useful compounds.
Nucleotide sequence accession number.
The nucleotide sequence encoding Nocardia CAR has been deposited in the GenBank-EMBL database under accession number AY495697.
Acknowledgments
A. He and T. Li are grateful for financial support through Center for Biocatalysis and Bioprocessing fellowships.
REFERENCES
- 1.Achterholt, S., H. Priefert, and A. Steinbuchel. 1998. Purification and characterization of the coniferyl aldehyde dehydrogenase from Pseudomonas sp. strain HR 199 and molecular characterization of the gene. J. Bacteriol. 180:4387-4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Arfman, H. A., and W. R. Abraham. 1993. Microbial reduction of aromatic carboxylic acids. Z. Naturforsch. Sect. C 48:52-57. [DOI] [PubMed] [Google Scholar]
- 4.Bachman, D. M., B. Dragoon, and S. John. 1960. Reduction of salicylate to saligenin by Neurospora. Arch. Biochem. Biophys. 91:326.. [DOI] [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Casey, J., and R. Dobb. 1992. Microbial routes to aromatic aldehydes. Enzyme Microbiol. Technol. 14:739-747. [Google Scholar]
- 7.Casqueiro, J., S. Guttierrez, O. Banuelos, F. Fierro, J. Velasco, and J. F. Martin. 1998. Characterization of the lys2 gene of Penicillium chrysogenum encoding α-aminoadipic acid reductase. Mol. Gen. Genet. 259:549-556. [DOI] [PubMed] [Google Scholar]
- 8.Chen, Y., and J. P. N. Rosazza. 1994. Microbial transformation of ibuprofen by a Nocardia species. Appl. Environ. Microbiol. 60:1292-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christner, J. E., M. J. Schlesinger, and M. J. Coon. 1964. Enzymatic activation of biotin: biotinyl adenylate formation. J. Biol. Chem. 239:3997-4005. [PubMed] [Google Scholar]
- 10.Cleland, W. W. 1979. Statistical analysis of enzyme kinetic data. Methods Enzymol. 63:103-138. [DOI] [PubMed] [Google Scholar]
- 11.Coque, J. J., M. Malumbres, J. F. Martin, and P. Liras. 1993. Analysis of the codon usage of the cephamycin C producer Nocardia lactamdurans. FEMS Microbiol. Lett. 110:91-96. [Google Scholar]
- 12.Drauz, K., and H. Waldmann (ed.). 2002. Enzyme catalysis in organic synthesis: a comprehensive handbook, 2nd ed. Wiley-VCH, New York, N.Y.
- 13.Ehmann, D. E., A. M. Gehring, and C. T. Walsh. 1999. Lysine biosynthesis in Saccharomyces cerevisiae: mechanism of α-aminoadipate reductase (Lys2) involves posttranslational phosphopantetheinylation by Lys5. Biochemistry 38:6171-6177. [DOI] [PubMed] [Google Scholar]
- 14.El-Sharkawy, S. H., W. Yang, L. Dostal, and J. P. N. Rosazza. 1992. Microbial oxidation of oleic acid. Appl. Environ. Microbiol. 58:2116-2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fersht, A. R., and M. M. Kaethner. 1976. Mechanism of aminoacylation of tRNA. Proof of the aminoacyl adenylate pathway for the isoleucyl- and tyrosyl-tRNA synthetases from Escherichia coli K12. Biochemistry 15:818-823. [DOI] [PubMed] [Google Scholar]
- 16.Gross, G. G. 1972. Formation and reduction of intermediate aryl-adenylate by aryl-aldehyde NADP oxidoreductase from Neurospora crassa. Eur. J. Biochem. 31:585-592. [DOI] [PubMed] [Google Scholar]
- 17.Gross, G. G., and M. H. Zenk. 1969. Reduktion aromatischer säuren zu Aldehyden und Alkoholen im zellfreien system. 1. Reinigung und Eigenschaften von Aryl-Aldehyde: NADP-Oxidoreduktase aus Neurospora crassa. Eur. J. Biochem. 8:413-419. [DOI] [PubMed] [Google Scholar]
- 18.Gross, G. G., and M. H. Zenk. 1969. Reduktion aromatischer säuren zu Aldehyden und Alkoholen im zellfreien system. 2. Reinigung und Eigenschaften von Aryl-Alkohol: NADP-Oxidoreduktase aus Neurospora crassa. Eur. J. Biochem. 8:420-425. [DOI] [PubMed] [Google Scholar]
- 19.Hijarrubia, M. J., J. F. Aparicio, J. Casqueiro, and J. F. Martin. 2001. Characterization of the lys2 gene of Acremonium chrysogenum encoding a functional α-aminoadipate activating and reducing enzyme. Mol. Gen. Genet. 264:755-762. [DOI] [PubMed] [Google Scholar]
- 20.Jezo, I., and J. Zemek. 1986. Enzymatische reduktion einiger aromatischer carboxysäuren. Chem. Pap. 40:279-281. [Google Scholar]
- 21.Kato, N., E. H. Joung, H. C. Yang, M. Masuda, M. Shimao, and H. Yanase. 1991. Purification and characterization of aromatic acid reductase from Nocardia asteroides JCM 3016. Agric. Biol. Chem. 55:757-762. [Google Scholar]
- 22.Kato, N., H. Konishi, K. Uda, M. Shimao, and C. Sakazawa. 1988. Microbial reduction of benzoate to benzyl alcohol. Agric. Biol. Chem. 52:1885-1886. [Google Scholar]
- 23.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 227:680-685. [DOI] [PubMed] [Google Scholar]
- 24.Li, T., and J. P. N. Rosazza. 1997. Purification, characterization, and properties of an aryl aldehyde oxidoreductase from Nocardia sp. strain NRRL 5646. J. Bacteriol. 179:3482-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, T., and J. P. N. Rosazza. 1998. NMR identification of an acyl-adenylate intermediate in the aryl-aldehyde oxidoreductase catalyzed reaction. J. Biol. Chem. 273:34230-34233. [DOI] [PubMed] [Google Scholar]
- 26.Li, T., and J. P. N. Rosazza. 2000. Biocatalytic synthesis of vanillin. Appl. Environ. Microbiol. 66:684-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mao, L.-F., D. S. Millington, and H. Schultz. 1992. Formation of a free acyl adenylate during the activation of 2-propylpentanoic acid. Valproyl-AMP: a novel cellular metabolite of valproic acid. J. Biol. Chem. 267:3143-3146. [PubMed] [Google Scholar]
- 28.Mulder, M. A., H. Zappe, and L. M. Steyn. 1997. Mycobacterial promoters. Tuber. Lung Dis. 78:211-223. [DOI] [PubMed] [Google Scholar]
- 29.Pelicic, V., M. Jackson, J. M. Reyrat, W. R. Jacobs, Jr., B. Gicquel, and C. Guilhot. 1997. Efficient allelic exchange and transposon mutagenesis in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 94:10955-10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raman, T. S., and E. R. B. Shanmugasundaram. 1962. Metabolism of some aromatic acids by Aspergillus niger. J. Bacteriol. 84:1340-1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodriguez, A., and E. Meighen. 1985. Fatty acyl-AMP as an intermediate in fatty acid reduction to aldehyde in luminescent bacteria. J. Biol. Chem. 260:771-774. [PubMed] [Google Scholar]
- 32.Rosazza, J. P. N., and T. Li. August. 1998. Carboxylic acid reductase and methods for use of the same. U.S. patent 5,795,759.
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34.Strohl, W. R. 1992. Compilation and analysis of DNA sequences associated with apparent streptomycete promoters. Nucleic Acids Res. 20:961-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-dependent gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsuda, Y., K. Kawai, and S. Naajima. 1984. Asymmetric reduction of 2-methyl-2-aryloxyacetic acids by Glomerella cingulata. Agric. Biol. Chem. 48:1373-1374. [Google Scholar]
- 37.Tsuda, Y., K. Kawai, and S. Nakajima. 1985. Microbial reduction of 2-phenylpropioic acid, 2-benzyloxypropionic acid and 2-(2-furfuryl) propionic acid. Chem. Pharm. Bull. 33:4657-4661. [Google Scholar]
- 38.van den Ban, E. C. D., H. M. Willemen, H. Wassink, C. Laane, and H. Haaker. 1999. Bioreduction of carboxylic acids by Pyrococcus furiosus in batch cultures. Enzyme Microbiol. Technol. 25:251-257. [Google Scholar]
- 39.Vignais, P. V., and I. Zabin. 1958. Synthesis and properties of palmityl adenylate, palmityl coenzyme A, and palmityl glutathione. Biochim. Biophys. Acta 29:263-269. [DOI] [PubMed] [Google Scholar]
- 40.White, H., G. Strobl, R. Feicht, and H. Simon. 1989. Carboxylic acid reductase: a new tungsten enzyme catalyses the reduction of non-activated carboxylic acids to aldehydes. Eur. J. Biochem. 184:89-96. [DOI] [PubMed] [Google Scholar]