Abstract
Papillomaviruses use rare codons with respect to the host. The reasons for this are incompletely understood but among the hypotheses is the concept that rare codons result in low protein production and this allows the virus to escape immune surveillance. We changed rare codons in the oncogenes E6 and E7 of the cottontail rabbit papillomavirus to make them more mammalian-like and tested the mutant genomes in our in vivo animal model. While the amino acid sequences of the proteins remained unchanged, the oncogenic potential of some of the altered genomes increased dramatically. In addition, increased immunogenicity, as measured by spontaneous regression, was observed as the numbers of codon changes increased. This work suggests that codon usage may modify protein production in ways that influence disease outcome and that evaluation of synonymous codons should be included in the analysis of genetic variants of infectious agents and their association with disease.
Keywords: Papillomavirus, Synonymous codons, Cancer, Immunogenicity, In vivo model, Codon usage, CRPV, E6, E7
BACKGROUND
Papillomaviruses are double stranded non-enveloped DNA tumor viruses with genomes of about 8Kb. These viruses infect mucosal and cutaneous tissues of many animal species and demonstrate high tissue and species specificities. Papillomaviruses infect the basal layer of the epidermis via micro abrasions in the skin and mucosa and are maintained at low copy number until the cells differentiate. At that point, copy number increases markedly and virion production ensues. The pathogens are dependent upon cellular differentiation for the completion of the life cycle and therefore the full life cycle is ideally studied in the intact host. The rabbit papillomavirus model is one of the best in vivo models for studying the course of papillomavirus disease from initiation of infection to malignant progression. For a review of the molecular biology of papillomaviruses see (Doorbar, 2006).
Papillomaviruses use rare codons relative to their hosts(Zhao, Liu et al., 2003;Zhou, Liu et al., 1999). The reasons for this bias are not well understood but it has been postulated that this is a mechanism to allow the virus to escape immune surveillance (Tindle, 2002). This theory suggests that the rare codons employed by the virus insure a low rate of translation resulting in protein levels sufficient to carry out viral functions but insufficient to trigger a significant immune response. In recent years, codon modifications that make the codons of many different genes more host-like have resulted in increased protein production in vitro(Mossadegh, Gissmann et al., 2004;Mechold, Gilbert et al., 2005;Gao, Li et al., 2003). Thus, in vitro studies support the hypothesis that papillomaviruses may use rare codons to limit protein production.
A second hypothesis is that papillomavirus codon usage may be tuned to the tRNA profile of the host and, specifically, to the cell in which the viral protein needs to be active as well as the stage of differentiation at which it needs to be expressed(Zhao, Gu et al., 2005;Ding, Doorbar et al., 2010;Gu, Li et al., 2004;Gu, Ding et al., 2007). We have shown that codon usage differs from gene to gene within a papillomavirus species as well as between a given gene for the alpha and beta papillomaviruses (Cladel, Bertotto et al., 2010). These observed differences are consistent with the hypothesis that tRNA profiles are related to location and timing of gene expression.
For many years, synonymous codons were thought to be redundant and were often ignored in the analysis of data. Recent work, however, has shown that synonymous codon variants may display marked phenotypic differences(Kallel, Rebai et al., 2009;Kimchi-Sarfaty, Oh et al., 2007;Capon, Allen et al., 2004;Duan, Wainwright et al., 2003;Nackley, Shabalina et al., 2006;Edwards, Hing et al., 2012;Bible, Mant et al., 2000;Hamano, Matsuo et al., 2007;Mueller, Papamichail et al., 2006;Burns, Shaw et al., 2006). The study of synonymous codon usage in both prokaryotes and eukaryotes is now an active field of investigation and has been correlated with 1) control of rate of translation (the speed at which translation occurs) ((Lavner & Kotlar, 2005;Lemm & Ross, 2002) 2) nucleosome positioning(Segal, Fondufe-Mittendorf et al., 2006) 3) tissue specificity requirements(Zhou, Liu, Peng, Sun, & Frazer, 1999;Gu, Li, Zhao, Fang, Bu, Frazer, & Zhao, 2004;Gu, Ding, Wang, de Kluyver, Saunders, Frazer, & Zhao, 2007) 4) mRNA stability and secondary structure(Capon, Allen, Ameen, Burden, Tillman, Barker, & Trembath, 2004;Duan, Wainwright, Comeron, Saitou, Sanders, Gelernter, & Gejman, 2003;Chamary & Hurst, 2005) 5) mRNA copy number(Akashi, 2003;Sharp & Cowe, 1991), 6) splicing(Fairbrother, Holste et al., 2004;Chamary, Parmley et al., 2006;Parmley & Hurst, 2007), 7) selection for translational efficiency (the rate at which properly folded protein is achieved) (Gingold & Pilpel, 2011; Chamary & Hurst, 2005), 8) selection for proper protein folding(Cortazzo, Cervenansky et al., 2002),9) regulation by microRNA(Zheng & Wang, 2011) and 10) selection for translational kinetics {“…is based on the right combination of codons( common and rare) that allows a regulated ribosome traffic rate ensuring the proper protein folding”} (Aragones, Guix et al., 2010). In a recent study, Chen et al(Chen, Davydov et al., 2010) concluded that synonymous SNPs (sSNPS) are just as likely as non-synonymous SNPS (nsSNPs) to be involved in disease mechanisms.
The E7 gene is one of the two major oncogenes of the cottontail rabbit papillomavirus (CRPV) and has been found to be poorly immunogenic(Han, Cladel et al., 1999). Since E7 is expressed in nearly all papillomavirus-associated malignancies, it is an important immunological target for therapeutic vaccines. In earlier work, our laboratory undertook the codon “optimization” of the CRPV oncogene E7 in the context of the wild type genome in an attempt to increase the protein production of this gene in vivo. The term “optimization” is defined in this context as the replacement of viral codons rarely used by the host with those commonly used by the host. We hypothesized that changing some of the codons to be more host-like would improve protein production and allow for better targeting of the viral oncoprotein by the immune system. We found that the codon modifications in the E7 gene resulted in both altered immune responses and papilloma growth rates relative to those found with the wild type virus(Cladel, Hu et al., 2008b). Enhanced protein production was noted following transient transfection of codon-modified E7 expression constructs. mRNA amounts and stabilities also increased under these conditions. To our knowledge this was the first use of codon optimization in the context of a pathogen to study host/pathogen interactions in vivo.
In the current study, the scope of our efforts was expanded to include the second CRPV oncogene, E6. E6 and E7 have been reported to work synergistically both in rabbit(Ganzenmueller, Matthaei et al., 2008) and in human(Moody & Laimins, 2010) papillomaviruses. We report here that codon optimizations in both oncogenes resulted in phenotypic changes, some of which were dramatic. Among these changes were 1) altered growth rates, 2) greatly reduced times to cancer 3) increased numbers of cancers and 4) enhanced immunogenicity. The results are novel and, to our knowledge, are the first to demonstrate an association between synonymous codons and malignant potential in vivo. The findings demonstrate the utility of the CRPV/rabbit model to elucidate the functions of synonymous codons at the same time that the biological complexities of the virus are being unraveled.
RESULTS
CRPV genomes with codon optimizations in the E6 gene showed phenotypic differences
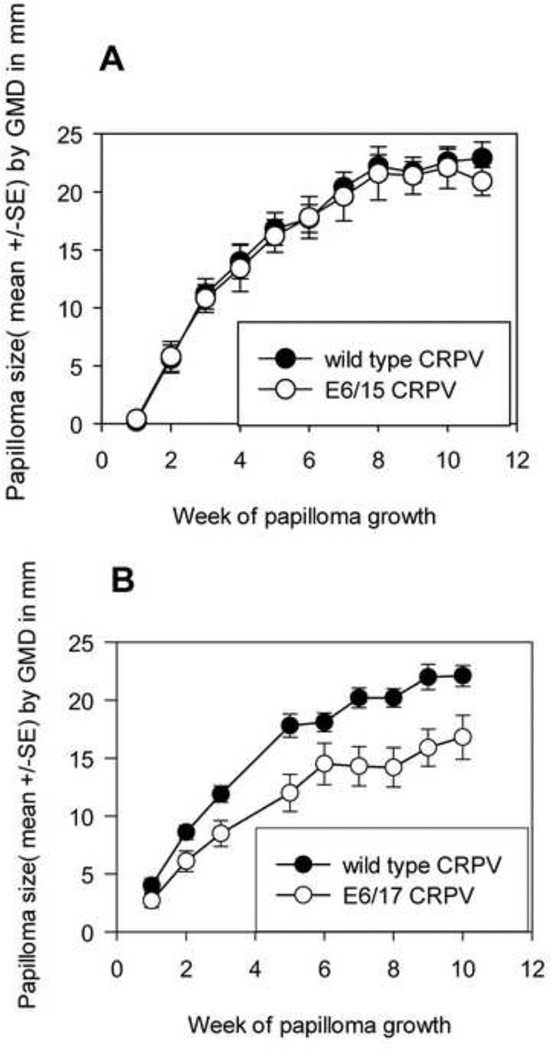
Data on codon optimizations in the E7 gene were reported in an earlier paper(Cladel, Hu, Balogh, & Christensen, 2008b)and are also shown in Supplementary figure #1. CRPV genomes with 15 (E6/15) and 17 (E6/17) synonymous codon changes in the N terminus of E6 were prepared for this study (Figure 1). CRPV has two E6 isoforms: long E6 (LE6) and short (SE6)(Barbosa & Wettstein, 1988). Long E6 was chosen for modification as we have shown that it is essential for papilloma formation whereas SE6 is not (Hu, Cladel et al., 2006). In addition, changes in short E6 would have involved concomitant changes in overlapping E8. All changes present in E6/15 were also present in E6/17. The changes were introduced within the 149 nucleotides upstream of the start of the overlapping E8 gene (CRPVnt 323) and downstream of the introduced Sac II cloning site (CRPVnt 174). Each modified E6 gene was substituted into the wild type Hershey CRPV (H. CRPV) backbone and the genomes were tested for function on a small number of rabbit skin sites. Both genomes produced papillomas. E6/15 grew at wild type rate whereas E6/17 grew more slowly (P <0.005, unpaired student t test, Figure 2A and B).
Fig. 1.
Codon optimizations in the 5’ region of E6 to create E6/15 and E6/17. Nucleotide changes were introduced into H. CRPV (Genbank accession number JF303889) Long E6 by site-directed mutagenesis. Changes for E6/15 and E6/17 are shown. Newly created codons are more commonly used in mammals than are the original codons. Nucleotides for the mutated portion of E6 are shown.
Fig. 2.
A and B. Papillomas generated from E6/15 and E6/17 grew at statistically different rates. Six animals were infected with wild type H. CRPV, H. CRPV with 15 codon changes in E6( E6/15) and H. CRPV with 17 changes in E6( E6/17) at four sites each for a total of 24 sites per genome. E6/15 papillomas grew at wild type rate and E6/17 papillomas grew more slowly than wild type (P<0.005 Student’s t- test).
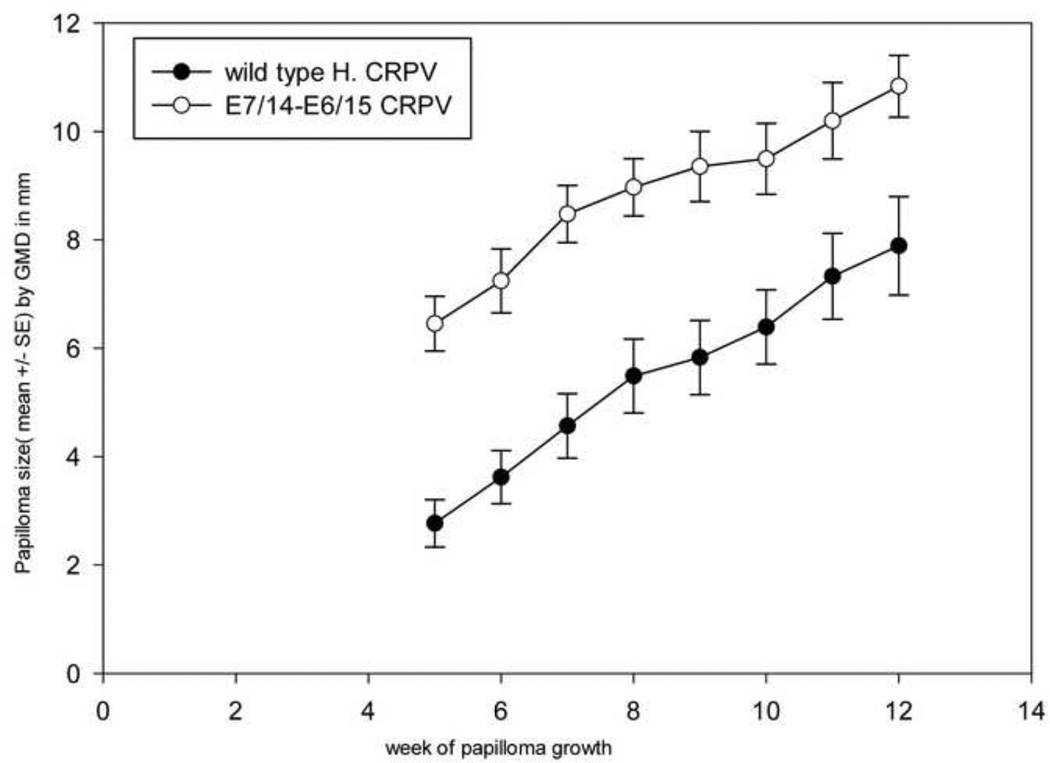
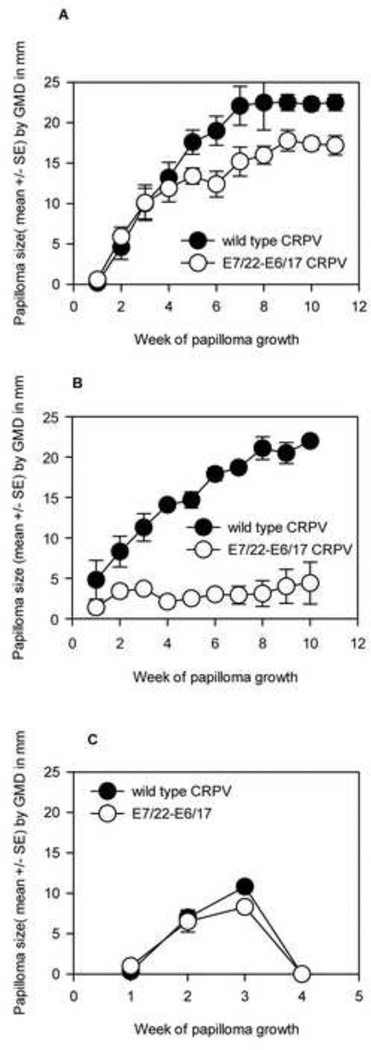
CRPV genomes with synonymous codon changes in both E6 and E7 displayed different phenotypes depending upon the combination of modified genes
Outbred New Zealand White (NZW) rabbits were used for all experiments. Growth curves for papillomas from CRPV with 14 changes in E7 and 15 changes in E6 (E7/14-E6/15) relative to wild type are shown in Figure 3. E7/14-E6/15 papillomas grew statistically faster than wild type. (P<0.01, unpaired Student t-test). Likewise E7/18-E6/17 induced papillomas that grew faster than wild type (P< 0.05, data not shown). CRPV E7/22-E6/17 papillomas followed three different courses: 1) progressive growth over time in four animals 2) markedly reduced, but persistent, growth relative to wild type in another four animals and 3) wild type growth followed by rapid regression in the remaining three animals (Figure 4A–C). In the third group, CRPV E7/22-E6/17 papillomas provoked immune responses that influenced the behavior of papillomas derived from the other genomes growing on the same rabbits. All papillomas on the regressing animals regressed including those initiated by wild type CRPV DNA (Figure 4C). E7/40-E6/17 was the genome with the greatest number of codon alterations. This genome generated papillomas at only a subset of sites and lesions were small. Detailed analysis of E7/40-E6/17 papillomas remains to be done due to inconsistent and slow growth rates.These data together demonstrated the influence of synonymous codons on progression and regression of papillomas and confirmed our earlier work(Cladel, Hu, Balogh, & Christensen, 2008b) that the immune system was responsive to synonymous changes in the codons.
Fig. 3.
Papillomas generated from E7/14-E6/15 grew statistically faster than papillomas from wild type CRPV. Six animals were infected with wild type CRPV at two sites and E7/14-E6/15 at two sites for a total of twelve sites per genome. E7/14-E6/15 grew statistically larger papillomas relative to wild type in outbred NZW rabbits (P<0.01, Student’s t-test).
Fig. 4.
A-C. Papillomas generated from E7/22-E6/17 displayed three different growth profiles. Twelve animals were infected at two sites each with wild type CRPV and with E7/22-E6/17 CRPV. One animal died before completion of the study. Papilloma growth displayed three different phenotypes. A) Papillomas on five animals grew steadily. There was no significant difference between wild type and E7/22-E6/17 growth rates at early time points in these animals; however at later time points the difference became significant (P=0.01, Student’s t-test) B) Papillomas on three other animals grew feebly but persistently. E7/22-E6/17 papillomas grew statistically more slowly in these animals relative to wild type (P=0.002) and statistically more slowly than the same papillomas in the progressor animals of group A (P<0.001, Student’s t-test). C )All papillomas on the remaining three animals rapidly regressed, including those of wild type controls.
Early cancers were detected in animals with papillomas from codon-optimized genomes
Forty nine animals were observed bearing papillomas initiated by both wild type Hershey CRPV (wt) and at least one of the codon-optimized genomes. By ten months post infection, 45% of these animals had developed at least one carcinoma originating from one of the codon-optimized genomes. None of the papillomas initiated by wt CRPV on the same animals became malignant during this time span [P=1.1×10−8, Fisher’s exact test, 2 tailed.
E7/14-E6/15 genomes produced 28 carcinomas out of 98 sites; cancers were detected from an early time point of 5 months (8 sites) to a late time point of 10 months (6 sites). There were no cancers at the 34 wild type sites on these rabbits (P=1.3×10−4, Fisher’s exact test, 2 tailed, Supplementary Table 1). A total of 17 animals were infected with E7/14-E6/15 and cancers were detected in 11 (65%) of these animals. Some regressions or partial regressions were noted in the remaining animals.
E7/18-E6/15 genomes produced 12 carcinomas out of 39 sites; cancers were detected from an early time point of 3 months (1site) to a late time point of 13 months (1site). There were no wild type cancers at the 38 sites during this time period (P= 1.8×10−4, Fisher’s exact test, 2 tailed, Supplementary Table 2). A total of 18 animals were infected with E7/18-E6/15 and cancers were detected in 9 (50%) of these animals. Some regressions or partial regressions were noted in the remaining animals.
Genomes E7/14, E6/15, E7/18-E6/17, E7/22-E6/17, and E7/17 all yielded early cancers although they were not studied in sufficient numbers to determine significance. Table 1 summarizes the cancer data for all of the genomes in this study. It is interesting to note that the genomes with the most codon changes progressed to cancer more rapidly. Thus cancers generated by both E7/18-E6/17 and E7/22-E6/17 occurred as early as 3 months post infection. These genomes will need to be tested in larger numbers to see if this trend holds up statistically.
Table 1.
Time to cancer for all codon-optimized genomes. Constructs were tested in the following numbers of animals: E7/8-E6/15, n=2; .E7/14-E6/15, n=17; E7/18-E6/15, n=18; E7/18-E6/17. n=22; E7/22-E6/17, n=l 1; E6/15. n=14; E7/14, n=4; E7/18, n=6. Each number in a box represents the number of cancers for a given genome(y axis) at a given month(x axis). Wild type cancers were from historical records as no wild type cancers were detected during the studies with the codon-optimized genomes.
| E7/8- E6/15 | 2 | 2 | |||||||||||||
| E7/14- E6/15 | 8 | 9 | 2 | 1 | 2 | 6 | |||||||||
| E7/18- E6/15 | 1 | 5 | 1 | 4 | 1 | ||||||||||
| E7/18- E6/17 | 2 | 6 | |||||||||||||
| E7/22- E6/17 | 2 | 2 | |||||||||||||
| E6/15 | 1 | 1 | 2 | 2 | |||||||||||
| E7/14 | 2 | 2 | 1 | 3 | 1 | ||||||||||
| E7/18 | 2 | ||||||||||||||
| WILD TYPE | 2 | 1 | 1 | ||||||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 |
Taken together, these data demonstrate the impact of synonymous codon modifications in the oncogenes of CRPV on the time of cancer development.
Reduction in growth rate and/or regressions were seen in animals bearing papillomas from codon-optimized genomes
Regressions of papillomas generated by wild type Hershey CRPV are rare. Regressions of lesions induced by the “regressor” strain, isolated both in this laboratory (“CRPVr”) and at the Pasteur Institute (“CRPVb”, Genbank accession number AJ243287) are common and occur early, within eight weeks of papilloma outgrowth(Salmon, Ramoz et al., 1997;Hu, Cladel et al., 2002). Clear evidence for host immunological mechanisms accounting for this regression is the observation that treatment with cyclosporine A results in persistent growth of papillomas initiated by the regressive strain of virus(Hu, Peng et al., 2005). In the animals bearing papillomas generated by our codon modified genomes, reduction in growth or outright regression was frequently observed. This regression was most clearly seen in some animals bearing papillomas generated by E7/22-E6/17, as noted above (figure 4 B and C) but was also observed with some of the other genomes. A total of 15 out of 49 animals experienced total regression or reduction in growth of their papillomas. In other unrelated experiments ongoing at the same time in our laboratory, no regressions were observed with wild type papillomas (n=53, P=4.5×10−5 ChiSquare with Yates Correction).
Two cancers that developed from papillomas from codon-altered genomes also displayed regression tendencies
Two cancers from papillomas generated by codon-altered genomes regressed and then later returned as aggressive cancers (data not shown).Q PCR on one of the samples showed abundant copies of CRPV DNA. This observation of cancer regression/resurgence has not been made in this laboratory with cancers derived from wild type H. CRPV nor has it been reported from other laboratories. Only two sites displayed this phenomenon, but it is worth noting that many of the sites that developed later (rather than earlier) cancers also displayed reduction in the growth of papillomas. (See supplementary tables 1 and 2). These observations would suggest that in such lesions there was competition between immunological and proliferative responses with the proliferative response generally gaining the advantage.
Synonymous changes were maintained in the DNA of the papillomas
DNA was extracted from selected papilloma biopsies and the E6 and E7 genes were amplified by PCR; the PCR products were subsequently sequenced. All modifications were maintained in the papilloma DNA. No other changes were noted.
Codon optimization did not influence mRNA levels in papillomas generated by codon-optimized genomes
Quantitative RT-PCR was used to determine baseline mRNA expression of wild type E6 and E7 in papillomas at week 8.5.We then analyzed mRNA levels from papillomas containing wild type E6 and E7, E7/14-E6/15 and E7/18-E6/15. There were six animals in this experiment and each animal was infected at two sites each with wt CRPV, E7/14-E6/15 and E7/18-E6/15. All papillomas yielded about the same amount of E7 mRNA at 8.5 weeks post infection. Likewise, wt E6 and E6/15 in both E7/14-E6/15 and E7/18-E6/15 yielded the same amount of E6 mRNA. (Figure 5). Thus, codon optimizations did not influence message in papillomas at 8.5 weeks post infection. The 8.5 week time point was chosen because at this point, papillomas have had time to develop substantial mass but have not yet progressed to cancer. Time-dependent variations in mRNA levels may occur.
Fig. 5.
Wild type and codon optimized E6 and E7 mRNA levels at 8.5 weeks post infection were not significantly different. Six animals were infected at two sites each with wild type CRPV, E7/14-E6/15 CRPV and E7/18-E6/15 CRPV. Wild type E7, E7/14, and E7/18 were all expressed at about the same level at 8.5 weeks post infection. Wild type E6 and E6/15 levels were the same at this time point. Thus codon optimization did not influence mRNA levels. E7 mRNA levels were statistically higher than E6 mRNA levels. P<0.0007, Student’s t-test.
M-Fold plots of codon-modified genes showed increased mRNA stabilities in all cases
mRNA stability has been associated with gene expression(Wei, Xiao et al., 2009;Edwards, Hing, Perry, Blaisdell, Kopelman, Fathke, Plum, Newell, Allen, S G, Shapiro, Okunji, Kosti, Shomron, Grigoryan, Przytycka, Sauna, Salari, Mandel-Gutfreund, Komar, & Kimchi-Sarfaty, 2012). We wanted to test whether the codon changes made to the E6 and E7 oncogenes would be predicted to alter mRNA stabilities. The Zuker M-Fold program(Zuker, 2003) was used to fold each full length E7 gene and the 5’ portions of the E6 genes in which the codon alterations were made. Table 4 shows optimal free energies for each mRNA. Each of the codon modified mRNAs with the exception of E7/18 was predicted to have enhanced stability (lower dG) over that of the wild type mRNA. These results show that mRNA stability could be a factor in the findings of this study. Note that in our earlier paper(Cladel, Hu, Balogh, & Christensen, 2008b) we showed increased stability of the codon-modified E7 mRNA in vitro.
Table 4.
E6 synonymous changes and resultant changes in frequency per thousand in rabbit. Synonymous changes were made to a total of nine different amino acids in E6 to convert codon usage from rare, as used in rabbit, to common, as used in rabbit. The codon changes and resultant frequency/1000 codons are shown for both E6/15 and E6/17.
| E6/15 | Transition | Frequency of virus codon/1000 in rabbit relative to frequency of modified codon/1000 in rabbit. |
|---|---|---|
| Ser | TCG>TCC | 5.7/19.4 |
| Gln | CAA>CAG | 9.2/33.0 |
| *Leu | TTA > CTA | 5.3/4.9 |
| Ile | ATA > ATC | 6.1/29.7 |
| Leu | TTG > CTG | 10.9/48.8 |
| Gly | GGT > GGC | 8.8/26.7 |
| Ile | ATA > ATC | 6.1/29.7 |
| Leu | TTG > CTG | 10.9/48.9 |
| Leu | TTG > CTG | 10.9/48.9 |
| Ala | GCA>GCC | 12.7/34.2 |
| Thr | ACG>ACC | 9.1 ,22.0 |
| Leu | CTA > CTG | 4.9/48.9 |
| Ile | ATT > ATC | 14.3 29.7 |
| Val | GTA > GTG | 4.8/33.3 |
| Pro | CCG > CCC | 8.7/20.8 |
| E6/17 | ||
| Leu | CTA > CTG | 4.9 / 48.9 |
| *Leu | CTA > CTG | 4.9/48.9 |
| Leu | TTG > CTG | 10.9/48.9 |
Not actually an Optimization but designed to be an intermediate to an optimized codon in E6/17.
The result of two nucleotide changes. The fust, in E6/15 above, is neutral.
Differences in proliferation markers were detected in a time course study comparing papillomas from wild type and E7/14-E6/15 infections
Eight rabbits were infected at three left sites with wild type CRPV and at three right sites with E7/14-E6/15. Measurements of papillomas were taken weekly. At weeks 5, 10, 15 and 20 post infection, two rabbits were sacrificed and sections of all papillomas were prepared. H and E analysis and CRPV E7, MCM7, and PHH3 staining were done on all sections except the time point 1(week 5) sections that were lost due to processor malfunction.
The steady state levels of E7 are low in most papillomas and have been challenging to detect immunohistochemically. We were able to detect the protein using our single monoclonal antibody to CRPV E7 by using antigen retrieval technology and found the protein to localize to the nucleus (Figure 6). We were not able, however, to confirm quantitative differences in protein production.
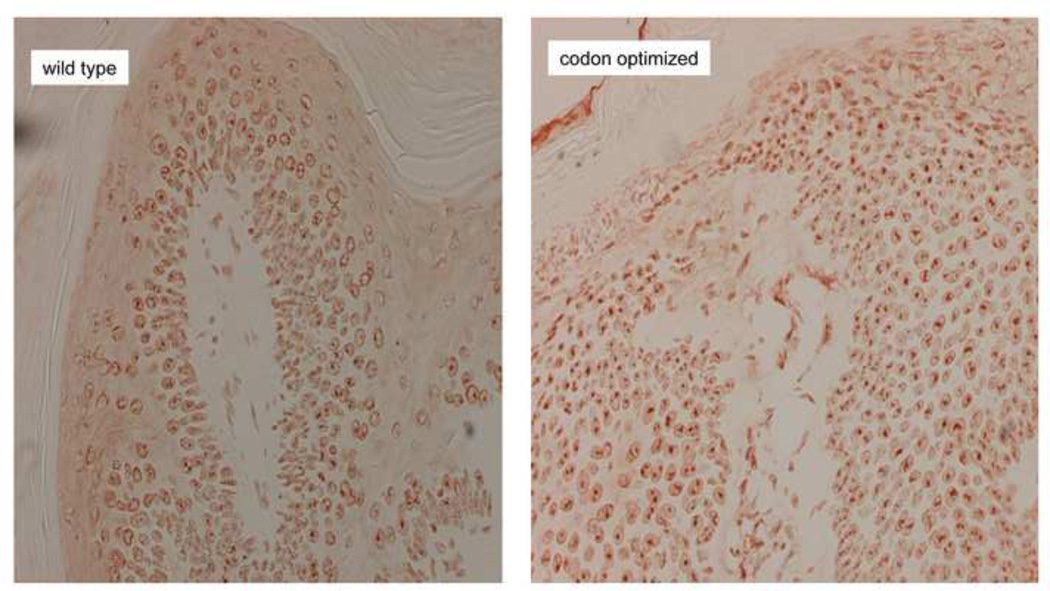
Fig. 6.
CRPV E7 could be detected in the nucleus. Papillomas from wild type and codon-optimized genomes were stained for CRPV E7 using an in house antibody raised to the E7 protein. E7 localized to the nucleus and appeared to be more pronounced in the codon-optimized sections. However, efforts to quantitate the differences were equivocal.
MCM7 is a commonly-accepted surrogate marker for E7 (Middleton, Peh et al., 2003). We therefore stained the time course sections for MCM7 protein. Over the five month period of the study, the intensities of the staining varied. We found that at time point 2(10 weeks), codon-modified sections stained more strongly; at time point 3(15 weeks), wild type staining had increased and codon-modified staining had decreased; by time point 4 (20 weeks) staining for both wild type and codon modified papillomas was lower than at the earlier time points. See Figure 7.
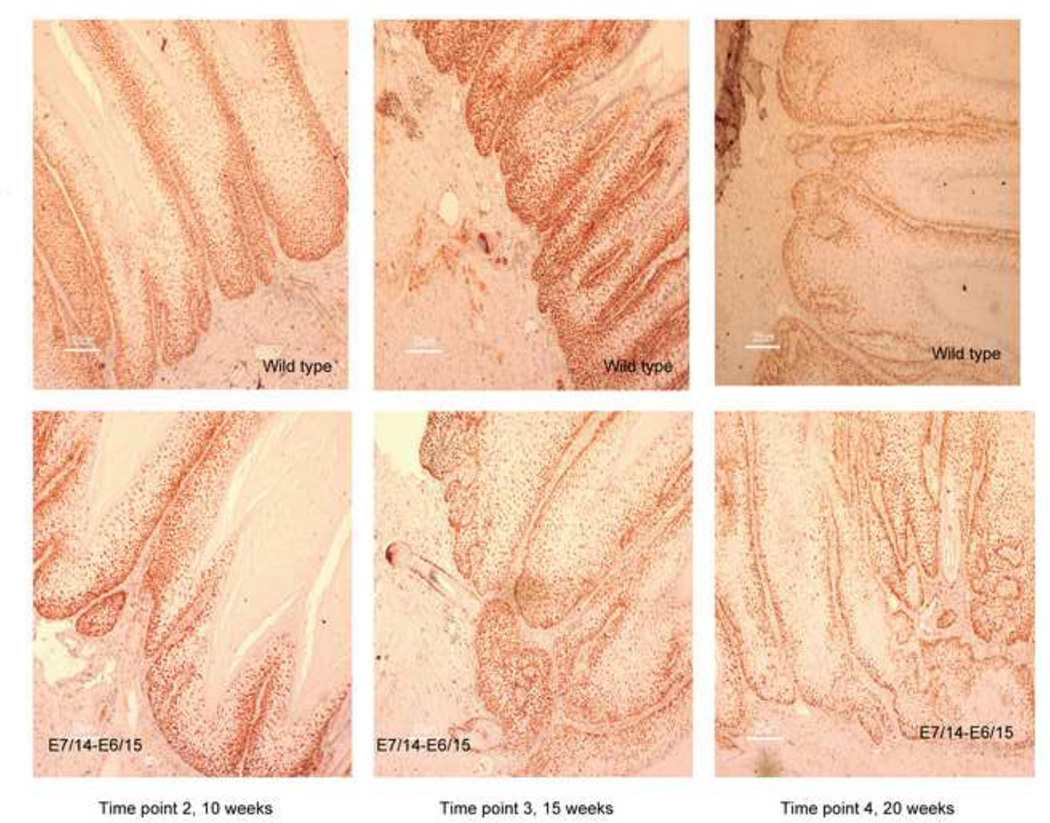
Fig. 7.
MCM7 staining patterns cchanged over time. A) At time point 2, staining of papillomas from E7/14-E6/15 genomes was stronger than wild type. B) At time point 3, the reverse was true and there was a reduction in staining of E7/14-E6/15 papillomas relative to time point 2. C) By time point 4, staining was reduced in papillomas from both wild type and E7/14-E6/15 genomes.
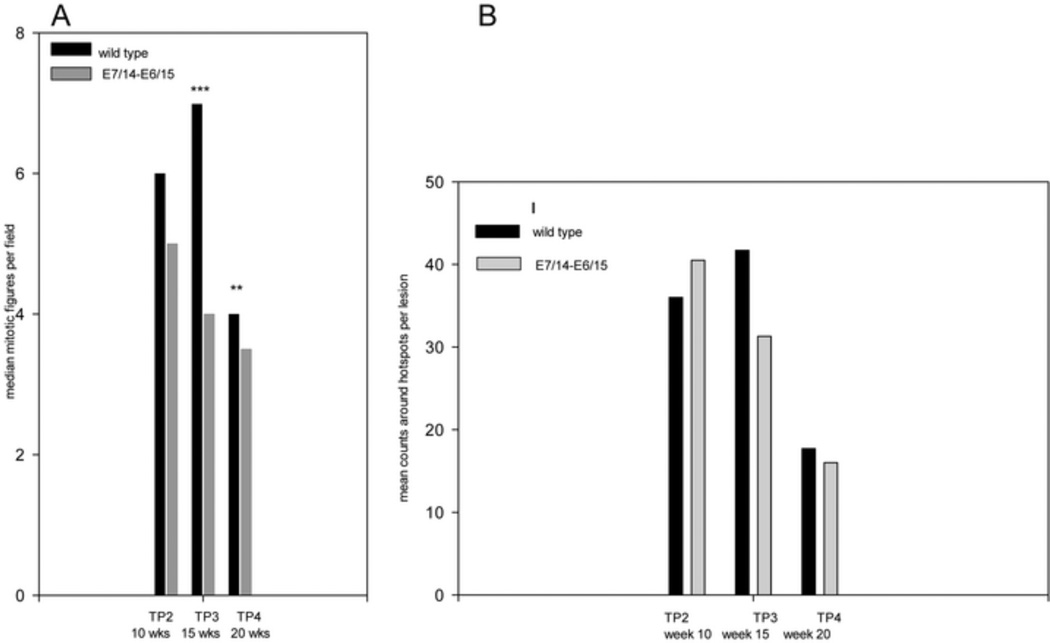
PhosphohistoneH3 (PHH3) is specifically expressed during mitosis but not at other points in the cell cycle(Juan, Traganos et al., 1998;Crosio, Fimia et al., 2002). We stained all sections with Rabbit anti-human PHH3 (Cell Signaling cat# 9701). We expected to see increased expression of protein in the codon-optimized papillomas based on their enhanced malignant potential. Instead, we detected a tendency for lower pHH3 positivity in the codon-optimized samples versus the wild type papillomas except at time point 2. In addition, in synchrony with the MCM7 observations, we observed decreased expression over time for both wild type and codon-modified papillomas. In this study, we quantitated the PHH3-positive cells in individual sections from time points 2, 3 and 4 (weeks 10, 15 and 20 respectively) using two different methods---hot spot analysis(Perry, Stafford et al., 1997) and counts per high powered field (Coupland, Campbell et al., 2008). Absolute numbers differed but with both computational methods, the trends were similar. The data for counts per high powered field are summarized in table 3 and graphed in figure 8. Figure 8 also includes a graph of the hot spot analysis for comparison. Using counts per high power field ,we found that codon-modified papillomas contained statistically fewer mitotic cells at time points 3( P<0.001 ) and 4( P=0.02) (Mann-whitney rank sum test) relative to wild type. In addition, over time there was a statistically significant reduction in counts in both codon-modified and wild type papillomas.
Table 3.
Statistical differences in PHH3 counts per high powered field between wild type (L) and E7/14-E6/15(R) papillomas at time points 2, 3, and 4 were noted, “n” represents the number of fields counted per site. Where statistical differences were noted for papillomas from the two genomes, there were always fewer mitoses in the codon-modified papillomas relative to wild type. Where significant differences were noted for the same genome at different time points, there were always lower counts at later time points.
| 2L | 2R | 3L | 3R | 4L | 4R | |
|---|---|---|---|---|---|---|
| 2 L(n=83) | ----- | P=0.56 | P=0.01 | ------ | P=0.01 | ------ |
| 2R(n=148) | P=0.56 | ------ | ------ | P<0.001 | ------ | P<0.001 |
| 3L(n=l04) | P=0.01 | ------ | ------ | P<0.001 | P<0.001 | ------ |
| 3R(n=137) | ------ | P<0.001 | P<0.001 | ------ | ------ | P=0.63 |
| 4L(n=96) | P=0.01 | ------ | P<0.001 | ------ | ------ | P=0.02 |
| 4R(n=114) | ------ | P<0.001 | ------------ | P=0.63 | P=0.02 | ------ |
Fig. 8.
PHH3 staining showed significantly fewer mitotic figures in sections of papillomas from the E7/14-E6/15 genome than from the wild type genome at time points 3 and 4. Left) Six sections from each time point for papillomas from both wild type CRPV and E7/14-E6/15 were examined for mitotic figures. Figures were counted per 20X high powered field(Coupland, Campbell, and Damato 1778–85) across the entire sections. Data were entered into Sigmaplot and significance was determined by the Mann-Whitney Rank Sum test. Table 5 shows statistics. Right) Mitotic figures were counted using Hot Spot analysis(Perry et al. 1455–65). In both cases, mitotic figures declined over time and were lower in papillomas from the codon-optimized genome for time points 3 and 4.
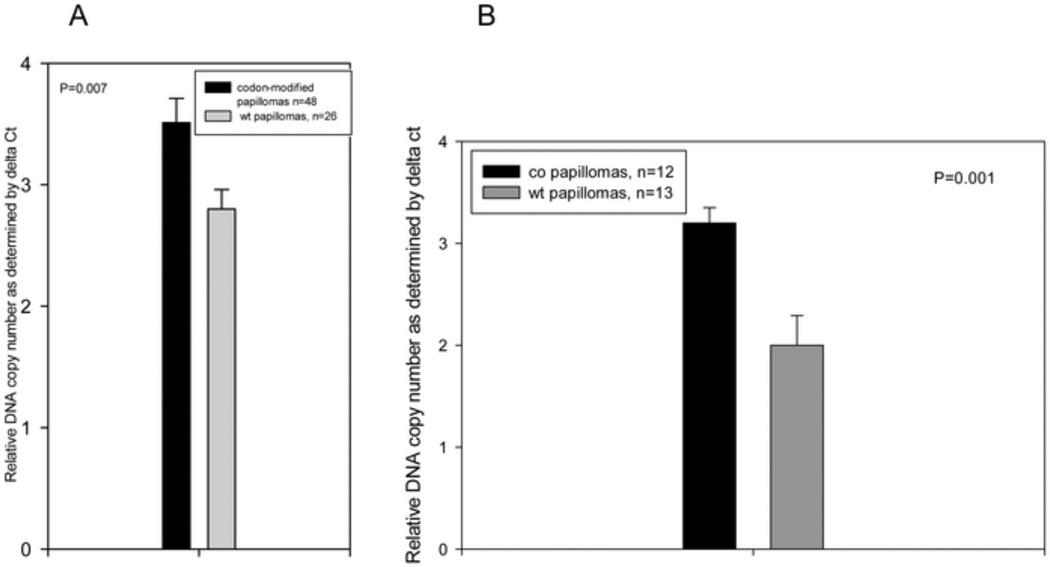
Papillomas generated from codon-optimized genomes contained a higher copy number of viral genomes
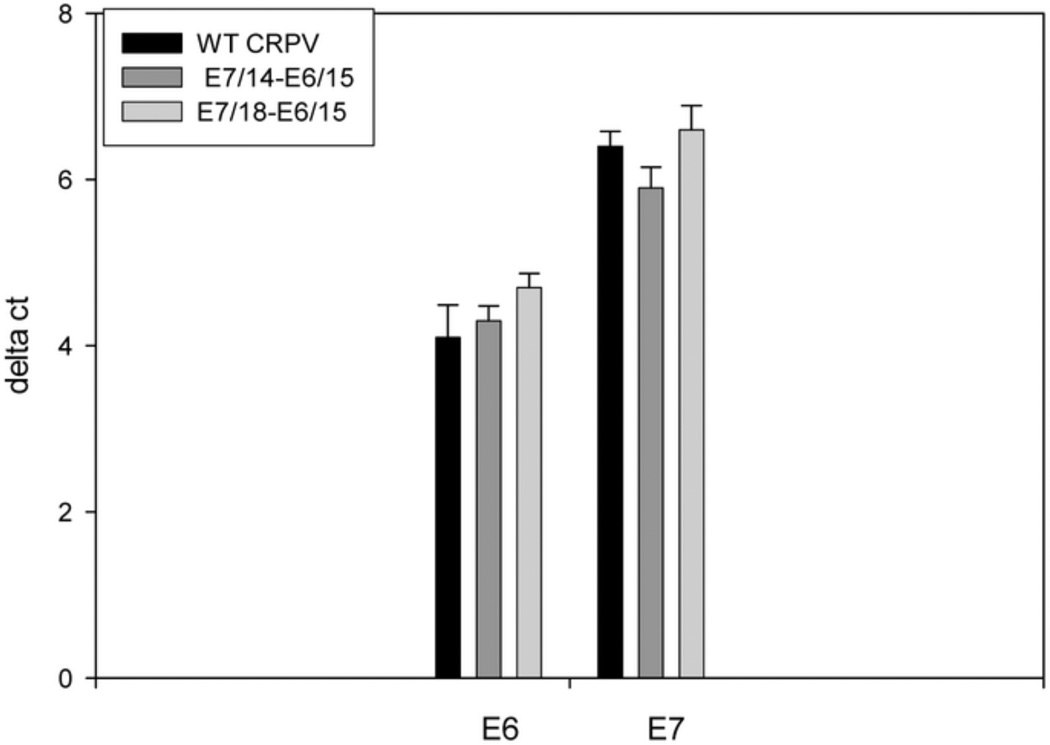
Quantitative PCR was used to determine relative viral DNA copy numbers in wild type and codon-optimized papillomas. Papillomas were excised at the time of sacrifice of the animals and most were between 6 and 10 months post infection. Wild type and codon-modified papillomas from the same animals were investigated in the same Q PCR reactions. In a comparison of 26 wild type and 44 codon-optimized papillomas, copy number was higher in the papillomas from codon-optimized genomes (P=0.007, unpaired Student’s t- test). Similar results (Figure 9) were obtained when copy numbers of wild type (n=15) and E7/14-E6/15 (n=22) genomes from the time course study were compared (P=0.001).
Fig. 9.
DNA copy number was higher in papillomas from codon-optimized genomes relative to wild type genome. A) DNA copy number was determined by Q PCR for wild type papillomas (n=26) and papillomas induced by codon-optimized genomes (n=48). Ribosomal RNA was used as internal control. Delta Ct values were entered into SigmaPlot and statistical significance was computed by Student’s t-test. P=0.007. B) The same analysis was done for DNAs from the time course study. P=0.001.
Dysplastic changes were noted histologically at very early time points in papillomas generated by codon-optimized genomes
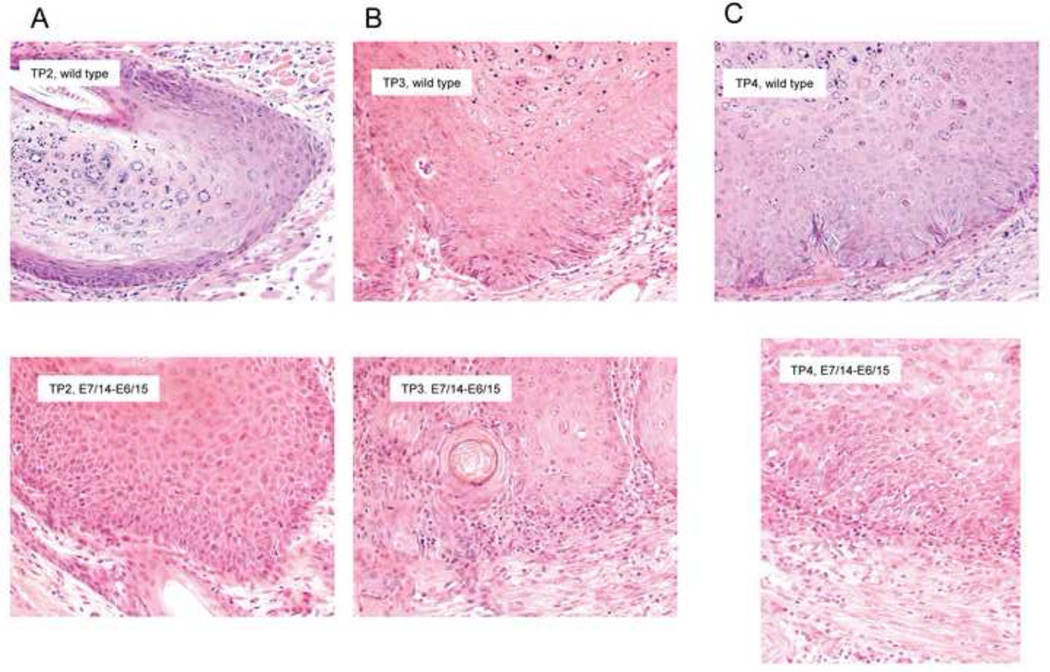
H and E sections from all of the papillomas from the time point study referenced above were prepared. As noted, in this experiment eight animals were infected with wild type DNA at three sites on the left back and with E7/14-E6/15 at three sites on the right back. At intervals of 4–5 weeks starting at week 5, two of the animals were sacrificed and the papillomas were excised. Evaluation of the sections showed rapid development of dysplasia in the papillomas; the dysplasia was most marked in papillomas from codon-modified genomes. Figure 10 shows representative wild type and codon-modified papillomas at time points 2, 3 and 4.
Fig. 10.
H and E analysis of papillomas at different time points revealed increased dysplasia in papillomas from the E7/14-E6/15 genome as well as more pronounced immune infiltrates. H and E sections were prepared from papillomas from the time course study. Representative sections from time points 2, 3 and 4 are shown. At each time point, the papillomas from codon-modified genomes were more disordered than the corresponding wild type papillomas as evidenced by disruption of the basement membrane and invasion into the dermis. Heavy immune infiltrates were also seen in the papillomas from E7/14-E6/15 at time points 3 and 4.
DISCUSSION
In this study, we have expanded on our earlier work(Cladel, Hu, Balogh, & Christensen, 2008b) to develop a robust animal model for the exploration of synonymous codon usage. This model was designed for two purposes: 1) to accelerate the appearance of phenotypic characteristics of the CRPV infection, thereby allowing for more expedient study of viral pathology and 2) to provide an in vivo mechanism for evaluating the findings on synonymous codon usage arising from the many in silico analyses currently being conducted. We demonstrate that it is possible to accomplish both goals simultaneously with this model and argue that the model will be a useful tool both for investigators interested in basic papillomavirus biology and for those wishing to test hypotheses of synonymous codon usage.
To carry out this work, we generated a panel of papillomavirus genomes with different numbers of synonymous codon “optimizations” in the oncogenes E6 and E7 and tested them in our rabbit model (Cladel, Hu et al., 2008a). We followed the papillomas over time for many months and observed altered growth rates relative to wild type, increased immunogenicity, as detected by complete or partial regressions, and, most strikingly, shortened times to and increasing numbers of cancers. These data affirmed the growing recognition that synonymous codons are not silent (Sauna & Kimchi-Sarfaty, 2011; Venetianer, 2012;Katsnelson, 2011;Edwards, Hing, Perry, Blaisdell, Kopelman, Fathke, Plum, Newell, Allen, S G, Shapiro, Okunji, Kosti, Shomron, Grigoryan, Przytycka, Sauna, Salari, Mandel-Gutfreund, Komar, & Kimchi-Sarfaty, 2012;Chen, Davydov, Sirota, & Butte, 2010).
We isolated the RNA from lesions in an experiment especially designed to compare RNAs generated from wild type and two codon-modified genomes. The animals were sacrificed at week 8.5, a time point at which the papillomas have reached a significant size but have not yet progressed to cancer. Interestingly, we did not see differences between wild type and codon-modified E6 or E7 mRNA levels. This finding was in contrast to our earlier in vitro studies that showed large increases in message with changes in codon usage(Cladel, Hu, Balogh, & Christensen, 2008b) and demonstrates the importance of an in vivo model to elucidate biologically meaningful outcomes.
Using an in vitro analysis, we showed in our earlier study, that codon-optimized E7 RNAs were more stable than wild type RNA. There are a very few assays to test RNA stability in vivo and they have been developed for specific circumstances(Hilgers, Pourquie et al., 2005;Lee, Bradley et al., 1998); we have not developed such an assay for the CRPV viral oncogene RNAs. Therefore, we used the Zuker M-Fold program, version 3.5, (Zuker, 2003) to check predicted stabilities of wild type and codon-modified genes, both E6 and E7. mRNAs from all codon-modified genes were predicted to be more stable than wild type with the exception of E7/18, which showed little change. We hypothesize that the codon changes may have conferred enhanced stability in vivo while at the same time influencing degradation such that steady state levels of mRNA did not appear to change(as observed in our Q RT PCR studies). For a review of mRNA stability and degradation, see Knapinska et al(Knapinska, Irizarry-Barreto et al., 2005). It is unclear how these factors would come together to affect the translation of the oncogenes.
Increased stability of mRNA is one mechanism that might be associated with more oncoprotein, as noted above. It is also possible that the codon changes may have influenced translation rate (Gingold & Pilpel, 2011) which, in turn, might have led to more properly folded protein. Synonymous codon usage has been associated with altered protein folding in numerous recent studies(Drummond & Wilke, 2008)
(Warnecke & Hurst, 2010;Zhou, Weems et al., 2009).
We analyzed E7 protein content using an in-house developed monoclonal antibody. Since we were looking for quantitative differences in protein production and since these differences were likely to be small, we chose to examine the E7 protein in histological sections rather than in papilloma homogenates. We found that the oncogene localized to the nucleus but we were not able to convincingly quantitate differences in E7 protein between the wild type and codon-optimized papillomas.
Our data show that cancers are much more common in the codon-optimized lesions and we hypothesized that proliferation markers would be more pronounced in these lesions. MCM7 staining was stronger in papillomas generated by codon-modified genome E7/14-E6/15 at the early time point but not at later time points. PHH3 positivity was less in papillomas from this genome for both time points 3 and 4. We know that cancer and the processes associated with its inception and progression are highly complex phenomena that are influenced by multiple factors. In a recent review (Hanahan & Weinberg, 2011), the authors note that excessive proliferation can trigger cellular senescence with resultant loss of proliferation markers. Eventually, selected cells may adapt to the stresses they are experiencing and proceed to renewed proliferation. These cells will have a competitive advantage and may progress to malignancy. Support for this sequence of events in breast cancer was recently published(Jerby, Wolf et al., 2012). Corroborating work on melanoma has also been reported(Hoek, Eichhoff et al., 2008). Davy and Doorbar in a 2007 review ((Davy & Doorbar, 2007)) report G2/M arrest induced by E6 and E7 expression in papillomavirus infections. We hypothesize that in the experiments reported here, time point 4 represents a period during which the cells are accommodating to the abundant stresses induced by earlier proliferation and that this leads to G2/M arrest and accounts for the reduction in proliferative signal seen with both MCM7 and PHH3 biomarkers. This period of relative senescence appears to be reached more quickly by the papillomas from codon-modified genomes.
We examined viral DNA copy number in our lesions and found statistically higher copy numbers of DNA in papillomas from codon-modified genomes relative to wild type. Davy and Doorbar in their review, () note that papillomaviruses are able to initiate a pseudo S phase to increase viral DNA synthesis following G2/M arrest.
Codon-modified papillomas showed a greater tendency for regression or reduced growth in this study. We have published several studies on regression rates of rabbit papillomas in the past(Hu, Cladel, Pickel, & Christensen, 2002;Wilgenburg, Budgeon et al., 2005;Okabayashi, Angell et al., 1993) and have shown that the process is associated with the infiltration of lymphocytes. Similar observations were also shown by Selvakumar et al (Selvakumar, Schmitt et al., 1997). For a review of the immunology of papillomaviruses see(Nicholls & Stanley, 2000). The wild type strain of CRPV in regular use in our laboratory rarely results in regressions. We hypothesize that the regression seen in papillomas initiated by codon-optimized DNAs is triggered by larger amounts of oncoproteins being produced in the lesions and subsequently targeted by the immune system. The genome that exhibited the greatest tendency to regression was the one we tested with the greatest number of codon-modifications, E7/22-E6/17. The differential response of the 11 animals in this experiment was undoubtedly due to the MHC differences in the outbred animals, differences that are not grossly obvious with wild type CRPV. .Note that very early cancers were seen in some of the progressing lesions from this genome. This evidence supports the concept of an interplay between malignant progression and immunological response. Grivennikov et al(Grivennikov, Greten et al., 2010) in a recent review on the connections between immunity, inflammation and cancer note that an in vivo model to study the influences of immunity and inflammation on cancer development is needed. Our model could serve this function.
Analysis of H and E sections from the time point experiment showed more abnormal pathology in the papillomas from codon-modified genomes at a given time point. Figure 10A–C are representative. Many of these sections showed high levels of lymphocytic infiltrates. Grivennikov et al (Grivennikov, Greten, & Karin, 2010) note that inflammation always accompanies cancer and that both tumor promoting inflammation and tumor controlling immunosurveillance can occur in the same lesion. This observation can help to explain the regressions and reductions in growth observed in this study as well as the behavior of the two papillomas that regressed and then returned to quickly become malignant.
The CRPV/rabbit model has considerable potential for the study of the significance of synonymous codon usage. The library of codon-modified CRPV genomes that we have created represents a small subset of the potential genomes possible. By altering the codons in targeted ways, it will be possible to test in vivo the observations accruing from in silico analyses. There are few other models available to conduct such studies. Carlini et. al successfully deoptimized leucines in the alcohol dehydrogenase (Adh) gene of Drosophila and showed decreasing enzymatic function with increasing numbers of altered codons. They concluded that the reduction in function was due to reduced translation of the protein and that each deoptimized codon contributed to a 2.13% decline in protein production as measured by reduction in enzymatic activity(Carlini, 2004). Recent work from the same group has shown that “optimizations” of the same leucines resulted in increased enzymatic activity in larvae but not in adult flies(Hense, Anderson et al., 2010). This study thus links stage of development as well as alterations in translation with codon usage. Several studies have investigated synonymous codon changes in the capsid protein of poliovirus. For example, Mueller et al(Mueller, Papamichail, Coleman, Skiena, & Wimmer, 2006) showed that codon deoptimization resulted in greatly attenuated virus and that the attenuation could be attributed to a modest reduction in rate of translation. This paper demonstrated that profound phenotypic differences could result from small changes in protein production. The work of Mueller et al was followed by a study by Coleman et al (Coleman, Papamichail et al., 2008) in the same laboratory. In this case, synonymous changes were made to codon pairs and similar observations were reported. A recent report from another laboratory in which CpG and TpA dinucleotides were increased in the capsid coding region also demonstrated viral attenuation with only a modest reduction in translation(Burns, Shaw, Campagnoli, Jorba, Vincent, Quay, & Kew, 2006).
The CRPV/rabbit model has distinct advantages to study the impact of synonymous codon changes in situ. The data presented in this paper demonstrate its utility. Among the attributes of our model that make it especially attractive for these studies are the following:
CRPV DNA is infectious. Modifications can be made to the small (about 8Kb) genome by common laboratory techniques and quickly tested in our well-established infection assay (Cladel, Hu, Balogh, Mejia, & Christensen, 2008a).
Because the genome is small, more extensive modifications can be made by direct synthesis of the codon-modified genome or part thereof.
Since this project was initiated, the rabbit genome has been sequenced (NCBI, Oryctolagus cuniculua, bio project PRJNA42933, PRJNA12819) and significant numbers of reagents are now available to investigators. The rabbit genome is closer to that of the human than is that of the mouse and many antibodies raised to human proteins cross-react with rabbit counterparts. We have made use of a number of these reagents in this study.
The CRPV genome demonstrates considerable plasticity as evidenced by work in our laboratory (Hu, Cladel, Balogh, Budgeon, & Christensen, 2006). Thus it can be expected to tolerate a wide range of synonymous codon changes as, indeed, we have shown with the genomes generated for this study. Phenotypic changes in outcome will help in the evaluation of the significance of the codon changes. Measurements as basic as size of the papillomas relative to those generated by wild type DNA will reveal much about the significance of the changes made.
The rabbit is a mammal with an immune system similar to that of humans and a propensity for some of the same types of infections. In this paper we have shown that codon changes in the CRPV oncogenes can influence both immunogenicity and oncogenicity of the virus. These observations pave the way for the study of the interplay between the two in a mammalian system akin to that of the human. To our knowledge, there is no better model available.
To date we have made selective changes to the E6 and E7 genes. However, changes to the other CRPV genes could also be revealing from both virological and codon usage perspectives. That is to say, basic properties of codon usage can be studied in this system and, as they are revealed, can be expected to shed light on properties of the virus itself. It is helpful that codon usage of the virus is very different from that of the host. This will allow for a wide latitude of changes. The need for a system that can couple the investigation of theories of codon usage with a model that provides a biologically meaningful readout has been articulated in the review of Plotkin et al (Plotkin & Kudla, 2011). The CRPV/rabbit model could serve this function well.
MATERIALS AND METHODS
Preparation of codon-altered genomes
The Hershey wild type cottontail rabbit papillomavirus (Hershey CRPV, wt H. CRPV, Genbank accession number JF303889) was used as the backbone for all constructs. The genome was cloned into PUC 19 at Sal1. The EcoR1 site was removed from the vector and a Cla1 site at 1383 just downstream of the E1 start and a Sac II site at 174 downstream of the E6 start were introduced into the CRPV genome by site-directed mutagenesis. Naturally occurring EcoR1 (1063) was used in conjunction with Cla1 to facilitate E7 replacements and EcoR1 and Sacll were used to facilitate E6 replacements. The genome with the engineered Sacll and Cla1 restriction sites was wild type in its growth characteristics and has been used as a backbone in many of our previous studies. Synonymous codon changes were introduced into E6 and E7 subclones by site-directed mutagenesis using the polymerase Pfu Turbo (Stratagene, LaJolla, CA) and a modification of the company’s Quick Change protocol as delineated in (Cladel, Hu, Balogh, & Christensen, 2008b). All products were sequenced in the core facility of the PA State University College of Medicine to verify the changes and to assure that additional alterations had not been introduced; the genes were then cloned into the wild type backbone in place of the wild type genes. Mutations in E6 are shown in Figure 1. Alterations in frequency of codon usage that resulted from the modifications are shown in Table 4 Mutations in E7 were reported in(Cladel, Hu, Balogh, & Christensen, 2008b) and are also shown in Supplementary Figure 1.
Codon changes were based on the work of Zhou et al(Zhou, Liu, Peng, Sun, & Frazer, 1999) and were done at random with the primary goal of changing a viral codon rare in mammals to one most common in mammals. Thus, even though the amino acids were chosen at random, those that would result in the most significant codon change were given priority. E6/15 changes involved 9 different amino acids. Leucine was most commonly altered both because of its frequency in the 5’ portion of the E6 gene and because the change from rare to common was most notable for this amino acid. This scenario was also true for E7 (data not shown). Each new construct contained all of the mutations from the previous one in addition to the new changes. Thus E7/14 contained all of the codon changes in E7/8 plus six more. E7/18 contained all of those in E7/14 plus four more and E7/22 contained all of the changes in E7/18 plus four additional changes. Since the time that this work was begun, enhanced sequence information has become available, including partial data for rabbit. Based on this information (http://www.kazusa.or.jp/codon/), Table 5 has been constructed showing relative codon usages for all amino acids for human, cow, rabbit and CRPV. These data do not differ markedly from the earlier data used by Zhou (Zhou, Liu, Peng, Sun, & Frazer, 1999) and support our decision to use those data as a foundation for our work. Chi Square analysis using the Stat Calc program freely available on the web (http://www.statpac.com/statistics-calculator/free-version.htm) was used to compare rabbit and CRPV codon usages and to determine that they are very different (P<0.00000000 for most amino acids).
Table 5.
Comparison of codon usage for mammals human, cow, and rabbit and for CRPV. Recent data from the Kazusa data base (http://www.kazusa.or.jp/codon/) were used to compile codon usage for three mammals, human, cow and rabbit, and for CRPV. Codon usage for the mammals is similar. However, codon usage between rabbit (host) and CRPV was computed to be very different for many of the codons. (StatCalc program (http://www.statpac.com/statistics-calculator/free-version.htm).
| Amino acid | codon | Human | Cow | Rabbit | CRPV |
|---|---|---|---|---|---|
| Arg | CGU | 4 5 | 4.6 | 3.7 | 3.3 |
| CGU | 10.4 | 11.1 | I3.0 | 8.2 | |
| CGA | 6.2 | 6.4 | 5.0 | 4.3 | |
| CGG | 11.4 | 12.5 | 11.4 | 4.3 | |
| AGA | 12.2 | 10.7 | 9.2 | 25.2 | |
| AGG | 12.0 | 11.4 | 10.6 | 12.7 | |
| Leu | UUA | 7.7 | 6.3 | 5.3 | 15.6 |
| UUG | 12.9 | 12.0 | 10.9 | 16.1 | |
| CUU | 13.2 | 11.9 | 10.1 | 18.1 | |
| cuC | 19.6 | 21.2 | 23.6 | 9.4 | |
| CUA | 7.2 | 6.1 | 4.9 | 16.4 | |
| CUG | 39.6 | 43.5 | 48.9 | 23.5 | |
| Ser | UCU | 15.2 | 13 1 | 10.4 | 15.1 |
| UCC | 17.7 | 17.3 | 19.4 | 8.2 | |
| UCA | 12.2 | 9.9 | 7.7 | 15.3 | |
| UCG | 4.4 | 5.0 | 5.7 | 3.7 | |
| AGU | 12.1 | 11.0 | 8.5 | 17.6 | |
| AGC | 19.5 | I9.3 | I9.3 | 17.3 | |
| Thr | ACU | 13.1 | 11.5 | 9.9 | 15.3 |
| ACC | 18.9 | 20.1 | 22.0 | 8.8 | |
| ACA | 15.1 | 13.0 | 11 7 | 27.2 | |
| ACG | 6.1 | 7.2 | 9 1 | 7.6 | |
| Pro | CCU | 17.5 | 15.8 | 12 6 | 16.6 |
| ccc | 19.8 | 20.4 | 20.8 | 15.0 | |
| CCA | 16.9 | 14.6 | 11.8 | 19.2 | |
| CCG | 6.9 | 7.8 | 8.7 | 13.2 | |
| Ala | GCU | 18.4 | 17.9 | 15.5 | 18.9 |
| GCC | 27.7 | 30.5 | 34.2 | 8 9 | |
| GCA | I5.8 | 14 3 | 12.7 | 25.1 | |
| GCG | 7.4 | 8.6 | 9.5 | 6.5 | |
| Gly | GGU | 10.8 | 10.8 | 8.8 | 16.7 |
| GGC | 22.2 | 24.4 | 26.7 | 10.8 | |
| GGA | 16.5 | 16 2 | 14.7 | 17.9 | |
| GGC | 16.5 | 16.8 | 17.0 | 22.5 | |
| Val | GUU | 11.0 | 10.1 | 8.7 | 15.1 |
| GUC | 14.5 | 15.9 | 18.0 | 13.5 | |
| GUA | 7.1 | 6.3 | 4.8 | 9.1 | |
| GUG | 28.1 | 30.8 | 33.3 | 20.4 | |
| Lys | AAA | 24.4 | 22.4 | 20.2 | 24.3 |
| AAG | 31.9 | 34.7 | 35.1 | 23.0 | |
| Asn | AAU | 17.0 | 14.7 | 13.4 | 16.8 |
| AAC | 19.1 | 21 4 | 24.2 | 18 7 | |
| Glu | CAA | 12.3 | 10 5 | 9.2 | 199 |
| CAG | 34.2 | 35 0 | 33.0 | 27.5 | |
| His | CAU | 10.9 | 9.4 | 7.3 | 11.2 |
| CAC | 15.1 | 15 5 | 16 0 | 6.9 | |
| Glu | GAA | 29.0 | 26.9 | 24.2 | 34.9 |
| GAG | 39.6 | 41.9 | 43.7 | 27.8 | |
| Asp | GAU | 21.8 | 20 5 | 17.6 | 34 3 |
| GAC | 23.l | 28.2 | 30.5 | 28.9 | |
| Tyr | UAU | 12.2 | 11.4 | 10.0 | 22.6 |
| UAC | 15.3 | 17.5 | 20.0 | 10.1 | |
| Cys | UGU | 10.6 | 9.3 | 8.2 | 17.9 |
| UGC | 12.6 | I2.6 | 13.6 | 19.2 | |
| Pbe | UUU | 17.6 | 16.4 | 16.4 | 25.8 |
| AUC | 20.3 | 22 3 | 28.4 | 14.5 | |
| Ile | AUU | 16.0 | 14.6 | 14.3 | 15.8 |
| AUC | 20.8 | 23.3 | 29.7 | 5.8 | |
| AUA | 7.5 | 6.7 | 6.1 | 16.3 |
FREQUENCY PER THOUSAND
DNA challenge and monitoring of tumors
The DNA constructs were prepared using the Qiagen maxiprep system (www.qiagen.com) and were purified by cesium chloride density ultracentrifugation and adjusted to 200µg/ml in 1X TE buffer for challenge on animals. New Zealand White (NZW) rabbits were maintained in the animal facility of the Pennsylvania State University College of Medicine. Most were outbred animals obtained from Robinson (Mocksville, NC); six were donated rabbits obtained from Millbrook, MA. The studies were approved by the Institutional Animal Care and Use Committee of the Pennsylvania State University (IACUC protocol #5, 97–202). For application of viral DNA, rabbits were sedated using ketamine/xylazine anesthesia. Back skin of the animals was scarified with a scalpel blade to create an abrasion. Three days later, again under anesthesia, the wounded sites were lightly scratched with a scalpel blade to introduce nicks into the scabs. Each site was then challenged with 5µgDNA in 50µl of 1×TE buffer and the DNA was scratched into the wound using a 21G needle(Cladel, Hu, Balogh, Mejia, & Christensen, 2008a). Monitoring of papilloma outgrowth began two weeks after infection and continued until time of sacrifice of the animals. In general, each animal was infected with wild type H. CRPV DNA at two sites and with experimental genomes at 8 sites for a total of 10 sites, five on each side of the back. In some experiments, the experimental genomes were the same and in others, they were mixed at different sites on the same animal. This allowed us to compare the behavior of different genomes on the same animal in some cases as well as to study individual genomes in isolation in other cases.
Immunohistochemical analysis of tumors
MCM7 (also known as cdc 47) staining was done on paraffin embedded, formalin-fixed tissues using NeoMarkers monoclonal antibody # MS-862-PO. Citrate buffer antigen retrieval at 90 degrees C was used before incubating the tissues at 1:150 dilution of the primary antibody for four hours. Slides were thoroughly rinsed in PBS followed by antibody detection using an HRP polymer conjugate (Zymed 87–9163) for 45 minutes. Amino ethyl carbazole (AEC) chromogen was applied for 20 minutes and the slides were washed and mounted in Crystal Mount.
For E7 staining, paraffin sections were dewaxed and rehydrated in water and washed in PBS. They were treated with 3% hydrogen peroxide for six minutes and washed well with three changes of PBS. The sections were incubated for 15 minutes in 4mg/ml pepsin dissolved in 0.1N HCI at 37– 45 degrees C. They were then washed with at least three changes of PBS. 3% BSA in PBS was applied for 30 minutes and then drained off without washing. The E7 MAb at 1:100 was applied for one hour and the slides were then washed with PBS. Super Picture Polymer Detection Agent (Invitrogen # 87–9163) was applied for 30 minutes and the slides were washed well with PBS. AEC chromogen was applied for 30 minutes. Slides were rinsed in water, allowed to air dry and coverslipped in aqueous media.
For PHH3 staining, five micron paraffin sections were dewaxed and rehydrated in distilled water. Antigen retrieval was performed using a Citrate Buffer pH 6.0 at 95°C for 12 minutes. Slides were allowed to cool in this solution for 30 minutes before washing in PBS. Non- specific binding was blocked by a 20 minute incubation in 3% BSA in PBS. Rabbit anti-human PHH3 (Cell Signaling cat# 9701) was diluted to 1:250 and allowed to incubate at RT for 2 hours. Primary antibody binding was detected using anti-Rabbit peroxidase conjugated secondary antibody (Vector ImmPRESS cat# MP-7401) for 40 minutes. Complexes were visualized with Nova Red substrate kit (Vector cat# SK4800). Slides were dehydrated through graded ethanols and coverslipped from xylene in a permanent mounting medium.
PHH3 quantitation
PHH3 was quantitated by two methods . In the first method, multiple fields from each section were scanned under a Nikon Eclipse (E600) microscope and the positive cells were counted and average total counts per 20X field were calculated (Coupland, Campbell, & Damato, 2008). Data for each animal were compared using the Mann-Whitney Rank Sum Test for uneven distributions.
In the second method, the “hot spots” and the fields adjacent to them were quantitated as per the protocol of(Perry, Stafford, Scheithauer, Suman, & Lohse, 1997). The Student’s t-test was used to compare counts for wild type and codon-modified papillomas.
Statistics
Papilloma size was determined by calculating the cubic root of the product of length × width × height of individual papillomas in millimeters to obtain a geometric mean diameter (GMD). Data were represented as the means (+ SEMs) of the GMDs for each test group. The significance of percentage of cancer sites/tumor sites induced by different constructs was determined by the Fisher’s exact test (P<0.05 was considered significant). Student’s t- test was used to determine significance of Q RT PCR and Q PCR results.
Fisher’s exact test was used to determine significance of the cancer and regression data.
Quantitative RTPCR analysis of total RNA isolated from papillomas
RNAs were isolated from flash frozen papilloma tissue using the TRIzol reagent (www.lnvitrogen.com). 10µg RNA was treated with Turbo DNA-free (product # AM 1907, www.invitrogen.com) to remove any contaminating DNA. RNAs were retrieved in a standard volume of 50 microliters. Reverse transcription and quantitative PCR were performed on 200ng of the RNA using Stratagene’s Brilliant II QRT-PCR one-step kit (# 600809). RNAs used in the study had 260/280 ratios of 1.9–2.0 as well as intact ribosomal bands as determined by agarose gel electrophoresis. Primers and probe were designed using Primer Express software© TATA-binding protein (TBP) amplicons, internal standards for the analyses, were generated using primers 5’ CACGGCACTGATTTTCAGTTCT 3’ (nt 627–648) and 5’TTCTTGCTGCCAGTCTGGACT 3’ (nt706–686) and used at final concentrations of 200nM. Probe for TBP was fluorogenic TaqMan probe 5’HEX TGTGCACAGGAGCCAAGAGTGAAGA BHQ-1 3’ and was used at a final concentration of 100nM. CRPV E7/14, 18 and 22 primers were 5’AAGCGCTGTAGGCAGACCA 3’ (16–35) and 5’TCGATTCAAGGTTCTGATGGC 3’ (84–105) and were used at a final concentration of 300nM. Probe was flurogenic TaqMan probe 5’FAM CAGCTTCGTCTGCGTCTGTGATCCA-BHQ-1 3’ (36–61) and was used at a final concentration of 100nM. Wild type CRPV E7 primers were 5’ CCATGTAAGCGCTGTAGGCAAA 3’ (10–32) and 5’GCAGTCGATTCAAGGTTCTTATGGC 3’ (85–109) and were used at a final concentration of 300nM. Probe was 5’FAM TCAGCTTCGTCTGCGTCTGTGATCC BHQ-1 3’ (35–60) and was used at a final concentration of 100nM. E6 primers for all genomes were 5’ GGGTCATTGCAGTTCTTG 3’ (233–250) and 5’ GGACGCAACACTCGTCACAT 3’ (314–333). These were used at a final concentration of 300nM. E6 probe was flurogenic TaqMan probe 5’FAM TCCCGGCAGTCGCTGCGG-BHQ −1 3’ used at a concentration of 100nM. Primers and probes for the E6 and E7 genes were synthesized by Biosearch Technologies (www.biosearchtech.com). Primers and probes were optimized prior to the running of E6/E7 analyses. A standard curve was run on each plate using 1:2 dilutions of one of the samples. This enabled efficiencies to be determined. All qRT-PCR reactions were performed using the Mx-3005P instrument (Agilent Technologies). Cycling conditions were 50°C for 30 min (reverse transcription) followed by 42 cycles of 94°C for 15 sec and 60°C for one min. Analysis was done using the delta delta Ct method (Pfaffl, 2001). Student’s t test was used to determine significance.
Compilation of Cancer data
The data in this study were collected over a period of several years. All animals in use in our laboratory and infected with wild type and /or codon-modified genomes were carefully monitored for papilloma growth and cancer development. Data were pooled to generate the cancer information. Cancer was first detected visually and then confirmed histologically.
Compilation of Regression data
The data in this study were collected over a period of several years. All animals in use in our laboratory and infected with wild type and /or codon-modified genomes were carefully monitored for papilloma growth. In those cases in which papillomas regressed or partially regressed, notations were made in the charts and regressions were followed. Data were pooled to generate the regression data.
Detection ofE6 and E7 sequences in papilloma DNA
DNAs were isolated from tissues using the Qiagen DNeasy system (product # 69506, www.qiagen.com). E6 and E7 genes were amplified by PCR and the products were sequenced in the core facility at the Pennsylvania State University College of Medicine.
Determination of relative copy numbers of viral DNA in papillomas
DNA was isolated from papilloma fragments using the DNeasy kit (www.Qiagen.com) and quantitated spectrophotometrically. 200 ng was subjected to quantitative PCR using the Sybr green kit #600828 from Agilent Technologies. For the amplification of a portion of CRPV E6, primers (5’GGA CGC AAC ACT CGT CAC AT) and (5’ GGG TCA TTG CAG TTC TTG CA) were used. As an internal control, 18S RNA was amplified in separate reactions on the same plate using primers (5’ CGC CGC TAG AGG TGA AAT TC) and (5’TTG GCA AAT GCT TTC GCT C). Primers were used at 300nM concentrations in each case after optimization of conditions. Standard curves were run on each plate for both E6 and 18S DNAs. Efficiencies were in the range of 90–110%.
The differences between the Cts for E6 for a given sample and 18S for the same sample were used to determine relative differences in DNA concentration. Each delta Ct represents a two-fold difference in concentration. Delta Cts for both wild type and codon-modified DNAs were entered into SigmaPlot version 11.0 and Student’s t -test was used to analyze results.
Analysis of RNA stabilities by mfold
Each wild type and codon-modified E6 and E7 gene was folded using the Zuker mfold program (version 3.5) freely available on the web (http://www.bioinfo.rpi.edu/applications/mfold). The program yields a number of possible folds listed in order of most stable (lowest free energy) to least stable. The most stable fold was chosen for each sequence. Results are shown in Table 2.
Table 2.
Gibb’s free energy (dG, kcal /mole) as computed by mfold for wild type and codon optimized oncogenes. The Zuker mfold program (http://www.bioinfo.rpi.edu/applications/mfold), version 3.5, was used to fold mRNAs of the mutated portion of E6 5’ of the E8 ATG and the entire E7. All optimized genes, with the exception of E7/18, were predicted to have higher stabilities (lower free energies) than the corresponding wild type gene.
| GENOME | dG, kcal/mole, calculated by mfold version 3.5 |
|---|---|
| E6, WILD TYPE, 5’ of E8 start | −29.96 |
| E6/15 5’of E8 start | −47.66 |
| E6/17 5’of E8 start | −57.70 |
| E7, WILD TYPE | −66.88 |
| E7/14 | −75.21 |
| E7/18 | −65.04 |
| E7/22 | −73.8 |
| E7/40 | −90.15 |
Supplementary Material
Highlights.
-
►
Papillomaviruses use rare codons with respect to the host.
-
►
Codons of the E6 and E7 oncogenes of CRPV were altered to be more mammalian-like.
-
►
Codon-modified genomes yielded more and earlier cancers.
-
►
Immunogenicity also increased.
-
►
The CRPV model can be used to study theories relating to synonymous codon usage.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Akashi H. Translational selection and yeast proteome evolution. Genetics. 2003;164:1291–1303. doi: 10.1093/genetics/164.4.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragones L, Guix S, Ribes E, Bosch A, Pinto RM. Fine-tuning translation kinetics selection as the driving force of codon usage bias in the hepatitis a virus capsid. PLoS Pathog. 2010;6:e1000797. doi: 10.1371/journal.ppat.1000797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa MS, Wettstein FO. The two proteins encoded by the cottontail rabbit papillomavirus E6 open reading frame differ with respect to localization and phosphorylation 2. J. Virol. 1988;62:1088–1092. doi: 10.1128/jvi.62.3.1088-1092.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bible JM, Mant C, Best JM, Kell B, Starkey WG, Raju KS, Seed P, Biswas C, Muir P, Banatvala JE, Cason J. Cervical lesions ave associated with human papillomavirus type 16 intratypic variants that have high transcriptional activity and increased usage of common mammalian codons 1. Journal of General Virology. 2000;81:1517–1527. doi: 10.1099/0022-1317-81-6-1517. [DOI] [PubMed] [Google Scholar]
- Burns CC, Shaw J, Campagnoli R, Jorba J, Vincent A, Quay J, Kew O. Modulation of poliovirus replicative fitness in HeLa cells by deoptimization of synonymous codon usage in the capsid region 1. Journal of Virology. 2006;80:3259–3272. doi: 10.1128/JVI.80.7.3259-3272.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capon F, Allen MH, Ameen M, Burden AD, Tillman D, Barker JN, Trembath RC. A synonymous SNP of the corneodesmosin gene leads to increased mRNA stability and demonstrates association with psoriasis across diverse ethnic groups. Human Molecular Genetics. 2004;13:2361–2368. doi: 10.1093/hmg/ddh273. [DOI] [PubMed] [Google Scholar]
- Carlini DB. Experimental reduction of codon bias in the Drosophila alcohol dehydrogenase gene results in decreased ethanol tolerance of adult flies. Journal of Evolutionary Biology. 2004;17:779–785. doi: 10.1111/j.1420-9101.2004.00725.x. [DOI] [PubMed] [Google Scholar]
- Chamary JV, Hurst LD. Evidence for selection on synonymous mutations affecting stability of mRNA secondary structure in mammals 1. Genome Biology. 2005;6 doi: 10.1186/gb-2005-6-9-r75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamary JV, Parmley JL, Hurst LD. Hearing silence: non-neutral evolution at synonymous sites in mammals 1. Nat.Rev.Genet. 2006;7:98–108. doi: 10.1038/nrg1770. [DOI] [PubMed] [Google Scholar]
- Chen R, Davydov EV, Sirota M, Butte AJ. Non-synonymous and synonymous coding SNPs show similar likelihood and effect size of human disease association. PLoS One. 2010;5:e13574. doi: 10.1371/journal.pone.0013574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cladel NM, Bertotto A, Christensen ND. Human alpha and beta papillomaviruses use different synonymous codon profiles. Virus Genes. 2010 doi: 10.1007/s11262-010-0451-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cladel NM, Hu J, Balogh K, Mejia A, Christensen ND. Wounding prior to challenge substantially improves infectivity of cottontail rabbit papillomavirus and allows for standardization of infection 1. J Virol.Methods. 2008a;148:34–39. doi: 10.1016/j.jviromet.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cladel NM, Hu J, Balogh KK, Christensen ND. CRPV genomes with synonymous codon optimizations in the CRPV E7 gene show phenotypic differences in growth and altered immunity upon E7 vaccination 1. PLoS ONE. 2008b;3:e2947. doi: 10.1371/journal.pone.0002947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman JR, Papamichail D, Skiena S, Futcher B, Wimmer E, Mueller S. Virus attenuation by genome-scale changes in codon pair bias. Science. 2008;320:1784–1787. doi: 10.1126/science.1155761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortazzo P, Cervenansky C, Marin M, Reiss C, Ehrlich R, Deana A. Silent mutations affect in vivo protein folding in Escherichia coli. Biochemical and Biophysical Research Communications. 2002;293:537–541. doi: 10.1016/S0006-291X(02)00226-7. [DOI] [PubMed] [Google Scholar]
- Coupland SE, Campbell I, Damato B. Routes of extraocular extension of uveal melanoma: risk factors and influence on survival probability. Ophthalmology. 2008;115:1778–1785. doi: 10.1016/j.ophtha.2008.04.025. [DOI] [PubMed] [Google Scholar]
- Crosio C, Fimia GM, Loury R, Kimura M, Okano Y, Zhou H, Sen S, Allis CD, Sassone-Corsi P. Mitotic phosphorylation of histone H3: spatio-temporal regulation by mammalian Aurora kinases. Mol.Cell Biol. 2002;22:874–885. doi: 10.1128/MCB.22.3.874-885.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davy C, Doorbar J. G2/M cell cycle arrest in the life cycle of viruses. Virology. 2007;368:219–226. doi: 10.1016/j.virol.2007.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J, Doorbar J, Li B, Zhou F, Gu W, Zhao L, Saunders NA, Frazer IH, Zhao KN. Expression of papillomavirus L1 proteins regulated by authentic gene codon usage is favoured in G2/M-like cells in differentiating keratinocytes. Virology. 2010;399:46–58. doi: 10.1016/j.virol.2009.12.021. [DOI] [PubMed] [Google Scholar]
- Doorbar J. Molecular biology of human papillomavirus infection and cervical cancer. Clinical Science. 2006;110:525–541. doi: 10.1042/CS20050369. [DOI] [PubMed] [Google Scholar]
- Drummond DA, Wilke CO. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution 1. Cell. 2008;134:341–352. doi: 10.1016/j.cell.2008.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan JB, Wainwright MS, Comeron JM, Saitou N, Sanders AR, Gelernter J, Gejman PV. Synonymous mutations in the human dopamine receptor D2 (DRD2) affect mRNA stability and synthesis of the receptor. Human Molecular Genetics. 2003;12:205–216. doi: 10.1093/hmg/ddg055. [DOI] [PubMed] [Google Scholar]
- Edwards NC, Hing ZA, Perry A, Blaisdell A, Kopelman DB, Fathke R, Plum W, Newell J, Allen CE, S G, Shapiro A, Okunji C, Kosti I, Shomron N, Grigoryan V, Przytycka TM, Sauna ZE, Salari R, Mandel-Gutfreund Y, Komar AA, Kimchi-Sarfaty C. Characterization of coding synonymous and non-synonymous variants in ADAMTS13 using ex vivo and in silico approaches. PLoS One. 2012;7:e38864. doi: 10.1371/journal.pone.0038864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairbrother WG, Holste D, Burge CB, Sharp PA. Single nucleotide polymorphism-based validation of exonic splicing enhancers 1. Plos Biology. 2004;2:1388–1395. doi: 10.1371/journal.pbio.0020268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganzenmueller T, Matthaei M, Muench P, Scheible M, Iftner A, Hiller T, Leiprecht N, Probst S, Stubenrauch F, Iftner T. The E7 protein of the cottontail rabbit papillomavirus immortalizes normal rabbit keratinocytes and reduces pRb levels, while E6 cooperates in immortalization but neither degrades p53 nor binds E6AP. Virology. 2008;372:313–324. doi: 10.1016/j.virol.2007.11.006. [DOI] [PubMed] [Google Scholar]
- Gao F, Li Y, Decker JM, Peyerl FW, Bibollet-Ruche F, Rodenburg CM, Chen Y, Shaw DR, Allen S, Musonda R, Shaw GM, Zajac AJ, Letvin N, Hahn BH. Codon usage optimization of HIV type 1 subtype C gag, pol, env, and nef genes: in vitro expression and immune responses in DNA-vaccinated mice 1. AIDS Res.Hum.Retroviruses. 2003;19:817–823. doi: 10.1089/088922203769232610. [DOI] [PubMed] [Google Scholar]
- Gingold H, Pilpel Y. Determinants of translation efficiency and accuracy. Molecular Systems Biology. 2011;7 doi: 10.1038/msb.2011.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Ding J, Wang X, de Kluyver RL, Saunders NA, Frazer IH, Zhao KN. Generalized substitution of isoencoding codons shortens the duration of papillomavirus L1 protein expression in transiently gene-transfected keratinocytes due to cell differentiation. Nucleic Acids Res. 2007;35:4820–4832. doi: 10.1093/nar/gkm496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Li M, Zhao WM, Fang NX, Bu S, Frazer IH, Zhao KN. tRNASer(CGA) differentially regulates expression of wild-type and codon-modified papillomavirus L1 genes. Nucleic Acids Res. 2004;32:4448–4461. doi: 10.1093/nar/gkh748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamano T, Matsuo K, Hibi Y, Victoriano AFB, Takahashi N, Mabuchi Y, Soji T, Irie S, Sawanpanyalert P, Yanai H, Hara T, Yamazaki S, Yamamoto N, Okamoto T. A single-nucleotide synonymous mutation in the gag gene controlling human immunodeficiency virus type 1 virion production. Journal of Virology. 2007;81:1528–1533. doi: 10.1128/JVI.01596-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han RC, Cladel NM, Reed CA, Peng XW, Christensen ND. Protection of rabbits from viral challenge by gene gun-based intracutaneous vaccination with a combination of cottontail rabbit papillomavirus E1, E2, E6, and E7 genes. Journal of Virology. 1999;73:7039–7043. doi: 10.1128/jvi.73.8.7039-7043.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hense W, Anderson N, Hutter S, Stephan W, Parsch J, Carlini DB. Experimentally increased codon bias in the Drosophila adh gene leads to an increase in larval, but not adult, alcohol dehydrogenase activity. Genetics. 2010;184:547–555. doi: 10.1534/genetics.109.111294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgers V, Pourquie O, Dubrulle J. In vivo analysis of mRNA stability using the Tet-Off system in the chicken embryo. Dev.Biol. 2005;284:292–300. doi: 10.1016/j.ydbio.2005.05.021. [DOI] [PubMed] [Google Scholar]
- Hoek KS, Eichhoff OM, Schlegel NC, Dobbeling U, Kobert N, Schaerer L, Hemmi S, Dummer R. In vivo switching of human melanoma cells between proliferative and invasive states. Cancer Res. 2008;68:650–656. doi: 10.1158/0008-5472.CAN-07-2491. [DOI] [PubMed] [Google Scholar]
- Hu J, Cladel NM, Balogh K, Budgeon L, Christensen ND. Impact of genetic changes to the CRPV genome and their application to the study of pathogenesis in vivo. Virology. 2006 doi: 10.1016/j.virol.2006.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Cladel NM, Pickel MD, Christensen ND. Amino acid residues in the carboxy-terminal region of cottontail rabbit papillomavirus E6 influence spontaneous regression of cutaneous papillomas. J Virol. 2002;76:11801–11808. doi: 10.1128/JVI.76.23.11801-11808.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Peng X, Cladel NM, Pickel MD, Christensen ND. Large cutaneous rabbit papillomas that persist during cyclosporin A treatment can regress spontaneously after cessation of immunosuppression. J.Gen.Virol. 2005;86:55–63. doi: 10.1099/vir.0.80448-0. [DOI] [PubMed] [Google Scholar]
- Jerby L, Wolf L, Denkert C, Stein GY, Hilvo M, Oresic M, Geiger T, Ruppin E. Metabolic associations of reduced proliferation and oxidative stress in advanced breast cancer. Cancer Res. 2012 doi: 10.1158/0008-5472.CAN-12-2215. [DOI] [PubMed] [Google Scholar]
- Juan G, Traganos F, James WM, Ray JM, Roberge M, Sauve DM, Anderson H, Darzynkiewicz Z. Histone H3 phosphorylation and expression of cyclins A and B1 measured in individual cells during their progression through G2 and mitosis. Cytometry. 1998;32:71–77. doi: 10.1002/(sici)1097-0320(19980601)32:2<71::aid-cyto1>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Kallel I, Rebai M, Khabir A, Farid NR, Rebai A. Genetic Polymorphisms in the EGFR (R521K) and Estrogen Receptor (T594T) Genes, EGFR and ErbB-2 Protein Expression, and Breast Cancer Risk in Tunisia. Journal of Biomedicine and Biotechnology. 2009 doi: 10.1155/2009/753683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsnelson A. Breaking the silence. Nat.Med. 2011;17:1536–1538. doi: 10.1038/nm1211-1536. [DOI] [PubMed] [Google Scholar]
- Kimchi-Sarfaty C, Oh JM, Kim IW, Sauna ZE, Calcagno AM, Ambudkar SV, Gottesman MM. A "silent" polymorphism in the MDR1 gene changes substrate specificity 1. Science. 2007;315:525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- Knapinska AM, Irizarry-Barreto P, Adusumalli S, Androulakis L, Brewer G. Molecular mechanisms regulating mRNA stability: Physiological and pathological significance. Current Genomics. 2005;6:471–486. [Google Scholar]
- Lavner Y, Kotlar D. Codon bias as a factor in regulating expression via translation rate in the human genome 1. Gene. 2005;345:127–138. doi: 10.1016/j.gene.2004.11.035. [DOI] [PubMed] [Google Scholar]
- Lee CH, Bradley G, Ling V. Increased P-glycoprotein messenger RNA stability in rat liver tumors in vivo. J.Cell Physiol. 1998;177:1–12. doi: 10.1002/(SICI)1097-4652(199810)177:1<1::AID-JCP1>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Lemm I, Ross J. Regulation of c-myc mRNA decay by translational pausing in a coding region instability determinant. Molecular and Cellular Biology. 2002;22:3959–3969. doi: 10.1128/MCB.22.12.3959-3969.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechold U, Gilbert C, Ogryzko V. Codon optimization of the BirA enzyme gene leads to higher expression and an improved efficiency of biotinylation of target proteins in mammalian cells. J Biotechnol. 2005;116:245–249. doi: 10.1016/j.jbiotec.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Middleton K, Peh W, Southern S, Griffin H, Sotlar K, Nakahara T, El Sherif A, Morris L, Seth R, Hibma M, Jenkins D, Lambert P, Coleman N, Doorbar J. Organization of human papillomavirus productive cycle during neoplastic progression provides a basis for selection of diagnostic markers. Journal of Virology. 2003;77:10186–10201. doi: 10.1128/JVI.77.19.10186-10201.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody CA, Laimins LA. Human papillomavirus oncoproteins: pathways to transformation. Nature Reviews Cancer. 2010;10:550–560. doi: 10.1038/nrc2886. [DOI] [PubMed] [Google Scholar]
- Mossadegh N, Gissmann L, Muller M, Zentgraf H, Alonso A, Tomakidi P. Codon optimization of the human papillomavirus 11 (HPV 11) L1 gene leads to increased gene expression and formation of virus-like particles in mammalian epithelial cells 1. Virology. 2004;326:57–66. doi: 10.1016/j.virol.2004.04.050. [DOI] [PubMed] [Google Scholar]
- Mueller S, Papamichail D, Coleman JR, Skiena S, Wimmer E. Reduction of the rate of poliovirus protein synthesis through large-scale codon deoptimization causes attenuation of viral virulence by lowering specific infectivity. Journal of Virology. 2006;80:9687–9696. doi: 10.1128/JVI.00738-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nackley AG, Shabalina SA, Tchivileva IE, Satterfield K, Korchynskyi O, Makarov SS, Maixner W, Diatchenko L. Human catechol-O-methyltransferase haplotypes modulate protein expression by altering mRNA secondary structure 1. Science. 2006;314:1930–1933. doi: 10.1126/science.1131262. [DOI] [PubMed] [Google Scholar]
- Nicholls PK, Stanley MA. The immunology of animal papillomaviruses. Vet lmmunol.Jmmunopathol. 2000;73:101–127. doi: 10.1016/s0165-2427(99)00165-8. [DOI] [PubMed] [Google Scholar]
- Okabayashi M, Angell MG, Budgeon LR, Kreider JW. Shope Papilloma Cell and Leukocyte Proliferation in Regressing and Progressing Lesions. American Journal of Pathology. 1993;142:489–496. [PMC free article] [PubMed] [Google Scholar]
- Parmley JL, Hurst LD. Exonic splicing regulatory elements Skew synonymous codon usage near intron-exon boundaries in mammals. Molecular Biology and Evolution. 2007;24:1600–1603. doi: 10.1093/molbev/msm104. [DOI] [PubMed] [Google Scholar]
- Perry Arie, Stafford Scott L, Scheithauer Bernd W, Suman Vera J, Lohse Cjhristine M. Meningioma Grading: An Analysis of Histologic Parameters. American Journal of Surgical Pathology. 1997;21(12):1455–1465. doi: 10.1097/00000478-199712000-00008. Ref Type: Generic. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotkin JB, Kudla G. Synonymous but not the same: the causes and consequences of codon bias. Nat.Rev.Genet. 2011;12:32–42. doi: 10.1038/nrg2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon J, Ramoz N, Cassonnet P, Orth G, Breitburd F. A cottontail rabbit papillomavirus strain (CRPVb) with strikingly divergent E6 and E7 oncoproteins: An insight in the evolution of papillomaviruses. Virology. 1997;235:228–234. doi: 10.1006/viro.1997.8680. [DOI] [PubMed] [Google Scholar]
- Sauna ZE, Kimchi-Sarfaty C. Understanding the contribution of synonymous mutations to human disease. Nat.Rev.Genet. 2011;12:683–691. doi: 10.1038/nrg3051. [DOI] [PubMed] [Google Scholar]
- Segal E, Fondufe-Mittendorf Y, Chen LY, Thastrom A, Field Y, Moore IK, Wang JPZ, Widom J. A genomic code for nucleosome positioning. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvakumar R, Schmitt A, Iftner T, Ahmed R, Wettstein FO. Regression of papillomas induced by cottontail rabbit papillomavirus is associated with infiltration of CD8+ cells and persistence of viral DNA after regression. Journal of Virology. 1997;71:5540–5548. doi: 10.1128/jvi.71.7.5540-5548.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM, Cowe E. Synonymous Codon Usage in Saccharomyces-Cerevisiae. Yeast. 1991;7:657–678. doi: 10.1002/yea.320070702. [DOI] [PubMed] [Google Scholar]
- Tindle RW. Immune evasion in human papillomavirus-associated cervical cancer 1. Nat.Rev.Cancer. 2002;2:59–65. doi: 10.1038/nrc700. [DOI] [PubMed] [Google Scholar]
- Venetianer Pal. Are synonymous codons indeed synonymous? Biomolecular Concepts. 2012;3(1):21–28. doi: 10.1515/bmc.2011.050. Ref Type: Generic. [DOI] [PubMed] [Google Scholar]
- Warnecke T, Hurst LD. GroEL dependency affects codon usage–support for a critical role of misfolding in gene evolution. Mol.Syst.Biol. 2010;6:340. doi: 10.1038/msb.2009.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Xiao Q, Zhang T, Mou Z, You J, Ma WJ. Differential regulation of mRNA stability controls the transient expression of genes encoding Drosophila antimicrobial peptide with distinct immune response characteristics. Nucleic Acids Res. 2009;37:6550–6561. doi: 10.1093/nar/gkp693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilgenburg BJ, Budgeon LR, Lang CM, Griffith JW, Christensen ND. Characterization of immune responses during regression of rabbit oral papillomavirus infections 1. Comparative Medicine. 2005;55:431–439. [PubMed] [Google Scholar]
- Zhao KN, Gu W, Fang NX, Saunders NA, Frazer IH. Gene codon composition determines differentiation-dependent expression of a viral capsid gene in keratinocytes in vitro and in vivo. Mol.Cell Biol. 2005;25:8643–8655. doi: 10.1128/MCB.25.19.8643-8655.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao KN, Liu WJ, Frazer IH. Codon usage bias and A+T content variation in human papillomavirus genomes. Virus Res. 2003;98:95–104. doi: 10.1016/j.virusres.2003.08.019. [DOI] [PubMed] [Google Scholar]
- Zheng ZM, Wang X. Regulation of cellular miRNA expression by human papillomaviruses 1. Biochim Biophys Acta. 2011;1809:668–677. doi: 10.1016/j.bbagrm.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Liu WJ, Peng SW, Sun XY, Frazer I. Papillomavirus capsid protein expression level depends on the match between codon usage and tRNA availability. Journal of Virology. 1999;73:4972–4982. doi: 10.1128/jvi.73.6.4972-4982.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Weems M, Wilke CO. Translationally optimal codons associate with structurally sensitive sites in proteins 1. Mol.Biol.Evol. 2009;26:1571–1580. doi: 10.1093/molbev/msp070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Research. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.