Abstract
The microbial populations responsible for anaerobic degradation of phthalate isomers were investigated by enrichment and isolation of those microbes from anaerobic sludge treating wastewater from the manufacturing of terephthalic acid. Primary enrichments were made with each of three phthalate isomers (ortho-, iso-, and terephthalate) as the sole energy source at 37°C with two sources of anaerobic sludge (both had been used to treat wastewater containing high concentrations of phthalate isomers) as the inoculum. Six methanogenic enrichment cultures were obtained which not only degraded the isomer used for the enrichment but also had the potential to degrade part of other phthalate isomers as well as benzoate with concomitant production of methane, presumably involving strictly syntrophic substrate degradation. Our 16S rRNA gene-cloning analysis combined with fluorescence in situ hybridization revealed that the predominant bacteria in the enrichment cultures were affiliated with a recently recognized non-sulfate-reducing subcluster (subcluster Ih) in the group ‘Desulfotomaculum lineage I' or a clone cluster (group TA) in the class delta-Proteobacteria. Several attempts were made to isolate these microbes, resulting in the isolation of a terephthalate-degrading bacterium, designated strain JT, in pure culture. A coculture of the strain with the hydrogenotrophic methanogen Methanospirillum hungatei converted terephthalate to acetate and methane within 3 months of incubation, whereas strain JT could not degrade terephthalate in pure culture. During the degradation of terephthalate, a small amount of benzoate was transiently accumulated as an intermediate, indicative of decarboxylation of terephthalate to benzoate as the initial step of the degradation. 16S rRNA gene sequence analysis revealed that the strain was a member of subcluster Ih of the group ‘Desulfotomaculum lineage I', but it was only distantly related to other known species.
To date, anaerobic (methanogenic) fermentation technology has been widely applied for the treatment of municipal and industrial wastes and wastewaters (2, 18). A number of anaerobic processes have been intensively developed over the past decades (31), and applications of these processes are now expanding to low-strength wastewaters (19), to wastes and wastewaters under extreme temperature conditions (16, 19, 35), and to more complex wastewaters containing anthropogenic compounds and/or compounds recalcitrant to biodegradation (22). Wastewaters with high concentrations of phthalate isomers (ortho-, meta-, and para-benzene dicarboxylic acid) are one of the complex wastewaters now being challenged by anaerobic processes. Phthalate isomers, which are primarily anthropogenic compounds, have been produced in massive amounts for use in manufacturing polyester resins, plastic bottles, plasticizers, polyester fibers, and other petroleum-based products in the world and are consequently eluted in the wastewater generated by the corresponding industries (25). From the economic and energetic aspects, anaerobic (methanogenic) processes have increasingly been introduced to treat those wastewaters, and as a consequence, more than 10 full-scale anaerobic bioreactors are currently in operation or under construction for the treatment of phthalate isomer-containing wastewaters (J. V. Duffel, presentation at the National Conference on Anaerobic Treatment of Complex Wastewaters, Breda, The Netherlands, 1993; H. Macarie and O. Monroy, presented at Journées Industrielles sur la Digestion Anaérobie, Narbonne, France, 1996; J. H. F. Pereboom, D. G. Man, and I. T. Su, presentation at the 7th International Symposium on Anaerobic Digestion, Cape Town, South Africa, 1994). However, all laboratory-, pilot-, and full-scale engineering studies have demonstrated that the processes require a long start-up time (long lag phase) for the removal of phthalate isomers and that the processes often stagnate during treatment (13, 20; Duffel, presentation, 1993). In particular, the long period of starting up, ranging from 1 to 3 months in batch studies (14, 15) to more than 1 year in full-scale reactors (Duffel, presentation, 1993; Pereboom et al., presentation, 1994), has been the most serious obstacle for further application and development of anaerobic technology for such wastewaters. Since these phenomena are attributed to the stability of the microbial populations responsible for phthalate isomer degradation under methanogenic conditions, attention has been paid to the phthalate isomer-degrading populations in those processes.
Under methanogenic conditions, phthalate degradation is thought to proceed by syntrophic association between different trophic groups of anaerobes. The current view is that the association may contain at least three groups of microbes, (i) phthalate-degrading, hydrogen (and/or formate)-producing, fermentative bacteria that form acetate, hydrogen (and/or formate), and carbon dioxide as end products, (ii) hydrogenotrophic methanogens, which scavenge hydrogen, and (iii) aceticlastic methanogens, which consume acetate. Since the reaction performed by the phthalate-degrading bacteria is energetically unfavorable under standard conditions, the presence of the last two microbial groups is thought to be essential to make the whole reaction energetically feasible. Due to these traits of syntrophic substrate-degrading bacteria, the isolation of such microbes has been considered difficult, and therefore, despite their importance in methanogenic environments, only limited members of such microbial groups have been successfully isolated and characterized so far. To date, only a few enrichment cultures that mineralize phthalate isomers under methanogenic conditions have been reported. Kleerebezem et al. recently reported three methanogenic consortia decomposing ortho-phthalate, isophthalate, or terephthalate (14). The physiological properties of the enrichment cultures, in particular the kinetics of degradation of phthalate isomers, were well described, but the cultures were not yet defined, and hence the phylogenetic positions of the microbial populations in the enrichment cultures were unknown.
In this paper, we report the enrichment, isolation, and partial characterization of phthalate isomer-degrading microbes under methanogenic conditions.
MATERIALS AND METHODS
Sources of methanogenic granules.
Anaerobic granular sludges were taken from two types (sludges I and II) of mesophilic (35°C) upflow anaerobic sludge blanket (UASB) reactors, both of which had been treating wastewater generated by the manufacturing of terephthalic acid. Sludge I was from a two-phase laboratory-scale system composed of an anaerobic fluidized bed reactor (pretreatment reactor; volume, 5,000 liters) and a UASB reactor (posttreatment reactor; volume, 5,000 liters) constructed in Taiwan. The first anaerobic fluidized bed reactor was used for the removal of organic substances which can be easily degraded, such as benzoic acid and acetic acid (both were present at 400 mg liter−1), while the second UASB reactor was employed for the removal of the remaining compounds, such as terephthalic acid (500 to 700 mg liter−1), ortho-phthalic acid and isophthalic acid (several milligrams per liter), and 4-methylbenzoic acid (600 to 700 mg liter−1). The granular sludge in the second UASB reactor was used for enrichment of phthalate isomer-degrading anaerobes (sludge I). Sludge II was the granular sludge from a full-scale, single-stage UASB reactor (volume, 8 × 105 liters) in Japan, in which wastewater containing terephthalic acid (200 to 300 mg liter−1), isophthalic acid (50 to 100 mg liter−1), ortho-phthalic acid (several milligrams per liter), acetic acid (1,000 mg liter−1), and benzoic acid (100 to 150 mg liter−1) had been treated. Granular sludges were taken from the two UASB reactors, immediately washed with phosphate buffer (10 mM, pH 7.2), and homogenized briefly for the primary enrichment cultures.
Microorganisms and cultivation.
The following organisms were used in this study. The terephthalate-degrading bacterium (strain JT) was isolated from sludge II in this study. Pelotomaculum thermopropionicum (DSM 13744), Desulfotomaculum thermobenzoicum (DSM 6193), Desulfotomaculum nigrificans (DSM 574), Desulfotomaculum thermosapovorans (DSM 6562), Syntrophobacter fumaroxidans (DSM 10017), Syntrophus gentianae (DSM 8423), Desulfovibrio vulgaris Marburg (DSM 2119), Methanospirillum hungatei (DSM 864), and Methanosaeta concilii (DSM 3671) were obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Braunschweig, Germany.
The basal medium used for enrichment and isolation was prepared as described previously (30). All cultivations were carried out anaerobically at 37°C in 50-ml serum vials containing 20 ml of medium (pH25°C, 7.0) under an atmosphere of N2-CO2 (80:20, vol/vol) without shaking. For enrichment of phthalate isomer-degrading anaerobes, either ortho-phthalate (1 mM), isophthalate (1 mM), or terephthalate (1 mM) was added to the basal medium as the sole energy source. For cultivation of a pure culture of strain JT, 10 mM crotonate and 0.02% yeast extract were used as the substrate. Methanospirillum hungatei was cultivated at 37°C in the medium as mentioned above except that hydrogen and acetate (5 mM) were added to the vials. For cocultivation, Methanospirillum hungatei and strain JT were inoculated into medium supplemented with terephthalate (1 mM) (inoculum size, 2.5 and 5%, respectively). Utilization of sulfate (sulfate concentration, 5 mM) by stain JT was tested in medium supplemented with 1 mM terephthalate or 5 mM benzoate as the electron donor and with exponential-phase culture as the inoculum. The purity of the strain isolated in this study was routinely examined by microscopy and incubation of the cultures with medium containing 0.1% yeast extract and a mixture of carbohydrates (sucrose, glucose, arabinose, and fructose; each at 2 mM) at 35 or 55°C.
Degradability experiments.
To test the degradability of phthalate isomers (1.5 mM each) and benzoate (5 mM) by enrichment cultures, batch experiments were carried out in 125-ml (liquid volume, 40 ml) serum vials with exponential-phase cultures as the inoculum at 37°C without shaking. A total volume of 400 ml of enrichment cultures (after 13 to 19 successive transfers over 2 years) was collected by centrifugation at 7,000 × g for 10 min at 35°C and resuspended in aliquots in 20 ml of medium without substrate. The cell suspension was then inoculated into five bottles containing medium supplemented with either (i) ortho-phthalate, (ii) isophthalate, or (iii) terephthalate, (iv) benzoate, or (v) no additional substrate as controls. The experiments were performed in duplicate. Substrate depletion and acetate and methane production were measured periodically. The degradability of each substance by the cultures was evaluated by percent substrate degradation in the 15-day incubation. In the calculation of electron recovery, the concentrations of fermentation products were corrected by subtracting the amounts of products formed in the control experiments, and the electron balance was calculated based solely on the amount of substrate consumed and the corrected amounts of acetate and methane formed.
Construction of 16S rRNA gene clone libraries from methanogenic phthalate isomer-degrading enrichment cultures.
DNA extraction, PCR amplification, cloning, and sequencing procedures for constructing 16S rRNA gene clone libraries were performed as previously reported (28) with slight modifications. For construction of the 16S rRNA gene clone library, we used the following primer set for PCR amplification of bacterial 16S rRNA genes: Bacteria-specific primer EUB8F (5′-AGAGTTTGATCCTGGCTCAG-3′; positions 8 to 27 in the Escherichia coli gene) and prokaryote-specific primer 1490R (5′-GGTTACCTTGTTACGACTT-3′, positions 1491 to 1509 in E. coli) (37). The PCR products were purified with a MicroSpin column (Amersham Pharmacia Biotech), followed by cloning into plasmids with the TA cloning kit (Novagen). For each enrichment culture, 10 clonal rRNA genes were randomly picked and screened by comparing restriction fragment length polymorphism (RFLP) patterns with HaeIII and HhaI restriction endonucleases. Representative clones having different RFLP patterns were then subjected to sequencing.
DNA extraction and amplification of 16S rRNA gene from a pure culture.
DNA from the pure culture was extracted by the method of Hiraishi (10). PCR amplification of bacterial 16S rRNA genes and purification of PCR products were carried out as described above. The purified PCR products were subjected to further analysis.
Sequencing of 16S rRNA gene and phylogenetic analysis.
Sequences of representative rRNA gene clones as well as the 16S rRNA gene of a pure culture were determined by dye terminator cycle sequencing with a Quick Start kit (Beckman Coulter) and an automated sequence analyzer (CEQ-2000XL; Beckman Coulter). Sequence data were aligned with the ARB program package (http://www.arb-home.de/), and the aligned data were manually corrected based on information about primary and secondary structures. The phylogenetic tree was constructed by the neighbor-joining method (26) implemented in the ARB program. Bootstrap resampling analysis (9) for 1,000 replicates was performed with the PAUP* 4.0 package (33) to estimate the confidence of tree topologies.
FISH.
For fluorescence in situ hybridization (FISH), fixation of cells in the enrichment cultures and subsequent whole-cell in situ hybridization were performed as described previously (29). The 16S rRNA-targeted oligonucleotide probes used in this study are listed in Table 1. For detection of the targeted bacteria in phthalate isomer-degrading enrichment cultures, we designed the following four probes: (i) JPIT74, specific for clones JP, JI, and JT (obtained from the phthalate isomer-degrading enrichment cultures from sludge II) (5′-TACAATTCGCAAGCTTCC-3′; E. coli positions 74 to 92); (ii) UP190, specific for clone UP (obtained from the ortho-phthalate-degrading enrichment culture from sludge I) (5′-TCCTTTCCTCATCCGTGC-3′; E. coli positions 190 to 208); (iii) UT62, specific for clones UT-1 and UT-2 (obtained from the terephthalate-degrading enrichment culture from sludge I) (5′-CATTGCAAACCCCGTTCG-3′; E. coli positions 62 to 80); and (iv) UI178, specific for clone UI (obtained from the isophthalate-degrading enrichment culture from sludge I) (5′-GTGTCGTGTGGTCTTATC-3′, E. coli positions 178 to 196).
TABLE 1.
Fluorescently labeled oligonucleotide probes used in this study
| Probe name | OPD namea | Target group | Probe sequence (5′ to 3′) | Fluorescent labelb | Reference |
|---|---|---|---|---|---|
| EUB338 | S-D-Bact-0338-a-A-18 | Bacteria | GCTGCCTCCCGTAGGAGT | Cy5 | 3 |
| MG1200 | S-O-Mmic-1200-a-A-21 | Methanomicrobiales | CGGATAATTCGGGGCATGCTG | Cy3 | 24 |
| MX825 | S-F-Msae-0825-a-A-23 | Methanosaetaceae | TCGCACCGTGGCCGACACCTAGC | Cy3 | 24 |
| JPIT74 | S-*JPIT-0074-a-A-18 | Clones JP, JI, and JT in ‘Desulfotomaculum lineage I’ | TACAATTCGCAAGCTTCC | Cy3 | This study |
| UP190 | S-*UP-0190-a-A-18 | Clone UP in ‘Desulfotomaculum lineage I’ | TCCTTTCCTCATCCGTGC | Cy3 | This study |
| UT62 | S-*UT-0062-a-A-18 | Clone UT in ‘Desulfotomaculum lineage I’ | CATTGCAAACCCCGTTCG | Cy3 | This study |
| UI178 | S-*UI-0178-a-A-18 | Clone UI in delta-Proteobacteria | GTGTCGTGTGGTCTTATC | Cy3 | This study |
OPD, oligonucleotide probe database (1).
Cy5, indodicarbocyanine; Cy3, indocarbocyanine.
To test the specificity of probes JPIT74, UP190, and UT62, the following species belonging to ‘Desulfotomaculum lineage I' (32) were used as reference strains: P. thermopropionicum (DSM 13744), Desulfotomaculum thermobenzoicum (DSM 6193), Desulfotomaculum nigrificans (DSM 574), and Desulfotomaculum thermosapovorans (DSM 6562). For evaluation of the specificity of probe UI178, the following organisms were employed as reference cells: Syntrophobacter fumaroxidans (DSM 10017), Syntrophus gentianae (DSM 8423), and Desulfovibrio vulgaris Marburg (DSM 2119). In addition, the specificity of each probe was also tested with cells in all the enrichment cultures established in this study. Hybridization stringency was adjusted by adding formamide to the hybridization buffer (0% [vol/vol] for JPIT74; 15% [vol/vol] for EUB338, UP190, UT62, and UI178). For double staining of the enrichment cultures, indodicarbocyanine- and indocarbocyanine-labeled probes were used simultaneously.
Microscopy and analytical methods.
An Olympus microscope equipped for epifluorescence was used for studies of cell morphology and epifluorescence (Olympus BX50F). Concentrations of phthalate isomers were analyzed by high-pressure liquid chromatography (HPLC) with a UV detector as described previously (23). Short-chain fatty acids, sulfate, alcohols, methane, hydrogen, carbon dioxide, and other intermediate substances such as succinate, malate, fumarate, and lactate were measured as described previously (11, 12).
Nucleotide sequence accession numbers.
Six 16S rRNA gene sequences of phthalate isomer-degrading clones as well as that of strain JT were deposited in the DNA databases under accession numbers AB091323 to AB091329.
RESULTS
Reactor performance.
For enrichment of methanogenic phthalate isomer-degrading consortia, sludges from two methane fermentation plants in Taiwan and Japan were used as the sources of inoculum. Both sludges (sludges I and II) had been treating wastewater from the manufacturing of terephthalate. The plant in Taiwan was composed of two-phase anaerobic bioreactors to enhance the removal of phthalate isomers (14, 21). As a whole system, the processes had exhibited good chemical oxygen demand (COD) removal efficiency (around 90%) and sufficient efficiency for phthalate isomer removal (85 to 90%) after 4 years of operation at the volumetric loading rate of 3 kg of COD m−3 day−1 and hydraulic retention time of 1 day. One of the reactors (the latter phase of the system, the UASB reactor) had been specifically used for the removal of phthalate isomers, and therefore a sludge sample (sludge I) was taken from the reactor for further analyses. On the other hand, the plant in Japan consisted of a full-scale, single-staged UASB reactor. Due to the relatively high COD loading rate at the sampling date (volumetric loading rate, 7.1 kg of COD m−3 day−1; hydraulic retention time, 6.4 h), the removal efficiency of COD and phthalate isomers was found to be lower than that of the system in Taiwan; COD and phthalate isomer removal was approximately 50 to 60% and 10 to 20%, respectively. We took the sludge from the reactor (sludge II) for further analyses.
Enrichment and isolation of phthalate isomer-degrading microbes.
To enrich for phthalate isomer-degrading microbes, primary enrichment was made with ortho-phthalate, isophthalate and terephthalate (1 mM each) as the sole carbon and energy source with the two different sludges as the inoculum. In total, six enrichment cultures were made: an enrichment with ortho-phthalate from sludge I, designated enrichment UP; an enrichment with isophthalate from sludge I, enrichment UI; an enrichment with terephthalate from sludge I, enrichment UT; an enrichment with ortho-phthalate from sludge II, enrichment JP; an enrichment with isophthalate from sludge II, enrichment JI; and an enrichment with terephthalate from sludge II, enrichment JT. The cultures were incubated anaerobically at 37°C.
In all six enrichment cultures, growth and phthalate isomer degradation were observed after 2 to 3 months of incubation. The cultures were successively transferred into fresh medium with 5 to 10% (vol/vol) inoculum when approximately 50% of the phthalate isomers had been degraded. The enrichment cultures always required a long time for complete mineralization of phthalate isomers, ranging from 20 to 50 days, and always formed methane (and acetate in some cases) as end products along with phthalate isomer degradation (Fig. 1). Additionally, the stagnation of growth and phthalate isomer degradation were found when we added 5 mM 2-bromoethanesulfonate, an inhibitor of methanogenesis, to active cultures, indicating strictly syntrophic phthalate isomer degradation, as previously reported by Kleerebezem et al. (14). In some cases, no growth occurred when we used the stationary-phase culture or a smaller inoculum (<5%) during the transfer.
FIG. 1.
Metabolism of phthalate isomers by enrichment cultures after more than 10 transfers. (a) Enrichment with ortho-phthalate from sludge I, enrichment UP; (b) enrichment with isophthalate from sludge I, enrichment UI; (c) enrichment with terephthalate from sludge I, enrichment UT; (d) enrichment with ortho-phthalate from sludge II, enrichment JP; (e) enrichment with isophthalate from sludge II, enrichment JI; (f) enrichment with terephthalate from sludge II, enrichment JT.
In the five enrichment cultures UP, UT, JP, JI, and JT, Methanospirillum-like F420-autofluorescent rods, Methanosaeta-like thick rods in some cases, and oval rods which seemed to be sporeformers were observed as the major morphotypes. The only exception was the isophthalate-degrading enrichment culture from sludge I (enrichment UI), which contained relatively thin, short, rod-shaped cells instead of the oval rods. FISH with Methanosaeta-specific probe MX825 (24) and Methanomicrobiales-specific probe MG1200 (24) revealed that in all the enrichment cultures, cells resembling Methanosaeta and Methanospirillum showed positive signals with the MX825 and MG1200 probes, respectively, suggesting that those cells were aceticlastic and hydrogenotrophic methanogens. When Bacteria-specific probe EUB338 was used, spore-forming oval rods (enrichments UP, UT, JP, JI, and JT) and thin short rods (enrichment UI) reacted with the probe, indicating that the cells belonged to the domain Bacteria and are likely to perform syntrophic phthalate isomer degradation. We therefore focused on those spore-forming oval rods and relatively thin short rods and attempted to establish defined cocultures with Methanospirillum hungatei or tricultures with Methanospirillum hungatei and Methanosaeta concilii.
Attempts were first made to isolate all of these bacteria in co- or tricultures on solid medium containing phthalate isomers; all tubes were inoculated beforehand with Methanospirillum hungatei (and Methanosaeta concilii in some trials). However, no visible colonies containing targeted cells could be found within 4 months of incubation. We then tried to isolate the targeted cells by replacing the substrate with other possible ones which were thought to support the growth of the targeted cells in pure culture, such as benzoate, crotonate, fumarate, and pyruvate. However, all of the substrates supported the growth of other microbes that showed no ability to metabolize phthalate isomers. We also tried to adapt the cultures with other external electron acceptors such as sulfate in phthalate isomer medium. However, cell growth was not observed on such media. Therefore, the enrichment cultures were further purified by serial dilution in liquid medium with the addition of Methanospirillum hungatei (and Methanosaeta concilii) cells. Co- and tricultures with Methanospirillum hungatei (and Methanosaeta concilii tricultures) showed similar growth properties; both cultures receiving 10−1 to 10−4 dilutions showed growth, phthalate isomer degradation, and methane production after 3 to 4 months of incubation for all enrichments. We therefore used only Methanospirillum hungatei cells for further dilution. This step was repeated several times over 15 months, and we obtained highly purified enrichment cultures, although they still contained contaminants that probably did not participate in phthalate isomer degradation.
Degradability of phthalate isomers by enrichment cultures.
To assess the degradability of each phthalate isomer as well as benzoate by all of the highly purified enrichment cultures, batch experiments were performed with dense cell suspensions. The degradability was evaluated by substrate degradation (as a percentage) in a 15-day incubation (Table 2). All six enrichment cultures degraded the phthalate isomer used for the primary enrichment within 7 to 15 days. One terephthalate enrichment culture (UT) could degrade only terephthalate within 15 days, but all of the other five enrichment cultures completely degraded 5 mM benzoate without a lag phase. Moreover, one ortho-phthalate enrichment culture (UP) could also degrade terephthalate completely within 15 days of incubation. For enrichment JT, this substrate range (i.e., terephthalate and benzoate utilization) was later confirmed for the principal component of the culture as a pure syntrophic coculture (strain JT; see below).
TABLE 2.
Degradability of benzoate and phthalate isomers by dense cells of phthalate isomer enrichment cultures within 15 days of incubationa
| Origin of enrichment cultures | Enrichment culture used | Substrate | Initial substrate concn (mM) | Final concn (mmol/liter)
|
% Degraded in 15 days | Electron recoveryb (%) | ||
|---|---|---|---|---|---|---|---|---|
| Substrate (mM) | Acetatea (mM) | Methanea (mmol/liter) | ||||||
| Sludge I | UP (enriched on ortho-phthalate medium) | ortho-Phthalate | 1.61 | 0 | 2.03 | 5.38 | 100 | 126 |
| Benzoate | 6.07 | 0 | 1.09 | 16.0 | 100 | 78 | ||
| Isophthalate | 1.53 | 1.53 | ND | ND | 0 | — | ||
| Terephthalate | 1.37 | 0 | 0 | 5.1 | 100 | 103 | ||
| UI (enriched on isophthalate medium) | Isophthalate | 1.25 | 0 | 0 | 5.37 | 100 | 119 | |
| Benzoate | 5.46 | 0 | 0 | 19.08 | 100 | 97 | ||
| ortho-Phthalate | 1.46 | 1.46 | ND | ND | 0 | — | ||
| Terephthalate | 1.24 | 1.24 | ND | ND | 0 | — | ||
| UT (enriched on terephthalate medium) | Terephthalate | 1.39 | 0 | 0.21 | 5.6 | 100 | 115 | |
| Benzoate | 5.21 | 5.21 | ND | ND | 0 | — | ||
| ortho-Phthalate | 1.28 | 1.28 | ND | ND | 0 | — | ||
| Isophthalate | 1.47 | 1.47 | ND | ND | 0 | — | ||
| Sludge II | JP (enriched on ortho-phthalate medium) | ortho-Phthalate | 1.61 | 0.14 | 4.24 | 0.93 | 91 | 95 |
| Benzoate | 6.48 | 0.26 | 11 | 3.61 | 96 | 65 | ||
| Isophthalate | 1.49 | 1.3 | 0.83 | 0.64 | 13 | 114 | ||
| Terephthalate | 1.38 | 0.92 | 1.34 | 0.25 | 33 | 98 | ||
| JI (enriched on isophthalate medium) | Isophthalate | 1.55 | 0 | 0 | 6.0 | 100 | 107 | |
| Benzoate | 5.49 | 0 | 0 | 16.1 | 100 | 81 | ||
| ortho-Phthalate | 1.54 | 1.46 | 0 | 1.60 | 5 | 123 | ||
| Terephthalate | 1.27 | 1.02 | 0 | 2.30 | 20 | 130 | ||
| JT (enriched on terephthalate medium) | Terephthalate | 1.27 | 0 | 4.4 | 0.96 | 100 | 113 | |
| Benzoate | 5.22 | 0 | 16.9 | 7.57 | 100 | 126 | ||
| ortho-Phthalate | 1.66 | 1.66 | ND | ND | 0 | — | ||
| Isophthalate | 1.56 | 1.56 | ND | ND | 0 | — | ||
The values shown were corrected by subtracting the amount of product formed in control experiments, to which substrates were not added. ND, not determined; —, not calculated.
Electron balance was calculated based only on substrate utilized, acetate formed, and methane formed.
Phylogenetic affiliation of bacterial populations in phthalate isomer enrichment cultures.
Since the isolation of targeted cells was difficult, we tried to identify the targeted bacteria by a full-cycle rRNA approach. Bacterial 16S rRNA genes from the highly enriched cultures after 1 year of successive transfers and serial dilutions were amplified and cloned into Escherichia coli, and RFLP analysis of clonal 16S rRNA genes with HaeIII and HhaI was performed on 10 randomly selected rRNA gene clones. From the analysis, one to five different RFLP patterns were found in each of the highly enriched cultures. The most abundant clones in the representative clone libraries for the five enrichment cultures UP, UT, JP, JI, and JT were affiliated with a recently recognized cluster (subcluster Ih) of the group Desulfotomaculum (‘Desulfotomaculum lineage I') (H. Imachi, Y. Qiu, Y. Sekiguchi, Y. Kamagata, A. Ohashi, and H. Harada, Abstr. 102nd Annu. Meet. Am. Soc. Microbiol. 2002, p. 260). These clones were most closely related to members of the genus Pelotomaculum, which is composed of syntrophic propionate-oxidizing bacteria such as Pelotomaculum thermopropionicum (Fig. 2A). In contrast, the clone library for enrichment culture UI showed only one RFLP pattern in 10 selected clones; the clone was affiliated with a recently recognized clone cluster (group TA) in the delta-Proteobacteria (Fig. 2B).
FIG. 2.
Phylogenetic trees among the group ‘Desulfotomaculum lineage I' (A) and the class delta-Proteobacteria (B) based on comparative analyses of 16S rRNA gene sequences, showing the phylogenetic positions of clones and an isolated bacterium (strain JT) obtained in this study (clones UP, UI, and UT were retrieved from enrichment cultures with sludge I as the inoculum, and clones JP, JI, and JT were obtained from enrichments with sludge II as the inoculum). The trees were calculated based on a distance matrix analysis of 16S rRNA gene sequences (neighbor-joining tree). The scale bars represent the number of nucleotide changes per sequence position. The symbols at nodes correlate with the bootstrap values (percent) obtained with 1,000 resamplings.
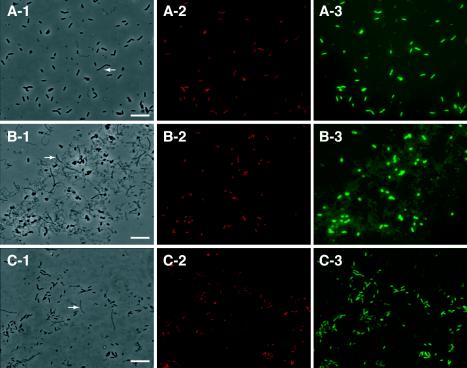
To confirm whether the 16S rRNA gene sequences that were obtained in the cloning analysis actually represented the dominant bacterial populations in the enrichment cultures, specific oligonucleotide probes were designed and applied to the enrichments. The specificity of the designed probes (JPIT74, UP190, and UT62 for clones in the group Pelotomaculum and UI178 for clone UI in the group TA) was first evaluated with reference organisms (see Materials and Methods) by FISH. All reference bacteria reacted with the Bacteria-specific probe EUB338 but not with probes JPIT74, UP190, UT62, or UI178 at any formamide concentration in the hybridization and washing buffers (data not shown). On the other hand, oval rods in the five enrichment cultures UP, UT, JP, JI, and JT hybridized with either the JPIT74, UP190, or UT62 probe (Fig. 3), i.e., rod-shaped cells in enrichments JP, JI, and JT hybridized with the JPIT74 probe (Fig. 3A), while rod-shaped cells in enrichments UP and UT reacted with the UP190 and UT62 probes (Fig. 3B). In addition, thin, short, rod-shaped cells in enrichment UI reacted with the UI178 probe (Fig. 3C). Each probe specifically detected cells in an enrichment-dependent manner; e.g., probe JPIT74 did not detect positive cells in enrichments UP, UT, and UI. These findings suggested that the probes constructed in this study were sufficiently specific for each clone and that each enrichment culture consisted mainly of a particular type of bacteria. Because these probes hybridized with most of the EUB338 probe-positive cells (Fig. 3), the majority of the bacterial cells in the enrichment cultures were actually assigned to either subcluster Ih of ‘Desulfotomaculum lineage I' (for enrichments UP, UT, JP, JI, and JT) or the clone clade (group TA) of the delta-Proteobacteria (for enrichment UI).
FIG. 3.
In situ hybridization of phthalate isomer-degrading enrichment cultures. The enrichment cultures were simultaneously hybridized with indodicarbocyanine-labeled Bacteria-specific probe EUB338 (green, panel 3) and indocarbocyanine-labeled clone-specific probes (red, panel 2). Phase-contrast micrographs (panel 1) and fluorescence micrographs of the same fields (panels 2 and 3) are shown. (A) JT culture (terephthalate culture with sludge II) hybridized with probe JPIT74, specific for clones JP, JI, and JT in subcluster Ih of ‘Desulfotomaculum lineage I'; (B) UT culture (terephthalate enrichment with sludge I) hybridized with probe UT62, specific for clone UT in subcluster Ih of ‘Desulfotomaculum lineage I'; (C) UI culture (isophthalate enrichment with sludge I) hybridized with the UI178 probe, specific for clone UI in the delta-Proteobacteria. Bars, 10 μm. Arrows indicate methanogen-like cells.
Isolation of phthalate isomer oxidizers in pure culture.
According to the results obtained by the rRNA approach, it was suggested that the five enrichment cultures UP, UT, JP, JI, and JT involved members of subcluster Ih of ‘Desulfotomaculum lineage I' as the major bacterial population. Since all members of ‘Desulfotomaculum lineage I' are spore-forming organisms, attempts were made to isolate these sporeformers by applying pasteurization to the enrichments. The five enrichments were pasteurized at 70°C for 40 min and then serially diluted into fresh medium previously inoculated with Methanospirillum hungatei cells. After 3 to 5 months of incubation, highly purified cocultures with the ability to degrade phthalate isomers were developed from the five enrichments, and cultures receiving 10−1 to 10−3 dilutions consisted almost solely of sporeformers and Methanospirillum hungatei. Therefore, the pasteurized enrichment cultures were further purified by repeated pasteurizations at 70°C for 40 min. Then the highly purified cocultures obtained through pasteurization were used for further attempts at isolation of sporeformers in pure culture. Several substrates, such as crotonate, lactate, fumarate, and pyruvate, that other known syntrophic bacteria can utilize in pure cultures were used for isolation. Of the compounds tested, crotonate (10 mM) supported the growth of the targeted cells from one terephthalate-degrading coculture (JT) after 1 month of incubation. Roll tube isolation was then conducted with medium containing 10 mM crotonate and 0.02% yeast extract, resulting in the formation of very small colonies that were light brown, lens shaped, and 0.1 to 0.15 mm in diameter after 1 month of incubation. This step was repeated several times until a pure culture was obtained. Strain JT (from enrichment JT) was reconstituted with Methanospirillum hungatei in terephthalate medium to check whether it was the targeted bacterium. Indeed, degradation of terephthalate occurred, with concomitant growth of the coculture (see Fig. 5). However, we were not able to find appropriate substrates that support other Pelotomaculum-like cells in the remaining four enrichments (UP, UT, JP, and JI). We were also not able to isolate the dominant bacterial cells in the isophthalate-degrading enrichment culture (UI), which contained cells belonging to the clone cluster (group TA) in the delta-Proteobacteria, since we could not obtain precise physiological information from phylogenetic analysis.
FIG. 5.
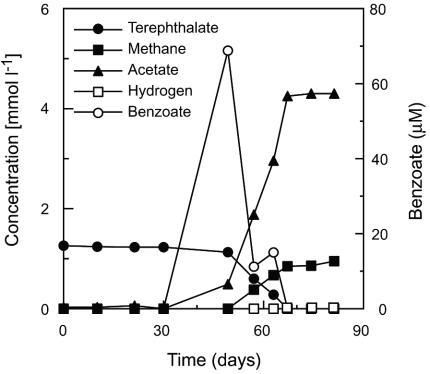
Terephthalate degradation by strain JT in coculture with Methanospirillum hungatei.
Partial characterization of strain JT.
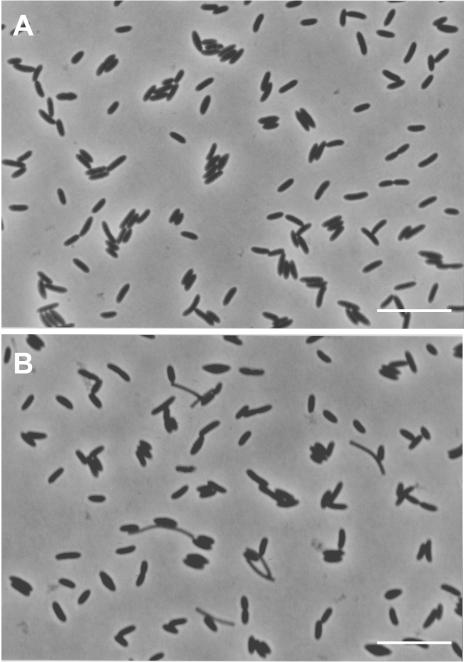
Strain JT was strictly anaerobic, since no growth occurred in medium under an atmosphere of N2 and O2 (80:20). The cells were nonmotile, rod-shaped, 0.8 to 1.0 μm wide, and 2.0 to 3.0 μm long (Fig. 4). Strain JT utilized terephthalate as well as benzoate in coculture with Methanospirillum hungatei (Fig. 5). However, growth and terephthalate (and benzoate) degradation were not observed in pure culture, indicating that strain JT is a syntrophic bacterium depending strictly on the presence of hydrogen (and/or formate)-consuming partner organisms to grow on terephthalate and benzoate. The coculture degraded terephthalate and produced 3.3 mol of acetate and 0.7 mol of methane per mol of terephthalate consumed (97% electron recovery). During terephthalate degradation, a small amount of benzoate (70 μM) was detected as a transiently excreted product. The isolate was able to grow on crotonate in pure culture. No growth of the strain was found on medium supplemented with sulfate as the electron acceptor in the presence of benzoate or terephthalate as electron donors. 16S rRNA gene sequence analysis revealed that the strain belongs to the genus Pelotomaculum. Organisms closely related to strain JT are P. thermopropionicum and “P. schinkii” (similarity values are 89% and 93%, respectively).
FIG. 4.
Phase-contrast micrographs showing the cell morphology of strain JT grown on crotonate (10 mM) (A) and on terephthalate (1 mM) in coculture with Methanospirillum hungatei (B). Bars, 10 μm.
DISCUSSION
Physiology of phthalate isomer enrichment cultures.
As with other known syntrophs, it took a long time (over 2 years) to establish highly enriched cultures as well as to isolate the microbes responsible for the degradation of phthalate isomers. One of the primary reasons for the difficulty in isolation was that the growth of the enrichment cultures on phthalate isomer medium was very slow. The specific growth rates (μmax, calculated based on methane production) of the six enrichment cultures on phthalate isomers medium were estimated to be 0.13 to 0.25 day−1. These values were almost in the same order of magnitude as those of poorly degradable substrates under methanogenic conditions, such as toluene (0.11 day−1) and o-xylene (0.07 day−1) (8). Another difficulty was that all enriched consortia were somewhat unstable, similar to the findings reported previously (14). Kleerebezem et al. suggested that one could no longer maintain stable growth of phthalate isomer-degrading consortia when (i) smaller amounts (<20%, vol/vol) of enrichment cultures were transferred to fresh medium or (ii) the cultures were transferred to fresh medium if inocula whose phthalate conversion rates were significantly lower than usual were used (14). In our cases, stable growth could normally be observed after transferring at least 5% of the active culture into fresh medium. But once we found the culture lost the ability to grow, we also needed to revive the culture by applying large amounts of active culture as an inoculum (15 to 20%, vol/vol).
The experiments testing the degradability of phthalate isomer with dense cell suspensions of each consortium suggested some important aspects of the physiology of the phthalate isomer degraders present in the consortia (Table 2). One of the most important findings was that almost all of the consortia except that in enrichment UT could degrade benzoate without a lag phase. As suggested by Kleerebezem et al. (14), this implied that the initial step in the degradation of phthalate isomers in methanogenic consortia is likely to be decarboxylation to benzoate, followed by degradation of benzoate presumably to carbon dioxide, acetate, and hydrogen (and/or formate). In fact, a small amount of benzoate (70 μM) was detected as a transiently excreted product during terephthalate degradation by strain JT in coculture with Methanospirillum hungatei. Another interesting finding was that some of the highly purified consortia could degrade other phthalate isomers besides the one used in the primary enrichment (Table 2). It cannot be ruled out whether one species of microbe actually degraded multiple forms of phthalate isomers and benzoate simultaneously in all the enrichment cultures, since the results were based on “highly purified” but not defined consortia. However, considering the fact that all of our enrichment cultures were found to contain homogeneous bacterial populations based on FISH analyses, it is very likely that two or three phthalate isomers as well as benzoate may be fermented by one species of organism under methanogenic conditions.
Kleerebezem et al. (14) reported that highly enriched methanogenic phthalate isomer-degrading cultures could only degrade the isomer used for enrichment but could degrade benzoate simultaneously. Therefore, they proposed that specific organisms were responsible for the degradation of each phthalate isomer. The discrepancy between the report by Kleerebezem et al. (14) and our data may be due simply to the physiological diversity of phthalate-degrading bacteria in methanogenic environments. However, our 16S rRNA gene-cloning experiments suggested that each of the consortia contained phylogenetically distinct bacteria as the major constituents, indicating that there may be specific types of microbes that are adapted to the degradation of different isomers.
Members of subcluster Ih (Pelotomaculum) of the group Desulfotomaculum.
Microscopic examinations of the enrichment cultures indicated that almost all of the consortia contained spore-forming oval rods as the major bacterial populations. This feature is consistent with the previous study (14), in which short fat rods and spore-forming microorganisms were presumed to be involved in the degradation of phthalate isomers under methanogenic conditions, although their phylogenetic positions were unknown. In our study, these spore-forming oval rods were phylogenetically identified as members of subcluster Ih in the spore-forming clade previously recognized as ‘Desulfotomaculum lineage I' (32). Currently, three genera are known to be involved in the clade, Desulfotomaculum, Sporotomaculum, and Pelotomaculum. Some Desulfotomaculum species can degrade aromatic compounds by sulfate reduction, but none was known to have the ability to metabolize such substances by syntrophic association (5, 34). The genus Sporotomaculum was recognized recently as a spore-forming, non-sulfate-reducing bacterial lineage of ‘Desulfotomaculum lineage I' (4). Two species within the genus have been described so far, of which one has the ability to grow on benzoate in syntrophic association with hydrogenotrophic methanogens (23). However, both are found to show no ability to grow on phthalate isomers under any culture conditions (4, 23). The genus Pelotomaculum was known to be a spore-forming, non-sulfate-reducing, syntrophic propionate-oxidizing lineage in the clade ‘Desulfotomaculum lineage I' (representing subcluster Ih) (12). Currently, two species are known as the members of the genus, but it is unclear whether they can metabolize phthalate isomers (6, 12).
A remarkable feature of our enrichment cultures is that they may not be able to utilize sulfate as an exogenous electron acceptor but grow by syntrophic association with hydrogenotrophic methanogens. Importantly, the methanogenic phthalate isomer-degrading syntrophic consortia reported by Kleerebezem et al. (14), in which sporeformers were observed as the major populations, were also shown to have no ability to utilize sulfate; these traits strongly suggest that the bacterial populations in the enrichments were likely to be similar to the ones we identified as members of subcluster Ih. Recently, strain 7, which is also a member of subcluster Ih in ‘Desulfotomaculum lineage I', was purified from a phenol-degrading anaerobic culture and showed the ability to metabolize phenol (17). In addition, a number of environmental clones belonging to subcluster Ih can be found in the public databases, some of which were retrieved from anaerobic petroleum-contaminated sites (7). These findings may imply that subcluster Ih is an important lineage of anaerobic microbes capable of degrading aromatic compounds.
Members of group TA in the class delta-Proteobacteria.
One enrichment culture (UI) was found to contain thin, rod-shaped cells belonging to a clone clade (group TA) (38) in the class delta-Proteobacteria as the major bacterial population. The clone lineage was recognized by recent culture-independent molecular studies on UASB sludges (28, 38), a contaminated aquifer site (7), an anaerobic trichlorobenzene-transforming consortium (36), and an anaerobic dichloropropane-dechlorinating consortium (27). More than 20 16S rRNA gene clonal sequences could be identified in the public databases as constituents of the group with no cultured representatives. Very importantly, the majority of such 16S rRNA gene clones were retrieved from the anaerobic sludge in a UASB reactor treating terephthalate-containing wastewater (38). Wu et al. demonstrated that the predominant bacterial populations in the anaerobic sludge community were affiliated with group TA (66.8% of the total bacterial clones) based on 16S rRNA gene-cloning analysis and subsequent FISH with a TA group-specific probe (38). Therefore, the members of the group were suggested to play significant roles in methanogenic terephthalate degradation. This finding, together with our enrichment UI, strongly indicates that certain members of group TA are responsible for the degradation of phthalate isomers.
Physiological properties of strain JT.
In the presence of the methanogen Methanospirillum hungatei, strain JT could degrade terephthalate as well as benzoate. The actual degradation and product formation by strain JT in coculture with Methanospirillum hungatei are nearly equivalent to the theoretical stoichiometry (14). In terms of phylogeny, the organisms most closely related to strain JT are members of the genus Pelotomaculum (6, 12). Strain JT shares some basic traits with the known species of Pelotomaculum, such as morphology, syntrophic growth, and inability to reduce sulfate. However, the strain may have differences in physiology, particularly in substrate range, from the known species. More detailed experiments with this unique isolate may elucidate the physiology and pathway of phthalate isomer degradation under methanogenic conditions.
In summary, we successfully identified and isolated the populations responsible for the mineralization of phthalate isomers under methanogenic conditions. Further molecular ecological studies are required to determine their abundance and spatial distribution in the original sludges treating actual terephthalate-containing wastewaters. In addition, the strains in the other enrichment cultures should be analyzed further to clarify the functions of the microbes. The isolation and more details on the growth and physiological properties of these microbes will be reported in the future.
Acknowledgments
We thank Tadashi Tagawa for information on the performance of UASB reactors.
This study was carried out as a part of the Project for Development of Technologies for Analyzing and Controlling the Mechanism of Biodegrading and Processing, which was entrusted to the New Energy and Industrial Technology Development Organization (NEDO), Japan, and financially supported by research grant 13355022 from the Grants-in-Aid for Scientific Research subsidized by the Japan Society for the Promotion of Science (JSPS).
REFERENCES
- 1.Alm, E. W., D. B. Oerther, N. Larsen, D. A. Stahl, and L. Raskin. 1996. The oligonucleotide probe database. Appl. Environ. Microbiol. 62:3557-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alvarez, J. M., S. Macé, and P. Llabrés. 2000. Anaerobic digestion of organic solid wastes. An overview of research achievements and perspectives. Bioresource Technol. 74:3-16. [Google Scholar]
- 3.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brauman, A., J. A. Müller, J. L. Garcia, A. Brune, and B. Schink. 1998. Fermentative degradation of 3-hydroxybenzoate in pure culture by a novel strictly anaerobic bacterium, Sporotomaculum hydroxybenzoicum gen. nov., sp. nov. Int. J. Syst. Bacteriol. 48:215-221. [DOI] [PubMed] [Google Scholar]
- 5.Cord-Ruwisch, R., and J. L. Garcia. 1985. Isolation and characterization of an anaerobic benzoate-degrading spore-forming sulfate-reducing bacterium, Desulfotomaculum sapomandens sp. nov. FEMS Microbiol. Lett. 29:325-330. [Google Scholar]
- 6.de Bok, F. A. M. 2002. Biochemistry and physiology of syntrophic propionate-oxidizing microbial consortia. Ph.D. thesis. Wageningen University, Wageningen, The Netherlands.
- 7.Dojka, M. A., P. Hugenholtz, S. K. Haack, and N. R. Pace. 1998. Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl. Environ. Microbiol. 64:3869-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edwards, E. A., and D. Grbic-Galic. 1994. Anaerobic degradation of toluene and o-xylene by a methanogenic consortium. Appl. Environ. Microbiol. 60:313-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Felsenstein, J. 1985. Confidence limits of phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 10.Hiraishi, A. 1992. Direct automated sequencing of 16S rDNA amplified by polymerase chain reaction from bacterial cultures without DNA purification. Lett. Appl. Microbiol. 15:210-213. [DOI] [PubMed] [Google Scholar]
- 11.Imachi, H., Y. Sekiguchi, Y. Kamagata, A. Ohashi, and H. Harada. 2000. Cultivation and in situ detection of a thermophilic bacterium capable of oxidizing propionate in syntrophic association with hydrogenotrophic methanogens in a thermophilic methanogenic granular sludge. Appl. Environ. Microbiol. 66:3608-3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Imachi, H., Y. Sekiguchi, Y. Kamagata, A. Ohashi, and H. Harada. 2002. Pelotomaculum thermopropionicum gen. nov., sp. nov., an anaerobic, thermophilic, syntrophic propionate-oxidizing bacterium. Int. J. Syst. Evol. Microbiol. 52:1729-1735. [DOI] [PubMed] [Google Scholar]
- 13.Kleerebezem, R., J. Mortier, L. W. Hulshoff Pol, and G. Lettinga. 1997. Anaerobic pre-treatment of petrochemical effluents: terephthalic acid wastewater. Water Sci. Technol. 36:237-248. [Google Scholar]
- 14.Kleerebezem, R., L. W. Hulshoff Pol, and G. Lettinga. 1999. Anaerobic degradation of phthalate isomers by methanogenic consortia. Appl. Environ. Microbiol. 65:1152-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kleerebezem, R., L. W. Hulshoff Pol, and G. Lettinga. 1999. Anaerobic biodegradability of phthalic acid isomers and related compounds. Biodegradation 10:63-73. [DOI] [PubMed] [Google Scholar]
- 16.Lepistö, R., and J. Rintala. 1999. Extreme thermophilic (70°C), VFA-fed UASB reactor, performance, temperature response, load potential and comparison with 35 and 55°C UASB reactors. Water Res. 33:3162-3170. [Google Scholar]
- 17.Letowski, J., P. Juteau, R. Villemur, M. F. Duckett, R. Beaudet, F. Lépine, and J. G. Bisaillon. 2001. Separation of a phenol carboxylating organism from a two-member, strict anaerobic co-culture. Can. J. Microbiol. 47:373-381. [PubMed] [Google Scholar]
- 18.Lettinga, G. 1995. Anaerobic digestion and wastewater treatment systems. Antonie Leeuwenhoek 67:3-28. [DOI] [PubMed] [Google Scholar]
- 19.Lettinga, G., S. Rebac, S. Parshina, A. Nozhevnikova, J. B. van Lier, and A. J. M. Stams. 1999. High-rate anaerobic treatment of wastewater at low temperatures. Appl. Environ. Microbiol. 65:1696-1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Macarie, H., A. Noyola, and J. P. Guyot. 1992. Anaerobic treatment of a petrochemical wastewater from a terephthalic acid plant. Water Sci. Technol. 25:223-235. [Google Scholar]
- 21.Macarie, H., and J. P. Guyot. 1992. Inhibition of the methanogenic fermentation of p-toluic acid (4-methylbenzoic acid) by acetate. Appl. Microbiol. Biotechnol. 38:398-402. [Google Scholar]
- 22.O'Neill, C., F. R. Hawkes, D. L. Hawkes, S. Esteves, and S. J. Wilcox. 2000. Anaerobic-aerobic biotreatment of simulated textile effluent containing varied ratios of starch and azo dye. Water Res. 34:2355-2361. [Google Scholar]
- 23.Qiu, Y. L., Y. Sekiguchi, H. Imachi, Y. Kamagata, I. C. Tseng, S. S. Cheng, A. Ohashi, and H. Harada. 2003. Sporotomaculum syntrophicum sp. nov., a novel anaerobic, syntrophic benzoate-degrading bacterium isolated from methanogenic sludge treating wastewater from terephthalate manufacturing. Arch. Microbiol. 179:242-249. [DOI] [PubMed] [Google Scholar]
- 24.Raskin, L., J. M. Stromley, B. E. Rittmann, and D. A. Stahl. 1994. Group-specific 16S rRNA hybridization probes to describe natural communities of methanogens. Appl. Environ. Microbiol. 60:1232-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ribbons, D. W., B. F. Taylor, R. W. Eaton, and B. N. Anderson. 1984. Microbial degradation of phthalates, p. 371-397. In D. T. Gibson (ed.), Microbial degradation of organic compounds. Marcel Dekker, New York, N.Y.
- 26.Saito, N., and M. Nei. 1987. The neighbor-joining method: a new method for constructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 27.Schlötelburg, C., F. von Wintzingerode, R. Hauck, W. Hegemann, and U. B. Göbel. 2000. Bacteria of an anaerobic 1,2-dichloropropane-dechlorinating mixed culture are phylogenetically related to those of other anaerobic dechlorinating consortia. Int. J. Syst. Evol. Microbiol. 50:1505-1511. [DOI] [PubMed] [Google Scholar]
- 28.Sekiguchi, Y., Y. Kamagata, K. Syutsubo, A. Ohashi, H. Harada, and K. Nakamura. 1998. Phylogenetic diversity of mesophilic and thermophilic granular sludges determined by 16S rRNA gene analysis. Microbiology 144:2655-2665. [DOI] [PubMed] [Google Scholar]
- 29.Sekiguchi, Y., Y. Kamagata, K. Nakamura, A. Ohashi, and H. Harada. 1999. Fluorescence in situ hybridization using 16S rRNA-targeted oligonucleotides reveals localization of methanogens and selected uncultured bacteria in mesophilic and thermophilic sludge granules. Appl. Environ. Microbiol. 65:1280-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sekiguchi, Y., Y. Kamagata, K. Nakamura, A. Ohashi, and H. Harada. 2000. Syntrophothermus lipocalidus gen. nov., sp. nov., a novel thermophilic, syntrophic, fatty-acid-oxidizing anaerobe which utilizes isobutyrate. Int. J. Syst. Evol. Microbiol. 50:771-779. [DOI] [PubMed] [Google Scholar]
- 31.Speece, R. E. 1996. Anaerobic biotechnology for industrial wastewaters. Archae Press, Nashville, Tenn.
- 32.Stackebrandt, E., C. Sproer, F. A. Rainey, J. Burghardt, O. Päuker, and H. Hippe. 1997. Phylogenetic analysis of the genus Desulfotomaculum: evidence for the misclassification of Desulfotomaculum guttoideum and description of Desulfotomaculum orientis as Desulfosporosinus orientis gen. nov., comb. nov. Int. J. Syst. Bacteriol. 47:1134-1139. [DOI] [PubMed] [Google Scholar]
- 33.Swofford, D. L. 2002. PAUP*: phylogenetic analysis using parsimony (*and other methods), version 4. Sinauer Associates, Sunderland, Mass.
- 34.Tasaki, M., Y. Kamagata, K. Nakamura, and E. Mikami. 1991. Isolation and characterization of a thermophilic benzoate-degrading, sulfate-reducing bacterium, Desulfotomaculum thermobenzoicum sp. nov. Arch. Microbiol. 155:348-352. [Google Scholar]
- 35.van Lier, J. B., S. Rebac, and G. Lettinga. 1997. High-rate anaerobic wastewater treatment under psychrophilic and thermophilic conditions. Water Sci. Tech. 35:199-206. [Google Scholar]
- 36.von Wintzingerode, F., B. Selent, W. Hegemann, and U. B. Göbel. 1999. Phylogenetic analysis of an anaerobic, trichlorobenzene-transforming microbial consortium. Appl. Environ. Microbiol. 65:283-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu, J. H., W. T. Liu, I. C. Tseng, and S. S. Cheng. 2001. Characterization of microbial consortia in a terephthalate-degrading anaerobic granular sludge system. Microbiology 147:373-382. [DOI] [PubMed] [Google Scholar]