Abstract
Investigations of the distribution and diversity of nitrogen-fixing microorganisms in natural environments have often relied on PCR amplification and sequence analysis of a portion of one of the key enzymes in nitrogen fixation, dinitrogenase reductase, encoded by nifH. Recent work has suggested that DNA macroarrays provide semiquantitative fingerprints of diversity within mixtures of nifH amplicons (G. F. Steward, B. D. Jenkins, B. B. Ward, and J. P. Zehr, Appl. Environ. Microbiol. 70:1455-1465, 2004). Here we report the application of macroarrays for a study in the Chesapeake Bay. Samples from different locations in the bay yielded distinct fingerprints. Analysis of replicates and samples from different locations by cluster analysis showed that replicates clustered together, whereas different samples formed distinct clusters. There was a correspondence between the hybridization pattern observed and that predicted from the distribution of sequence types in a corresponding clone library. Some discrepancies between the methods were observed which are likely a result of the high nifH sequence diversity in the Chesapeake Bay and the limited number of sequences represented on this version of the array. Analyses of sequences in the clone library indicate that the Chesapeake Bay harbors unique, phylogenetically diverse diazotrophs. The macroarray hybridization patterns suggest that there are spatially variable communities of diazotrophs, which have been confirmed by quantitative PCR methods (S. M. Short, B. D. Jenkins, and J. P. Zehr, Appl. Environ. Microbiol., in press). The results show that DNA macroarrays have great potential for mapping the spatial and temporal variability of functional gene diversity in the environment.
Microbial communities play important roles in aquatic biogeochemical cycles and food webs. The discovery that most aquatic bacteria are not represented by culture collections (13, 24) has spawned many studies that describe the composition and diversity of aquatic communities with molecular phylogenetic methods (21, 39). However, the biochemical capacities or functions of individual microbes in environmental communities are not well understood. The functions of microbes are defined by the genetic potential contained in their genomes, and it is the expression of specific genes in relation to physical and chemical factors that makes individual species competitive in the environment. The factors controlling the distribution of microbes and specific genes in aquatic environments are not well known.
Estuaries are complex hydrodynamic environments that exhibit strong gradients in oxygen, nutrients, organic matter, and salinity. Relatively little is known about the dynamics of microbial assemblages in estuaries (8, 9, 23). Many estuaries receive high concentrations of anthropogenic nutrients (26), which can support high productivity rates and biomass. Estuaries and coastal waters are often nutrient limited as a result of the transition from fresh to salt water and depletion of nutrients by phytoplankton blooms. In the Chesapeake Bay, N limitation increases with salinity as a result of the difference between the N/P ratios of freshwater and seawater inputs (11, 12, 20). There is seasonality to nutrient inputs, such that N limitation is pronounced during the summer (11, 12, 20).
N2-fixing microorganisms are one component of aquatic microbial communities that can be important in providing fixed nitrogen in nitrogen-limited systems. In nitrogen-limited estuaries, blooms of nitrogen (N2)-fixing cyanobacteria can occur (4, 22). Diverse heterotrophic N2-fixing bacteria are found in estuaries (1), but the roles that these microorganisms play in estuarine nutrient dynamics and the factors that determine the distribution of N2-fixing microorganisms (36) have not been defined.
N2 fixation is catalyzed by the enzyme nitrogenase, which is composed of two multisubunit proteins encoded by the nif genes. N2 fixation is an energetically expensive process, and approximately 20 gene products are involved in the maturation and assembly of nitrogenase (16). Therefore, N2 fixation capacity represents a substantial genomic component (approximately 20 kb of DNA). Gene mutation occurs at high frequencies in microbial populations, and genes are deleted and transferred prior to being fixed in the population. Neutral or nearly neutral genes should not be fixed in a population due to random mutation events (3); therefore, the presence of specific genes in natural populations suggests that the gene products confer a selective advantage in the environment. The presence of nitrogenase genes in natural populations should indicate a selective advantage, yet diverse N2-fixing microorganisms have been found in aquatic environments where N2 fixation has not been detected, such as in hypersaline Mono Lake (29). In this study, we sampled the genetic diversity of nifH-containing microbial populations along the freshwater-to-saltwater gradient in the Chesapeake Bay and one of its subestuaries, the Choptank River, in order to determine how N2-fixing microorganisms are distributed and to begin to identify the factors that contribute to the selection and distribution of N2 fixation genes.
DNA arrays have been developed and applied to study the dynamics of microbial communities and genes encoding enzymes involved in specific biogeochemical processes (“functional genes”) (7, 10, 14, 18, 25, 30, 32-34), including nitrogenase (32, 28). We used a recently developed DNA macroarray approach, which is a simple-to-construct membrane spotted with PCR products (28) to compare the distribution of nifH gene phylotypes in samples from a transect of surface water samples from the Chesapeake Bay.
MATERIALS AND METHODS
Chesapeake Bay and Choptank River sample collection.
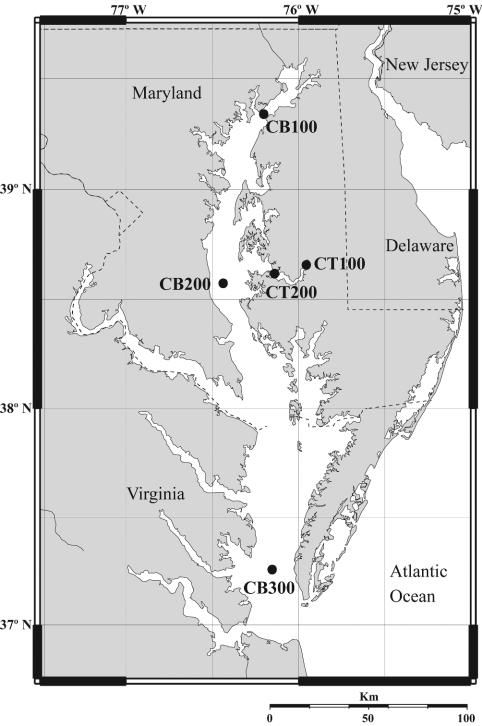
Water samples were collected from three stations in the Chesapeake Bay on cruises aboard the R/V Cape Henlopen with Niskin bottles (General Oceanics, Miami, Fla.) mounted on a rosette (Fig. 1). Choptank River samples from two stations (Fig. 1) were collected with a peristaltic pump with tubing attached to a conductivity, temperature, and depth meter. The dates on which samples were obtained, the depths at which samples were obtained, and the names by which the samples are referenced are indicated in Table 1. Samples were filtered onto 0.22-μm Sterivex-GV capsules (Millipore, Billerica, Mass.) with a peristaltic pump until flow diminished, resulting in filtration volumes of 180 to 1,500 ml within approximately 10 to 20 min. Following filtration, water was purged from the filters, and the filters were frozen immediately in liquid nitrogen and then stored at −80°C prior to extraction.
FIG. 1.
Map of the Chesapeake Bay (CB) and Choptank River (CT), indicating the sampling stations surveyed in this study. Station CB100 is located at 39°21′N, 76°11′W; CB200 is located at 38°34′N, 76°27′W; CB300 is located at 37°18′N, 76°09′W; CT100 is located at 38°48′N, 75°55′W; and CT200 is located at 38°37′N, 76°08′W.
TABLE 1.
Samples extracted and amplified for clone library construction and macroarray experimentsa
| Location | Date (mo/day/yr) | No. of clones sequenced | Depth (m) | Temp (°C) | Salinity (PSU) | NH4 (μM) | NO3 (μM) | NO2 (μM) | Array data |
|---|---|---|---|---|---|---|---|---|---|
| CB100 | 4/4/2001 | 39 | 1.8 | 7.3 | 0.4 | 8.4 | 88.7 | 0.8 | Yes |
| 4/4/2001 | 7 | 4.8 | 7.2 | 0.5 | N/A | N/A | N/A | Yes | |
| 4/4/2001 | 9 | 9.7 | 6.7 | 4.3 | 9.9 | 77.0 | 0.6 | ||
| CB200 | 4/5/2001 | 73 | 1.8 | 8.3 | 10.1 | 2.5 | 43.0 | 0.6 | Yes |
| 4/5/2001 | 7 | 11.2 | 7.4 | 16.5 | N/A | N/A | N/A | Yes | |
| 4/5/2001 | 8 | 17.6 | 7.2 | 18.6 | 4.5 | 2.4 | 0.2 | ||
| CB300 | 4/6/2001 | 10 | 1.9 | 9.3 | 23.5 | 0.4 | 1.5 | 0.1 | Yes |
| 4/6/2001 | 9 | 8.3 | 8.8 | 23.1 | N/A | N/A | N/A | Yes | |
| 4/6/2001 | 8 | 11.3 | 8.7 | 24.1 | 2.0 | 1.1 | 0.1 | ||
| CT100 | 4/3/2001 | 9 | 1 | N/A | N/A | 15.5 | 69.0 | 0.9 | |
| 7/11/2000 | 9 | 0.9 | N/A | N/A | 1.2 | 35.7 | 0.2 | ||
| CT200 | 7/11/2000 | 9 | 0.9 | N/A | N/A | 2.9 | 0.0 | 0.1 | |
| 7/11/2000 | 10 | 7.9 | N/A | N/A | 10.4 | 0.7 | 0.1 |
PSU, practical salinity units; N/A, data not available. Array data means the sample was subjected to analysis by DNA macroarrays shown in Fig. 3.
Genomic DNA extraction.
STE buffer (1.8 ml; 20% sucrose, 50 mM Tris-HCl, 50 mM EDTA) containing lysozyme (5 mg/ml) was added directly to the filters. The filters were incubated at 25°C for 1 h, followed by addition of proteinase K (2 mg/ml, final concentration) and sodium dodecyl sulfate (SDS) (1%, final concentration) and incubation at 60°C for 1 h. Liquid was removed from the filter with a syringe and transferred to a Corex (Corning) tube. Filters were rinsed with 1 ml of TE (10 mM Tris-HCl, 1.0 mM EDTA, pH 8.0), and the rinses were pooled with their corresponding samples. RNase cocktail (Ambion, Austin, Tex.; 2.5 U/ml, final concentration) was added, and the samples were incubated for 10 min at room temperature. Ammonium acetate was added to 2 M final concentration, and samples were centrifuged at 14,800 × g for 6 min to precipitate proteins. The supernatant was transferred to a new tube, followed by the addition of 2 volumes of ethanol. Nucleic acids were precipitated at −20°C for 30 min, followed by centrifugation at 12,000 × g for 35 min at 4°C. Pellets were washed with 70% ethanol and resuspended in 500 μl of TE. Samples were extracted with an equal volume of phenol-chloroform-isoamyl alcohol, 25:24:1, followed by an equal volume of chloroform. The extracted supernatant was precipitated with 100% ethanol and 0.3 M sodium acetate. Samples were further purified with the DNeasy mini-kit (Qiagen, Valencia, Calif.) following the manufacturer's instructions.
PCR amplification, cloning, and sequencing of nifH genes.
A fragment of the nifH gene (approximately 360 bp) was amplified with a nested PCR strategy. First-round reactions were performed with the primers described previously (29). The genomic DNA extract (15 to 40 ng) was added to PCR mixtures containing 5 μl of 10× ExTaq buffer (Takara, Madison, Wis.), 4 μl of a mix of deoxynucleoside triphosphates (2.5 mM each), 37.5 μl of water, 0.5 μl of 100 μM nifH32F (5′-TGAGACAGATAGCTATYTAYGGHAA-3′), 0.5 μl of 100 μM nifH623R (5′-GATGTTCGCGCGGCACGAADTRNATSA-3′), and 0.5 μl of ExTaq DNA polymerase (5 U/μl; Takara, Madison, Wis.). The reaction mixtures were amplified for one denaturation step (5 min at 94°C), followed by 30 cycles of 94°C for 1 min, 50°C for 1 min, and 72°C for 1 min, and one final 7-min extension cycle at 72°C. One microliter of this reaction product was transferred into a second 50-μl reaction mixture containing the same reagent mixture with an additional 1 μl of water and the internal nifH primers designed previously (37): 0.5 μl of 100 μM nifH1 (TGYGAYCCNAARGCNGA) and 0.5 μl of 100 μM nifH2 (ADNGCCATCATYTCNCC).
Second-round reactions were amplified with one denaturation step of 5 min at 94°C, 30 cycles of 94°C for 1 min, 57°C for 1 min, and 72°C for 1 min, and a final 7-min extension step at 72°C. PCR products from this reaction were gel purified with the QIAquick gel extraction kit (Qiagen, Valencia, Calif.) and cloned into a pGEM T-system II vector (Promega, Madison, Wis.). Plasmids were purified with the QIAprep spin miniprep kit (Qiagen, Valencia, Calif.), and nifH fragments were cycle sequenced in both directions with T7 and Sp6 primers and BigDye version 3 chemistry on an ABI Prism 310 or 3100 sequencer (Applied Biosystems, Foster City, Calif.).
Phylogenetic analysis and selection of sequences for array construction.
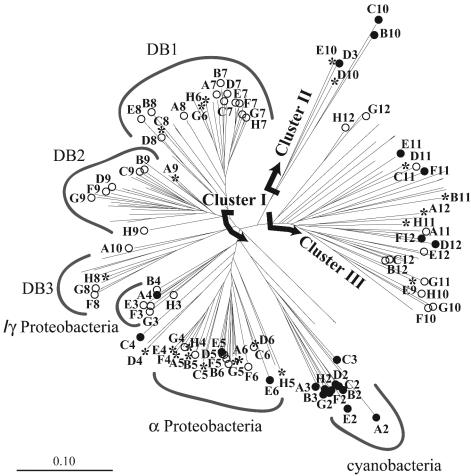
The Chesapeake Bay and Choptank River sequences were aligned with other nifH sequences from GenBank with a nifH Pfam and the HMMER algorithm (29). The output from the Hammer alignment was brought into an ARB database (www.arb-home.de/), and a neighbor-joining tree was generated (Fig. 2). Sixty-five clones from the Chesapeake Bay and Choptank River libraries that represented the range of diversity found in those environments were identified. Clones were chosen so that on average each shared 87% sequence identity with the next nearest selected clone (range, 73 to 96%). An array was prepared with the 65 Chesapeake and Choptank environmental clones, two Pacific Ocean environmental clones representing cyanobacteria (38), and clones from 21 cultivated microorganisms (28) (Table 2).
FIG. 2.
nifH phylogenetic tree constructed with all nifH sequences recovered from the Chesapeake Bay and Choptank River clone libraries and sequences from cultivated microorganisms spotted on the macroarray. Letters indicate the array coordinates of the sequences printed on the DNA macroarray. Solid circles indicate sequences from cultivated microorganisms, open circles indicate sequences from the Chesapeake Bay, and asterisks indicate sequences from the Choptank River. Deeply branching phylogenetic groups that contain no sequences from cultivated microorganisms are indicated (DB1, DB2, and DB3). The neighbor-joining tree was constructed with aligned ≈330-bp nifH DNA fragments and the ARB software package (www.arb-home.de/).
TABLE 2.
Positions, names, and GenBank accession numbers of sequences printed on the macroarraya
| Row | Column
|
|||||
|---|---|---|---|---|---|---|
| 1a | 2 | 3 | 4 | 5 | 6 | |
| A | 2.500 | Arcobacter nitrofigilis (DSMZ 7299), AY221825 | Oscillatoria sancta (PCC 7515), AY221815 | Azotobacter chroococcum (NRRL B-14637), AY351672 | CB916H1, AY224034 | u_CB916H2, AY224035 |
| B | 0.833 | Cyanobacterial isolate WH001, AY221818 | Symploca sp. (PCC 8802), AY221816 | CB894H8, AY223950 | CB895H8, AY223957 | u_CB907H6, AY223986 |
| C | 0.278 | Cyanobacterial isolate WH8501, AY221821 | Vibrio diazotrophicus (DSMZ 2604), AY221828 | Frankia sp. (DSMZ 43829), AY351671 | CB891H8, AY223911 | u_CB912H5, AY224023 |
| D | 0.093 | HT1903, AF299422 | Paenibacillus azotofixans (DSMZ 5976), AY221826 | CB911H3, AY224015 | CB907H2, AY223968 | Chlorobium limicola (DSMZ 245), AY221831 |
| E | 0.031 | HT1902, AF299420 | CB894H2, AY223945 | CB891H1, AY223907 | CB907H9, AY223997 | u_CB909H2, AY224001 |
| F | 0.010 | Anabaena cylindrica (UTEX 629) AY221813 | CB909H9, AY224005 | CB891H9, AY223909 | Xanthobacter flavus (NRRL B-14838), AY221812 | Pelodictyon lutcolum (DSMZ 273), AY22183 |
| G | 0.003 | Tolypothrix sp. (PCC 7101), AY221817 | Klebsiella oxytoca (NRRL B-199), AY221827 | CB894H10, AY223944 | CB891H3, AY223908 | u_CB910H2, AY224007 |
| H | 0.000 | Nostoc muscorum (UTEX 486), AY221814 | CB909H5, AY224002 | CB914H1, AY224026 | CB921H3, AY224040 | u_CB912H1, AY224020 |
Nanograms of biotinylated λ DNA printed at each position (A1 to H1).
Table 2a.
| Column | |||||
|---|---|---|---|---|---|
| 7 | 8 | 9 | 10 | 11 | 12 |
| CB910H3, AY224008 | CB909H8, AY224004 | CB891H5, AY223910 | CB895H3, AY223954 | u_CB910H5, AY224010 | u_CB916H2, AY224035 |
| CB910H8, AY224012 | CB911H1, AY224014 | CB894H6, AY223948 | Methanothermobacter thermoautotrophicum (DSMZ 1850), AY221829 | u_CB914H8, AY224032 | u_CB907H6, AY223986 |
| CB910H4, AY224009 | CB914H6, AY224030 | CB895H2, AY223953 | Methanococcus vannielli (DSMZ 1224), AY221830 | u_CB909H1, AY224000 | u_CB912H5, AY224023 |
| CB910H6, AY224011 | CB907H10, AY223960 | CB895H4, AY223955 | CB921H10, AY224038 | u_CB914H3, AY224028 | Chlorobium limicola (DSMZ 245), AY221831 |
| CB907H5, AY223979 | CB907H4, AY223976 | CB914H7, AY224031 | CB921H8, AY224043 | Desulfobacter latus (DSMZ 3381), AY221822 | u_CB909H2, AY224001 |
| CB912H4, AY224022 | CB894H4, AY223946 | CB895H9, AY223958 | CB911H9, AY224019 | Desulfotomaculum mgrificans (DSMZ 574), AY221823 | Pelodictyon lutcolum (DSMZ 273), AY22183 |
| CB910H10, AY224006 | CB895H1, AY223952 | CB895H10, AY223951 | CB912H2, AY224021 | u_CB912H9, AY224025 | u_CB910H2, AY224007 |
| CB911H10, AY224013 | CB921H1, AY224039 | CB894H5, AY223947 | CB911H7, AY224018 | u_CB921H4, AY224041 | u_CB912H1, AY224020 |
Macroarray construction and target hybridization.
nifH sequences used as array probes were amplified from the cloning site of the pGEM-T vector with the Sp6 and T7 primers in a 50-μl PCR containing 1 μl of miniprep DNA, 1 μl of each primer (SP6 and T7) at 25 μM, 5 μl of 10× ExTaq buffer, 4 μl of deoxynucleoside triphosphate mix (2.5 mM each), 0.5 μl of ExTaq DNA polymerase (Takara), and 38 μl of water. The reactions were amplified with a denaturation step of 5 min at 94°C, followed by 30 cycles of 94°C for 30 s, 42°C for 30 s, and 72°C for 30 s, and a final 3-min extension cycle at 72°C. PCR products were gel purified with the QIAquick gel extraction kit, and DNA was quantified with PicoGreen (Molecular Probes, Eugene, Oreg.) and a Cary spectrofluorometer (Varian, Palo Alto, Calif.). DNA for array probes was diluted in TE to a concentration of 2.5 ng/μl. Control DNA was prepared with biotinylated lambda phage DNA (see below) serially diluted in both TE and unlabeled lambda DNA so that the DNA concentration for each standard remained constant at 2.5 ng/μl and the concentration of the labeled portion varied (2.5, 0.83, 0.28, 0.09, 0.03, 0.01, 0.003, and 0 ng/μl). Probes and control DNA were spotted onto nylon membranes (SuPerCharge; Schleicher & Schuell, Keene, N.H.) with a hand-operated, 96-pin tool that delivers 1 μl per pin (V&P Scientific, San Diego, Calif.). Control DNA (biotinylated phage lambda) was spotted to serve as a positive control for signal detection. Spotted membranes were denatured by incubating the membrane (printed side up) for 10 min on sheets of blotting paper soaked with 3 M NaCl and 0.4 N NaOH. Membranes were neutralized by placing them on blotting paper soaked with 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 5 min, followed by paper soaked with 1× SSC for 1 min. Wet membranes were placed face down on plastic wrap and exposed face down on a UV transilluminator for 2 min. Membranes were air dried and stored at room temperature before hybridization.
Biotin labeling of target and control DNA.
Target DNA for hybridization was amplified from genomic DNA with the nested PCR strategy described above. For a few selected samples (CB100 1.8 m, CB200 1.8 m, and CB300 1.9 m), we took extra precautions to minimize variation between independent PCRs from each sample. In these cases, the first-round reactions for each sample were conducted in triplicate and then pooled; 1 μl of the pooled reaction product was transferred into each of triplicate second-round reactions, and the products of these reactions were pooled. In all amplifications, the second-round reaction products were gel purified with the QIAquick gel extraction kit (Qiagen). DNA was quantified with PicoGreen (Molecular Probes) and a Cary spectrofluorometer (Varian). Target DNA was labeled with biotin at a concentration of 10 ng/μl in a 20-μl volume with the BrightStar psoralen-biotin kit (Ambion). Targets were heat denatured in a thermocycler for 12 min at 99°C, placed directly on ice, and protected from light. Psoralen-biotin reagent (4 μl) was added, and the mixture was UV irradiated at 365 nm for 45 min in the dark. TE was added to a total volume of 100 μl, and unincorporated biotin was removed with two extractions of 200 μl of water-saturated butanol. Control lambda DNA was labeled with the same method at an initial concentration of 50 ng/μl.
Array hybridization and quantification of relative signal intensity.
Membranes were prehybridized in a thermally sealed bag with 5 ml of hybridization buffer (1 mM EDTA, 6% SDS, 250 mM sodium phosphate [pH 7.2], and 40% formamide) and mixed gently for 1 h at 60°C. Biotinylated targets were heat denatured for 12 min at 99°C, cooled directly in an ice-water bath, and added to 5 ml of 60°C hybridization buffer at a final concentration of 10 ng/ml. Prehybridization solution was removed from the bag and replaced with the hybridization solution containing the labeled target. Blots were hybridized overnight (10 to 12 h) with rocking, washed twice (2× SSC-1% SDS) for 5 min and twice (0.1× SSC and 1% SDS) for 15 min at 60°C, and twice (1× SSC) at room temperature for 5 min. Blocking and detection reagents were part of the Southern-Star chemiluminescence detection system (Applied Biosystems, Foster City, Calif.). Blots were rinsed twice for 5 min in blocking buffer (1× phosphate-buffered saline, 0.2% I-Block reagent, 0.5% SDS) and then incubated with a 1:2,000 dilution of AvidX-AP in blocking buffer for 20 min. Blots were then washed once in blocking buffer for 5 min, three times in 1× PBS-0.5% SDS for 5 min, and twice in assay buffer for 2 min. CDP-Star (2.5 ml) was added to each blot. Blots were incubated for 30 min prior to exposure to X-ray film (Biomax Light; Kodak). Film images were scanned on a flatbed scanner and saved as TIFF images in Adobe Photoshop. Pixel intensity was quantified with the ImageQuant version 5.0 software package (Amersham Biosciences, Piscataway, N.J.). Signal for each spot was expressed as a percentage of the most intense spot on each macroarray, since the intensity of the standard curve on each blot varied between experiments. The normalized signal intensity data from the blots were analyzed by cluster analysis based on Euclidean distances with single-linkage clustering (Data Desk version 6; Data Description Inc., Ithaca, N.Y.) to determine the similarity of replicates in comparison to differences between stations.
Nucleotide sequence accession numbers.
The sequences from the Chesapeake Bay and Choptank River nifH clone libraries were submitted to GenBank (accession numbers AY223907 to AY224045).
RESULTS
We recovered nifH sequence types in three major phylogenetic groups (clusters I, II, and III) from different stations and depths in the Chesapeake Bay and the Choptank River (Fig. 1). The majority of Chesapeake Bay and Choptank River sequences are in cluster I (Fig. 2), a large phylogenetic group containing sequences from cultivated proteobacteria, cyanobacteria, and Firmicutes and sequences from a variety of aquatic and terrestrial habitats (36). There are also Chesapeake Bay nifH sequences in cluster III, a phylogenetic group containing sequences from anaerobic bacteria such as sulfate reducers and clostridia (Fig. 2). Some Chesapeake Bay sequences in this cluster are similar to sequences recovered from Chesapeake Bay sediment samples (6) and, in one case, nearly identical (99% identity). Only one Choptank River station (CT200) had sequences in cluster III. Chesapeake Bay and Choptank River cluster III nifH sequences were less than 81% identical to sequences obtained from cultivated organisms (Table 3). A few Choptank River nifH sequences grouped in cluster II (Fig. 2), a phylogenetic cluster that contains sequences from archaea and the second alternative, non-vanadium-, non-molybdenum-containing Fe protein genes (anfH). The Neuse River is the only other aquatic environment with nifH sequences represented in this cluster.
TABLE 3.
Phylogenetic identity of probes that hybridized to Chesapeake Bay targets based on genetic distances to sequences from cultivated microorganisms
| Array position | Probe | GenBank accession no. | Blots with signal for corresponding probe | Closest cultivated relatives | Bacterial group | Accession no. | Genetic distance from probe |
|---|---|---|---|---|---|---|---|
| F3 | CB909H9 | AY224005 | CB200, CB300 | Azotobacter vinelandii | γ | M20568 | 0.13 |
| Pseudomonas stutzeri (A15b) | γ | AJ297529 | 0.14 | ||||
| H3 | CB909H5 | AY224002 | CB100, CB200, CB300 | Marichromatium purpuratum (95Carr100a) | γ | AF059648 | 0.10 |
| Pseudomonas stutzeri (A15) | γ | AJ297529 | 0.11 | ||||
| A5 | CB916H1 | AY224034 | CB100 | Burkholderia sp. strain STM678 | β | AJ302315 | 0.12 |
| Herbaspirillum seropedicae Z78 | β | Z54207 | 0.13 | ||||
| B5 | CB895H8 | AY223957 | CB100 | Herbaspirillum seropedicae Z78 | β | Z54207 | 0.14 |
| C5 | CB891H8 | AY223911 | CB100, CB200 | Methylosinus sp. strain LW3 | α | AF378720 | 0.15 |
| Herbaspirillum seropedicae Z78 | β | Z54207 | 0.15 | ||||
| E4 | CB891H1 | AY223907 | CB100 | Methylosinus sp. strain LW4 | α | AF378721 | 0.14 |
| Methylosinus sp. strain LW3 | α | AF378720 | 0.15 | ||||
| G4 | CB894H10 | AY223944 | CB100 | Xanthobacter flavus NRRL B-14838 | α | AY221812 | 0.15 |
| H4 | CB914H1 | AY224026 | CB100 | Xanthobacter flavus NRRL B-14838 | α | AY221812 | 0.12 |
| Rhizobium sp. strain ORS571 | α | M16710 | 0.13 | ||||
| D5 | CB907H2 | AY223968 | CB200 | Rhizobium sp. strain ORS571 | α | M16710 | 0.12 |
| Xanthobacter flavus NRRL B-14838 | α | AY221812 | 0.13 | ||||
| E5 | CB907H9 | AY224005 | CB100, CB200 | Methylocystis sp. LW5 | α | AF378719 | 0.12 |
| Methylosinus trichosporium OB3 | α | AF378724 | 0.12 | ||||
| Rhizobium sp. strain ORS571 | α | M16710 | 0.12 | ||||
| Xanthobacter flavus NRRL B-14838 | α | AY221812 | 0.12 | ||||
| G5 | CB891H3 | AY223908 | CB100, CB200, CB300 | Methylosinus trichosporium OB3b | α | AF378724 | 0.07 |
| Methylocystis sp. strain LW5 | α | AF378719 | 0.08 | ||||
| H5 | CB921H3 | AY224040 | CB100, CB200 | Methylocystis sp. strain LW5 | α | AF378719 | 0.14 |
| A6 | CB921H7 | AY224042 | CB100, CB200, CB300 | Methylocystis sp. strain LW5 | α | AF378719 | 0.10 |
| Methylosinus trichosporium OB3b | α | AF378724 | 0.10 | ||||
| B6 | CB909H7 | AY224003 | CB100, CB200 | Beijerinckia indica subsp. indica KS1-7 | α | AF296350 | 0.08 |
| Beijerinckia indica subsp. indica KS1-7 | α | AF296353 | 0.08 | ||||
| C6 | CB895H7 | AY223956 | CB100 | Rhodobacter sphaeroides 16PHC | α | AF031817 | 0.13 |
| Rhodospirillum rubrum | α | M33774 | 0.12 | ||||
| D6 | CB914H5 | AY224029 | CB100, CB200 | Rhodobacter sphaeroides 16PHC | α | AF031817 | 0.09 |
| Azospirillum lipoferum Sp59b | α | AF216882 | 0.10 | ||||
| F6 | CB908H5 | AY223998 | CB100, CB200 | Azospirillum lipoferum Sp59b | α | AF216882 | 0.12 |
| Gluconacetobacter diazotrophicus PAL5 | α | AF030414 | 0.12 |
DNA macroarray analysis of nifH sequence diversity in surface water samples along a Chesapeake Bay transect.
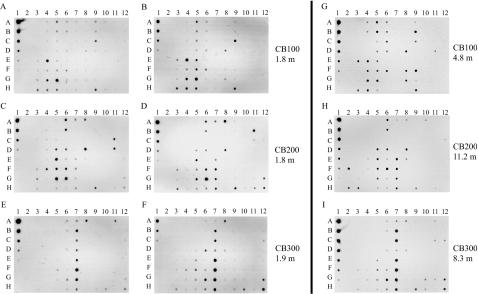
Arrays were hybridized in duplicate with targets amplified from three surface water samples (Fig. 3A to F) to test the repeatability of the procedure. The results for each pair of blots represent completely independent analyses by two different researchers, including independent amplification and labeling of the targets and independent hybridizations. For these comparisons, the targets were prepared from pooled triplicate PCR amplifications as described in Materials and Methods. In general, hybridization patterns were similar between duplicate target samples and clustered together when the array data were analyzed by cluster analysis (Fig. 4D), though some array-to-array differences in overall signal intensity are evident from the differences in the intensity of the standard dilution series (Fig. 3A to F, column 1, A to H).
FIG. 3.
Hybridization of nifH macroarray to targets generated from filtered water samples collected at stations CB100 (A and B, sample from 1.8 m; G, sample from 4.8 m), CB200 (C and D, sample from 1.8 m; H, sample from 11.2 m), and CB300 (E and F, sample from 1.9 m; I, sample from 8.3 m). The duplicate blots for each surface water station (e.g., panels A and B) represent independent replicate PCR amplifications of the target samples and independent blot hybridizations. The targets shown in panels A to F were from three pooled amplification products. The targets shown in panels G to I were from individual amplification products.
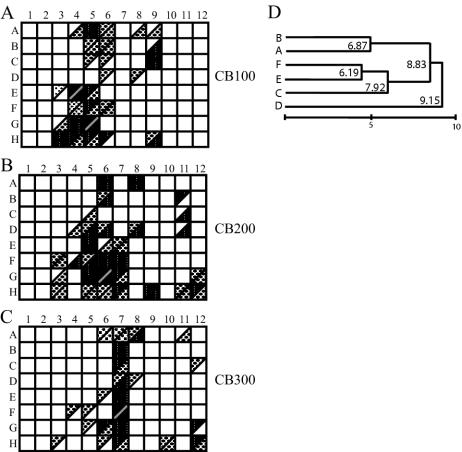
FIG. 4.
Quantification of signal intensity from macroarray hybridizations. Relative signal intensity (calculated from the experiments shown in Fig. 3A to F) was expressed as a percentage of the most intense spot on each array. Probes with 80 to 100% of the intensity of the most intense signal are indicated with solid triangles. Probes with 20 to 80% of the intensity of the most intense signal are indicated with medium gray triangles. Probes with 9 to 20% of the intensity of the most intense signal are indicated with light gray triangles. Boxes with equivalent shading represent similar signal levels on the duplicate blots. Divided squares indicate that different signal intensities were obtained from the duplicate blots. (D) Cluster analysis of the raw data from replicated surface sample blots, showing clustering of replicates (A and B, CB100; C and D, CB200; and E and F, CB300) and differences between samples. Replicates from the mid-depth station were the most variable and did not cluster together. Euclidian distances to the cluster node at the right end of the scale bar are indicated.
The observed trend in hybridization pattern at each sample site was consistent between the replicated surface water samples and samples collected deeper in the water column on the same date (Fig. 3). Although the targets from the deeper samples were not pooled as were the surface water samples, the overall hybridization pattern was very similar (Fig. 3). Therefore, the trends with sampling station appear to be robust for both sample replicates and independent samples collected at the same site.
The blots replicated hybridization intensity fairly well. In order to simplify the analysis of the macroarray data, the spot intensity of the blot images was quantified (shown in Fig. 3A to F). The signal for individual spots was expressed as a percentage of that of the most intense spot on each macroarray, since the standard curve on each blot did not control for hybridization kinetics and the exposure intensity varied between experiments. Figure 4 shows a graphic comparison of the quantified signal intensity between the duplicate experiments at each station, binned for hybridization intensity into low, medium, and high signals. Boxes with the same shading in each half indicate that signals were consistent between blots. For example, the signal intensity was similar for probes A5 and A6 between replicate arrays hybridized with CB100 1.8 m (Fig. 4A). A number of probes had hybridization signals at different relative intensities on the duplicate blots, indicated by different shading in each half of the divided squares (Fig. 4). For example, probe H9 had a stronger hybridization signal on one of the CB100 replicate blots than on the other (Fig. 4A). Although the signal from each probe was not always the same binned intensity between duplicate experiments, the general hybridization levels were similar, which was confirmed by cluster analysis of the raw unbinned data (Fig. 4D). Probe signals in the highest ranges are comparable between blots, and the general trend of probes that hybridized was similar in the duplicate experiments.
The most intense hybridization signals from surface water samples at the head of the estuary (station CB100 1.8 m) were to sequence types that clustered near cultivated alpha-proteobacteria (positions E4 and G5, Fig. 3A, B; Fig. 4A; Table 3). There was also some hybridization to probes that are similar to cultivated beta and gamma Proteobacteria (positions H3 and A5, Fig. 3A and B; Fig. 4A; Table 3). Hybridization was also detected for some of the probes (column 9, positions A to C and G to H, Fig. 3A and B; Fig. 4A) from a group of uncultivated cluster I sequence types, designated DB1 in Fig. 2. Some of these hybridization signals were detected at station CB100 at both 1.8 and 4.8 m (Fig. 3A, B, and G; Fig. 4A, column 9, A to C) and not the other stations. The community composition was different at station CB200, located at the middle of the estuary, adjacent to the outflow from the Choptank River. The arrays indicate that sequences in a group of uncultivated cluster I sequence types designated DB1 in Fig. 2 are the majority at CB200 1.8 m (position G6, column 7 E to H, A8, and D8 in Fig. 3C and D; Fig. 4B) as well as sequences in the alpha-proteobacteria group (column 5, D to H, and column 6, A, B, and D to F in Fig. 3C and D; Fig. 4B; Table 3). There were also a few beta- and gamma-proteobacteria sequence types detected on the CB200 1.8 m arrays (positions F3 and H3, Fig. 3C and D; Fig. 4B; Table 3). In addition, the CB200 1.8-m samples hybridized to cluster III sequences on the array (column 11, B to D, and column 12, G to H, in Fig. 3C and D; Fig. 4B). The macroarray hybridizations indicate that there may be a substantially different diazotroph community at station CB300 compared to CB200 and CB100. Most of the CB300 sample hybridization at 1.9 m and 8.3 m was to probes from uncultivated cluster I sequence types indicated as DB1 on Fig. 2 (column 6, G and H, and column 7, A to H, position A8, in Fig. 3E, F, and I; Fig. 4C), as well as some cluster III sequence types (positions H10, A11, and H12, Fig. 3E, F, and I; Fig. 4C).
The arrays indicate shifts in relative abundance of sequence types within phylogenetic groups between stations, based on the hybridization intensity of the PCR-amplified target. In particular, the alpha-proteobacteria probes that hybridized to CB100 (column 4, E to H, positions A5 and B5 in Fig. 3A, B, and G; Fig. 4A) are much less abundant at CB200 and CB300 (Fig. 3C, D, E, F, H, and I; Fig. 4B and C). In contrast, alpha-proteobacteria probe sequences (positions A6, B6, and D6) are more abundant at CB200 than at CB100 or CB300 (Fig. 3C, D, and H; Fig. 4B).
Comparison of clone library composition with DNA macroarray results.
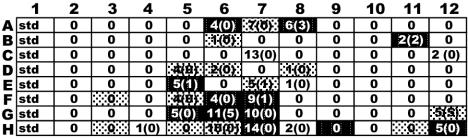
In order to assess how well DNA macroarrays assay community composition compared to nifH clone library sequence analysis, 63 clones from a nifH library were sequenced from the CB200 target that was hybridized to the array (target hybridized in Fig. 3D). Preliminary experiments showed that sequences with greater than ≈85% DNA sequence identity hybridized to probes on the macroarray under the stringency conditions used in the experiments (28). Figure 5 shows the number of CB200 clone library sequences predicted to hybridize to each array probe at two different levels of sequence identity (85% and 95%), based on distances calculated between CB200 clone library sequences and the probe sequences on the macroarray. It should be noted that, although sequences will hybridize at 85% identity, the signal is substantially weaker than that generated by sequences of greater than 95% identity (28). The grayscale shading in Fig. 5 indicates the range of signal intensity measured for each probe hybridized to the CB200 target.
FIG. 5.
Comparison of array signal intensity with sequence composition of nifH clone library from station CB200. Shading corresponds to the levels in Fig. 4 (low, light gray; medium, dark gray; and high, black). The numbers at each array position are the number of CB200 nifH sequences in the clone library that were ≥85% identical to the probe sequences. Numbers in parentheses indicate the number of CB200 clones that were 95% or greater identical to the probe sequences.
There is good agreement between the predicted and observed probe hybridization signals. In a few instances, probes were predicted to hybridize with target sequences at 85% to 95% identity and there was no detectable array signal (Fig. 5, white boxes with numbers). When target sequences were greater than 95% identical to the probe, there was always a corresponding hybridization signal (Fig. 5, positions E5, G6, E7, D7, A8, B2, and G12). There was less agreement between the abundance of individual sequence types predicted to hybridize to a probe and the signal intensity observed for that probe (Fig. 5). For example, the most intense array hybridization signal corresponded to position G6, and 5 of 62 sequences from the clone library were predicted to hybridize to the probe at 95% identity. In contrast, 5 of 62 sequences from the clone library were predicted to hybridize to probe G11 at 95% identity, but only weak hybridization was observed. A large fraction (40%) of the clones obtained in the library were less than 85% identical to the probes on the array and would not be predicted to generate any hybridization signal.
DISCUSSION
nifH sequence diversity in the Chesapeake Bay and Choptank River.
The Chesapeake Bay and Choptank River clone libraries generated from water column samples show that there is a high diversity of potential N2-fixing microorganisms in the estuary. In general, the nifH phylotypes detected in the Chesapeake and Choptank have also been reported from other aquatic environments, including estuaries, freshwater lakes, hypersaline lakes, salt marshes, and microbial mats (1, 17, 19, 27, 35). The distribution of sequence types from the Chesapeake Bay is very similar to that found in the Neuse River estuary (1). Many Chesapeake and Choptank River sequences group in cluster I (Fig. 2, DB1, DB2, and DB3), which contains only sequences recovered from environmental studies of estuaries, lakes, and marshes and associated with sea grasses and rice roots (clusters 1A and 1C [36]). Other sequences group with sequences from cultivated proteobacteria that have only thus far been reported from saline aquatic environments and marshes (cluster 1N [36]). Some Chesapeake Bay and Choptank River sequences grouped with sequences found in both fresh and saline aquatic environments and in microbial mats, marshes, and soils (clusters 1J and 1K [36]). Additionally, there are a number of Chesapeake Bay sequences that group with cluster III nifH sequence types that have not been reported from the open ocean but are found in freshwater aquatic environments, marshes, and microbial mats and are associated with sea grasses (36). The different clusters may represent organisms that are autochthonous to the water column or allochthonous from terrestrial runoff.
Interestingly, no cyanobacterial nifH sequence types were found in the Chesapeake Bay or the Choptank River sequence libraries. The Chesapeake Bay is a shallow estuary (the bottom at the deepest station CB200 is 18.6 m), and our clone libraries include samples taken 1 m from the bottom at all stations. Thus, the water column N2-fixing communities may represent both organisms selected in the water column and those transported or resuspended from benthic sediments. In this study, particle-attached versus free-living assemblages were not differentiated, and it could be that the different clusters also occupy different microhabitats in the water column.
The high diversity of nifH sequences in the Chesapeake Bay makes it very difficult to use clone library data to compare diazotroph community composition between stations, depths in the water column, or seasons. Clone libraries would have to be large and exhaustively sequenced in order to provide reproducible, statistically meaningful comparative analyses. We used clone library data from different stations and depths to construct a DNA macroarray with probes that represent the major groups of phylogenetic diversity in the clone libraries. Our nifH database, although large (over 2,000 sequences) at the beginning of this study, had few sequences that were >85% identical to the clones recovered in the Chesapeake Bay clone libraries. Therefore, new libraries are necessary to develop array probes that can detect the most abundant nifH sequences from the Chesapeake Bay.
Limitations of DNA macroarrays as a fingerprinting technique.
Any probing technique based on PCR amplification is subject to the effects of PCR bias (31), but the sensitivity provided by amplification via PCR or reverse transcription-PCR is necessary for many environmental applications. Regardless of potential bias in amplification, the array hybridization was generally reproducible when PCR products from multiple reactions were pooled. The arrays with PCR products as probes can distinguish sequence types within approximately 85% identity (28, 34), making it possible to detect target sequences in the environment even if they have not been sequenced previously. However, hybridization of target samples to the probes provides only relative phylogenetic information, since the probes cross-hybridize to target sequences within 85% identity (28). We have found the arrays to be semiquantitative (28), since sequences that are very similar to the probe will generate stronger signals than less similar sequences.
It is difficult to comprehensively analyze the contributions to signal intensity from cross-hybridization because the signal intensity generated from abundant, low-similarity sequences cannot necessarily be resolved from less abundant but highly similar sequence types. In some cases, the range of sequence types that can cross-hybridize to a specific probe will not necessarily precisely define the phylogenetic affiliation of an environmental nifH sequence. For example, it is difficult to distinguish some beta-proteobacterial from gamma-proteobacterial nifH sequences, since sequences from some cultivated isolates in these groups are more than 85% identical. Since sequences within 15% sequence identity cannot be resolved with the probes, microheterogeneity within populations cannot be detected. However, the arrays are useful for detecting patterns of phylogenetic association (within approximately 85% identity) and are useful for comparing community composition between samples. It is not likely to be possible to create arrays that include specific probes that cover the entire diversity of nifH, particularly in environments that have not yet been characterized by intensive sequencing efforts. Thus, the cross-hybridization at 85% can actually facilitate the characterization of patterns of diversity and is one of the benefits of the macroarrays with longer PCR products as probes.
Comparison of array hybridization pattern to clone library composition.
To compare how well the array hybridization patterns and clone library data correspond, a clone library from one of the target samples (CB200) was extensively sequenced. The fact that 40% of the sequences in this library would not be detected by the array highlights the importance of obtaining sequence information from the environment of interest prior to constructing the array. Furthermore, it is unlikely that an array made from sequences from cultivated isolates or one environment will perform well when hybridized to samples from other environments. Although there was good correlation between the composition and diversity of the nifH clone libraries and the array hybridization patterns, there were discrepancies between the number of sequences predicted to hybridize to each array probe and the observed hybridization signal intensity. These discrepancies could be due to several factors, including G+C content and the positions of mismatches within the sequence (28). We obtained 43 unique nifH sequence types (sequences with less than 98% identity) from the CB200 library containing 62 sequences. Rarefaction analysis of these data demonstrates that we did not saturate the possible numbers of unique sequences that could be obtained in this library (data not shown). Therefore, the numbers of sequences predicted to hybridize to each probe may not reflect overall distribution in the clone library.
Macroarray analysis reveals shifts in diazotroph community composition along the Chesapeake Bay estuary.
Array hybridization patterns indicated that there might be differences in overall diazotroph diversity among the sampling stations in the Chesapeake Bay. More probes hybridized to the CB100 and CB200 samples (both at surface and at mid-depth; Fig. 3A, B, and G and Fig. 3C, D, and H, respectively) in comparison to the CB300 samples (Fig. 3E, F, and I). It may be possible that CB300 sequence diversity was less well represented on the array even though the arrays were constructed with phylogenetic information from equal numbers of sequences from CB100, CB200, and CB300 clone libraries (28).
The macroarrays detected shifts in specific diazotroph populations among the sampling stations in duplicate PCR and hybridization experiments from surface water samples (Fig. 3A to F; Fig. 4). The arrays detected a shift from gamma-, beta-, and alpha-proteobacteria subclusters at station CB100 and CB200 to a deeply branching group of cluster I sequences at CB300. The phylogenetic affiliations of these deeply branching nitrogenase genes to cultivated microorganisms cannot be inferred, and they may represent novel lineages or lineages from which nitrogenase genes have yet to be sequenced.
Cluster III nifH probes hybridized to surface water samples from stations CB200 and CB300. Cluster III sequence types were also found in both the Chesapeake Bay and Choptank River nifH clone libraries. The cluster III array probes are closely related only to sequences recovered from the Chesapeake Bay estuary and are less than 85% identical to cluster III sequences reported from other environments. Thus, hybridization to these probes indicates the presence of sequence types that have thus far only been reported from the Chesapeake Bay. This highlights the importance of making arrays tailored to the environment being targeted, as otherwise these sequences may not have been detected by the array.
The array results indicate shifts in the relative abundance of specific phylotypes among stations. The differences in hybridization patterns between stations indicate shifts of species within phylogenetic groups, such as the alpha-proteobacteria, as well as shifts between groups (e.g., Deep Branch 1 and Deep Branch 2 in cluster III). The overall pattern of hybridization from CB100 to CB300 was reflected in experiments with independent samples from the same station but at different depths (Fig. 3G to I). Even though these additional samples were not replicated in PCR or hybridization, they show the same general hybridization pattern and indicate the same overall shifts in diazotroph community composition as the surface water samples. This is consistent with similar water column properties in the surface water and mid-depth samples hybridized in Fig. 3 (compare temperature and salinity in Table 1). Thus, the array hybridization patterns reflect a distinct change in nifH genes along the transect.
The macroarrays are useful for elucidating patterns and identifying individual phylotypes that are present at different relative abundances along the estuary. The probe F7 nifH phylotype (Fig. 3 and 4) is abundant at the mouth of the estuary (CB300), less abundant at CB200, and undetectable at the top of the estuary (CB100). The same trend was found for the absolute abundance of the F7 nifH sequence type between these stations by quantitative PCR (26a).
The high diversity of nifH genes is interesting because the Chesapeake Bay is only nitrogen limited seasonally (11, 12, 20) and N2 fixation in the water column has not been detected (T. Kana, personal communication). The high diversity of nifH sequences in the Chesapeake Bay may indicate that there are microorganisms that persist in the estuary that contain the nifH gene and may not fix nitrogen (36). Alternatively, there may be regions of local N limitation that require organisms to fix N2 at rates that are not currently detectable in bottle experiments. In order to understand the relationships between distributions of microorganisms and the physical-chemical environment, the unifying concept of biocomplexity, the distribution of microorganisms and environmental factors, needs to be investigated over large temporal and spatial scales (2). The shifts in diazotroph populations observed with the arrays may correspond to differences in temperature, salinity, and fixed inorganic nitrogen concentrations between stations (Table 1). Salinity and nutrients are correlated parameters; the freshwater at the northern extreme of the Chesapeake Bay (CB100) has the highest nutrient load and the lowest salinity, while low-nutrient water is found at the southern end of the Chesapeake Bay (CB300) (Table 1). It is unclear which environmental factors cause shifts in estuarine microbial communities, but recent studies in the Choptank River indicate shifts in bacterial communities that correlate with changes in salinity and dissolved oxygen concentration (5, 15).
The apparent correlation in community composition and relative nifH abundance with salinity and nutrients could be caused by either selection or environmental factors or by differences in sources and transport of bacteria from freshwater inputs or sediment resuspension. With arrays in hand, it is now possible to probe estuary water samples with sufficient temporal and spatial resolution to determine the sources and fates of specific diazotroph populations. Furthermore, specific nifH phylotypes can be targeted by quantitative PCR to define more precisely their distribution and potentially even their growth rates and gene expression levels.
The DNA macroarray for nitrogenase was found to be useful for fingerprinting nifH sequence diversity and suggests spatial trends in phylotype distributions in the Chesapeake Bay. Due to the diversity of nifH sequences, arrays need to be tailored to the sequence types found in the environment of interest. Although they cannot provide information about sequence types not represented on the array, they allow rapid comparison of the relative abundance and diversity of environmental sequences. Ultimately, the application of unamplified DNA to arrays, currently not possible due to the detection limits of arrays, will avoid amplification biases and be an even more powerful approach for characterizing microbial functional diversity and ecosystem biocomplexity (2). However, arrays probed with amplification products are particularly useful for identifying specific phylotypes as targets for more quantitative methods, such as quantitative PCR (26a).
Acknowledgments
We thank George Jackson for creating and maintaining the biocomplexity web site (http://snow.tamu.edu/), Pat Glibert and Todd Kana for providing us with nutrient data, and W. Boicourt for CTD data.
This research was supported by NSF biocomplexity grants to J.P.Z. (OCE 9981437) and B.B.W. (OCE 9981482).
REFERENCES
- 1.Affourtit, J., J. P. Zehr, and H. W. Paerl. 2001. Distribution of nitrogen-fixing microorganisms along the Neuse River Estuary, North Carolina. Microb. Ecol. 41:114-123. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, O. R. 2003. A model of biocomplexity and its application to the analysis of some terrestrial and marsh eukaryotic microbial communities with an emphasis on amoeboid protists. J. Eukaryot. Microbiol. 50:86-91. [DOI] [PubMed] [Google Scholar]
- 3.Berg, O. G., and C. G. Kurland. 2002. Evolution of microbial genomes: Sequence acquisition and loss. Mol. Biol. Evol. 19:2265-2276. [DOI] [PubMed] [Google Scholar]
- 4.Bianchi, T. S., E. Engelhaupt, P. Westman, T. Andren, C. Rolff, and R. Elmgren. 2000. Cyanobacterial blooms in the Baltic Sea: natural or human-induced? Limnol. Oceanogr. 45:716-726. [Google Scholar]
- 5.Bouvier, T. C., and P. A. del Giorgio. 2002. Compositional changes in free-living bacterial communities along a salinity gradient in two temperate estuaries. Limnol. Oceanogr. 47:453-470. [Google Scholar]
- 6.Burns, J. A., J. P. Zehr, and D. G. Capone. 2002. Nitrogen fixing phylotypes of Chesapeake Bay and Neuse River Estuary sediments. Microb. Ecol. 44:336-343. [DOI] [PubMed] [Google Scholar]
- 7.Cho, J. C., and J. M. Tiedje. 2002. Quantitative detection of microbial genes by using DNA microarrays. Appl. Environ. Microbiol. 68:1425-1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crump, B. C., E. V. Armbrust, and J. A. Baross. 1999. Phylogenetic analysis of particle-attached and free-living bacterial communities in the Columbia River, its estuary, and the adjacent coastal ocean. Appl. Environ. Microbiol. 65:3192-3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.del Giorgio, P. A., and T. C. Bouvier. 2002. Linking the physiologic and phylogenetic successions in free-living bacterial communities along an estuarine salinity gradient. Limnol. Oceanogr. 47:471-486. [Google Scholar]
- 10.Dennis, P., E. A. Edwards, S. N. Liss, and R. Fulthorpe. 2003. Monitoring gene expression in mixed microbial communities by using DNA microarrays. Appl. Environ. Microbiol. 69:769-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fisher, T. R., A. B. Gustafson, K. Sellner, R. Lacouture, L. W. Haas, R. L. Wetzel, R. Magnien, D. Everitt, B. Michaels, and R. Karrh. 1999. Spatial and temporal variation of resource limitation in Chesapeake Bay. Mar. Biol. 133:763-778. [Google Scholar]
- 12.Fisher, T. R., E. R. Peele, J. W. Ammerman, and L. W. Harding. 1992. Nutrient limitation of phytoplankton in Chesapeake Bay. Mar. Ecol. Prog. Ser. 82:51-63. [Google Scholar]
- 13.Giovannoni, S. J., T. B. Britschgi, C. L. Moyer, and K. G. Field. 1990. Genetic diversity in Sargasso Sea bacterioplankton. Nature (London) 344:60-63. [DOI] [PubMed] [Google Scholar]
- 14.Guschin, D. Y., B. K. Mobarry, D. Proudnikov, D. A. Stahl, B. E. Rittmann, and A. D. Mirzabekov. 1997. Oligonucleotide microchips as genosensors for determinative and environmental studies in microbiology. Appl. Environ. Microbiol. 63:2397-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heidelberg, J. F., K. B. Heidelberg, and R. R. Colwell. 2002. Seasonality of Chesapeake Bay bacterioplankton species. Appl. Environ. Microbiol. 68:5488-5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Howard, J. B., and D. C. Rees. 1996. Structural basis of biological nitrogen fixation. Chem. Rev. 96:2965-2982. [DOI] [PubMed] [Google Scholar]
- 17.Lovell, C. R., M. J. Friez, J. W. Longshore, and C. E. Bagwell. 2001. Recovery and phylogenetic analysis of nifH sequences from diazotrophic bacteria associated with dead aboveground biomass of Spartina alterniflora. Appl. Environ. Microbiol. 67:5308-5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loy, A., A. Lehner, N. Lee, J. Adamczyk, H. Meier, J. Ernst, K. H. Schleifer, and M. Wagner. 2002. Oligonucleotide microarray for 16S rRNA gene-based detection of all recognized lineages of sulfate-reducing prokaryotes in the environment. Appl. Environ. Microbiol. 68:5064-5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.MacGregor, B. J., B. Van Mooy, B. J. Baker, M. Mellon, P. H. Moisander, H. W. Paerl, J. Zehr, D. Hollander, and D. A. Stahl. 2001. Microbiological, molecular biological and stable isotopic evidence for nitrogen fixation in the open waters of Lake Michigan. Environ. Microbiol. 3:205-219. [DOI] [PubMed] [Google Scholar]
- 20.Malone, T. C., D. J. Conley, T. R. Fisher, P. M. Glibert, L. W. Harding, and K. G. Sellner. 1996. Scales of nutrient-limited phytoplankton productivity in Chesapeake Bay. Estuaries 19:371-385. [Google Scholar]
- 21.Methe, B. A., W. D. Hiorns, and J. P. Zehr. 1998. Contrasts between marine and freshwater bacterial community composition: analyses of communities in Lake George, N. Y. and six other Adirondack lakes. Limnol. Oceanogr. 43:368-374. [Google Scholar]
- 22.Moisander, P. H., and H. W. Paerl. 2000. Growth, primary productivity, and nitrogen fixation potential of Nodularia spp. (Cyanophyceae) in water from a subtropical estuary in the United States. J. Phycol. 36:645-658. [DOI] [PubMed] [Google Scholar]
- 23.Murray, A. E., J. T. Hollibaugh, and C. Orrego. 1996. Phylogenetic compositions of bacterioplankton from two California estuaries compared by denaturing gradient gel electrophoresis of 16S rDNA fragments. Appl. Environ. Microbiol. 62:2676-2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pace, N. R. 1997. A molecular view of microbial diversity and the biosphere. Science 276:734-740. [DOI] [PubMed] [Google Scholar]
- 25.Peplies, J., F. O. Glockner, and R. Amann. 2003. Optimization strategies for DNA microarray-based detection of bacteria with 16S rRNA-targeting oligonucleotide probes. Appl. Environ. Microbiol. 69:1397-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pinckney, J. L., H. W. Paerl, P. Tester, and T. L. Richardson. 2001. The role of nutrient loading and eutrophication in estuarine ecology. Environ. Health Perspect. 109:699-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26a.Short, S. M., B. D. Jenkins, and J. P. Zehr. The spatial and seasonal distribution of two diazotrophic bacteria in the Chesapeake Bay. Appl. Environ. Microbiol. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steppe, T. F., J. L. Pinckney, J. Dyble, and H. W. Paerl. 2001. Diazotrophy in modern marine Bahamian stromatolites. Microb. Ecol. 41:36-44. [DOI] [PubMed] [Google Scholar]
- 28.Steward, G. F., B. D. Jenkins, B. B. Ward, and J. P. Zehr. Development and testing of a DNA macroarray to assess nitrogenase (nifH) gene diversity. Appl. Environ. Microbiol., in press. [DOI] [PMC free article] [PubMed]
- 29.Steward, G. F., J. P. Zehr, R. P. Jellison, J. P. Montoya, and J. T. Hollibaugh. Vertical distribution of nitrogen-fixing phylotypes in a meromictic, hypersaline lake. Microb. Ecol., in press. [DOI] [PubMed]
- 30.Stine, O. C., A. Carnahan, R. Singh, J. Powell, J. P. Furuno, A. Dorsey, E. Silbergeld, H. N. Williams, and J. G. Morris. 2003. Characterization of microbial communities from coastal waters using microarrays. Environ. Monit. Assess. 81:327-336. [PubMed] [Google Scholar]
- 31.Suzuki, M., M. S. Rappe, and S. J. Giovannoni. 1998. Kinetic bias in estimates of coastal picoplankton community structure obtained by measurements of small-subunit rRNA gene PCR amplicon length heterogeneity. Appl. Environ. Microbiol. 64:4522-4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taroncher-Oldenburg, G., E. M. Griner, C. A. Francis, and B. B. Ward. 2003. Oligonucleotide microarray for the study of functional gene diversity in the nitrogen cycle in the environment. Appl. Environ. Microbiol. 69:1159-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilson, K. H., W. J. Wilson, J. L. Radosevich, T. Z. DeSantis, V. S. Viswanathan, T. A. Kuczmarski, and G. L. Andersen. 2002. High-density microarray of small-subunit ribosomal DNA probes. Appl. Environ. Microbiol. 68:2535-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu, L. Y., D. K. Thompson, G. S. Li, R. A. Hurt, J. M. Tiedje, and J. Z. Zhou. 2001. Development and evaluation of functional gene arrays for detection of selected genes in the environment. Appl. Environ. Microbiol. 67:5780-5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zani, S., M. T. Mellon, J. L. Collier, and J. P. Zehr. 2000. Expression of nifH genes in natural microbial assemblages in Lake George, N.Y., detected with reverse transcription-PCR. Appl. Environ. Microbiol. 66:3119-3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zehr, J. P., B. D. Jenkins, S. M. Short, and G. F. Steward. 2003. Nitrogenase gene diversity and microbial community structure: a cross-system comparison. Environ. Microbiol. 5:539-554. [DOI] [PubMed] [Google Scholar]
- 37.Zehr, J. P., and L. A. McReynolds. 1989. Use of degenerate oligonucleotides for amplification of the nifH gene from the marine cyanobacterium Trichodesmium spp. Appl. Environ. Microbiol. 55:2522-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zehr, J. P., J. B. Waterbury, P. J. Turner, J. P. Montoya, E. Omoregie, G. F. Steward, A. Hansen, and D. M. Karl. 2001. Unicellular cyanobacteria fix N-2 in the subtropical North Pacific Ocean. Nature 412:635-638. [DOI] [PubMed] [Google Scholar]
- 39.Zwart, G., W. D. Hiorns, B. A. Methe', M. P. Agterveld, R. Huismans, S. C. Nold, J. P. Zehr, and H. J. Laanbroek. 1998. Near-identical 16S rRNA sequences recovered from lakes in North America and Europe indicate the existence of clades of freshwater bacteria with global distribution. Syst. Appl. Microbiol. 21:546-556. [DOI] [PubMed] [Google Scholar]