Abstract
Study of stem cells may reveal promising treatment for diseases. The fate and function of transplanted stem cells remain poorly defined. Recent studies demonstrate that reporter genes can monitor real-time survival of transplanted stem cells in living subjects. We examined the effects of a novel and versatile triple fusion (TF) reporter gene construction on embryonic stem (ES) cell function by proteomic analysis. Murine ES cells were stably transduced with a self-inactivating lentiviral vector containing fluorescence (firefly luciferase; Fluc), bioluminescence (monomeric red fluorescence protein; mRFP), and positron emission tomography (herpes simplex virus type 1 truncated thymidine kinase; tTK) reporter genes. Fluorescence-activated cell sorting (FACS) analysis isolated stably transduced populations. TF reporter gene effects on cellular function were evaluated by quantitative proteomic profiling of control ES cells versus ES cells stably expressing the TF construct (ES-TF). Overall, no significant changes in protein quantity were observed. TF reporter gene expression had no effect on ES cell viability, proliferation, and differentiation capability. Molecular imaging studies tracked ES-TF cell survival and proliferation in living animals. In summary, this is the first proteomic study, demonstrating the unique potential of reporter gene imaging for tracking ES cell transplantation non-invasively, repetitively, and quantitatively.
Keywords: Embryonic stem cells, Mass spectrometry, Molecular imaging, Protein quantification
1 Introduction
Stem cells are derived from adult or embryonic sources and are capable of self-renewal. The therapeutic potential of stem cells has been demonstrated for a number of disorders including ischemic heart disease [1], diabetes [2], liver failure [3], neurodegenerative disorders [4], and hematological diseases [5]. Despite the recent success of this therapeutic strategy, the fate of these cells following injection is not well defined. Thus, the specific contribution of stem cell therapy to the observed functional changes is still poorly understood in many disease models. To investigate the mechanism of stem cell fate, novel reporter gene-imaging techniques have been developed in recent years to track cellular events in vivo. Although imaging studies on cell trafficking have been reported, few have specifically addressed the potential effects of reporter genes on stem cell function, a wholly critical aspect that cannot be overlooked [6, 7].
Proteomic analysis is a tremendous tool that is ideal for the study of proteins and their effects in complex biological samples. In particular, the final stages of a biological event that manifest in some activity are often a consequence of protein interactions and associated with changes in protein abundance and modification [8, 9]. Although microarray gene analysis provides a large amount of quantitative transcriptional data from comparative populations of cells or tissues, this approach presents some problems. These include (i) rapid and non-steady-state changes in mRNA levels, which make it difficult to observe consistent results; (ii) reproducibility of results among different laboratories; (iii) data mining and analysis, which remain significant hurdles; and, (iv) the daunting number of genes altered, such that a comprehensive screening to identify functional targets relevant to an outcome becomes more labor intensive than looking at the changes that occur on the protein level [10–13]. Furthermore, many genes can change in their transcription levels, but this change is often not reflected in changes in protein levels [8, 14–18]. Thus, methods that can pare down the list of possible candidates for specific function(s) will greatly facilitate the identification of a reasonable number of targets.
The analysis of protein abundance alterations will provide a feasible number of candidates that can be assayed for functional significance. The existence of many transcriptional products in cells can serve to facilitate the rapid production of proteins in tightly regulated responses, in such conditions as stress and apoptosis [19], which are reflected by changes in protein levels. Not all proteins are essential for a given activity in response to a trigger or stimulus. A select subpopulation of transcripts that code for a specific subset of proteins may be all that are required for the event. This point is illustrated by the redundancy of biological regulatory mechanisms that govern the need for rapid protein expression to maintain critical events such as cell viability, as exemplified in multidrug resistance (MDR) [20]. Here, multiple stimuli activate pathways that are all dedicated to the activation of MDR1. Therefore, changes in protein levels in response to specific stimuli may more accurately reflect those proteins directly linked to a cellular event and provide a clearer representation of critical participants compared to micro-array analysis.
In this study, we assessed the potential effects of triple fusion (TF) reporter gene expression on embryonic stem (ES) cell function. To accomplish this, we used isotopic labeling of peptides with 16O/18O [21, 22] and observed no significant changes by quantitative protein analysis. Likewise, the TF did not adversely affect ES cell viability, proliferation, and differentiation. Finally, we demonstrated the potential of molecular imaging to track ES cell survival and proliferation in living subjects.
2 Materials and methods
2.1 Culture of undifferentiated ES cells
The murine ES-D3 cell line (CRL-1934, ATCC) was originally isolated from day-4 blastocytes of 129/Sv mice [23]. ES cells were kept in an undifferentiated, pluripotent state by using 1000 IU/mL leukemia inhibitory factor (LIF; Chemicon) and grown on top of murine embryonic fibroblasts feeder layer inactivated by 10 μg/mL of mitomycin C (Sigma, St. Louis, MO). ES cells were cultured on 0.1% gelatin-coated plastic dishes in ES medium containing DMEM supplemented with 15% fetal calf serum, 0.1 mM β-mercaptoethanol, 2 mM glutamine, and 0.1 mM nonessential amino acids as described [24]. To confirm their undifferentiated state, ES cells were labeled with 4′,6-diamidino-2-phenylindole (DAPI) for nuclear marker, Oct-4 antibodies (Sigma) for ES cell transcriptional factor and E-cadherin (Invitrogen) for ES cell surface receptor marker. Images were obtained using a ZEISS Axiovert microscope (Sutter Instrument Company, Novato, CA).
2.2 Construction of the pUb-Fluc-mRFP-tTK triple fusion reporter gene
PCR amplification and standard cloning techniques were used to insert firefly luciferase (Fluc) and monomeric red fluorescence protein (mRFP) genes from plasmid pCDNA 3.1-CMV-Fluc (Promega, Madison, WI) and pCDNA3.1-CMV-mRFP in frame with the truncated thymidine kinase (tTK) gene into the pCDNA3.1-HSV-tTK as described [25]. This TF reporter gene fragment (3.3 kbp) was released from the plasmid with Not1 and BamH1 restriction enzymes before blunt-end ligation into the multiple cloning site of lentiviral transfer vector, FUG, driven by the human ubiquitin-C promoter [26].
2.3 Lentiviral production and infection of ES cells
SIN lentivirus was prepared by transient transfection of 293T cells as described [27]. Briefly, pFUG-TF containing the TF reporter gene was co-transfected into 293T cells with HIV-1 packaging vector (δ8.9) and vesicular stomatitis virus G glycoprotein-pseudotyped envelop vector (pVSVG). Lentivirus supernatant was concentrated by sediment centrifugation using SW29 rotor at 50 000 × g for 2 h. Concentrated virus was titered on 293T cells. Murine ES cells were transduced with LV-pUb-Fluc-mRFP-tTK at a multiplicity of infection (MOI) of 10. The infectivity was determined by mRFP expression as analyzed by fluorescence-activated cell sorting using FACscan (Becton Dickinson, San Jose, CA).
2.4 Determination of TF-induced changes in ES cells by quantitative analysis using MS
Twenty million murine ES cells transduced with and without the TF reporter gene were analyzed for quantitative protein changes in the first data set containing three trials, and 70 million cells were used in the second set of data containing five trials. A flow diagram illustrates the process used (Fig. 1A). Preparation of cell cytosol and plasma membrane preparation were previously described [28–30]. In brief, pelleted cells were resuspended in hypotonic lysis buffer containing 20 mM Tris pH 7.4, 5 mM EDTA, 5 mM EGTA, 5 mM DTT, and 5 mM Na3VO4. Lysis was performed by sonication on ice, followed by microcentrifugation at 4500 rpm, and the supernatant was saved. The remaining material was resuspended in PBS, centrifuged at 126 000×g, and the supernatant was saved. The pellet fraction was lysed in hypotonic buffer containing 1% Triton X-100 or 1% NP40, and spun at 126 000×g. The fractions were pooled prior to labeling. The 16O/18O isotopic labeling of peptides was done as described previously [21, 22]. This involved labeling individual samples, mixing the samples at a 1:1 ratio and peptide separation by RP chromatography and robotic fractionation by a Probot (Dionex, Sunnyvale, CA) onto a target plate for analysis by MALDI TOF/TOF (Ultraflex, Bruker Daltonics, Bremen, Germany). The sample was initially passed over a salt trap C18 PepMap 100 column (1 mm id×15 mm, 5 μm, 100 A) (Dionex, Sunnyvale, CA). Peptide separation was done using Vydac C18 RP column (150 mm×75 μm, 3 μm, 300 A) (Western Analytical, Wildomar, CA), and applying a linear gradient RP A, 0.1% TFA/5% ACN/95% H20; RP B, 0.1% TFA/20% H20/80% ACN; 5%A:100%B for 40 min, for a total run time of 80 min at a flow rate of 250 nL/min (LC Packings Nano LC System, Dionex Corporation, Sunnyvale, CA). Protein and peptide identifications were made by PMF and MS/MS using MASCOT (Matrix Science, London, UK). Search criteria include peptide tolerance of 0.5 Da and MS/MS tolerance of 1.2 Da. MS and MS/MS peaks from a combination of spots are considered (positions A1/A2/A3, A2/A3/A4, etc.) such that the MS peaks from these spots are submitted for PMF searching and the product ion spectra from these peaks are submitted for the corresponding MS/MS searches. The Stanford Proteomics and Integrative Research Facility has developed a software application that takes into account the peptide identifications from both the MS/MS and PMF, and assigns combined scores to provide a more accurate protein identification. PMF is important, as all the peaks that show a quantitative change might not have undergone MS/MS. Quantitative analysis was performed using this software, which applies a modified algorithm for determining peak pairs and looks at the ratio of the peaks to determine fold change (manuscript in preparation, provisional filed) (Fig. 1B). The application is an extension of the algorithm previously described [31] and incorporates an additional mass-dependent isotopic pattern point. The original algorithm calculates two additional isotopic cluster points beyond the monoisoptopic peak, based on the monoisotopic peak mass to charge ratio and intensity. This application uses a Gaussian fit over the monoisotopic peak profile. By incorporating all the isotopic peaks in addition to the two described in the original algorithm, we can obtain an improved deconvolution estimate. A baseline correction is done before the deconvolution followed by peak comparison where the relative peak intensities are compared for a given pair. As a control, we performed analyses on samples where we included either BSA or human lactotransferrin at a 5:1, 1:5, and 1:1 ratio in the background of the murine ES cell lysates (Table 1). We see the relative fold change that is expected in all cases, which serves as a control for the detection capability of the analysis process and software.
Figure 1.

(A) Schematic representation of quantitative proteomic profiling process. The sample preparation phase involves cell lysis, trypsin cleavage of the protein mixture, and 16O/18O isotopic labeling. Samples are mixed, separated by 1-D LCMS, robotically spotted using a Probot (Dionex Corporation, Sunnyvale, CA) onto a sample target plate and subjected to MS/MS for identification of peptides through a MALDI TOF-TOF (Bruker Daltonics, Bremen, Germany). Quantitative analysis was completed using proprietary software (Stanford Proteomics and Integrative Research Facility) and other methods were used to provide comprehensive quantitative data. (B) Quantitative analysis and processing of data. The analysis portion involves comparison of files generated to look for matches, peak identification, and deconvolution followed by normalization of data to provide the data (manuscript in preparation).
Table 1.
BSA or human lactotransferrin added at a 5:1, 1:5 and 1:1 ratios, respectively. Either BSA or human lactotransferrin or both are added to the murine ES cell lysates and the ratio for each situation is established. The appropriate fold change can be detected, as the ratio of these marker proteins is known. Shown are the identifications, mass of protein, peptide number, percent of protein coverage, fold change, and mass of peptide
| Accession | Name | Mass | Peptides | % Coverage | Fold change | Peptide |
|---|---|---|---|---|---|---|
| 5:1 | ||||||
| gi|640213 | Chain C, legume isolectin Ii (Lol Ii) complexed with lactotransferrin (N2 Fragment) | 17 509 | 1 | 5 | 5.0388 DOWN | 1047.984 |
| gi|16198357 | Lactotransferrin [Homo sapiens] | 78 279 | 1 | 1 | 5.0388 DOWN | 1047.984 |
| gi|16198359 | Lactotransferrin [Homo sapiens] | 78 295 | 1 | 1 | 5.0388 DOWN | 1047.984 |
| gi|18490850 | Lactotransferrin [Homo sapiens] | 78 278 | 1 | 1 | 5.0388 DOWN | 1047.984 |
| gi|54607120 | Lactotransferrin [Homo sapiens] | 78 132 | 1 | 1 | 5.0388 DOWN | 1047.984 |
| gi|640213 | Chain C, legume isolectin Ii (Lol Ii) complexed with lactotransferrin (N2 Fragment) | 17 509 | 1 | 5 | 5.0901 DOWN | 1047.501 |
| gi|16198357 | Lactotransferrin [Homo sapiens] | 78 279 | 1 | 1 | 5.0901 DOWN | 1047.501 |
| gi|16198359 | Lactotransferrin [Homo sapiens] | 78 295 | 1 | 1 | 5.0901 DOWN | 1047.501 |
| gi|18490850 | Lactotransferrin [Homo sapiens] | 78 278 | 1 | 1 | 5.0901 DOWN | 1047.501 |
| gi|20151212 | Chain B, crystal structure of a domain-opened mutant (R121d) of the human lactoferrin N-lobe refine | 36 849 | 1 | 2 | 5.0901 DOWN | 1047.501 |
| gi|54607120 | Lactotransferrin [Homo sapiens] | 78 132 | 1 | 1 | 5.0901 DOWN | 1047.501 |
| gi|162648 | Albumin [Bos taurus] | 69 248 | 1 | 1 | 5.9942 DOWN | 1163.598 |
| gi|229552 | Albumin | 66 088 | 1 | 1 | 5.9942 DOWN | 1163.598 |
| gi|30794280 | Albumin [Bos taurus] | 69 278 | 1 | 1 | 5.9942 DOWN | 1163.598 |
| 1:5 | ||||||
| gi|7428767 | Lactotransferrin [Homo sapiens] | 78 320 | 1 | 1 | 5.0235 UP | 1578.824 |
| gi|16198357 | Lactotransferrin [Homo sapiens] | 78 279 | 1 | 1 | 5.0235 UP | 1578.824 |
| gi|16198359 | Lactotransferrin [Homo sapiens] | 78 295 | 1 | 1 | 5.0235 UP | 1578.824 |
| gi|18490850 | Lactotransferrin [Homo sapiens] | 78 278 | 1 | 1 | 5.0235 UP | 1578.824 |
| gi|54607120 | Lactotransferrin [Homo sapiens] | 78 132 | 1 | 1 | 5.0235 UP | 1578.824 |
| 1:1 | ||||||
| gi|16198357 | Lactotransferrin [Homo sapiens] | 78 279 | 1 | 1 | 2.3144 DOWN | 1616.128 |
| gi|16198359 | Lactotransferrin [Homo sapiens] | 78 295 | 1 | 1 | 2.3144 DOWN | 1616.128 |
| gi|18490850 | Lactotransferrin [Homo sapiens] | 78 278 | 1 | 1 | 2.3144 DOWN | 1616.128 |
| gi|54607120 | Lactotransferrin [Homo sapiens] | 78 132 | 1 | 1 | 2.3144 DOWN | 1616.128 |
2.5 Determination of in vitro reporter enzyme activity
Equal number (1×106) of control ES cells, unsorted ES-TF cells (before FACS), and sorted ES-TF cells (after FACS) were treated with Passive Lysis Buffer (Promega, Madison, WI). Cells underwent freeze-thawing three times at −80°C for 15 min each. The homogenate was centrifuged at 14 000 rpm for 15 min and the Fluc enzyme activity was assessed using 20 μL of supernatant with 100 μL of Luciferase Assay Reagent (Promega, Madison, WI). The results were normalized to relative light unit per microgram of protein (RLU/μg) as described [32]. The same samples were assayed for thymidine kinase enzyme activity using [3H]-penciclovir as the substrate as described [33].
2.6 Effect of reporter genes and reporter probes on ES cell viability and proliferation
Cells were plated uniformly in 96-well plates at a density of 5000 cells/well. The trypan blue exclusion assay was used to assess for viability at 24-, 48-, and 72-h time points. The microplate spectrofluorometer (Gemini EM, Sunnyvale, CA) was used to measure the CyQuant cell proliferation assay (Molecular Probes, Eugene, OR) at the 24-, 48-, and 72-h time points. For these assays, results of six samples were averaged.
2.7 RT-PCR analysis of embryonic and tissue-specific transcripts
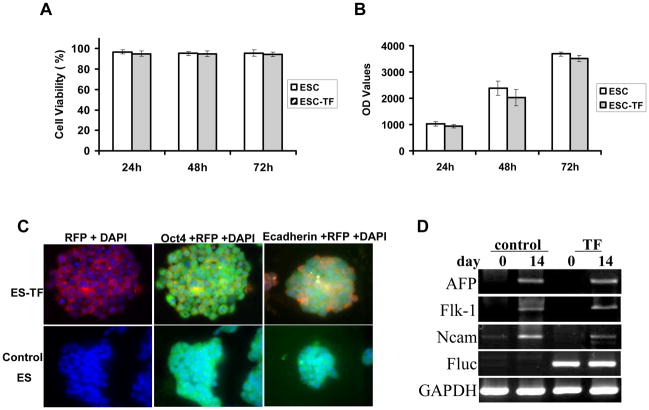
RT-PCR was used to compare the expression of ectodermal-specific protein neural cell adhesion molecule (NCAM), mesodermal-specific protein FMS-like tyrosine kinase 1 (Flk-1), endodermal-specific protein alphafetoprotein (AFP), and reporter gene (Fluc) between control nontransduced ES and ES-TF cells. Total RNA was prepared from ES-TF cells using Trizol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. The primer sets used in the amplification reaction are as follow: AFP forward primer, TATCAGCCACTGCTGCAACT, reverse primer, GTTCAGGCTTTTGCTTCACC; Flk-1 forward primer, CACCTGGCACTCTCCACCTTC, reverse primer, GATTTCATCCCACTA CCGAAAG; Ncam forward primer, GGAAGGGAACCAAGTGAACA, reverse primer, ACGGTGTGTCTGCTTGAACA; Fluc forward primer, ATCTACTGGTCTGCCTAAAG, reverse primer, CAGCTCTTCTTCAAATCTATAC; and control GAPDH forward primer, TTCACCACCATGGAGAAGGC, reverse primer, GGCATGGACTGTGGTCATGA. PCR products were separated on 1% agarose gel electrophoresis and quantified by using Labworks 4.6 Image Acquisition and analysis software (UVP Bio-imaging systems, Upland, CA).
2.8 Subcutaneous transplantation of ES cells onto mice shoulders
Adult nude mice (weighing 20–25 g) received isoflurane (2%) for general anesthesia. Animals (n = 8) were injected subcutaneously in the right shoulder with 1×106 of ES-TF cells and in the left shoulder with 1×106 of control untransduced ES cells in 50 μL of PBS. The same animals underwent longitudinal bioluminescence and positron emission tomography (PET) imaging protocols as described below.
2.9 Optical bioluminescence imaging of ES cell transplantation
Bioluminescence imaging was performed using the Xenogen In Vivo Imaging System as described [32]. Briefly, after intraperitoneal injection of the reporter probe D-Luciferin (125 mg/kg body weight), animals were imaged for 30 min using 1-min acquisition intervals. The same mice were scanned every 1–3 days for 2 weeks. Bioluminescence was quantified in units of maximum photons per second per centimeter square per steridian (p/s/cm2/sr).
2.10 Small animal PET imaging of ES cell transplantation
PET imaging was acquired using the P4 Concorde MicroPET system (Knoxville, TN). Briefly, animals were injected with the reporter probe [18F]-FHBG (171 ± 12 μCi) and images from 60–75 min post-injection were reconstructed by filtered back projection algorithm as described [33]. Regions of interest (ROI) were drawn over the shoulder region. The ROI counts/mL/min were converted to counts/g/min (assuming a tissue density of 1 g/mL) and divided by the injected dose to obtain an image ROI-derived [18F]-FHBG percentage of injected dose per gram of heart (% ID/g) using the ASIPro VM software as previously described [33].
2.11 Postmortem immunohistochemical staining
After imaging, all animals were sacrificed using protocols approved by the Stanford Animal Research Committee. Samples from explanted shoulder regions were fixed with 10% neutral buffered formalin, processed, embedded in paraffin, sectioned, and stained with hematoxylin and eosin (H&E). Slides were interpreted by an expert pathologist blinded to the study (AJC).
2.12 Data analysis
Data are given as mean ± SD. For statistical analysis, the two-tailed Student’s t–test was used. Differences were considered significant at P < 0.05.
3 Results
3.1 ES cells expressing bioluminescence, fluorescence, and PET reporter genes
To develop a multi-modality imaging approach, we used a TF reporter gene bearing Fluc, mRFP, and tTK reporter gene driven by a constitutive ubiquitin promoter in a lentiviral vector (Fig. 2A). The reporter genes were joined by a 14-(LENSHASAGYQAST) and 8-amino acid (TAGPGSAT) long linker, respectively. FACS analysis showed 29 ± 5% of ES cells with red fluorescence after lentiviral transduction (Fig. 2B). As expected, the unsorted ES-TF cells had significantly higher Fluc and tTK activities compared to control ES cells (*p < 0.05) (Figs. 2C and D). The sorted ES-TF cells had significantly higher Fluc and tTK activities compared to control ES cells as well as unsorted ES-TF cells (**p < 0.05).
Figure 2.
Isolation and characterization of ES-TF cells. (A) Map of the TF reporter gene containing firefly luciferase (Fluc), monomeric red fluorescence protein (mRFP), and herpes simplex virus type 1 truncated thymidine kinase (tTK) separated by 14- and 8-amino acid-long linkers, respectively. (B) After lentiviral transduction (MOI = 10), 29 ± 5% of these cells were positive for red fluorescence. (C) Sorted ES-TF cells showed significantly higher levels of Fluc enzyme activity compared to unsorted ES-TF cells and control ES cells. (D) Sorted ES-TF cells showed significantly higher levels of tTK enzyme activity compared to unsorted ES-TF cells and control ES cells. *p < 0.05 vs. control ES cells. **p < 0.05 vs. unsorted ES cells.
3.2 Proteomic profiling of control ES versus ES-TF cells
To determine if the TF reporter gene had any effects on ES protein expression, quantitative analysis using 16O/18O-labeled samples were repeated in two sets. The first is a trial that was repeated three times using 20 million cells per trial (Set 1.1, Set 1.2, Set 1.3) (Figs. 3A–C) and the second is a trial that was repeated five times using more than three times the number of cells (70 million) as in the first set (Set 2.1, Set 2.2, Set 2.3, Set 2.4, Set 2.5) (Figs. 3D–H). The purpose of this strategy is to determine if any protein(s) consistently change in abundance, which would indicate a potential influence of the TF reporter gene expression on cellular function. Moreover, we tested the reliability of our method by using ratios of well-defined protein mixtures (5:1, 1:5, and 1:1) to confirm expected changes when the proportion of protein present is known. Table 1 lists the results of the 5:1, 1:5 and 1.1 control samples where lysates were spiked with either BSA or human lactotransferrin at these ratios. We selected a background cutoff of threefold to ensure that changes are significant in the context of a biological system. The background threshold is indicated in each panel of Fig. 3 by red lines, which mark the threefold cut-off, and where points occurring above or below the red line are proteins that exhibit a significant increase or decrease, respectively. A point within the two lines is deemed a protein with insignificant change. A list of proteins for each trial from both sets of data with peptide number and protein coverage can be found at http://spir.stanford.edu/research.htm. The proteins that changed by more than threefold in any trial are also shown in Table 2, where changes were observed in only Set 2.1, and Set 2.5, and none of these proteins were consistently present in other trials with the exception of one protein that occurred in two trials, but did not meet the confidence criteria. As there were two experimental sets that differed in cell number and time of assay, we compared the findings within an experimental set. We do see proteins that did not change, which are identified in more than one trial. The summary table of the proteins that were identified per trial can be viewed at http://spir.stanford.edu/research.htm. Collectively, these results support the conclusion that the transduction of the TF construct does not induce any significant changes in protein levels in murine ES cells.
Figure 3.

Quantitative profiling. (A–C) Three trials of quantitative proteomic profiling were performed comparing control ES versus TF-ES cells, where 20 million cells were used for each trial. Each trial is represented in the plot where the red lines denote the background and the peptides identified are displayed based on m/z versus fold change. (D–H) Five trials of quantitative proteomic profiling comparing ES versus TF-ES cells, where 70 million cells were used per trial. Proteins that changed in abundance that exceeded background are shown in Table 2.
Table 2.
Proteins identified that exhibited change in abundance. The table shows a list of proteins that changed in abundance, which exceeded the background threshold for the experiment. Fold changes are shown where UP indicates a fold increase and DOWN represents a fold decrease in protein abundance. Peptide mass is also shown. Each protein identified exhibited a changed in only one trial.
| Accession | Description | Fold change | ||
|---|---|---|---|---|
| 2.5 | MSMS | PMF | ||
| gi|1125030 | Meltrin gamma [Mus musculus] | 9.2737 DOWN | X | X |
| gi|22799041 | TGFB-inducible early growth response protein 3 [Mus musculus] | 5.7747 UP | X | X |
| gi|14789893 | Ubiquitin-like 4 [Mus musculus] | 5.9809 DOWN | X | X |
| gi|202071 | Thymidine kinase [Mus musculus] | 3.0437 UP | X | X |
| gi|13542680 | Tubulin, beta, 2 [Mus musculus] | 6.0689 DOWN | X | X |
| gi|20988232 | Myosin binding protein C, fast-type [Mus musculus] | 3.5752 UP | X | X |
| 2.1 | ||||
| gi|896143 | Ig kappa chain | 5.041 DOWN | X | X |
To assess the detection capability of this approach, we looked for proteins that are expressed in undifferentiated ES cells and are species-specific, as differences exist between human and murine proteins [34, 35]. Table 3 is a list of proteins expressed in undifferentiated murine ES cells that were identified through this approach [34–36]. Finally, the thymidine kinase reporter gene (tTK) used in our TF reporter construct is a truncated version of the mutant herpes simplex virus thymidine kinase (HSV1-sr39tk), whereby the nuclear localization signal has been deleted in order to enhance tTK cytoplasmic accumulation and to decrease cellular toxicity [25]. Of the thymidine kinase proteins identified, one protein is the predicted HSV1-thymidine kinase protein (Table 4). We attribute the identification of thymidine kinase proteins that are not an exact match to the changes that were introduced into the original mutant HSV1-sr39tk. The MASCOT server utilizing the National Center for Bioinformatics non-redundant (NCBInr) database is currently undergoing modification of the entries. To date, these are the proteins that matched the peptides identified.
Table 3.
Proteins expressed in murine ES cells. The table is a list of the proteins or protein families that are known to be expressed in murine ES cells and were identified. Also shown are the mass of the protein, number of peptides, and percentage of protein coverage
| Accession | Name | Mass | Peptides | Coverage |
|---|---|---|---|---|
| Mitochondrial ribosomal protein | ||||
| gi|30519921 | Mitochondrial ribosomal protein L50 [Mus musculus] | 18 202 | 2 | 8 |
| gi|58331153 | Mitochondrial ribosomal protein S33 isoform 2 [Mus musculus] | 3 859 | 1 | 35 |
| Pleckstrin | ||||
| gi|10120815 | Chain B, structure of the pleckstrin homology domain from Grp1 in complex with inositol(1,3,4,5,6)pentakisphosphate | 15 186 | 5 | 20 |
| gi|82895324 | PREDICTED: pleckstrin homology domain containing, family M (with RUN domain) member 2 isoform 3 [Mus musculus] | 45 605 | 2 | 3 |
| gi|82895346 | PREDICTED: similar to pleckstrin homology domain containing, family M (with RUN domain) member 1 [Mus musculus] | 45 186 | 6 | 12 |
| SWI/SNF | ||||
| gi|82954481 | PREDICTED: similar to SWI/SNF related matrix-associated actin-dependent regulator of chromatin subfamily C member I | 24 114 | 3 | 8 |
| T-cell activation protein | ||||
| gi|198767 | lck Protein | 1 140 | 2 | 100 |
| gi|22671463 | Platelet and T cell activation antigen 1 isoform 2; PTA1; CD226 [Mus musculus] | 25 024 | 4 | 14 |
| Polycomb | ||||
| gi|82887063 | PREDICTED: similar to putative polycomb group protein ASXL1 (additional sex combs-like protein 1) isoform 1 | 16 2574 | 7 | 2 |
| gi|82887065 | PREDICTED: similar to putative polycomb group protein ASXL1 (additional sex combs-like protein 1) isoform 2 | 13 6912 | 6 | 2 |
| gi|82887488 | PREDICTED: similar to putative polycomb group protein ASXL1 (additional sex combs-like protein 1) [Mus musculus] | 157 370 | 7 | 3 |
| Homeobox | ||||
| gi|193981 | Homeobox protein | 2 872 | 1 | 32 |
| gi|309310 | Homeobox protein | 3 300 | 1 | 25 |
| gi|1196549 | Homeobox protein | 10 367 | 2 | 30 |
| gi|2765440 | Iroquois homeobox protein 1 [Mus musculus] | 8 555 | 2 | 21 |
| gi|17066573 | PEM homeobox [Mus musculus] | 2 258 | 2 | 45 |
| gi|80479165 | Ladybird homeobox homolog 2 [Mus musculus] | 20 894 | 6 | 22 |
| gi|82994021 | PREDICTED: similar to oocyte specific homeobox 5 [Mus musculus] | 11 301 | 4 | 23 |
| gi|82994039 | PREDICTED: similar to oocyte specific homeobox 1 [Mus musculus] | 16 998 | 5 | 17 |
| gi|83000379 | PREDICTED: similar to ES cell derived homeobox [Mus musculus] | 30 970 | 3 | 8 |
| gi|83000396 | PREDICTED: similar to reproductive homeobox on chromosome X, 4 [Mus musculus] | 31 342 | 2 | 5 |
| gi|83000443 | PREDICTED: similar to reproductive homeobox on X chromosome, 12 [Mus musculus] | 19 205 | 2 | 8 |
| Misc | ||||
| gi|9501362 | Ankyrin repeat domain 2 [Mus musculus] | 36 684 | 3 | 9 |
| gi|26992098 | Annexin II [Mus sp.] | 1 774 | 2 | 100 |
| gi|833814 | Putative serine/threonine kinase | 2 265 | 1 | 63 |
| gi|82895727 | PREDICTED: similar to serine/threonine-protein kinase tousled-like 2 (Tousled-like kinase 2) [Mus musculus] | 4 300 | 2 | 21 |
| gi|82934049 | PREDICTED: similar to RAC-beta serine/threonine-protein kinase (RAC-PK-beta) (Protein kinase Akt-2) | 43 321 | 3 | 6 |
| gi|83004885 | PREDICTED: similar to serine/threonine protein phosphatase with EF-hands-1 (PPEF-1) (Protein phosphatase with EF calcium binding domain) | 25 236 | 3 | 9 |
| gi|22002066 | 60S ribosomal protein L27a (L29) | 16 579 | 6 | 20 |
| gi|38082564 | PREDICTED: similar to 60S ribosomal protein L11 [Mus musculus] | 22 694 | 7 | 19 |
| gi|63501803 | PREDICTED: similar to 60S ribosomal protein L3 (L4) [Mus musculus] | 22 553 | 6 | 13 |
| gi|82801893 | PREDICTED: similar to 60S ribosomal protein L11 [Mus musculus] | 18 083 | 3 | 8 |
| gi|82884758 | PREDICTED: similar to 60S ribosomal protein L39 [Mus musculus] | 6 377 | 1 | 13 |
| gi|82895010 | PREDICTED: similar to 60S ribosomal protein L29 [Mus musculus] | 8 275 | 2 | 28 |
| gi|82899647 | PREDICTED: similar to 60S ribosomal protein L35 [Mus musculus] | 14 123 | 3 | 20 |
| gi|82904934 | PREDICTED: similar to 60S ribosomal protein L38 [Mus musculus] | 6 486 | 2 | 13 |
| gi|82914505 | PREDICTED: similar to 60S ribosomal protein L39 [Mus musculus] | 6 403 | 1 | 13 |
| gi|82923199 | PREDICTED: similar to 60S ribosomal protein L5 [Mus musculus] | 13 262 | 4 | 26 |
| gi|82930144 | PREDICTED: similar to 60S ribosomal protein L11 isoform 2 [Mus musculus] | 19 014 | 5 | 17 |
| gi|82932018 | PREDICTED: similar to 60S ribosomal protein L23 [Mus musculus] | 11 158 | 2 | 16 |
| gi|82935692 | PREDICTED: similar to 60S ribosomal protein L9 [Mus musculus] | 31 668 | 3 | 9 |
| gi|82951993 | PREDICTED: similar to 60S ribosomal protein L27a (L29) [Mus musculus] | 16 648 | 6 | 20 |
| gi|82952413 | PREDICTED: similar to 60S ribosomal protein L3 (L4) [Mus musculus] | 13 397 | 5 | 30 |
| gi|82954930 | PREDICTED: similar to 60S ribosomal protein L17 (L23) (amino acid starvation-induced protein) (ASI) | 16 495 | 3 | 15 |
| gi|82959035 | PREDICTED: similar to 60S ribosomal protein L29 (P23) [Mus musculus] | 16 159 | 3 | 16 |
| gi|82959254 | PREDICTED: similar to 60S ribosomal protein L26 [Mus musculus] | 4 400 | 1 | 18 |
| gi|82986593 | PREDICTED: similar to 60S ribosomal protein L7a [Mus musculus] | 15 621 | 2 | 11 |
| gi|82995359 | PREDICTED: similar to 60S ribosomal protein L9 [Mus musculus] | 4 881 | 1 | 16 |
| gi|82997503 | PREDICTED: similar to 60S ribosomal protein L17 (L23) (amino acid starvation-induced protein) (ASI) | 42 578 | 5 | 8 |
| gi|83000571 | PREDICTED: similar to 60S ribosomal protein L35 [Mus musculus] | 8 151 | 4 | 43 |
| gi|83002858 | PREDICTED: similar to 60S ribosomal protein L12 [Mus musculus] | 14 789 | 2 | 10 |
| gi|83003174 | PREDICTED: similar to 60S ribosomal protein L12 [Mus musculus] | 12 377 | 2 | 12 |
| gi|83014023 | PREDICTED: similar to 60S ribosomal protein L23a [Mus musculus] | 7 313 | 2 | 26 |
| gi|18077587 | Cyclin B3 [Mus musculus] | 4 104 | 1 | 20 |
| gi|50613 | Cyclin B2 [Mus musculus] | 45 423 | 4 | 5 |
| gi|563900 | Murine cyclin H [Mus musculus] | 2 104 | 1 | 61 |
| gi|6684129 | Cyclin D1 [Mus musculus] | 2 746 | 1 | 60 |
| gi|82898104 | PREDICTED: similar to cyclin-dependent kinase 6 [Mus musculus] | 7 636 | 5 | 37 |
| gi|82955419 | PREDICTED: similar to cyclin-dependent kinase 4 [Mus musculus] | 20 198 | 3 | 17 |
| gi|82956097 | PREDICTED: similar to cell division protein kinase 4 (cyclin-dependent kinase 4) (PSK-J3) (CRK3) [Mus musculus] | 27 339 | 4 | 19 |
| gi|14714757 | Procollagen (type III) N-endopeptidase [Mus musculus] | 21 594 | 4 | 21 |
| gi|1777336 | Alpha2(XI) collagen [Mus musculus] | 1 020 | 1 | 62 |
| gi|4103062 | Alpha 1 type IX collagen short form [Mus musculus] | 2 730 | 1 | 69 |
| gi|49614260 | Alpha1(V) collagen [Mus musculus] | 3 401 | 1 | 23 |
| gi|553856 | Alpha-1 type IX collagen | 3 364 | 1 | 34 |
| gi|553882 | Alpha-2 type I collagen | 2 470 | 2 | 34 |
| gi|21618685 | P450 (cytochrome) oxidoreductase [Mus musculus] | 76 995 | 10 | 10 |
| gi|296469 | Cytochrome P450 [Mus musculus] | 4 789 | 2 | 30 |
| gi|553905 | Cytochrome p-450b | 2 978 | 2 | 48 |
| gi|56541056 | Ubiquinol-cytochrome c reductase binding protein [Mus musculus] | 13 553 | 3 | 27 |
| gi|6680986 | Cytochrome c oxidase, subunit Va [Mus musculus] | 16 020 | 3 | 10 |
| gi|6753580 | Cytochrome P450, family 2. subfamily c, polypeptide 37 [Mus musculus] | 55 478 | 4 | 5 |
| gi|80478978 | Cytochrome P450, family 24, subfamily a, polypeptide 1 [Mus musculus] | 59 415 | 6 | 8 |
| gi|82801364 | PREDICTED: similar to NADPH cytochrome B5 oxidoreductase [Mus musculus] | 18 063 | 3 | 28 |
| gi|82988418 | PREDICTED: similar to ubiquinol-cytochrome c reductase binding protein [Mus musculus] | 8 721 | 2 | 16 |
| gi|82993194 | PREDICTED: similar to cytochrome P450 XXI (steroid 21-hydroxylase) (21-OHase) (P450-C21) (P-450c21) | 6 885 | 2 | 21 |
| gi|82882433 | PREDICTED: similar to DEAD (Asp-Glu-Ala-Asp) box polypeptide 18 [Mus musculus] | 23 425 | 4 | 22 |
| gi|82930835 | PREDICTED: similar to DnaJ (Hsp40) homolog, subfamily C, member 13 isoform 4 [Mus musculus] | 181 891 | 7 | 3 |
| gi|2689718 | DNA (cytosine-5)-methyltransferase [Mus musculus] | 2 850 | 1 | 33 |
| gi|14250182 | Eukaryotic translation initiation factor 3, subunit 4 (delta) [Mus musculus] | 35 616 | 5 | 10 |
| gi|82796315 | PREDICTED: similar to eukaryotic translation initiation factor 1A, Y-linked [Mus musculus] | 10 939 | 2 | 13 |
| gi|82998188 | PREDICTED: similar to eukaryotic translation initiation factor 4A, isoform 1 isoform 2 [Mus musculus] | 39 297 | 6 | 13 |
| gi|82998190 | PREDICTED: similar to eukaryotic translation initiation factor 4A, isoform 1 isoform 1 [Mus musculus] | 37 370 | 7 | 16 |
| gi|82998245 | PREDICTED: similar to eukaryotic translation initiation factor 4A, isoform 1 isoform 2 [Mus musculus] | 22 109 | 4 | 16 |
| gi|54607022 | F-box and leucine-rich repeat protein 10 isoform 1 [Mus musculus] | 149 638 | 11 | 6 |
| gi|56800427 | Novel F-box domain containing protein [Mus musculus] | 58 363 | 5 | 5 |
| gi|6456100 | F-box protein FBL10 [Mus musculus] | 34 150 | 3 | 7 |
| gi|82801364 | PREDICTED: similar to NADPH cytochrome B5 oxidoreductase [Mus musculus] | 18 063 | 3 | 28 |
| gi|13177608 | GTP-binding protein RAB12 [Mus musculus] | 5 519 | 2 | 32 |
| gi|13177612 | GTP-binding protein RAB16 [Mus musculus] | 7 487 | 2 | 29 |
| gi|83013653 | PREDICTED: similar to Rho-related GTP-binding protein RhoN (Rho family GTPase 2) (Rnd2) (Rho7) [Mus musculus] | 9 564 | 4 | 41 |
| gi|9971156 | GTP-binding like protein 2 [Mus musculus] | 56 436 | 8 | 13 |
| gi|14582204 | Huntingtin-interacting protein-1 protein interactor [Mus musculus] | 48 725 | 7 | 13 |
| gi|3378243 | Integrin alpha 7 [Mus musculus] | 126 637 | 3 | 1 |
| gi|3925800 | Mcm4(cdc21) [Mus musculus] | 4 541 | 2 | 50 |
| gi|13384946 | NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1 [Mus musculus] | 8 620 | 4 | 35 |
| gi|82881464 | PREDICTED: similar to NADH-ubiquinone oxidoreductase MWFE subunit (complex I-MWFE) (CI-MWFE) [Mus musculus] | 3 861 | 1 | 20 |
| gi|82942594 | PREDICTED: similar to NADH-ubiquinone oxidoreductase B9 subunit (complex I-B9) (CI-B9) [Mus musculus] | 7 542 | 2 | 13 |
| gi|83020260 | PREDICTED: similar to NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 7 [Mus musculus] | 11 476 | 3 | 15 |
| gi|11999273 | Group XII-2 phospholipase A2 [Mus musculus] | 17 188 | 4 | 18 |
| gi|20070853 | Group XII-1 phospholipase A2, isoform 1 [Mus musculus] | 21 275 | 6 | 25 |
| gi|82953351 | PREDICTED: similar to DNA-directed RNA polymerase III largest subunit (RPC155) (RPC1) isoform 9 [Mus musculus] | 29 679 | 4 | 13 |
| gi|13278483 | Peptidylprolyl isomerase F [Mus musculus] | 21 723 | 6 | 23 |
| gi|53237015 | Peptidylprolyl isomerase A [Mus musculus] | 17 960 | 4 | 20 |
| gi|63702001 | PREDICTED: similar to peptidyl prolyl isomerase H [Mus musculus] | 22 976 | 7 | 14 |
| gi|82891759 | PREDICTED: similar to peptidyl-prolyl cis-trans isomerase NIMA-interacting 4 (Rotamase Pin4) (PPIase Pin4) | 14 579 | 3 | 35 |
| gi|82914090 | PREDICTED: similar to peptidyl-prolyl cis-trans isomerase-like 2 (PPIase) (Rotamase) (Cyclophilin-60) | 14 298 | 3 | 14 |
| gi|82932250 | PREDICTED: peptidylprolyl isomerase (cyclophilin)-like 6 isoform 1 [Mus musculus] | 35 539 | 4 | 10 |
| gi|82949793 | PREDICTED: similar to peptidylprolyl isomerase D [Mus musculus] | 17 922 | 6 | 17 |
| gi|18043908 | THO complex 3 [Mus musculus] | 38 713 | 2 | 3 |
| gi|21312090 | THO complex 3 [Mus musculus] | 38 608 | 2 | 3 |
| gi|83007150 | PREDICTED: similar to THO complex subunit 4 (Tho4) (RNA and export factor binding protein 1) (REF1-I) | 13 717 | 2 | 11 |
| gi|63102314 | Hepatocellular carcinoma-associated antigen 127 [Mus musculus] | 26 255 | 1 | 6 |
| gi|83002662 | PREDICTED: similar to hepatocellular carcinoma-associated antigen 127 [Mus musculus] | 16 626 | 1 | 10 |
| gi|37194893 | U2AF homology motif (UHM) kinase 1 [Mus musculus] | 46 459 | 5 | 12 |
| gi|68534509 | U2 small nuclear ribonucleoprotein auxiliary factor (U2AF) 1, related sequence 2, isoform 2 [Mus musculus] | 17 097 | 4 | 19 |
| gi|82897422 | PREDICTED: similar to ubiquitin-conjugating enzyme E2 variant 1 [Mus musculus] | 5 404 | 2 | 25 |
| gi|82933724 | PREDICTED: similar to ubiquitin-conjugating enzyme E2 L3 (ubiquitin-protein ligase L3) (ubiquitin carrier protein L3) | 11 360 | 2 | 12 |
| gi|82949698 | PREDICTED: similar to ubiquitin-conjugating enzyme E2 L3 (ubiquitin-protein ligase L3) (Ubiquitin carrier protein L3) | 18 032 | 3 | 11 |
| gi|82950415 | PREDICTED: similar to ubiquitin-conjugating enzyme E2 H (ubiquitin-protein ligase H) (Ubiquitin carrier protein L3) | 9 213 | 3 | 31 |
| gi|82957240 | PREDICTED: similar to ubiquitin-conjugating enzyme E2 C (ubiquitin-protein ligase C) (Ubiquitin carrier protein C) | 10 696 | 2 | 15 |
| gi|85662417 | Ubiquitin-conjugating enzyme E2, J2 homolog isoform a [Mus musculus] | 30 273 | 4 | 11 |
| gi|82951721 | PREDICTED: similar to nucleolar transcription factor 1 (upstream binding factor 1) (UBF-1) [Mus musculus] | 18 775 | 3 | 15 |
| gi|6678517 | Uroporphyrinogen decarboxylase [Mus musculus] | 40 572 | 4 | 6 |
| gi|82881456 | PREDICTED: similar to ubiquitin carboxyl-terminal hydrolase CYLD [Mus musculus] | 13 052 | 2 | 11 |
| gi|20809864 | WD repeat domain 78 [Mus musculus] | 68 621 | 5 | 7 |
| gi|47059149 | WD repeat domain 51B [Mus musculus] | 53 470 | 6 | 9 |
| gi|82879175 | PREDICTED: similar to WD repeat and SOCS box containing protein 2 (WSB-2) (SOCS box-containing WD protein SWiP-2) | 5 894 | 4 | 27 |
Table 4.
Identification of HSV1 thymidine kinase. To confirm that the reporter proteins can be detected, a search for the thymidine kinase protein was done. The tTK reporter gene within the TF construct is a truncated version of the mutant HSV1-sr39tk reporter gene. Protein mass, number of peptides and percentage of protein coverage are shown. This modification, which truncates the already modified protein, may limit the identification of the protein, ase fewer peptides may be matched in the database.
| Accession | Name | Mass | Peptides | Coverage |
|---|---|---|---|---|
| gi|471721 | ICP18.5 homolog of HSV-1 | 10 141 | 3 | 23 |
| gi|34732881 | Thymidine kinase; Tk [Bacteriophage 44RR2.8t] | 21 630 | 2 | 6 |
| gi|32453595 | Tk thymidine kinase [Enterobacteria phage RB69] | 21 403 | 3 | 11 |
| gi|34732881 | Thymidine kinase; Tk [Bacteriophage 44RR2.8t] | 21 630 | 3 | 6 |
| gi|60099578 | Thymidine kinase [Duck enteritis virus] | 40 680 | 1 | 2 |
| gi|62115016 | Thymidine kinase [Duck enteritis virus] | 40 564 | 1 | 2 |
| gi|33324244 | Truncated thymidine kinase [African swine fever virus] | 21 085 | 1 | 7 |
| gi|33324248 | Truncated thymidine kinase [African swine fever virus] | 21 055 | 1 | 7 |
| gi|52551092 | UL24 [Fibropapilloma-associated turtle herpesvirus] | 33 753 | 1 | 2 |
| gi|3769435 | Thymidine kinase [macropodid herpesvirus type 1] | 36 654 | 1 | 2 |
| gi|83722592 | UL24 [Cercopithecine herpesvirus 16] | 28 065 | 4 | 19 |
3.3 Effects of reporter genes on ES cell function
Although none of the proteins examined showed significant changes compared to background levels, we decided to further address the question of whether ES cell function was adversely affected. Figure 4A shows that there was no significant difference in the viability between control ES cell and ES-TF cells. Likewise, the cell proliferation was not significantly altered at the 24-, 48-, and 72-h time points (Fig. 4B). Immunofluorescence staining showed similar expression pattern of Oct-4 transcriptional factor and E-cadherin surface marker on undifferentiated ES cells (Fig. 4C). To assess whether ES cell differentiation was affected, we compared embryoid body formation from days 0 and 14. RT-PCR analysis showed that the expression patterns of the endodermal (AFP), mesodermal (Flk-1), and ectodermal (NCAM) specific proteins were also similar between the two groups. These additional data suggest that the expression of TF reporter gene had no significant effects on ES cell viability, proliferation, and differentiation, a demonstration which is necessary before proceeding to in vivo imaging of ES cell transplantation in living animals.
Figure 4.
Effects of TF reporter gene expression on ES cell function. (A, B) Trypan blue cell viability and CyQuant cell proliferation assay showed no significant difference between control ES and ES-TF cells at various time points. (C) Both cell types also showed similar morphology on brightfield microscopy as well as staining patterns of stem cell Oct-4 transcriptional factor and E-cadherin cell surface marker on fluorescence microscopy. DAPI staining is used as a nuclear marker. (D) RT-PCR analysis shows the in vitro differentiation capacity into ectoderm-, mesoderm-, and endoderm-specific phenotypes is also similar between ES and ES-TF cells. Fluc is present only in ES-TF cells as expected. GAPDH is a loading control.
3.4 Bioluminescence and PET imaging ES cell survival and proliferation
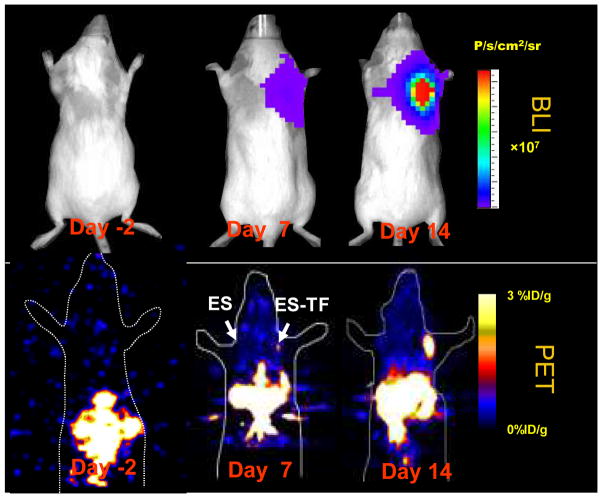
The in vitro replication capacities of human and murine ES cells have been documented in previous studies [37, 38]. However, their in vivo survival and proliferation kinetics have not yet been well defined. To address these issues, control ES and ES-TF cells were injected subcutaneously into the left and right shoulders, respectively, of adult nude mice. Imaging was performed over the next 2 weeks (Fig. 5). At day 7, the bioluminescence and PET signals in the right shoulder were 5.4×107±1.3×106 photons/s/cm2/sr and 0.75± 0.06%ID/g, respectively, compared to background signals of 4.3×105 ± 2.8×104 photons/s/cm2/sr and 0.044.3× 105 ±0.01%ID/g in the left shoulder (p < 0.05). At day 14, the bioluminescence and PET signals in the right shoulder increased to 4.3×109±1.1×108 photons/s/cm2/sr and 2.37±0.50%ID/g, respectively, compared to background signals of 1.1×106±3.7×104 photons/s/cm2/sr and 0.05±0.01%ID/g in the left shoulder (p < 0.05). Because the Fluc and tTK genes are fused together, quantification of both imaging signals showed a strong correlation in vivo (r2 = 0.89). After imaging, all animals were sacrificed and ES cell presence was confirmed by ex vivo enzyme assays and conventional histology. There was a robust correlation (r2 = 0.85 for Fluc and r2 = 0.88 for tTK) between ex vivo enzyme assays of ES-TF cells explanted from the right shoulders with in vivo imaging signals (data not shown). Finally, to assess if the reporter genes could negatively hamper differentiation of ES cells into different cell lineages in vivo, histological analysis was performed. Both control ES and ES-TF cells showed similar patterns of ectoderm, mesoderm, and endoderm tissue formation in teratomas arising from ES cells (Fig. 6). The teratomas included large areas of immature cells in rest and sheets, intermixed with foci with development to immature glands, respiratory epithelium, immature neural tissue, brain-like neuronal tissue, squamous epithelium, and combined osteochondroid development. Focal necrosis and hemorrhage was also seen in teratomas from both groups.
Figure 5.
Noninvasive imaging of ES cell transplant in living subjects. Bioluminescence (top row) and PET imaging (bottom row) of a representative animal before (day −2) and after ES cell transplantation (days 7 and 14). The survival of control ES cells in the left shoulder could not be assessed because they lack reporter genes. In contrast, ES-TF cells transplanted into the right shoulder showed progressive increases in bioluminescence and PET signals at days 7 and 14, reflecting both cell survival and cell proliferation during this period. The bioluminescence activity is quantified as units of photons per second per centimeter square per steridian (p/s/cm2/sr). The PET activity is quantified as percentage of injected dose of [18F]-FHBG per gram of tissue (% ID/g). Note significant PET activity is also seen in the gut region due to the natural excretion and clearance route of [18F]-FHBG.
Figure 6.

Postmortem histological analysis of explanted shoulder tissues. H&E staining of teratomas derived from either control ES cells (A–D) or ES-TF cells (E–H) show similar patterns of in vivo differentiation between the two groups. (A) and (D) osteochondroid formation; (B) and (E) squamous cell differentiation with keratin pearl; (C) and (F) respiratory epithelium with ciliated columnar and mucin producing goblet cells; (D) and (F) immature brain-like neural cells. All slides are shown at 400x magnification.
4 Discussion
In recent years, significant research has focused the use of stem cell transplantation for tissue repair. This potential has already been explored in several clinical arenas [39]. Hematopoietic stem cells obtained from the bone marrow or peripheral blood were first used for treatment of patients with leukemia in the early 1970s and have now been expanded to lymphomas, solid tumors, and congenital disorders [40]. More recently, cardiovascular cell therapies based on skeletal myoblasts, endothelial progenitor cells, or bone marrow stem cells have yielded significant improvements in myocardial function of patients, and these successes have generated an enormous growth in basic research and clinical trials [41]. The possibility of isolating mouse [37] and human [38] ES cells, which can then differentiate into neurons, cardiomyocytes, β-islet cells, osteoblasts, hepatocytes, and hematopoietic progenitors indicates the huge promise of this line of work [39]. However, many challenges lie ahead, not the least of which is the current state of imperfect understanding of many fundamental aspects of stem cell biology and physiology in living subjects.
To better understand stem cell fate in vivo, novel techniques that allow noninvasive, repetitive, and quantitative assessment of underlying stem cell biology are clearly needed. The ideal imaging platform should (i) be biocompatible, safe, and nontoxic; (ii) allow detection at the single cell level; (iii) serve as reliable indicator of cell survival and proliferation; and (iv) expedite studies in basic research as well as clinical trials. At present, no single imaging modality possesses all of these parameters [42]. Taking advantage of the strengths and weaknesses of different modalities, we used a novel TF reporter gene with the following logic. The fluorescence reporter gene (mRFP) allows imaging at the single-cell level by fluorescence microscopy and isolation of stable clone populations by FACS. The bioluminescence reporter gene (Fluc) can be used for high-throughput analysis of stem cell transplant in small animals at relatively low cost per scan [32]. However, both fluorescence and bioluminescence imaging rely on low energy light photons that become attenuated within deep tissues. Thus, the PET reporter gene (tTK) was added for pre-clinical validation in living subjects. Compared to computed tomography (CT) or magnetic resonance (MR) imaging, the detection threshold of PET is 6–8 log-order more sensitive [7]. This is one of the main reasons why PET imaging of reporter gene expression has now been adapted into clinical studies involving patients with recurrent glioblastoma [43] and hepatocellular carcinoma [44].
A critical question with regard to reporter genes is whether they would affect cellular traits that might hinder future clinical applications. To address this issue, we comparatively profiled protein levels in control ES and ES-TF cells. Our strategy employed labeling proteins with 16O/18O, a method that effectively and stably labels nearly all of the peptides [45]. The subsequent MS analysis is done using the co-eluted heavy and light peptides [46]. In addition, the analysis software developed by the Stanford Proteomic and Integrative Research Facility (SPIR) (Fig. 1B) is able to identify the quantitative changes using peak picking criteria and link these peaks with peptide identification through MS/MS from MASCOT, as shown in Fig. 3 and in link http://spir.stanford.edu/research.htm. The assay was done in one trial in triplicate and repeated five times in a second trial, and no significant changes that consistently occurred were observed in either data set, which is consistent with the results for cell viability, proliferation, and function as shown in Fig. 4.
While gene microarrays can be used to evaluate changes in transcriptional activity, the interactions between increased or decreased transcriptional regulation and protein expression do not always correlate [15–18]. We previously observed the expected lack of coordinate changes between the studies from genomic microarray analysis performed on these same cells and under the same experimental conditions [47]. For the first trial (Set 1), we observed 2127 unique protein entries. Upon comparison of these entries with a gene database, 1522 original entries that did not undergo any changes in protein level were identified using the National Center for Biotechnology Information non-redundant database (National Institutes of Health/National Library of Medicine, Bethesda MD). Of these, 1115 were matched to genes through batch Entrez database (National Institutes of Health/National Library of Medicine, Bethesda, MD) analysis. Of these, 680 matched the National Institutes of Aging clone set, which consists of some 22 000 gene probes, out of which 25 unique genes that were observed to change in transcription levels were found to match proteins that did not change. In the second trial (Set 2), we observed the identification of proteins that range in number from 831 to 3134 per trial, where the collective total number of unique protein entries is 6225. We found a greater number of proteins identified in Set 2 as compared to Set 1, which is consistent with the increase in cell number used for Set 2. In addition, 856 proteins are common to both sets. These results are consistent with the lack of correlation between transcriptional and translational levels. Furthermore, of those proteins observed to change above background levels, their occurrence was unique for a given trial (Table 2 and http://spir.stanford.edu/research.htm). These results indicate that the introduction of TF has no significant negative influence on the cellular function of the ES cells studied here.
Our studies show that proteomic profiling on a quantitative level can reliably be used to observe changes that may directly influence cellular function. A lack of change in transcript abundance, even with no change in protein abundance, can still have a functional outcome by modifications to the protein [48]. We also identified proteins that were phosphorylated among the list. If there is an effect of those modifications, one might expect to see some influence on the outcome of the viability and proliferation studies as well as some up- or down-regulation of kinase or phosphatase activity, but none was observed. This suggests that the modifications on these proteins are probably part of their normal or resting state in the cell. In cellular situations such as stress and apoptosis, translational regulation is a more expedient means of responding to a critical cellular state [19]. The amount of data generated from assessing transcriptional changes can be quite large, as transcriptional regulation produces many forms of RNA with varying rates of stability and noise [47, 49, 50]. Although transcriptional regulation can influence protein expression, other factors that can mediate protein processing, cannot be directly observed by quantitative changes on the transcriptional level. Moreover, translational regulation is involved in the rapid phenotypic responses required by a biological system [12, 13]. Therefore, through the acquisition of data that assesses quantitative changes in protein levels, we provide a smaller and potentially relevant list of proteins that will serve as an indicator of whether the introduction of reporter gene constructs has an influence on cellular events.
In conclusion, we reported the first quantitative proteomic profile for determining the effects of fluorescence, bioluminescence, and PET reporter genes on stably transduced ES cells. The use of enzymatic labeling strategies to ensure more uniformly labeled samples, coupled with analysis applications following collection of MS data, is an efficient method to provide a rapid survey of protein level changes in complex mixtures. Finally, we demonstrated the use of this novel TF reporter gene construct for noninvasive imaging of ES cell survival and proliferation in living animals.
Acknowledgments
The authors would like to thank Joshua Spin for his helpful discussions. This work was supported in part by grants from the AHA, ARA, and NHLBI (JCW); NCI ICMIC-P50CA114747, NHLBI R01 HL078632, and NCI SAIRP (SSG); and funding from the Department of Pathology, Stanford University School of Medicine (EWW).
Abbreviations
- ES
embryonic stem
- FACS
fluorescence-activated cell sorting
- mRFP
monomeric red fluorescence protein
- PET
positron emission tomography
- TF
triple fusion
References
- 1.Orlic D, Kajstura J, Chimenti S, Jakoniuk I, et al. Nature. 2001;410:701–705. doi: 10.1038/35070587. [DOI] [PubMed] [Google Scholar]
- 2.Ramiya VK, Maraist M, Arfors KE, Schatz DA, et al. Nat Med. 2000;6:278–282. doi: 10.1038/73128. [DOI] [PubMed] [Google Scholar]
- 3.Kobayashi N, Fujiwara T, Westerman KA, Inoue Y, et al. Science. 2000;287:1258–1262. doi: 10.1126/science.287.5456.1258. [DOI] [PubMed] [Google Scholar]
- 4.Lindvall O, Kokaia Z, Martinez-Serrano A. Nat Med. 2004;10(Suppl):S42–50. doi: 10.1038/nm1064. [DOI] [PubMed] [Google Scholar]
- 5.Wright DE, Wagers AJ, Gulati AP, Johnson FL, Weissman IL. Science. 2001;294:1933–1936. doi: 10.1126/science.1064081. [DOI] [PubMed] [Google Scholar]
- 6.Blasberg RG, Tjuvajev JG. J Clin Invest. 2003;111:1620–1629. doi: 10.1172/JCI18855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Massoud TF, Gambhir SS. Genes Dev. 2003;17:545–580. doi: 10.1101/gad.1047403. [DOI] [PubMed] [Google Scholar]
- 8.Feder ME, Walser JC. J Evol Biol. 2005;18:901–910. doi: 10.1111/j.1420-9101.2005.00921.x. [DOI] [PubMed] [Google Scholar]
- 9.Monti M, Orru S, Pagnozzi D, Pucci P. Biosci Rep. 2005;25:45–56. doi: 10.1007/s10540-005-2847-z. [DOI] [PubMed] [Google Scholar]
- 10.Allison DB, Cui X, Page GP, Sabripour M. Nat Rev Genet. 2006;7:55–65. doi: 10.1038/nrg1749. [DOI] [PubMed] [Google Scholar]
- 11.Ewis AA, Zhelev Z, Bakalova R, Fukuoka S, et al. Expert Rev Mol Diagn. 2005;5:315–328. doi: 10.1586/14737159.5.3.315. [DOI] [PubMed] [Google Scholar]
- 12.Ozbudak EM, Thattai M, Kurtser I, Grossman AD, van Oudenaarden A. Nat Genet. 2002;31:69–73. doi: 10.1038/ng869. [DOI] [PubMed] [Google Scholar]
- 13.Thattai M, van Oudenaarden A. Proc Natl Acad Sci USA. 2001;98:8614–8619. doi: 10.1073/pnas.151588598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen G, Gharib TG, Huang CC, Taylor JM, et al. Mol Cell Proteomics. 2002;1:304–313. doi: 10.1074/mcp.m200008-mcp200. [DOI] [PubMed] [Google Scholar]
- 15.Greenbaum D, Colangelo C, Williams K, Gerstein M. Genome Biol. 2003;4:117. doi: 10.1186/gb-2003-4-9-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gygi SP, Rochon Y, Franza BR, Aebersold R. Mol Cell Biol. 1999;19:1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Izzotti A, Bagnasco M, Cartiglia C, Longobardi M, De Flora S. Int J Oncol. 2004;24:1513–1522. [PubMed] [Google Scholar]
- 18.Lorenz P, Ruschpler P, Koczan D, Stiehl P, Thiesen HJ. Proteomics. 2003;3:991–1002. doi: 10.1002/pmic.200300412. [DOI] [PubMed] [Google Scholar]
- 19.Holcik M, Sonenberg N. Nat Rev Mol Cell Biol. 2005;6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- 20.Shtil AA, Azare J. Int Rev Cytol. 2005;246:1–29. doi: 10.1016/S0074-7696(05)46001-5. [DOI] [PubMed] [Google Scholar]
- 21.Mirgorodskaya OA, Kozmin YP, Titov MI, Korner R, et al. Rapid Commun Mass Spectrom. 2000;14:1226–1232. doi: 10.1002/1097-0231(20000730)14:14<1226::AID-RCM14>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 22.Schnolzer M, Jedrzejewski P, Lehmann WD. Electrophoresis. 1996;17:945–953. doi: 10.1002/elps.1150170517. [DOI] [PubMed] [Google Scholar]
- 23.Doetschman TC, Eistetter H, Katz M, Schmidt W, Kemler R. J Embryol Exp Morphol. 1985;87:27–45. [PubMed] [Google Scholar]
- 24.Boheler KR, Czyz J, Tweedie D, Yang HT, et al. Circ Res. 2002;91:189–201. doi: 10.1161/01.res.0000027865.61704.32. [DOI] [PubMed] [Google Scholar]
- 25.Ray P, De A, Min JJ, Tsien RY, Gambhir SS. Cancer Res. 2004;64:1323–1330. doi: 10.1158/0008-5472.can-03-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D. Science. 2002;295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- 27.De A, Lewis XZ, Gambhir SS. Mol Ther. 2003;7:681–691. doi: 10.1016/s1525-0016(03)00070-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Damen JE, Ware MD, Kalesnikoff J, Hughes MR, Krystal G. Blood. 2001;97:1343–1351. doi: 10.1182/blood.v97.5.1343. [DOI] [PubMed] [Google Scholar]
- 29.Liu L, Damen JE, Cutler RL, Krystal G. Mol Cell Biol. 1994;14:6926–6935. doi: 10.1128/mcb.14.10.6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miura K, MacGlashan DW. Blood. 2000;96:2199–2205. [PubMed] [Google Scholar]
- 31.Wehofsky M, Hoffman R, Hubert M, Spengler B. Eur J Mass Spectrom. 2001;7:39–46. [Google Scholar]
- 32.Wu JC, Inubushi M, Sundaresan G, Schelbert HR, Gambhir SS. Circulation. 2002;105:1631–1634. doi: 10.1161/01.cir.0000014984.95520.ad. [DOI] [PubMed] [Google Scholar]
- 33.Wu JC, Inubushi M, Sundaresan G, Schelbert HR, Gambhir SS. Circulation. 2002;106:180–183. doi: 10.1161/01.cir.0000023620.59633.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Valdimarsdottir G, Mummery C. Apmis. 2005;113:773–789. doi: 10.1111/j.1600-0463.2005.apm_3181.x. [DOI] [PubMed] [Google Scholar]
- 35.Van Hoof D, Passier R, Ward-Van Oostwaard D, Pinkse MW, et al. Mol Cell Proteomics. 2006 doi: 10.1074/mcp.M500405-MCP200. [DOI] [PubMed] [Google Scholar]
- 36.Lee TI, Jenner RG, Boyer LA, Guenther MG, et al. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Evans MJ, Kaufman MH. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 38.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, et al. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 39.Vats A, Bielby RC, Tolley NS, Nerem R, Polak JM. Lancet. 2005;366:592–602. doi: 10.1016/S0140-6736(05)66879-1. [DOI] [PubMed] [Google Scholar]
- 40.Jansen J, Hanks S, Thompson JM, Dugan MJ, Akard LP. J Cell Mol Med. 2005;9:37–50. doi: 10.1111/j.1582-4934.2005.tb00335.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wollert KC, Drexler H. Circ Res. 2005;96:151–163. doi: 10.1161/01.RES.0000155333.69009.63. [DOI] [PubMed] [Google Scholar]
- 42.Frangioni JV, Hajjar RJ. Circulation. 2004;110:3378–3383. doi: 10.1161/01.CIR.0000149840.46523.FC. [DOI] [PubMed] [Google Scholar]
- 43.Jacobs A, Voges J, Reszka R, Lercher M, et al. Lancet. 2001;358:727–729. doi: 10.1016/s0140-6736(01)05904-9. [DOI] [PubMed] [Google Scholar]
- 44.Penuelas I, Mazzolini G, Boan JF, Sangro B, et al. Gastroenterology. 2005;128:1787–1795. doi: 10.1053/j.gastro.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 45.Murphy RC, Clay KL. Biomed Mass Spectrom. 1979;6:309–314. doi: 10.1002/bms.1200060709. [DOI] [PubMed] [Google Scholar]
- 46.Zhang R, Sioma CS, Wang S, Regnier FE. Anal Chem. 2001;73:5142–5149. doi: 10.1021/ac010583a. [DOI] [PubMed] [Google Scholar]
- 47.Mendell JT, Sharifi NA, Meyers JL, Martinez–Murillo F, Dietz HC. Nat Genet. 2004;36:1073–1078. doi: 10.1038/ng1429. [DOI] [PubMed] [Google Scholar]
- 48.Czupalla C, Mansukoski H, Pursche T, Krause E, Hoflack B. Proteomics. 2005;5:3868–3875. doi: 10.1002/pmic.200402059. [DOI] [PubMed] [Google Scholar]
- 49.Arias AM, Hayward P. Nat Rev Genet. 2006;7:34–44. doi: 10.1038/nrg1750. [DOI] [PubMed] [Google Scholar]
- 50.Stamm S, Ben-Ari S, Rafalska I, Tang Y, et al. Gene. 2005;344:1–20. doi: 10.1016/j.gene.2004.10.022. [DOI] [PubMed] [Google Scholar]