Abstract
The BCL-2 family protein BAK is responsible for mitochondrial outer membrane permeabilization (MOMP), which leads to apoptosis. The BCL-2 homology (BH) 3-only protein BID activates BAK to perform this function. We report the NMR solution structure of the human BID BH3–BAK complex, which identified the activation site at the canonical BH3-binding groove of BAK. Mutating the BAK BH1 in the groove prevented activation and MOMP but not the binding of BID. BAK BH3 mutations allowed BID binding and activation but blunted function by blocking BAK oligomerization. BAK activation follows a “hit-and-run” mechanism whereby BID dissociates from the trigger site allowing BAK oligomerization at an overlapping interface. In contrast, the BH3-only proteins NOXA and BAD are predicted to clash with the trigger site, and are not activators of BAK. These findings provide insights into the early stages of BAK activation.
INTRODUCTION
The interaction of pro- and anti-apoptotic BCL-2 proteins determines mitochondrial outer membrane permeabilization (MOMP), a critical decision point in the mitochondrial pathway of apoptosis1. If MOMP occurs, proteins of the mitochondrial intermembrane space, including cytochrome c (cyt c), trigger caspase activation and apoptosis2. Even if caspase activity is blocked or disrupted, the cell usually succumbs to bioenergetic catastrophe as a consequence of MOMP3. Therefore, the control of MOMP by BCL-2 proteins is central to our understanding of this major pathway of cell death.
During MOMP, the pro-apoptotic BCL-2 effector proteins, BAK and BAX, are responsible for the permeabilization event4, and without these neither MOMP nor the engagement of the mitochondrial pathway of apoptosis occurs5,6. BAK and BAX reside in cells in inactive forms and upon activation, oligomerize and insert into the membrane, causing permeabilization and thus apoptosis. One step in this oligomerization appears to involve the exposure of one of the BCL-2 homology (BH) domains, BH3, which then binds into the canonical hydrophobic groove of another BAK or BAX molecule8,9. This groove, present in the anti-apoptotic and pro-apoptotic effector BCL-2 proteins, binds BH3 and often the C-terminal helix of the protein and is called the BC groove7. Anti-apoptotic BCL-2 proteins, such as BCL-2, BCL-xL, and MCL-1, can bind the BH3 of active BAK or BAX, and this is one way in which they block MOMP and apoptosis7,10.
Pro-apoptotic effector and anti-apoptotic BCL-2 proteins are regulated by a subfamily of proteins that share only the BH3 region (“BH3-only proteins”)11. Some of these, particularly BID and BIM, are capable of activating BAK and BAX12,13 and are “direct activators”14. All of the BH3-only proteins bind to BCL-2, BCL-xL, or MCL-17, but the sequestration of BID and BIM is another way in which anti-apoptotic BCL-2 proteins block apoptosis7,10.
Little is known regarding how BID and BIM trigger the activation of BAK and BAX. Peptides corresponding to the BH3 regions can be modified by internal cross-links (the BH3 stabilized alpha-helices of BCL-2 family proteins: SAHB15), and these show enhanced binding to and activation of pro-apoptotic effectors16,17. A BIM or BAX SAHB can bind to a “trigger site” in BAX that was topologically opposite the BC groove, suggesting that this interaction causes the downstream conformational changes that displace the BAX C-terminal helix from the BC groove, promoting MOMP16,17.
To gain insights into the direct activation of BAK during apoptosis, here we characterize the structure of a recombinant, soluble BAK18 bound via its BC groove to a BH3 SAHB of BID, and describe conformational changes that accompany this binding.
RESULTS
BID binding at the BC groove of BAK
To characterize the BID SAHB–BAK interaction, we used a truncated, soluble, human recombinant BAK (cBAK, residues 15–186)18. For the SAHB of BID BH3, we positioned the stabilizing bridge at sites along the hydrophilic side of the amphipathic BID BH3 helix to minimally interfere with the effector-binding interface (Fig. 1a, Supplementary Fig 1a, Supplementary Table 1)15,19,20. Unlike conventional BH3 peptides, BID SAHBs were helical as indicated by circular dichroism (Supplementary Fig. 1c), as described15. Positioning the staple within the C-terminal half of BID BH3 (SAHBa) enhanced BAK binding by ~10-fold over that of the unstapled counterpart, producing a 1:1 complex, as assessed by sedimentation velocity analytical ultracentrifugation (AUC) analysis (Supplementary Tables 1, 2, Fig. 1a). The addition of SAHBa to enriched mitochondria induced the release of cyt c in a BAK-dependent manner (Fig. 1b, Supplementary Fig. 1b; wt mitochondria contain BAK but not BAX). The TROSY spectra of 15N-cBAK in complex with SAHBa or its unstapled counterpart were very similar, showing that the bridge did not induce significant changes in cBAK conformation but instead improved the affinity of the interaction (Fig. 1c, Supplementary Fig. 1d, e).
Figure 1. BID BH3 binds the BC groove to directly activate BAK.
(a) Unstapled and stapled BID peptides. The stabilizing peptide chemical bridge involves the positions indicated by X occupied by pentenylalanines. B is occupied by norleucine. Conserved BH3 residues are colored.
(b) MOMP assays measured cyt c release from purified B6 mouse liver mitochondria after 45 min incubations with the peptides at 5, 0.5 and 0.05 μM. Cyt c in the supernatant (S) and pellet (P) was assessed by Western blotting.
(c) [15N-1H] TROSY spectra of 150 μM 15N-cBAK ± 8× or 1.5× excess BH3a or SAHBa, respectively.
(d) 15N-1H chemical shift perturbations (CSPs) plotted for the SAHBa–cBAK complex in (c) as a function of BAK residue number. Residues with significant CSPs above the calculated threshold (orange line) are located at the BC groove of BAK.
(e) 15N-1H CSPs mapped onto the “closed” apo cBAK structure (PDB ID 2IMT) shown as surface representation. Minimum to maximum CSPs are color coded from white to red. Select side chains of residues within and peripheral to the BC groove are illustrated to help identify the CSPs. (See also Supplementary Fig. 1.)
We characterized the SAHBa–cBAK complex in solution by nuclear magnetic resonance (NMR) spectroscopy. A comparison of chemical shift values for apo cBAK18 and those for the SAHBa–cBAK complex suggested that SAHBa bound within the BC groove of BAK (Fig. 1d, e). Additional SAHBa binding-induced chemical shift perturbations mapped to residues on either side of the BC groove in helices α1 and α6 that did not contact the peptide, probably as a result of changes in the chemical environment as the groove opened to accommodate the incoming SAHBa (Fig. 1c–e). These observations were confirmed through determination of the solution structure of 13C/15N-labeled cBAK bound to unlabeled SAHBa. An ensemble of the 20 lowest-energy structures exhibited backbone atom root mean square deviation of 0.53 Å (Table 1, Supplementary Fig. 2a, b). The lowest energy conformer is used to represent the 3D structure of the SAHBa–cBAK complex in all the figures.
Table 1.
NMR and refinement statistics
| cBAK (SAHBa) | |
|---|---|
| NMR distance and dihedral constraints | |
| Distance constraints | |
| Total NOE | 2702 (324) |
| Intra-residue | 992 |
| Inter-residue | |
| Sequential (|i – j| = 1) | 632 |
| Medium-range (|i – j| < 4) | 511 |
| Long-range (|i – j| > 5) | 567 |
| Intermolecular | 162 |
| Hydrogen bonds | 126 (14) |
| Total dihedral angle restraints | 218 (24) |
| φ | 109(12) |
| ψ | 109(12) |
| Structure statistics | |
| Violations (mean and s.d.) | |
| Distance constraints (Å) | 0.015 ± 0.002 |
| Dihedral angle constraints (°) | 0.352 ± 0.114 |
| Max. dihedral angle violation (°) | 0.632 ± 0.255 |
| Max. distance constraint violation (Å) | 0.019 ± 0.160 |
| Deviations from idealized geometry | |
| Bond lengths (Å) | 0.021 |
| Bond angles (°) | 0.223 |
| Impropers (°) | 1.73 |
| Average pairwise r.m.s. deviation** (Å) | |
| Heavy | 0.92 ± 0.08 |
| Backbone | 0.53 ± 0.10 |
Pairwise backbone and heavy atom r.m.s. deviations were obtained by superimposing residues 23–45 and 65–187 of BAK and 80–101 of SAHBa peptide among 20 lowest-energy refined structures.
BID induces opening of the occluded BC groove of BAK
The SAHBa peptide adopts a conformation with 6 helical turns upon binding within an opened BC groove on cBAK (Fig. 2, Supplementary Fig. 2). The SAHBa peptide binds the BC groove in an orientation seen previously in structures of complexes between BH3 peptides and anti-apoptotic BCL-2 proteins7. In all of these complexes, the hydrophobic faces of the amphipathic BH3 helices bind within the BC groove (Fig. 2a, c, Supplementary Fig. 2e, f). In the SAHBa–cBAK complex, this allowed the bridge-containing hydrophilic residues of SAHBa to face the solvent and limit contact with residues of the BC groove in cBAK (Supplementary Fig. 2b). Further, the loop between BAK helix α1 and α2 exhibited a disordered conformation compared with that of the BAK crystal structure18, although other studies have suggested similar disorder of this loop in apo BAK (Supplementary Fig. 2a)21,22.
Figure 2. BID induces opening of the occluded BC groove in BAK.
The first high-resolution NMR structure for a complex between BAK and a direct activator BH3 peptide is represented in (a) and (c). Surface charge, cartoon, and mixed representations of SAHBa–cBAK (a, c) and apo cBAK (b, d; 2IMT). Images were produced in Pymol (pymol.sourceforge.net). Calpain-susceptible sites exposed upon BAK activation in the presence of membranes or detergents (red and green arrowheads in (c)). (See also Supplementary Fig. 2.)
The SAHBa–cBAK structure displays interactions for the highly conserved Leu90 and Asp95 of BID with a hydrophobic groove pocket and a charged patch in the BH1 of BAK, respectively (Fig. 2a, Supplementary Fig. 2c, g). The side chain of Tyr89 at the end of BAK BH3, positioned centrally to occlude the BC groove in the apo BAK structure (Fig. 2d)18, is displaced by SAHBa, which, in the complex, positions four hydrophobic residues deep and central within the groove—I86, L90, V93, and N97 (Fig. 2c). On the opposite side of the groove from Tyr89, BAK BH1 Arg127 and Asn124 form hydrogen bonds with Asp95 and Asp98 of BID, respectively. Additional details of the binding of SAHBa to the cBAK BC groove are shown in Supplementary Fig. 2 and Supplementary Movies 1 and 2. The SAHBa–cBAK structure suggests that BID binding to the occluded BC groove is a primary event during BAK activation, producing a stable conformer in membrane-free environments.
BID-induced collapse of BAK N-terminal bundle in CHAPS
To study active BAK in cells undergoing apoptosis, we previously used an assay in which conformational changes alter its sensitivity to calpain proteolysis10. In the absence of apoptotic stimulation, native cellular BAK is fully resistant to calpain. Upon activation, BAK becomes susceptible to proteolysis at two positions, at the end of the BH4 and within the BH3 region, therefore producing two stable fragments that stretch to the C-terminus (Fig. 3a). Non-ionic detergents induce the active conformation of BAK in the absence of direct activators, which manifests as sensitivity to calpain and the appearance of two stable fragments10. The small-micelle detergent CHAPS did not induce this conformational change10, and we therefore probed the conformation of the BID–cBAK complexes in CHAPS (Fig. 3, Supplementary Fig. 3). In the absence of detergent, the BID–cBAK complexes, including SAHBa–cBAK, were fully resistant to proteolysis by calpain (Supplementary Fig. 3a). In contrast, in the presence of CHAPS the BH3a–cBAK and SAHBa–cBAK complexes were susceptible to proteolysis by calpain (Supplementary Fig. 3a, 3b).
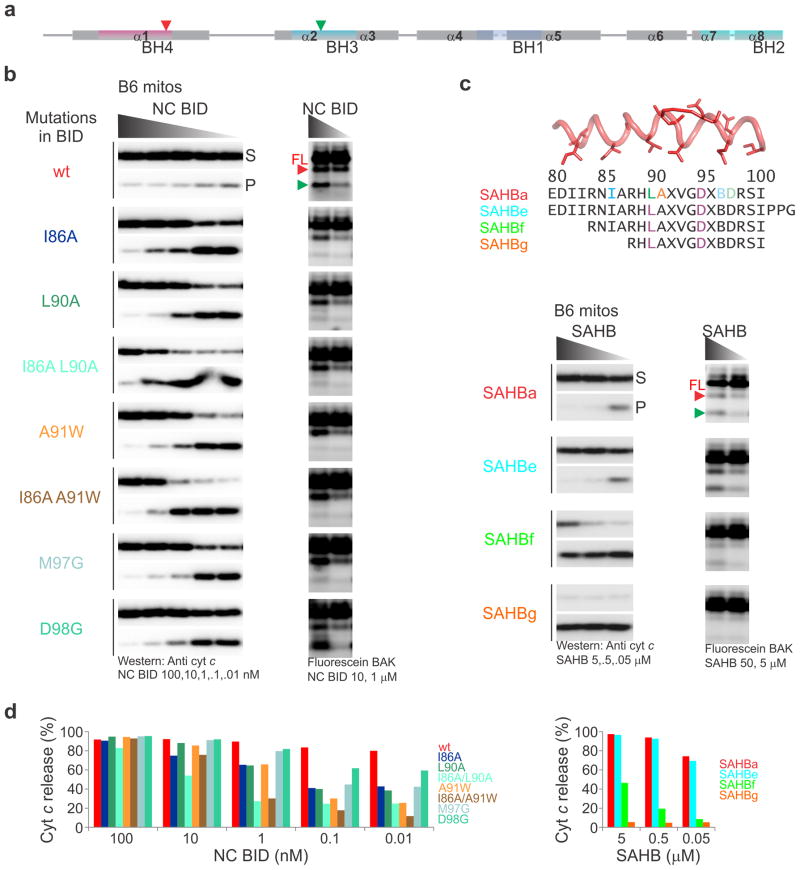
Figure 3. Molecular determinants for BAK direct activation extend throughout BID BH3.
(a) Calpain-susceptible sites exposed during BAK activation (red and green arrowheads).
(b) MOMP (left panels) and calpain sensitivity assays using 1.75 μM cBAK (right panels) were performed as described in Fig. 1b and S3b, respectively, with the indicated purified NC BID as direct activator. Full-length cBAK (FL).
(c) MOMP (left panels) and protease sensitivity assays (right panels) were performed as in (b) with the indicated SAHB as direct activator. DMSO or SAHB (5 μM) alone did not permeabilize bak−/−mitochondria.
(d) Bands from the MOMP images in (b) and (c) were integrated by densitometry, and cyt c release was calculated and represented as histograms. The MOMP and protease sensitivity assays were performed at the same time for all BID ligands. Representative profiles were extracted from larger images. (See also Supplementary Fig. 3.)
Using this approach, we analyzed structure-activity relationships for BID. The opening of the BC groove of cBAK to accommodate SAHBa suggests that the hydrophobic face of the BID BH3 helix is important for BAK activation. We performed extensive mutagenesis of BID BH3 in the context of active, full-length, cleaved, recombinant BID, referred to as NC BID (Fig. 3b, d; BID is activated through proteolysis within its disordered loop between helix α1 and the BH3-containing helix α2)23, and SAHBa (Supplementary Fig. 3c, f, Supplementary Table 1). BID BH3 hydrophobic residue replacements by Ala or Gly, including I86A, L90A, and M97G, resulted in impaired BAK activation and increased the minimal BID concentration required for BAK-mediated MOMP by almost two orders of magnitude (Fig. 3b, d, Supplementary Fig. 3c, f). Equally potent in disrupting BID function was the A91W substitution on the hydrophilic face of BID BH3, likely through clashes of the large Trp side chain with the BH1 of BAK. The combined double substitutions, I86A L90A and I86A A91W, showed an additive effect on BAK-mediated MOMP and active BAK conformation, further disabling BID activation of BAK (Fig. 3b, d, Supplementary Fig. 3c, f). In contrast, the A98G substitution on the hydrophilic side of the BID BH3, predicted to interact with a charged patch in the BH1 of BAK, less dramatically affected function than the hydrophobic residue substitutions (Fig. 3b, d, Supplementary Fig. 3c, f).
We tested deletions of residues at the N terminus and C-terminal extension in the BID SAHB (Fig. 3c, d, Supplementary Table 1). N-terminal deletion of four and eight residues sequentially diminished the ability of SAHB to activate BAK (Fig. 3c, d right panel). Accordingly, deletion of the first 4 residues ablated BAK binding as measured by NMR, while weak binding and activity were detectable by AUC and functional assays, respectively (Supplementary Tables 1, 2). Further deletion completely blocked binding and activity (Supplementary Tables 1, 2). Addition of three native residues at the C-terminus did not significantly improve SAHB binding or activity (Fig. 3c, d right panel). Our combined mutagenesis and truncation analysis suggests that effective activation of BAK by BID requires critical hydrophobic contacts by BID at the four most central hydrophobic residues, I86, L90, V93, and M97.
Structural basis for a “hit-and-run” activation mechanism
In contrast to the high-affinity interactions between anti-apoptotic BCL-2 proteins and their BH3 ligands (Kd ~10 nM)24,25, BAK direct activation may be governed by a transient “hit-and-run” mechanism11,26. Accordingly, direct activator BH3 ligands transiently engage the BC groove of BAK with moderate affinity (Kd ~10 μM) to induce the conformational changes seen in the SAHBa–cBAK structure. In the presence of biological membranes, direct activator BH3-only proteins dissociate from BAK26. We tested the hit-and-run model using engineered direct activator–effector complexes produced through disulfide bonding of BID and BAK. We introduced single cysteines in BID BH3 at I83, I86, M97, D98, and I101. Given their minimal effect on NC BID and SAHBa function (Supplementary Fig. 3c, e, f), these cysteines provided disulfide bond “trapping” sites in BID. Guided by the SAHBa–cBAK structure, we engineered single cysteines in the BH2 region, proximal to BID Ile101, and produced them in BAK containing serine substitutions at the naturally occurring cysteines (C14S C166S, Fig. 4a, b, Supplementary Fig. 4a, b).
Figure 4. Activator–effector complexes support a “hit-and-run” activation mechanism.
(a) Schematic representation of the SAHB–BAK complexes highlighting the engineered disulfide “traps” between I101C SAHBq and the BH2 or helix α6 of cBAK. For BH2 mutants of BAK Cys14 and Cys166 were replaced by Ser.
(b) Cartoon representation of the contacts Ile101 of BID makes at the BC groove of BAK.
(c) Protocol flowchart for the production and analysis of SAHBq–cBAK complexes.
(d) Oxidized SAHBq–cBAK complexes (orange arrowheads) migrate more slowly than their reduced counterparts (red arrowheads), as shown by SDS-PAGE.
(e) Protease sensitivity assays were performed at 24°C in 50 mM HEPES pH 7.5, 1 μM m-calpain, 0.5 mM CaCl2, 1% CHAPS, and the oxidized complexes in (d) ± 50 μM SAHBa in the absence of reducing agent. After 80 min, the reaction was quenched in SDS sample buffer ± reducing agent. Schematic representation of conformational changes and protease susceptibility are illustrated for each condition.
(f) MOMP assays tested cyt c release from purified bak−/− mouse liver mitochondria after 1 h incubations of the proteins in (d, lower panel) in the absence or presence of NC BID ± ABT-737. Cyt c in the supernatant (S) and pellet (P) was assessed by Western blotting. The MOMP and protease sensitivity assays were performed at the same time for all BID–BAK complexes. Representative profiles were extracted from larger images. (See also Supplementary Fig. 4.)
The efficiency of cross-linking to a BID SAHB containing the I101C substitution (SAHBq, Fig. 4c, d, Supplementary Fig. 4c, Supplementary Table 1) was proportional to the amount of SAHB. At a 2.5 molar excess of SAHBq to cBAK (20 μM), L183C and G184C (in cBAK) cross-linked to I101C in SAHBq with ~40–80% efficiency upon oxidation with copper/1,10-phenanthroline (Cu/Phe) (Fig. 4d; the orientation of SAHBq at the BC groove in these complexes is illustrated in Fig. 4a, b). We also produced an I101C SAHBq adduct to cBAK Cys166 with ~20% efficiency. This “trap” site, on the face opposite the BC groove, was expected to allow free motion of the SAHB about the disulfide bond without perturbing the access to the BC groove (Fig. 4a). The SAHBq–cBAK oxidized complexes migrated more slowly than their reduced counterparts during SDS PAGE (Fig. 4d).
We tested calpain sensitivity of SAHBq–cBAK oxidized complexes in CHAPS with or without excess SAHBa to assess their activation conformation. The disulfide-bonded SAHBq–cBAK BH2 adducts, but not the Cys166-targeted adduct, were completely resistant to calpain proteolysis (Fig. 4e). Adding excess SAHBa activated both of the SAHBq–cBAK BH2 complexes (Fig. 4e). These results suggested that SAHBq–cBAK BH2 complexes potently “trapped” SAHBq at the BC groove, preventing the conformational change associated with calpain sensitivity, whereas the Cys166-targeted complex activated BAK in trans, supporting the “hit-and-run” model. The SAHBq–cBAK BH2 complexes required excess SAHBa to partially displace the “trapped” SAHBq by competition at the BC groove, as seen in the SAHBa–cBAK structure. This is likely facilitated by the ability of the BAK BH2 to undergo the conformational changes associated with activation and move away from the BH3 (Fig. 4e).
Oxidation at 5× molar excess of peptide to cBAK (100 μM) resulted in efficiencies of cross-linking for I101C SAHBq to L183C and G184C cBAK of >95% and to Cys166 cBAK of ~50% (Supplementary Fig. 4c). The efficiently oxidized SAHBq–cBAK complexes were tested by gel filtration chromatography to assess oligomerization. The L183C and G184C SAHBq–cBAK traps were monomeric and remained so even when a 4× excess SAHBa was supplemented (Supplementary Fig. 4d, e). In contrast, the Cys166 trap displayed a monomeric to oligomeric chromatography profile eluting as species of ~22 kDa to >670 kDa. To probe the accessibility of SAHBq in the Leu183Cys and Gly184Cys traps, we applied the gel filtration samples to an Ni2+-NTA agarose affinity column preloaded with BCL-xL containing a C-terminus His6 tag (Supplementary Fig. 4f). While the Cys166 trap was largely retained by the BCL-xL affinity column, both BH2 trapped proteins were not (Fig. S4f). This supports the notion that neither the SAHBq nor the BAK BH3 were accessible to BCL-xL in the L183C and G184C traps.
We next tested the activity of purified SAHBq–cBAK traps in MOMP assays. We originally showed that cBAK displays synergism with NC BID during MOMP of mitochondria from bak−/− MEFs, albeit in a narrow range of concentrations (5–0.5 μM cBAK), likely due to poor membrane targeting in the absence of the transmembrane region18. To accurately assess the activity of the SAHBq–cBAK traps in MOMP assays, we aimed to produce traps devoid of contaminating cBAK species. The Cys166 trap preparations consistently co-purified with contaminating Cys166–Cys166 cBAK dimer (Fig. 4d lower panel). It has therefore remained impossible to accurately assess activity of the Cys166 trap although it consistently displayed more activity than Cys166 cBAK (Fig. 4f, compare C166 and C166 trap). The L183C and G184C traps that flowed through BCL-xL affinity columns provided largely homogeneous preparations (Fig. 4d, Supplementary Fig. 4f). Consistent with data presented in Fig. 4e, the BH2 tethered SAHBq traps were inactive in effecting MOMP in response to NC BID, whereas at similar concentrations the untethered Leu183Cys and Gly184Cys cBAK showed robust release of cyt c (Fig. 4f, Supplementary Fig. 4h). Moreover, addition of the BCL-2 and BCL-xL antagonist ABT-737 (ref. 27) failed to enhance the activity of the BH2-tethered traps, but augmented that of the untethered cBAK mutants (Fig. 4f), excluding the possibility that contaminating BCL-xL may have prevented their activation. Our structure-based disulfide-bonded SAHBq–cBAK traps support the hit-and-run BAK activation mechanism, which relies on transient engagement of the BC groove by BID.
BAK activation resolved by structure-guided mutagenesis
We generated structure-guided mutants of BAK and examined direct activation, oligomerization, and cell death (Fig. 5a, b). To test function in a cellular system using a variety of apoptotic stimuli, we stably expressed BAK mutants in bak−/− bax−/− MEFs (Fig. 5b, c). Consistent with other studies8,28, mutation of the BH1 residue Gly126 to alanine rendered BAK inactive for apoptosis (Fig. 5c, Supplementary Fig. 5), cyt c release (Fig. 5d), and oligomerization (Fig. 5e). The G126A mutant was expressed at higher levels than wt in DKO MEFs and displayed reduced protease sensitivity in enriched mitochondria activated by NC BID (Fig. 5b, f). Similarly, G126A substitution prevented the expected conformational change in recombinant cBAK as assessed by SAHBa-induced protease sensitivity in CHAPS (Fig. 5g). While the affinity of the BID–BAK interaction is an important determinant for BAK direct activation, BID binding to the G126A cBAK, as detected by various methods including AUC and NMR (Supplementary Table 1), was not sufficient to induce the specific conformational changes seen for SAHBa–wt cBAK detected by NMR and the protease sensitivity assay.
Figure 5. Direct activation and oligomerization separated by point mutations in BAK.
(a) Cartoon representation of the mutated BC groove residues to test direct activation, oligomerization, and BAK function.
(b) Protein levels of BAK reconstituted in DKO MEFs was assessed by Western blotting.
(c) Cell death of wt and BAK mutants stably reconstituted in DKO MEFs was induced with 1 ng ml−1mouse TNF and 0.5 μg ml−1 CHX, 5 mJ UV or 0.5 μM ActD. After 24 h the cells were prepared for FACS analysis to monitor the extent of apoptosis as measured by annexin V positivity. Data are represented as mean ± SD (n=3).
(d) MOMP assays tested cyt c release from enriched mitochondria purified from bak−/− bax−/− MEFs reconstituted with the corresponding BAK mutants after 45 min incubations with 10, 1, 0.1, 0.01, or 0 nM NC BID. Cyt c in the supernatant (S) and pellet (P) was assessed by Western blotting.
(e and f) BMH cross-linking and protease sensitivity assays of the mitochondrial pellets in (d) were performed for an additional 30 min and 45 min, respectively, at 24°C. BAK (FL) in the pellet (P) was assessed by Western blotting with N-terminus (e) and internal targeted antibodies (f).
(g) Protease sensitivity assays probed the conformation of purified wt and mutant cBAK (FL) at 24°C in 50 mM HEPES, pH 7.5, 10 mM dithiothreitol (DTT), 1 μM m-calpain, 0.5 mM CaCl2, and 1% CHAPS ± 50 μM SAHBa. (See also Supplementary Fig. 5)
In contrast, while an I81A mutation in the BH3 of BAK impaired BAK oligomerization (Fig. 5e) and apoptosis in reconstituted DKO MEFs (Fig. 5c, Supplementary Fig. 5), it showed potent NC BID- and SAHBa-induced conformational changes, as assessed by protease sensitivity (Fig. 5f, g). Although Ile81 of BAK makes direct contact with Ile101 of BID, it appears to be dispensable for BID-induced activation of BAK, as it does not contribute significantly to binding or the conformational changes detected by protease sensitivity in CHAPS (Supplementary Table 1, Fig. 5f, g). We suggest that upon activation, I81A BAK exposes its BH3 (ref. 10) but may fail to engage BH3-to-BC groove-mediated oligomerization8, although a structural basis for this is not provided in our study.
The lack of apoptotic activity of both G126A and I81A correlated with their poor ability to mediate MOMP in enriched mitochondria from bak−/− bax−/− MEFs reconstituted with the respective mutants (Fig. 5d). It is noteworthy that wt BAK, purified with native mitochondrial membranes from reconstituted bak−/− bax−/− MEFs, showed less accumulation of the smaller calpain-resistant fragment upon activation with NC BID than did the I81A mutant (Fig. 5f). This is a fragment produced by proteolysis within the BH3. In contrast, upon NC BID activation, wt BAK mostly accumulated the larger BH3-containing calpain-resistant fragment, possibly because the BH3 was efficiently buried and therefore protected in the oligomer, consistent with the postulated BH3-to-BC groove oligomerization interface8. The decreased oligomerization in the I81A mutant correlated with a higher level of the smaller fragment upon proteolysis, suggesting that this region remained exposed in the activated mutant molecule.
Our testing of these and other BAK mutants reconstituted in DKO MEFs using a diverse array of apoptotic conditions, including TNF/cycloheximide, UV, and actinomycin D (Fig. 5a–c, Supplementary Fig. 5), suggested that BAK-mediated apoptosis followed a similar activation route regardless of the specific activation mechanisms engaged. Receptor-mediated apoptosis activated BID to engage the mitochondrial pathway29, but the other apoptotic stimuli tested did not, relying instead on additional direct activator BH3-only proteins such as BIM, which therefore are likely to trigger BAK activation by also engaging the BC groove3.
NOXA and BAD are not direct BAK activators
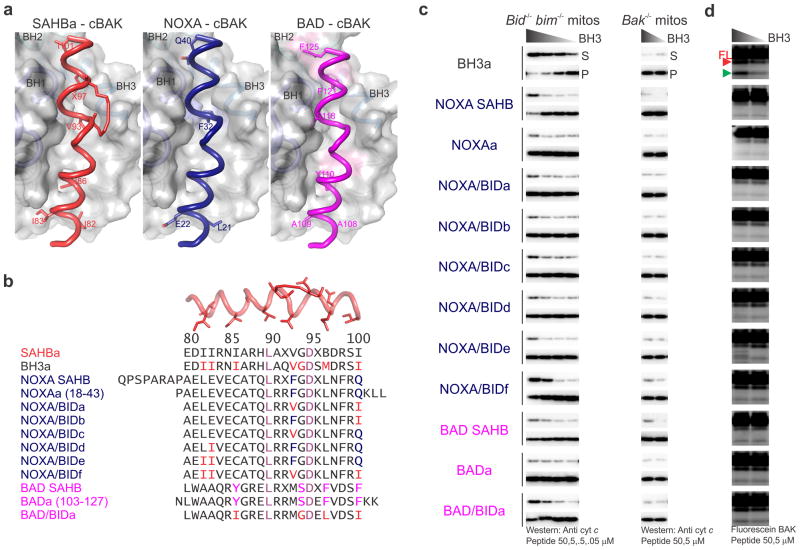
A long-standing issue in the BCL-2 family regulation of MOMP has been the unpredictable specificity of BH3-only direct activators for the effectors BAK and BAX. To date, BID and BIM have remained the two most potent direct activators of both BAK and BAX7,25, although reports have potentially implicated PUMA30,31 and NOXA28,32 as direct activators of the effectors. Our structural analysis provided an opportunity to test the direct activation function based on the specificity of BID BH3 for the trigger site in BAK. Based on modeling of the BH3 region of BAD and NOXA over that of BID in the SAHBa–cBAK complex, we predicted that BAD and NOXA BH3 would produce significant clashes with the BC groove of BAK (Fig. 6a). The BAD BH3 would project four residues corresponding to I86(W), G94(S), M97(F), and I101(F) within 2Å of BC groove residues in BAK; the first three N-terminal residues represent the minimal BH3 core required for potent direct activation by BID (Fig. 6a, b). Similarly, NOXA BH3 would project two residues within 2Å of the BC groove corresponding to V93(F) and I101(Q), the latter representing a hydrophobic-to-polar substitution (Fig. 6a, b). We therefore tested BAD and NOXA BH3 peptides in MOMP assays using enriched mitochondrial fractions from bid−/− bim−/− liver (Fig. 6, Supplementary Fig. 6). In contrast to BID BH3a, NOXAa and BADa peptides were largely inactive even at 50 μM (Fig. 6c). Furthermore, we found that NOXAa and BADa peptides were unable to activate cBAK in the protease sensitivity assays (Fig. 6d). Moreover, the stapled NOXA SAHB and BAD SAHB were unable to robustly activate BAK, although NOXA SAHB induced weak BAK-mediated MOMP at the highest concentration tested (Fig. 6b, c). In other studies, membrane disruption by NOXA BH3 peptide was observed at non-physiologically high concentrations of cBAK (5 μM), although NOXA BH3 did not induce MOMP in digitonin-permeabilized bid−/− bim−/− MEFs32. We systematically engineered NOXA and BAD BH3s to contain BID substitutions at the positions predicted to clash at the BC groove of BAK. By replacing at least 4 residues to those of BID (NOXA/BIDf and BAD/BIDa in Fig. 6b) both peptides were weakly active in triggering BAK-mediated MOMP and induced enhanced protease sensitivity of purified cBAK in CHAPS associated with direct activation (Fig. 6b, c). In addition, we tested N-and C-terminal extensions in NOXA that share the minimal BH3 core and observed no evidence of direct BAK activation by any (Supplementary Fig. 6). We therefore conclude that BAD and NOXA BH3 are not direct activators of BAK.
Figure 6. NOXA and BAD are not direct activators of BAK.
(a) Human NOXA (blue) and human BAD BH3 (magenta) were modeled over BID (red) in the SAHBa–cBAK complex. The BC grooves of BAK found within 2Å of NOXA and BAD BH3 are colored light blue and pink, respectively.
(b) BH3 peptide alignment. The NOXAa and BADa BH3 peptides were previously shown to be inactive at inducing MOMP in digitonin permeabilized bid−/− bim−/− MEFs32.
(c) MOMP assays tested cyt c release from purified bid−/− bim−/− and bak−/− mouse liver mitochondria after 1 h incubations with peptide. Cyt c in the supernatant (S) and pellet (P) was assessed by Western blotting.
(d) Protease sensitivity assays with the respective peptides were performed as in Fig. S3b. The MOMP and protease sensitivity assays were performed at the same time for all BH3–BAK complexes, with the exception of NOXA SAHB and BAD SAHB complexes, which were performed at the same time with the peptides in Supplementary Fig. 6. Representative profiles were extracted from larger images. (See also Supplementary Fig. 6.)
Discussion
When the pro-apoptotic effector proteins, BAX and BAK, bind to the BH3 of BID or BIM, they undergo conformational changes, followed by oligomerization and insertion into the outer mitochondrial membranes to effect MOMP. This is the pivotal step in the life-death decision of the mitochondrial pathway of apoptosis, the primary way in which vertebrate cells die. The solution structure of the BID BH3 bound to BAK provides insights into this key activation process. BID BH3 opens a canonical BC groove in BAK in a manner that is strikingly similar to that of other structures of BH3 binding to anti-apoptotic BCL-2 family proteins7 and evidently to BAX33. We validated the activation site of BAK at its BC groove in several structure-function assays in vitro, and in cell culture by extensive mutagenesis of both the effector and the direct activator, consistent with studies performed with truncated28 and full-length forms of BAK34. Previously unresolved aspects of the role of BID as direct activator28,34 emerged from our analysis, including the minimal helical BH3 region required for strong activation of BAK, defined as residues 80–101, which displays extensive interactions at the BC groove of BAK, dominated by the four central conserved hydrophobic residues I86, L90, V93, and M97.
Direct activation of BAK by BID in the absence of membranes induced significant rearrangements in the BH3, which opened the occluded BC groove by destabilizing contacts between the BH1 and BH3 observed in free BAK18. Although BH3–BCL-2 family complexes have revealed details of the interactions between BH3 peptides and BC grooves of anti-apoptotic BCL-2 proteins, the effectors manifest multi-level binding-induced conformational changes that are significantly augmented by membranes, their activation being poorly correlated with BH3 binding in membrane-free environments, although fully dependent on it33. The BID-induced BAK activation fully manifested in the presence of membranes, which support the shedding of the N-terminal bundle of BAK (N terminus to BH3) from the C-terminal bundle (BH3 to C terminus), as indicated by the protease sensitivity assay in CHAPS detergent. We found that Gly126 mutations in the BH1 of BAK prevented binding-induced conformational changes by BID, and thereby prevented all activation events. In contrast, I81A in the BAK BH3 did not prevent these conformational changes resulting in protease susceptibility (exposure of the BH3), but prevented subsequent BAK–BAK oligomerization, MOMP, and apoptosis. Therefore, through analysis of these different BAK mutants, critical individual steps in BAK activation were separated: BID BH3 binding, exposure of its BH3, displacement of the BID BH3 by a neighboring BAK BH3, and oligomerization. Our study, while providing no structural insights into oligomerization, is consistent with the proposed activation mechanism of BAX by BID33.
In our current understanding of BAK effector activation, BAK is constitutively targeted to the outer mitochondrial membrane via a C-terminal hydrophobic tail, which does not modulate the pro-apoptotic activity of BAK beyond targeting the membrane28,34,35. The membrane-tethered BCL-2-like core of BAK contains the necessary structural components associated with MOMP activity (Fig. 7). BAK is dormant in the absence of apoptotic stimulation, identified by resistance to calpain proteolysis. The direct activators BID and BIM, but probably not NOXA or BAD, transiently engage the BCL-2 core of BAK in the manner of the SAHBa–cBAK complex during apoptosis. BH3-to-BC groove binding destabilizes the BCL-2 core by unwinding the BH3 and restructuring the BH1–BH3 hydrophobic core. This creates a deep cavity that potentially weakens the interactions of the long BH4-containing helix α1 at the bottom of the BC groove. Further unwinding of the BCL-2 core is disfavored in membrane-free environments. In native BAK, proximity to the membranes facilitates membrane insertion: BAK’s C-terminal bundle (BH3 to C terminus) probably inserts, while the N-terminal bundle (N terminus to BH3) unfolds, becoming sensitive to calpain proteolysis. This is facilitated by the “hit-and-run” nature of the activator BH3, which in the presence of membranes dissociates from the effector BC groove, freeing it for interaction with an adjacent active BAK monomer that supplies its exposed BH3 for a BH3-to-BC groove oligomerization interface8,36. If hydrophobic residues that may be exposed in active BAK are critical for disruption of the membrane, we can then hypothesize that consolidation of such disruption events may be necessary for MOMP. This, then, may be the function of additional BAK–BAK interactions, such as that described as α6–α6 (ref. 37). By bringing together local areas of lipid disruption, a “hole” may develop. Studies have suggested that while permeabilization of a membrane by four molecules of the effector protein BAX is sufficient to release small molecules, approximately 20 activated BAX molecules are needed to release proteins38. The precise nature of the interactions that permit such large oligomers to manifest remains unknown.
Figure 7. Direct activation triggers BAK-mediated MOMP.

BAK-mediate MOMP follows a multistep activation–oligomerization mechanism. Dormant BAK is protease resistant, soluble, monomeric and is not kept in check by anti-apoptotic BCL-2 proteins10,18. During direct activation in the absence of membranes, BID engages BAK as a soluble, stable 1:1 BH3-to-BC groove complex that remains resistant to proteolysis. In the presence of the outer mitochondrial membrane (OMM) or CHAPS the 1:1 BID–BAK active complex opens up as it associates with the lipidic milieu, becoming protease sensitive at the BH4 and the BH3. Poorly understood membrane interactions likely facilitate the dissociation of BID from the BC groove, conforming to the “hit-and-run” activation mechanism7,10. The BH1 Gly126 mutants block binding-induced conformational changes associated with direct activation. Oligomeric BAK represents the end-stage of BAK activation, being membrane associated and presumed to be mediated by two oligomerization interfaces as shown8,36,37. As the BH3 participates in the BH3-to-BC groove interface, it becomes buried and resistant to proteolysis, and only the BH4 remains accessible in oligomeric BAK. The BH3 Ile81 mutants block oligomerization by preventing the BH3-to-BC groove, although inhibitory mutations have not yet been identified at the alternate interface.
In contrast to BAK, activation of the effector BAX during MOMP involves an additional step of targeting it from the cytosol to the outer mitochondrial membrane. This step is reversed by BCL-xL and potentially by other anti-apoptotic BCL-2 proteins, which help retrotranslocate BAX from the outer mitochondrial membrane to the cytoplasm39. Whether mitochondrial translocation requires binding of the BAX trigger site opposite the BC groove16,17 or the BC groove itself, as seen for SAHBa–cBAK, has not been clearly resolved, but the binding of the BID BH3 to the BC groove of BAX, in a manner similar to BAK, has been resolved at high resolution33. Potentially, BAX activation is governed by a hit-and-run mechanism involving sequential transient engagement of both trigger sites, as supported by a recent study that compared the BH3 binding interface of the full-length isoforms of BAX and BAK34. BAX triggering also exhibits similarities to BAK, in that engagement of the BAX BC groove by BH3 activators in lipidic milieus alters BAX conformation40, including a straightening of hydrophobic helices α5 and α6 (ref. 33), which we did not observe in our structure of BID SAHBa–cBAK. Further, a structural basis for BH3-to-groove BAX oligomerization has potentially been elucidated33. Our study with BAK and the recent efforts with BAX33 have thus provided missing insights into the nature of the initial events that precipitate the activation of BCL-2 effectors during apoptosis.
Online Methods
Mammalian and bacterial protein expression, purification, and site directed mutagenesis
The human BAK and BID constructs and mutants thereof used in this study were engineered for bacterial or mammalian expression as follows. Full-length BAK was expressed from pMX-IRES-GFP retroviral vector41. The retroviral vectors were transiently transfected using Lipofectamine 2000 in Phoenix Amphotropic or Ecotropic virus packaging helper cell lines. After overnight recovery in 10% fetal calf serum (FCS) and Dubelco modified Eagle’s medium (DMEM) supernatants containing virus were used to infect bak−/− bax−/− mouse embryonic fibroblasts (MEFs). Stable GFP-positive MEFs were sorted three times by fluorescence activated cell sorting (FACS) to enrich for the high-expressing BAK population. BAK expression in reconstituted DKO MEFs was assessed by immunoblotting with an anti-BAK polyclonal (G23 Santa Cruz; 1:1000 dilution) or a monoclonal antibody (Ab-1 EMD Millipore; 1:1000 dilution). Protein loading was determined by immunoblotting with the anti-actin monoclonal antibody (C4 Millipore; 1:100000 dilution). Secondary anti-mouse or anti-rabbit HRP-conjugated antibodies were used at 1:20000 dilution (Millipore).
The bacterial expression of BAK was based on our previous study in which the C terminus-truncated BAK-ΔTM was engineered with a four-residue leader to improve protein expression levels (MEAS-A2SGQGP … LNLGNG186LEHHHHHH)18. The bacterial expression of BID was based on pGEX-4T-1 vector producing a GST-BID fusion protein containing a thrombin cleavage site between the GST and BID. C terminus-truncated mouse BCL-xL-ΔTM, referred to in the text as BCL-xL or BCL-xL-His6, was expressed from the pET29d vector42. Rat m-calpain and μ-calpain protease core, μI-II, were expressed as previously reported43,44. All His-tagged proteins were purified using multiple chromatography steps. Batch Ni2+-NTA affinity chromatography (Qiagen) was followed by fast protein liquid chromatography (FPLC) using S100 or S200 gel filtration and Q-Sepharose columns (GE Healthcare). Purified MEAS-BAK-ΔTM was subjected to overnight μI-II proteolysis to remove the N-terminus (the MEAS leader and the first 14 residues) and the C-terminal His tag, followed by additional Q-Sepharose FPLC purification to give rise to calpain-proteolyzed cBAK (residues 15–186), essentially the stable, soluble BCL-2-like core of BAK18. We also modified cBAK with the thiol-reactive fluorescein-5 maleimide tag (F150, LifeTechnologies), which reacted with the remaining Cys166 in cBAK (the other, Cys14, is absent in cBAK). We refer to this fluorescein conjugate as cBAK-F150, which was further purified by S100 and Q-Sepharose FPLC to remove excess unreacted F150. GST-BID was batch purified on GSH resin, and tag proteolysis by thrombin was performed overnight directly on the resin. The digested material is NC BID, as thrombin also digests the intrinsically disordered loop in BID. NC BID was subsequently purified by Q-Sepharose FPLC.
Site directed mutagenesis was performed using the QuikChange II and II XL protocols (Agilent Technologies). The mutagenesis primers were designed using the QuikChange primer design tool (www.genomics.agilent.com).
Synthesis, hydrocarbon stapling, and characterization of BID BH3 peptides
Peptide synthesis and hydrocarbon stapling of BID BH3 were performed at the Hartwell Center for Bioinformatics and Biotechnology at St. Jude Children’s Research Hospital. All peptides were N-terminal acetylated and C-terminal amidated. In the hydrocarbon-stapled versions, Met97 was replaced with norleucine (Nle). Ruthenium-mediated ring-closing olefin metathesis (RCM) was performed for the stapled peptides, all of which were synthesized with the alanine analogue, (S)-N-Fmoc-2-(4′-pentenyl) alanine using the previously described procedure15,19,45. All the peptides were HPLC purified to greater than 90% homogeneity, and their molecular weights were confirmed by mass spectrometry.
For circular dichroism (CD) analysis, BID BH3a and SAHBa were dissolved in 10 mM phosphate buffer (pH 7.0) at 25 μM. CD spectra were obtained at 25°C on an AVIV model 202-01 spectropolarimeter; four scans were averaged using 1 nm bandwidth and a 1 mm path length quartz cell (Hellma). Molar ellipticity was calculated as CD/[number of amino acids × concentration (mol/L) × path length (0.1 cm) × 10 (conversion factor to decimoles)].
NMR titrations and structure determination
Uniformly 13C/15N-labeled or 15N-labeled BAK was produced in the MEAS-BAK-ΔTM-expressing BL21* cells according to the standard minimal M9 medium bacterial growth protocols supplemented with 15NH4Cl and with 13C-glucose as appropriate46. Labeled BAK was then purified, processed with calpain and repurified as with all our unlabeled BAK proteins to obtain highly purified, uniformly labeled cBAK. We previously assigned ~90% of the backbone resonances for free cBAK, residues 15–186 (ref. 18). [15N/1H] TROSY NMR titrations of 15N-labeled cBAK with unlabeled wt and mutant BID BH3 and BID SAHB peptide ligands were performed at 30°C in a buffer containing 10% D2O and either 20 mM HEPES pH 6.8 or 20 mM phosphate buffer pH 6.8. BID peptide stocks were prepared at 100 mM in deuterated dimethylsulfoxide (DMSO-d6) (Cambridge Isotope). 15N-1H chemical shift perturbations were calculated as CSPs = [(Δ1H ppm)2 + (Δ15N ppm/5)2]1/2.
Using the best-behaved BID SAHBa, which showed the highest affinity for cBAK in solution, we determined the structure of its complex with cBAK by NMR spectroscopy using 13C/15N-labeled cBAK and unlabeled BID SAHBa in a 20 mM phosphate buffer pH 6.8 and 10% D2O at 30°C. This sample was stable during the entire period of data collection required for structure determination47. We collected the NMR spectra using Bruker Avance 600 MHz or 800 MHz spectrometers equipped with a cryogenically cooled triple resonance z-gradient probe. Data processing was performed in TopSpin (Bruker BioSpin) and spectral analysis in Cara48. For backbone assignment of cBAK in the SAHBa–cBAK complex we performed three triple-resonance experiments including HNCA, HNCACB, CBCA(CO)NH, which were analyzed in conjunction with a [15N-1H] TROSY spectrum. HA and HB assignments for BAK were made using HBHA(CBCACO)NH spectra aided by 15N-edited [1H-1H] NOESY. The remaining aliphatic carbon and hydrogen resonances were assigned using 13C-detected HCC-TOCSY and 1H HCCH-TOCSY along with [13C-1H]-HSQC spectra. The aromatic [13C-1H]-TROSY and 13C-edited [1H-1H] aromatic NOESY spectra were used in obtaining the side chain resonances of aromatic residues. We assigned the SAHBa peptide resonances from 2D-[15N-13C] filtered TOCSY and NOESY experiments. The intermolecular cBAK–SAHBa NOEs were obtained from 3D spectra of 13C/15N-half-filtered spectra of 15N-edited NOESY and 13C-edited NOESY experiments for the complex. Structure calculations were performed originally with automatic NOE peak picking in UNIO49 and CYANA50 (Table 1). Manual structure refinement was performed in CYANA. The CYANA library files for the unnatural amino acids pentenylalanine, which forms the stabilizing bridge, and for norleucine were generated and used in the structure calculations. Ramachandran analysis of the lowest energy SAHBa–cBAK structures identified 87.8, 10.8, 0.9 and 0.5% of residues in allowed, additionally allowed, generously allowed and disallowed regions, respectively.
Analytical ultracentrifugation
Experiments were carried out in a ProteomeLab XL-I analytical ultracentrifuge with an eight-hole rotor (Beckman An-50Ti) and cells containing sapphire or quartz windows and charcoal-filled Epon double-sector centre pieces (Beckman Coulter, Fullerton, CA). AUC was performed in buffer 20 mM HEPES pH 7.0, 100 mM NaCl, 0.5 mM DTT. The density and viscosity of the ultracentrifugation buffer at 20°C were measured with a DMA 5000M density meter and an AMVn viscometer (Anton Paar USA, Ashland, VA). The partial specific volume at 20°C and the molecular weights of the proteins and peptides were calculated based on their amino acid composition using the software SEDNTERP51. All samples were dialyzed against the ultracentrifugation buffer and this buffer was used as optical reference.
For the sedimentation velocity experiments the loading volume of 400 μl was identical for the reference and sample chambers of the double-sector centrepiece. Fringe displacement data at time intervals of 1 min were collected with the Rayleigh interference system for 12 h at a rotor speed of 50k rpm and analysed with SEDFIT software (www.analyticalultracentrifugation.com) using the model for continuous sedimentation coefficient distribution c(s) with deconvolution of diffusional effects52,53.
The sedimentation coefficient distribution c(s) was calculated with maximum entropy regularization at a confidence level of p = 0.68 and at a resolution of sedimentation coefficients of n = 100. The positions of the meniscus and bottom, as well as time-invariant and radial noises, were fitted. The velocity data were also fitted to the single-site hetero-association model (A+B↔AB, with the peptide species A and cBAK species B) using the SEDPHAT software (www.analyticalultracentrifugation.com).
Sedimentation equilibrium was attained at a rotor temperature of 20°C at increasing speeds of 16k rpm (28 h), 20k rpm (24 h) and 30k rpm (24 h)54. Protein at concentrations of between 3.4 and 14.1 μM (130 μL) were loaded into double-sector centrepieces and absorbance distributions recorded at 280 in 0.001 cm radial intervals with 20 replicates for each point. Global least squares modelling were performed at multiple rotor speeds with the software SEDPHAT using a single species model as well as the single-site hetero-association model (A+B↔AB, with the peptide species A and cBAK species B.)54.
Supplementary Material
Acknowledgments
We thank C. Dillon, L. McCormick and M. Young for managing the mouse colony, R. Cassell and P. Rodriques of the St. Jude Hartwell Center for producing the peptides, and R. Cross and G. Lennon of the St. Jude Flow Cytometry facility for cell sorting. This work was supported by US National Institute of Health grants AI40646, GM52735, and GM096208 (D.R.G.), by R01CA082491 and 1R01GM083159 (R.W.K.), and by the American Lebanese Syrian Associated Charities.
Footnotes
Accession Codes
Protein Data Bank: The NMR solution structure of SAHBa–cBAK has been deposited under accession codes 2M5B. Biomagnetic Resonance Bank: Related BMRB entry to PDB 2M5B has been deposited under accession code 19045.
Author Contributions
T.M. and C.R.G. contributed the NMR analyses, T.M., F.L. and P.F. the biochemical analyses, and T.M. and A.N. the AUC analyses. All authors contributed conceptually to various aspects of the project. T.M., R.W.K., and D.R.G wrote the manuscript.
References
- 1.Green DR. Apoptotic pathways: ten minutes to dead. Cell. 2005;121:671–4. doi: 10.1016/j.cell.2005.05.019. [DOI] [PubMed] [Google Scholar]
- 2.Jiang X, Wang X. Cytochrome C-mediated apoptosis. Annu Rev Biochem. 2004;73:87–106. doi: 10.1146/annurev.biochem.73.011303.073706. [DOI] [PubMed] [Google Scholar]
- 3.Tait SW, Green DR. Mitochondria and cell death: outer membrane permeabilization and beyond. Nat Rev Mol Cell Biol. 2010;11:621–32. doi: 10.1038/nrm2952. [DOI] [PubMed] [Google Scholar]
- 4.Kuwana T, et al. Bid, Bax, and lipids cooperate to form supramolecular openings in the outer mitochondrial membrane. Cell. 2002;111:331–42. doi: 10.1016/s0092-8674(02)01036-x. [DOI] [PubMed] [Google Scholar]
- 5.Lindsten T, et al. The combined functions of proapoptotic Bcl-2 family members bak and bax are essential for normal development of multiple tissues. Mol Cell. 2000;6:1389–99. doi: 10.1016/s1097-2765(00)00136-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wei MC, et al. Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death. Science. 2001;292:727–30. doi: 10.1126/science.1059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chipuk JE, Moldoveanu T, Llambi F, Parsons MJ, Green DR. The BCL-2 family reunion. Mol Cell. 2010;37:299–310. doi: 10.1016/j.molcel.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dewson G, et al. To trigger apoptosis, Bak exposes its BH3 domain and homodimerizes via BH3:groove interactions. Mol Cell. 2008;30:369–80. doi: 10.1016/j.molcel.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 9.Dewson G, et al. Bax dimerizes via a symmetric BH3:groove interface during apoptosis. Cell Death Differ. 2012;19:661–70. doi: 10.1038/cdd.2011.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Llambi F, et al. A unified model of mammalian BCL-2 protein family interactions at the mitochondria. Mol Cell. 2011;44:517–31. doi: 10.1016/j.molcel.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chipuk JE, Green DR. How do BCL-2 proteins induce mitochondrial outer membrane permeabilization? Trends Cell Biol. 2008;18:157–64. doi: 10.1016/j.tcb.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walensky LD, Gavathiotis E. BAX unleashed: the biochemical transformation of an inactive cytosolic monomer into a toxic mitochondrial pore. Trends Biochem Sci. 2011;36:642–52. doi: 10.1016/j.tibs.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Westphal D, Dewson G, Czabotar PE, Kluck RM. Molecular biology of Bax and Bak activation and action. Biochim Biophys Acta. 2011;1813:521–31. doi: 10.1016/j.bbamcr.2010.12.019. [DOI] [PubMed] [Google Scholar]
- 14.Certo M, et al. Mitochondria primed by death signals determine cellular addiction to antiapoptotic BCL-2 family members. Cancer Cell. 2006;9:351–65. doi: 10.1016/j.ccr.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 15.Bird GH, Bernal F, Pitter K, Walensky LD. Synthesis and biophysical characterization of stabilized alpha-helices of BCL-2 domains. Methods Enzymol. 2008;446:369–86. doi: 10.1016/S0076-6879(08)01622-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gavathiotis E, Reyna DE, Davis ML, Bird GH, Walensky LD. BH3-triggered structural reorganization drives the activation of proapoptotic BAX. Mol Cell. 2010;40:481–92. doi: 10.1016/j.molcel.2010.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gavathiotis E, et al. BAX activation is initiated at a novel interaction site. Nature. 2008;455:1076–81. doi: 10.1038/nature07396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moldoveanu T, et al. The X-ray structure of a BAK homodimer reveals an inhibitory zinc binding site. Mol Cell. 2006;24:677–88. doi: 10.1016/j.molcel.2006.10.014. [DOI] [PubMed] [Google Scholar]
- 19.Kim YW, Grossmann TN, Verdine GL. Synthesis of all-hydrocarbon stapled alpha-helical peptides by ring-closing olefin metathesis. Nat Protoc. 2011;6:761–71. doi: 10.1038/nprot.2011.324. [DOI] [PubMed] [Google Scholar]
- 20.Walensky LD, et al. Activation of apoptosis in vivo by a hydrocarbon-stapled BH3 helix. Science. 2004;305:1466–70. doi: 10.1126/science.1099191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Graslund S, et al. The use of systematic N- and C-terminal deletions to promote production and structural studies of recombinant proteins. Protein Expr Purif. 2008;58:210–21. doi: 10.1016/j.pep.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 22.Wang H, et al. Novel dimerization mode of the human Bcl-2 family protein Bak, a mitochondrial apoptosis regulator. J Struct Biol. 2009;166:32–7. doi: 10.1016/j.jsb.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 23.Gross A, et al. Caspase cleaved BID targets mitochondria and is required for cytochrome c release, while BCL-XL prevents this release but not tumor necrosis factor-R1/Fas death. J Biol Chem. 1999;274:1156–63. doi: 10.1074/jbc.274.2.1156. [DOI] [PubMed] [Google Scholar]
- 24.Ku B, Liang C, Jung JU, Oh BH. Evidence that inhibition of BAX activation by BCL-2 involves its tight and preferential interaction with the BH3 domain of BAX. Cell Res. 2011;21:627–41. doi: 10.1038/cr.2010.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Letai AG. Diagnosing and exploiting cancer’s addiction to blocks in apoptosis. Nat Rev Cancer. 2008;8:121–32. doi: 10.1038/nrc2297. [DOI] [PubMed] [Google Scholar]
- 26.Wei MC, et al. tBID, a membrane-targeted death ligand, oligomerizes BAK to release cytochrome c. Genes Dev. 2000;14:2060–71. [PMC free article] [PubMed] [Google Scholar]
- 27.Oltersdorf T, et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–81. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 28.Dai H, et al. Transient binding of an activator BH3 domain to the Bak BH3-binding groove initiates Bak oligomerization. J Cell Biol. 2011;194:39–48. doi: 10.1083/jcb.201102027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strasser A, Jost PJ, Nagata S. The many roles of FAS receptor signaling in the immune system. Immunity. 2009;30:180–92. doi: 10.1016/j.immuni.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim H, et al. Stepwise activation of BAX and BAK by tBID, BIM, and PUMA initiates mitochondrial apoptosis. Mol Cell. 2009;36:487–99. doi: 10.1016/j.molcel.2009.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ren D, et al. BID, BIM, and PUMA are essential for activation of the BAX- and BAK-dependent cell death program. Science. 2010;330:1390–3. doi: 10.1126/science.1190217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Du H, et al. BH3 domains other than Bim and Bid can directly activate Bax/Bak. J Biol Chem. 2011;286:491–501. doi: 10.1074/jbc.M110.167148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Czabotar PE, et al. Bax Crystal Structures Reveal How BH3 Domains Activate Bax and Nucleate Its Oligomerization to Induce Apoptosis. Cell. 2013;152:519–31. doi: 10.1016/j.cell.2012.12.031. [DOI] [PubMed] [Google Scholar]
- 34.Leshchiner ES, Braun CR, Bird GH, Walensky LD. Direct activation of full-length proapoptotic BAK. Proc Natl Acad Sci. 2013 doi: 10.1073/pnas.1214313110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ferrer PE, Frederick P, Gulbis JM, Dewson G, Kluck RM. Translocation of a Bak C-terminus mutant from cytosol to mitochondria to mediate cytochrome C release: implications for Bak and Bax apoptotic function. PLoS One. 2012;7:e31510. doi: 10.1371/journal.pone.0031510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oh KJ, et al. Conformational changes in BAK, a pore-forming proapoptotic Bcl-2 family member, upon membrane insertion and direct evidence for the existence of BH3-BH3 contact interface in BAK homo-oligomers. J Biol Chem. 2010;285:28924–37. doi: 10.1074/jbc.M110.135293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dewson G, et al. Bak activation for apoptosis involves oligomerization of dimers via their alpha6 helices. Mol Cell. 2009;36:696–703. doi: 10.1016/j.molcel.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 38.Lovell JF, et al. Membrane binding by tBid initiates an ordered series of events culminating in membrane permeabilization by Bax. Cell. 2008;135:1074–84. doi: 10.1016/j.cell.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 39.Edlich F, et al. Bcl-xL retrotranslocates Bax from the mitochondria into the cytosol. Cell. 2011;145:104–16. doi: 10.1016/j.cell.2011.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Montessuit S, et al. Membrane remodeling induced by the dynamin-related protein Drp1 stimulates Bax oligomerization. Cell. 2010;142:889–901. doi: 10.1016/j.cell.2010.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only references
- 41.Liu X, et al. Generation of mammalian cells stably expressing multiple genes at predetermined levels. Anal Biochem. 2000;280:20–8. doi: 10.1006/abio.2000.4478. [DOI] [PubMed] [Google Scholar]
- 42.Denisov AY, et al. Structural model of the BCL-w-BID peptide complex and its interactions with phospholipid micelles. Biochemistry. 2006;45:2250–6. doi: 10.1021/bi052332s. [DOI] [PubMed] [Google Scholar]
- 43.Moldoveanu T, Gehring K, Green DR. Concerted multi-pronged attack by calpastatin to occlude the catalytic cleft of heterodimeric calpains. Nature. 2008;456:404–8. doi: 10.1038/nature07353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moldoveanu T, et al. A Ca2+ switch aligns the active site of calpain. Cell. 2002;108:649–60. doi: 10.1016/s0092-8674(02)00659-1. [DOI] [PubMed] [Google Scholar]
- 45.Verdine GL, Hilinski GJ. Stapled peptides for intracellular drug targets. Methods Enzymol. 2012;503:3–33. doi: 10.1016/B978-0-12-396962-0.00001-X. [DOI] [PubMed] [Google Scholar]
- 46.Hill JM. NMR screening for rapid protein characterization in structural proteomics. Methods Mol Biol. 2008;426:437–46. doi: 10.1007/978-1-60327-058-8_29. [DOI] [PubMed] [Google Scholar]
- 47.Sattler M, Schleucher J, Griesinger C. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog Nucl Magn Reson Spectrosc. 1999;34:93–158. [Google Scholar]
- 48.Keller R. The computer aided resonance assignment tutorial. CANTINA Verlag; Goldau, Switzerland: 2004. [Google Scholar]
- 49.Guerry P, Herrmann T. Comprehensive automation for NMR structure determination of proteins. Methods Mol Biol. 2012;831:429–51. doi: 10.1007/978-1-61779-480-3_22. [DOI] [PubMed] [Google Scholar]
- 50.Guntert P, Mumenthaler C, Wuthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J Mol Biol. 1997;273:283–98. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 51.Laue TM, Shah BD, Ridgeway TM, Pelletier SL The Royal Society of Chemistry, C. Analytical Ultracentrifugation in Biochemistry and Polymer Science. The Royal Society of Chemistry. 1992:90–125. [Google Scholar]
- 52.Brown PH, Balbo A, Schuck P. Characterizing protein-protein interactions by sedimentation velocity analytical ultracentrifugation. Curr Protoc Immunol. 2008;Chapter 18(Unit 18):15. doi: 10.1002/0471142735.im1815s81. [DOI] [PubMed] [Google Scholar]
- 53.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J. 2000;78:1606–19. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Balbo A, Brown PH, Braswell EH, Schuck P. Measuring protein-protein interactions by equilibrium sedimentation. Curr Protoc Immunol. 2007;Chapter 18(Unit 18):8. doi: 10.1002/0471142735.im1808s79. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.